Abstract
Context:
Fatigue is the most common symptom associated with cancer and its treatment. Investigation of molecular mechanisms associated with fatigue may identify new therapeutic targets.
Objective:
The objective of this pilot study was to evaluate the relationships between gene expression and methylation status and evening fatigue severity in women with breast cancer who received chemotherapy.
Methods:
Latent class analysis (LCA) was used to identify evening fatigue phenotypes. In this analysis, the lowest (i.e., moderate, n = 7) and highest (i.e., very high, n = 29) fatigue-severity classes identified using LCA were analyzed via two stages. First, a total of 32,609 transcripts from whole blood were evaluated for differences in expression levels between the classes. Next, 637 methylation sites located within the putative transcription factor binding sites for those genes demonstrating differential expression were evaluated for differential methylation state between the classes.
Results:
A total of 89 transcripts in 75 unique genes were differentially expressed between the moderate (the lowest fatigue-severity class identified) and very high evening fatigue classes. In addition, 23 differentially methylated probes and three differentially methylated regions were found between the moderate and very high evening fatigue classes.
Conclusions:
Using a multistaged integrated analysis of gene expression and methylation, differential methylation was identified in the regulatory regions of genes associated with previously hypothesized mechanisms for fatigue, including inflammation, immune function, neurotransmission, circadian rhythm, skeletal muscle energy, carbohydrate metabolism, and renal function as well as core biological processes including gene transcription and the cell-cycle regulation.
Keywords: fatigue, breast cancer, chemotherapy, gene expression, methylation, integrated genomic analysis
Fatigue is the most common symptom associated with cancer and its treatment (Berger et al., 2015). Severe fatigue has a negative impact on patients’ ability to tolerate treatments as well as on their quality of life (Bower & Ganz, 2015). Given the high occurrence rates and significant negative impact of fatigue on patients’ lives, it is vital that effective treatments be developed for this devastating symptom.
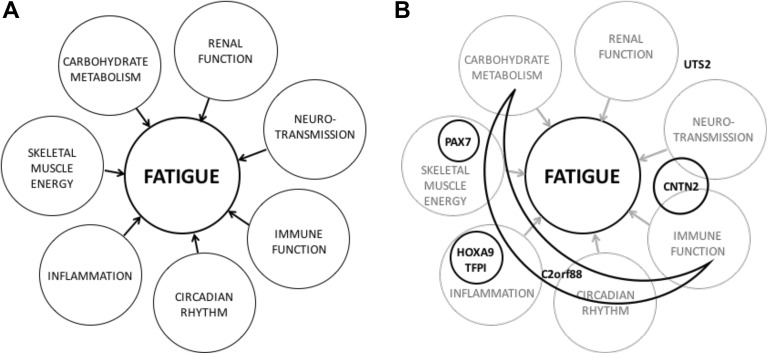
One of the major reasons for the paucity of efficacious interventions to treat this disabling symptom is lack of knowledge of the underlying mechanisms. Findings from several studies suggest that fatigue is associated with alterations in inflammation/immune function, energy metabolism, neurotransmission, and circadian rhythm (Barsevick et al., 2010; Bower, 2014; Kober, Dunn et al., 2016; Landmark-Hoyvik et al., 2010; Saligan & Kim, 2012; Saligan et al., 2015). Findings from our prior study suggest that energy metabolism could be differentiated into skeletal muscle energy and carbohydrate metabolism and that alterations in renal function may contribute to fatigue severity (Figure 1; Flowers et al., 2018).
Figure 1.
Hypothesized mechanisms to explain fatigue severity (Panel A; Flowers et al., 2018). In Panel B, the potential relationships between differentially methylated genes identified in this study and hypothesized mechanisms of fatigue are shown in bold font and shapes. The interrelationships between components related to fatigue in Panel A are not shown.
An evaluation of changes in gene expression can be used to identify mechanisms that underlie differences in fatigue severity. Gene expression produces the functional gene products of various genetic and epigenetic processes. In our previous study (Kober, Dunn et al., 2016), we identified differences in the expression of 12 genes that were associated with evening fatigue severity in breast cancer patients who were categorized into low (n = 19) and high (n = 25) fatigue-severity classes. In a longitudinal study using a more refined phenotype (Flowers et al., 2018), we identified 29 genes that were differentially expressed in oncology patients with moderate (n = 65, the lowest level of fatigue observed) versus very high (n = 195) levels of evening fatigue. In these two studies, evening fatigue severity was evaluated using Lee Fatigue Scale (LFS) scores that were categorized using a clinically meaningful cut point (i.e., >5.6 on a 0–10 scale; Fletcher et al., 2008). Findings from these studies were consistent with previous reports that evaluated the relationship between fatigue severity and changes in gene expression in oncology patients (Hsiao, Reddy, Chen, & Saligan, 2016; Landmark-Hoyvik et al., 2009; Saligan et al., 2013).
Gene expression is regulated by epigenetic mechanisms (e.g., methylation; Razin & Riggs, 1980). Methylation of DNA affects the initiation or inactivation of transcription through attachment of a methyl group to a cytosine nucleotide (Portela & Esteller, 2010). While an evaluation of the epigenetic modifications in genes associated with fatigue may explain its persistence in cancer survivors, we only identified one study that explored associations between DNA methylation status and fatigue severity in breast cancer patients who did and did not receive chemotherapy (CTX) following radiation therapy (RT; Smith et al., 2014). In that study, higher plasma concentrations of soluble tumor necrosis factor receptor 2 (sTNFR2) and interleukin-6 (IL-6) were associated with more severe fatigue. Changes in methylation status between pretreatment and 6 months post-RT were inversely correlated with changes in the concentrations of sTNFR2 and IL-6. However, the investigators found no associations between methylation status and fatigue severity. A number of study limitations warrant consideration. First, the adjustments the authors made for the large number of comparisons (they tested 484,496 methylation loci) to control for Type I error can lead to an inflation of Type II error. Second, the researchers measured fatigue at only two time points, which may not be representative of the more persistent phenotype. Third, they did not evaluate diurnal variations in fatigue. Finally, the study did not address the potential for heterogeneity in methylation patterns due to blood-cell type.
To our knowledge, no studies have evaluated the association between regulation of gene expression by methylation and fatigue severity. In this pilot study, we performed a multistage integrated analysis of gene expression and methylation status (Buescher & Driggers, 2016; Ritchie, Holzinger, Li, Pendergrass, & Kim, 2015) in a sample of breast cancer patients who received CTX and were phenotyped for longitudinal patterns of evening fatigue severity. In the first stage, we performed a whole-transcriptome analysis to evaluate for differences in gene expression associated with evening fatigue severity in order to identify candidate genes that demonstrated functional differences between the two fatigue-severity classes. In the second stage, we performed an exploratory candidate gene analysis to evaluate for associations between fatigue severity and methylation state in the candidate genes that we identified in Stage 1. Using this approach, we could relate potential loci for regulation of gene expression (i.e., methylation sites) with functional differences in gene expression. Specifically, we interrogated only those methylation loci located in putative transcription factor binding site (TFBS) regions of the candidate genes. For these analyses, we evaluated for changes in gene expression and methylation in patients with breast cancer who were classified into moderate (n = 7, the lowest) versus very high (n = 29, the highest) evening fatigue latent classes.
Materials and Methods
Patients and Settings
For this study, we leveraged data from a previously completed longitudinal study of the symptom experience of oncology outpatients receiving CTX (Wright et al., 2015b). Eligible patients were ≥18 years of age; had a diagnosis of breast, gastrointestinal, gynecological, or lung cancer; had received CTX within the preceding 4 weeks; were scheduled to receive at least two additional cycles of CTX; were able to read, write, and understand English; and gave written informed consent. Of the 582 patients included in the original study, 36 women with breast cancer (n = 7 moderate, n = 29 very high levels of evening fatigue) had both gene expression and methylation data available and were included in this study.
Instruments
A demographic questionnaire was used to obtain information on age, gender, ethnicity, marital status, living arrangements, education, employment status, income, and past medical history. The Karnofsky Performance Status Scale was used to evaluate patients’ functional status (Karnofsky, Abelmann, Craver, & Burchenal, 1948). The Self-Administered Comorbidity Questionnaire (SCQ) was used to evaluate occurrence of, treatment for, and functional impact of common comorbid conditions (e.g., diabetes, osteoarthritis, renal disease; Sangha, Stucki, Liang, Fossel, & Katz, 2003). Total SCQ score can range from 0 to 39, and the instrument has well-established validity and reliability (Olomu, Corser, Stommel, Xie, & Holmes-Rovner, 2012).
The 13-item LFS was used to assess physical fatigue (Lee, Hicks, & Nino-Murcia, 1991). Patients rated each item using a 0–10 numeric rating scale based on how they felt prior to going to bed each night over the previous week (i.e., evening fatigue). Higher LFS scores indicate greater fatigue severity. A score of ≥5.6 indicates a clinically meaningful level of evening fatigue (Fletcher et al., 2008). The LFS has well-established validity and reliability (Lee et al., 1991).
Study Procedures
The Committee on Human Research at the University of California, San Francisco, and each of the study sites approved the study. Research staff members approached patients in the infusion unit where CTX was administered to discuss participation in the study. All patients signed written informed consents. All patients completed questionnaires in their homes a total of 6 times over two cycles of CTX (i.e., prior to the next CTX administration, approximately 1 week after CTX administration, approximately 2 weeks after CTX administration). Blood for RNA and DNA was collected at a single time point prior to the administration of the next cycle of CTX. Medical records were reviewed for disease and treatment information.
