Abstract
Five genes encode the five human signal peptide peptidases (SPPs), which are intramembrane-cleaving aspartyl proteases (aspartyl I-CLiPs). SPPs have been conserved through evolution with family members found in higher eukaryotes, fungi, protozoa, arachea, and plants. SPPs are related to the presenilin family of aspartyl I-CLiPs but differ in several key aspects. Presenilins (PSENs) and SPPs both cleave the trans-membrane region of membrane proteins; however, PSENs cleave type 1 membrane proteins whereas SPPs cleave type 2 membrane proteins. Though the overall homology between SPPs and PSENs is minimal, they are multipass membrane proteins that contain two conserved active site motifs YD and GxGD in adjacent membrane-spanning domains and a conserved PAL motif of unknown function near their COOH-termini. They differ in that the active site YD and GxGD containing transmembrane domains of SPPs are inverted relative to PSENs, thus, orienting the active site in a consistent topology relative to the substrate. At least two of the human SPPs (SPP and SPPL3) appear to function without additional cofactors, but PSENs function as a protease, called γ-secretase, only when complexed with Nicastrin, APH-1 and Pen-2. The biological roles of SPP are largely unknown, and only a few endogenous substrates for SPPs have been identified. Nevertheless there is emerging evidence that SPP family members are highly druggable and may regulate both essential physiologic and pathophysiologic processes. Further study of the SPP family is needed in order to understand their biological roles and their potential as therapeutic targets.
Keywords: Intramembrane-cleaving proteases (I-CLiPs), Signal peptide peptidase (SPP), Gamma-secretase, Presenilin, Signal peptide peptidase-like (SPPL)
1. Introduction: intramembrane-cleaving proteases
Until recently, proteases had been shown to carry out catalysis in an aqueous, cytoplasmic lumenal, or extracellular environment. The necessity of a water molecule in the catalytic mechanism of peptide bond cleavage (“hydrolysis”) indicates that water must have access to the catalytic site. Indeed, “classic” proteases have catalytic domains that are present in a domain with free access to an aqueous environment. Recently, three new families of proteases have been discovered that possess the ability to carry out peptide bond hydrolysis of transmembrane domains (TMDs); these three families of proteases are referred to generically as intramembrane-cleaving proteases (I-CLiPs). I-CLiPs are polytopic membrane proteases that carry out peptide bond hydrolysis in the hydrophobic plane of the membrane. The core catalytic residues for each of the three classes are the same catalytic residues used by classical representative water-soluble enzymes. Consistent with use of conserved catalytic mechanisms, initial structural studies suggest that the polytopic I-CLiPs form a pore like structure within the membrane; thus, permitting water access to the active site [1–3]. Examples of the three types of intramembrane-cleaving proteases now can be found in almost all organisms studied: (i) the S2P-M50 super family of site 2 proteases, which are zinc metalloprotease Paragraph integrity is hard to read here due to justified alignment selected. I-CLiPs http://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?ascbin=8&maxaln=10&seltype=2&uid=127338 [4,5], (ii) the cl10483 Rhomboid superfamily http://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?uid=cl10483, which are serine I-CLiPs [6,7], and (iii) the cl01342 Peptidase_A22B superfamily http://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?ascbin=8&maxaln=10&seltype=2&uid=120578&querygi=60218981&aln=1,0,7,135, which includes the presenilins (PSENs) [8,9] and the signal peptide peptidases (SPPs) [10], which are aspartyl I-CLiPs.
2. SPPs and γ-secretase: similarities and differences
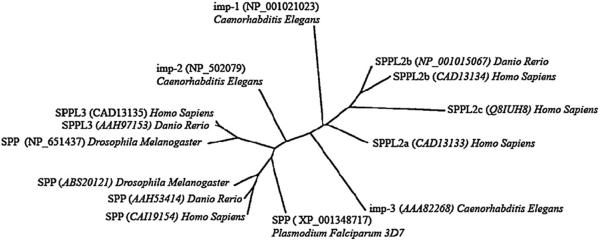
Five human SPPs and numerous SPPs in other species were originally identified through bioinformatic methods [11], and in the absence of functional data, they were named “presenilin homologs.” Shortly thereafter, one of the human SPPs was shown to catalyze the intramembrane proteolytic processing of signal peptides of major histocompatability complex class I molecules after cleavage by signal peptidase [12]. Thus, despite several disparate initially proposed nomenclatures, the field has adopted the nomenclature of signal peptide peptidase and signal peptide peptidase-like (SPPL), stemming from the original functional studies [11–13]. The human genes encoding SPPs and other studied othologs can be divided into two branches based on homology and initial functional studies. One branch consists of the SPP and SPPL3 proteins. The second branch consists of the SPPL2 proteins (SPPL2a,b,c) (Fig. 1) [12,14–17].
Fig. 1.
A phylogenetic tree of the various SPPs that have been studied to date is shown. This tree was created using the multiple alignment sequence program found at http://3w.molgen.mpg.de/. Protein sequences used to generate the alignment and the phylogenetic tree are shown in parenthesis. Note that such an alignment suggests that one of the Drosophila Melanogaster SPPs (NP 651437) should probably be renamed SPPL3. The phylogenetic tree demonstrates that the SPP and SPPL3 orthologs are more closely related then the SPPL2 family members.
