Summary
Background
The Memorial Sloan Kettering Cancer Center (MSKCC) risk model is an established prognostic tool for metastatic renal-cell carcinoma that integrates clinical and laboratory data, but is agnostic to tumour genomics. Several mutations, including BAP1 and PBRM1, have prognostic value in renal-cell carcinoma. Using two independent clinical trial datasets of patients with metastatic renal-cell carcinoma, we aimed to study whether the addition of the mutation status for several candidate prognostic genes to the MSKCC model could improve the model’s prognostic performance.
Methods
In this retrospective cohort study, we used available formalin-fixed paraffin-embedded tumour tissue and clinical outcome data from patients with metastatic renal-cell carcinoma assigned to treatment with tyrosine kinase inhibitors in the COMPARZ trial (training cohort; n=357) and RECORD-3 trial (validation cohort; n=258). Eligible patients in both trials were treatment-naive; had histologically confirmed, advanced, or metastatic renal-cell carcinoma; and a Karnofsky performance status score of at least 70. For each cohort, data from patients in all treatment groups (sunitinib and pazopanib in the training cohort, and everolimus and sunitinib in the validation cohort) were pooled for this analysis. In the training cohort, tumour tissue was used to evaluate somatic mutations by next-generation sequencing, and the association between cancer-specific outcomes (overall survival, progressionfree survival, and overall response) and the mutation status of six genes of interest (BAP1, PBRM1, TP53, TERT, KDM5C, and SETD2) was tested. Only those genes with prognostic value in this setting were added to the MSKCC risk model to create a genomically annotated version. The validation cohort was used to independently test the prognostic value of the annotated model compared with the original MSKCC risk model.
Findings
357 (32%) of 1110 patients assigned to protocol treatment in the COMPARZ study between August, 2008, and September, 2011, were evaluable for mutation status and clinical outcomes in the training cohort. The independent validation cohort included 258 (55%) of 471 evaluable patients, enrolled between October, 2009, and June, 2011, on the RECORD-3 study. In the training cohort, the presence of any mutation in BAP1 or TP53, or both, and absence of any mutation in PBRM1 were prognostic in terms of overall survival (TP53wt/BAP1mut, TP53mut/BAP1wt or TP53mut/BAP1mut vs TP53wt/BAP1wt hazard ratio [HR] 1·57, 95% CI 1·21–2·04; p=0·0008; PBRM1wt vs PBRMmut, HR 1·58, 1·16–2·14; p=0·0035). The mutation status for these three prognostic genes were added to the original MSKCC risk model to create a genomically annotated version. Distribution of participants in the training cohort into the three risk groups of the original MSKCC model changed from 87 (24%) of 357 patients deemed at favourable risk, 217 (61%) at intermediate risk, and 53 (15%) at poor risk, to distribution across four risk groups in the genomically annotated risk model, with 36 (10%) of 357 deemed at favourable risk, 77 (22%) at good risk, 108 (30%) at intermediate risk, and 136 (38%) at poor risk. Addition of genomic information improved model performance for predicting overall survival (C-index: original model, 0·595 [95% CI 0·557–0·634] vs new model, 0·637 [0·595–0·679]) and progression-free survival (0·567 [95% CI 0·529–0·604] vs 0·602 [0·560–0·643]) with adequate discrimination of the proportion of patients who achieved an objective response (Cochran-Armitage one-sided p=0·0014). Analyses in the validation cohort confirmed the superiority of the genomically annotated risk model over the original version.
Interpretation
The mutation status of BAP1, PBRM1, and TP53 has independent prognostic value in patients with advanced or metastatic renal-cell carcinoma treated with first-line tyrosine kinase inhibitors. Improved stratification of patients across risk groups by use of a genomically annotated model including the mutational status of these three genes warrants further investigation in prospective trials and could be of use as a model to stratify patients with metastatic renal-cell carcinoma in clinical trials.
Funding
Novartis Pharmaceuticals Corporation, MSKCC Support Grant/Core Grant, and the J Randall & Kathleen L MacDonald Research Fund.
Introduction
Clear-cell renal-cell carcinoma is the most common form of kidney cancer and, in the majority of cases, is driven by mutation events that results in loss of function in the von Hippel-Lindau tumour suppressor (VHL) gene at chromosome 3p25.1 VHL mutations occur as a result of the loss of chromosome 3p, are found in over 90% of cases of clear-cell renal-cell carcinoma, and predispose individuals to secondary mutation events in other genes at chromosome 3, including the chromatin-modifying tumour-suppressor genes PBRM1 and BAP1.1 PBRM1 is the second most frequently mutated gene after VHL,1 and the mutations acquired in this gene largely do not overlap with loss-of-function events in BAP1, with alterations across both genes affecting more than 50% of patients with clear-cell renal-cell carcinoma.2 A growing number of analyses in the non-metastatic setting have suggested that the mutation status of BAP1 and PBRM1 are prognostic for cancer-specific outcomes.3–5 Similarly, retrospective studies suggest that acquired mutations in several other genes that are recurrently mutated, such as SETD2, KDM5C, TP53, and TERT, might have prognostic implications in patients with metastatic disease who are given standard drugs.2,6 VHL status has not been shown to be a useful prognostic marker for several reasons, including that gene function is almost uniformly lost, and high frequencies of mutation or epigenetic silencing restrict its use for genomic classification. Additionally, various features of the VHL locus itself have posed technical challenges for sequencing, including heavy guanine-cytosine (GC) content, a small coding region, and frequent occurrences of large indels.
The established standards for prognostication and stratification of patients with metastatic renal-cell carcinoma receiving systemic therapy are models that integrate clinical and laboratory data,7,8 yet molecular—and specifically genomic—biomarkers have not previously been integrated successfully. The Memorial Sloan Kettering Cancer Center (MSKCC) risk model is one of the most commonly applied stratification tools used in general clinical practice and research settings. The model allows stratification of treatment-naive patients with metastatic disease into one of three categories (favourable, intermediate, or poor prognosis) when they intiate first-line therapy.7 It estimates progression-free survival and overall survival on the basis of five clinical and laboratory parameters (hypercalcaemia, anaemia, Karnofsky performance status, lactate dehydrogenase concentration, and time from diagnosis to start of therapy).7 One of the main limitations of this model is uneven distribution across its three risk groups, with most patients typically being classified as intermediate risk.
Here, we used two clinical trial datasets of patients receiving approved targeted drugs for the treatment of metastatic renal-cell carcinoma and correlated patients’ outcomes with their mutation status regarding six genes frequently mutated and previously proposed to have prognostic relevance:2,6 BAP1, PBRM1, SETD2, KDM5C, TP53, and TERT. The genes confirmed to have prognostic value on univariate testing and deemed to be independently prognostic on multivariate testing were to be integrated into the MSKCC risk model to build a new, genomically annotated model to improve prognostication.
Methods
Study design and participants
In this population-based, retrospective cohort study, we used formalin-fixed paraffin-embedded tumour tissue collected at baseline and clinical outcomes data from patients with metastatic renal-cell carcinoma enrolled in the COMPARZ and RECORD-3 trials. MSKCC was the lead centre for both trials and treated patients on the studies. Integrated Mutation Profiling of Actionable Cancer Targets (IMPACT; using Illumina HiSeq 2500 (Illumina Inc, San Diego, CA, USA) assay testing for all patients presented here (those treated at this institution or others) was done at MSKCC. In this study, we included all participants with adequate archival tumour tissue for DNA extraction (quality of tissues were decided on a case-bycase basis) and testing of acquired somatic mutations by next-generation sequencing.