Procedures for RNA Processing to Obtain Gene Expression and Methylation Data
RNA sample processing, microarray hybridization, and gene expression data preprocessing and quality control followed the protocol we described previously (Kober, Dunn et al., 2016) and again in detail in the Supplementary Methods (see https://doi.org/10.5281/zenodo.1190578). DNA sample processing, microarray hybridization, and methylation data processing and quality control are also described in detail in the Supplementary Methods (https://doi.org/10.5281/zenodo.1190578). Briefly, differentially expressed candidate genes were located on the reference human genome. Then, because methylation can inhibit transcription through alterations in TFBS (Jones, 2012), we limited our selection of probes to those that were located in these regions. Probes identified from the putative TFBS region for each candidate gene and the microarray control probes were included in the analyses.
Data Analyses
Latent profile analysis (LPA)
We described in detail the method used to identify the evening fatigue latent classes in a previous publication (Kober, Cooper et al., 2016). Briefly, we used LPA to identify subgroups of patients (i.e., latent classes) with distinct evening fatigue trajectories over two cycles of CTX. Of the 582 patients included in the LPA, 20.0% (n = 116) were in the moderate, 21.8% (n = 127) were in the high, and 58.2% (n = 339) were in the very high evening fatigue classes. For this article, we made comparisons between the patients with breast cancer in the moderate (n = 7) and very high (n = 29) evening fatigue classes who had both gene expression and methylation data available.
Demographic and clinical data
Demographic and clinical data were analyzed using SPSS, Version 23 (IBM, Armonk, NY). Independent sample t tests, Mann–Whitney U tests, χ2 tests, and Fisher’s exact tests were used to evaluate for differences in demographic and clinical characteristics between the moderate and very high evening fatigue classes.
Differential gene expression
Differential expression of genes between the latent classes was examined with an estimation of gene-by-gene variance using the LIMMA package for R (Version 3.30.13; Ritchie, Phipson, et al., 2015). Demographic and clinical characteristics and surrogate variables were evaluated as potential covariates in the models. Adjustment for multiple comparisons was performed using the Benjamini and Hochberg method (Hochberg & Benjamini, 1990) with a false-discovery rate cutoff of 10%.
Differential methylation
Tests for differentially methylated probes (DMPs) between classes were done using an F test implemented in the minfi package for R (Version 1.20.2; Aryee et al., 2014). Differences between the latent classes in the differentially methylated regions (DMRs) were evaluated using bumpHunter for R (Version 1.14.0; Jaffe et al., 2012). Demographic and clinical characteristics and surrogate variables were evaluated for inclusion as potential covariates in the models. The Type I error threshold for this exploratory study of differential methylation for both DMPs and DMRs was set at .05.
Results
Demographic and Clinical Characteristics
As shown in Table 1, at the enrollment assessment (i.e., prior to the next dose of CTX), mean fatigue score in the very high evening fatigue class was 6.4 ± 1.6 compared to 3.5 ± 2.2 in the moderate fatigue class (p < .0001). We found no additional statistically significant differences between the evening fatigue classes. Therefore, we included no demographic or clinical characteristics as covariates in our models that evaluated for differences in gene expression and methylation.
Table 1.
Differences in Demographic and Clinical Characteristics Between Moderate and Very High Evening Fatigue Classes.
| Characteristic | Moderate Evening Fatigue (n = 7), Mean ± SD | Very High Evening Fatigue (n = 29), Mean ± SD | Statistics |
|---|---|---|---|
| Age (years) | 53.8 ± 15.6 | 55.6 ± 8.6 | t = −0.43, p = .671 |
| Education (years) | 15.6 ± 2.3 | 17.1 ± 2.2 | t = −1.69, p = .101 |
| Body mass index (kg/m2) | 27.0 ± 4.9 | 26.7 ± 5.4 | t = 0.12, p = .905 |
| Lee Fatigue Scale evening fatigue score | 3.5 ± 2.2 | 6.4 ± 1.6 | t = −3.93, p = .000 |
| Karnofsky Performance Status score | 82.9 ± 16.0 | 80.7 ± 13.6 | t = 0.37, p = .717 |
| Self-Administered Comorbidity Questionnaire score | 2.6 ± 2.4 | 2.2 ± 1.3 | t = 0.51, p = .615 |
| Time since diagnosis (years) | 8.3 ± 13.6 | 4.0 ± 6.1 | t = 1.28, p = .211 |
| Hemoglobin (g/dl) | 11.5 ± 1.7 | 11.4 ± 1.4 | t = 0.07, p = .945 |
| Hematocrit (%) | 34.7 ± 4.8 | 34.0 ± 4.2 | t = 0.40, p = .694 |
| n (%) | n (%) | ||
| Self-reported ethnicity | χ2 = 4.88, p = .181 | ||
| White | 4 (57.1) | 23 (79.3) | |
| Asian/Pacific Islander | 1 (14.3) | 4 (13.8) | |
| Black | 0 (0.0) | 1 (3.4) | |
| Hispanic/Mixed/Other | 2 (28.6) | 1 (3.4) | |
| Married or partnered (yes) | 3 (42.9) | 23 (79.3) | FE, p = .076 |
| Lives alone (yes) | 2 (28.6) | 4 (13.8) | FE, p = .573 |
| Currently employed (yes) | 1 (14.3) | 12 (41.4) | FE, p = .382 |
| Annual household income | U, p = .061 | ||
| <US$30,000 | 0 (0.0) | 1 (3.8) | |
| US$30,000–US$70,000 | 2 (50.0) | 2 (7.7) | |
| US$70,000–US$100,000 | 1 (25.0) | 0 (0.0) | |
| >US$100,000 | 1 (25.0) | 23 (88.5) | |
| Childcare responsibilities (yes) | 2 (33.3) | 7 (26.9) | FE, p = 1.000 |
| Eldercare responsibilities (yes) | 1 (20.0) | 3 (11.5) | FE, p = .525 |
| Exercise on a regular basis (yes) | 5 (71.4) | 25 (86.2) | FE, p = .573 |
| Prior cancer treatment | U, p = .586 | ||
| None | 1 (14.3) | 3 (10.7) | |
| Only CTX, surgery, or RT | 2 (28.6) | 14 (50.0) | |
| CTX and surgery, or surgery and RT, or CTX and RT | 1 (14.3) | 3 (10.7) | |
| CTX, surgery, and RT | 3 (42.9) | 8 (28.6) | |
| Metastatic sites | χ2 = 3.35, p = .340 | ||
| No metastasis | 1 (14.3) | 12 (41.4) | |
| Only one lymph node metastasis | 2 (28.6) | 4 (13.8) | |
| Only metastatic disease in other sites | 0 (0.0) | 3 (10.3) | |
| Metastatic disease in lymph nodes and other sites | 4 (57.1) | 10 (34.5) |
Note. CTX = chemotherapy; FE = Fisher’s exact test; RT = radiation therapy; U = Mann–Whitney U test.
Gene Expression
Expression measurements of 32,609 probes were included in the analyses. No surrogate variables in the gene expression data were identified using Surrogate Variable Analysis (SVA). Therefore, no additional covariates (i.e., demographic, clinical, or surrogate variables) were included in the model that tested for differences in gene expression. Differential expression of 89 probes was found in the very high compared to moderate evening fatigue classes (Supplemental Table S1, https://doi.org/10.5281/zenodo.1190578). These 89 probes were annotated to their unique gene symbols (Supplemental Table S2, https://doi.org/10.5281/zenodo.1190578).
Methylation
For the 89 probes that were differentially expressed between the moderate and very high evening fatigue classes, we mapped 75 RefSeq gene identifiers. For these 75 genes, 637 individual methylation probes were found in TFBS regions. These probes were included in the differential methylation analysis (Supplemental Table S3, https://doi.org/10.5281/zenodo.1190578). No surrogate variables were identified in the methylation data using SVA. Therefore, no additional covariates (i.e., demographic, clinical, or surrogate variables) were included in the model that was used to test for differential methylation.
We identified 23 significant DMPs (Table 2; Supplemental Table S4, https://doi.org/10.5281/zenodo.1190578). Of the 23 genes, 2 (i.e., collagen type XI alpha 1 chain [COL11A1] and chromosome 2 open reading frame 88 [C2orf88]) contained two DMPs each, and each of the remaining 21 genes contained one DMP each. One DMP (cg14210694) was associated with differential expression of the gene in which the probe was located (C2orf88; ρ = .42, p = .01069).
Table 2.
