Abstract
Background
Progression of breast cancer involves both genetic and epigenetic factors. Parkin gene has been identified as a tumor suppressor gene in the pathogenesis of various cancers. Nevertheless, the putative role of Parkin in breast cancer remains largely unknown. Therefore, we evaluated the regulation of Parkin through both genetic and epigenetic mechanisms in breast carcinoma.
Method
A total of 156 breast carcinoma and their normal adjacent tissue samples were included for mutational analysis through SSCP, and sequencing. MS-PCR was employed for methylation study whereas Parkin protein expression was evaluated using immunohistochemistry and western blotting. For the survival analysis, Kaplan–Meier curve and Cox’s proportional hazard model were used.
Results
In expression analysis, Parkin protein expression was found to be absent in 68% cases of breast cancer. We found that aberrant promoter methylation of Parkin gene is a frequent incident in breast cancer tumors and cell lines. Our MS-PCR result showed that Parkin promoter methylation has a significant role (p = 0.0001) in reducing the expression of Parkin protein. Consistently, expression of Parkin was rectified by treatment with 5-aza-2-deoxycytidine. We also found significant associations of both Parkin negative expression and Parkin promoter methylation with the clinical variables. Furthermore, we found a very low frequency (5.7%) of Parkin mutation with no clinical significance. In survival analysis, patients having Parkin methylation and Parkin loss had a worse outcome compared to those harboring none of these events.
Conclusion
Overall, these results suggested that promoter methylation-mediated loss of Parkin expression could be used as a prognostic marker for the survival of breast cancer.
Electronic supplementary material
The online version of this article (10.1186/s12885-019-6013-6) contains supplementary material, which is available to authorized users.
Keywords: PARK-2 gene, Mutation, Methylation, Expression, Breast cancer
Background
Globally, breast cancer is the most fatal malignancy in women and a second major cause of cancer-related deaths among females [1]. In India, the incidence of breast cancer cases has overtaken cervical cancer as the most commonly diagnosed cancer among women, witnessing a rapid rise and more likely to increase in the future [2].
Parkin (PARK2 or PRKN) gene which spans more than 1.38 Mb is one of the largest human genes maps to chromosome 6q25.2-q27 [3]. Parkin gene lies within FRA6E region, the third most fragile site which is prone to rearrangement and breakage in tumors [3]. Alterations in Parkin, an E3 ubiquitin-protein ligase are mainly associated with Parkinson’s disease [4]. Nevertheless, accumulating pieces of evidence have highlighted its tumor-suppressive role in addition to the one-sided view of its ubiquitin ligase activity [4, 5]. Several studies have demonstrated Parkin gene alterations in a wide array of cancers including brain, breast, liver, pancreas, kidney, ovarian, cervical, and colorectal cancer [6–12]. Numerous groups have reported lack of Parkin expression due to mutation and hypermethylation in a variety of cancers [8, 13–15]. Besides, down-regulation and copy number loss of the Parkin are common events in pancreatic cancers [11]. Moreover, Parkin is found to regulate energy metabolism namely Warburg effect thereby suppressing tumorigenesis [16]. Recently, Parkin has been suggested as a key player involved in different hallmarks of cancer cell [17]. Amazingly, a functional interplay has been reported between the Parkin and p53, a well-established tumor suppressor [16, 18]. Whereas another study indicated the role of Parkin in the metastasis through interaction with HIF-1α (hypoxia-inducible factor 1α) thus highlighted the pivotal role of Parkin in tumor suppression [19]. Collectively, the aforementioned studies emphasized that downregulation of Parkin may promote cancer however the precise mechanism of Parkin inactivation remains unexplored mainly in breast cancer.
Aberrant promoter methylation is a widespread mechanism in cancer. It is an emerging molecular marker which raises the hopes for the development of novel therapeutics in combating cancer [20, 21]. Recent studies have reported aberrant methylation at Parkin promoter among acute lymphoid leukemia (ALL), chronic granulocytic leukemia (CGL) [15], nasopharyngeal carcinoma [22] and cervical cancer [9]. Although the precise genetic and epigenetic mechanisms contributing to Parkin loss in breast cancer remain elusive, this prompted us to investigate the possible mechanisms as well as the potential role of Parkin gene in breast cancer.
Methods
Ethical approval
The present study was approved by the Ethics Committee and Institutional Review Board (ECIRB) of Jamia Millia Islamia, New Delhi and Rajiv Gandhi Cancer Institute and Research Centre, New Delhi, India. Each participating patient signed informed written consent.
Tumor specimens and cell lines
The study comprised of 156 pairs of histologically confirmed breast carcinoma and their adjacent normal tissue samples (without any tumor cell infiltration) from sporadic breast cancer patients undergoing biopsies at Rajiv Gandhi Cancer Research Institute and Research Centre, New Delhi, from 2013 to 2017. After surgery, all samples were instantly put in liquid nitrogen and kept in − 80 °C until further use. Clinicopathological parameters of the breast cancer patients were obtained from the hospital database (Additional file 1: Table S1). The exclusion criteria for the study were metastasized cases from other organs, cases having a prior history of any cancer and prior exposure to chemotherapy and radiation. Three breast cancer cell lines; MCF-7, MDA-MB-231, MDA-MB-468, and one normal HEK-293 (Human embryonic kidney) cells were procured from National Centre for Cell Sciences (NCCS) Pune, India. The cells were grown as a monolayer culture in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum (Gibco, Thermoscientific, South American origin) and antibiotics (100 U penicillin and, 100 mg L−1streptomycin) at 37 °C in a humidified atmosphere of 5% CO2 and were subcultured twice a week [23]. Stock culture of the cell lines was maintained in the exponential growth phase by passaging as monolayer culture, the dislodged cells were suspended and reseeded routinely in complete medium.
Nucleic acid extraction
Genomic DNA was extracted using proteinase K/phenol-chloroform protocol from a total of 156 breast cancer (confirmed by a pathologist) and adjacent normal tissues, and also from four cell lines (MCF-7, MDA-MB-231, MDA-MB-468, and HEK-293). Besides, a total RNA was isolated by TRIzol Reagent from cell lines (Invitrogen) according to the manufacturer’s instructions.
PCR–SSCP and sequencing
A total of 12 exons were amplified to reveal any somatic mutation of Parkin by single-stranded conformational polymorphism (SSCP). Extracted DNA was used for PCR amplification using primers (Additional file 1: Table S2) and amplification conditions as described earlier [9]. The amplified products were visualized by electrophoresis using 2% agarose gel and stained with ethidium bromide. SSCP protocol was followed as mentioned earlier [9]. Samples that demonstrated differences in band-shifts with respect to the wild-type bands were categorized as mutants. To confirm mutations those samples were re-amplified in 40 μL reactions for DNA sequencing using forward and reverse primers. For each sample sequencing was repeated to minimize sequencing artifacts and to confirm mutations. The BLAST tool was employed for pair-wise nucleotide sequence alignment.
TCGA (the Cancer genome atlas) and COSMIC (catalogue of somatic mutations in Cancer) analysis
To analyze the Parkin genetic alterations among breast tumors in TCGA (The Cancer Genome Atlas) we used the cBioPortal database (available at www.cbioportal.org). TCGA (http://cancergenome.nih.gov/) is a publically available reservoir containing 33 types of cancer having more than eleven thousand human tumors with their clinical and molecular phenotypes [24, 25]. The COSMIC (Catalog of Somatic Mutations in Cancer) database (https://cancer.sanger.ac.uk/cosmic) analysis was done to figure out mutations of Parkin (PARK2). Pie charts showing distribution and substitutions of Parkin mutations in breast cancer were obtained.
