Abstract
Background
The methylation status of the O6-methylguanine-DNA methyltransferase (MGMT) promoter has emerged as a favorable independent prognostic and predictive biomarker in glioma. This study aimed to build a radiomics signature based on 18F-fluorodeoxyglucose (FDG) positron emission tomography (PET) for noninvasive measurement of the MGMT promoter methylation status in glioma.
Methods
One hundred and seven pathology-confirmed primary diffuse glioma patients were retrospectively included and randomly assigned to the primary (n = 71) or validation cohort (n = 36). The MGMT promoter methylation status was measured by pyrosequencing. A total of 1561 radiomics features were extracted from the three-dimensional region of interest (ROI) on the standard uptake value (SUV) maps that were generated from the original 18F-FDG PET data. A radiomics signature, a clinical signature and a fusion signature that combined the clinical and radiomics features together were generated. The performance of the three signatures was evaluated by receiver operating characteristic (ROC) curve analysis, and the patient prognosis was stratified based on the MGMT promoter methylation status and the signature with the best performance.
Results
Five radiomics features were selected to construct the radiomics signature, and displayed the best performance with area under the receiver operating characteristic (ROC) curve (AUC) reaching 0.94 and 0.86 in the primary and validation cohorts, respectively, which outweigh the performances of clinical signature and fusion signature. With a median follow-up time of 32.4 months, the radiomics signature stratified the glioma patients into two risk groups with significantly different prognoses (p = 0.04).
Conclusions
18F-FDG-PET-based radiomics is a promising approach for preoperatively evaluating the MGMT promoter methylation status in glioma and predicting the prognosis of glioma patients noninvasively.
Keywords: Radiomics, FDG PET, MGMT promoter methylation, Glioma, Prognosis
Background
Glioma is one of the most malignant central nervous system (CNS) tumors, with an annual incidence of 5.26 per 100,000 individuals [1]. Alkylating agents, such as temozolomide (TMZ), induce guanine-alkyl groups to the DNA and trigger tumor cell death, and have been widely utilized in the treatment of glioma [2, 3]. This methylation damage to DNA can be remedied by a DNA repair enzyme, O6-methylguanine-DNA methyltransferase (MGMT), which can be epigenetically silenced according to its promoter methylation status, making the MGMT promoter methylation status a strong prognostic and predictive biomarker in glioma [3–5] that is routinely measured in the clinical evaluation of glioma patients. However, the MGMT status is mainly assessed based on tumor samples by pyrosequencing, methylation-specific polymerase chain reaction (PCR) or methylation chip analysis [6–8], and these methods are restricted by comparatively long detection periods and high detection costs, the existence of intratumor heterogeneity, and the unattainability of tumor samples through surgery or biopsy. Therefore, noninvasive measurement of the MGMT promoter methylation status has great clinical significance to precisely guide treatment and predict prognosis.
Radiomics, a recently emerging technique for quantifying tumor characteristics with high-throughput radiomics features, allows prediction of the tumor phenotype through mathematic models that are built with selected radiomics features [9]. Current radiomics studies in the glioma field have shown promising results in demonstrating correlations between magnetic resonance imaging (MRI) features and clinical manifestations [10], WHO grades [11], molecular characteristics [12–15], and prognoses [16]. Specifically, Li et al. and Xi et al. predicted the MGMT promoter methylation status in glioblastoma [13, 14] and Wei et al. investigated the imaging features of WHO grade II-IV astrocytoma [15] using radiomics, suggesting the efficacy of using radiomics to predict the MGMT promoter methylation status.
18F-fluorodeoxyglucose (FDG) positron emission tomography (PET) is an alternative molecular imaging technique that has been applied to tumor grading [17], surgical planning [18], recurrence identification [19], and prognosis prediction [20] in glioma. In particular, Choi et al. found that MGMT-methylated WHO grade III and IV gliomas had a significantly higher maximum tumor-to-normal tissue uptake ratio (TNR) and identified a trend of higher mean TNRs in MGMT-methylated gliomas than in MGMT-unmethylated gliomas [21]. In addition, Colavolpe et al. reported a case of multicentric glioblastoma in which the lesion showed higher MGMT expression and intense 18F-FDG uptake [22], suggesting a potential correlation between the 18F-FDG-PET results and the MGMT promoter methylation status in glioma. However, to the best of our knowledge, no studies have focused on predicting the MGMT promoter methylation status using an 18F-FDG-PET-based radiomics approach. Since the MGMT promoter methylation status has been proven to be an independent prognostic and predictive marker in glioma regardless of the WHO classification or chemotherapy regimen [3–5, 23, 24], prediction of the MGMT promoter methylation status using 18F-FDG-PET radiomics may have great clinical potential.
This study retrospectively investigated the radiomics characteristics of gliomas by 18F-FDG-PET to build a conceivable model for predicting the MGMT promoter methylation status and patient prognosis noninvasively.
Methods
Patients
Patients who were pathologically diagnosed with primary glioma and underwent an 18F-FDG-PET/CT examination between March 2010 and May 2018 at Peking Union Medical College Hospital were retrospectively reviewed. The inclusion criteria included the following: 1) adults with histopathologically confirmed WHO grade II-IV primary diffuse glioma without a previous history of CNS tumors; 2) preoperative 18F-FDG PET/CT examination of the brain; 3) sufficient paraffin-embedded tumor tissue for measurement of the MGMT promoter methylation status; and 4) no chemotherapy or radiotherapy delivered before 18F-FDG PET/CT acquisition and surgery. The study design was approved by the Institutional Review Board, and all patients provided informed consent. A total of 107 patients met the inclusion criteria and were randomly assigned to the primary cohort (n = 71) or the validation cohort (n = 36). The patient recruitment pathway is displayed in Fig. 1.