Differential Methylation Between Moderate and Very High Evening Fatigue Classes.
| Gene Symbol | Name | Entrez ID | Probe ID or Regiona | logFCb | Δβc | p Valued | Mechanism(s)e |
|---|---|---|---|---|---|---|---|
| Differentially methylated probes | |||||||
| CCDC93 | Coiled-coil domain containing 93 | 54520 | cg16326819 | .57 | .02 | F = 4.54, p < .040 | T |
| CCNJ | Cyclin J | 54619 | cg11930665 | −.30 | −.01 | F = 4.92, p < .033 | T, CC |
| CNTN2 | Conactin 2 | 6900 | cg12269394 | −.25 | −.05 | F = 13.77, p < .001 | NT |
| COL11A1 | Collagen type XI alpha 1 chain | 1301 | cg00172849 cg26256793 |
−0.19 | −.06 −.07 |
F = 5.73, p < .022 F = 5.70, p < .023 |
U |
| C2orf88 | Chromosome 2 open reading frame 88 | 84281 | cg22003435 cg14210694 |
−.48 | −.02 .01 |
F = 5.55, p < .024 F = 5.43, p < .026 |
I, E, NT |
| HHIPL1 | HIP-like 1 | 84439 | cg07456670 | −.20 | .05 | F = 7.60, p < .010 | I |
| HIST1H4E | Histone cluster 1 H4 family member E | 8367 | cg02656667 | −.46 | .01 | F = 4.94, p < .033 | T |
| HOXA9 | Homeobox A9 | 3205 | cg26521404 | −.22 | −.04 | F = 6.65, p < .015 | I |
| LRRC15 | Leucine-rich repeat containing 15 | 131578 | cg18766468 | −.57 | .06 | F = 4.43, p < .043 | I |
| KMT2E | Lysine methyltransferase 2E | 55904 | cg14527213 | .79 | .01 | F = 4.67, p < .038 | T, CC |
| PAPOLG | Poly(A) polymerase gamma | 64895 | cg08129754 | −.24 | <.01 | F = 6.46, p < .016 | T |
| PARP16 | Poly(ADP-ribose) polymerase family member 16 | 54956 | cg11718315 | −.46 | .02 | F = 4.53, p < .041 | T |
| PAX7 | paired box 7 | 5081 | cg12601945 | −.23 | −.03 | F = 8.35, p < .007 | E |
| SNORD1A | Small nucleolar RNA, C/D box 1A | 677848 | cg03742250 | −.33 | −.02 | F = 4.20, p < .049 | I, T |
| SNORD1C | Small nucleolar RNA, C/D box 1C | 677850 | cg27048959 | −.33 | −.01 | F = 5.07, p < .031 | T |
| TAF1C | TATA-box-binding protein–associated factor, RNA polymerase I subunit C | 9013 | cg20315643 | .66 | .01 | F = 6.10, p < .019 | T |
| TFIP11 | Tuftelin interacting protein 11 | 24144 | cg10311833 | .93 | −.01 | F = 4.50, p < .041 | T |
| TFPI | Tissue factor pathway inhibitor | 7035 | cg27342837 | −.24 | −.03 | F = 4.76, p < .036 | I |
| TRMT2A | tRNA methyltransferase 2 homolog A | 27037 | cg16982658 | −.26 | .02 | F = 14.37, p < .001 | T |
| CEP41 | Centrosomal protein 41 | 95681 | cg26966001 | −.29 | .01 | F = 6.48, p < .016 | CC |
| UTS2 | Urotensin 2 | 10911 | cg09938876 | −.81 | .01 | F = 4.23, p < .048 | R |
| VPS41 | VPS41, HOPS complex subunit | 27072 | cg07273388 | .72 | −.02 | F = 4.31, p < .046 | U |
| ZNF607 | Zinc finger protein 607 | 84775 | cg16046616 | −.42 | −.02 | F = 4.26, p < .047 | T |
| Differentially methylated regions | |||||||
| DEFB124 | Defensin beta 124 | chr20:30060822–30063038 | −.22 | p < .030 | I | ||
| PRM1 | Protamine 1 | chr16:11374090–11376309 | −.23 | p < .013 | U | ||
| TAF1C | TATA-box-binding protein–associated factor, RNA polymerase I subunit C | chr16:84223440–84225080 | .66 | p < .004 | T, CC | ||
Note. tRNA = transfer RNA.
a Chromosome: region start–region end.
b Log fold change in gene expression between patients in the very high and moderate evening fatigue classes. A positive value denotes higher gene expression in the very high compared to the Moderate evening fatigue class.
c Difference in methylation state between patients in the very high and moderate evening fatigue classes. A positive value denotes a higher methylation in the very high compared to the moderate evening fatigue class.
d p value of the F test for differentially methylated probe; area p value for differentially methylated region.
e Mechanism: I = inflammation, E = skeletal muscle energy, NT = neurotransmission, R = renal function, T = transcription, CC = cell-cycle regulation, U = unknown.
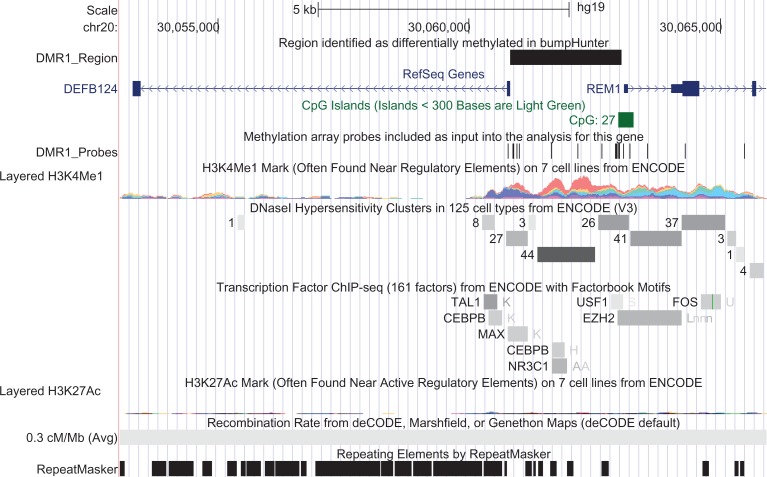
The bumpHunter algorithm identified 61 regions (i.e., bumps) that conferred three significant DMRs in TFBS regions of genes that were differentially expressed between the moderate and very high fatigue classes (Table 2; Supplemental Table S5, https://doi.org/10.5281/zenodo.1190578). The first DMR (chr20:30060822–30063038) was located upstream of the TFBS for defensin beta 124 (DEFB124; Figure 2). A total of 19 probes were included as input into the analysis for this gene, and 12 reside within the DMR. The second DMR (chr16:84223440–84225080) was located upstream of the TFBS for TATA-box-binding protein–associated factor, RNA polymerase I subunit C (TAF1C). A total of 18 probes were included as input for the analysis of this gene. The third DMR (chr16:11374090–11376309) was located upstream of the TFBS for protamine 1 (PRM1). A total of nine probes were included as input for the analysis of this gene.
Figure 2.
Screenshot of the University of California Santa Cruz Genome browser displaying the region on chromosome 20 of the hg19 (genome reference consortium Version 37) assembly of the human genome that includes the defensin beta 124 and RRAD and GEM-Like GTPase 1 genes, the region identified in this study as differentially methylated, and the locations of the methylation probe included in the analysis. Assembly tracks show scale, chromosome, position of the region, and gaps in the assembly. The gene models are provided by the RefSeq. The gene models depict exons as solid blocks connected by lines in introns with arrows showing the direction of transcription. Putative regulatory regions are identified by the layered H3K4Me1, layered H3K27Ac, transcription factor Chromatin Immunoprecipitation Sequencing (ChIP-seq), DNase hypersensitivity clusters, and CpG island (i.e., 5'—C—phosphate—G—3' linear DNA sequence) tracks. Predicted recombination rates and DNA repeat elements in the region are displayed in the final tracks.
Discussion
Differential Gene Expression Identified Candidate Genes for the Methylation Analyses
To our knowledge, this study is the first to use a multistaged integrated analysis of gene expression and methylation to provide preliminary evidence for potential mechanisms of evening fatigue in women receiving CTX for breast cancer. In the first stage of this data-integrated approach (Buescher & Driggers, 2016; Ritchie, Holzinger et al., 2015), differences in gene expression between the moderate and very high evening fatigue classes were evaluated. In the second stage, a candidate gene approach was used to evaluate for differences in methylation in the subset of genes that were differentially expressed. Methylation of cytosine, particularly in TFBS regions, is one epigenetic mechanism that underlies the regulation of gene expression (Stephens, Miaskowski, Levine, Pullinger, & Aouizerat, 2013). The use of this methodology focused the methylation analysis on only the subset of genes that exhibited functional differences in gene expression in this sample.
While evidence exists to support a relationship between methylation and gene expression, the patterns of these associations can vary (Jones, 2012). For example, higher gene expression can be associated with increased methylation in the promoter (Kass, Landsberger, & Wolffe, 1997) and with decreased methylation within the gene (Jones, 1999). These associations vary with the distance from the promoter (Schultz et al., 2015) as well as between individuals and across tissues (Wagner et al., 2014). Our findings are consistent with previously observed variability in associations in that some loci exhibit increased and some loci exhibit decreased methylation between the moderate and very high evening fatigue classes.
Previous studies by our group (Flowers et al., 2018; Kober, Dunn et al., 2016) and others (Wang, 2008; Wang & Woodruff, 2015) identified a number of mechanisms that could influence fatigue severity in oncology patients receiving CTX (Figure 1, Panel A). Findings from the current study provide additional support for many of these mechanisms, including inflammation, neurotransmission, carbohydrate metabolism, renal function, immune function, and skeletal muscle energy (Figure 1, Panel B), which we discuss below in detail. In addition, we found evidence for multiple loci that can contribute to two or more of these mechanisms simultaneously.
Inflammation
A total of five differentially expressed and methylated genes are solely associated with inflammation (Table 2). Previous studies have identified a large number of inflammatory genes associated with higher levels of fatigue (Doong et al., 2015; Illi et al., 2012; Kober, Smoot et al., 2016; McCann et al., 2012; Wang, Yin, Miller, & Xiao, 2017). While the exact relationship between inflammation and fatigue is not completely understood, it is likely that multiple putative genes and gene products interact and are regulated by epigenetic processes (e.g., methylation). Homeobox (HOX) genes are involved in normal hematopoiesis (Argiropoulos & Humphries, 2007). Expression of homeobox A9 (HOXA9) can be inhibited by inflammatory signals, including nuclear factor kappa-light-chain-enhancer of activated B cells (NFκ-β), which controls cytokine production (Trivedi, Patel, & Patel, 2008). Consistent with our previous study that found that differential perturbation of the NFκ-β pathway was associated with fatigue severity (Flowers et al., 2018), expression of HOXA9 was decreased in the very high fatigue class.
