Abstract
Colorectal cancer (CRC) is one of the most common and deadly malignancies in the world. In China, the morbidity rate of CRC has increased during the period 2000 to 2011. Biomarker detection for early CRC diagnosis can effectively reduce the mortality of patients with CRC. To explore the underlying mechanisms of effective biomarkers and identify more of them, we performed weighted correlation network analysis (WGCNA) on a GSE68468 dataset generated from 378 CRC tissue samples. We screened the gene set (module), which was significantly associated with CRC histology, and analyzed the hub genes. The key genes were identified by obtaining six colorectal raw data (i.e., GSE25070, GSE44076, GSE44861, GSE21510, GSE9348, and GSE21815) from the GEO database (https://www.ncbi.nlm.nih.gov/geo). The robust differentially expressed genes (DEGs) in all six datasets were calculated and obtained using the library “RobustRankAggreg” package in R 3.5.1. An integrated analysis of CRC based on the top 50 downregulated DEGs and hub genes in the red module from WGCNA was conducted, and the intersecting genes were screened. The Kaplan–Meier plot was further analyzed, and the genes associated with CRC prognosis based on patients from the TCGA database were determined. Finally, we validated the candidate gene in our clinical CRC specimens. We postulated that the candidate genes screened from the database and verified by our clinical pathological data may contribute to understanding the molecular mechanisms of tumorigenesis and may serve as potential biomarkers for CRC diagnosis and treatment.
1. Introduction
Colorectal cancer (CRC) is a malignant tumor that ranks third in terms of incidence and second in terms of mortality worldwide [1]. Similarly, the incidence and mortality of CRC rank fifth in China [2]. Despite dramatic reduction in the overall CRC incidence and mortality [3, 4], the morbidity rate in China is still rising from 2000 to 2011 [2]. Hence, further research is desperately needed to elucidate the causes for the increasing burden of CRC and to advance treatments for tumor subtypes with low response rates to current therapies. Treatment according to the distinctive tumor biology is an effective means to reduce the mortality of patients with CRC, as well as the detection of biomarkers that enable the stratification of patients with CRC into different prognostic subgroups and in relation therapeutic response [5]. Advances in RNA sequencing technologies and bioinformatics analysis provide novel potential biomarkers and drug targets for tumor treatment [6]. Weighted gene coexpression network analysis (WGCNA), as a systems biology algorithm, can enable the identification of highly coexpressed gene clusters (modules) [7]. Then, such interest modules and hub genes related to clinical traits can be screened out and used to identify candidate biomarkers [8]. The robust rank aggregation (RRA) method can be used to integrate multiple sets of chip data gene lists and to perform comprehensive reordering to find the most significant difference genes [9]. RRA prevents cross-platform standardization processing, and the number of samples per chip has no strict limit, which is of great significance for the effective evaluation of the results of different gene expression profiles [10].
In our study, we performed WGCNA on GSE68468 and screened out the key gene modules and hub genes significantly associated with the CRC histology. Additionally, we conducted RRA on six raw data (i.e., GSE25070, GSE44076, GSE44861, GSE21510, GSE9348, and GSE21815) and calculated the top 50 robust DEGs in all the six data by using the library “RobustRankAggreg” package. We analyzed the integrated genes between DEGs and hub genes in the key module by using Kaplan–Meier analysis in the TCGA database and obtained the candidate genes associated with OS. Finally, we validated the candidate gene in our clinical CRC specimens. The candidate genes screened from the database and verified using our clinical pathological data might have significant clinical implications for CRC diagnosis, treatment, and prognosis prediction.
2. Results
2.1. Weighted Gene Coexpression Network Construction and Module Detection
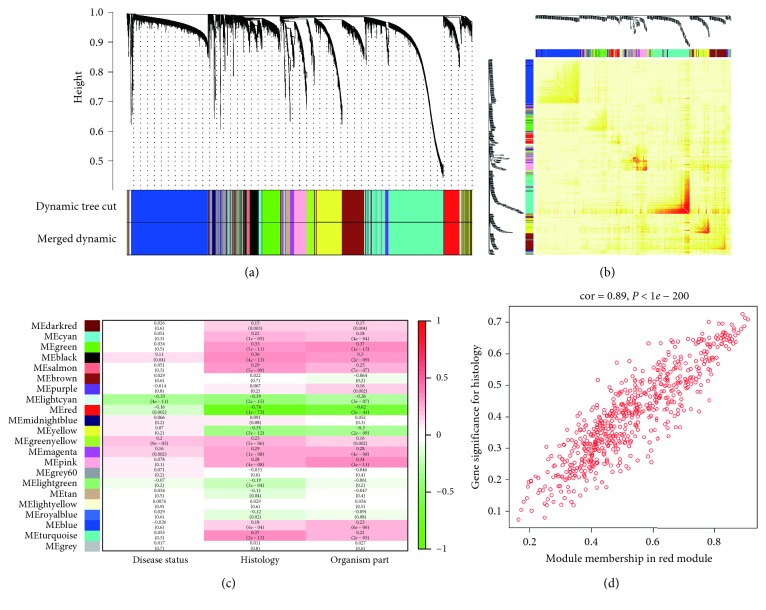
We performed WGCNA to find the highly correlated genes. The sample dendrogram and trait heatmap are shown in Figure S1A. As shown in Figure S1B, power value 4 was set to guarantee the high-scale independence and low mean connectivity of 13515 genes. We set the dissimilarity as 0.25 to merge similar modules (Figure S1C), and 22 modules were generated (Figure 1(a)). Furthermore, the interaction relationship network of 22 modules was plotted (Figure 1(b)). From those obtained modules, the red module had the deepest association with tumor histology (cor = −0.76, P=1E − 73), which was selected for further analysis (Figure 1(c)). Additionally, the module memberships in the red module and the gene significance had a high correlation (0.89) and a high P value (<1e − 200) (Figure 1(d)), suggesting its suitability for identifying the hub genes associated with CRC occurrence and metastasis.
Figure 1.
Modules defined by the WGCNA. (a) Dendrogram tree represents the clustering dendrogram of genes in GSE68468, and the bottom colorful bands represent 22 modules assigned by genes. (b) A network heatmap plot of 400 genes. The intensity of the red color indicates the strength of the correlation between pairs of modules on a linear scale. (c) Correlation between modules and clinicopathologic features. The correlation coefficient and corresponding P value of each module are shown in each cell. (d) Correlation (Pearson) between module membership and gene significance for the red module.
2.2. Coexpression Network Construction and Hub Gene Identification
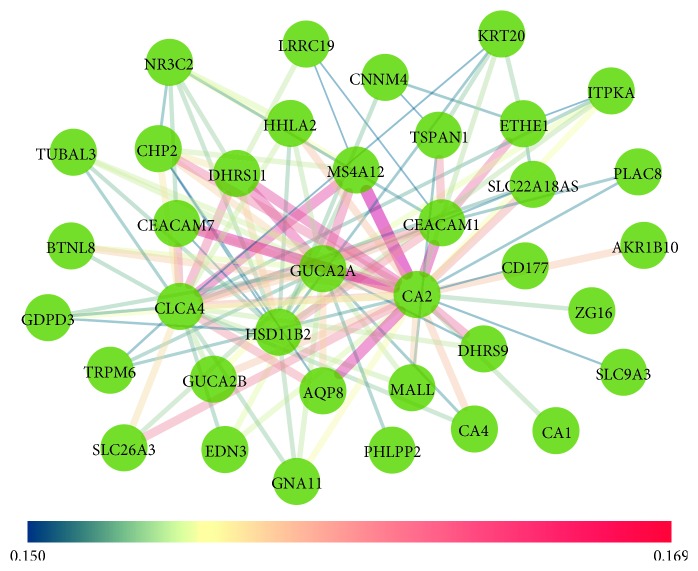
In our study, we obtained 616 genes in the red modules. The hub genes in the red module were filtered by a condition of the weight value being greater than 0.15, and a total of 37 edges were obtained and are visualized in Figure 2. The top 10 hub genes were CA2, MS4A12, DHRS11, GUCA2A, ETHE1, CLCA4, TSPAN1, HSD11B2, AQP8, and CHP2.
Figure 2.
Coexpression network of hub genes. The hub genes in the red modules were filtered by a condition of the weight value (>0.15) and visualized using Cytoscape 3.6.0. The size and color of the edge represent the intensity of weight interaction in the network.
2.3. RRA Analysis
We calculated DEGs expressed between the cancerous and adjacent tissues in each dataset and displayed the results by using volcano maps (Figures S2a–f). Then, DEGs in all six data were recalculated and reorganized using the library “RobustRankAggreg” package. A total of 464 robust DEGs were identified, including 176 upregulated genes and 288 downregulated genes (Table 1), by using adjusted P value < 0.01 and |logFC| ≥ 1 as the cutoff criteria. The 50 most prominent up- and downregulated genes were screened and are visualized in Figures 3(a) and 3(b).
Table 1.