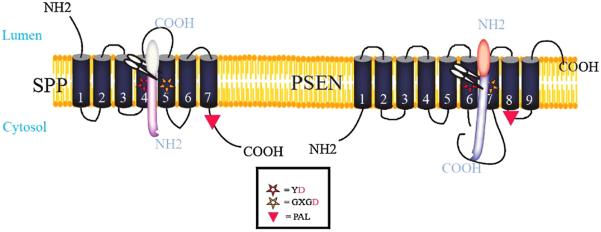
Despite limited areas of direct sequence homology, human PSENs and human SPPs are membrane proteins whose amino acid sequences can be aligned almost throughout their entire lengths [11]. These proteins share identical active site motifs, YD and GxGD [10,12,15,18]. In addition, they contain a third conserved motif, PAL, near their COOH-termini [11,19] (Fig. 2). The YD and GxGD motifs, which in PSENs and SPP contain the catalytic aspartate residues, are unusual in that they are present within predicted adjacent and opposing transmembrane regions [3,10]. However, topology studies demonstrate that the orientation of these transmembrane regions in all examined human SPP family members is inverted relative to PSENs [12,15,16,20]. This inversion of the active site appears to have functional consequences; SPPs cleave type 2 membrane proteins whereas PSENs cleave type 1 membrane proteins.
Fig. 2.
A schematic of SPP and PSEN show that they are multipass membrane protein that contains two catalytic aspartate residues indicated by the stars. These aspartates are present in opposing and adjacent transmembrane domains. A third conserved motif in is the PAL motif near the carboxyl-terminus (red triangle). As depicted, SPP and its homologs cleave type 2 membrane proteins. Althougha7TMD model is shown, other models suggest additional TMDs. For either model the exact topology of SPP has not been experimentally validated. PSEN exhibit an inverted topology relative to SPP. The schematic shows the 9 TMD topology model for PSEN which has been extensively validated using genetic and biochemical methods [59].
The similarities between PSENs and SPP, especially within the active site YD and GxGD motifs, point to a common catalytic mechanism. As in PSENs, mutation of the conserved aspartates in the YD and GxGD motif abolishes proteolytic activity [12,21,22]. In addition, a subset of γ-secretase inhibitors (GSIs) that bind PSEN also inhibit SPP and conversely certain SPP inhibitors inhibit PSEN dependent γ-secretase activity [1,20,21]. In a very interesting study, Iben and colleagues have recently shown that the most abundant cellular target of a γ-secretase transition-state inhibitor, L-685,458, is in fact SPPs and not γ-secretase [23]. Indeed, though this GSI binds and inhibits both γ-secretase and SPPs, SPPs are typically found at much higher levels than PSEN/γ-secretase in cells, thus, accounting for the majority of GSI binding. Given the sequence divergence outside of the conserved active site motifs, one would expect that not all γ-secretase inhibitors would inhibit SPP and vice versa. This appears to be the case. For example, the SPP inhibitor Z-LL2 ketone shows marked selectivity for inhibition of SPP over γ-secretase. LY-411,575 preferentially inhibits γ-secretase but does inhibit SPP at higher concentration, whereas the γ-secretase inhibitor DAPT that is similar in structure to LY-411,575 does not inhibit SPP [1,20]. More recently, helical peptide inhibitors have been reported that inhibit both SPP and γ-secretase but are more potent against SPP [24]. Because γ-secretase and SPP family members are potential targets for therapeutic intervention, PSENs for the treatment of AD and cancer and SPPs for HCV and malaria, it will be imperative in the future to identify compounds with drug-like properties that are highly selective for each. Indeed, the recent identification of novel Notch-sparing APP-selective γ-secretase inhibitors suggests that it may be possible to fine tune the specificity of aspartyl I-CLiP inhibitors in unexpected ways [25].
It is well established that γ-secretase cleaves the TMD of substrates at multiple points [8,26]. Though many models of this cleavage have been put forth, most data converge on mechanisms whereby the initial cleavage occurs near the cytoplasmic end of the TMD (the ε-cleavage site in APP and the S3 cleavage site in Notch) followed by sequential cleavages that result in liberation of the N-termini of the substrate with the final cleavages representing the γ-cleavage of APP or the S4-cleavage of Notch. Recent studies on SPP and SPPL2b indicate that, like γ-secretase, these enzymes may cleave the substrate at multiple sites within the TMD and suggest that, as with γ-secretase, this processing may be sequential [18,21]. Such studies further reinforce the similarities in catalysis of peptide bonds carried out by SPPs and PSENs. Nevertheless, it should be stated that at present the precise mechanism whereby γ-secretase or SPPs catalyze multiple cleavages of a single substrate is not known.
A major difference between SPP and PSEN is that SPP can catalyze proteolysis of substrate without additional co-factors [12,21,27]. Whether other SPP family members (SPPL3 and SPPL2a,b) function without co-factors remains an open question. In contrast, PSENs function as the catalytic protease core of γ-secretase only when complexed with three additional proteins [2,26]. Though Nicastrin has been suggested to serve as the substrate recognition factor in the γ-secretase complex [28], recent data suggest that this may not be the case [29]. Several elegant studies do suggest that the substrate binding site and the active site are distinct in both γ-secretase and SPP [24,30]. SPPs and γ-secretase preferentially catalyze the cleavage of transmembrane proteins, that either have (i) short extracellular/lumenal domains, or (ii) have been previously cleaved in a process referred to as ectodomain shedding to release the extracellular/luminal domain to generate a short membrane bound “stub” [2,3]. In the case of γ-secretase, this ectodomain shedding is largely attributed to α-and β-secretase activities. For SPP, the classical signal peptidase cleavage performs this “shedding” [12,31], and for SPPL2 family members, it appears that some substrates can be cleaved by metalloprotease disintegrins (ADAMs) [15–17,32,33]. Again, why these “membrane-stubs” are preferentially cleaved and how they are recognized by these aspartyl I-CLiPs remains an area of debate, and one worthy of further study.