The training set comprised patient data from the randomised, open-label, phase 3 COMPARZ study9 comparing pazopanib versus sunitinib in treatmentnaive patients with histologically confirmed, advanced or metastatic renal-cell carcinoma with a clear-cell component, a Karnofsky performance status score of at least 70, and measurable disease per Response Evaluation Criteria in Solid Tumors (RECIST; version 1.1) enrolled at sites in 14 countries in North America, Europe, Australia, and Asia. Pazopanib was given at 800 mg, orally, once daily on a continuous schedule; sunitinib was given at 50 mg, orally, once daily on a schedule of 4 weeks on and 2 weeks off. The sunitinib and pazopanib treatment groups were analysed together for this study.
The validation cohort similarly included treatmentnaive patients with advanced or metastatic renal-cell carcinoma, measurable disease as per RECIST, and a Karnofsky performance status score of at least 70 enrolled in the randomised, phase 2, RECORD-3 trial at 95 sites in 19 countries in Australia, Europe, North America, South America, and Asia.10 The trial compared first-line everolimus (10 mg, orally, once daily on a continuous schedule) with sunitinib (50 mg, orally, once daily, on a schedule of 4 weeks on and 2 weeks off). Data from both treatment groups were pooled for this study.
The original study protocols had been reviewed and approved by institutional ethical review boards at participating centres and included consent for tissue acquisition and biomarker investigation, and all participants provided written consent.
Procedures
We separated tumour and normal tissue by manual review and microsection of formalin-fixed paraffin-embedded tissue samples collected at baseline in both trials. We extracted and purified DNA from tumour and normal tissue and did deep sequencing using the MSKCC IMPACT assay, as previously described.11 IMPACT is designed to detect single nucleotide variants, short copy number aberrations, and structural rearrangements, with deep coverage across all exonic sequences, plus select introns of more than 340 cancer-related genes of interest, with germline comparison as previously described.11
The original MSKCC risk model for treatment-naive patients with advanced renal cell carcinoma7 estimates cancer-specific outcomes on the basis of five clinical and laboratory parameters (Karnofsky performance status score, <80; lactate dehydrogenase, >1·5 times the upper limit of normal, low haemoglobin (anaemia), corrected calcium >10 mg/dL, and time from diagnosis to start of first-line therapy <1 year). For each of the parameters, one point is assigned when patients meet the specified criteria (table 1). Patients are then separated into one of three risk categories: favourable (zero points), intermediate (one to two points), or poor (three to five points).
Table 1:
MSKCC original model and MSKCC annotated risk model
| Original MSKCC model | Genomically annotated MSKCC model | |
|---|---|---|
| Parameters | ||
| Karnofsky performance status score | <80 | <80 |
| Lactate dehydrogenase concentration | >1.5 × ULN | >1.5 × ULN |
| Haemoglobin concentration | Low | Low |
| Corrected calcium concentration | >10 mg/dL | >10 mg/dL |
| Time from diagnosis | <1 year | <1 year |
| TP53 or BAP1* | NA | One or more mutations in either gene |
| PBRM1* | NA | Wild type or mutation with concurrent mutation in BAP1 and TP53 |
| Groups | ||
| Favourable risk | 0 | 0 |
| Good risk | NA | 1 |
| Intermediate risk | 1–2 | 2 |
| Poor risk | 3 or more | 3 or more |
One point is assigned for each parameter, with the grouping being the sum of the points scored. A higher score equates to a poorer outcome.
One and only one point can be added for each of the two genomic questions. MSKCC=Memorial Sloan Kettering Cancer Center. NA=not applicable. ULN=upper limit of normal.
Using a training and validation cohort, we set to improve the risk stratification performance of the model by incorporating genomic variants.
We first aimed to test the association of cancer-specific outcomes (progression-free survival, overall survival, and objective response) with the mutation status of the following genes of interest: PBRM1, SETD2, KDM5C, BAP1, TP53, and TERT. Genomic data were correlated with outcome by univariate analysis and subsequent multivariate testing, integrating genomics with clinical and laboratory data. The mutation status of independently prognostic genes were then integrated as a novel factor in the annotated MSKCC risk model.
Outcomes
Overall survival was defined as the time interval between randomisation and date of patient death because of any cause or censoring on the day of the last follow-up visit. Progression-free survival was defined as the time interval between the date of randomisation and the earliest date of either disease progression or death because of any cause. The proportion of participants with an objective response was defined as the combined proportion of participants with complete responses and partial responses.
Statistical analysis
We studied the association between the mutation status of genes of interest and patient oncological outcomes with the aim of seeing if they were independent of the previously established prognostic factors of the original MSKCC model in separate analyses. We considered the genes that were deemed to be candidate prognostic markers using univariate testing for survival endpoints (Kaplan-Meier method and log-rank comparisons) and objective response (Fisher’s exact test) and integrated those confirmed to be independently prognostic on multivariate testing into the annotated risk model. This multivariate analysis (Cox proportional hazards regression) of overall survival integrated the original MSKCC clinical and laboratory parameters and the presence of any mutation of interest. A two-sided p value of less than 0·05 was considered statistically significant for the Cox multivariate analysis.
Contingent on confirmation of the independent prognostic value of genomic variables, we developed a new MSKCC risk model by assigning points for original and genomic features. Effect sizes from the multivariate model informed the weighing of point scores for individual parameters. We did Kaplan-Meier analyses of individual subgroups of patients based on integrated scores. Log-rank comparison across all groups was chosen in lieu of paired testing of individual risk groups because of small sample sizes and to avoid multiple testing. We did not correct for multiple comparisons in this step, and instead of pair-wise results we determined significance across all groups. We manually reviewed the Kaplan-Meier curves and did cut-point analyses to optimise new cutoffs for the updated risk groups in the new model.
Once the annotated model was created, we used Harrell’s concordance index (C-index)12 to test discrimination for overall survival and progression-free survival in the original and new MSKCC risk models. We assessed the proportion of patients who achieved an objective response within each risk group as determined by the two different MSKCC risk models by the Cochran-Armitage trend test.13 We did not use any subjective weights, but the Cochran-Armitage trend test was adjusted to the sample sizes of each group. This test provided one-sided p values at a significance level of 0·05.
Finally, the new MSKCC risk model was validated in a cohort of patients from the RECORD-3 trial (ie, the validation cohort), including C-statistics, Kaplan-Meier analysis, and log-rank comparison for overall survival in the original and annotated model. We also used the Cochran-Armitage trend test to determine correlation between risk status and the proportion of patients who achieved an objective response as per RECIST in the validation cohort. We tested the calibration performance of the model separately in the training and validation cohorts using the Hosmer-Lemeshow test.14 For this goodness-of-fit test, a p value of less than 0·05 indicates evidence of poor fit, and a p value of 0·05 or higher indicates that a model is correctly specified. We did all analyses using SAS version 9.2
The cohorts used in this retrospective study were registered with ClinicalTrials.gov, COMPARZ with number and RECORD-3 with number .