Whole DEGs (176 upregulated genes and 288 downregulated genes) identified in the six microarray data from the GEO database by using adjusted P value < 0.01 and |logFC| ≥ 1 as the cutoff criteria.
| Name | P value | Adjusted P value | LogFC |
|---|---|---|---|
| Upregulated DEGs | |||
| MMP7 | 1.04E − 16 | 2.71E − 12 | 3.717319 |
| THBS2 | 3.32E − 16 | 8.67E − 12 | 2.821997 |
| TGFBI | 7.96E − 16 | 2.08E − 11 | 2.584099 |
| MMP3 | 1.59E − 15 | 4.15E − 11 | 2.998933 |
| DPEP1 | 4.59E − 15 | 1.20E − 10 | 3.250292 |
| CDH3 | 4.60E − 14 | 1.20E − 09 | 3.266416 |
| KRT23 | 1.55E − 13 | 4.05E − 09 | 3.658825 |
| NFE2L3 | 1.32E − 12 | 3.45E − 08 | 2.149137 |
| GZMB | 1.36E − 12 | 3.55E − 08 | 2.078558 |
| COL11A1 | 3.42E − 12 | 8.95E − 08 | 2.728714 |
| CXCL1 | 3.70E − 12 | 9.65E − 08 | 2.210194 |
| CEMIP | 4.96E − 12 | 1.30E − 07 | 2.894223 |
| FAP | 5.19E − 12 | 1.36E − 07 | 2.449763 |
| SLCO1B3 | 6.55E − 12 | 1.71E − 07 | 2.197776 |
| CLDN1 | 7.48E − 12 | 1.95E − 07 | 2.916603 |
| MYC | 1.15E − 11 | 2.99E − 07 | 1.831720 |
| IFITM1 | 1.15E − 11 | 2.99E − 07 | 1.625712 |
| TESC | 2.53E − 11 | 6.61E − 07 | 2.108654 |
| CXCL11 | 2.95E − 11 | 7.70E − 07 | 1.989137 |
| EDNRA | 3.02E − 11 | 7.89E − 07 | 1.496142 |
| MMP1 | 3.29E − 11 | 8.60E − 07 | 2.399938 |
| FOXQ1 | 5.40E − 11 | 1.41E − 06 | 2.749133 |
| TPX2 | 5.42E − 11 | 1.41E − 06 | 1.634694 |
| COL4A1 | 5.77E − 11 | 1.51E − 06 | 1.630033 |
| TCN1 | 7.96E − 11 | 2.08E − 06 | 1.926601 |
| EGFL6 | 9.09E − 11 | 2.38E − 06 | 1.752245 |
| TRIP13 | 9.50E − 11 | 2.48E − 06 | 1.729510 |
| PMAIP1 | 1.01E − 10 | 2.63E − 06 | 1.621771 |
| CXCL3 | 1.19E − 10 | 3.10E − 06 | 1.933844 |
| PSAT1 | 1.33E − 10 | 3.48E − 06 | 1.749995 |
| CTHRC1 | 1.36E − 10 | 3.56E − 06 | 2.547903 |
| PLS3 | 1.76E − 10 | 4.61E − 06 | 1.561012 |
| INHBA | 1.82E − 10 | 4.76E − 06 | 2.470231 |
| IFITM2 | 2.22E − 10 | 5.81E − 06 | 1.477155 |
| PHLDA1 | 2.77E − 10 | 7.25E − 06 | 2.025229 |
| CSE1L | 2.77E − 10 | 7.25E − 06 | 1.509054 |
| BMP4 | 4.28E − 10 | 1.12E − 05 | 1.311002 |
| CRNDE | 4.93E − 10 | 1.29E − 05 | 2.196882 |
| S100A9 | 5.05E − 10 | 1.32E − 05 | 1.793278 |
| CDK4 | 6.20E − 10 | 1.62E − 05 | 1.268501 |
| XPOT | 6.54E − 10 | 1.71E − 05 | 1.350678 |
| LY6E | 6.54E − 10 | 1.71E − 05 | 1.773015 |
| CCL20 | 8.45E − 10 | 2.21E − 05 | 1.681642 |
| TSPAN5 | 8.63E − 10 | 2.25E − 05 | 1.300671 |
| CDC25B | 9.62E − 10 | 2.51E − 05 | 1.477785 |
| LGR5 | 1.03E − 09 | 2.69E − 05 | 1.765688 |
| ASCL2 | 1.06E − 09 | 2.78E − 05 | 2.270850 |
| IFITM3 | 1.12E − 09 | 2.94E − 05 | 1.535829 |
| PLAU | 1.35E − 09 | 3.52E − 05 | 1.449998 |
| RIPK2 | 1.40E − 09 | 3.65E − 05 | 1.304180 |
| COL5A2 | 1.42E − 09 | 3.72E − 05 | 1.709418 |
| CXCL10 | 1.45E − 09 | 3.78E − 05 | 1.832510 |
| DUSP14 | 1.49E − 09 | 3.89E − 05 | 1.604280 |
| TRIB3 | 1.52E − 09 | 3.97E − 05 | 2.009118 |
| CKMT2 | 1.71E − 09 | 4.46E − 05 | 2.047320 |
| CEL | 1.77E − 09 | 4.62E − 05 | 1.921534 |
| TGIF2 | 1.82E − 09 | 4.74E − 05 | 1.594622 |
| HOMER1 | 1.89E − 09 | 4.94E − 05 | 1.595741 |
| SRPX2 | 1.97E − 09 | 5.15E − 05 | 1.759897 |
| SLC7A5 | 2.05E − 09 | 5.37E − 05 | 1.907805 |
| UBD | 2.06E − 09 | 5.38E − 05 | 1.848031 |
| GPSM2 | 2.90E − 09 | 7.58E − 05 | 1.310352 |
| AZGP1 | 3.52E − 09 | 9.20E − 05 | 2.058214 |
| AURKA | 3.81E − 09 | 9.96E − 05 | 1.248453 |
| STC2 | 3.90E − 09 | 0.000102 | 1.838371 |
| SULF1 | 3.90E − 09 | 0.000102 | 2.003268 |
| ATAD2 | 4.09E − 09 | 0.000107 | 1.500415 |
| TMEM158 | 4.11E − 09 | 0.000107 | 1.212075 |
| PPAT | 4.12E − 09 | 0.000108 | 1.305250 |
| SPP1 | 4.13E − 09 | 0.000108 | 2.205887 |
| CXCL2 | 4.36E − 09 | 0.000114 | 1.826375 |
| COL1A2 | 4.44E − 09 | 0.000116 | 1.882105 |
| SLC7A11 | 5.29E − 09 | 0.000138 | 1.208023 |
| TIMP1 | 5.69E − 09 | 0.000149 | 1.661168 |
| MMP12 | 6.08E − 09 | 0.000159 | 1.933314 |
| SCD | 6.11E − 09 | 0.000160 | 1.564105 |
| SOX9 | 6.43E − 09 | 0.000168 | 1.352557 |
| CXCL9 | 6.92E − 09 | 0.000181 | 1.461166 |
| HSPH1 | 8.01E − 09 | 0.000209 | 1.238503 |
| GDPD5 | 8.12E − 09 | 0.000212 | 1.308392 |
| CXCL8 | 8.27E − 09 | 0.000216 | 2.588362 |
| COL1A1 | 8.38E − 09 | 0.000219 | 1.769908 |
| FABP6 | 8.52E − 09 | 0.000223 | 2.079844 |
| PMEPA1 | 8.52E − 09 | 0.000223 | 1.333566 |
| ANXA9 | 9.83E − 09 | 0.000257 | 1.608179 |
| TDGF1P3 | 1.15E − 08 | 0.000301 | 1.841176 |
| CEP55 | 1.22E − 08 | 0.000319 | 1.470764 |
| COL10A1 | 1.27E − 08 | 0.000331 | 1.780470 |
| MCM10 | 1.37E − 08 | 0.000358 | 1.211513 |
| MPP6 | 1.51E − 08 | 0.000396 | 1.432320 |
| PROX1 | 1.58E − 08 | 0.000413 | 1.364230 |
| TEAD4 | 1.65E − 08 | 0.000431 | 1.447381 |
| CLDN2 | 1.72E − 08 | 0.000449 | 1.698260 |
| ODAM | 1.79E − 08 | 0.000468 | 1.471032 |
| MTHFD2 | 1.83E − 08 | 0.000478 | 1.111261 |
| KRT6B | 1.85E − 08 | 0.000484 | 1.485079 |
| HILPDA | 1.87E − 08 | 0.000488 | 1.488635 |
| DACH1 | 2.14E − 08 | 0.000559 | 1.610398 |
| ESM1 | 2.17E − 08 | 0.000568 | 1.708858 |
| CYP4X1 | 2.22E − 08 | 0.000581 | 1.382114 |
| GNG4 | 2.25E − 08 | 0.000588 | 1.178933 |
| ARID3A | 2.35E − 08 | 0.000613 | 1.275140 |
| PPBP | 2.38E − 08 | 0.000621 | 1.748166 |
| EPHX4 | 2.42E − 08 | 0.000633 | 2.381989 |
| LRP8 | 2.67E − 08 | 0.000697 | 1.522085 |
| PTP4A3 | 2.67E − 08 | 0.000697 | 1.183443 |
| SERPINE2 | 2.67E − 08 | 0.000697 | 1.154912 |
| AGT | 2.81E − 08 | 0.000734 | 1.405610 |
| C2 | 2.84E − 08 | 0.000743 | 1.372973 |
| RRM2 | 2.86E − 08 | 0.000748 | 1.196973 |
| NEBL | 3.49E − 08 | 0.000912 | 1.521795 |
| RPP40 | 3.49E − 08 | 0.000912 | 1.285132 |
| ARNTL2 | 3.77E − 08 | 0.000984 | 1.419157 |