3. SPP, substrates and function
Based on both subcellular fractionation studies and immunocytochemical localization, it appears that SPP and SPPL3 are predominantly localized to the endoplasmic reticulum (ER) [15,16,34–38]. Originally, SPP was functionally identified as an ER protease responsible for the generation of self-peptides presented by HLA-E [12]. These initial functional studies used a SPP-directed photolabile inhibitor that blocked the cleavage and presentation of MHC class I signal peptides by HLA-E [1], indicating that one major role of SPP may be to regulate normal immunologic surveil-lance [2,39]. In the ER, MHC class I molecules are first processed by the signal peptidase, followed by SPP catalyzed cleavage within the TMD of the residual signal peptide [1]. Once generated, the MHC derived peptides bind to HLA-E inducing its cell-surface expression. It is thought that the presentation of such self-peptides by HLA-E on the cell surface protects the cell from natural killer cell attack, though this has not been proven in vivo.
SPP may also play a role in membrane protein dislocation from the ER during human cytomegalovirus infection [40]. Dislocatio, which is more commonly referred to as retro-translocation, refers to the process by which misfolded proteins in the ER are exported from the ER into the cytoplasm, where they are degraded by the ubiquitin proteasome system. SPP associates with US2, a protein that plays an essential role in retro-translocation of MHC class I heavy chains (HC) [40]. SPP knockdown by RNA silencing decreased US2-mediated retro-translocation of MHC class I HC. Intriguingly, a requirement for SPP mediated proteolysis was not reported in the retro-translocation pathway. Thus, there remains only a tantalizing link between the proven proteolytic function of SPP mediated proteolysis of Class I molecules and HLA-E loading of the Class I peptides and a potential role of SPP-mediated proteolysis in US2 mediated MHC class HC retro-translocation.
Other identified substrates of SPP include the eosinophil cationic protein (ECP), IgSF1 (p120/inhibin-binding protein), and preprocalcitonin. SPP-mediated liberation of the N-terminus of ECP signal peptide is reported to result in increased transforming-growth factor alpha expression [41]. Cleavage of IgSF1 (p120/inhibin-binding protein), an immunoglobulin (Ig) domain protein that is highly expressed in the pituitary and brain, by SPP results in the generation of two separate Ig-containing proteins [42]. Similar to the cleavage of MHC Class I signal peptide, SPP-catalyzed cleavage of the signal peptide of preprocalcitonin appears to generate a peptide that is then presented by HLA-A2. Overexpression of preprocalcitonin results in enhanced presentation that can then be recognized by a cytotoxic lymphocyte clone [43].
Hepatitis C virus (HCV) core protein and GB virus B core protein are also known substrates of SPP [22,34,44–50]. In HCV infected cells, the immature HCV core protein is transiently anchored in the ER membrane via a C-terminal, signal peptide-like sequence. Intramembrane proteolysis of the signal peptide in the HCV core protein by SPP promotes the final processing of core protein and its release from the ER membrane into the cytosol where it can then bind to lipid droplets. SPP plays a similar role in cleavage of the GB virus core protein [46]. Although the literature is somewhat unclear on the absolute requirement for SPP-catalyzed cleavage of the HCV core protein and GB virus with respect to the virus life cycle [45–49], these studies have implicated SPP as a potential target for anti-HCV viral therapy as blocking SPP-mediated cleavage of HCV will, at a minimum, slow viral production [46].
4. SPPL3, function and substrates
The function of the closest homolog of SPP, SPPL3 remains unknown. Although, SPPL3 is only ~25% identical to SPP and ~45% homologous, based on cleavage of several substrates and its cellular location within the ER, it is likely that it has an overlapping function with SPP [14,15,36]. However, study of SPPL3 has been hampered by the difficulties with stable overexpression of this protein in mammalian cell culture. Notably based on a variety of web-based resources to examine gene expression including the Allen Brian atlas and the GNF symAtlas, SPPL3 mRNA appears to be fairly widely and abundantly expressed in a number of tissues. Thus, further studies will be needed to understand why its overexpression is problematic in mammalian cell culture.
5. SPPL2a and b, function and substrates
The SPPL2 branch of the SPP family consists of three human proteins, SPPL2a,b,c. SPPL2a and b are approximately 50% identical and 70% homologous to each other. Both SPPL2a and b appear to be most highly expressed at the mRNA level in various immune cells (see Biogps at https://biogps.gnf.org/). SPPL2c mRNA is detectable in tissues, but unlike the other SPP family members the gene encoding it contains no introns and is highly polymorphic [11]. To date SPPL2c has not been detected at the protein level, and based on its gene structure, low level of expression, and high degree of sequence polymorphism, it may to represent a recently derived pseudogene.
Based on initial subcellular localization studies that show that SPPL2b localizes to the later secretory pathway, such as endosomes and lysosomes, it was postulated that SPPL2b was likely to have non-overlapping functions with SPP and SPPL3 [15]. Indeed, the recent identification of three substrates of SPPL2a/b suggests that they have distinct cellular functions as compared to SPPL3/SPP. The first substrate of SPPL2a/b to be identified was tumor necrosis factor alpha (TNF-α). Following cleavage of TNF-α by a disintegrin metalloprotease (ADAM), the membrane stub is cleaved in its TMD by SPP2a/b. The SPPL2a/b cleavage of TNF-α liberates the cytoplasmic N-terminus, which can translocate to the nucleus and trigger interleukin-12 production. The cleavage also generates a small C-terminal peptide that is released into the extracellular space [16,17]. Similarly SPPl2a/b processes the Fas ligand (FasL), which is a type II transmembrane protein belonging to the tumour necrosis factor family. FasL binding to the cognate Fas receptor triggers the apoptosis that plays a pivotal role in the maintenance of immune system homeostasis. Again, following cleavage of the extracellular domain by an ADAM protease, the FasL membrane remnant is further processed by SPPL2a/b to liberate a small and unstable fragment containing the intracellular FasL domain. This intracellular FasL domain can translocate to the nucleus and is capable of inhibiting gene transcription [51]. A final substrate for SPPL2a/b is the BRI2 protein [32]. This type 2 membrane protein, when mutated, can cause a CNS amyloidosis and dementia, but its normal function is unknown. In any case, like TNF-α and FasL, BRI2 is cleaved sequentially by an ADAM protease and SPPL2a/b. In this case, no function has been assigned to the TMD cleavage of BRI2 [32].