Role of the funding source
The study was designed by academic investigators and representatives of the funder, who were involved in data collection, data analyses, data interpretation, and writing of the report. All authors had full access to the data and contributed to the development and approval of the manuscript. The corresponding author had full access to the data throughout the study and had final responsibility for the decision to submit for publication.
Results
Of 1110 patients assigned to treatment in the COMPARZ trial between August, 2008, and September, 2011,9 377 (34%) had available tissue samples for DNA extraction and were assigned to protocol treatment (IMPACT next-generation sequencing), of whom 357 (95%) had available data for all five MSKCC risk factors so we could recalculate the MSKCC scores. Therefore, for this study the training cohort comprised 357 participants with complete data available for analysis, because 20 (5%) of 377 were excluded from the MSKCC risk score calculation because they did not have data for at least one of the five MSKCC risk factors. Of these 357 participants, 175 (49%) were assigned to sunitinib and 182 (51%) were assigned to pazopanib in the original study.
471 participants were assigned to treatment in RECORD-3 between October, 2009, and June, 2011, 258 (55%) of whom had successful DNA extractions and IMPACT next-generation sequencing analysis done and comprised the validation cohort.
Baseline characteristics for all patients in the COMPAR2 training cohort are summarised in table 2. The cohorts included in this study seem to be representative of the overall study population enrolled in each trial (table 2). Outcomes for the trials have previously been reported.9,10 In accordance with similarities in patient characteristics, we found similar cancer-specific outcomes between the study cohorts and full study populations (appendix p 1).
Table 2:
Baseline characteristics for COMPARZ and RECORD-3 cohorts
| COMPARZ biomarker cohort (n=377)* | COMPARZ total (n=927) | RECORD-3 biomarker cohort (n=258)* | RECORD-3 total (n=471) | |
|---|---|---|---|---|
| Age, years | 61 (30–86) | 62 (18–88) | 62 (20–89) | 62 (20–89) |
| Sex | ||||
| Male | 279 (74%) | 671 (72%) | 195 (76%) | 342 (73%) |
| Female | 98 (26%) | 256 (28%) | 63 (24%) | 129 (27%) |
| MSKCC risk group† | ||||
| Favourable | 87 (24%) | 212 (24%) | 82 (32%) | 140 (30%) |
| Intermediate | 217 (61%) | 508 (58%) | 145 (56%) | 264 (56%) |
| Poor | 53 (15%) | 157 (18%) | 31 (12%) | 67 (14%) |
| Unknown | 20 | 50 | 0 | 0 |
| Previous nephrectomy | 362 (96%) | 779 (84%) | 258 (100%) | 471 (100%) |
| Number of metastatic sites | ||||
| ≤2 metastatic sites at baseline | 249 (66%) | 603 (65%) | 150 (58%) | 262 (56%) |
| >2 metastatic sites at baseline | 128 (34%) | 324 (35%) | 108 (42%) | 209 (44%) |
| First-line therapy | ||||
| Sunitinib | 188 (50%) | 463 (50%) | 130 (50%) | 233 (50%) |
| Pazopanib | 189 (50%) | 464 (50%) | NA | NA |
| Everolimus | NA | NA | 128 (50%) | 238 (51%) |
Data are median (range), n (%), or n. MSKCC=Memorial Sloan Kettering Cancer Center. NA=not available.
Biomarker cohort is all the patients from intention-to-treat population that have available next-generation sequencing data.
Denominator for risk groups is total cohort minus unknown samples.
For each of the six genes investigated, all available data for patients from COMPARZ and RECORD-3 were combined and categorised as mutant or wild type. Mutation frequencies and distributions are summarised in the (pp 9,10).
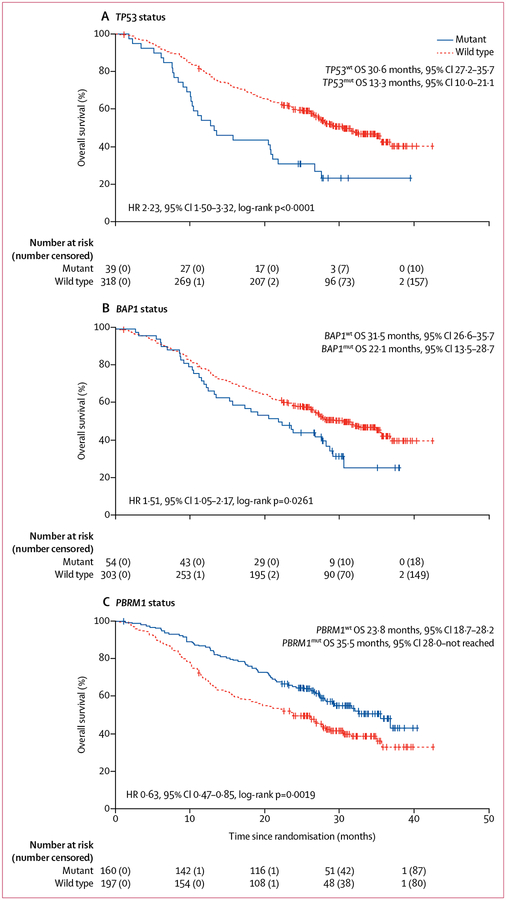
For the training cohort, we correlated the mutation status (mutant vs wild type) for each of the six genes of interest with outcomes on first-line tyrosine kinase inhibitor therapy in COMPARZ. Univariate analyses showed that the mutation status for BAP1, TP53, and PBRM1 was prognostic for overall survival, but no correlations were found for the mutation status of KDM5C, SETD2, and TERT (table 3). Patients with mutations in BAP1 or TP53, or both, had worse overall survival than those with wild-type copies of these genes, while those with acquired mutations in PBRM1 had improved overall survival (figure 1).