| AHCY | 3.84E − 08 | 0.001004 | 1.237807 |
| GTF2IRD1 | 3.95E − 08 | 0.001032 | 1.371062 |
| WNT2 | 4.00E − 08 | 0.001044 | 1.452032 |
| TOP2A | 4.04E − 08 | 0.001057 | 1.372624 |
| PUS7 | 4.09E − 08 | 0.001069 | 1.249772 |
| NMU | 4.30E − 08 | 0.001122 | 1.284463 |
| COL6A3 | 4.44E − 08 | 0.001160 | 1.270615 |
| RNF43 | 4.57E − 08 | 0.001195 | 1.397306 |
| COL5A1 | 4.69E − 08 | 0.001226 | 1.191502 |
| CDK1 | 4.70E − 08 | 0.001228 | 1.317782 |
| ETV4 | 4.86E − 08 | 0.001271 | 2.204983 |
| PTGS2 | 5.04E − 08 | 0.001317 | 1.233879 |
| CHEK1 | 5.20E − 08 | 0.001358 | 1.126745 |
| MMP10 | 5.41E − 08 | 0.001414 | 1.195256 |
| FZD3 | 5.52E − 08 | 0.001442 | 1.089222 |
| UBE2C | 5.81E − 08 | 0.001518 | 1.241558 |
| RNASEH2A | 6.16E − 08 | 0.001609 | 1.071208 |
| ENC1 | 6.27E − 08 | 0.001637 | 1.375197 |
| IGF2BP3 | 6.37E − 08 | 0.001665 | 1.082776 |
| VSNL1 | 6.75E − 08 | 0.001764 | 1.786437 |
| GALNT6 | 6.79E − 08 | 0.001774 | 1.266153 |
| ERP27 | 6.97E − 08 | 0.001821 | 1.218536 |
| PROCR | 7.73E − 08 | 0.002020 | 1.376152 |
| PAFAH1B3 | 7.88E − 08 | 0.002058 | 1.100418 |
| RFC3 | 8.14E − 08 | 0.002126 | 1.372173 |
| NKRF | 8.14E − 08 | 0.002127 | 1.043072 |
| CXCL5 | 9.69E − 08 | 0.002531 | 1.294918 |
| SQLE | 1.03E − 07 | 0.002700 | 1.308167 |
| EXOSC5 | 1.08E − 07 | 0.002828 | 1.132422 |
| S100P | 1.13E − 07 | 0.002944 | 1.551099 |
| ELOVL5 | 1.27E − 07 | 0.003331 | 1.176539 |
| ZAK | 1.29E − 07 | 0.003360 | 1.541699 |
| TCFL5 | 1.45E − 07 | 0.003786 | 1.270140 |
| LAPTM4B | 1.67E − 07 | 0.004376 | 1.026874 |
| CEBPB | 1.70E − 07 | 0.004431 | 1.333258 |
| SLCO4A1 | 1.83E − 07 | 0.004790 | 1.606904 |
| TTK | 1.89E − 07 | 0.004934 | 1.386804 |
| TMEM97 | 1.98E − 07 | 0.005179 | 1.082142 |
| CHI3L1 | 2.01E − 07 | 0.005239 | 1.621308 |
| PPM1H | 2.10E − 07 | 0.005475 | 1.353835 |
| SORD | 2.14E − 07 | 0.005591 | 1.037586 |
| CST1 | 2.20E − 07 | 0.005747 | 1.894531 |
| CENPF | 2.46E − 07 | 0.006422 | 1.024242 |
| GTF3A | 2.47E − 07 | 0.006451 | 1.215577 |
| APCDD1 | 2.59E − 07 | 0.006772 | 1.580775 |
| KIF4A | 2.59E − 07 | 0.006772 | 1.267447 |
| MFAP2 | 2.65E − 07 | 0.006914 | 1.397671 |
| CCNB1 | 2.77E − 07 | 0.007241 | 1.203637 |
| GDF15 | 2.88E − 07 | 0.007514 | 1.563948 |
| NXT1 | 2.93E − 07 | 0.007667 | 1.046948 |
| SHMT2 | 2.95E − 07 | 0.007697 | 1.002641 |
| SNTB1 | 2.96E − 07 | 0.007726 | 1.291950 |
| TACSTD2 | 2.97E − 07 | 0.007754 | 1.640872 |
| SNX10 | 2.99E − 07 | 0.007814 | 1.021758 |
| SLC12A2 | 3.04E − 07 | 0.007933 | 1.134681 |
| TNFRSF12A | 3.13E − 07 | 0.008190 | 1.332447 |
| SLC5A6 | 3.25E − 07 | 0.008488 | 1.064032 |
| RUVBL1 | 3.26E − 07 | 0.008513 | 1.066507 |
| FXYD5 | 3.32E − 07 | 0.008680 | 1.172860 |
| ZNF239 | 3.35E − 07 | 0.008745 | 1.158161 |
| REG1A | 3.40E − 07 | 0.008876 | 1.168719 |
| OLFML2B | 3.57E − 07 | 0.009326 | 1.332885 |
| ERCC6L | 3.63E − 07 | 0.009472 | 1.325393 |
|
| |||
| Downregulated DEGs | |||
| GUCA2B | 4.94E − 21 | 1.29E − 16 | −5.47325 |
| GUCA2A | 5.63E − 20 | 1.47E − 15 | −4.98756 |
| CA4 | 9.10E − 20 | 2.38E − 15 | −4.93587 |
| MS4A12 | 2.15E − 19 | 5.61E − 15 | −5.22647 |
| CA2 | 3.16E − 19 | 8.27E − 15 | −4.64843 |
| AQP8 | 7.18E − 19 | 1.88E − 14 | −5.22336 |
| CLCA1 | 1.67E − 17 | 4.37E − 13 | −4.41598 |
| CLCA4 | 2.30E − 17 | 6.01E − 13 | −5.78604 |
| AKR1B10 | 3.47E − 17 | 9.06E − 13 | −3.66480 |
| CA1 | 1.37E − 16 | 3.58E − 12 | −4.85127 |
| HSD17B2 | 1.42E − 15 | 3.72E − 11 | −3.65299 |
| GCG | 1.86E − 15 | 4.87E − 11 | −4.24507 |
| CWH43 | 3.10E − 15 | 8.09E − 11 | −3.35700 |
| MT1M | 6.63E − 15 | 1.73E − 10 | −3.74527 |
| CHP2 | 6.63E − 15 | 1.73E − 10 | −3.59700 |
| FCGBP | 9.37E − 15 | 2.45E − 10 | −3.35836 |
| ADH1C | 1.89E − 14 | 4.93E − 10 | −3.66678 |
| CD177 | 7.18E − 14 | 1.88E − 09 | −3.56989 |
| CA12 | 1.04E − 13 | 2.73E − 09 | −2.48416 |
| CHGA | 1.04E − 13 | 2.73E − 09 | −3.21194 |
| SLC26A3 | 1.09E − 13 | 2.85E − 09 | −3.26689 |
| CEACAM7 | 1.16E − 13 | 3.02E − 09 | −3.26188 |
| PLAC8 | 1.75E − 13 | 4.58E − 09 | −2.93747 |
| CLDN8 | 1.79E − 13 | 4.67E − 09 | −4.07661 |
| PCK1 | 3.95E − 13 | 1.03E − 08 | −2.41432 |
| BEST2 | 5.80E − 13 | 1.52E − 08 | −2.88517 |
| GBA3 | 6.08E − 13 | 1.59E − 08 | −3.15495 |
| MT1H | 7.81E − 13 | 2.04E − 08 | −2.45079 |
| HMGCS2 | 1.28E − 12 | 3.35E − 08 | −2.63704 |
| SI | 1.53E − 12 | 3.99E − 08 | −3.18335 |
| GCNT3 | 1.86E − 12 | 4.87E − 08 | −2.55705 |
| EDN3 | 2.02E − 12 | 5.29E − 08 | −2.36855 |
| SLC26A2 | 2.58E − 12 | 6.74E − 08 | −2.74311 |
| TSPAN1 | 3.02E − 12 | 7.89E − 08 | −2.43173 |
| UGT2B17 | 3.26E − 12 | 8.51E − 08 | −2.68448 |
| BCAS1 | 3.43E − 12 | 8.96E − 08 | −2.14870 |
| LRRC19 | 4.61E − 12 | 1.20E − 07 | −2.60514 |
| LGALS2 | 5.96E − 12 | 1.56E − 07 | −2.50281 |
| ANPEP | 8.52E − 12 | 2.23E − 07 | −2.96251 |
| VSIG2 | 8.52E − 12 | 2.23E − 07 | −2.90829 |
| MT1E | 9.09E − 12 | 2.37E − 07 | −2.30341 |
| MT1G | 9.49E − 12 | 2.48E − 07 | −2.13170 |
| NXPE4 | 9.66E − 12 | 2.53E − 07 | −2.93788 |
| KRT20 | 9.69E − 12 | 2.53E − 07 | −2.12657 |
| CA7 | 1.29E − 11 | 3.38E − 07 | −2.72816 |
| MT1F | 1.32E − 11 | 3.45E − 07 | −2.15911 |
| KLF4 | 1.38E − 11 | 3.59E − 07 | −2.38010 |
| AHCYL2 | 1.43E − 11 | 3.74E − 07 | −2.24169 |
| ITLN1 | 1.74E − 11 | 4.55E − 07 | −3.42798 |
| HSD11B2 | 2.10E − 11 | 5.49E − 07 | −2.34259 |
| NR3C2 | 2.53E − 11 | 6.60E − 07 | −2.37317 |
| EPB41L3 | 2.57E − 11 | 6.72E − 07 | −2.39867 |
| ITM2C | 2.67E − 11 | 6.97E − 07 | −2.12884 |
| MMP28 | 2.86E − 11 | 7.48E − 07 | −1.93771 |
| ZG16 | 3.09E − 11 | 8.06E − 07 | −4.23120 |
| ABCG2 | 3.75E − 11 | 9.80E − 07 | −3.20141 |