6. What regulates SPP and SPPL2 proteolysis?
Using various substrates and mutant versions of these substrates, several groups have attempted to define the specificity of SPP cleavage [1,20,22]. Though these studies all reinforce the idea that SPP cleavage appears to be most efficient following “shedding” of the lumenal portion of the substrate by signal peptide cleavage, the results of these studies are somewhat divergent. Such differences may be attributable to the different methods of evaluating cleavage. Using substrates produced by in vitro translation reactions, it was demonstrated that helix destabilising residues are required for SPP cleavage, but that flanking sequences can affect the process [1]. A subsequent study showed that a reporter substrate based on a transmembrane domain sequence resistant to SPP proteolysis following in vitro translation could be cleaved by SPP when transfected into cells [20]. With only a handful of substrates identified, the study of the specificity of SPP is clearly in its infancy. However, as with γ-secretase, it is unclear whether identifying additional substrates will identify signature motifs that direct SPP cleavage, as studies of presenilin dependent γ-secretase activity show promiscuous sequence specificity [14].
Though the subcellular localizations of SPP and SPPL2b are distinct [15,16], when analyzed by sucrose flotation density gradients, both proteins fractionate into buoyant membrane microdomains which are often referred to as lipid rafts or detergent-resistant membranes [14]. This localization to lipid rafts is quite similar to the localization of γ-secretase in rafts [52,53]. Though lipid composition of the membrane clearly regulates γ-secretase cleavage, the extent to which the raft localization is required for SPP activity is uncertain. As raft microdomains appear to play important roles with respect to signalling cascades, it is likely that the clustering of SPPs within such domains is important. Perhaps one way SPP cleavage is regulated is through control of substrate entry into the local membrane microdomain in which SPP is localized.
7. In vivo SPP knockout and knockdown suggest an essential role in development
Knockout or siRNA-mediated knockdown studies of SPP family members have only been performed in D. melanogaster, C. elegans, and D. rerio (Zebrafish) [15,35,54]. No mouse knockouts have been reported. The studies in model organisms suggest that SPP and its homologs have vital functional roles in development. In C. elegans, deficiency of ce-imp-2 (a SPP-like gene), causes a severe developmental phenotype [54]. The effect of genetically disrupting the two other C. elegans SPP homologs has not been reported. Drosophila deficient in one of two SPP genes, CG11840, had defective trachea and died as larvae [35]. In Zebrafish, when either the SPP or SPPL3 homologs were knocked down, a similar embryonic lethal phenotype was observed, with a prominent effect on nervous system development noted [15]. Knockdown of the SPPL2b homolog resulted in a distinct developmental phenotype with an enlarged caudal vein [15]. Such genetic studies show that SPPs and its homologs have important, and in some cases distinct, physiologic roles essential during normal development. The molecular determinants underlying these phenotypes have not been identified and it is unlikely that SPP cleavage of the known substrates can account for the phenotypes associated with SPP deficiency in these models organisms.
8. A role in protein turnover?
In some cases, γ-secretase cleavage of substrates plays a role in signal transduction and in other cases, γ-secretase cleavage has been shown to terminate a signal of a transmembrane protein receptor [2]. Whether all cleavages of transmembrane protein substrates modulate signalling events is not known. γ-Secretase cleavage may also play a role more analogous to the proteasome, digesting type 1 transmembrane proteins to generate smaller soluble fragments that can then be broken down by other proteolytic systems in the cell [55]. Given the abundance of SPP and SPPL3 ad their location in the ER and the tentative link to ER dislocation, it is interesting to speculate that these I-CLiPs may play a normal role in cleavage of both misfolded and normally folded transmembrane proteins [40]. Furthermore, it is also possible that the SPPL2 proteins may also play dual roles in signalling and protein turnover. Given that SPPl2a and b are present in the endosomal and lysosomal system, it would seem highly plausible that in some cases they play a role in protein turnover.
9. Is SPP a monomer, dimer or both?
Monomeric SPP is a ~45 kDa N-linked glycoprotein [12]. However, solubilized SPP often appears as two bands following SDS-PAGE and Western Blotting, one at ~42 kDa, and one at ~95 kDa [13,20,56]. Under mild lysis conditions, SPP is primarily detected s a ~95 kDa homodimer [56], but the SDS-stable homodimer is dissociable to a monomer by heating in the presence of SDS and reducing agent [56]. In addition, an NH2-terminally FLAG epitope tagged SPP co-purifies with a COOH-terminally V5 epitope tagged SPP [56]. These biochemical studies suggest that SPP is likely to exist as a dimer in vivo. Indeed, fluorescence imaging studies suggest that SPP does exist as a dimer in cells. However, in vitro studies demonstrate that detergent-solubilized monomeric SPP is capable of cleaving exogenous synthetic peptide substrates [21], and further in vitro studies suggest that even a fragment of SPP can catalyze proteolysis as a monomer in vitro. Although the bulk of evidence would suggest that the majority of SPP and its closest homolog, SPPL3 [57], are present within the cell as homodimers, it is unclear whether the monomer or dimer represent the active form. Efforts to obtain more structural information on SPP may provide additional insights into the nature of its active form.