Tissue factor pathway inhibitor (TFPI) inhibits the coagulation cascade, which is part of a cytokine-mediated inflammatory response (Esmon, 2005; Wood et al., 2013). In the present study, the increase in methylation that we observed in the regulatory region of TFPI in the very high fatigue class may underlie the decreased expression of TFPI. This decrease in TFPI expression may result in decreased inhibition of coagulation, which is seen in inflammatory states (Esmon, 2005). This decreased inhibition may be one mechanism by which the inflammatory response is activated, resulting in higher levels of fatigue.
The human hedgehog interacting protein-like 1 (HHIPL1) gene is a member of the glucose/sorbosone dehydrogenase family that is expressed in many tissues but has the highest levels of expression in brain tissue (Fagerberg et al., 2014). In addition, it is a member of the scavenger receptor cysteine-rich domain family (HGNC:1404), which is a highly conserved module of the innate immune system (Sarrias et al., 2004). We observed lower gene expression levels and higher methylation states of HHIPL1 in our very high fatigue class. This inverse relationship is similar to the pattern researchers have observed for the human hedgehog interacting protein HHIP in human hepatoma cells (Tada et al., 2008), pancreatic cancer cells (Martin et al., 2005), and gastrointestinal cancer cells (Taniguchi et al., 2007). These results support a relationship between the methylation status of and gene expression in HHIPL1 and suggest an interesting target for future research, as HHIPL1 expression may be restored by epigenetic modifier drugs.
DEFB124 is a member of the β-defensin family that is involved in innate host defense mechanisms (Patil, Cai, Sang, Blecha, & Zhang, 2005). This DMR overlaps with an annotated CpG island and lies in a region with strong evidence of nearby regulatory DNA elements (Figure 2). These elements include DNase I hypersensitivity clusters, TFBSs, and sites susceptible to histone modifications (i.e., H3K4Me1). NFκ-β increases the expression of DEFB124 and is associated with increased production of chemokines and cytokines (Kim, Lee, Han, & Myung, 2014). However, we observed a decrease in gene expression in DEFB124 in the very high evening fatigue class. In light of this finding, as well as the decreased expression in a paralog defensing (defensin beta 103, DEF103B) in a previous study (Kober, Dunn et al., 2016), additional research is needed to evaluate the associations between members of the β-defensin family and the severity of evening fatigue.
Leucine-rich repeat containing 15 is a protein-coding gene involved in collagen and laminin binding. It is a member of the cytokine-mediated signaling pathway (GO:0019221), which is activated as part of immune regulation (Bezbradica & Medzhitov, 2009). It can be upregulated in the presence of pro-inflammatory cytokines (Satoh, Hata, & Yokota, 2002). However, we observed a decrease in the expression of this gene in the very high evening fatigue class relative to the moderate class. This finding warrants further exploration in a future study.
Renal Function
Urotensin 2 (UTS2) is a highly conserved gene whose product has both potent vasoconstrictor and vasodilatory activity that is widely expressed across many tissues (Ames et al., 1999; Langham & Kelly, 2013; Ong, Lam, & Cheung, 2005). We observed a decrease in gene expression of UTS2 in the very high evening fatigue class. Previous studies found associations between genetic polymorphisms in UTS2 and plasma levels of UTS2 and hypertension (He et al., 2016; Zoccali et al., 2008), as well as complications from hypertension (e.g., renal disease; Hursitoglu et al., 2012). Of note, patients with renal disease report high levels of fatigue (Amro et al., 2016). In addition, in our previous study (Flowers et al., 2018), increases in fatigue severity were associated with differential expression of genes associated with renal function.
Skeletal Muscle Energy
Alterations in skeletal muscle energy are another hypothesized mechanism for fatigue (Wang, 2008). In the present study, we identified differential expression and methylation of the paired box 7 (PAX7) gene. The protein product of this gene facilitates myogenesis (McKinnell et al., 2008), supports myoblast proliferation (Riuzzi et al., 2014), and mediates skeletal muscle atrophy (Joung et al., 2014). Polymorphisms in this gene are associated with rhabdomyosarcoma (Du et al., 2005). We observed a decreased level of PAX7 expression in the very high fatigue class, which supports the hypothesis that fatigue may be related to changes in myoblast stability.
Neurotransmission
The relative role of central versus peripheral mechanisms in the development and maintenance of fatigue remains an area of active research (Yavuzsen et al., 2009). Peripheral changes can contribute to a reciprocal regulation between the innate immune and neural systems (Cole, Hawkley, Arevalo, & Cacioppo, 2011; Irwin & Cole, 2011; Powell, Mays, Bailey, Hanke, & Sheridan, 2011). Pro-inflammatory cytokines cross the blood–brain barrier and can lead to increased synthesis of cytokines in the brain (Quan & Banks, 2007) that can influence neural and endocrine activity (Dantzer, O’Connor, Freund, Johnson, & Kelley, 2008). Increasing evidence suggests that peripheral gene expression reflects system-wide biology (Liew, Ma, Tang, Zheng, & Dempsey, 2006) and can provide insights into the activities of both central and peripheral mechanisms (Cole, 2013).
Findings from previous studies suggest that the contactin 2 gene (CNTN2; also known as transient axonal glycoprotein-1) is part of the immunoglobulin superfamily of adhesion molecules and is associated with the development of neurons in the cerebellum (Derfuss et al., 2009; Iijima, Koike, Katsuno, & Sobue, 2011; Pang et al., 2012; Rickman et al., 2001; Stogmann et al., 2013; Yan & Jiang, 2016). We observed a decrease in gene expression of CNTN2 in the very high evening fatigue class. Polymorphisms in this gene are associated with chronic inflammatory demyelinating polyneuropathy and responses to immunoglobulin treatment for that condition (Iijima et al., 2011; Pang et al., 2012).
Overlapping Hypothesized Mechanisms
Any given gene product may act as a part of multiple pathways. Genes can have different effects depending on where their products lie in the various pathways. In the present study, we found candidate loci that overlap in multiple processes associated with several mechanisms for fatigue (i.e., inflammation, circadian rhythm, hypothalamic–pituitary–adrenal [HPA] axis, and gluconeogenesis). These findings suggest that changes within seemingly disparate mechanisms may be related to changes in a single or small number of loci.
Specifically, we found that two noncoding small nucleolar RNAs (snoRNAs) were differentially expressed and differentially methylated (i.e., snoRNA, C/D box 1A SNORD1A [i.e., R38A, snR38A] and snoRNA, C/D box 1C SNORD1C [i.e., R38C, snR38C]). While snoRNAs primarily control methylation of ribosomal RNA, they also regulate some pre-messenger RNA processing (Falaleeva et al., 2016). In addition, some miRNAs that regulate gene expression across multiple levels of transcription and translation originate from genomic regions that code for snoRNAs (Brameier, Herwig, Reinhardt, Walter, & Gruber, 2011; Ender et al., 2008). Results from one study support SNORD1A involvement in multiple inflammatory pathways (Liang, Patil, & Nakai, 2015). Because the regulatory impact of changes in expression of snoRNAs are broad, associations between expression of their regulatory elements and fatigue severity warrant additional investigation.
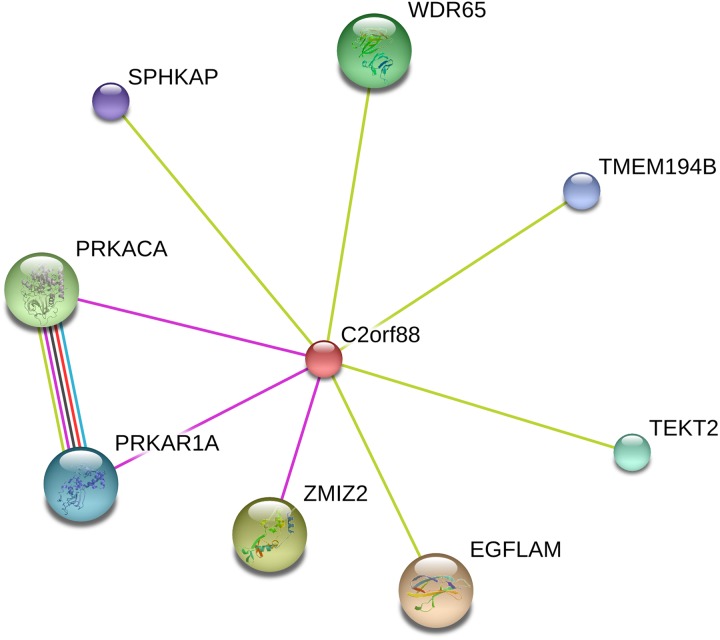
Differential expression of C2orf88 was associated with differential methylation of a probe annotated to this gene region in the present study. C2orf88 is involved in an interaction network with the protein kinase cyclic AMP-dependent type 1 regulatory subunit alpha (PRKAR1A) and protein kinase C alpha (PRKCA) genes (Figure 3). PRKAR1A and PRKCA are subunits of the protein kinase A gene, which regulates secretion of cortisol (Berthon, Szarek, & Stratakis, 2015). We observed decreased expression of C2orf88 in the very high evening fatigue class as well as a significant correlation between the methylation state of cg14210694 and expression levels of C2orf88. Changes in levels of cortisol can influence inflammatory pathways that contribute to fatigue severity (Gleeson et al., 2011; Steptoe, Hamer, & Chida, 2007) as well as activity of the HPA axis (Tomas, Newton, & Watson, 2013), which can also contribute to fatigue severity.
Figure 3.