| SLC4A4 | 4.13E − 11 | 1.08E − 06 | −3.28247 |
| SCGB2A1 | 4.33E − 11 | 1.13E − 06 | −2.48958 |
| MALL | 4.61E − 11 | 1.20E − 06 | −2.29999 |
| TUBAL3 | 4.92E − 11 | 1.29E − 06 | −2.39086 |
| RUNDC3B | 5.33E − 11 | 1.39E − 06 | −1.89913 |
| NR1H4 | 5.50E − 11 | 1.44E − 06 | −2.46055 |
| SLC30A10 | 5.68E − 11 | 1.48E − 06 | −3.22414 |
| PIGR | 6.05E − 11 | 1.58E − 06 | −2.18018 |
| MUC2 | 6.24E − 11 | 1.63E − 06 | −2.62907 |
| HPGD | 6.74E − 11 | 1.76E − 06 | −2.64497 |
| TSPAN7 | 6.84E − 11 | 1.79E − 06 | −2.30396 |
| PAPSS2 | 1.30E − 10 | 3.39E − 06 | −1.78856 |
| SLC17A4 | 1.31E − 10 | 3.43E − 06 | −2.31184 |
| DHRS9 | 1.36E − 10 | 3.56E − 06 | −2.74003 |
| CAPN9 | 1.65E − 10 | 4.32E − 06 | −1.77713 |
| PTPRH | 1.70E − 10 | 4.43E − 06 | −1.93474 |
| SST | 1.81E − 10 | 4.73E − 06 | −2.35448 |
| HEPACAM2 | 1.82E − 10 | 4.76E − 06 | −2.85009 |
| SMPDL3A | 2.03E − 10 | 5.31E − 06 | −1.85516 |
| UGT2A3 | 2.16E − 10 | 5.65E − 06 | −2.42200 |
| TRPM6 | 2.42E − 10 | 6.34E − 06 | −2.40017 |
| CES2 | 2.58E − 10 | 6.74E − 06 | −1.80950 |
| NR5A2 | 2.64E − 10 | 6.91E − 06 | −1.65550 |
| DHRS11 | 2.77E − 10 | 7.25E − 06 | −2.40964 |
| ENTPD5 | 2.88E − 10 | 7.52E − 06 | −1.91140 |
| PLCL2 | 2.98E − 10 | 7.79E − 06 | −1.71267 |
| MYO1A | 2.98E − 10 | 7.79E − 06 | −1.82841 |
| BTNL8 | 3.28E − 10 | 8.58E − 06 | −2.68136 |
| PLCD1 | 3.40E − 10 | 8.88E − 06 | −1.69423 |
| ADTRP | 3.71E − 10 | 9.69E − 06 | −2.66899 |
| ARL14 | 4.37E − 10 | 1.14E − 05 | −2.10661 |
| PDZD3 | 4.78E − 10 | 1.25E − 05 | −1.73340 |
| SCNN1B | 5.25E − 10 | 1.37E − 05 | −2.51853 |
| MEP1A | 5.25E − 10 | 1.37E − 05 | −2.15953 |
| C2orf88 | 5.40E − 10 | 1.41E − 05 | −2.60606 |
| GPA33 | 5.69E − 10 | 1.49E − 05 | −1.85470 |
| SCIN | 6.20E − 10 | 1.62E − 05 | −2.19971 |
| MAOA | 6.61E − 10 | 1.73E − 05 | −1.79573 |
| PIGZ | 6.89E − 10 | 1.80E − 05 | −1.67717 |
| PYY | 6.96E − 10 | 1.82E − 05 | −2.53788 |
| C4orf19 | 7.26E − 10 | 1.90E − 05 | −1.57357 |
| TNFRSF17 | 7.47E − 10 | 1.95E − 05 | −2.36650 |
| ADAMDEC1 | 7.47E − 10 | 1.95E − 05 | −2.39500 |
| TEX11 | 8.71E − 10 | 2.28E − 05 | −2.24440 |
| CDHR2 | 9.44E − 10 | 2.47E − 05 | −1.71114 |
| TMEM171 | 9.64E − 10 | 2.52E − 05 | −2.11860 |
| STMN2 | 1.28E − 09 | 3.36E − 05 | −2.05581 |
| PKIB | 1.45E − 09 | 3.78E − 05 | −2.98037 |
| CLIC5 | 1.57E − 09 | 4.11E − 05 | −1.55858 |
| BMP2 | 1.60E − 09 | 4.19E − 05 | −1.76798 |
| RETSAT | 1.63E − 09 | 4.26E − 05 | −1.68389 |
| PADI2 | 1.69E − 09 | 4.42E − 05 | −2.10023 |
| SLC9A2 | 1.80E − 09 | 4.70E − 05 | −1.55732 |
| CYP2C18 | 2.16E − 09 | 5.65E − 05 | −1.71017 |
| C1orf115 | 2.32E − 09 | 6.05E − 05 | −1.74460 |
| CEACAM1 | 2.42E − 09 | 6.31E − 05 | −1.84011 |
| PDE9A | 2.44E − 09 | 6.37E − 05 | −2.05665 |
| SLCO2A1 | 2.58E − 09 | 6.75E − 05 | −1.53024 |
| BCAR3 | 2.63E − 09 | 6.87E − 05 | −1.50716 |
| METTL7A | 2.65E − 09 | 6.92E − 05 | −1.62515 |
| DSC2 | 2.65E − 09 | 6.92E − 05 | −1.44448 |
| IL1R2 | 2.75E − 09 | 7.19E − 05 | −2.16852 |
| CCDC68 | 2.75E − 09 | 7.19E − 05 | −1.89156 |
| GDPD3 | 2.78E − 09 | 7.28E − 05 | −2.08667 |
| PDE6A | 2.81E − 09 | 7.34E − 05 | −1.93721 |
| EPHX2 | 3.12E − 09 | 8.16E − 05 | −1.49186 |
| SPIB | 3.35E − 09 | 8.76E − 05 | −2.27898 |
| ITPKA | 3.41E − 09 | 8.91E − 05 | −1.62863 |
| SCUBE2 | 3.66E − 09 | 9.57E − 05 | −1.51582 |
| AMPD1 | 4.13E − 09 | 0.000108 | −1.81548 |
| SELENBP1 | 4.61E − 09 | 0.000120 | −2.04099 |
| TFCP2L1 | 5.10E − 09 | 0.000133 | −1.16817 |
| BTNL3 | 5.13E − 09 | 0.000134 | −2.10512 |
| VIPR1 | 5.45E − 09 | 0.000142 | −1.57631 |
| ZZEF1 | 5.53E − 09 | 0.000144 | −1.27744 |
| NAT2 | 5.61E − 09 | 0.000147 | −1.61058 |
| CDHR5 | 5.81E − 09 | 0.000152 | −1.97788 |
| PTGDR | 6.44E − 09 | 0.000168 | −2.08658 |
| HHLA2 | 7.58E − 09 | 0.000198 | −1.91952 |
| PLCE1 | 7.60E − 09 | 0.000199 | −1.63459 |
| BEST4 | 7.87E − 09 | 0.000206 | −2.37175 |
| FUCA1 | 8.12E − 09 | 0.000212 | −1.41919 |
| FGFBP1 | 8.12E − 09 | 0.000212 | −1.45128 |
| MT1X | 8.24E − 09 | 0.000215 | −1.87572 |
| SLC16A9 | 8.52E − 09 | 0.000223 | −1.97426 |
| SEMA6A | 8.52E − 09 | 0.000223 | −1.66469 |
| FXYD3 | 8.58E − 09 | 0.000224 | −1.39744 |
| DEFB1 | 8.94E − 09 | 0.000233 | −1.76340 |
| ACADS | 9.22E − 09 | 0.000241 | −1.70299 |
| UGDH | 9.50E − 09 | 0.000248 | −1.45862 |
| HRASLS2 | 1.06E − 08 | 0.000276 | −1.48365 |
| CYP4F12 | 1.09E − 08 | 0.000284 | −1.26493 |
| JCHAIN | 1.10E − 08 | 0.000288 | −2.33454 |
| ABHD3 | 1.13E − 08 | 0.000295 | −1.54939 |
| VWA5A | 1.23E − 08 | 0.000321 | −1.12976 |
| LDHD | 1.27E − 08 | 0.000332 | −2.10259 |
| XDH | 1.28E − 08 | 0.000334 | −1.53406 |
| INSL5 | 1.29E − 08 | 0.000337 | −2.42430 |
| SPINK5 | 1.38E − 08 | 0.000359 | −2.43812 |
| VILL | 1.50E − 08 | 0.000391 | −1.41689 |
| SQRDL | 1.51E − 08 | 0.000396 | −1.38225 |
| FABP2 | 1.52E − 08 | 0.000398 | −1.88374 |
| LGALS4 | 1.61E − 08 | 0.000421 | −1.42830 |
| DNASE1L3 | 1.65E − 08 | 0.000431 | −1.82008 |
| SLC44A4 | 1.66E − 08 | 0.000434 | −1.59136 |
| GOLM1 | 1.68E − 08 | 0.000439 | −1.20581 |
| RNF186 | 1.72E − 08 | 0.000450 | −1.21896 |
| GDPD2 | 1.85E − 08 | 0.000484 | −1.90186 |
| SLC22A18AS | 1.85E − 08 | 0.000484 | −1.62665 |
| MGLL | 1.91E − 08 | 0.000499 | −1.32867 |
| B3GNT6 | 1.92E − 08 | 0.000501 | −1.59112 |
| IL6R | 1.92E − 08 | 0.000502 | −1.86011 |
| RAPGEFL1 | 1.92E − 08 | 0.000502 | −1.32000 |
| TRPM4 | 1.94E − 08 | 0.000508 | −1.34414 |
| ETFDH | 2.00E − 08 | 0.000522 | −1.52397 |
| TMPRSS2 | 2.05E − 08 | 0.000536 | −1.39816 |
| APOBR | 2.08E − 08 | 0.000544 | −1.69871 |