Fig. 1.
Patient recruitment pathway. A total of 168 patients were screened, and 107 patients were included in the current study. Patients were randomly assigned to the primary or validation cohort
MGMT promoter methylation status measurement
The methylation status of the MGMT promoter was measured by pyrosequencing, as previously described [25]. Briefly, DNA was extracted from formalin-fixed, paraffin-embedded tumor samples with a Simplex OUP® FFPE DNA Extraction Kit (TIB, China) and quantified by spectrophotometry with a NanoDrop 2000 system (Thermo Fisher, US). Bisulfate modification was performed with an EpiTect Bisulfite Kit (Qiagen, Germany), and PCR was carried out with a DRR007 Kit (Takara, Japan) using a Verity 96-Well Thermal Cycler (Thermo Fisher, US). Pyrosequencing was subsequently performed in 10 CpG island regions within the MGMT promoter using the PyroMark Q96 system (Qiagen, Germany). Gliomas were defined as having a methylated MGMT promoter if the average methylation rate of the CpG regions was greater than or equal to 8%; gliomas were defined as having an unmethylated MGMT promoter if the average methylation rate was less than 8% [25].
18F-FDG-PET/CT data acquisition
18F-FDG was produced using an RDS-111 Cyclotron (CTI, US). A dose of 5.55 MBq (0.15 mCi) 18F-FDG per kilogram of body weight was intravenously administered after the patient had fasted at least 4 h and their blood glucose level was determined not to exceed the normal limit (6.4 mM). The patient underwent 18F-FDG-PET/CT on a Biograph 64 TruePoint TrueV PET/CT system (Siemens Medical Solutions, Germany) after a 40–60 min time lag in standardized conditions (quiet, dimly lit room with patient’s eyes closed), and acquired 148 axial slides with an interslice spacing of 3 mm.
Tumor segmentation
The three-dimensional region of interest (ROI) was segmented by two experienced neurosurgeons for the 18F-FDG-PET data on the merged PET/CT images using ITK-SNAP software (http://www.itksnap.org/pmwiki/pmwiki.php), with patients’ contrast-enhanced T1-weighted images (for contrast-enhanced tumors) and T2-weighted fluid attenuated inversion recovery (FLAIR) images (for non-contrast-enhanced tumors) as anatomical reference. The ROIs were subsequently reviewed by a senior nuclear medical scientist blinded to the patients’ information. If there was a discrepancy of less than 5% between the ROIs placed by the two neurosurgeons, the final ROI was defined as the region of overlap, and if the discrepancy was greater than or equal to 5%, the nuclear medical scientist made the final decision.
Radiomics feature extraction and selection
Standard uptake value (SUV) maps were generated from the original 18F-FDG-PET DICOM data using MATLAB version R2015b (Math Works, US). A total of 1561 radiomics features, including 13 shape and size features, 18 first-order features, 68 texture features, 688 wavelet features and 680 further filtered (logarithm, square, exponential, gradient, square root, lbp-2D, lbp-3D) features were extracted using PyRadiomics (https://github.com/Radiomics/pyradiomics) [26]. The radiomics features were normalized to the interval of 0 to 1.
The radiomics features were reduced and selected through sequential application of the Wilcoxon rank-sum test and multivariate linear logistic regression with the L1 penalty.
Clinical feature evaluation
Five clinical features, respectively, age, sex, metabolic pattern (cystic or solid), SUVmax and SUVmean, were also evaluated. Cystic metabolic tumor was defined as a lesion with visible marginal 18F-FDG update but significant low central radioactivity, and solid metabolic tumor was defined as a lesion without a significant low metabolic necrosis or cysts inside the ROI [27, 28]. SUVmax and SUVmean were defined as radiomics feature ‘First order_Maximum’ and ‘First order_Mean’ that extracted from the ROI.
Signature construction, validation and evaluation
Three predictive signatures, namely, a radiomics signature, clinical signature, and fusion signature, were constructed. The radiomics signature was generated with the radiomics features that were previously selected with a support vector machine (SVM). The clinical signature was generated with 5 clinical features using the logistic regression after selection by the Akaike information criterion (AIC). The selected clinical features and selected radiomics features were combined to generate the fusion signature using the logistic regression. The 3 signatures were independently validated in the validation cohort.
The signatures were evaluated in terms of the area under the receiver operating characteristic (ROC) curve (AUC), accuracy, sensitivity, specificity, and positive and negative predictive values. Decision curve analysis was applied to reflect the clinical utility of the model [29, 30], and the Delong test was utilized to evaluate the difference in the performance of the models.
Prognosis analysis
The overall survival (OS) of patients was evaluated up to May 31, 2018. Kaplan-Meier curves were plotted based on the MGMT promoter methylation status and the signature with the best performance in stratifying the OS of patients. The log-rank test was utilized to determine differences in survival between the groups.
Statistical analysis
Statistical analysis was performed with SPSS Statistics software, version 18.0 (Chicago, US) and R software, version 3.4.1 (https://www.r-project.org/). Statistically significant differences were defined by a two-tailed threshold of p < 0.05.