Oncomine database and UALCAN analysis
Oncomine database was exploited to investigate the mRNA expression of Parkin in breast cancer using the criteria of p-value less than 10–4 and fold change more than 1.5 [26]. The top 10% of the resulted lists were examined (https://www.oncomine.org/). The Parkin mRNA expression was analyzed in TCGA breast and Compendia cell lines datasets. Moreover, UALCAN tool (http://ualcan.path.uab.edu/index.html) was also used to correlate the clinicopathological parameters among breast cancer in TCGA data.
Bisulphite-modification and MSP (methylation specific polymerase chain reaction)
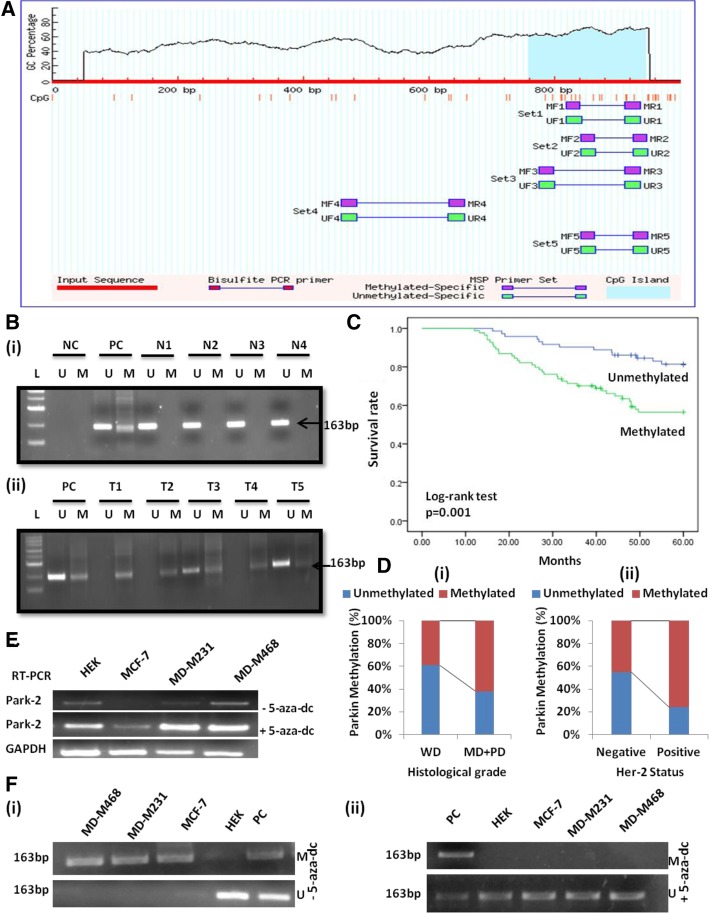
DNA isolated from the tissues was employed in bisulfite modifications by using EZ DNA Methylation-Gold TM kit (Zymo Research, USA) according to manufacturer’s instruction. In Parkin promoter region one CpG island (187 bp) was found just before the transcription site through the Methprimer tool (https://www.urogene.org/methprimer/) (Fig. 1a). MSP reaction was carried out using the unmethylated and methylated primers in a final volume of 25 μL (Additional file 1: Table S2). PCR reaction conditions were, an initial denaturation at 95 °C for 5 min, after that 40 cycles of denaturation at 95 °C for 45 s, followed by annealing step at 63 °C for 35 s, and extension was done at 72 °C for 45 s, a last terminating cycle of final extension was carried out at 72 °C for 7 min. Commercially available methylated and unmethylated bisulfite converted human genomic DNA (Zymo Research Corp., Orange, CA) were used as positive controls of methylated and unmethylated alleles. As a negative control double distilled water (ddH2O) was used in each PCR reaction. Amplified PCR products were then visualized on 2% agarose gels with 100 bp DNA ladder as a standard reference and photographed using Gel Doc (Bio-Rad laboratories, CA, USA) under UV (ultraviolet) illumination.
Fig. 1.
Parkin (PARK-2) methylation analysis. a Graphical representation of CpG islands (187) in the Parkin promoter region taken from MethPrimer (https://www.urogene.org/methprimer/); Criteria used: Island size > 100, GC Percent > 50.0, Obs/Exp > 0.60 b MS-PCR gel pictures, representing methylation of Parkin in (i) normal breast tissues & (ii) breast carcinoma tissues, PC-Positive control, NC-Negative control & L- Ladder (100 bp). c The survival curve was analyzed according to promoter methylation of Parkin protein (SPSS version 17.0). d Frequency distribution of Parkin methylation in (i) Histological grade (ii) Her-2 status (p < 0.05). Demethylating treatment with 5-aza-dC restored Parkin expression and unmethylated status in breast cancer cell lines. e Parkin mRNA expression through RT-PCR, showing that demethylating treatment with 5-aza-dC restored Parkin expression in MCF-7, MDA-MB-231, and MDA-MB-468 cell lines. HEK a non-tumor derived cell line was used as a positive control. GAPDH was amplified as an internal control. f Methylation-specific PCR of Parkin promoter in breast cancer cell lines MCF-7, MD-M231 & MD-M468 before (i) and after (ii) 5-aza-dC treatment. HEK was used as a positive control. PC- Positive control totally methylated and unmethylated bisulfite converted human genomic DNA
5-Aza-2-deoxycytidine treatment
Three breast cancer (i.e. MCF-7, MDA-MB-231, and MD-MB-468) and one non-tumor derived HEK cell lines were seeded at a density of 2 × 105 cells into six-well plates. Next day, Iscove’s modified Dulbecco’s medium containing 10 μmol/L 5-aza-dC (Sigma, USA) was added and changed every 24 h. After 96 h, RNA and DNA were isolated for RT-PCR and MSP analysis respectively.
RT-PCR (reverse transcriptase PCR)
RNA extracted from the cell lines were then used for the synthesis of complementary DNA (cDNA) using iScript™ Reverse Transcription Reagents (Bio-Rad Laboratories, Inc.) and were stored at − 80 °C. PCR reaction was carried out in a final volume of 25 μl, using 2 μl of cDNA, 1 U AmpliTaq Gold DNA Polymerase (Applied Biosystems, Foster City, CA) 0.2 mM dNTPs, 1.5 mM MgCl2, and 20 pmol of primers as listed in Additional file 1: Table S2. PCR conditions were, an initiation of 94 °C for 10 min, followed by 35 cycles of denaturation at 94 °C for 1 min, annealing at 58 °C for 45 s and 7 min extension at 72 °C followed by a final extension of 10 min at 72 °C. Additionally, in each reaction, a set of primers specific for the GAPDH gene (Applied Biosystems) was included as an internal control. Amplified aliquots were then visualized on 2.0% agarose gels. For semi-quantitative analysis, Quantity One v 4.4.0 software (Bio-Rad, USA) was used.
Immunohistochemistry (IHC)
Immunohistochemical staining was carried out on formalin-fixed paraffin-embedded tissue blocks of each sample. The 3–4 μm thin tissue sections were then taken on Poly-L-lysine coated slides. The protocol was followed as described previously [9]. The slides were incubated in a humidified chamber with 1:100 dilution of anti-PARK-2 antibody (cat #ab15954, Abcam) at 4 °C for 24 h. After washing with PBS thrice, slides were next incubated with biotinylated secondary antibody and with an avidin-horseradish peroxidase for 25–30 min. To visualize antigen-antibody reaction 3, 3′-diaminobenzidine (DAB) substrate (DAB substrate kit, Vector Laboratories) was added followed by a counterstaining with hematoxylin dye. Normal adjacent breast tissues were used as positive controls.