| APPL2 | 2.11E − 08 | 0.000551 | −1.46951 |
| IQGAP2 | 2.17E − 08 | 0.000566 | −1.55628 |
| FAM83E | 2.17E − 08 | 0.000568 | −1.17487 |
| ETHE1 | 2.29E − 08 | 0.000599 | −1.61458 |
| FLVCR2 | 2.39E − 08 | 0.000624 | −1.01211 |
| ALDH1A1 | 2.41E − 08 | 0.000631 | −1.28928 |
| DENND2A | 2.51E − 08 | 0.000657 | −1.32793 |
| HRCT1 | 2.57E − 08 | 0.000671 | −1.58787 |
| ATP8B1 | 2.69E − 08 | 0.000703 | −1.00964 |
| A1CF | 2.74E − 08 | 0.000715 | −1.36877 |
| ELOVL6 | 2.96E − 08 | 0.000773 | −1.08274 |
| C11orf86 | 3.02E − 08 | 0.000789 | −2.11210 |
| TMEM100 | 3.14E − 08 | 0.000821 | −1.69604 |
| ASAP3 | 3.27E − 08 | 0.000854 | −1.44474 |
| S100A14 | 3.29E − 08 | 0.000859 | −1.21353 |
| PHLPP2 | 3.42E − 08 | 0.000892 | −1.45264 |
| LIMA1 | 3.47E − 08 | 0.000907 | −1.20588 |
| FABP1 | 3.51E − 08 | 0.000916 | −1.62316 |
| FMO5 | 3.62E − 08 | 0.000947 | −1.41152 |
| LRMP | 3.64E − 08 | 0.000950 | −1.84646 |
| UGP2 | 3.68E − 08 | 0.000961 | −1.28750 |
| ATP2A3 | 3.74E − 08 | 0.000978 | −1.51001 |
| SLC22A5 | 3.74E − 08 | 0.000978 | −1.30616 |
| FAM46C | 3.78E − 08 | 0.000989 | −1.41521 |
| LXN | 3.97E − 08 | 0.001037 | −1.10935 |
| CDKN2B | 3.98E − 08 | 0.001039 | −2.23550 |
| HMOX1 | 3.99E − 08 | 0.001043 | −1.23547 |
| NEDD4L | 4.03E − 08 | 0.001054 | −1.23680 |
| CLDN7 | 4.18E − 08 | 0.001093 | −1.29905 |
| PARM1 | 4.21E − 08 | 0.001099 | −1.52095 |
| HSD3B2 | 4.29E − 08 | 0.001120 | −2.26639 |
| KIAA0513 | 4.39E − 08 | 0.001146 | −1.22322 |
| ACADVL | 4.50E − 08 | 0.001176 | −1.23429 |
| SYTL2 | 4.74E − 08 | 0.001238 | −1.07640 |
| SLC1A1 | 4.79E − 08 | 0.001251 | −1.40176 |
| CNTN3 | 4.81E − 08 | 0.001256 | −2.13362 |
| CNNM4 | 4.97E − 08 | 0.001300 | −1.36807 |
| CASP7 | 5.04E − 08 | 0.001317 | −1.34691 |
| GLTP | 5.30E − 08 | 0.001385 | −1.40471 |
| PGM1 | 5.49E − 08 | 0.001435 | −1.23037 |
| ERN2 | 5.60E − 08 | 0.001464 | −1.10843 |
| NAAA | 5.72E − 08 | 0.001494 | −1.45647 |
| GNA11 | 6.10E − 08 | 0.001593 | −1.40416 |
| CCL23 | 6.37E − 08 | 0.001665 | −1.50079 |
| STYK1 | 6.40E − 08 | 0.001673 | −1.56848 |
| CFD | 6.56E − 08 | 0.001715 | −1.80374 |
| C15orf48 | 6.82E − 08 | 0.001783 | −1.45026 |
| OASL | 6.89E − 08 | 0.001800 | −1.13508 |
| GPT | 6.99E − 08 | 0.001827 | −1.44289 |
| CLDN23 | 7.12E − 08 | 0.001859 | −1.93577 |
| MB | 7.62E − 08 | 0.001991 | −1.42542 |
| HIGD1A | 7.65E − 08 | 0.002000 | −1.40813 |
| EMP1 | 8.37E − 08 | 0.002188 | −1.46200 |
| ADH1B | 8.46E − 08 | 0.002211 | −2.01430 |
| LPAR1 | 8.49E − 08 | 0.002219 | −1.26559 |
| GPAT3 | 8.70E − 08 | 0.002272 | −2.01808 |
| CHGB | 8.90E − 08 | 0.002324 | −1.55043 |
| P3H2 | 9.09E − 08 | 0.002376 | −1.48288 |
| PRKACB | 9.28E − 08 | 0.002424 | −1.46492 |
| OSBPL1A | 9.84E − 08 | 0.002572 | −1.16940 |
| SPINK4 | 1.04E − 07 | 0.002716 | −1.69181 |
| MT2A | 1.04E − 07 | 0.002726 | −1.55590 |
| SULT1B1 | 1.07E − 07 | 0.002807 | −1.66731 |
| NAT1 | 1.08E − 07 | 0.002828 | −1.17391 |
| ADRA2A | 1.11E − 07 | 0.002892 | −1.54370 |
| MEP1B | 1.13E − 07 | 0.002960 | −2.16734 |
| LPCAT4 | 1.18E − 07 | 0.003077 | −1.02702 |
| PCSK7 | 1.18E − 07 | 0.003077 | −1.00185 |
| ST6GALNAC1 | 1.18E − 07 | 0.003088 | −1.65355 |
| SGK2 | 1.21E − 07 | 0.003160 | −1.41158 |
| GRAMD3 | 1.26E − 07 | 0.003287 | −1.25518 |
| RIOK3 | 1.27E − 07 | 0.003331 | −1.21281 |
| CITED2 | 1.29E − 07 | 0.003374 | −1.11561 |
| CXCL12 | 1.41E − 07 | 0.003684 | −1.54414 |
| ITM2A | 1.45E − 07 | 0.003786 | −1.47880 |
| SEPP1 | 1.51E − 07 | 0.003956 | −1.10723 |
| KBTBD11 | 1.53E − 07 | 0.004006 | −1.05544 |
| RHOF | 1.55E − 07 | 0.004040 | −1.11757 |
| GSTA1 | 1.57E − 07 | 0.004105 | −1.34829 |
| GALNT12 | 1.61E − 07 | 0.004214 | −1.23449 |
| BCHE | 1.70E − 07 | 0.004446 | −1.57410 |
| HGD | 1.78E − 07 | 0.004657 | −1.24750 |
| HOXD1 | 1.83E − 07 | 0.004793 | −1.79058 |
| FGL2 | 1.90E − 07 | 0.004977 | −1.31339 |
| ATP8A1 | 2.03E − 07 | 0.005307 | −1.05015 |
| HIST1H1C | 2.06E − 07 | 0.005393 | −1.25170 |
| MYOT | 2.10E − 07 | 0.005480 | −1.72660 |
| PBLD | 2.11E − 07 | 0.005502 | −1.58127 |
| RBM47 | 2.15E − 07 | 0.005614 | −1.08864 |
| CES3 | 2.18E − 07 | 0.005700 | −1.45856 |
| CD1D | 2.18E − 07 | 0.005704 | −1.29631 |
| FAM150B | 2.19E − 07 | 0.005719 | −1.50655 |
| RARRES1 | 2.23E − 07 | 0.005819 | −1.46770 |
| LGALS9 | 2.24E − 07 | 0.005842 | −1.01981 |
| KLK1 | 2.27E − 07 | 0.005936 | −1.29450 |
| TOX | 2.27E − 07 | 0.005942 | −1.29132 |
| ACVRL1 | 2.36E − 07 | 0.006175 | −1.13083 |
| CPT2 | 2.44E − 07 | 0.006372 | −1.22566 |
| SGK1 | 2.57E − 07 | 0.006718 | −1.64587 |
| ALPI | 2.62E − 07 | 0.006834 | −1.09455 |
| SLC51B | 2.65E − 07 | 0.006919 | −2.27581 |
| SLC20A1 | 2.71E − 07 | 0.007076 | −1.03315 |
| SCGN | 2.74E − 07 | 0.007164 | −1.86108 |
| ASPA | 2.75E − 07 | 0.007186 | −1.45497 |
| FAM107B | 2.81E − 07 | 0.007337 | −1.24898 |
| AGR3 | 2.82E − 07 | 0.007358 | −1.80558 |
| HOXA5 | 2.85E − 07 | 0.007437 | −1.09160 |
| NPY1R | 2.97E − 07 | 0.007754 | −1.92966 |
| TNFSF10 | 2.97E − 07 | 0.007754 | −1.20956 |
| FAM107A | 3.18E − 07 | 0.008318 | −1.40373 |
| MUC4 | 3.26E − 07 | 0.008520 | −1.83234 |
| TST | 3.31E − 07 | 0.008645 | −1.30505 |
| NXPE1 | 3.57E − 07 | 0.009326 | −1.90526 |
| SPON1 | 3.60E − 07 | 0.009400 | −1.78810 |
| ST6GALNAC6 | 3.78E − 07 | 0.009874 | −1.79857 |
FC: fold change.
Figure 3.
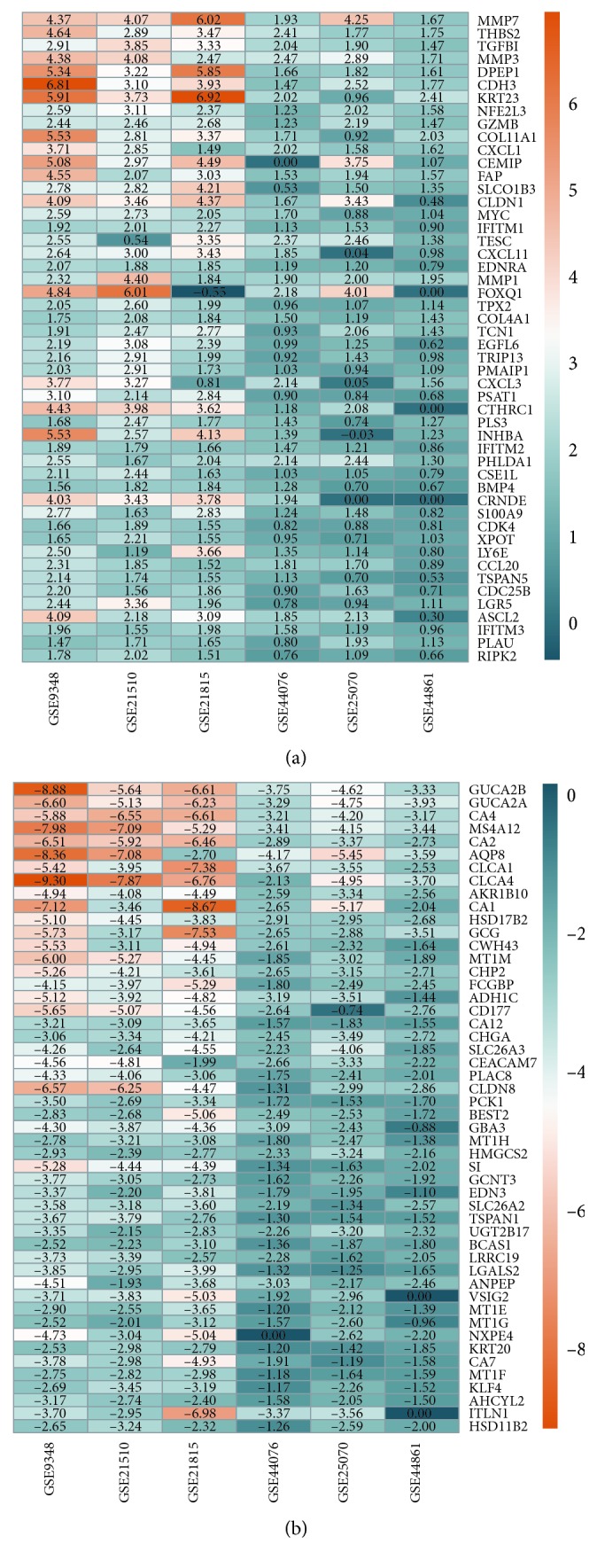
Top 50 robust DEGs identified in the six microarray data (i.e., GSE25070, GSE44076, GSE44861, GSE21510, GSE9348, and GSE21815) from the GEO database: (a) upregulated genes; (b)downregulated genes. These genes were ranked by adjusted P value.