Results
Clinical characteristics
The clinical characteristics of patients in the primary and validation cohorts are summarized in Table 1. The MGMT methylation rate in the primary and validation cohorts was 54.9 and 55.6%, respectively. There were no significant interclass differences in age, sex, body weight, metabolic pattern, WHO grade, SUVmax or SUVmean among the included patients (p = 0.11–0.84). However, tumors with MGMT promoter methylation tend to have a higher rate for cystic metabolic pattern, and the difference of metabolic pattern for MGMT methylated and MGMT unmethylated patients reached statistical significance in the validation cohort (p = 0.20 and 0.02 in the primary and validation cohort, respectively).
Table 1.
Patients’ Characteristics of Primary and Validation Cohorts
| Characteristics | Primary cohort (n = 71) | Validation cohort (n = 36) | P | ||||
|---|---|---|---|---|---|---|---|
| Methylated (n = 39) | Unmethylated (n = 32) | P | Methylated (n = 20) | Unmethylated (n = 16) | P | ||
| Age (mean ± SD, years) | 50.72 ± 14.01 | 50.50 ± 14.82 | 0.95 | 46.70 ± 12.45 | 58.33 ± 11.95 | 0.08 | 0.65 |
| Gender | 0.97 | 0.45 | 0.84 | ||||
| Male | 23 | 19 | 10 | 10 | |||
| Female | 16 | 13 | 10 | 6 | |||
| Weight (mean ± SD, kg) | 67.24 ± 12.36 | 64.20 ± 10.11 | 0.27 | 69.05 ± 14.74 | 66.28 ± 9.56 | 0.50 | 0.45 |
| Metabolic Pattern | 0.20 | 0.02 | 0.34 | ||||
| Cystic | 23 | 14 | 15 | 6 | |||
| Solid | 16 | 18 | 5 | 10 | |||
| WHO Grading | 0.05 | 0.08 | 0.11 | ||||
| Low Grade Glioma | 13 | 2 | 11 | 2 | |||
| High Grade Glioma | 26 | 30 | 9 | 14 | |||
| SUVmax | 9.18 ± 4.05 | 10.51 ± 4.45 | 0.20 | 9.89 ± 4.11 | 7.84 ± 3.28 | 0.11 | 0.33 |
| SUVmean | 4.00 ± 2.10 | 4.60 ± 1.86 | 0.22 | 4.31 ± 2.01 | 3.40 ± 1.59 | 0.14 | 0.36 |
Abbreviations: SD Standard deviation, WHO World Health Organization, SUV Standard uptake value
Note: Chi-Square or Fisher Exact tests, as appropriate, were used to compare the differences in categorical variables, while the independent sample t-test was used to compare the differences in age
Feature selection and signature construction
Among the 1561 extracted radiomics features, 1543 redundant features were reduced through the Wilcoxon rank-sum test, and 5 final features were selected by logistic regression with the L1 penalty to build the radiomics signature. Only the metabolic pattern was selected by the AIC to build the clinical signature, and the fusion signature was built based on the radiomics signature and metabolic pattern. The selected radiomics features are shown in Table 2.
Table 2.
Selected Features in the Radiomics Signature
| Feature Name | Matrix | Filter |
|---|---|---|
| Skewness | First Order | Logarithm |
| 90 Percentile | First Order | Logarithm |
| Median | First Order | Logarithm |
| Joint Average | GLCM | Logarithm |
| Maximum 2D Diameter Slice | Shape | Original |
Abbreviations: GLCM, Gray-Level Co-occurrence Matrix; 2D, two-dimensional
Diagnostic performance of the three signatures
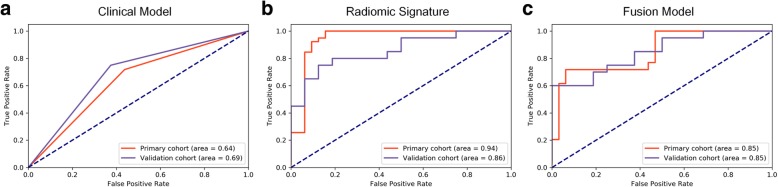
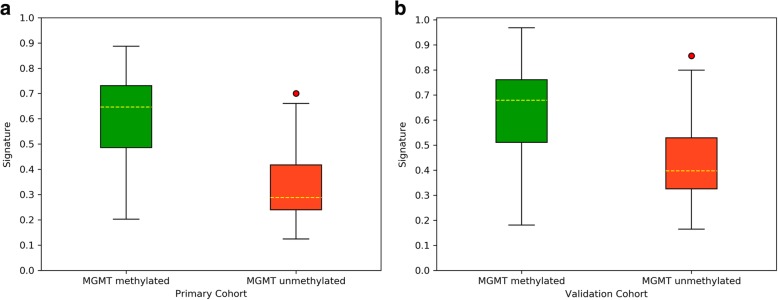
The radiomics signature performed the best among the three signatures in predicting the MGMT promoter methylation status, reaching an AUC of 0.94 in the primary cohort and 0.86 in the validation cohort. The clinical signature demonstrated a moderate predictive value and reached an AUC of 0.64 and 0.69 in the primary and validation cohorts, respectively. The fusion signature performed better than the clinical signature but poorer than the radiomics signature, with an AUC of 0.85 both in the primary and validation cohorts. The Delong test demonstrated that the radiomics signature performed significantly better than the clinical and fusion signatures in the primary cohort (p < 0.0001 and p = 0.036, respectively), but the differences in the validation cohort were not significant (p = 0.115 and 0.900, respectively) due to the limited number of patients. The decision curve reflecting the benefit of the radiomics signature showed a net benefit outweighing both schemes at any threshold probability in the primary cohort. The performance of the radiomics, clinical and fusion signatures is summarized in Table 3. The ROC curves are displayed in Fig. 2, and the box plots are demonstrated in Fig. 3. The decision curve is shown in Fig. 4 (a).