Staining interpretation
Stained slides were evaluated by two expert histopathologists at 100X and 400X magnifications under the light microscope. At least three tissue cores from each case were evaluated. The staining < 5% was considered as negative expression and more than 5% were measured as positive.
Western blotting
Protein was extracted from the breast cancer cell lines and tissues with a RIPA lysis buffer (150 mM NaCl, 1% sodium deoxycholate, 1% Triton X-100, 0.1% SDS, 50mMTris_HCl, pH 7.5/2mMEDTA) and was quantified by using the BCA kit (Pierce). Samples of 40 μg of protein were loaded per well in SDS-PAGE followed by transfer onto nitrocellulose membranes (Bio-Rad). Non-specific binding was blocked using 5% bovine serum albumin (BSA) containing 0.05% Tween-20 for 1 h. Subsequently, incubation with primary antibodies, anti-PARK-2 (1:1000; cat #ab15954, Abcam) and GAPDH (1:1000 dilution; Santa Cruz Biotechnology) was done at 4 °C for overnight. Following washing with PBST (phosphate-buffered saline with Tween 20), membranes were re-incubated with secondary antibody conjugated with horseradish peroxidase (HRP) anti-rabbit for PARK-2 and anti-mouse for GAPDH for 2 h at room temperature. The bands were then developed in the darkroom on photographic films through luminol method [27]. The Western blots were quantified by densitometry analysis using ImageJ software 1.46r version.
Statistical analysis
All Statistical evaluations of data were done through the SPSS (Statistical Package for the Social Sciences), version 17.0 for the window. Fisher’s exact test was used for all the comparisons to evaluate the statistical significance with P values < 0.05 and the confidence intervals were quoted at 95% level. Overall survival (OS) was analyzed from the date of surgery to date of the event. Besides, Univariate analysis of time to death (as a result of cancer) was done using the Kaplan-Meier method, and the log-rank test was used to compare the survival times. Univariate and multivariate analysis of the prognostic factors were examined by Cox’s proportional hazard model to identify independent variables predictive of OS. The P value < 0.05 was considered statistically significant for all methods.
Results
Mutational analysis of Parkin gene
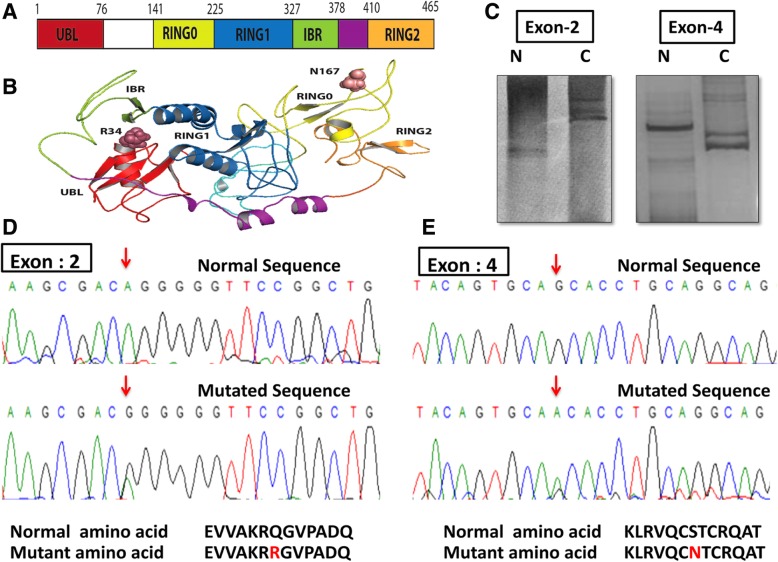
In the mutational analysis, we found that only 5.7% (9/156) breast cancer cases have somatic mutations in exon 2 and exon 4 of Parkin gene (Fig. 2d & e), the same were absent in the normal adjacent tissues. Out of these nine cases, six samples demonstrated A to G transition at nucleotide position 235 leading to conversion of glutamine to arginine at codon 34 in exon 2, while other three samples showed the transition of G to A at nucleotide position 634 leading to conversion of serine to asparagine at codon 167 in exon 4 (Additional file 1: Table S3, Fig. 2a-e). We for the first time reported these two novel somatic mutations; Glu34Arg and Ser167Asp, which were not found in the available list of COSMIC (Additional file 2: Figure S2). Markedly, all mutated cases coincided with the loss of Parkin protein, although we did not find any significant association of these mutations (p > 0.05). Our result is consistent with the TCGA data (cBioPortal: http://www.cbioportal.org/) and COSMIC databases showing that the mutation frequency of Parkin is very low (2.3%) in breast cancer (Additional file 2: Figures S1, S2 and S3).
Fig. 2.
Representation of Parkin (PARK-2) somatic mutations. a Illustration of Parkin 2D structure showing different domains (b) a ribbon representation of the Parkin conformation highlighting somatically mutated residues taken from PyMOL (version 1.7.4.5 Edu.) c Gel pictures of SSCP showing band shift, N-Normal Adjacent tissue, C-Cancerous tissue. d Sequencing histogram of Exon 2 with transition A → G at nucleotide position 235 leading to the conversion of glutamine (Q) to arginine (R) at codon 34 & (e) Exon 4 showing transition G → A at nucleotide position 634 leading to the conversion of Serine (S) to asparagine (N) at codon 167
Parkin protein expression is frequently absent in breast tumors
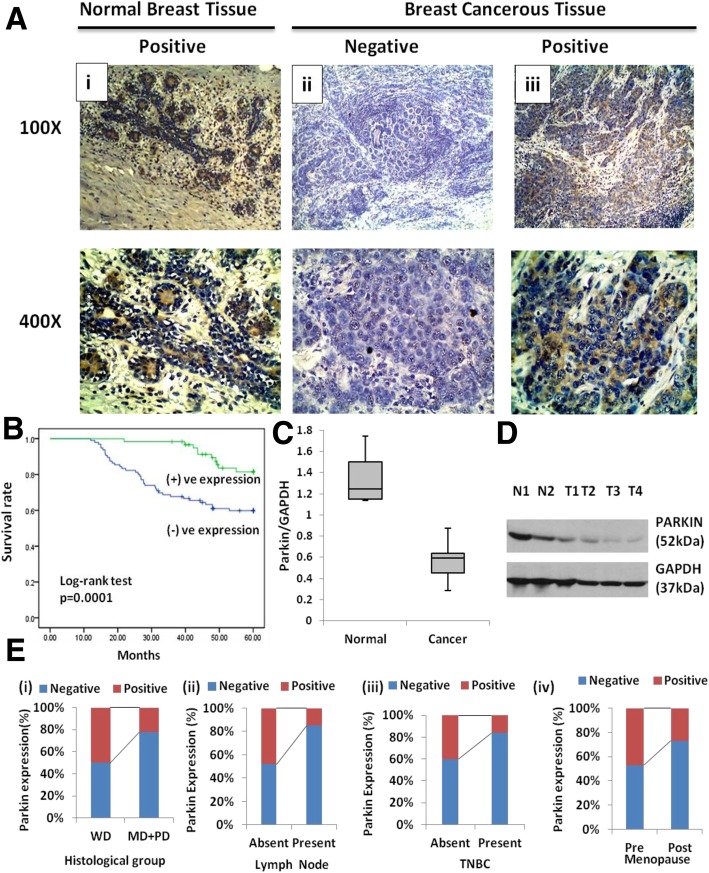
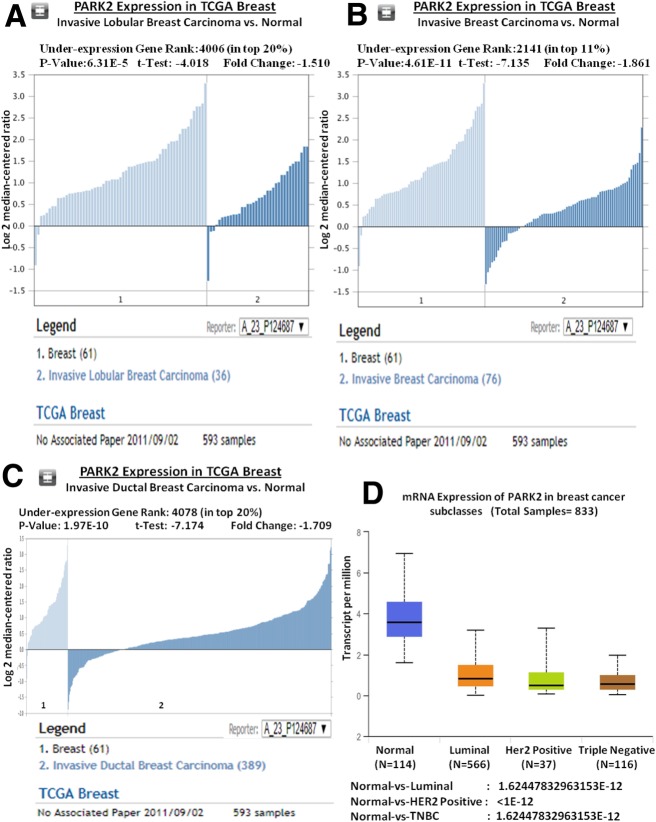
Immunohistochemical study showed predominant cytoplasmic expression of Parkin protein in normal breast tissues (Fig. 3a). The IHC results revealed Parkin protein to be frequently absent in 68% (106/156) cases (Fig. 3a (i), (ii) & (iii)). Interestingly, results of western blot were found to be very well corroborated for the Parkin protein expression where we also found a lower level of Parkin protein in cancer tissues in contrast to normal tissues (Fig. 3c & d). Oncomine study demonstrated that expression of Parkin mRNA is low in different breast cancer types in comparison to normal tissues (Fig. 4a-c). The Compendia cell lines dataset also showed lower expression of Parkin mRNA in breast cancer cell lines in comparison to most of the other cancer cell lines (Additional file 2: Figure S4).