Protein–protein interaction network of predicted functional partners for the chromosome 2 open reading frame 88 gene (C2orf88). Network interaction representation for C2orf88 was generated by the STRING database (Szklarczyk et al., 2017). Nodes represent all proteins produced by a single protein-coding gene locus. Edges represent specific or meaningful associations. Node size: protein of unknown 3D structure (small), protein of known or predicted 3D structure (large). Color of the edges connecting the nodes represents the types of evidence supporting the connections (colors visible in online version of article only): predicted gene neighborhood (green), predicted gene fusions (red), known interactions from experimental evidence (pink), co-expression (black), and text mining (green).
Transcription and Regulation
As noted above, single genes may operate simultaneously within multiple biological pathways. In addition, single or multiple genes may influence core biological processes that propagate out and have effects on many phenotypes. In the present study, we found 12 candidate loci that have functions within the core biological mechanisms of transcription, translation, posttranslational modification, and cell-cycle regulation (Table 2). The functions of these genes are summarized in Table 3. Because this study is the first to identify changes in the expression of these genes that are involved in fundamental biological processes, they warrant confirmation in future studies of fatigue in oncology patients.
Table 3.
Differentially Expressed Genes Involved in Transcription and Cell-Cycle Regulation.
| Symbol | Name | ID | Description |
|---|---|---|---|
| CCDC93 | Coiled-coil domain containing 93 | 54520 | Involved in protein binding and transport. Expressed widely across multiple tissues (Fagerberg et al., 2014). Although the specific function of CCDC93 is unclear, it is found to interact with ubiquitin (Danielsen et al., 2011). The covalent attachment of proteins to ubiquitin is known to be involved in regulation of many cellular functions through posttranslational modifications (Yang, 2005). |
| CCNJ | Cyclin J | 54619 | Expressed in the nucleus and widely expressed across multiple tissues (Fagerberg et al., 2014). However, its specific function is not known. More broadly, the cyclin family of proteins controls the progression through the cell cycle and transcription by activating cyclin-dependent kinase enzymes (Malumbres et al., 2009). CCNJ may be involved in regulation of one or more pathways directly via transcription or less directly via cell-cycle control (e.g., expansion of cell lineages in inflammatory cell types). |
| CEP41 | Centrosomal protein 41 | 95681 | Also known as TGSA14. A gene that encodes a protein that binds to the centrosome and microtubules and is involved in the cell cycle (Andersen et al., 2003). Mutations in CEP genes alter miRNA binding sites, which may affect the cell cycle (Gopalakrishnan, Kamaraj, & Purohit, 2014). |
| COL11A1 | Collagen type XI alpha 1 chain | 1301 | Codes for a protein involved in protein digestion and absorption. These protein products are expressed in blood and bone marrow tissue. |
| HIST1H4E | Histone cluster 1 H4 family member E | 8367 | Codes for a basic nuclear protein involved in the nucleosome structure of the chromosomal fiber and instrumental in regulating fundamental cellular processes including transcription (Vardabasso et al., 2014). |
| HOXA9 | Homeobox A9 | 3205 | A protein-coding gene in the class of transcription factors that regulate gene expression. Member of the “transcriptional misregulation in cancer” pathway (KEGG hsa05202). Recent studies found an inverse relationship between methylation status of cg26521404 and expression levels of HOXA9 in samples of patients with breast cancer (Park, Choi, Bae, Choi, & Kim, 2017) and adenocarcinoma (Yan et al., 2017). |
| KMT2E | Lysine methyltransferase 2E | 55904 | Also known as NKp44L and MLL5. A protein-coding gene involved in the regulation of transcription, DNA methylation, cell-cycle mediation and survival, and hematopoiesis (Zhang, Novera, Zhang, & Deng, 2017). Expressed widely across major tissues. Binds to histone H3K4me3 and serves as a transcriptional regulatory switch (Ali et al., 2013). Overexpression inhibits cell-cycle progression (Zhang et al., 2017). |
| PAPOLG | Poly(A) polymerase gamma | 64895 | A protein-coding gene that catalyzes the posttranscriptional adenylation of the 3′ end of mRNA precursors and small RNAs (Topalian et al., 2001). |
| PARP16 | Poly(ADP-ribose) polymerase family member 16 | 54956 | Catalyzes the posttranslational modification of proteins. |
| PRM1 | Protamine 1 | 5619 | Known to affect DNA compaction during spermatogenesis and expressed primarily in testes (Fagerberg et al., 2014). |
| SNORD1A | Small nucleolar RNA, C/D box 1A | 677848 | An RNA gene affiliated with the snoRNA class that is involved in site-specific rRNA modification. |
| SNORD1C | Small nucleolar RNA, C/D box 1C | 677850 | An RNA gene affiliated with the snoRNA class that is involved in site-specific rRNA modification. |
| TAF1C | TATA-box binding protein associated factor, RNA polymerase I subunit C | 9013 | A component of the transcription factor selectivity factor/transcription initiation factor-IB complex (SL1/TIF-1B). Codes for one of three TATA box-binding proteins necessary for the formation of the complex required by RNA polymerase I to initiate transcription (Di Pietro et al., 2000). Knockdown of TAF1C decreases the expression of HOX genes (including HOXA9) and other genes necessary for initiation of RNA polymerase II–dependent translation (Okuda, Kanai, Ito, Matsui, & Yokoyama, 2015). p53 interacts with TAF1C to inhibit transcription by RNA polymerase I in tumor tissue (Oh, An, Yoo, & Lee, 2015). A mutation in TAF1C may cause a loss of interaction with p53 (Oh et al., 2015). |
| TFIP11 | Tuftelin interacting protein 11 | 24144 | Also known as Spp382p. A protein-coding gene that is a component of the spliceosome involved in late-stage splicing of pre-mRNA (Herrmann et al., 2007) and plays a key role in cell survival (Okuda et al., 2015). |
| TRMT2A | tRNA methyltransferase 2 homolog A | 27037 | A protein-coding gene of unknown function. However, experimental evidence (Szklarczyk et al., 2017; Wan et al., 2015) suggests chemical interactions with histone-lysine N-methyltransferase (SMYD3), which specifically methylates histone H3 and is involved in transcriptional activation (Van Aller et al., 2012). Expressed across major tissues, with expression varying over the cell cycle (Whitfield et al., 2002). |
| VPS41 | VPS41, HOPS complex subunit | 27072 | Codes for a protein that is involved in the lysosomal system (Harrington, Yacoubian, Slone, Caldwell, & Caldwell, 2012). |
| ZNF607 | Zinc finger protein 607 | 84775 | Codes for a protein that is involved in gene expression and transcription factor activity. |
Note. mRNA = messenger RNA; miRNA = micro RNA; snoRNA = small nucleolar RNA.
Overlapping Loci
We found two loci that demonstrated differential methylation and resided in the putative TFBSs of multiple genes. In both cases, we had gene expression information for only the differentially expressed (DE) loci. First, the region of TAF1C overlaps a potential TFBS for a nearby gene, adenosine deaminase domain containing 2 (ADAD2, RefSeq NM_001145400, Entrez Gene 161931). However, because the transcript probes for ADAD2 did not pass our quality-control filters, we could not compare expression measurements of TAF1C and ADAD2. ADAD2 is expressed in the testes, brain, esophagus, prostate, skin, and spleen (Fagerberg et al., 2014; McKee et al., 2005). While the function of ADAD2 is not well characterized, it is related to the RNA adenosine deaminases family, which is involved in RNA editing (Nishikura, 2016).
Second, we observed a DMR in DEFB124 that overlaps with the potential TFBS for RRAD and GEM like GTPase 1 (REM1, RefSeq NM_014012.5, Entrez Gene 28954). In our differential-expression analysis (Stage 1), the transcript probes for REM1 did not pass our prespecified quality-control filters. Therefore, we were not able to compare the expression measurements of DEFB124 and REM1. Although the functions of REM1 are not well understood, results from in vitro studies suggest that the protein product of REM1 is expressed in skeletal muscle myocytes and inhibits L-calcium channel activity (Finlin, Crump, Satin, & Andres, 2003). These findings support a possible association between alterations in skeletal muscle energy and fatigue severity.
Limitations
Several limitations warrant consideration. While this pilot study was focused on methylation, other factors that contribute to regulation of gene expression (e.g., miRNAs, genetic variations) need to be evaluated. Our sample size was small. The group sizes were unbalanced, which may limit the effectiveness of the DE analysis (Kerr, 2009). Despite the small sample size, we identified statistically significant differences in a number of loci. Finally, because only female patients with breast cancer were included in these analyses, our findings may not generalize to male patients or to patients with other cancer diagnoses.
Conclusion
This pilot study was the first to use a multistaged integrated analysis of gene expression and methylation to identify genes with functional differences using an unbiased approach and then interrogate these findings in a hypothesis-driven analysis of differential methylation in the TFBS regions of the candidate genes. We identified 23 differentially methylated sites and 3 differentially methylated regions that were associated with membership in the very high evening fatigue class. Some of these methylated sites are located in genes that are involved in the hypothesized mechanisms for fatigue. Of note, we identified differential methylation of PAX7 and C2orf88, which both have potential roles in skeletal muscle function. These associations may provide support for the mechanisms that underlie the benefits of exercise for fatigue (Mustian et al., 2017; Wright et al., 2015a). Finally, we found candidate loci involved in core biological processes or multiple overlapping pathways, which may simultaneously impact multiple pathways. These findings are particularly interesting because they suggest that a small number of loci could influence multiple molecular mechanisms. Additional studies are needed to confirm these findings and elucidate additional relationships among methylation, changes in gene expression, and fatigue severity. While this study demonstrated the benefits of integrating multiple types of omics data, future research should include heritable information (i.e., genetics) in the analysis.