Table 3.
The Performances of the Three Predictive Models
| Models | AUC (95%CI) | ACC (95%CI) | SEN (95%CI) | SPE (95%CI) | PPV (95%CI) | NPV (95% CI) |
|---|---|---|---|---|---|---|
| Radiomics model | ||||||
| Primary cohort | 0.94 (0.93, 0.96) | 91.3% (89.8, 93.3) | 94.9% (93.1, 96.6) | 87.5% (84.4, 90.7) | 90.2% (87.8, 92.8) | 93.3% (91.1, 95.6) |
| Validation cohort | 0.86 (0.83, 0.88) | 77.8% (75.2, 80.3) | 75.0% (71.5, 78.6) | 81.3% (77.5, 84.9) | 83.3% (80.0, 86.7) | 72.2% (68.2, 76.1) |
| Clinical model | ||||||
| Primary cohort | 0.64 (0.61, 0.67) | 64.8% (61.9, 67.9) | 71.8% (68.1, 75.7) | 56.3% (51.6, 61.1) | 66.7% (62.9, 70.6) | 62.1% (57.3, 67.2) |
| Validation cohort | 0.69 (0.66, 0.72) | 66.4% (66.6, 72.3) | 75.0% (71.3, 78.5) | 62.5% (58.1, 67.0) | 71.4% (67.9, 75.1) | 66.7% (61.9, 71.1) |
| Fusion model | ||||||
| Primary cohort | 0.85 (0.83, 0.87) | 64.8% (62.0, 67.7) | 71.8% (68.1, 75.5) | 56.3% (51.6, 60.9) | 66.7% (63.0, 70.4) | 62.1% (57.5, 66.7) |
| Validation cohort | 0.85 (0.82, 0.87) | 72.7% (78.1, 82.9) | 80.0% (76.6, 83.5) | 62.5% (58.0, 67.0) | 72.7% (69.3, 76.3) | 71.4% (66.7, 76.1) |
Abbreviations: CI Confidence interval, AUC Area under receiver-operating characteristic curve, ACC Accuracy, SEN Sensitivity, SPE Specificity, PPV Positive predictive value, NPV Negative predictive value
Fig. 2.
Receiver operating characteristic (ROC) curves of the prediction models. ROC curve of the clinical (a), radiomics (b), and fusion (c) predictive models in both the primary and validation cohorts
Fig. 3.
Box plots of the radiomics signature. Box plots of the radiomics signature in the primary (a) and validation cohorts (b). The signature displayed a higher value for the patients with MGMT-methylated tumors in both cohorts
Fig. 4.
Clinical utility of the radiomics signature. The decision curve of the radiomics signature in the primary cohort (a). The x axis represented the threshold probability, where the expected benefit of treatment as MGMT methylated is equal to the expected benefit of treatment as MGMT unmethylated (the threshold probability varies from patient to patient). The y axis indicated the net benefit for the treatment which considered the benefit of true positive and loss of false positive, and higher net benefit value indicates better model. The net benefit of the radiomics signature is further compared with the default strategies, which we treat all patients as MGMT methylated (red line) or as MGMT unmethylated (black line). The current prediction model outweigh both default strategies at any threshold probability, suggesting the clinical value of our model at all circumstances. Kaplan-Meier curves revealed the prognosis-based groups stratified by the MGMT promoter methylation status and the radiomics signature (b)
Prognostic performance of the Radiomics signature
Among the 107 included patients, 100 patients who were known to survive to the closing date or to have an exact time of death were included in the prognosis analysis, and the median follow-up time is 32.4 months. Both the MGMT promoter methylation status and the radiomics signature stratified the glioma patients into a high-risk group and a low-risk group (p = 0.0002 and 0.04, respectively), and the differences within the high- and low-risk groups did not reach statistical significance. The Kaplan-Meier curves are shown in Fig. 4 (b).
Discussion
In this study, 18F-FDG-PET radiomics features were extracted, selected and analyzed, and three prediction signatures, respectively, and a radiomics signature, a clinical signature, and a fusion signature, were built to predict the MGMT promoter methylation status. The radiomics signature displayed the best performance, with an accuracy of 91.3% and an AUC of 0.94 in the primary cohort, and an accuracy of 77.8% and an AUC of 0.86 in the validation cohort, respectively. The clinical value of the radiomics signature was further demonstrated by the prognosis analysis. These results suggest that 18F-FDG-PET-based radiomics is a promising method for predicting the MGMT promoter methylation status and prognosis noninvasively, demonstrating strong potential for clinical application.