10. SPPs as therapeutic targets
The intense focus on PSENs/γ-secretase as a therapeutic target in Alzheimer's disease resulted in the development of extremely potent GSIs targeting both the active site aspartates and allosteric sites of PSENs. Several GSIs have entered human clinical trials. These studies demonstrate that the aspartyl I-CLiPs are, from a pharmacologic point of view, viable drug targets. Selective high affinity inhibitors with good pharmacokinetic properties have been identified that target γ-secretase in vivo. Unfortunately, due to mechanism-based toxicity largely mediated by inhibition of Notch signalling, there are concerns that long-term administration of a γ-secretase inhibitor will not be well-tolerated in humans. Such concerns resulted in the search for Notch-sparing GSIs. As previously mentioned previously several Notch-sparing GSIs have been reported. Whether these selective GSIs inhibit SPPs has not been reported [25].
Early inhibitor studies also provided “proof of concept” that it is possible to selectively target the activities of SPP and γ-secretase. Based on their role in HCV core protein processing, human SPPs have been postulated to be a potential target for anti-HCV therapy [22,34,44–50]. To date, no studies prove that SPP is a realistic target in the setting of HCV infection. As HCV infection and the resulting chronic hepatitis is a major cause of liver failure, further study of SPP, SPP inhibitors, and inhibition of HCV virus are certainly needed. However, given the uncertainty of the normal functions of SPP, it is not clear whether such treatment will be tolerated. Furthermore, given the possible functional overlap between SPP and SPPL3; it may be important to develop both non-selective and selective SPP and SPPL3 inhibitors.
Further complicating matters, is the fact that all SPP inhibitors identified to date seem to inhibit SPPL2s as well as SPP and SPPL3. Given the recent studies implicating SPPL2 in potentially modifying immune signalling pathways [16,17,51], one has to wonder whether SPPL2a and b are also potential targets for immune modulatory therapy, and whether selective inhibitors of these can be identified as well. Though SPP is an intriguing drug target for HCV infection a number of questions probably need to be answered before considering a major effort at developing human SPP inhibitors. Such questions include: What are the consequences to the organism of inhibiting HLA-E antigen presentation? Does SPP cleavage play a role in autoimmune disease? Would inhibition of SPP function be useful to modulate immune system function? Clearly, the study of SPPs is in its infancy and the answers to such questions will require additional tools (e.g. selective potent in vivo inhibitors and SPP knockout mice) to help find answers to these interesting but speculative questions.
We have recently proposed that the SPP homologs present in several major human protozoal pathogens might represent novel drug targets [14]. Multiple human protozoal organisms have only a single aspartyl I-Clip that is more homologous to SPP then to SPPL2a/b or PSENs. Indeed, it appears that the SPP (mSPP) from the malarial parasite, Plasmodium falciparum does possess proteolytic activity and can be inhibited by several established SPP inhibitors [14,58]. If the mSPP is a critical gene for parasite development, then the pharmacological inhibition of mSPP may be lethal to the parasite. Given that the mSPP is capable of being targeted by drug-like compounds, it may be possible to develop selective inhibitors of the mSPP that do not inhibit human SPP or γ-secretase. Furthermore, if it can be shown that mSPP is a good drug target, similar techniques and reagents could be developed to target multiple human parasitic pathogens.
11. Summary
SPPs are a newly recognized family of proteases. Though no direct link to disease process has been forthcoming, additional study of SPPs will likely reveal novel physiologic and pathophysiologic functions in humans. An understanding of SPP function in other organisms may also shed light on its normal physiologic role and provide insight into whether SPP like proteins indeed represent novel therapeutic targets in certain parasitic disease. Further study of SPP and SPP homologs will, no doubt, provide substantial insight into how aspartyl I-CLiPs recognize and catalyze the cleavage of transmembrane proteins.
Acknowledgments
This work was supported by grants from the NIH (NS39072 to T.E.G. and GM79555 to M.S.W.).
