Abstract
Understanding the evolution of biodiversity on Earth is a central aim in biology. Currently, various disciplines of science contribute to unravel evolution at all levels of life, from individual organisms to species and higher ranks, using different approaches and specific terminologies. The search for common origin, traditionally called homology, is a connecting paradigm of all studies related to evolution. However, it is not always sufficiently taken into account that defining homology depends on the hierarchical level studied (organism, population, and species), which can cause confusion. Therefore, we propose a framework to define homologies making use of existing terms, which refer to homology in different fields, but restricting them to an unambiguous meaning and a particular hierarchical level. We propose to use the overarching term “homology” only when “morphological homology,” “vertical gene transfer,” and “phylogenetic homology” are confirmed. Consequently, neither phylogenetic nor morphological homology is equal to homology. This article is intended for readers with different research backgrounds. We challenge their traditional approaches, inviting them to consider the proposed framework and offering them a new perspective for their own research.
Keywords: Analogy, character, common ancestry, genealogy, homology, homoplasy, orthology, paralogy
Homology is a central concept in biology, and it is used at all hierarchical levels of life. Minelli and Fusco (2013) published an extensive review of the long trajectory across centuries of the concept(s) of homology. Nevertheless, the current most common definition of homology is similarity due to common ancestry and mainly refers to the species level, implying a phylogenetic context (e.g., Darwin 1859; Wake et al. 2011; Nixon and Carpenter 2012). This of course implies fixation of at least some characters. Homology assessments, which are the subject of this text, are based on such fixed characters. We do not deny the relevance of character polymorphism (different phenotypes within a same species, see e.g., Fusco and Minelli 2010) in evolution and such cases may obscure homology assessments (e.g., Shubin et al. 2009) with subsequent methodological problems such as divergent time estimations (see e.g., Charlesworth 2010). Similarity which is not based on common ancestry is often called homoplasy (Lankester 1870; Hennig 1950, 1966; see also Sanderson and Hufford 1996; Scotland 2010) or analogy (Remane 1956). Although these definitions appear to be clear and unambiguous, we detected several causes for confusion leading to the misidentification of presumed homologies or homoplasies. Some of these confusions result from:
Historical change of the meaning of the term homology, which originally had an exclusively morphological context (Owen 1843, 1848). Later, the homology concept became accepted in a phylogenetic context (Haeckel 1866), but both approaches are coexisting until today (Kaplan 1984).
Using homology in different contexts, for example, in a morphological versus a phylogenetic context (Remane 1956). This results in a partial overlap of terms such as analogy and homology sensu Owen (1848), or leads to the creation of new terms such like homonomy, homonymy, homotypy, and homodynamy (Remane 1956). Recently, Vogt (2017), following Brigandt (2003) and Assis (2015), resumed the context-sensitive aspect of homology: the historical approach of homology assumes that homologs have a common evolutionary origin. Complementary, the “mechanistic account of homology” assumes that homologs are caused by a same developmental module (Wagner 1996 in Vogt 2017). The “nonevolutionary comparative account of homology” refers to the pre-Darwinistic recognition of homology from Belon (1555) to Owen(1843–1849).
Restricting the term homology to a narrow and specific context, for example equating it to synapomorphy (Patterson 1982), or to developmental pathways (Sattler 1966, 1994).
Adopting different terms for similar kinds of homology, such as serial homology versus iterative homology versus homonomy (Patterson 1982); homoiology sensu Plato versus homologous analogy sensu Mivart (cited by Remane 1956).
Mixing reference frameworks such as position and development, for instance in “partial homology” (Sattler and Rutishauser 1992).
The use of homology at different hierarchical levels, the so-called “subclasses” of homology (Fitch 1970). Confusion may result when the considered hierarchical level is not explicitly cited. For example, “transformational homology” is related to character and character state definitions at the organism level while “taxic homology” refers to character evolution at the species level (Patterson 1982). In another example at the molecular level, new terms were introduced such as orthology, which refers to “the relationship among DNA sequences that diversify through speciation” (Fitch 1970), thus originating from the same ancestral sequence. In addition, paralogy, which is “the relationship of DNA sequences that diverge through gene duplications” (Fitch 1970), refers to duplicates of a same ancestral DNA sequence. Xenology is “the relationship among DNA sequences that are horizontally transferred” (Gray and Fitch 1983), referring to the relation between the DNA of a donor and an acceptor species which do not share common origin.