Previous studies on radiological evaluation of the MGMT promoter methylation status have mainly focused on the visual features, quantitative parameters or high-throughput radiomics features [13–15, 31–34] of gliomas (mostly glioblastomas) based on multimodal MRI and have reported accuracies ranging from 0.58–0.89 and AUCs ranging from 0.75–0.92 (without distinguishing training and validation data). Our prediction model demonstrated comparable accuracy and AUC values, suggesting the capability of 18F-FDG-PET radiomics to predict the MGMT promoter methylation status. However, most previous studies on imaging-based prediction of the MGMT promoter methylation status have mainly focused on glioblastomas, and limited studies have included less aggressive gliomas (e.g., lower grade gliomas, such as WHO grade II and III gliomas), in which the MGMT promoter status also has prognostic and predictive value [3–5, 23, 24]. Although there may be discriminative imaging characteristics, our 18F-FDG-PET-based radiomics signature can predict the MGMT promoter methylation status regardless of the WHO grade (e.g., in lower grade gliomas and glioblastomas) or pathological information (e.g., in astrocytomas and oligodendrogliomas), suggesting the capability of noninvasive prediction without previous knowledge based on tumor samples.
Unlike MRI, which displays the structural characteristics of tumors, PET is a highly sensitive molecular imaging technique that reflects the altered tumor metabolism that is ubiquitous among cancer cells. Malignant brain tumors usually exhibit an altered glucose metabolism, in which glucose is converted to pyruvate and further into lactate instead of entering mitochondria and the citric acid cycle [35]. 18F-FDG, a glucose analogue, can be taken up by cells but not further catabolized through glycolysis, making it a reliable radiotracer for measuring cancer cell metabolism. Considering the relationship between the glucose metabolism and oncogenic reprogramming [36], radiogenomic analysis based on 18F-FDG-PET may reflect certain molecular processes through imaging data, which is the theoretical basis of our study. However, compared with anatomical imaging modalities (e.g., CT and MRI), 18F-FDG-PET has a relatively low spatial resolution, which limits the stability and accuracy of certain features, especially in lesions with a relatively small volume [37].
Feature selection is a core step in radiomics studies since most features have little relevance to the MGMT promoter methylation status and may overwhelm the distinguishable features if they cannot be effectively reduced. The number of final selected features also needs to be balanced according to the patient cohort size because the addition of relevant features may increase performance in the primary cohort but may also result in overfitting of the radiomics signature. In our study, the Wilcoxon rank-sum test removed 1543 of the 1561 radiomics features that were irrelevant to the MGMT promoter methylation status, and logistic regression with the L1 penalty diluted the weights, allowing selection of the final 5 radiomics features to construct the radiomics signature. Although the selected radiomics features are not visually available to nuclear medicine physicians (though they are mathematically easy to comprehend), the radiomics signature did provide additional assistance to physicians in the noninvasive molecular diagnosis of glioma (Fig. 5).
Fig. 5.
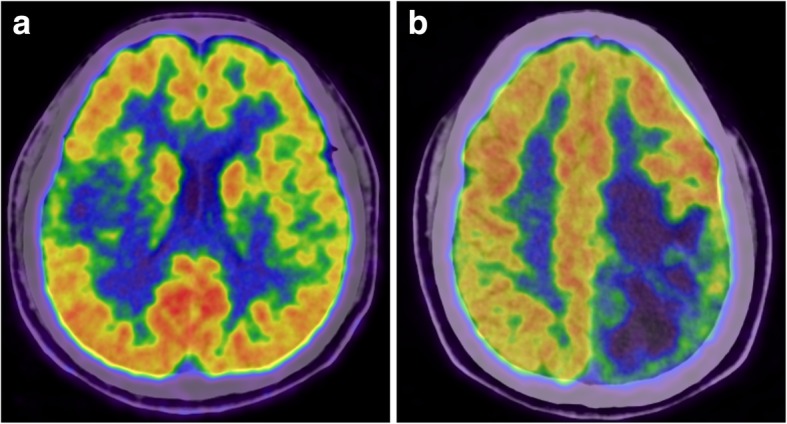
Examples of using the radiomics signature to evaluate the MGMT promoter methylation status noninvasively. A 37/M was histopathologically diagnosed with anaplastic astrocytoma with a methylated MGMT promoter (a), and a 44/M was histopathologically diagnosed with anaplastic astrocytoma with an unmethylated MGMT promoter (b). Determination of the MGMT promoter methylation status is difficult based on clinical and visually assessed imaging characteristics, but the radiomics signature demonstrated values of 0.84 (a) and 0.27 (b) in these two patients and successfully predicted their MGMT status (the cutoff value of the radiomics signature was 0.50)
Three signatures were built in our study to predict the MGMT promoter methylation status. In addition to the radiomics signature, the clinical signature was built with visualized imaging features (e.g., metabolic pattern), and the fusion signature was built with the 5 selected radiomics features and the metabolic pattern. However, the radiomics signature demonstrated the best performance and outweighed the clinical signature in both the primary and validation cohorts, suggesting that the selected radiomics features are more reliable than the clinically assessed imaging features in differentiating tumors based on the MGMT promotor methylation status. Objective clinical features (e.g., age and sex) and the most frequently used quantitative imaging parameters (e.g., SUVmax and SUVmean) were excluded by the AIC when building the clinical signature, although some of these features are the only references for physicians in noninvasively evaluating the MGMT promoter methylation status without radiomics. Moreover, the addition of the clinical feature (i.e., metabolic pattern) to the set of radiomics features decreased the AUC of the prediction model, indicating a potential disturbance to the signature with the addition of features with less relevance. Thus, clinical features may not be integrated into the noninvasive radiomics evaluation of the MGMT promoter methylation status.