Footnotes
Authors’ Note: The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Author Contributions: Elena Flowers, Jon Levine, Steven Paul, Christine Miaskowski, and Kord Kober contributed to conception and design. Elena Flowers, Annesa Flentje, Christine Miaskowski, and Kord Kober contributed to acquisition, analysis, and interpretation; drafted the manuscript; critically revised the manuscript; gave final approval; and agrees to be accountable for all aspects of work ensuring integrity and accuracy. Jon Levine, Adam Olshen, Marilyn Hammer, Steven Paul, and Yvette Conley contributed to analysis and interpretation, critically revised the manuscript, gave final approval, and agrees to be accountable for all aspects of work ensuring integrity and accuracy.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Research reported in this publication obtained grants from the National Cancer Institute (CA134900, CA168960), the National Center for Advancing Translational Sciences of the National Institutes of Health (NIH, TR000143), and a UCSF School of Nursing Intramural Research Award. Dr. Miaskowski is an American Cancer Society Clinical Research Professor.
References
- Ali M. Rincon-Arano H. Zhao W. Rothbart S. B. Tong Q. Parkhurst S. M.…Kutateladze T. G. (2013). Molecular basis for chromatin binding and regulation of MLL5. Proceedings of the National Academy of Sciences USA, 110, 11296–11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames R. S. Sarau H. M. Chambers J. K. Willette R. N. Aiyar N. V. Romanic A. M.…Douglas S. A. (1999). Human urotensin-II is a potent vasoconstrictor and agonist for the orphan receptor GPR14. Nature, 401, 282–286. [DOI] [PubMed] [Google Scholar]
- Amro A., Waldum-Grevbo B., von der Lippe N., Brekke F. B., Miaskowski C., Os I. (2016). Symptom clusters from dialysis to renal transplantation: A five-year longitudinal study. Journal of Pain and Symptom Management, 51, 512–519. [DOI] [PubMed] [Google Scholar]
- Andersen J. S., Wilkinson C. J., Mayor T., Mortensen P., Nigg E. A., Mann M. (2003). Proteomic characterization of the human centrosome by protein correlation profiling. Nature, 426, 570–574. [DOI] [PubMed] [Google Scholar]
- Argiropoulos B., Humphries R. K. (2007). Hox genes in hematopoiesis and leukemogenesis. Oncogene, 26, 6766. [DOI] [PubMed] [Google Scholar]
- Aryee M. J., Jaffe A. E., Corrada-Bravo H., Ladd-Acosta C., Feinberg A. P., Hansen K. D., Irizarry R. A. (2014). Minfi: A flexible and comprehensive bioconductor package for the analysis of infinium DNA methylation microarrays. Bioinformatics, 30, 1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsevick A., Frost M., Zwinderman A., Hall P., Halyard M., Consortium G. (2010). I’m so tired: Biological and genetic mechanisms of cancer-related fatigue. Quality of Life Research, 19, 1419–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger A. M. Mooney K. Alvarez-Perez A. Breitbart W. S. Carpenter K. M. Cella D.…Smith C. (2015). Cancer-related fatigue, version 2.2015. Journal of National Comprehensive Cancer Networks, 13, 1012–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthon A. S., Szarek E., Stratakis C. A. (2015). PRKACA: The catalytic subunit of protein kinase A and adrenocortical tumors. Frontiers in Cellular and Developmental Biology, 3, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezbradica J. S., Medzhitov R. (2009). Integration of cytokine and heterologous receptor signaling pathways. Nature Immunology, 10, 333–339. [DOI] [PubMed] [Google Scholar]
- Bower J. E. (2014). Cancer-related fatigue—Mechanisms, risk factors, and treatments. Nature Reviews Clinical Oncology, 11, 597–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower J. E., Ganz P. A. (2015). Symptoms: Fatigue and cognitive dysfunction. Advances in Experimental Medicine and Biology, 862, 53–75. [DOI] [PubMed] [Google Scholar]
- Brameier M., Herwig A., Reinhardt R., Walter L., Gruber J. (2011). Human box C/D snoRNAs with miRNA like functions: Expanding the range of regulatory RNAs. Nucleic Acids Research, 39, 675–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buescher J. M., Driggers E. M. (2016). Integration of omics: More than the sum of its parts. Cancer & Metabolism, 4, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole S. W. (2013). Nervous system regulation of the cancer genome. Brain, Behavior, and Immunity, 30, S10–S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole S. W., Hawkley L. C., Arevalo J. M., Cacioppo J. T. (2011). Transcript origin analysis identifies antigen-presenting cells as primary targets of socially regulated gene expression in leukocytes. Proceedings of the National Academy of Sciences USA, 108, 3080–3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielsen J. M. Sylvestersen K. B. Bekker-Jensen S. Szklarczyk D. Poulsen J. W. Horn H.…Nielsen M. L. (2011). Mass spectrometric analysis of lysine ubiquitylation reveals promiscuity at site level. Molecular and Cellular Proteomics, 10, M110.003590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantzer R., O’Connor J. C., Freund G. G., Johnson R. W., Kelley K. W. (2008). From inflammation to sickness and depression: When the immune system subjugates the brain. Nature Reviews Neuroscience, 9, 46–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derfuss T. Parikh K. Velhin S. Braun M. Mathey E. Krumbholz M.…Meinl E. (2009). Contactin-2/TAG-1-directed autoimmunity is identified in multiple sclerosis patients and mediates gray matter pathology in animals. Proceedings of the National Academy of Sciences USA, 106, 8302–8307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pietro C. Rapisarda A. Amico V. Bonaiuto C. Viola A. Scalia M.…Purrello M. (2000). Genomic localization of the human genes TAF1A, TAF1B and TAF1C, encoding TAF(I)48, TAF(I)63 and TAF(I)110 subunits of class I general transcription initiation factor SL1. Cytogenetics and Cell Genetics, 89, 133–136. [DOI] [PubMed] [Google Scholar]
- Doong S. H. Dhruva A. Dunn L. B. West C. Paul S. M. Cooper B. A.…Miaskowski C. (2015). Associations between cytokine genes and a symptom cluster of pain, fatigue, sleep disturbance, and depression in patients prior to breast cancer surgery. Biological Research for Nursing, 17, 237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du S., Lawrence E. J., Strzelecki D., Rajput P., Xia S. J., Gottesman D. M., Barr F. G. (2005). Co-expression of alternatively spliced forms of PAX3, PAX7, PAX3-FKHR and PAX7-FKHR with distinct DNA binding and transactivation properties in rhabdomyosarcoma. International Journal of Cancer, 115, 85–92. [DOI] [PubMed] [Google Scholar]
- Ender C. Krek A. Friedlander M. R. Beitzinger M. Weinmann L. Chen W.…Meister G. (2008). A human snoRNA with microRNA-like functions. Molecular Cell, 32, 519–528. [DOI] [PubMed] [Google Scholar]
- Esmon C. T. (2005). The interactions between inflammation and coagulation. British Journal of Haematology, 131, 417–430. [DOI] [PubMed] [Google Scholar]
- Fagerberg L. Hallstrom B. M. Oksvold P. Kampf C. Djureinovic D. Odeberg J.…Uhlen M. (2014). Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Molecular and Cellular Proteomics, 13, 397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falaleeva M. Pages A. Matuszek Z. Hidmi S. Agranat-Tamir L. Korotkov K.…Stamm S. (2016). Dual function of C/D box small nucleolar RNAs in rRNA modification and alternative pre-mRNA splicing. Proceedings of the National Academy of Sciences USA, 113, E1625–E1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlin B. S., Crump S. M., Satin J., Andres D. A. (2003). Regulation of voltage-gated calcium channel activity by the Rem and Rad GTPases. Proceedings of the National Academy of Sciences USA, 100, 14469–14474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher B. S. Paul S. M. Dodd M. J. Schumacher K. West C. Cooper B.…Miaskowski C. (2008). Prevalence, severity, and impact of symptoms on female family caregivers of patients at the initiation of radiation therapy for prostate cancer. Journal of Clinical Oncology, 26, 599–605. [DOI] [PubMed] [Google Scholar]
- Flowers E. Miaskowski C. Conley Y. Hammer M. J. Levine J. Mastick J.…Kober K. M. (2018). Differential expression of genes and differentially perturbed pathways associated with very high evening fatigue in oncology patients receiving chemotherapy. Supportive Care in Cancer, 26, 739–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleeson M., Bishop N. C., Stensel D. J., Lindley M. R., Mastana S. S., Nimmo M. A. (2011). The anti-inflammatory effects of exercise: Mechanisms and implications for the prevention and treatment of disease. Nature Reviews Immunology, 11, 607–615. [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan C., Kamaraj B., Purohit R. (2014). Mutations in microRNA binding sites of CEP genes involved in cancer. Cell Biochemistry and Biophysics, 70, 1933–1942. [DOI] [PubMed] [Google Scholar]
- Harrington A. J., Yacoubian T. A., Slone S. R., Caldwell K. A., Caldwell G. A. (2012). Functional analysis of VPS41-mediated neuroprotection in Caenorhabditis elegans and mammalian models of Parkinson’s disease. Journal of Neuroscience, 32, 2142–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W. Y., Chen G. J., Lai X., Wu F., Tang C. S., Zhang A. H. (2016). Expression levels of urotensin II are associated with endoplasmic reticulum stress in patients with severe preeclampsia. Journal of Human Hypertension, 30, 129–135. [DOI] [PubMed] [Google Scholar]