Table 3:
Univariate analysis of mutations of interest and clinical outcomes in the training cohort
| Frequency of Mutation (n=357) | Overall Response (n=329) | Overall survival (n=357) | Progression-free survival (n=357) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n (%) | n (%) | Median, months (95% CI) | Event/total | HR (95% CI) | p value | Median, months (95% CI) | Event/Total | HR (95% CI) | p value | |
| BAP1 | ||||||||||
| Wild type | 303 (85%) | 94 (34%) | 31.5 (26.6–35.7) | 152/303 | ref | .. | 10.7 (8.3–11.1) | 186/303 | ref | .. |
| Mutant | 54 (15%) | 17 (32%) | 22.1 (13.5–28.7) | 36/54 | 1.51 (1.05–2.17) | 0.0261 | 5.5 (4.2–10.9) | 35/54 | 1.38 (0.96–1.98) | 0.083 |
| KDM5C | ||||||||||
| Wild type | 315 (88%) | 101 (35%) | 27.7 (25.2–32.6) | 167/315 | ref | .. | 9.5 (8.3–11.0) | 194/315 | ref | .. |
| Mutant | 42 (12%) | 10 (25%) | 31.5 (18.9-not reached) | 21/42 | 0.94 (0.60–1.48) | 0.780 | 8.0 (5.6–14.1) | 27/42 | 1.00 (0.67–1.50) | 0.984 |
| PBRM1 | ||||||||||
| Wild type | 197 (55%) | 51 (28%) | 23.8 (18.7–28.2) | 116/197 | ref | .. | 8.2 (5.5–8.4) | 134/197 | ref | .. |
| Mutant | 160 (45%) | 60 | 35.5 (28.0-not reached) | 72/160 | 0.63 (0.47–0.85) | 0.0019 | 11.1 (9.5–16.5) | 87/160 | 0.67 (0.51–0.88) | 0.004 |
| SETD2 | ||||||||||
| Wild type | 266 (75%) | 83 (34%) | 27.6 (24.2–31.6) | 144/266 | ref | .. | 10.0 (8.3–11.1) | 169/266 | ref | .. |
| Mutant | 91 (25%) | 28 (34%) | 32.0 (19.6-not reached) | 44/91 | 0.86 (0.62–1.21) | 0.395 | 9.0 (8.1–11.2) | 52/91 | 0.92 (0.67–1.25) | 0.582 |
| TERT | ||||||||||
| Wild type | 315 (88%) | 102 (35%) | 28.2 (26.2–34.9) | 162/315 | ref | .. | 9.5 (8.3–11.0) | 197/315 | ref | .. |
| Mutant | 42 (12%) | 9 (26%) | 25.2 (13.7–36.8) | 26/42 | 1.26 (0.83–1.91) | 0.274 | 8.3 (2.7–13.8) | 24/42 | 1.16 (0.76–1.77) | 0.501 |
| TP53 | ||||||||||
| Wild type | 318 (89%) | 102 (35%) | 30.6 (27.2–35.7) | 159/318 | ref | .. | 10.6 (8.3–11.1) | 193/318 | ref | .. |
| Mutant | 39 (11%) | 9 (27%) | 13.3 (10.0–21.1) | 29/39 | 2.23 (1.50–3.32) | <0.0001 | 5.5 (2.7–10.8) | 28/39 | 1.42 (0.95–2.10) | 0.09 |
All 357 evaluable patients in the training cohort were included. HR=hazard ratio.
Figure 1: Median overall survival by mutation status in the training cohort.
OS=overall survival. HR=hazard ratio.
For progression-free survival, only the prognostic effect of PBRM1 status was confirmed (table 3). Best radiographic overall response versus lack of response was favourably associated with mutations in PBRM1 (proportion of patients with an overall response: wild type 28%, mutant 41%; Fisher’s exact test p=0·0070), but showed no significant associations with mutation status in BAP1 (p=0·43) and TP53 (p=0·06; table 3).
27 (8%) of 357 patients had concurrent mutations in more than one of the three genes that were prognostic for overall survival on univariate analysis: eight (2%) of 357 had PBRM1/BAP1, 18 (5%) had PBRM1/TP53, 11 (3%) had BAP1/TP53, and five (1%) had BAP1/PBRM1/TP53. Concurrent mutations in BAP1 and TP53 were associated with adverse outcome, with similar effects observed for acquired alterations in either of the two genes alone (appendix p 11). Furthermore, among patients with either BAP1 or TP53 mutations, concurrent acquired alterations in PBRM1 seemed to be associated with improved overall survival compared with alterations of TP53 or BAP1 alone (appendix p 12). However, the prognostic effect of PBRM1 status was reversed in patients with mutations in all three genes (TP53, BAP1, and PBRM1), who had worse overall survival compared with those whose tumours had mutant BAP1 and TP53, and wild-type PBRM1 (appendix p 12).
The multivariate model of overall survival confirmed that both the presence of any mutation in BAP1 or TP53, or both, and the absence of mutations in PBRM1, to be independently associated with adverse outcomes in the training cohort (TP53wt/BAP1mut vs TP53wt/BAP1wt or TP53mut/BAP1wt vs TP53wt/BAP1wt or TP53mut/BAP1mut vs TP53wt/BAP1wt, hazard ratio [HR] 1·57, 95% CI 1·21–2·04; p=0·0008; PBRM1wt vs PBRMmut, HR 1·58, 1·16–2·14; p=0·0035; appendix p 2). Effect sizes were similar for individual clinical variables and genomic variables, supporting the construction of a new model that weighs clinical and genomic factors equally for an integrated risk stratification tool that combines clinical, laboratory, and genomic features for individual patients.
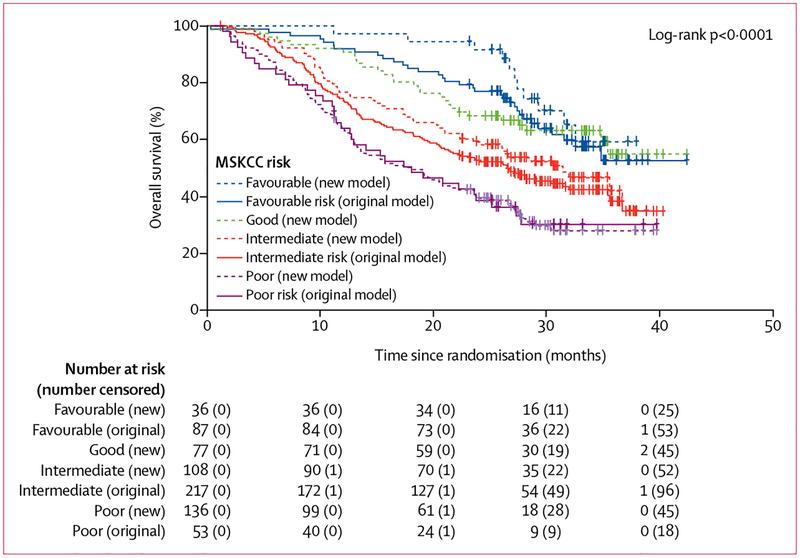
To construct the molecularly annotated model, we assigned points for the same features as in the original MSKCC risk model at the same thresholds for the clinical categories, and we asked two questions regarding tumour genomics with one additional point for each. First, the mutation status of TP53 and BAP1 is determined and one point is added for the presence of one or more mutations in BAP1 or TP53, or both. Then the mutation status of PBRM1 is determined, and if PBRM1 is wild type, or if the patient has concurrent mutations in all three genes (BAP1, TP53, and PBRM1), one point is added (table 1). With addition of up to two points, patients could have a score between zero and seven. We defined the new group assignments as follows, introducing a fourth group: favourable risk (zero points), good risk (one point), intermediate risk (two points), and poor risk (three or more points). The decision to proceed with these four groups was based on a review of individual Kaplan-Meier curves per number of total points assigned (appendix p 13) and added cut-point analyses (data not shown).
Differences in the distribution of risk categories based on the original versus the new genomically annotated model in the training cohort are in table 1. We applied the original MSKCC model to the training cohort and found a good separation in terms of overall survival for the three risk groups defined (table 4). Overall survival differed significantly (p<0·0001) between patients classified as favourable, intermediate, and poor risk. By use of the annotated MSKCC model and intrapatient comparison, individual scores changed for 218 (61%) of 357 patients in the training cohort and overall risk status changed for 174 (49%) participants, leading to substantial changes in the distribution of participants in each risk group (table 4). A detailed overview of redistribution across risk groups by both models in the training cohort is in the appendix (p 3).