2.4. Survival Analysis in the TCGA Database and Validation in the GEO Database
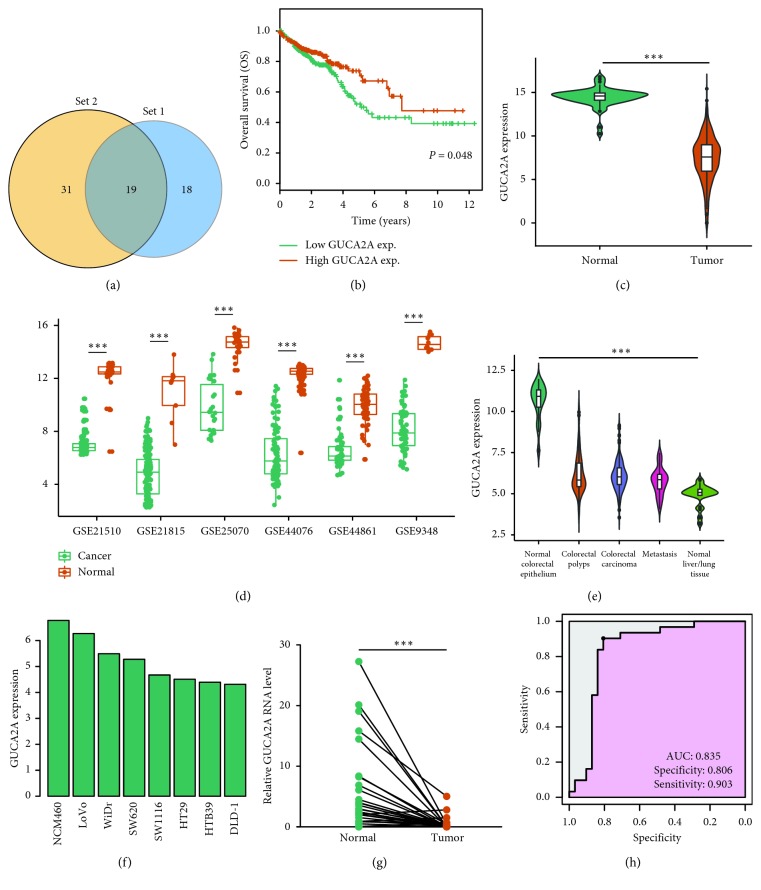
By taking the intersection of the 50 downregulated DEGs and hub genes in the red module from WGCNA (Figure 4(a)), we obtained 19 interacting genes.
Figure 4.
Aberrant expression of GUCA2A can be a candidate prognostic biomarker in CRC. (a) Venn diagram of the robust DEGs and genes in the red module revealed 19 interacting genes. Set 1: hub genes in the red module; Set 2: downregulated DEGs. (b) Five-year OS of CRC patients from the TCGA database (P=0.048). Patients with a low expression of GUCA2A have a significantly shorter OS. (c) GUCA2A expression in the TCGA database. (d) GUAC2A expression between CRC tissues and normal tissues in the six datasets. (e) GUAC2A expression in different tissue histology. (f) CRC cells in GSE68468. (g) GUCA2A expression levels in human CRC and their matched adjacent normal tissues. (h) ROC curves of tissue GUCA2A expression for differentiating CRC tissue from normal tissue. ∗∗∗ P < 0.001.
K-M analysis was conducted to evaluate the relationship between gene expression and the overall survival (OS) of CRC, and only GUCA2A was clearly related to the OS of CRC patients in the TCGA database. Patients with a lower GUCA2A expression had a significantly shorter OS than those with a higher expression (P < 0.05) (Figure 4(b)). Obviously, GUCA2A was abnormally expressed in tumor tissues and was significantly different in TCGA and GEO databases (Figures 4(c) and 4(d)). We further validated the aberrant expression of GUCA2A in GSE68468, which contains both CRC tissues and cellular RNA-Seq data. Compared with adjacent normal tissues, the expression of GUCA2A in tumor and metastatic tissues was significantly low, and the normal liver and lung tissues had the lowest expression value (P < 0.05) (Figure 4(e)). The colonic epithelial cell (NCM460) has the highest GUCA2A expression compared with other CRC cells (Figure 4(f)).
2.5. Human Tissue Samples and Quantitative Real-Time PCR
We performed real-time PCR to examine the levels of GUCA2A in 31 paired CRC/adjacent tissues to further validate the dysregulated expression of GUCA2A (Figure 4(g)). Then, we further evaluated the diagnostic value of GUCA2A for CRC tissues and normal counterparts using ROC curve analyses. The results showed that the area under the ROC curve (AUC) was 0.835 (P < 0.001; sensitivity: 0.806; specificity: 0.903). These results suggest that GUCA2A downregulation may play an important role in colorectal tumorigenesis and has a potential diagnostic value for CRC patients.
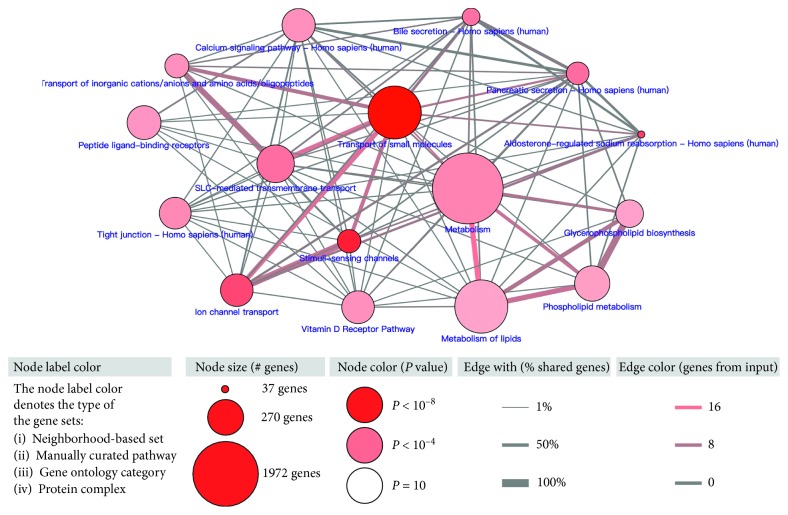
2.6. Pathway Analysis
The pathway enrichment analysis of GUCA2A coexpressed genes was conducted in the ConsensusPathDB human database. From the 16 pathways shown in Figure 5, the transport of small molecules and metabolism are prominent.
Figure 5.
Pathway enrichment analysis for GUCA2A coexpressed genes.
3. Discussion
Early detection and complete resection before metastasis are considered the only curative therapy for CRC [11]. The five-year survival rate of CRC patients was obviously better when the localized disease was detected at an early stage than that of CRC patients with distant metastasis. Cancer is a molecularly heterogeneous disease, and none of the currently identified biomarkers are sensitive and specific enough for reliable CRC screening in the clinical setting. Thus, identifying novel molecular biomarkers has significant clinical benefits.
In our study, we first performed WGCNA for GSE68468 and screened the pathologically related hub genes. We conducted RRA analysis for six datasets and screened the top 100 robust DEGs, which had a high or low expression in all expression profiles. By taking the intersection, we obtained 19 candidate genes, and only the expression of GUCA2A was associated with the OS of CRC patients in the TCGA database. We found that GUCA2A was prominent and top-ranking in both the hub gene network (Figure 2) and robust DEGs (Figure 3(b)), indicating its value in tumorigenesis.
Guanylin (GUCA2A) and uroguanylin (GUCA2B) are two peptide hormones that function as paracrine endogenous ligands for the guanylate cyclase-C (GUCY2C) receptor [12]. A previous study indicated the role of GUCY2C signaling in facilitating mucosal wounding and inflammation mediation, in part, through the control of resistin-like molecule β production [13]. GUCA2A, GUCA2B, and GUCY2C are downregulated in inflammatory bowel disease [14], which may have implications in inflammatory bowel disease pathogenesis. A recent study suggests that GUCY2C can suppress tumor progress [15] in the intestine, and the loss of the GUCY2C signaling cascade increases CRC susceptibility [16]. Intestinal homeostasis disruption and CRC tumorigenesis are associated with a fairly common loss of GUCA2A and GUCA2B [17–19]. Bashir et al. revealed the possibility of GUCA2A loss silencing GUCY2C, which leads to microsatellite instability tumor [20].
Consistent with the expression of GUCA2A in our study, the expression of GUCA2B was significantly downregulated in CRC tissues and had a relative high weight in the red module, which further indicates that GUCA2A and GUCA2B may play a consistent role in CRC neoplasia.
In conclusion, we revealed that GUCA2A was downregulated in CRC tissues. Aberrantly expressed GUCA2A can be a candidate marker of poor prognosis in patients with CRC, which may be a therapeutic target for precision medicine in cervical cancer. However, further studies are still needed to explore the underlying molecular mechanism through which GUCA2A plays a role in CRC. However, future in vivo and in vitro experiments are still required to explore the mechanisms underlying the roles of GUCA2A in CRC.
4. Methods
4.1. WGCNA Construction and Module Detection
We performed WGCNA to microarray data GSE68468 generated from 378 CRC tissue samples, and the “WGCNA” package in R 3.5.1 was used to construct a coexpression network. WGCNA seeks to identify modules of densely interconnected genes by searching for genes with similar patterns of connection strengths or high topological overlap. For each dataset, Pearson correlation coefficients were calculated for all pairwise comparisons of expressed genes across all samples. Genes with similar expression profiles were classified into modules based on the TOM dissimilarity with a minimum size of 30 for the gene dendrogram and visualized via hierarchical clustering [7]. Then, the modules whose eigengenes were highly correlated (correlation above 0.7) were merged. The gene network was visualized with randomly selected 400 genes.