Fig. 3.
Parkin (PARK-2) expressional analysis. a Immunohistochemical analysis of Parkin protein in breast tissue at 100X and 400X magnifications. (i) Parkin positive expression in normal tissue, (ii) Parkin negative expression, (iii) Parkin positive expression in breast carcinoma respectively (Scale bar: 1000 μm). b The Survival curve was analyzed according to the expression status of Parkin protein (SPSS version 17.0). c Parkin protein expression in breast cancer tissues, N-Normal, T-Tumor. d Frequency distribution of Parkin expression with (i) Lymph node, (ii) histological grade and (iii) TNBC cases (p < 0.05). e Distribution of the Parkin/GAPDH ratio in normal and breast cancer tissues
Fig. 4.
Oncomine analysis showing loss of Parkin (PARK-2) expression in different types of breast carcinoma in TCGA. a Invasive lobular breast carcinoma vs. normal tissues, b Invasive breast carcinoma vs. normal tissues, and c Invasive ductal breast carcinoma vs. normal tissues using TCGA breast data. d Relation of Parkin mRNA expression among different subgroups of breast cancer such as Normal, Luminal, Her2 Positive and Triple-negative breast cancers. Oncomine database (https://www.oncomine.org/); TCGA: The Cancer Genome Atlas (Significant value: P < 0.05)
Hypermethylation of Parkin in breast cancer tumors
Promoter methylation of Parkin was detected in 54% (84/156) cases of breast tumors (Fig. 1b; (i) and (ii)). Interestingly, we found a strong correlation between the Parkin methylation and Parkin loss (p = 0.0001). The results showed that methylation frequency coincided with the lower expression of Parkin protein, as evident in 82% (69/84) cases (Table 1).
Table 1.
Correlation of Parkin promoter methylation with Parkin protein expression
| Parkin Promoter | Parkin expressiona | Total (%) | p Valueb | OR (95%CI) | |
|---|---|---|---|---|---|
| Negative | Positive | ||||
| Unmethylated | 37 (51) | 35 (49) | 72 (46) | 0.0001* | 0.2298 (0.1113–0.4744) |
| Methylated | 69 (82) | 15 (18) | 84 (54) | ||
| Total (%) | 106 (68) | 50 (32) | 156 (100) | ||
aProtein expression through IHC (Immunohistochemistry)
bFisher’s Exact test, *Significant Correlation (P < 0.05)
Parkin promoter methylation and loss of Parkin expression
To verify the role of Parkin promoter methylation in the loss of Parkin protein expression, the mRNA level of Parkin was checked in three breast cancer cell lines: MCF-7, MDA-MB-231, and MDA-MB-468. The MCF-7 was found to be Parkin negative, while MDA-MB-231 & MDA-MB-468 have shown very low expression of Parkin (Fig. 1e). Additionally, these cell lines were then treated with 5-aza-dC, a demethylating agent. Following the treatment, restoration of Parkin mRNA expression was witnessed in MCF-7 cells while MDA-MB-231 & MDA-MB-468 cell lines showed relatively increased expression (Fig. 3e). Furthermore, to confirm the result MSP was performed and found only unmethylated bands for all the cell lines (Fig. 3f (i) & (ii)).
Correlation of Parkin protein expression with clinicopathological parameters and patient survival
While finding the statistical correlation of Parkin protein expression with clinical parameters of the patients, we observed a significant correlation of Parkin negative expression with the histological grade (p = 0.0001), Lymph node (p < 0.0001), Menopause (p = 0.028) and TNBC status (p = 0.003) (Table 2) (Fig. 3e (i), (ii) & (iii)). While examining the TCGA breast dataset using UALCAN tool, we speculated that the expression of Parkin mRNA is significantly correlated with the TNBC cases as revealed by our data. On contrary, TCGA data also demonstrated a significant association between the Parkin mRNA expression and Her2 positivity which we failed to get in our study that might be due to differences in population or sample size. Furthermore, Kaplan-Meier survival curve demonstrated that Parkin positive expression has a better mean survival time of 58.2 months than the negative expression 46.6 months (p = 0.0001)(Fig. 3b). In a stratified univariate analysis, the prognostic value of Parkin expression became even more pronounced for OS along with the clinical stage, histological grade and lymph node thereby chosen as the factors to be included in the same Cox regression model. A multivariate analysis also validated that clinical stage (relative risk, 1.476; 95% CI: 1.094–1.990, p = 0.011), histological grade (relative risk, 3.198; 95% CI: 1.412–7.245, p = 0.005), lymph node (relative risk, 2.194; 95% CI: 1.150–4.186, p = 0.017) and Parkin expression (relative risk, 0.057; 95% CI: 0.008–0.418, p = 0.005) are independent prognostic factors for OS (Table 3).
Table 2.