In this article, we present a conceptual framework addressing all evolutionary disciplines and levels. This way, different approaches to homology are integrated into a single, concise, and hierarchical model. At the same time, we propose a simple and logical terminology, mostly based on existing though clearly defined terms (see Appendix 1). This is particularly relevant in a time when morphological and molecular approaches are being integrated and when terminology is being revised in an ontological context (e.g., www.plantontology.org).
Conceptual Framework
Basic Assumptions
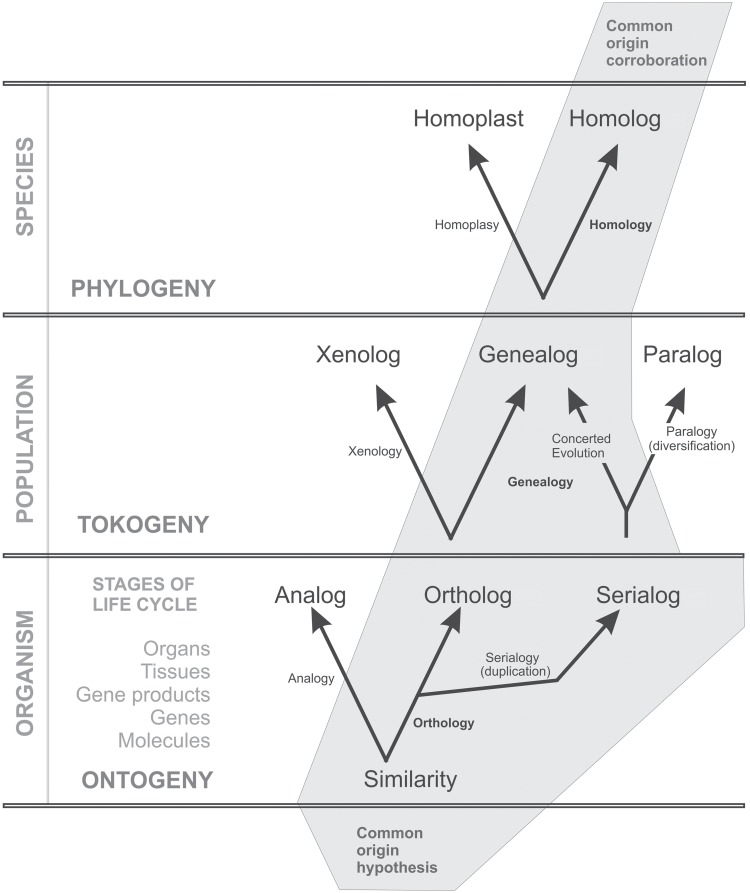
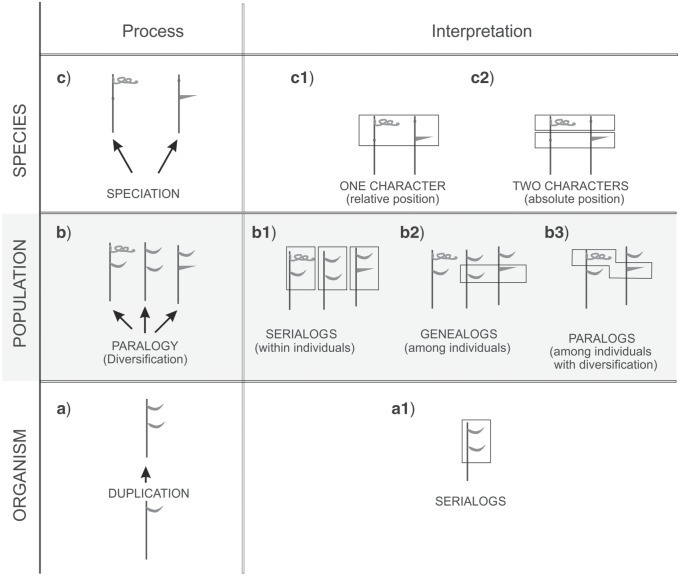
Fundamentally, evolution is studied at three hierarchical levels (Fig. 1), the organism level (ontogeny), the population level (tokogeny), and the species level (phylogeny).
Figure 1.
Conceptual framework for the study of homology. Three hierarchical levels (organism, population, and species) with their respective processes (ontogeny, tokogeny, and phylogeny) need to be studied to assess common origin at each level. Homology requires common origin at all levels: genealogy at the population level and orthology at the organism level. The organism level includes different sublevels and ontogenetic life stages.