The resulting Pearson correlation matrix was transformed into a matrix of connection strengths (that is, an adjacency matrix) by using a power function (connection). All the modules were assigned to the corresponding color. The relationships of modules and clinical traits (i.e., disease status, histology, and organism part) were calculated. Among these clinical traits, pathology, including normal mucosa, polyps, CRC tissue, CRC with metastases, and normal lung/liver tissues, can reflect the occurrence and metastasis of CRC. The associations of individual genes with the clinical trait, namely, gene significance (GS), and the module eigengenes, namely, module membership (MM), were evaluated. Then, the correlation between GS and MM was calculated, and the highly correlated interest module can be used to construct the coexpression network and identify the hub genes.
4.2. Coexpression Network Construction and Hub Gene Identification
The genes in the key modules screened from WGCNA were further analyzed.
We filtered the hub genes by a condition of the weight value (>0.15) and visualized them using Cytoscape 3.6.0 [21].
4.3. Robust Rank Aggregation (RRA) Analysis
To further increase the reliability of the results and screen out the ideal candidate, we enrolled six published colorectal microarray data (i.e., GSE25070, GSE44076, GSE44861, GSE21510, GSE9348, and GSE21815) [22–31] from the GEO database, which have 530 CRC tissues and 50 normal tissues in our study (Table 2). After screening DEGs (adjusted P value < 0.05 and |logFC| ≥ 2) in each dataset by using the “limma” package [32], the RRA method was used to identify significantly robust DEGs by using the library “RobustRankAggreg” package in R 3.5.1. The statistically significant DEGs were defined to have adjusted P value < 0.01 and |logFC| ≥ 1. Finally, the top 50 up- and downregulated DEGs were selected and visualized by using a heatmap.
Table 2.
Detailed information on the six microarrays from the GEO database.
| Reference | Tissue | GEO | Platform | Normal | Tumor |
|---|---|---|---|---|---|
| Hinoue et al. [22] | CRC | GSE25070 | GPL6883 | 51 | 51 |
| Cordero et al. [23–26] | CRC | GSE44076 | GPL13667 | 148 | 98 |
| Ryan et al. [27] | CRC | GSE44861 | GPL3921 | 55 | 56 |
| Tsukamoto et al. [28] | CRC | GSE21510 | GPL570 | 25 | 123 |
| Hong et al. [29] | CRC | GSE9348 | GPL570 | 12 | 70 |
| Mimori et al. [30, 31] | CRC | GSE21815 | GPL6480 | 9 | 132 |
4.4. Survival Analysis in the TCGA Database
In order to increase the reliability of the results, the intersection of hub genes in the red module from WGCNA and 50 downregulated robust DEGs was performed and analyzed. Kaplan–Meier (K-M) analysis was conducted to evaluate the relationship between gene expression and the overall survival (OS) of 617 CRC patients in The Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/). Patients were classified into high- or low-expression groups according to the median value. Then, genes associated with CRC survival were screened.
4.5. Human Tissue Samples and Quantitative Real-Time PCR
Overall, the 31 CRC and adjacent normal tissues obtained at Jiangsu Cancer Hospital (Nanjing, China) were frozen immediately after surgical resection and kept at 80°C until further analysis. Tumor histopathology was classified according to the World Health Organization Classification of Tumors system. The present study was done with the approval of the local ethics committee.
RNA isolation (Takara, Dalian, China) was performed according to the manufacturer's instructions. Reverse transcription was conducted with a PrimeScript RT reagent kit (Takara, Kusatsu City, Shiga, Japan) and the fluorescent quantitative experiment with ABI qPCR 7300 (Torrance, CA). The PCR reactant mix consisted of 2 µl cDNA solution, 10 μl 2× PowerUp™ SYBR™ Green Master Mix (Thermo Fisher Scientific, Carlsbad, CA), 0.5 μl of 10 μM forward and reverse PCR primer, and 7 μl DNA template Nuclease-Free Water. The PCR conditions were set as follows: denaturation at 50°C for 2 min, 95°C for 2 min, 95°C for 15 s, and 60°C for 1 min with 40 cycles. A GAPDH primer set was used as an internal control.
4.6. Pathway Analysis
We performed pathway enrichment analysis for the coexpressed genes to explore the possible mechanism of the candidate gene in CRC. The coexpressed genes were obtained from the TCGA database, and the top 100 genes with the highest Pearson correlation coefficient were considered to be significantly coexpressed (Table 3). Pathway enrichment analysis was performed by using the ConsensusPathDB human database (http://cpdb.molgen.mpg.de/) [33]. The overrepresentation gene set analysis was utilized, and the following pathway databases were enrolled in our analysis: Manual upload, NetPath, SignaLink, PID, EHMN, HumanCyc, INOH, KEGG, BioCarta, WikiPathways, SMPDB, and PharmGKB. Minimum overlap input list >5 and P value cutoff <0.01 were considered significant enrichment.
Table 3.
GUCA2A coexpressed genes in the TCGA database.
| Genes | Correlation | P value |
|---|---|---|
| GUCA2A | 1 | 0 |
| TMIGD1 | 0.81034948 | 6.78E − 145 |
| CA4 | 0.78988642 | 1.02E − 132 |
| GUCA2B | 0.78006796 | 2.39E − 127 |
| SCNN1B | 0.76922102 | 9.92E − 122 |
| CDKN2B-AS1 | 0.75976020 | 4.48E − 117 |
| LINC00974 | 0.72826962 | 5.10E − 103 |
| CA1 | 0.72589778 | 4.85E − 102 |
| OTOP2 | 0.72524636 | 8.98E − 102 |
| B3GNT7 | 0.69874083 | 1.59E − 91 |
| CA2 | 0.69872813 | 1.60E − 91 |
| CLDN8 | 0.69162255 | 5.79E − 89 |
| CLCA4 | 0.68724381 | 2.01E − 87 |
| MS4A12 | 0.67084612 | 6.81E − 82 |
| CEACAM7 | 0.65697702 | 1.74E − 77 |
| BTNL3 | 0.64672606 | 2.24E − 74 |
| SLC25A34 | 0.64457072 | 9.77E − 74 |
| C11orf86 | 0.64075941 | 1.28E − 72 |
| CLDN23 | 0.64014863 | 1.93E − 72 |
| EDN3 | 0.62755226 | 7.21E − 69 |
| RP11-35P15.1 | 0.62358770 | 8.90E − 68 |
| B3GALT5-AS1 | 0.62060619 | 5.75E − 67 |
| SULT1A2 | 0.61591517 | 1.04E − 65 |
| PYY | 0.61445537 | 2.53E − 65 |
| ZG16 | 0.61434490 | 2.71E − 65 |
| ALPI | 0.61109844 | 1.93E − 64 |
| SLC25A47P1 | 0.60742616 | 1.73E − 63 |
| BTNL8 | 0.60619878 | 3.58E − 63 |
| BEST4 | 0.59344211 | 5.70E − 60 |
| CD177 | 0.59322686 | 6.44E − 60 |
| ST6GALNAC6 | 0.59074429 | 2.60E − 59 |
| CDHR5 | 0.58538873 | 5.07E − 58 |
| TRPM6 | 0.57770739 | 3.26E − 56 |
| SEPP1 | 0.57757017 | 3.51E − 56 |
| KRTAP13-2 | 0.57186209 | 7.21E − 55 |
| SLC4A4 | 0.56853673 | 4.08E − 54 |
| RP11-203J24.9 | 0.55828248 | 7.58E − 52 |
| MYPN | 0.55589062 | 2.50E − 51 |
| RP11-202A13.1 | 0.54993651 | 4.66E − 50 |
| SLC30A10 | 0.54640858 | 2.57E − 49 |
| SLC26A3 | 0.54279335 | 1.45E − 48 |
| SMPD1 | 0.54142083 | 2.77E − 48 |
| CDKN2B-AS | 0.53902904 | 8.55E − 48 |
| RN7SKP127 | 0.53769807 | 1.59E − 47 |
| HHLA2 | 0.53644205 | 2.86E − 47 |
| TMEM82 | 0.53029307 | 4.85E − 46 |
| LYPD8 | 0.52880179 | 9.56E − 46 |
| CD177P1 | 0.52733551 | 1.85E − 45 |
| SCNN1G | 0.52521670 | 4.81E − 45 |
| ABCC13 | 0.52358277 | 9.98E − 45 |
| GDPD2 | 0.51942104 | 6.29E − 44 |
| TTC22 | 0.51932599 | 6.56E − 44 |
| B3GALT5 | 0.51777531 | 1.29E − 43 |
| C2orf88 | 0.51755217 | 1.43E − 43 |
| DHRS9 | 0.51539770 | 3.64E − 43 |
| TSPAN1 | 0.51536862 | 3.69E − 43 |
| INSL5 | 0.51280701 | 1.11E − 42 |
| TMEM253 | 0.50785220 | 9.21E − 42 |
| SDCBP2 | 0.50717257 | 1.23E − 41 |
| SLC17A4 | 0.50515764 | 2.86E − 41 |
| PLAC8 | 0.50131456 | 1.42E − 40 |
| MMP28 | 0.49830422 | 4.90E − 40 |
| PPY | 0.49796769 | 5.62E − 40 |
| PLA2G10 | 0.49555028 | 1.51E − 39 |
| GLRA4 | 0.49493081 | 1.94E − 39 |
| ITM2C | 0.49231037 | 5.57E − 39 |
| TRIM40 | 0.49190614 | 6.55E − 39 |
| SMPDL3A | 0.49188755 | 6.60E − 39 |
| MALL | 0.49120642 | 8.68E − 39 |
| BCAS1 | 0.49019232 | 1.30E − 38 |
| CDKN2B | 0.48925853 | 1.89E − 38 |
| SLC51B | 0.48708976 | 4.46E − 38 |
| BMP3 | 0.48536366 | 8.79E − 38 |
| AMPD1 | 0.48202823 | 3.23E − 37 |
| ENTPD5 | 0.47054777 | 2.56E − 35 |
| PTPRH | 0.46882533 | 4.87E − 35 |
| RPL12P14 | 0.46759668 | 7.68E − 35 |
| RP11-92A5.2 | 0.46652136 | 1.14E − 34 |
| AC106869.2 | 0.46640139 | 1.19E − 34 |
| MUC12 | 0.46578524 | 1.50E − 34 |
| CASC18 | 0.46570335 | 1.55E − 34 |
| PEX26 | 0.46071845 | 9.52E − 34 |
| TMEM72 | 0.46026123 | 1.12E − 33 |
| GLDN | 0.45995677 | 1.25E − 33 |
| TEX11 | 0.45994953 | 1.26E − 33 |
| NAAA | 0.45741196 | 3.13E − 33 |
| TRANK1 | 0.45665719 | 4.10E − 33 |
| SMIM6 | 0.45572985 | 5.71E − 33 |
| LINC00507 | 0.44872641 | 6.70E − 32 |
| AOC1 | 0.44821442 | 8.01E − 32 |
| CDHR2 | 0.44663931 | 1.38E − 31 |
| APPL2 | 0.44091022 | 9.81E − 31 |
| TMPRSS2 | 0.43862005 | 2.12E − 30 |
| CCDC152 | 0.43855903 | 2.17E − 30 |
| TRPV3 | 0.43605403 | 5.02E − 30 |
| NPY2R | 0.43603750 | 5.04E − 30 |
| GNA11 | 0.43389107 | 1.03E − 29 |
| ACVRL1 | 0.43021941 | 3.44E − 29 |
| SEMA6D | 0.42947338 | 4.39E − 29 |
| GPR15 | 0.42871749 | 5.62E − 29 |
| RP1-117B12.4 | 0.40855340 | 3.19E − 26 |
Acknowledgments
This study was supported in part by grants from the Jiangsu Provincial Key Research Development Program (BE2016794 to Jifeng Feng and BE2016795 to Jianzhong Wu).