Correlation of Parkin protein expression with clinical parameters among breast cancer patients
| S. No. | Variables | Total | Parkin Expressiona | P valueb | OR (95%CI) | ||
|---|---|---|---|---|---|---|---|
| Positive | Negative | ||||||
| 1 | Age (Years) | < 50 | 30 | 9 (30) | 21 (70) | 0.832 | 1.1255(0.4738–2.6737) |
| ≥50 | 126 | 41 (33) | 85 (67) | ||||
| 2 | Weight (Kg) | < 60 | 75 | 26 (35) | 49 (65) | 0.607 | 0.7935 (0.4046–1.5562) |
| ≥60 | 81 | 24 (30) | 57 (70) | ||||
| 3 | Tumour size (cm) | < 4’ | 73 | 29 (40) | 44 (60) | 0.060 | 0.5139 (0. 2599–1.0161) |
| ≥4 | 83 | 21 (25) | 62 (75) | ||||
| 4 | Clinical Stage | I + II | 88 | 29 (33) | 59 (67) | 0.863 | 0.9090 (0.4606–1.7941) |
| III + IV | 68 | 21 (31) | 47 (69) | ||||
| 5 | Histologicalc grade | WD | 54 | 27 (50) | 27 (50) | 0.0001* | 0.2911 (0.1435–0.5906) |
| MD + PD | 102 | 23 (22) | 79 (78) | ||||
| 6 | Lymph Node | Negative | 81 | 39 (48) | 42 (52) | < 0.0001* | 0.1851 (0.0854–0.4014) |
| Positive | 75 | 11 (15) | 64 (85) | ||||
| 7 | Menopause | Pre | 38 | 18 (47) | 20 (53) | 0.028* | 0.4134 (0.1943–0.8797) |
| Post | 118 | 32 (27) | 86 (73) | ||||
| 8 | ERd | Negative | 81 | 23 (28) | 58 (72) | 0.391 | 1.9438 (0.0190–3.7081) |
| Positive | 75 | 27 (36) | 48 (64) | ||||
| 9 | PRe | Negative | 103 | 32 (31) | 71 (69) | 0.721 | 1.1411 (0.5637–2.3099) |
| Positive | 53 | 18 (34) | 35 (66) | ||||
| 10 | Her-2f | Negative | 111 | 36 (32) | 75 (68) | 1.000 | 0.9409 (0.4463–1.9835) |
| Positive | 45 | 14 (31) | 31 (69) | ||||
| 11 | TNBCg | No | 106 | 42 (40) | 64 (60) | 0.003* | 0.2902 (0.1240–0.6793) |
| Yes | 50 | 8 (16) | 42 (84) | ||||
a Protein expression through IHC (Immunohistochemistry)
b Fisher’s exact test, *Significant Correlation (P < 0.05)
c WD Well differentiated, MD Moderately differentiated, PD Poorly differentiated
d Estrogen receptor
e Progesterone receptor
f human epidermal growth factor receptor 2
g Triple Negative breast cancer
Table 3.
Univariate and multivariate overall survival analysis of different prognostic variables and Parkin expression in breast cancer patients by cox proportional hazard model
| Variables | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| Hazard ratio | 95% CI | P Value | Hazard ratio | 95% CI | P Value | |
| Age (< 50 yr. vs. ≥50 yr.) | 1.535 | 0.688–3.422 | 0.295 | – | – | – |
| Weight (< 60 kg vs. ≥60 kg) | 0.720 | 0.408–1.270 | 0.295 | – | – | – |
| Tumour size (< 4 cm vs. ≥4 cm) | 1.322 | 0.745–2.347 | 0.340 | – | – | – |
| Clinical Stage (I vs. II vs. III vs. IV) | 1.409 | 1.028–1.930 | 0.033* | 1.476 | 1.094–1.990 | 0.011* |
| Histologicala grade (WD vs. MD + PD) | 3.659 | 1.641–8.162 | 0.002* | 3.198 | 1.412–7.245 | 0.005* |
| Lymph Node (Neg. vs. Pos.) | 3.427 | 1.812–6.483 | < 0.0001* | 2.194 | 1.150–4.186 | 0.017* |
| Menopause (Pre vs. Post) | 2.184 | 0.979–.4.870 | 0.056 | – | – | – |
| ERb (Neg. vs. Pos.) | 0.843 | 0.476–1.491 | 0.557 | – | – | – |
| PRc (Neg. vs. Pos.) | 0.717 | 0.380–1.356 | 0.306 | – | – | – |
| Her-2d (Neg. vs. Pos.) | 1.784 | 0.999–3.184 | 0.050* | – | – | – |
| TNBCe (No vs. Yes) | 0.839 | 0.450–1.563 | 0.579 | – | – | – |
| Parkin (Neg. vs. Pos.) | 0.035 | 0.005–0.253 | 0.001* | 0.057 | 0.008–0.418 | 0.005* |
a WD Well differentiated, MD Moderately differentiated, PD Poorly differentiated
b Estrogen receptor
c Progesterone receptor
d human epidermal growth factor receptor 2
e Triple Negative breast cancer, *Significant Correlation (P < 0.05)
Correlation of Parkin methylation with clinicopathological parameters and patient survival
In the descriptive analysis, Parkin promoter methylation was correlated with different clinicopathological variables of breast cancer patients. We observed a significant correlation of Parkin methylation with the histological grade (p = 0.007) and Her-2 status (p = 0.001) (Fig. 1d; (i) & (ii)) (Table 4). However, no other clinical parameter showed a significant correlation with Parkin promoter methylation (p > 0.05). In survival analysis, Kaplan-Meier method also illustrated that the patients with methylated Parkin promoter had a mean survival time of 46.7 months as compared with 55.3 months for patients with the unmethylated Parkin promoter (p = 0.001)(Fig. 1c). The clinical parameters; Clinical stage, histological grade, and lymph node were found significant prognostic indicators for OS in univariate analysis, thus included as the parameters in the same Cox regression model. Results of multivariate analysis further provided the evidence that Parkin promoter methylation (relative risk, 2.286; 95% CI: 1.190–4.389, p = 0.013) is an independent prognostic factor for OS (Table 5).
Table 4.
Correlation of PARK-2 promoter methylation with clinical parameters among breast cancer patients
| S No. | Variables | Total | Parkin Promoter | P valuea | OR (95%CI) | ||
|---|---|---|---|---|---|---|---|
| Unmethylated | Methylated | ||||||
| 1 | Age (Years) | < 50 | 30 | 14 (47) | 16 (53) | 1.000 | 1.0259 (0.4617–2.2791) |
| ≥50 | 126 | 58 (46) | 68 (54) | ||||
| 2 | Weight (Kg) | < 60 | 75 | 36 (48) | 39 (52) | 0.748 | 1.1538 (0.6143–2.1672) |
| ≥60 | 81 | 36 (44) | 45 (56) | ||||
| 3 | Tumour size (cm) | < 4’ | 73 | 37 (51) | 36 (49) | 0.335 | 1.4095 (0.7487–2.6537) |
| ≥4 | 83 | 35 (42) | 48 (58) | ||||
| 4 | Clinical Stage | I + II | 88 | 36 (41) | 52 (59) | 0.148 | 0.6154 (0.3251–1.1650) |
| III + IV | 68 | 36 (53) | 32 (47) | ||||
| 5 | Histological gradeb | WD | 54 | 33 (61) | 21 (39) | 0.007* | 2.5385 (1.2894–4.9974) |
| MD + PD | 102 | 39 (38) | 63 (62) | ||||
| 6 | Lymph Node | Negative | 81 | 42 (52) | 39 (48) | 0.151 | 1.6154 (0.8559–3.0487) |
| Positive | 75 | 30 (46) | 45 (54) | ||||
| 7 | Menopause | Pre | 38 | 23 (61) | 15 (39) | 0.061 | 2.1592 (1.0235–4.5549) |
| Post | 118 | 49 (42) | 69 (58) | ||||
| 8 | ERc | Negative | 81 | 41 (51) | 40 (49) | 0.264 | 1.4548 (0.7723–2.7404) |
| Positive | 75 | 31 (41) | 44 (59) | ||||
| 9 | PRd | Negative | 103 | 49 (48) | 54 (52) | 0.735 | 1.1836 (0.6076–2.3056) |
| Positive | 53 | 23 (43) | 30 (57) | ||||
| 10 | Her-2e | Negative | 111 | 61 (55) | 50 (45) | 0.001* | 3.7709 (1.7351–8.1925) |
| Positive | 45 | 11 (24) | 34 (76) | ||||
| 11 | TNBCf | No | 106 | 49 (46) | 57 (54) | 1.000 | 1.0092 (0.5140–1.9812) |
| Yes | 50 | 23 (46) | 27 (54) | ||||
a Fisher’s exact test, *Significant Correlation (P < 0.05)
b WD Well differentiated, MD Moderately differentiated, PD Poorly differentiated
c Estrogen receptor
d Progesterone receptor
e human epidermal growth factor receptor 2
f Triple negative breast cancer
Table 5.