Table 4:
Overall survival in the training cohort by risk model
| Original MSKCC risk model (n=357) | Annotated MSKCC risk model (n=357) | |||||
|---|---|---|---|---|---|---|
| n (%) | Median overall survival, months (95% CI) | Overall survival, months (25th percentile) | n (%) | Median overall survival, months (95% CI) | Overall survival, months (25th percentile) | |
| Favourable risk | 87 (24%) | Not reached (30.6-not reached)* | 26 | 36 (10%) | Not reached (31.6-not reached)* | 28 |
| Good risk | 0 | NA | NA | 77 (22%) | Not reached (34.8-not reached)* | 21 |
| Intermediate risk | 217 (61%) | 26.6 (21.0–32.0)* | 12 | 108 (30%) | 30.6 (22.4–36.8)* | 13 |
| Poor risk | 53 (15%) | 18.1 (11.9–25.2)* | 10 | 136 (38%) | 17.4 (13.0–23.7)* | 9 |
| C-index for overall survival (95% CI) | NA | 0.595 (0.557–0.634) | NA | NA | 0.637 (0.595–0.679) | NA |
Adapted MSKCC model includes the laboratory and clinical parameters of the original model, and the mutation status of BAP1, TP53, and PBRM1. MSKCC=Memorial Sloan Kettering Cancer Center. NA=not applicable.
p value for overall survival between categories was <0.0001; p values were calculated from log-rank tests of each model.
The updated model stratified patients with significant differences in overall survival in differentiated risk groups (p<0·0001; table 4; figure 2). Median overall survival was not reached for patients with favourable and good risk, but the median overall survival for patients at intermediate risk with the genomically annotated model was 30·6 months (95% CI 22·4–36·8) and for those at poor risk was 17·4 months (13·0–23·7).
Figure 2: Overall survival by risk status and model in the training cohort.
MSKCC=Memorial Sloan Kettering Cancer Center.
When compared with the original model, the updated model showed a better fit with prediction of overall survival in the training cohort, with the C-index increasing from 0·595 (95% CI 0·557–0·634) to 0·637 (0·595–0·679; table 4). Similarly, the C-index for progression-free survival increased from 0·567 (95% CI 0·529–0·604) in the original model to 0·602 (0·560–0·643) in the annotated model, with adequate discrimination of the proportion of patients who achieved an objective response (Cochran-Armitage one-sided p=0·0014). Using Hosmer-Lemeshow testing to evaluate the overall goodness of fit of the models, we found satisfactory p values for the training cohort for both the original and annotated risk models (p=0·066 and p=0·17; appendix p 4).
Comparing the redistributed risk groups, we found that a large proportion of the training cohort were at intermediate risk using the original model (217 [61%] of 357 participants), with a median overall survival of 26·6 months (95% CI 21·0–32·0). Of these participants, 40 (18%) were reclassified as at good risk and had a numerically improved median overall survival of 35·5 months (95% CI 21·6–not evaluable; appendix p 5) after redistribution. Furthermore, the annotated model reclassified 83 (38%) of 217 intermediate risk participants as at poor risk. These 83 patients had worse median overall survival, estimated at 16·5 months (95% CI 12·8–23·8, appendix p 5), than the original poor risk group (18·1 months, 11·9–25·2). Similar trends were also observed for reclassification of favourable risk participants by the original model to the good and intermediate risk groups; and similar effects were seen for progression-free survival, as summarised in the appendix (p 5).
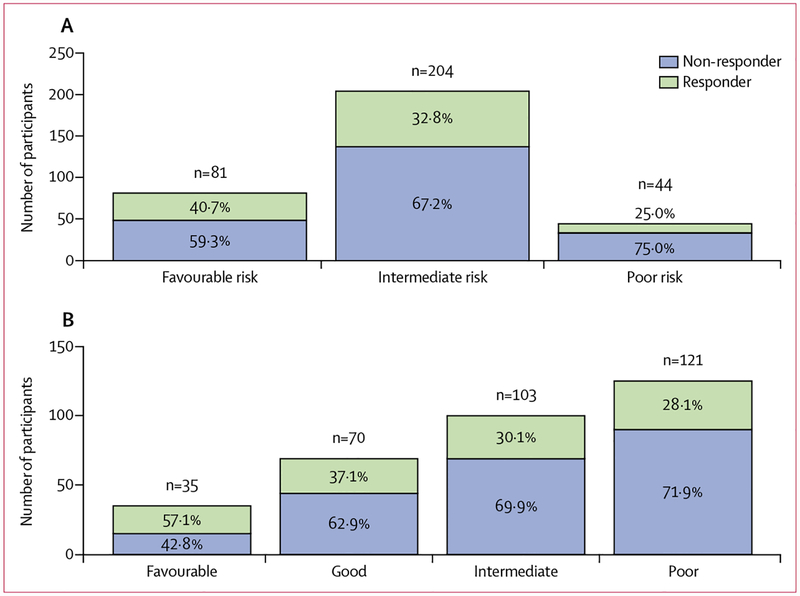
In the training cohort, 329 (92%) of 357 were evaluable for overall response after treatment in the original trial. Cochran-Armitage trend tests of best response for the original and the genomically annotated MSKCC models showed significant correlation between risk groups and objective responses, with an evident Cochran-Armitage one-sided p value of 0·0014 for the new model and a p value of 0·03 for the original model.
The distribution of patients who achieved an objective response among risk groups for the original and genomically annotated models is shown in figure 3. 20 (57%) of 35 patients classified as favourable risk by the genomically annotated model were responders, compared with 26 (37%) of 70 classified as good risk, 31 (30%) of 103 classified as intermediate risk, and only 34 (28%) of 121 classified as poor risk.
Figure 3: Proportions of patients in the training cohort who achieved an objective response, stratified by model.
All patients who achieved an overall response in the training cohort (329 [92%] of 357) are shown. Patients were stratified into risk categories with the original risk prediction model (A) and the genomically annotated model (B).
Next, we sought to validate the new MSKCC model in an independent dataset from the RECORD-3 trial (validation cohort; n=258).2,10 Each patient was categorised into MSKCC risk groups using the original and new models (appendix pp 6, 14). Additional points for genomic annotation were added for 168 (65%) of 258 patients, and on intrapatient comparison with the original MSKCC risk model, risk categories were altered for 145 (56%) patients. The four risk groups in the new model differeed significantly with regards to overall survival (p<0·appendix p 14). The C-index for overall survival with first-line therapy by use of the new model was superior to that of the original MSKCC model (original model C-index 0·658, 95% CI 0·616–0·699; genomically annotated model C-index 0·670, 0·625–0·715; appendix p 6). Our Kaplan-Meier analysis of overall survival for the training cohort, in which patients were grouped by their scores from the original MSKCC model, showed that curves for patients with one to three points overlap at various points (appendi p 15). Similarly to the training cohort, Hosmer-Lemeshow testing for the validation cohort showed overall goodness-of-fit for both the original model (p=0·47) and the new model (p=0·43; appendix p 4). Also similar to the training cohort, redistribution of participants on the basis of their score from the new model was reflected in changes in the median over all survival—ie, the median overall survival of the original model intermediate-risk group w 23·1 months (95% CI 19·2–31·4). For patients redistributed from the intermediate-risk group to the good-risk group, median overall survival was 34·9 months (27·7–46·1). For those remaining in the intermediate-risk group per the new model, median overall survival was 27·7 months (17·7–37·2) and, for those to the redistributed from the intermediate-risk group to the poor-risk group, this value was 16·9 months (9·8–20·4; appendix p 7). Cochran-Armitage testing to discern trends in the proportion of participants who achieved an objective response by RECIST remained significant in the validation cohort for the genomically annotated model (original model, p=0·0007; annotated model, p<0·0001; appendix p 8).