The MGMT promoter has proven to be a strong prognostic biomarker in glioma. The retrospective investigation of the EORTC 26981/22981 trial demonstrated that the MGMT promoter methylation status is a favorable independent prognostic biomarker in glioblastoma [5, 6]; the NOA-04 trial and the EORTC 26951/26053/22054 trial demonstrated its prognostic value in anaplastic glioma regardless of the histopathological classification and treatment strategy [3, 23, 24]. The recently reported RTOG 0424 trial also suggests that the MGMT promotor methylation status can predict the prognosis of patients with low-grade glioma treated with radiotherapy and TMZ [4]. In accordance with previous evidences, patients with MGMT promoter methylation displayed significantly longer OS in our research. The clinical use of a radiomics signature can be further supported if the signature not only detects the MGMT promotor methylation status noninvasively but also predicts the patients’ prognosis before treatment. In our study, the radiomics signature could stratify patients into two significantly different groups based on the prognosis, suggesting the feasibility of using the radiomics signature to predict prognosis in addition to distinguishing molecular features. Moreover, the differences between the MGMT promotor methylation status-predicted and radiomics signature-predicted prognosis within each risk group were nonsignificant, even with population discrepancies within each risk group (e.g., a 20% difference in the composition of the low-risk group), indicating that the radiomics signature can serve to evaluate the prognosis aside from the MGMT promoter methylation status. Despite the results from the EORTC 26981/22981/26053/22054 and NOA-04 trials suggesting that the MGMT promoter methylation status is a predictive biomarker that can be used to evaluate whether a patient will benefit from TMZ [3, 5, 6, 24], chemotherapy strategies were not integrated into the prognosis analysis due to their diversity and the retrospective nature of this study.
The current study has several limitations. First, this was a single-center, retrospective study with a limited sample size, and the validation cohort is particularly restricted. Further prospective, multicenter studies with large patient cohorts may be essential for improving the generality and performance of the prediction model. Second, there may be a selection bias of the included patients since 18F-FDG-PET examination was not mandatorily performed. The necessity of differential diagnosis of the intracranial lesion or the evaluation of the extracranial situation were the major consideration to suggest an 18F-FDG-PET scan. Third, the radiomics model was constructed without subclassification of the metabolic pattern (i.e., solid or cystic) and therefore may not include distinguishable features for determining the MGMT promoter methylation status in each subclassification. Fourth, more than half of the patients did not reach the endpoint of the prognosis analysis, which may have introduced bias to the prognosis data. Further studies with long-term follow-up periods may be needed to eliminate such imbalances. Finally, in addition to 18F-FDG-PET data, multimodality imaging data (e.g., data from MRI and PET with alternative tracers) may be further integrated into the radiomics model for predicting the MGMT promoter methylation status in glioma.
Conclusions
18F-FDG-PET-based radiomics is a promising method for preoperatively evaluating the MGMT promoter methylation status in glioma and has the potential to guide the treatment and predict the prognosis of glioma patients noninvasively.
Acknowledgements
The authors thank American Journal Experts for providing language help. The authors thank the Clinical Biobank of Peking Union Medical College Hospital for the storage and supply of tumor samples.
Abbreviations
- AIC
Akaike information criterion
- AUC
Area under the ROC curve
- CNS
Central nervous system
- FDG
Fluorodeoxyglucose
- MGMT
O6-methylguanine-DNA methyltransferase
- MRI
Magnetic resonance imaging
- OS
overall survival
- PCR
Polymerase chain reaction
- PET
Positron emission tomography
- RFE
Recursive feature elimination
- ROC
Receiver operating characteristic
- ROI
Region of interest
- SVM
Support vector machine
- TMZ
Temozolomide
- TNR
Tumor-to-normal tissue uptake ratio
Authors’ contributions
Conception: Xin Cheng, Jie Tian, Wenbin Ma; design of the work: Xin Cheng, Jie Tian, Zhenyu Liu, Yu Wang; acquisition of data: Xin Cheng, Ziren Kong, Delin Liu, Xuying Qin; analysis of data: Ziren Kong, Yusong Lin, Chendan Jiang, Longfei Li, Zehua Liu; interpretation of data: Ziren Kong, Chendan Jiang, Yuekun Wang, Congxin Dai; drafted the work: Ziren Kong, Yusong Lin, Chendan Jiang, Longfei Li, Zehua Liu, Yuekun Wang, Congxin Dai, Delin Liu, Xuying Qin; substantively revised the work: Xin Cheng, Jie Tian, Wenbin Ma, Zhenyu Liu, Yu Wang. All authors have approved the submitted version, and agreed both to be personally accountable for the author’s own contributions and to ensure that questions related to the accuracy or integrity of any part of the work.
Funding
This work was supported by the Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences [grant numbers 2016-I2M-2-001, 2018-I2M-3-001], the Fundamental Research Funds for the Central Universities [grant number 3332018029], the National Natural Science Foundation of China [grant numbers 81772009, 81772012], the Scientific and Technological Research Project of Henan Province [grant number 182102310162], the Beijing Natural Science Foundation [grant number 7182109], the Chinese Academy of Sciences [grant numbers GJJSTD20170004 and QYZDJ-SSW-JSC005], and the Youth Innovation Promotion Association of Chinese Academy of Sciences [grant number 2019136].
Availability of data and materials
The datasets used and analyzed in the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The study design was approved by the Institutional Review Board, and all patients provided informed consent.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ziren Kong, Yusong Lin and Chendan Jiang are Co-first author.