Univariate and multivariate overall survival analysis of different prognostic variables and Parkin methylation in breast cancer patients by cox proportional hazard model
| Variables | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| Hazard ratio | 95% CI | P Value | Hazard ratio | 95% CI | P Value | |
| Age (< 50 yr. vs. ≥50 yr.) | 1.535 | 0.688–3.422 | 0.295 | – | – | – |
| Weight (< 60 kg vs. ≥60 kg) | 0.720 | 0.408–1.270 | 0.295 | – | – | – |
| Tumour size (< 4 cm vs. ≥4 cm) | 1.322 | 0.745–2.347 | 0.340 | – | – | – |
| Clinical Stage (I + II vs. III + IV) | 1.409 | 1.028–1.930 | 0.033* | 1.541 | 1.139–2.083 | 0.005* |
| Histologicala grade (WD vs. MD + PD) | 3.659 | 1.641–8.162 | 0.002* | 3.316 | 1.462–7.524 | 0.004* |
| Lymph Node (Neg. vs. Pos.) | 3.427 | 1.812–6.483 | < 0.0001* | 2.769 | 1.453–5.277 | 0.002* |
| Menopause (Pre vs. Post) | 2.184 | 0.979–.4.870 | 0.056 | – | – | – |
| ERb (Neg. vs. Pos.) | 0.843 | 0.476–1.491 | 0.557 | – | – | – |
| PRc (Neg. vs. Pos.) | 0.717 | 0.380–1.356 | 0.306 | – | – | – |
| Her-2d (Neg. vs. Pos.) | 1.784 | 0.999–3.184 | 0.050 | – | – | – |
| TNBCe (No vs. Yes) | 0.839 | 0.450–1.563 | 0.579 | – | – | – |
| Methylation (Meth. vs. Unmeth.) | 2.892 | 1.528–5.474 | 0.001* | 2.286 | 1.190–4.389 | 0.013* |
a WD Well differentiated, MD Moderately differentiated, PD Poorly differentiated
b Estrogen receptor
c Progesterone receptor
d human epidermal growth factor receptor 2
e Triple Negative breast cancer, *Significant Correlation (P < 0.05)
Discussion
The expression of Parkin gene is frequently downregulated in a wide spectrum of tumors and cancer cell lines [4], while exogenous expression of Parkin protein inhibits cell proliferation and tumor growth in breast cancer [12]. Furthermore, the break at the Parkin gene/ FRA6E site have been linked with poor overall survival in breast carcinoma [28]. However, the molecular mechanism by which Parkin expression is down-regulated in tumors remains unclear. This was an important breakthrough for further investigations in the regulatory mechanism of Parkin gene expression and its pivotal contribution in the prognosis of breast cancer.
Loss of Parkin expression in a cell could persuade growth-promoting effect as a result of the failure of pro-apoptotic and cell cycle-suppressive regulations [29–31]. Our study indicated that in breast cancer, expression of Parkin was significantly lower/absent which confirms the results of earlier studies [4, 7–10, 28, 32]. Interestingly, unlike the previous study [28], we found a significant correlation of Parkin loss with the poorly/moderately differentiated grade of breast cancer, which highlights its relevance to breast carcinoma. We also found a statistically significant link between Parkin down-regulation and increased lymph node metastasis. This link suggests that Parkin gene has a metastasis suppressive role that is essential to prevent malignant progression in breast cancer as proposed by an earlier study in case of pancreatic cancer [11]. A previous report demonstrated no link between the Parkin expression and breast cancer subtypes [19]. In our study, we, however, found that the absence of Parkin expression appears to have a more profound significance in TNBC (Triple-negative breast cancer) cases which was also supported by the TCGA data. Another study has reported a short metastasis-free survival of patients with Parkin break but not with the loss of Parkin expression [28]. Our study, in contrast, demonstrates that a low Parkin expression is linked with a worse prognosis of breast cancer. Thus, in the present study reduced expression of Parkin and its correlation with an aggressive subtype like TNBC suggests a tumor-suppressive function of Parkin.
Reports have demonstrated somatic mutations and intragenic deletions of Parkin in colon cancer, glioblastoma, in addition to lung cancer [5]. Remarkably, Parkin mutations sometimes occur in the same domains or even at the same amino acids in cancer, which, when found in germline, causes neurodegeneration [5]. Here, we reported two novel somatic mutations/variants of Parkin; Q34R (exon 2) and S167 N (exon 4) which were absent in normal breast tissues. Interestingly, the mutation Q34R lies in the UBL (ubiquitin-like region) domain that is involved in substrate ubiquitination [33, 34]. The variant S167 N was found to be situated in the RING0 domain, that plays a crucial role in Parkin inactivation [35]. The mutation Q34R has been reported yet, neither in cancer nor in PD while the status of S167 N variant does not have any known significance in case of Parkinson’s disease [36, 37]. Hence, it is possible to hypothesize that variant S167 N in Parkinson’s disease could be included in those genotypes which may influence the occurrence of Parkin somatic mutations at the same residue in cancer as indicated by an earlier study [5]. Notably, all mutated cases coincided with the loss of Parkin protein, however, we failed to get any significant association of these mutations. This observation indicates that Parkin mutation is a rare and not a predetermining factor for breast cancer as reported by an earlier study [38].
Addressing the molecular cause for Parkin loss, next we focused on its promoter methylation. It is reported that the Parkin promoter sequence contains distinct CpG islands that are frequently methylated in other cancers [15, 22]. As far as the authors are aware, we provide the first evidence that promoter methylation is a major event for the Parkin inactivation in breast cancer. To validate our hypothesis that Parkin is epigenetically regulated, we have further shown that the treatment of 5′-azacytidine, a methyltransferase inhibitor restores Parkin mRNA expression in different breast cancer cell lines. In this context, methylation at the Parkin promoter should not be viewed as a surrogate biomarker for the loss of Parkin but as a frequently identifiable attribute of breast cancer. Another substantial finding of the present study is that both Parkin promoter methylation and its expressional loss are associated with the advanced histological grade in breast cancer. Thus indicating towards a predominant link of methylation-mediated Parkin loss with poor pathophenotype in Indian breast cancer patients. We also observed that Parkin methylation but not its expression is linked with Her-2 positivity. The observed disparity in correlations provides a hint that Parkin methylation might be an early event with more therapeutic options whereas; Parkin loss may involve some other mechanisms besides the methylation, like transcriptional regulation. The clinical significance of molecular markers and their correlation with survival may carry a more favorable prognosis and increased chemotherapy sensitivity. Remarkably, in the present study, multivariate analysis indicated that the Parkin expression and methylation both are independent prognostic molecular markers of OS in breast cancer patients. Our results also illustrated that breast cancer patients with Parkin methylation exhibited poor overall survival while those with Parkin positive expression having a better 5-year patient survival.