- Herrmann G. Kais S. Hoffbauer J. Shah-Hosseini K. Bruggenolte N. Schober H.…Schar P. (2007). Conserved interactions of the splicing factor Ntr1/Spp382 with proteins involved in DNA double-strand break repair and telomere metabolism. Nucleic Acids Research, 35, 2321–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg Y., Benjamini Y. (1990). More powerful procedures for multiple significance testing. Statistics in Medicine, 9, 811–818. [DOI] [PubMed] [Google Scholar]
- Hsiao C. P., Reddy S. Y., Chen M. K., Saligan L. N. (2016). Genomic profile of fatigued men receiving localized radiation therapy. Biological Research for Nursing, 18, 281–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hursitoglu M. Tukek T. Cikrikcioglu M. A. Kara O. Kazancioglu R. Ozkan O.…Erek A. (2012). Urotensin II levels in patients with chronic kidney disease and kidney transplants. Upsala Journal of Medical Sciences, 117, 22–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima M., Koike H., Katsuno M., Sobue G. (2011). Polymorphism of transient axonal glycoprotein-1 in chronic inflammatory demyelinating polyneuropathy. Journal of the Peripheral Nervous System, 16, 52–55. [DOI] [PubMed] [Google Scholar]
- Illi J. Miaskowski C. Cooper B. Levine J. D. Dunn L. West C.…Aouizerat B. E. (2012). Association between pro- and anti-inflammatory cytokine genes and a symptom cluster of pain, fatigue, sleep disturbance, and depression. Cytokine, 58, 437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin M. R., Cole S. W. (2011). Reciprocal regulation of the neural and innate immune systems. Nature Reviews Immunology, 11, 625–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe A. E., Murakami P., Lee H., Leek J. T., Fallin M. D., Feinberg A. P., Irizarry R. A. (2012). Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. International Journal of Epidemiology, 41, 200–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P. A. (1999). The DNA methylation paradox. Trends in Genetics, 15, 34–37. [DOI] [PubMed] [Google Scholar]
- Jones P. A. (2012). Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nature Reviews Genetics, 13, 484–492. [DOI] [PubMed] [Google Scholar]
- Joung H. Eom G. H. Choe N. Lee H. M. Ko J. H. Kwon D. H.…Kook H. (2014). Ret finger protein mediates Pax7-induced ubiquitination of MyoD in skeletal muscle atrophy. Cellular Signaling, 26, 2240–2248. [DOI] [PubMed] [Google Scholar]
- Karnofsky D. A., Abelmann W. H., Craver L. V., Burchenal J. H. (1948). The use of nitrogen mustards in the palliative treatment of carcinoma. Cancer, 1, 634–656. [Google Scholar]
- Kass S. U., Landsberger N., Wolffe A. P. (1997). DNA methylation directs a time-dependent repression of transcription initiation. Current Biology, 7, 157–165. [DOI] [PubMed] [Google Scholar]
- Kerr K. F. (2009). Comments on the analysis of unbalanced microarray data. Bioinformatics, 25, 2035–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. H., Lee J., Han J. H., Myung S. C. (2014). Beta-defensin 124 is required for efficient innate immune responses in prostate epithelial RWPE-1 cells. Korean Journal of Urology, 55, 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kober K. M. Cooper B. A. Paul S. M. Dunn L. B. Levine J. D. Wright F.…Miaskowski C. (2016). Subgroups of chemotherapy patients with distinct morning and evening fatigue trajectories. Supportive Care in Cancer, 24, 1473–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kober K. M. Dunn L. Mastick J. Cooper B. Langford D. Melisko M.…Miaskowski C. (2016). Gene expression profiling of evening fatigue in women undergoing chemotherapy for breast cancer. Biological Research for Nursing, 18, 370–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kober K. M., Smoot B., Paul S. M., Cooper B. A., Levine J. D., Miaskowski C. (2016). Polymorphisms in cytokine genes are associated with higher levels of fatigue and lower levels of energy in women after breast cancer surgery. Journal of Pain and Symptom Management, 52, 695–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landmark-Hoyvik H., Reinertsen K. V., Loge J. H., Fossa S. D., Borresen-Dale A. L., Dumeaux V. (2009). Alterations of gene expression in blood cells associated with chronic fatigue in breast cancer survivors. Pharmacogenomics Journal, 9, 333–340. [DOI] [PubMed] [Google Scholar]
- Landmark-Hoyvik H. Reinertsen K. V. Loge J. H. Kristensen V. N. Dumeaux V. Fossa S. D.…Edvardsen H. (2010). The genetics and epigenetics of fatigue. Physical Medicine and Rehabilitation, 2, 456–465. [DOI] [PubMed] [Google Scholar]
- Langham R. G., Kelly D. J. (2013). Urotensin II and the kidney. Current Opinions in Nephrology and Hypertension, 22, 107–112. [DOI] [PubMed] [Google Scholar]
- Lee K. A., Hicks G., Nino-Murcia G. (1991). Validity and reliability of a scale to assess fatigue. Psychiatry Research, 36, 291–298. [DOI] [PubMed] [Google Scholar]
- Liang K. C., Patil A., Nakai K. (2015). Discovery of intermediary genes between pathways using sparse regression. PLoS One, 10, e0137222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew C. C., Ma J., Tang H. C., Zheng R., Dempsey A. A. (2006). The peripheral blood transcriptome dynamically reflects system wide biology: A potential diagnostic tool. Journal of Laboratory and Clinical Medicine, 147, 126–132. [DOI] [PubMed] [Google Scholar]
- Malumbres M. Harlow E. Hunt T. Hunter T. Lahti J. M. Manning G.…Wolgemuth D. J. (2009). Cyclin-dependent kinases: A family portrait. Nature Cell Biology, 11, 1275–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S. T. Sato N. Dhara S. Chang R. Hustinx S. R. Abe T.…Goggins M. (2005). Aberrant methylation of the Human Hedgehog Interacting Protein (HHIP) gene in pancreatic neoplasms. Cancer Biology & Therapy, 4, 728–733. [DOI] [PubMed] [Google Scholar]
- McCann B. Miaskowski C. Koetters T. Baggott C. West C. Levine J. D.…Miaskowski C. (2012). Associations between pro- and anti-inflammatory cytokine genes and breast pain in women prior to breast cancer surgery. Journal of Pain, 13, 425–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee A. E., Minet E., Stern C., Riahi S., Stiles C. D., Silver P. A. (2005). A genome-wide in situ hybridization map of RNA-binding proteins reveals anatomically restricted expression in the developing mouse brain. BMC Developmental Biology, 5, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnell I. W. Ishibashi J. Le Grand F. Punch V. G. Addicks G. C. Greenblatt J. F.…Rudnicki M. A. (2008). Pax7 activates myogenic genes by recruitment of a histone methyltransferase complex. Nature Cell Biology, 10, 77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustian K. M. Alfano C. M. Heckler C. Kleckner A. S. Kleckner I. R. Leach C. R.…Miller S. M. (2017). Comparison of pharmaceutical, psychological, and exercise treatments for cancer-related fatigue: A meta-analysis. Journal of American Medical Association Oncology, 3, 961–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K. (2016). A-to-I editing of coding and non-coding RNAs by ADARs. Nature Reviews Molecular Cell Biology, 17, 83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H. R., An C. H., Yoo N. J., Lee S. H. (2015). Frameshift mutations of TAF1C gene, a core component for transcription by RNA polymerase I, and its regional heterogeneity in gastric and colorectal cancers. Pathology, 47, 101–104. [DOI] [PubMed] [Google Scholar]
- Okuda H., Kanai A., Ito S., Matsui H., Yokoyama A. (2015). AF4 uses the SL1 components of RNAP1 machinery to initiate MLL fusion- and AEP-dependent transcription. Nature Communications, 6, 8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olomu A. B., Corser W. D., Stommel M., Xie Y., Holmes-Rovner M. (2012). Do self-report and medical record comorbidity data predict longitudinal functional capacity and quality of life health outcomes similarly? BMC Health Services Research, 12, 398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong K. L., Lam K. S., Cheung B. M. (2005). Urotensin II: Its function in health and its role in disease. Cardiovascular Drugs and Therapy, 19, 65–75. [DOI] [PubMed] [Google Scholar]
- Pang S. Y. Chan K. H. Mak W. W. Kung M. H. Lee C. N. Tsoi T. H.…Ho S. L. (2012). Single-nucleotide polymorphism of transient axonal glycoprotein-1 and its correlation with clinical features and prognosis in chronic inflammatory demyelinating polyneuropathy. Journal of the Peripheral Nervous System, 17, 72–75. [DOI] [PubMed] [Google Scholar]
- Park S. M., Choi E. Y., Bae M., Choi J. K., Kim Y. J. (2017). A long-range interactive DNA methylation marker panel for the promoters of HOXA9 and HOXA10 predicts survival in breast cancer patients. Clinical Epigenetics, 9, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil A. A., Cai Y., Sang Y., Blecha F., Zhang G. (2005). Cross-species analysis of the mammalian beta-defensin gene family: Presence of syntenic gene clusters and preferential expression in the male reproductive tract. Physiological Genomics, 23, 5–17. [DOI] [PubMed] [Google Scholar]
- Portela A., Esteller M. (2010). Epigenetic modifications and human disease. Nature Biotechnology, 28, 1057–1068. [DOI] [PubMed] [Google Scholar]
- Powell N. D., Mays J. W., Bailey M. T., Hanke M. L., Sheridan J. F. (2011). Immunogenic dendritic cells primed by social defeat enhance adaptive immunity to influenza A virus. Brain, Behavior, and Immunity, 25, 46–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan N., Banks W. A. (2007). Brain-immune communication pathways. Brain, Behavior, and Immunity, 21, 727–735. [DOI] [PubMed] [Google Scholar]