References
- [1].Lemberg MK, Martoglio B. Requirements for signal peptide peptidase-catalyzed intramembrane proteolysis. Mol Cell. 2002;10(4):735–44. doi: 10.1016/s1097-2765(02)00655-x. [DOI] [PubMed] [Google Scholar]
- [2].Wolfe MS, Kopan R. Intramembrane proteolysis: theme and variations. Science. 2004;305(5687):1119–23. doi: 10.1126/science.1096187. [DOI] [PubMed] [Google Scholar]
- [3].Golde TE, Eckman CB. Physiologic and pathologic events mediated by intramembranous and juxtamembranous proteolysis. Sci STKE. 2003;2003(172):RE4. doi: 10.1126/stke.2003.172.re4. [DOI] [PubMed] [Google Scholar]
- [4].Brown MS, Ye J, Rawson RB, Goldstein JL. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell. 2000;100(4):391–8. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- [5].Rudner DZ, Fawcett P, Losick R. A family of membrane-embedded metalloproteases involved in regulated proteolysis of membrane-associated transcription factors. Proc Natl Acad Sci USA. 1999;96(26):14765–70. doi: 10.1073/pnas.96.26.14765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Urban S, Lee JR, Freeman M. A family of Rhomboid intramembrane proteases activates all Drosophila membrane-tethered EGF ligands. Embo J. 2002;21(16):4277–86. doi: 10.1093/emboj/cdf434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Urban S, Lee JR, Freeman M. Drosophila rhomboid-1 defines a family of putative intramembrane serine proteases. Cell. 2001;107(2):173–82. doi: 10.1016/s0092-8674(01)00525-6. [DOI] [PubMed] [Google Scholar]
- [8].Wolfe MS, Selkoe DJ. Biochemistry. Intramembrane proteases—mixing oil and water. Science. 2002;296(5576):2156–7. doi: 10.1126/science.1073844. [DOI] [PubMed] [Google Scholar]
- [9].Selkoe D, Kopan R. Notch and Presenilin: regulated intramembrane proteolysis links development and degeneration. Annu Rev Neurosci. 2003;26:565–97. doi: 10.1146/annurev.neuro.26.041002.131334. [DOI] [PubMed] [Google Scholar]
- [10].Martoglio B, Golde TE. Intramembrane-cleaving aspartic proteases and disease: presenilins, signal peptide peptidase and their homologs. Hum Mol Genet. 2003;2(Spec No 2):R201–206. doi: 10.1093/hmg/ddg303. [DOI] [PubMed] [Google Scholar]
- [11].Ponting CP, Hutton M, Nyborg A, Baker M, Jansen K, Golde TE. Identification of a novel family of presenilin homologues. Hum Mol Genet. 2002;11(9):1037–44. doi: 10.1093/hmg/11.9.1037. [DOI] [PubMed] [Google Scholar]
- [12].Weihofen A, Binns K, Lemberg MK, Ashman K, Martoglio B. Identification of signal peptide peptidase, a presenilin-type aspartic protease. Science. 2002;296(5576):2215–8. doi: 10.1126/science.1070925. [DOI] [PubMed] [Google Scholar]
- [13].Grigorenko AP, Moliaka YK, Korovaitseva GI, Rogaev EI. Novel class of polytopic proteins with domains associated with putative protease activity. Biochemistry (Mosc) 2002;67(7):826–35. doi: 10.1023/a:1016365227942. [DOI] [PubMed] [Google Scholar]
- [14].Nyborg AC, Ladd TB, Jansen K, Kukar T, Golde TE. Intramembrane proteolytic cleavage by human signal peptide peptidase like 3 and malaria signal peptide peptidase. Faseb J. 2006;20(10):1671–9. doi: 10.1096/fj.06-5762com. [DOI] [PubMed] [Google Scholar]
- [15].Krawitz P, Haffner C, Fluhrer R, Steiner H, Schmid B, Haass C. Differential localization and identification of a critical aspartate suggest non-redundant proteolytic functions of the presenilin homologues SPPL2b and SPPL3. J Biol Chem. 2005;280(47):39515–23. doi: 10.1074/jbc.M501645200. [DOI] [PubMed] [Google Scholar]
- [16].Friedmann E, Hauben E, Maylandt K, Schleeger S, Vreugde S, Lichtenthaler SF, et al. SPPL2a and SPPL2b promote intramembrane proteolysis of TNFalpha in activated dendritic cells to trigger IL-12 production. Nat Cell Biol. 2006;8(8):843–8. doi: 10.1038/ncb1440. [DOI] [PubMed] [Google Scholar]
- [17].Fluhrer R, Grammer G, Israel L, Condron MM, Haffner C, Friedmann E, et al. A gamma-secretase-like intramembrane cleavage of TNFalpha by the GxGD aspartyl protease SPPL2b. Nat Cell Biol. 2006;8(8):894–6. doi: 10.1038/ncb1450. [DOI] [PubMed] [Google Scholar]
- [18].Fluhrer R, Fukumori A, Martin L, Grammer G, Haug-Kroper M, Klier B, et al. Intramembrane proteolysis of GXGD-type aspartyl proteases is slowed by a familial Alzheimer disease-like mutation. J Biol Chem. 2008;283(44):30121–8. doi: 10.1074/jbc.M806092200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wang J, Beher D, Nyborg AC, Shearman MS, Golde TE, Goate A. C-terminal PAL motif of presenilin and presenilin homologues required for normal active site conformation. J Neurochem. 2006;96(1):218–27. doi: 10.1111/j.1471-4159.2005.03548.x. [DOI] [PubMed] [Google Scholar]
- [20].Nyborg AC, Jansen K, Ladd TB, Fauq A, Golde TE. A signal peptide peptidase (SPP) reporter activity assay based on the cleavage of type II membrane protein substrates provides further evidence for an inverted orientation of the SPP active site relative to presenilin. J Biol Chem. 2004;279(41):43148–56. doi: 10.1074/jbc.M405879200. [DOI] [PubMed] [Google Scholar]
- [21].Sato T, Nyborg AC, Iwata N, Diehl TS, Saido TC, Golde TE, et al. Signal peptide peptidase: biochemical properties and modulation by nonsteroidal antiinflammatory drugs. Biochemistry. 2006;45(28):8649–56. doi: 10.1021/bi060597g. [DOI] [PubMed] [Google Scholar]