Ziren Kong, Yusong Lin and Chendan Jiang contributed equally to this work.
Contributor Information
Ziren Kong, Email: kongziren@pumc.edu.cn.
Yusong Lin, Email: yslin@ha.edu.cn.
Chendan Jiang, Email: jiangchendan@gmail.com.
Longfei Li, Email: lfli@ha.edu.cn.
Zehua Liu, Email: zhliu2@ha.edu.cn.
Yuekun Wang, Email: skafear@yeah.net.
Congxin Dai, Email: daicongxin@163.com.
Delin Liu, Email: liudl@student.pumc.edu.cn.
Xuying Qin, Email: qxy5758@163.com.
Yu Wang, Email: ywang@pumch.cn.
Zhenyu Liu, Email: zhenyu.liu@ia.ac.cn.
Xin Cheng, Phone: +8618611674487, Email: pumch_chengxin@126.com.
Jie Tian, Phone: +8601082618465, Email: jie.tian@ia.ac.cn.
Wenbin Ma, Phone: +8613701364566, Email: mawb2001@hotmail.com.
References
- 1.Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. Jama. 2013;310(17):1842–1850. doi: 10.1001/jama.2013.280319. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 3.Wick W, Hartmann C, Engel C, et al. NOA-04 randomized phase III trial of sequential radiochemotherapy of anaplastic glioma with procarbazine, lomustine, and vincristine or temozolomide. J Clin Oncol. 2009;27(35):5874–5880. doi: 10.1200/JCO.2009.23.6497. [DOI] [PubMed] [Google Scholar]
- 4.Bell EH, Zhang P, Fisher BJ, et al. Association of MGMT promoter methylation status with survival outcomes in patients with high-risk glioma treated with radiotherapy and Temozolomide: an analysis from the NRG oncology/RTOG 0424 trial. JAMA Oncol. 2018. [DOI] [PMC free article] [PubMed]
- 5.Gorlia T, van den Bent MJ, Hegi ME, et al. Nomograms for predicting survival of patients with newly diagnosed glioblastoma: prognostic factor analysis of EORTC and NCIC trial 26981-22981/CE.3. Lancet Oncol. 2008;9(1):29–38. doi: 10.1016/S1470-2045(07)70384-4. [DOI] [PubMed] [Google Scholar]
- 6.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 7.Quillien V, Lavenu A, Sanson M, et al. Outcome-based determination of optimal pyrosequencing assay for MGMT methylation detection in glioblastoma patients. J Neuro-Oncol. 2014;116(3):487–496. doi: 10.1007/s11060-013-1332-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bady P, Sciuscio D, Diserens AC, et al. MGMT methylation analysis of glioblastoma on the Infinium methylation BeadChip identifies two distinct CpG regions associated with gene silencing and outcome, yielding a prediction model for comparisons across datasets, tumor grades, and CIMP-status. Acta Neuropathol. 2012;124(4):547–560. doi: 10.1007/s00401-012-1016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14(12):749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 10.Liu Z, Wang Y, Liu X, et al. Radiomics analysis allows for precise prediction of epilepsy in patients with low-grade gliomas. Neuroimage Clin. 2018;19:271–278. doi: 10.1016/j.nicl.2018.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hwan-Ho C, Hyunjin P. Classification of low-grade and high-grade glioma using multi-modal image radiomics features. Conf Proc IEEE Eng Med Biol Soc. 2017;2017:3081–3084. doi: 10.1109/EMBC.2017.8037508. [DOI] [PubMed] [Google Scholar]
- 12.Han Y, Xie Z, Zang Y, et al. Non-invasive genotype prediction of chromosome 1p/19q co-deletion by development and validation of an MRI-based radiomics signature in lower-grade gliomas. J Neuro-Oncol. 2018;140(2):297–306. doi: 10.1007/s11060-018-2953-y. [DOI] [PubMed] [Google Scholar]
- 13.Li ZC, Bai H, Sun Q, et al. Multiregional radiomics features from multiparametric MRI for prediction of MGMT methylation status in glioblastoma multiforme: a multicentre study. Eur Radiol. 2018;28(9):3640–3650. doi: 10.1007/s00330-017-5302-1. [DOI] [PubMed] [Google Scholar]
- 14.Xi YB, Guo F, Xu ZL, et al. Radiomics signature: a potential biomarker for the prediction of MGMT promoter methylation in glioblastoma. J Magn Reson Imaging. 2018;47(5):1380–1387. doi: 10.1002/jmri.25860. [DOI] [PubMed] [Google Scholar]
- 15.Wei J, Yang G. Hao X, et al. Eur Radiol: A multi-sequence and habitat-based MRI radiomics signature for preoperative prediction of MGMT promoter methylation in astrocytomas with prognostic implication; 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kickingereder P, Neuberger U, Bonekamp D, et al. Radiomic subtyping improves disease stratification beyond key molecular, clinical, and standard imaging characteristics in patients with glioblastoma. Neuro-Oncology. 2018;20(6):848–857. doi: 10.1093/neuonc/nox188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoon JH, Kim JH, Kang WJ, et al. Grading of cerebral glioma with multiparametric MR imaging and 18F-FDG-PET: concordance and accuracy. Eur Radiol. 2014;24(2):380–389. doi: 10.1007/s00330-013-3019-3. [DOI] [PubMed] [Google Scholar]
- 18.Ideguchi M, Nishizaki T, Ikeda N, et al. A surgical strategy using a fusion image constructed from 11C-methionine PET, 18F-FDG-PET and MRI for glioma with no or minimum contrast enhancement. J Neuro-Oncol. 2018. [DOI] [PubMed]