Discussion
We here present an updated MSKCC risk model that, through addition of the mutation status of three genes (BAP1, PBRM1, and TP53) and a fourth risk group, shows improved risk stratification of patients with metastatic renal-cell carcinoma who are initiating first-line therapy. Compared with the original MSKCC risk model, our genomically annotated tool altered risk grouping for about half of participants analysed from two independent cohorts, with dedicated analyses suggesting improved correlation with overall survival, progression-free survival, and the proportion of patients who achieved an objective response.
Clear-cell renal-cell carcinomas have substantial interpatient variability in clinical behaviour, even in the metastatic setting. This heterogeneity in aggressiveness has necessitated the development of broadly applicable tools that facilitate prognostication and enable stratification of patients who are enrolled in clinical trials. The MSKCC risk model was originally developed from a large dataset of 463 patients with advanced renal-cell carcinoma treated in clinical trials of interferon-α-based regimens.7 A candidate list of clinical variables (sites of metastatic disease, hallmarks of disease kinetics, and performance status) and biochemical parameters were tested by use of univariate analysis, and subsequent multivariate testing. The model has been validated in several settings, including independent cohorts of patients treated with cytokines and VEGF-targeted drugs,15 and it has also successfully stratified according to prognosis patients being treated with checkpoint inhibitors.16 The MSKCC model is one of the most commonly applied stratification tools to minimise imbalances between treatment groups in contemporary randomised renal-cell carcinoma trials—eg, the trials that led to the approval of nivolumab () and cabozantinib ().
Other models have taken similar approaches to stratify patients with advanced disease into risk groups, integrating clinical and laboratory features and grouping patients into three risk categories, including the International Metastatic Database Consortium (IMDC)8 and the International Kidney Cancer Working Group (IKCWG).17 Because they overlap substantially in their development and composition, these tools perform similarly when directly compared.18 Furthermore, they share important limit ations. First, they do not account for individual difference in tumour biology. Next-generation sequence testing was not broadly feasible when these models were developed, and understanding of the mutational landscape of renal-cell carcinoma was insufficient. During the conception of the IKCWG model, tumour grade and histology were investigated but did not reach prognostic significance, even in univariate analysis.17 Clinical behaviour is the end product of tumour and host biology, and the need to integrate molecular features, which are now easier to investigate and are better understood, was what motivated this study. Second, the existing models result in quite an uneven distribution of patients. All models quoted here categorise patients into three groups, all typically classifying 50% or more of patients as intermediate risk. Such poor distribution limits the utility of these models for the stratification of patients in randomised trials. Various molecular features have been investigated as candidate biomarkers to forecast the risks for patients with advanced renal-cell carcinoma receiving tyrosine kinase inhibitor therapy, including circulating angiogenic factors or cytokines, RNA-based expression signatures, immunohistochemistry, germline single-nucleotide polymorphisms, and somatic mutations.2,19–21 Other investigators have made efforts to improve established clinical models by adding molecular data. de Velasco and colleagues22 integrated the IMDC model and ClearCode34—a 34-gene expression signature—and showed improved accuracy in a cohort of 54 patients who were given tyrosine kinase inhibitors and whose tumours had been analysed as part of The Cancer Genome Atlas project. Similarly, Zurita and colleagues23 analysed data for 343 patients from the placebo-controlled pazopanib registration study and created a novel model integrating three circulating markers (osteopontin, interleukin 6, and metalloproteinase inhibitor 1) and five clinical variables that performed with higher prognostic accuracy than the IMDC model did.23 Although neither of the approaches have been validated in independent datasets, these reports support the hypothesis of incorporating molecular variables in stratification tools.
The broader application of these approaches in routine practice is difficult to imagine, given the challenges of implementing the assays beyond the academic setting. However, panel testing for mutation profiling is already available to many clinicians through institutional or commercial assays, and genotyping using next-generation sequencing is increasingly being done outside the academic setting. Although genomic literacy varies among physicians, confidence is growing among treating oncologists.24 Hence, the addition of defined mutation signatures could be adapted by many physicians in routine care, although the financial burden associated with such testing is still a substantial limitation. With steadily decreasing costs of next-generation sequencing and increasingly easy access to commercial assays, the genomic information we used here could soon be available for most newly diagnosed patients.
We selected the six genes investigated here because of their relevance for renal-cell carcinoma pathobiology and their mutation frequency in the setting of metastatic disease.2,6 Previous studies have suggested the prognostic value for each of the candidates, but we only found three to be independently associated with overall survival in the training cohort. Notably, we did not consider mutations in phosphoinositide 3-kinase pathway components, such as serine/threonine-protein kinase (mTOR), in this setting because previous investigations have not detected associations between outcomes for treatment with mTOR tyrosine kinase inhibitors (rapamycin and its analogues) and the classes of drugs given to the cohorts included here.2 We found the presence of mutations in BAP1 or TP53, or both, conferred similar adverse effects on overall survival, whereas the absence of mutations in PBRM1 resulted in worse outcomes than when the gene had mutations. These findings are incorporated into the new, genomically annotated MSKCC model. Subgroup analyses of treatment-related outcomes for patients who were reclassified by use of the new model indicated that the new classifications more accurately reflected their actual outcomes than the original model. Overall, the annotated model discriminated overall survival via log-rank testing across individual scores and the four risk groups. Because of the small sample size of subgroups and to avoid multiple testing, we did not do paired comparisons between individual scores. Harrell’s concordance indices for overall survival and progression-free survival, the two endpoints analysed for development of the original MSKCC model, were improved with the genomically annotated model. Additionally, we tested if the prognostic utility of the new model extended to the discrimination of patients who achieved an objective response according to RECIST assessment, another clinically relevant outcome measure for the prognostication of patients in the clinical setting. A stepwise change in the proportion of patients who achieved an objective response was apparent across risk groups and was confirmed by formal trend testing.
One question of interest is whether introduction of a fourth risk group, with redistribution of the intermediate risk patients, would add similar prognostic value to the original model without the introduction of genomic data. In our Kaplan-Meier analysis of overall survival for the training cohort, with patients grouped by their scores from the original MSKCC model, curves for patients with one to three points overlap at various points. Hence, the addition of another group is unlikely to result in as successful a redistribution of risk groups for this dataset as with our new genomic model.
We substantiated our analyses using an independent validation cohort and compared the two risk models and again found improved distribution across the risk groups with the new model compared with the original. Again, we determined concordance for overall survival going from the original to the annotated MSKCC model and consistent trends for the proportion of patients who achieved an objective response across the four groups. However, validation analyses were restricted to testing the model as developed in the training cohort. Univariate and multivariate analyses leading to the conception of the new model itself were not repeated in the validation cohort.