- [22].Okamoto K, Moriishi K, Miyamura T, Matsuura Y. Intramembrane proteolysis and endoplasmic reticulum retention of hepatitis C virus core protein. J Virol. 2004;78(12):6370–80. doi: 10.1128/JVI.78.12.6370-6380.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Iben LG, Olson RE, Balanda LA, Jayachandra S, Robertson BJ, Hay V, et al. Signal peptide peptidase and gamma-secretase share equivalent inhibitor binding pharmacology. J Biol Chem. 2007;282(51):36829–36. doi: 10.1074/jbc.M707002200. [DOI] [PubMed] [Google Scholar]
- [24].Sato T, Ananda K, Cheng CI, Suh EJ, Narayanan S, Wolfe MS. Distinct pharmacological effects of inhibitors of signal peptide peptidase and gamma-secretase. J Biol Chem. 2008;283(48):33287–95. doi: 10.1074/jbc.M805670200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Mayer SC, Kreft AF, Harrison B, Abou-Gharbia M, Antane M, Aschmies S, et al. Discovery of Begacestat, a Notch-1-Sparing gamma-secretase inhibitor for the treatment of Alzheimer's Disease. J Med Chem. 2008;51(23):7348–51. doi: 10.1021/jm801252w. [DOI] [PubMed] [Google Scholar]
- [26].Steiner H, Fluhrer R, Haass C. Intramembrane proteolysis by gamma-secretase. J Biol Chem. 2008;283(44):29627–31. doi: 10.1074/jbc.R800010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Narayanan S, Sato T, Wolfe MS. A C-terminal region of signal peptide peptidase defines a functional domain for intramembrane aspartic protease catalysis. J Biol Chem. 2007;282(28):20172–9. doi: 10.1074/jbc.M701536200. [DOI] [PubMed] [Google Scholar]
- [28].Shah S, Lee SF, Tabuchi K, Hao YH, Yu C, LaPlant Q, et al. Nicastrin functions as a gamma-secretase-substrate receptor. Cell. 2005;122(3):435–47. doi: 10.1016/j.cell.2005.05.022. [DOI] [PubMed] [Google Scholar]
- [29].Chavez-Gutierrez L, Tolia A, Maes E, Li T, Wong PC, de Strooper B. Glu(332) in the Nicastrin ectodomain is essential for gamma-secretase complex maturation but not for its activity. J Biol Chem. 2008;283(29):20096–105. doi: 10.1074/jbc.M803040200. [DOI] [PubMed] [Google Scholar]
- [30].Berezovska O, Randya P, Skoch J, Wolfe MS, Bacskai B, Hyman BT. Amyloid precursor protein associates with a nicastrin-dependent docking site on the presenilin 1-gamma-secretase complex in cells demonstrated by fluorescence lifetime imaging. J Neurosci. 2003;23(11):4560–6. doi: 10.1523/JNEUROSCI.23-11-04560.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Weihofen A, Martoglio B. Intramembrane-cleaving proteases: controlled liberation of proteins and bioactive peptides. Trends Cell Biol. 2003;13(2):71–8. doi: 10.1016/s0962-8924(02)00041-7. [DOI] [PubMed] [Google Scholar]
- [32].Martin L, Fluhrer R, Reiss K, Kremmer E, Saftig P, Haass C. Regulated intramembrane proteolysis of Bri2 (Itm2b) by ADAM10 and SPPL2a/SPPL2b. J Biol Chem. 2008;283(3):1644–52. doi: 10.1074/jbc.M706661200. [DOI] [PubMed] [Google Scholar]
- [33].Haffner C, Haass C. Cellular functions of gamma-secretase-related proteins. Neurodegen Dis. 2006;3(4–5):284–9. doi: 10.1159/000095268. [DOI] [PubMed] [Google Scholar]
- [34].Dev KK, Chatterjee S, Osinde M, Stauffer D, Morgan H, Kobialko M, et al. Signal peptide peptidase dependent cleavage of type II transmembrane substrates releases intracellular and extracellular signals. Eur J Pharmacol. 2006;540(1–3):10–7. doi: 10.1016/j.ejphar.2006.04.011. [DOI] [PubMed] [Google Scholar]
- [35].Casso DJ, Tanda S, Biehs B, Martoglio B, Kornberg TB. Drosophila signal peptide peptidase is an essential protease for larval development. Genetics. 2005;170(1):139–48. doi: 10.1534/genetics.104.039933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Friedmann E, Lemberg MK, Weihofen A, Dev KK, Dengler U, Rovelli G, et al. Consensus analysis of signal peptide peptidase and homologous human aspartic proteases reveals opposite topology of catalytic domains compared with presenilins. J Biol Chem. 2004;279(49):50790–8. doi: 10.1074/jbc.M407898200. [DOI] [PubMed] [Google Scholar]
- [37].Urny J, Hermans-Borgmeyer I, Schaller HC. Cell-surface expression of a new splice variant of the mouse signal peptide peptidase. Biochim Biophys Acta. 2006;1759(3–4):159–65. doi: 10.1016/j.bbaexp.2006.02.007. [DOI] [PubMed] [Google Scholar]
- [38].Urny J, Hermans-Borgmeyer I, Gercken G, Schaller HC. Expression of the presenilin-like signal peptide peptidase (SPP) in mouse adult brain and during development. Gene Expr Patterns. 2003;3(5):685–91. doi: 10.1016/s1567-133x(03)00094-2. [DOI] [PubMed] [Google Scholar]
- [39].Lemberg MK, Bland FA, Weihofen A, Braud VM, Martoglio B. Intramembrane proteolysis of signal peptides: an essential step in the generation of HLA-E epitopes. J Immunol. 2001;167(11):6441–6. doi: 10.4049/jimmunol.167.11.6441. [DOI] [PubMed] [Google Scholar]
- [40].Loureiro J, Lilley BN, Spooner E, Noriega V, Tortorella D, Ploegh HL. Signal peptide peptidase is required for dislocation from the endoplasmic reticulum. Nature. 2006;441(7095):894–7. doi: 10.1038/nature04830. [DOI] [PubMed] [Google Scholar]