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Supplementary Materials
S1: sample dendrogram and soft-thresholding value estimation. (A) Sample dendrogram and trait heatmap. The three traits correspond to the disease status, histology, and organism part, respectively. (B) Scale independence and mean connectivity of soft-thresholding values (β). (C) Clustering of module eigengenes. The dissimilarity was set as 0.25 to merge the similar modules. S2: volcano plots of DEGs between cancerous and adjacent tissues in six microarray data.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Chen W., Zheng R., Baade P. D., et al. Cancer statistics in China, 2015. CA: A Cancer Journal for Clinicians. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Daly M., Paquette I. Surveillance, epidemiology, and end results (SEER) and SEER-medicare databases: use in clinical research for improving colorectal cancer outcomes. Clinics in Colon and Rectal Surgery. 2019;32(01):61–68. doi: 10.1055/s-0038-1673355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siegel R. L., Miller K. D., Jemal A. Cancer statistics, 2017. CA: A Cancer Journal for Clinicians. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 5.Punt C. J. A., Koopman M., Vermeulen L. From tumour heterogeneity to advances in precision treatment of colorectal cancer. Nature Reviews Clinical Oncology. 2017;14(4):235–246. doi: 10.1038/nrclinonc.2016.171. [DOI] [PubMed] [Google Scholar]
- 6.Baltimore D. Our genome unveiled. Nature. 2001;409(6822):814–816. doi: 10.1038/35057267. [DOI] [PubMed] [Google Scholar]
- 7.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9(1) doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xue Z., Huang K., Cai C., et al. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature. 2013;500(7464):593–597. doi: 10.1038/nature12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kolde R., Laur S., Adler P., Vilo J. Robust rank aggregation for gene list integration and meta-analysis. Bioinformatics. 2012;28(4):573–580. doi: 10.1093/bioinformatics/btr709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Võsa U., Kolde R., Vilo J., Metspalu A., Annilo T. RNA Mapping. Vol. 1182. New York, NY, USA: Humana Press; 2014. Comprehensive meta-analysis of microRNA expression using a robust rank aggregation approach; pp. 361–373. [DOI] [PubMed] [Google Scholar]
- 11.Hollis M., Nair K., Vyas A., Chaturvedi L. S., Gambhir S., Vyas D. MicroRNAs potential utility in colon cancer: early detection, prognosis, and chemosensitivity. World Journal of Gastroenterology. 2015;21(27):8284–8292. doi: 10.3748/wjg.v21.i27.8284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuhn M. Molecular physiology of membrane guanylyl cyclase receptors. Physiological Reviews. 2016;96(2):751–804. doi: 10.1152/physrev.00022.2015. [DOI] [PubMed] [Google Scholar]
- 13.Steinbrecher K. A., Harmel-Laws E., Garin-Laflam M. P., et al. Murine guanylate cyclase C regulates colonic injury and inflammation. The Journal of Immunology. 2011;186(12):7205–7214. doi: 10.4049/jimmunol.1002469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brenna Ø., Bruland T., Furnes M. W., et al. The guanylate cyclase-C signaling pathway is down-regulated in inflammatory bowel disease. Scandinavian Journal of Gastroenterology. 2015;50(10):1241–1252. doi: 10.3109/00365521.2015.1038849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pattison A. M., Merlino D. J., Blomain E. S., Waldman S. A. Guanylyl cyclase C signaling axis and colon cancer prevention. World Journal of Gastroenterology. 2016;22(36):8070–8077. doi: 10.3748/wjg.v22.i36.8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blomain E. S., Lin J. E., Kraft C. L., et al. Translating colorectal cancer prevention through the guanylyl cyclase C signaling axis. Expert Review of Clinical Pharmacology. 2013;6(5):557–564. doi: 10.1586/17512433.2013.827406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Basu N., Arshad N., Visweswariah S. S. Receptor guanylyl cyclase C (GC-C): regulation and signal transduction. Molecular and Cellular Biochemistry. 2010;334(1-2):67–80. doi: 10.1007/s11010-009-0324-x. [DOI] [PubMed] [Google Scholar]
- 18.Li P., Schulz S., Bombonati A., et al. Guanylyl cyclase C suppresses intestinal tumorigenesis by restricting proliferation and maintaining genomic integrity. Gastroenterology. 2007;133(2):599–607. doi: 10.1053/j.gastro.2007.05.052. [DOI] [PubMed] [Google Scholar]
- 19.Lin J. E., Li P., Snook A. E., et al. The hormone receptor GUCY2C suppresses intestinal tumor formation by inhibiting AKT signaling. Gastroenterology. 2010;138(1):241–254. doi: 10.1053/j.gastro.2009.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bashir B., Merlino D. J., Rappaport J. A., et al. Silencing the GUCA2A-GUCY2C tumor suppressor axis in CIN, serrated, and MSI colorectal neoplasia. Human Pathology. 2019;87(19):103–114. doi: 10.1016/j.humpath.2018.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su G., Morris J. H., Demchak B., Bader G. D. Biological network exploration with cytoscape 3. Current Protocols in Bioinformatics. 2014;47(1):8.13.1–8.13.24. doi: 10.1002/0471250953.bi0813s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hinoue T., Weisenberger D. J., Lange C. P. E., et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Research. 2012;22(2):271–282. doi: 10.1101/gr.117523.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Solé X., Crous-Bou M., Cordero D., et al. Discovery and validation of new potential biomarkers for early detection of colon cancer. PLoS One. 2014;9(9) doi: 10.1371/journal.pone.0106748.e106748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cordero D., Solé X., Crous-Bou M., et al. Large differences in global transcriptional regulatory programs of normal and tumor colon cells. BMC Cancer. 2014;14(1) doi: 10.1186/1471-2407-14-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanz-Pamplona R., Berenguer A., Cordero D., et al. Aberrant gene expression in mucosa adjacent to tumor reveals a molecular crosstalk in colon cancer. Molecular Cancer. 2014;13(1):p. 46. doi: 10.1186/1476-4598-13-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Closa A., Cordero D., Sanz-Pamplona R., et al. Identification of candidate susceptibility genes for colorectal cancer through eQTL analysis. Carcinogenesis. 2014;35(9):2039–2046. doi: 10.1093/carcin/bgu092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ryan B. M., Zanetti K. A., Robles A. I., et al. Germline variation inNCF4, an innate immunity gene, is associated with an increased risk of colorectal cancer. International Journal of Cancer. 2014;134(6):1399–1407. doi: 10.1002/ijc.28457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsukamoto S., Ishikawa T., Iida S., et al. Clinical significance of osteoprotegerin expression in human colorectal cancer. Clinical Cancer Research. 2011;17(8):2444–2450. doi: 10.1158/1078-0432.ccr-10-2884. [DOI] [PubMed] [Google Scholar]
- 29.Hong Y., Downey T., Eu K. W., Koh P. K., Cheah P. Y. A ‘metastasis-prone’ signature for early-stage mismatch-repair proficient sporadic colorectal cancer patients and its implications for possible therapeutics. Clinical & Experimental Metastasis. 2010;27(2):83–90. doi: 10.1007/s10585-010-9305-4. [DOI] [PubMed] [Google Scholar]
- 30.Kogo R., Shimamura T., Mimori K., et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Research. 2011;71(20):6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 31.Iwaya T., Yokobori T., Nishida N., et al. Downregulation of miR-144 is associated with colorectal cancer progression via activation of mTOR signaling pathway. Carcinogenesis. 2012;33(12):2391–2397. doi: 10.1093/carcin/bgs288. [DOI] [PubMed] [Google Scholar]
- 32.Ritchie M. E., Phipson B., Wu D., et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research. 2015;43(7):p. e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamburov A., Wierling C., Lehrach H., Herwig R. ConsensusPathDB-a database for integrating human functional interaction networks. Nucleic Acids Research. 2009;37(1):D623–D628. doi: 10.1093/nar/gkn698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S1: sample dendrogram and soft-thresholding value estimation. (A) Sample dendrogram and trait heatmap. The three traits correspond to the disease status, histology, and organism part, respectively. (B) Scale independence and mean connectivity of soft-thresholding values (β). (C) Clustering of module eigengenes. The dissimilarity was set as 0.25 to merge the similar modules. S2: volcano plots of DEGs between cancerous and adjacent tissues in six microarray data.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.