Conclusions
In conclusion, we demonstrated that the breast cancer patients have a hugely varied course for the regulation of Parkin expression, where promoter methylation seems to play a major role while Parkin mutation otherwise may have a somewhat little contribution. The low frequency of two mutations in the studied samples indicates that these might be passenger mutations although their damaging effect cannot be overruled. This study also speculates that Parkin methylation may have a negative prognostic effect on breast cancer. Moreover, our results also strongly support the notion that Parkin expression is a potential prognostic factor for advanced and triple-negative breast cancer and can be used as a biomarker. However, investigation of Parkin status with more attention towards the molecular subtypes like TNBC, which has a relatively worse prognosis and currently lacks sufficient attention, is warranted.
Additional files
Table S1 Clinicopathological characteristics of breast cancer patients included for the study (N = 156). Table S2 Representative table of primer sequence for PCR and their respective temperature 558 and amplicon size. Table S3 Summary of mutations obtained along with codon and nucleotide alteration in breast tumor samples. (DOCX 24 kb)
Figure S1 PARK2 alterations. Figure S2 PARK2 mutations. Figure S3 PARK2 mutations in COSMIC. Figure S4 A. Oncomine analysis showed loss of Parkin expression in different types of breast carcinoma cell line in comparison to other cancer cell lines using compendia cell lines data (61 cell lines) (https://www.oncomine.org/). (DOCX 925 kb)
Acknowledgments
Authors would like to thank Department of Science and Technology for providing INSPIRE- Fellowship (IF140532) to KW and India Council of Medical Research (ICMR) for providing Senior Research Fellowship to AP.
Abbreviations
- 5′-azadC
5-Aza-2-deoxycytidine
- COSMIC
Catalogue of Somatic Mutations in Cancer analysis
- DAB
3, 3′-Diaminobenzidine
- ER
Estrogen receptor
- HER-2
Human epidermal growth factor receptor 2
- IHC
Immunohistochemistry
- MSP
Methylation-specific polymerase chain reaction
- OS
Overall Survival
- PD
Parkinson’s disease
- PR
Progesterone receptor
- Q34R
Glu34Arg
- RT-PCR
Reverse transcriptase PCR
- S167N
Ser167Asp
- SSCP
Single-stranded conformational polymorphism
- TCGA
The Cancer Genome Atlas
- TNBC
Triple-negative breast cancer
- UBL
Ubiquitin-like region
Authors’ contributions
MMAR was the principal investigator of the work. KW designed the study, performed the experiments, compiled the data, and contributed to the writing. AP performed in vitro experiments, data analysis and contributed to the writing. ZIB helped in drafting the manuscript. SK and AM provided valuable clinical input and reviewed the manuscript. All the authors read and approved the final manuscript.
Funding
None
Availability of data and materials
It is confirmed that all the data generated or analyzed during this study are included in this published article.
Ethics approval and consent to participate
The study was approved by the Ethics Committee and Institutional Review Board of Jamia Millia Islamia, New Delhi and Rajiv Gandhi Cancer Institute and Research Centre, New Delhi, India. All participating patients have signed a prior informed written consent.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Khushnuma Wahabi, Email: alikhushiw@gmail.com.
Ahmad Perwez, Email: ahmad86.delhi@gmail.com.
Shabeena Kamarudheen, Email: shabinkamar@gmail.com.
Zafar Iqbal Bhat, Email: zafari598@gmail.com.
Anurag Mehta, Email: anumehta11@gmail.com.
M. Moshahid A. Rizvi, Email: rizvijmi@gmail.com
References
- 1.DeSantis CE, Fedewa SA, Goding Sauer A, Kramer JL, Smith RA, Jemal A. Breast cancer statistics, 2015: convergence of incidence rates between black and white women. CA Cancer J Clin. 2016;66:31–42. doi: 10.3322/caac.21320. [DOI] [PubMed] [Google Scholar]
- 2.Gangane N, Anshu, Manvatkar S, Ng N, Hurtig A-K, Sebastián MS. Prevalence and risk factors for patient delay among women with breast Cancer in rural India. Asia Pac J Public Health. 2016;28:72–82. doi: 10.1177/1010539515620630. [DOI] [PubMed] [Google Scholar]
- 3.Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 4.Cesari R, Martin ES, Calin GA, Pentimalli F, Bichi R, McAdams H, et al. Parkin, a gene implicated in autosomal recessive juvenile parkinsonism, is a candidate tumor suppressor gene on chromosome 6q25-q27. Proc Natl Acad Sci U S A. 2003;100:5956–5961. doi: 10.1073/pnas.0931262100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Veeriah S, Taylor BS, Meng S, Fang F, Yilmaz E, Vivanco I, et al. Somatic mutations of the Parkinson’s disease-associated gene PARK2 in glioblastoma and other human malignancies. Nat Genet. 2010;42:77–82. doi: 10.1038/ng.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yeo CWS, Ng FSL, Chai C, Tan JMM, Koh GRH, Chong YK, et al. Parkin pathway activation mitigates glioma cell proliferation and predicts patient survival. Cancer Res. 2012;72:2543–2553. doi: 10.1158/0008-5472.CAN-11-3060. [DOI] [PubMed] [Google Scholar]
- 7.Lee K, Lee MH, Kang YW, Rhee K-J, Kim TU, Kim YS. Parkin induces apoptotic cell death in TNF-α-treated cervical cancer cells. BMB Rep. 2012;45:526–531. doi: 10.5483/BMBRep.2012.45.9.104. [DOI] [PubMed] [Google Scholar]
- 8.Poulogiannis G, McIntyre RE, Dimitriadi M, Apps JR, Wilson CH, Ichimura K, et al. PARK2 deletions occur frequently in sporadic colorectal cancer and accelerate adenoma development in Apc mutant mice. Proc Natl Acad Sci U S A. 2010;107:15145–15150. doi: 10.1073/pnas.1009941107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naseem A, Bhat ZI, Kalaiarasan P, Kumar B, Gandhi G, Rizvi MMA. Genetic and epigenetic alterations affecting PARK-2 expression in cervical neoplasm among north Indian patients. Tumor Biol. 2017;39:101042831770363. doi: 10.1177/1010428317703635. [DOI] [PubMed] [Google Scholar]
- 10.Mehdi SJ, Ali A, Rizvi MMA. Parkin gene alterations in ovarian carcinoma from northern Indian population. Pathol Oncol Res. 2011;17:579–586. doi: 10.1007/s12253-010-9351-x. [DOI] [PubMed] [Google Scholar]
- 11.Sun X, Liu M, Hao J, Li D, Luo Y, Wang X, et al. Parkin deficiency contributes to pancreatic tumorigenesis by inducing spindle multipolarity and misorientation. Cell cycle (Georgetown, Tex) 2013;12:1133–1141. doi: 10.4161/cc.24215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tay S-P, Yeo CWS, Chai C, Chua P-J, Tan H-M, Ang AXY, et al. Parkin enhances the expression of cyclin-dependent kinase 6 and negatively regulates the proliferation of breast cancer cells. J Biol Chem. 2010;285:29231–29238. doi: 10.1074/jbc.M110.108241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denison SR, Wang F, Becker NA, Schüle B, Kock N, Phillips LA, et al. Alterations in the common fragile site gene Parkin in ovarian and other cancers. Oncogene. 2003;22:8370–8378. doi: 10.1038/sj.onc.1207072. [DOI] [PubMed] [Google Scholar]