- [41].Chang HT, Kao YL, Wu CM, Fan TC, Lai YK, Huang KL, et al. Signal peptide of eosinophil cationic protein upregulates transforming growth factor-alpha expression in human cells. J Cell Biochem. 2007;100(5):1266–75. doi: 10.1002/jcb.21120. [DOI] [PubMed] [Google Scholar]
- [42].Robakis T, Bak B, Lin SH, Bernard DJ, Scheiffele P. An internal signal sequence directs intramembrane proteolysis of a cellular immunoglobulin domain protein. J Biol Chem. 2008;283(52):36369–76. doi: 10.1074/jbc.M807527200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].El Hage F, Stroobant V, Vergnon I, Baurain JF, Echchakir H, Lazar V, et al. Preprocalcitonin signal peptide generates a cytotoxic T lymphocyte-defined tumor epitope processed by a proteasome-independent pathway. Proc Natl Acad Sci USA. 2008;105(29):10119–24. doi: 10.1073/pnas.0802753105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].McLauchlan J, Lemberg MK, Hope G, Martoglio B. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. Embo J. 2002;21(15):3980–8. doi: 10.1093/emboj/cdf414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Ma HC, Ku YY, Hsieh YC, Lo SY. Characterization of the cleavage of signal peptide at the C-terminus of hepatitis C virus core protein by signal peptide peptidase. J Biomed Sci. 2007;14(1):31–41. doi: 10.1007/s11373-006-9127-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Targett-Adams P, Schaller T, Hope G, Lanford RE, Lemon SM, Martin A, et al. Signal peptide peptidase cleavage of GB virus B core protein is required for productive infection in vivo. J Biol Chem. 2006;281(39):29221–7. doi: 10.1074/jbc.M605373200. [DOI] [PubMed] [Google Scholar]
- [47].Vauloup-Fellous C, Pene V, Garaud-Aunis J, Harper F, Bardin S, Suire Y, et al. Signal peptide peptidase-catalyzed cleavage of hepatitis C virus core protein is dispensable for virus budding but destabilizes the viral capsid. J Biol Chem. 2006;281(38):27679–92. doi: 10.1074/jbc.M602587200. [DOI] [PubMed] [Google Scholar]
- [48].Ait-Goughoulte M, Hourioux C, Patient R, Trassard S, Brand D, Roingeard P. Core protein cleavage by signal peptide peptidase is required for hepatitis C virus-like particle assembly. J Gen Virol. 2006;87(Pt 4):855–60. doi: 10.1099/vir.0.81664-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Hope RG, McElwee MJ, McLauchlan J. Efficient cleavage by signal peptide peptidase requires residues within the signal peptide between the core and E1 proteins of hepatitis C virus strain J1. J Gen Virol. 2006;87(Pt 3):623–7. doi: 10.1099/vir.0.81371-0. [DOI] [PubMed] [Google Scholar]
- [50].Majeau N, Gagne V, Bolduc M, Leclerc D. Signal peptide peptidase promotes the formation of hepatitis C virus non-enveloped particles and is captured on the viral membrane during assembly. J Gen Virol. 2005;86(Pt 11):3055–64. doi: 10.1099/vir.0.81174-0. [DOI] [PubMed] [Google Scholar]
- [51].Kirkin V, Cahuzac N, Guardiola-Serrano F, Huault S, Luckerath K, Friedmann E, et al. The Fas ligand intracellular domain is released by ADAM10 and SPPL2a cleavage in T-cells. Cell Death Differ. 2007;14(9):1678–87. doi: 10.1038/sj.cdd.4402175. [DOI] [PubMed] [Google Scholar]
- [52].Wahrle S, Das P, Nyborg AC, McLendon C, Shoji M, Kawarabayashi T, et al. Cholesterol-dependent gamma-secretase activity in buoyant cholesterol-rich membrane microdomains. Neurobiol Dis. 2002;9(1):11–23. doi: 10.1006/nbdi.2001.0470. [DOI] [PubMed] [Google Scholar]
- [53].Lee SJ, Liyanage U, Bickel PE, Xia W, Lansbury PT, Jr, Kosik KS. A detergent-insoluble membrane compartment contains A beta in vivo. Nat Med. 1998;4(6):730–4. doi: 10.1038/nm0698-730. [DOI] [PubMed] [Google Scholar]
- [54].Grigorenko AP, Moliaka YK, Soto MC, Mello CC, Rogaev EI. The Caenorhabditis elegans IMPAS gene, imp-2, is essential for development and is functionally distinct from related presenilins. Proc Natl Acad Sci USA. 2004;101(41):14955–60. doi: 10.1073/pnas.0406462101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Kopan R, Ilagan MX. Gamma-secretase: proteasome of the membrane? Nat Rev Mol Cell Biol. 2004;5(6):499–504. doi: 10.1038/nrm1406. [DOI] [PubMed] [Google Scholar]
- [56].Nyborg AC, Kornilova AY, Jansen K, Ladd TB, Wolfe MS, Golde TE. Signal peptide peptidase forms a homodimer that is labeled by an active site-directed gamma-secretase inhibitor. J Biol Chem. 2004;279(15):15153–60. doi: 10.1074/jbc.M309305200. [DOI] [PubMed] [Google Scholar]
- [57].Nyborg AC, Herl L, Berezovska O, Thomas AV, Ladd TB, Jansen K, et al. Signal peptide peptidase (SPP) dimer formation as assessed by fluorescence lifetime imaging microscopy (FLIM) in intact cells. Mol Neurodegen. 2006;1:16. doi: 10.1186/1750-1326-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Li X, Chen H, Oh SS, Chishti AH. A Presenilin-like protease associated with Plasmodium falciparum micronemes is involved in erythrocyte invasion. Mol Biochem Parasitol. 2008;158(1):22–31. doi: 10.1016/j.molbiopara.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Spasic D, Tolia A, Dillen K, Baert V, De Strooper B, Vrijens S, et al. Presenilin-1 maintains a nine-transmembrane topology throughout the secretory pathway. J Biol Chem. 2006;281(36):26569–77. doi: 10.1074/jbc.M600592200. [DOI] [PubMed] [Google Scholar]