Our study had several limitations. 50% of patients in the validation cohort received first-line treatment with everolimus. Although this treatment has relevant implications for progression-free survival assessments, recognising that first-line progression-free survival differed for sunitinib and everolimus on RECORD-3,2 it is relevant to note that overall survival was similar between the two treatment groups.25 This similarity could be explained by the crossover design of RECORD-3 and the broad availability of VEGF-directed drugs outside of the investigational setting at the time the trial was held. Presumably, most patients who were randomly assigned to the first-line everolimus group on RECORD-3 went on to receive a VEGF tyrosine kinase inhibitor after progression (either sunitinib or another drug outside of the trial). Assessment of overall survival, as done in our confirmation analyses here, considers the composite of all systemic drugs received, not only the first-line drug. Progression-free survival was not non-inferior for everolimus in RECORD-3, as per the study’s primary endpoint analysis.2 Accordingly, we did not undertake progression-free survival investigations for the validation cohort.
Future work could include extending our investigations to include the IMDC model,8 the second of the two most commonly applied tools for prognostication and stratification in the metastatic space, which would have been of high interest, but was not possible since additional details to calculate this score were not available for the validation cohort.
In our approach, we did not scrutinise the functional effects of individual mutations; instead, the model assumes that all mutations in the genes in question confer loss of function. Although this approach could be optimised in research settings by investigating the functional effects of individual missense mutations, such an undertaking would not be feasible in the clinical setting. Clonal evolution and tumour heterogeneity are commonly cited as challenges in tissue-based biomarker development for renal-cell carcinoma.26 Panel testing is typically done using archival nephrectomy specimens, which are frequently collected long before assessment, and often before metastatic recurrence of the disease. This approach poses the question of whether somatic mutations that are detected in specimens are reflective of the same genotype present in the metastatic sites. This possibility has been confirmed in a retrospective cohort,27 with the degree of variance differing between the individual genes of interest.27 This variance is probably a function of clonal evolution, with the understanding that mutations occurring early in the development of a cancer are likely to be common across sites—ie, shared between primary tumours and metastases. One study28 of 349 unmatched primary tumours and 229 metastases showed that clear-cell renal-cell carcinoma primary tumours and metastases encompass a uniform distribution of common genomic alterations, as tested by next-generation sequencing panels. However, variability between matched primary tumours and metastases, or the change in genomic alterations over time and after sequential systemic therapies, was not addressed.28 For PBRM1, sampling is unlikely to be an issue considering that acquired somatic mutations are understood to be an early event in the evolution of most renal-cell carcinomas.29 Like PBRM1, BAP1 is located on chromosome 3p (together with VHL), which suffers heterozygous chromosomal loss almost uniformly in clear-cell renal-cell carcinoma. Similar to PBRM1, this situation makes loss of BAP1 more likely to occur early in disease evolution. The mutations in TP53, the least commonly mutated of the three genes studied here, are likely to be later events,29 and hence might be more prone to heterogeneity and sampling errors. The independent association confirmed on multivariate testing in this study, and the improvements in concordance seen and confirmed by use of the new model, suggest that such challenges can be overcome with the current approach.
In summary, to our knowledge this study is the largest to date that uses contemporary next-generation sequence genomic testing to prognosticate outcomes for patients with metastatic renal cell carcinoma treated with tyrosine kinase inhibitors receiving standard tyrosine kinase inhibitor treatment in the first-line setting, combining genomic parameters with established clinical and laboratory features. More than half of patients included in our analysis had mutations in one or more of three genes of interest, which were confirmed to have prognostic relevance, emphasising the applicability of this approach in broad practice. We showed the improved prognostic accuracy of an integrated model and we confirmed our findings in an independent cohort of patients treated in a separate trial. The novel genomically annotated MSKCC risk model can be used to project outcomes for patients with metastatic renal-cell carcinoma treated with tyrosine kinase inhibitors and should be considered for future clinical trial design.
Supplementary Material
Research in context.
Evidence before this study
We searched PubMed for publications from database inception to Feb 23, 2018, for studies in English determining the prognostic value of genomic markers in advanced renal-cell carcinoma and molecular annotation of tools for risk stratification in the metastatic setting. We used the search terms “genomic risk model RCC”, “prognostic somatic mutations renal cell carcinoma“, “mutational biomarkers VEGF targeted therapy”, “MSKCC risk stratification renal cell carcinoma”, and “molecular biomarkers advanced renal cell carcinoma” with dedicated attention to studies that assessed the prognostic role of acquired somatic mutations in metastatic renal-cell carcinoma or annotation of existing risk stratification tools with molecular tumour features, or both. Although other molecular parameters have been considered for addition into existing models by some authors, we found no other publications that integrated standard risk profiling with mutation testing in the metastatic setting.
Added value of this study
This study investigated the prognostic value of common tumour mutations for patients with metastatic renal-cell carcinoma receiving first-line therapy in, to our knowledge, the largest cohort so far. We made use of the information in a discovery cohort to improve the accuracy of the Memorial Sloan Kettering Cancer Center (MSKCC) risk model by incorporating the mutational status of three genes with prognostic value into the model. The genomically annotated model was validated in an independent cohort of patients with similar baseline characteristics. Mutations in BAP1, PBRM1, or TP53 occurred in more than 50% of cases in this setting, and consideration of this information altered the risk categorisation for about half the patients across two cohorts. We showed that outcomes for patients who are re-categorised are better reflected by their new group assignments and confirmed this finding in a large independent cohort. Furthermore, cohort-wide discrimination and calibration testing in discovery and confirmation sets support the utility of the genomically annotated model.
Implications of all the available evidence
The MSKCC risk model has been updated to integrate relevant individual genomic information. This annotated risk stratification tool improves the prognostic accuracy of the commonly applied MSKCC model, and is the first validated model to integrate moelcular features with clinical and laboratory parameters in the metaststic setting. It has been independently validated and could be applied for prognostication of individual patients in future clinical trials.
Acknowledgments
We thank Anuradha Bandaru for medical writing and editorial support. We thank Almedina Redzematovic for her supervision of specimen handling at Memorial Sloan Kettering Cancer Center (MSKCC) and for technical support. The study was designed by academic investigators and representatives of the funder (Novartis Pharmaceuticals Corporation, East Hanover, NJ, USA). Patients treated at MSKCC were supported in part by a MSKCC Support Grant/Core Grant (P30 CA008748). Additional funding was provided through the J Randall & Kathleen L MacDonald Research Fund.
Declaration of interests
MHV reports personal fees from GlaxoSmithKline, Calithera, Novartis, Exelixis, Pfizer, Alexion, Eisai, and Corvus; and non-financial support from Takeda outside of the submitted work. AR, YC, PP, and MM are employees of Novartis. YC owns stock in Novartis. TKC reports consulting and advisory fees from AstraZeneca, Bayer, Bristol-Myers Squibb, Cerulean, Eisai, Foundation Medicine, Exelixis, Genentech, Roche, GlaxoSmithKline, Merck, Novartis, Peloton, Pfizer, Prometheus Labs, Corvus, and Ipsen; and research grants from AstraZeneca, Bristol-Myers Squibb, Exelixis, Genentech, GlaxoSmithKline, Merck, Novartis, Peloton, Pfizer, Roche, Tracon, and Eisai. JJH reports grants and personal fees from Novartis during the conduct of the study; grants and personal fees from Eisai; personal fees from Chugai; and grants from Cancer Genetics outside of the submitted work. RJM reports grants, personal fees, and clinical trial support paid to his employer from Novartis during the conduct of the study; personal fees and clinical trial support paid to his employer from Pfizer and Genetech/Roche; and clinical trial support paid to his employer from Bristol-Myers Squibb and Merck outside of the submitted work. All other authors declare no competing interests.