- 14.Fujiwara M, Marusawa H, Wang H-Q, Iwai A, Ikeuchi K, Imai Y, et al. Parkin as a tumor suppressor gene for hepatocellular carcinoma. Oncogene. 2008;27:6002–6011. doi: 10.1038/onc.2008.199. [DOI] [PubMed] [Google Scholar]
- 15.Agirre X, Román-Gómez J, Vázquez I, Jiménez-Velasco A, Garate L, Montiel-Duarte C, et al. Abnormal methylation of the common PARK2 and PACRG promoter is associated with downregulation of gene expression in acute lymphoblastic leukemia and chronic myeloid leukemia. Int J Cancer. 2006;118:1945–1953. doi: 10.1002/ijc.21584. [DOI] [PubMed] [Google Scholar]
- 16.Zhang C, Lin M, Wu R, Wang X, Yang B, Levine AJ, et al. Parkin, a p53 target gene, mediates the role of p53 in glucose metabolism and the Warburg effect. Proc Natl Acad Sci U S A. 2011;108:16259–16264. doi: 10.1073/pnas.1113884108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wahabi K, Perwez A, Rizvi MA. Parkin in Parkinson’s disease and Cancer: a double-edged sword. Mol Neurobiol. 2018;55(8):6788–6800. doi: 10.1007/s12035-018-0879-1. [DOI] [PubMed] [Google Scholar]
- 18.Checler F, Alves da Costa C. Interplay between parkin and p53 governs a physiological homeostasis that is disrupted in Parkinson’s disease and cerebral cancer. Neuro-degener Dis. 2014;13:118–121. doi: 10.1159/000354075. [DOI] [PubMed] [Google Scholar]
- 19.Liu J, Zhang C, Zhao Y, Yue X, Wu H, Huang S, et al. Parkin targets HIF-1α for ubiquitination and degradation to inhibit breast tumor progression. Nat Commun. 2017;8:1823. doi: 10.1038/s41467-017-01947-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laird PW. Early detection: the power and the promise of DNA methylation markers. Nat Rev Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 21.Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov. 2006;5:37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- 22.Ni H, Zhou Z, Jiang B, Yuan X, Cao X, Huang G, et al. Inactivation of parkin by promoter methylation correlated with lymph node metastasis and genomic instability in nasopharyngeal carcinoma. Tumor Biol. 2017;39:101042831769502. doi: 10.1177/1010428317695025. [DOI] [PubMed] [Google Scholar]
- 23.Qureshi R, Arora H, Biswas S, Perwez A, Naseem A, Wajid S, et al. Mutation analysis of EGFR and its correlation with the HPV in Indian cervical cancer patients. Tumor Biol. 2016;37:9089–9098. doi: 10.1007/s13277-016-4789-4. [DOI] [PubMed] [Google Scholar]
- 24.Tomczak K, Czerwińska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Poznan, Poland) 2015;19:A68–A77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: a Cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khan P, Idrees D, Moxley MA, Corbett JA, Ahmad F, von Figura G, et al. Luminol-based chemiluminescent signals: clinical and non-clinical application and future uses. Appl Biochem Biotechnol. 2014;173:333–355. doi: 10.1007/s12010-014-0850-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Letessier A, Garrido-Urbani S, Ginestier C, Fournier G, Esterni B, Monville F, et al. Correlated break at PARK2/FRA6E and loss of AF-6/Afadin protein expression are associated with poor outcome in breast cancer. Oncogene. 2007;26:298–307. doi: 10.1038/sj.onc.1209772. [DOI] [PubMed] [Google Scholar]
- 29.Gong Y, Zack TI, Morris LGT, Lin K, Hukkelhoven E, Raheja R, et al. Pan-cancer genetic analysis identifies PARK2 as a master regulator of G1/S cyclins. Nat Genet. 2014;46:588–594. doi: 10.1038/ng.2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gong Y, Schumacher SE, Wu WH, Tang F, Beroukhim R, Chan TA. Pan-Cancer analysis links PARK2 to BCL-XL-dependent control of apoptosis. Neoplasia. 2017;19:75–83. doi: 10.1016/j.neo.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee MH, Cho Y, Jung BC, Kim SH, Kang YW, Pan C-H, et al. Parkin induces G2/M cell cycle arrest in TNF-α-treated HeLa cells. Biochem Biophys Res Commun. 2015;464:63–69. doi: 10.1016/j.bbrc.2015.05.101. [DOI] [PubMed] [Google Scholar]
- 32.Picchio MC, Martin ES, Cesari R, Calin GA, Yendamuri S, Kuroki T, et al. Alterations of the tumor suppressor gene Parkin in non-small cell lung cancer. Clin Cancer Res : an official journal of the American Association for Cancer Research. 2004;10:2720–2724. doi: 10.1158/1078-0432.ccr-03-0086. [DOI] [PubMed] [Google Scholar]
- 33.Shimura H, Hattori N, Kubo SI, Mizuno Y, Asakawa S, Minoshima S, et al. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat Genet. 2000;25:302–305. doi: 10.1038/77060. [DOI] [PubMed] [Google Scholar]
- 34.Finney N, Walther F, Mantel P-Y, Stauffer D, Rovelli G, Dev KK. The cellular protein level of Parkin is regulated by its ubiquitin-like domain. J Biol Chem. 2003;278:16054–16058. doi: 10.1074/jbc.C300051200. [DOI] [PubMed] [Google Scholar]
- 35.Wauer T, Komander D. Structure of the human Parkin ligase domain in an autoinhibited state. EMBO J. 2013;32:2099–2112. doi: 10.1038/emboj.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mata IF, Alvarez V, García-Moreira V, Guisasola LM, Ribacoba R, Salvador C, et al. Single-nucleotide polymorphisms in the promoter region of the PARKIN gene and Parkinson’s disease. Neurosci Lett. 2002;329:149–152. doi: 10.1016/S0304-3940(02)00619-5. [DOI] [PubMed] [Google Scholar]
- 37.Wu R-M, Bounds R, Lincoln S, Hulihan M, Lin C-H, Hwu W-L, et al. Parkin Mutations and Early-Onset Parkinsonism in a Taiwanese Cohort. Arch Neurol. 2005;62:82. doi: 10.1001/archneur.62.1.82. [DOI] [PubMed] [Google Scholar]
- 38.Je EM, Yoo NJ, Lee SH. Somatic mutation of PARK2 tumor suppressor gene is not common in common solid cancers. Pathol & Oncol Res. 2013;19:393–395. doi: 10.1007/s12253-012-9591-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Clinicopathological characteristics of breast cancer patients included for the study (N = 156). Table S2 Representative table of primer sequence for PCR and their respective temperature 558 and amplicon size. Table S3 Summary of mutations obtained along with codon and nucleotide alteration in breast tumor samples. (DOCX 24 kb)
Figure S1 PARK2 alterations. Figure S2 PARK2 mutations. Figure S3 PARK2 mutations in COSMIC. Figure S4 A. Oncomine analysis showed loss of Parkin expression in different types of breast carcinoma cell line in comparison to other cancer cell lines using compendia cell lines data (61 cell lines) (https://www.oncomine.org/). (DOCX 925 kb)
Data Availability Statement
It is confirmed that all the data generated or analyzed during this study are included in this published article.