- 19.Dankbaar JW, Snijders TJ, Robe PA, et al. The use of 18F-FDG PET to differentiate progressive disease from treatment induced necrosis in high grade glioma. J Neuro-Oncol. 2015;125(1):167–175. doi: 10.1007/s11060-015-1883-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Santra A, Kumar R, Sharma P, Bal C, Julka PK, Malhotra A. F-18 FDG PET-CT for predicting survival in patients with recurrent glioma: a prospective study. Neuroradiology. 2011;53(12):1017–1024. doi: 10.1007/s00234-011-0898-3. [DOI] [PubMed] [Google Scholar]
- 21.Choi H, Bang JI, Cheon GJ, et al. (1)(8)F-fluorodeoxyglucose and (1)(1)C-methionine positron emission tomography in relation to methyl-guanine methyltransferase promoter methylation in high-grade gliomas. Nucl Med Commun. 2015;36(3):211–218. doi: 10.1097/MNM.0000000000000236. [DOI] [PubMed] [Google Scholar]
- 22.Colavolpe C, Guedj E, Metellus P, et al. FDG-PET to predict different patterns of progression in multicentric glioblastoma: a case report. J Neuro-Oncol. 2008;90(1):47–51. doi: 10.1007/s11060-008-9629-y. [DOI] [PubMed] [Google Scholar]
- 23.van den Bent MJ, Dubbink HJ, Sanson M, et al. MGMT promoter methylation is prognostic but not predictive for outcome to adjuvant PCV chemotherapy in anaplastic oligodendroglial tumors: a report from EORTC brain tumor group study 26951. J Clin Oncol. 2009;27(35):5881–5886. doi: 10.1200/JCO.2009.24.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van den Bent MJ, Baumert B, Erridge SC, et al. Interim results from the CATNON trial (EORTC study 26053-22054) of treatment with concurrent and adjuvant temozolomide for 1p/19q non-co-deleted anaplastic glioma: a phase 3, randomised, open-label intergroup study. Lancet. 2017;390(10103):1645–1653. doi: 10.1016/S0140-6736(17)31442-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reifenberger G, Hentschel B, Felsberg J, et al. Predictive impact of MGMT promoter methylation in glioblastoma of the elderly. Int J Cancer. 2012;131(6):1342–1350. doi: 10.1002/ijc.27385. [DOI] [PubMed] [Google Scholar]
- 26.van Griethuysen JJM, Fedorov A, Parmar C, et al. Computational Radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77(21):e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suh HB, Choi YS, Bae S, et al. Primary central nervous system lymphoma and atypical glioblastoma: differentiation using radiomics approach. Eur Radiol. 2018;28(9):3832–3839. doi: 10.1007/s00330-018-5368-4. [DOI] [PubMed] [Google Scholar]
- 28.Kong Z, Jiang C, Zhu R, et al. 18F-FDG-PET-based radiomics features to distinguish primary central nervous system lymphoma from glioblastoma. NeuroImage: Clinical. 2019;101912. [DOI] [PMC free article] [PubMed]
- 29.Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Mak. 2006;26(6):565–574. doi: 10.1177/0272989X06295361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Z, Rousson V, Lee WC, et al. Decision curve analysis: a technical note. Ann Transl Med. 2018;6(15):308. doi: 10.21037/atm.2018.07.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Drabycz S, Roldan G, de Robles P, et al. An analysis of image texture, tumor location, and MGMT promoter methylation in glioblastoma using magnetic resonance imaging. Neuroimage. 2010;49(2):1398–1405. doi: 10.1016/j.neuroimage.2009.09.049. [DOI] [PubMed] [Google Scholar]
- 32.Carrillo JA, Lai A, Nghiemphu PL, et al. Relationship between tumor enhancement, edema, IDH1 mutational status, MGMT promoter methylation, and survival in glioblastoma. AJNR Am J Neuroradiol. 2012;33(7):1349–1355. doi: 10.3174/ajnr.A2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Romano A, Calabria LF, Tavanti F, et al. Apparent diffusion coefficient obtained by magnetic resonance imaging as a prognostic marker in glioblastomas: correlation with MGMT promoter methylation status. Eur Radiol. 2013;23(2):513–520. doi: 10.1007/s00330-012-2601-4. [DOI] [PubMed] [Google Scholar]
- 34.Han Y, Yan L-F, Wang X-B, et al. Structural and advanced imaging in predicting MGMT promoter methylation of primary glioblastoma: a region of interest based analysis. BMC Cancer. 2018;18(1):215. doi: 10.1186/s12885-018-4114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maher EA, Marin-Valencia I, Bachoo RM, et al. Metabolism of [U-13 C]glucose in human brain tumors in vivo. NMR Biomed. 2012;25(11):1234–1244. doi: 10.1002/nbm.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim MM, Parolia A, Dunphy MP, Venneti S. Non-invasive metabolic imaging of brain tumours in the era of precision medicine. Nat Rev Clin Oncol. 2016;13(12):725–739. doi: 10.1038/nrclinonc.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee JW, Lee SM. Radiomics in oncological PET/CT: clinical applications. Nucl Med Mol Imaging. 2018;52(3):170–189. doi: 10.1007/s13139-017-0500-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed in the current study are available from the corresponding author on reasonable request.