Footnotes
Data sharing
Novartis is committed to sharing access to patient-level data and supporting clinical documents from eligible studies with qualified external researchers. These requests are reviewed and approved by an independent review panel on the basis of scientific merit. All data provided are anonymised to respect the privacy of patients who have participated in the trial in line with applicable laws and regulations. This trial data availability is according to the criteria and process described on www.clinicalstudydatarequest.com.
References
- 1.Sato Y, Yoshizato T, Shiraishi Y, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet 2013; 45: 860–67. [DOI] [PubMed] [Google Scholar]
- 2.Hsieh JJ, Chen D, Wang PI, et al. Genomic biomarkers of a randomized trial comparing first-line everolimus and sunitinib in patients with metastatic renal cell carcinoma. Eur Urol 2017; 71: 405–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Joseph RW, Kapur P, Serie DJ, et al. Clear cell renal cell carcinoma subtypes identified by BAP1 and PBRM1 expression. J Urol 2016; 195: 180–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kapur P, Peña-Llopis S, Christie A, et al. Effects on survival of BAP1 and PBRM1 mutations in sporadic clear-cell renal-cell carcinoma: a retrospective analysis with independent validation. Lancet Oncol 2013; 14: 159–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hakimi AA, Ostrovnaya I, Reva B, et al. Adverse outcomes in clear cell renal cell carcinoma with mutations of 3p21 epigenetic regulators BAP1 and SETD2: a report by MSKCC and the KIRC TCGA research network. Clin Cancer Res 2013; 19: 3259–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlo MI, Manley B, Patil S, et al. Genomic alterations and outcomes with VEGF-targeted therapy in patients with clear cell renal cell carcinoma. Kidney Cancer 2017; 1: 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Motzer RJ, Bacik J, Murphy BA, Russo P, Mazumdar M. Interferon-alfa as a comparative treatment for clinical trials of new therapies against advanced renal cell carcinoma. J Clin Oncol 2002; 20: 289–96. [DOI] [PubMed] [Google Scholar]
- 8.Heng DY, Xie W, Regan MM, et al. Prognostic factors for overall survival in patients with metastatic renal cell carcinoma treated with vascular endothelial growth factor-targeted agents: results from a large, multicenter study. J Clin Oncol 2009; 27: 5794–99. [DOI] [PubMed] [Google Scholar]
- 9.Motzer RJ, Hutson TE, Cella D, et al. Pazopanib versus sunitinib in metastatic renal-cell carcinoma. N Engl J Med 2013; 369: 722–31. [DOI] [PubMed] [Google Scholar]
- 10.Motzer RJ, Barrios CH, Kim TM, et al. Phase II randomized trial comparing sequential first-line everolimus and second-line sunitinib versus first-line sunitinib and second-line everolimus in patients with metastatic renal cell carcinoma. J Clin Oncol 2014; 32: 2765–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn 2015; 17: 251–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harrell FE Jr, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med 1996; 15: 361–87. [DOI] [PubMed] [Google Scholar]
- 13.Liu Hui. Paper SP05: Cochran-Armitage trend test using SAS. Rahway, NJ: Merck Research Labs, Merck & Co, 2007. https://www.lexjansen.com/pharmasug/2007/sp/SP05.pdf (accessed Oct 17, 2018). [Google Scholar]
- 14.Taktak AFG, Eleuteri A, Lake SP, Fisher AC. Evaluation of prognostic models: discrimination and calibration performance. 2007. https://pdfs.semanticscholar.org/fdb0/6f1262a41c3639bed0f06ffab9e2ba067992.pdf?_ga=2.251755145.1611769987.1539786819-1783829099.1539786819 (accessed Oct 17, 2018).
- 15.Patil S, Figlin RA, Hutson TE, et al. Prognostic factors for progression-free and overall survival with sunitinib targeted therapy and with cytokine as first-line therapy in patients with metastatic renal cell carcinoma. Ann Oncol 2011; 22: 295–300. [DOI] [PubMed] [Google Scholar]
- 16.Motzer RJ, Rini BI, McDermott DF, et al. Nivolumab for metastatic renal cell carcinoma: results of a randomized phase II trial. J Clin Oncol 2015; 33: 1430–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manola J, Royston P, Elson P, et al. Prognostic model for survival in patients with metastatic renal cell carcinoma: results from the international kidney cancer working group. Clin Cancer Res 2011; 17: 5443–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heng DYC, Xie W, Regan MM, et al. External validation and comparison with other models of the International Metastatic Renal-Cell Carcinoma Database Consortium prognostic model: a population-based study. Lancet Oncol 2013; 14: 141–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Voss MH, Chen D, Marker M, et al. Circulating biomarkers and outcome from a randomised phase II trial of sunitinib vs everolimus for patients with metastatic renal cell carcinoma. Br J Cancer 2016; 114: 642–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choueiri TK, Figueroa DJ, Fay AP, et al. Correlation of PD-L1 tumor expression and treatment outcomes in patients with renal cell carcinoma receiving sunitinib or pazopanib: results from COMPARZ, a randomized controlled trial. Clin Cancer Res 2015; 21: 1071–77. [DOI] [PubMed] [Google Scholar]
- 21.Garcia-Donas J, Esteban E, Leandro-García LJ, et al. Single nucleotide polymorphism associations with response and toxic effects in patients with advanced renal-cell carcinoma treated with first-line sunitinib: a multicentre, observational, prospective study. Lancet Oncol 2011; 12: 1143–50. [DOI] [PubMed] [Google Scholar]
- 22.de Velasco G, Culhane AC, Fay AP, et al. Molecular subtypes improve prognostic value of international metastatic renal cell carcinoma database consortium prognostic model. Oncologist 2017; 22: 286–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zurita AJ, Gagnon RC, Liu Y, et al. Integrating cytokines and angiogenic factors and tumour bulk with selected clinical criteria improves determination of prognosis in advanced renal cell carcinoma. Br J Cancer 2017; 117: 478–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gray SW, Hicks-Courant K, Cronin A, Rollins BJ, Weeks JC. Physicians’ attitudes about multiplex tumor genomic testing. J Clin Oncol 2014; 32: 1317–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knox JJ, Barrios CH, Kim TM, et al. Final overall survival analysis for the phase II RECORD-3 study of first-line everolimus followed by sunitinib versus first-line sunitinib followed by everolimus in metastatic RCC. Ann Oncol 2017; 28: 1339–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sankin A, Hakimi AA, Mikkilineni N, et al. The impact of genetic heterogeneity on biomarker development in kidney cancer assessed by multiregional sampling. Cancer Med 2014; 3: 1485–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Becerra MF, Reznik E, Redzematovic A, et al. Comparative genomic profiling of matched primary and metastatic tumors in renal cell carcinoma. Eur Urol Focus 2017; published online Oct 20. DOI: 10.1016/j.euf.2017.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Velasco G, Wankowicz SA, Madison R, et al. Targeted genomic landscape of metastases compared to primary tumours in clear cell metastatic renal cell carcinoma. Br J Cancer 2018; 118: 1238–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerlinger M, Horswell S, Larkin J, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet 2014; 46: 225–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.