- Razin A., Riggs A. D. (1980). DNA methylation and gene function. Science, 210, 604–610. [DOI] [PubMed] [Google Scholar]
- Rickman D. S. Tyagi R. Zhu X. X. Bobek M. P. Song S. Blaivas M.…Hanash S. M. (2001). The gene for the axonal cell adhesion molecule TAX-1 is amplified and aberrantly expressed in malignant gliomas. Cancer Research, 61, 2162–2168. [PubMed] [Google Scholar]
- Ritchie M. D., Holzinger E. R., Li R., Pendergrass S. A., Kim D. (2015). Methods of integrating data to uncover genotype-phenotype interactions. Nature Reviews Genetics, 16, 85–97. [DOI] [PubMed] [Google Scholar]
- Ritchie M. E., Phipson B., Wu D., Hu Y., Law C. W., Shi W., Smyth G. K. (2015). LIMMA powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research, 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riuzzi F., Sorci G., Sagheddu R., Sidoni A., Alaggio R., Ninfo V., Donato R. (2014). RAGE signaling deficiency in rhabdomyosarcoma cells causes upregulation of PAX7 and uncontrolled proliferation. Journal of Cell Science, 127, 1699–1711. [DOI] [PubMed] [Google Scholar]
- Saligan L. N. Hsiao C. P. Wang D. Wang X. M. St John L. Kaushal A.…Dionne R. A. (2013). Upregulation of alpha-synuclein during localized radiation therapy signals the association of cancer-related fatigue with the activation of inflammatory and neuroprotective pathways. Brain, Behavior, and Immunity, 27, 63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saligan L. N., Kim H. S. (2012). A systematic review of the association between immunogenomic markers and cancer-related fatigue. Brain, Behavior, and Immunity, 26, 830–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saligan L. N. Olson K. Filler K. Larkin D. Cramp F. Yennurajalingam S.…Mustian K. (2015). The biology of cancer-related fatigue: A review of the literature. Supportive Care in Cancer, 23, 2461–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangha O., Stucki G., Liang M. H., Fossel A. H., Katz J. N. (2003). The self-administered comorbidity questionnaire: A new method to assess comorbidity for clinical and health services research. Arthritis and Rheumatology, 49, 156–163. [DOI] [PubMed] [Google Scholar]
- Sarrias M. R., Gronlund J., Padilla O., Madsen J., Holmskov U., Lozano F. (2004). The Scavenger Receptor Cysteine-Rich (SRCR) domain: An ancient and highly conserved protein module of the innate immune system. Critical Reviews in Immunology, 24, 1–37. [DOI] [PubMed] [Google Scholar]
- Satoh K., Hata M., Yokota H. (2002). A novel member of the leucine-rich repeat superfamily induced in rat astrocytes by beta-amyloid. Biochemical and Biophysical Research Communications, 290, 756–762. [DOI] [PubMed] [Google Scholar]
- Schultz M. D. He Y. Whitaker J. W. Hariharan M. Mukamel E. A. Leung D.…Ecker J. R. (2015). Human body epigenome maps reveal noncanonical DNA methylation variation. Nature, 523, 212–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. K. Conneely K. N. Pace T. W. Mister D. Felger J. C. Kilaru V.…Torres M. A. (2014). Epigenetic changes associated with inflammation in breast cancer patients treated with chemotherapy. Brain, Behavior, and Immunity, 38, 227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens K. E., Miaskowski C. A., Levine J. D., Pullinger C. R., Aouizerat B. E. (2013). Epigenetic regulation and measurement of epigenetic changes. Biological Research for Nursing, 15, 373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steptoe A., Hamer M., Chida Y. (2007). The effects of acute psychological stress on circulating inflammatory factors in humans: A review and meta-analysis. Brain, Behavior, and Immunity, 21, 901–912. [DOI] [PubMed] [Google Scholar]
- Stogmann E. Reinthaler E. Eltawil S. El Etribi M. A. Hemeda M. El Nahhas N.…Zimprich A. (2013). Autosomal recessive cortical myoclonic tremor and epilepsy: Association with a mutation in the potassium channel associated gene CNTN2. Brain, 136, 1155–1160. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D. Morris J. H. Cook H. Kuhn M. Wyder S. Simonovic M.…von Mering C. (2017). The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Research, 45, D362–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada M. Kanai F. Tanaka Y. Tateishi K. Ohta M. Asaoka Y.…Omata M. (2008). Down-regulation of hedgehog-interacting protein through genetic and epigenetic alterations in human hepatocellular carcinoma. Clinical Cancer Research, 14, 3768–3776. [DOI] [PubMed] [Google Scholar]
- Taniguchi H. Yamamoto H. Akutsu N. Nosho K. Adachi Y. Imai K.…Shinomura Y. (2007). Transcriptional silencing of hedgehog-interacting protein by CpG hypermethylation and chromatic structure in human gastrointestinal cancer. Journal of Pathology, 213, 131–139. [DOI] [PubMed] [Google Scholar]
- Tomas C., Newton J., Watson S. (2013). A review of hypothalamic-pituitary-adrenal axis function in chronic fatigue syndrome. International Scholarly Research Notices Neuroscience, 2013, 784520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalian S. L., Kaneko S., Gonzales M. I., Bond G. L., Ward Y., Manley J. L. (2001). Identification and functional characterization of neo-poly(A) polymerase, an RNA processing enzyme overexpressed in human tumors. Molecular Cell Biology, 21, 5614–5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi C. M., Patel R. C., Patel C. V. (2008). Differential regulation of HOXA9 expression by nuclear factor kappa B (NF-kappaB) and HOXA9. Gene, 408, 187–195. [DOI] [PubMed] [Google Scholar]
- Van Aller G. S. Reynoird N. Barbash O. Huddleston M. Liu S. Zmoos A. F.…Kruger R. G. (2012). Smyd3 regulates cancer cell phenotypes and catalyzes histone H4 lysine 5 methylation. Epigenetics, 7, 340–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardabasso C., Hasson D., Ratnakumar K., Chung C. Y., Duarte L. F., Bernstein E. (2014). Histone variants: Emerging players in cancer biology. Cellular and Molecular Life Sciences, 71, 379–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner J. R., Busche S., Ge B., Kwan T., Pastinen T., Blanchette M. (2014). The relationship between DNA methylation, genetic and expression inter-individual variation in untransformed human fibroblasts. Genome Biology, 15, R37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan C. Borgeson B. Phanse S. Tu F. Drew K. Clark G.…Emili A. (2015). Panorama of ancient metazoan macromolecular complexes. Nature, 525, 339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Yin J., Miller A. H., Xiao C. (2017). A systematic review of the association between fatigue and genetic polymorphisms. Brain, Behavior, and Immunity, 62, 230–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. S. (2008). Pathophysiology of cancer-related fatigue. Clinical Journal of Oncology Nursing, 12, 11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. S., Woodruff J. F. (2015). Cancer-related and treatment-related fatigue. Gynecological Oncology, 136, 446–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield M. L. Sherlock G. Saldanha A. J. Murray J. I. Ball C. A. Alexander K. E.…Botstein D. (2002). Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Molecular Biology of the Cell, 13, 1977–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood J. P., Bunce M. W., Maroney S. A., Tracy P. B., Camire R. M., Mast A. E. (2013). Tissue factor pathway inhibitor-alpha inhibits prothrombinase during the initiation of blood coagulation. Proceedings of the National Academy of Sciences USA, 110, 17838–17843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright F. D’Eramo Melkus G. Hammer M. Schmidt B. L. Knobf M. T. Paul S. M.…Miaskowski C. (2015. a). Predictors and trajectories of morning fatigue are distinct from evening fatigue. Journal of Pain and Symptom Management, 50, 176–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright F. D’Eramo Melkus G. Hammer M. Schmidt B. L. Knobf M. T. Paul S. M.…Miaskowski C. (2015. b). Trajectories of evening fatigue in oncology outpatients receiving chemotherapy. Journal of Pain and Symptom Management, 50, 163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H. Guan Q. He J. Lin Y. Zhang J. Li H.…He F. (2017). Individualized analysis reveals CpG sites with methylation aberrations in almost all lung adenocarcinoma tissues. Journal of Translational Medicine, 15, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Jiang Y. (2016). RACK1 affects glioma cell growth and differentiation through the CNTN2-mediated RTK/Ras/MAPK pathway. International Journal of Molecular Medicine, 37, 251–257. [DOI] [PubMed] [Google Scholar]
- Yang X. J. (2005). Multisite protein modification and intramolecular signaling. Oncogene, 24, 1653–1662. [DOI] [PubMed] [Google Scholar]
- Yavuzsen T. Davis M. P. Ranganathan V. K. Walsh D. Siemionow V. Kirkova J.…Yue G. H. (2009). Cancer-related fatigue: Central or peripheral? Journal of Pain and Symptom Management, 38, 587–596. [DOI] [PubMed] [Google Scholar]
- Zhang X., Novera W., Zhang Y., Deng L. W. (2017). MLL5 (KMT2E): Structure, function, and clinical relevance. Cellular and Molecular Life Sciences, 74, 2333–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoccali C., Mallamaci F., Benedetto F. A., Tripepi G., Pizzini P., Cutrupi S., Malatino L. (2008). Urotensin II and cardiomyopathy in end-stage renal disease. Hypertension, 51, 326–333. [DOI] [PubMed] [Google Scholar]