Ontogeny refers to the development within an individual organism. All life stages (“semaphoronts” of an individual sensu Hennig 1966, p. 6–7; Prashant et al. 2017; Sharma et al. 2017) of an organism are studied to identify characters. These identified characters are then used for homology assessments at the species level. Consequently, comparison of structures must be done among corresponding life stages, for example, structures occurring in larvae should not be compared with structures in imagos, or root hairs of sporophytes should not be compared to rhizoids of gametophytes (see Scotland 2010).
The term tokogeny was introduced by Hennig (1966), referring to the reticulated, nonhierarchical relationships among individuals of a same species (horizontal gene transfer within species, see also Sanderson and Hufford 1996). In contrast, phylogeny refers to the evolutionary history of species, that is, to the vertical gene transfer from an ancestral species to its descendants. The vertical gene transfer from the ancestor species to its descendants results in hierarchical relationships among them: one ancestor can have several descendants, but not inversely. In tokogeny, there are no such hierarchical relationships with descendants having more than one ancestor.
Each hypothesis of common ancestry is primarily based on common origin of structures at the organism level (same ontogenetic origin), common origin at the tokogenetic level (vertical gene transfer), and common origin at the phylogenetic level (shared ancestry or a same phylogenetic origin). Consequently, we propose that common ancestry should be assessed at each of these three levels (Fig. 1).
Central in our framework is that we distinguish “morphological homology” (see below under “orthologs and analogs”) for the organism level, “genealogical homology” (see p. 6) for the population level, and “phylogenetic homology” (see p. 12) for the species level. Common origin at the population level (genealogical homology) implies common origin at the organism level (morphological homology). Common origin at the species level (phylogenetic homology) implies genealogical homology and morphological homology (Fig. 1). Only when common origin is agreed on each level, homology is corroborated (Figs. 1 and 6).
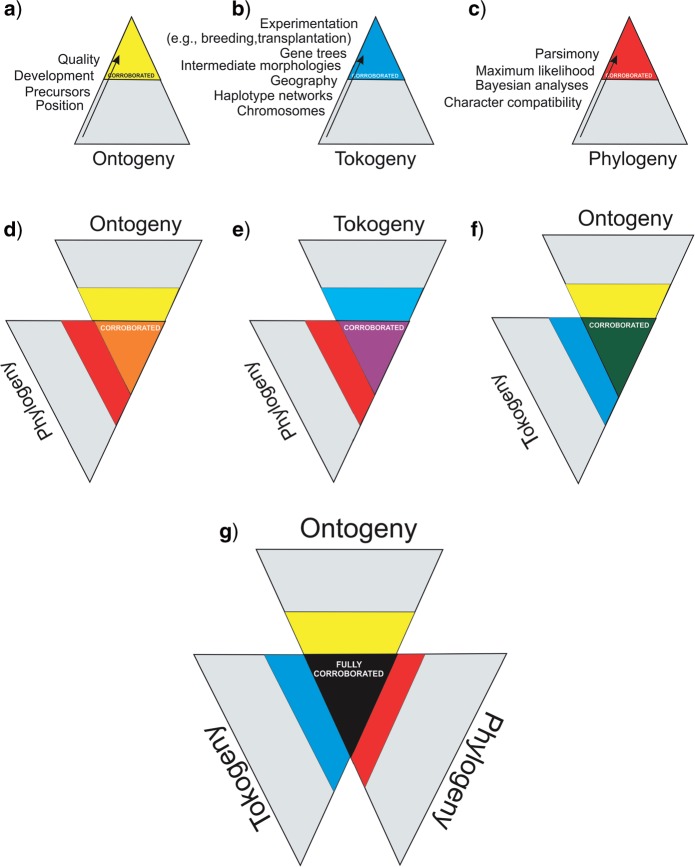
Figure 6.
Methodological approach to fully corroborate homology. Each triangle represents one of the three hierarchical levels, respectively the organism level, the population level, and the species level. Tests exist at each hierarchical level (a–c) that reduce the initial number of possible homology hypotheses to a limited set of corroborated hypotheses at each hierarchical level. The area of each triangle reflects the relative number of homology hypotheses before (full area) and after the tests (grey shades area). Corroboration at more than one level (d–f) increases confidence for the initial homology hypothesis. When the three levels have been tested (g), the initial homology hypothesis is fully corroborated. This figure appears in color in the online version of the article.
Organism Level
Orthologs and analogs
At the organism level, common origin is tested by morphological, anatomical/histological, physiological, and genetic investigations at the corresponding hierarchical sublevels of organs, tissues, gene products, genes, and molecules (Fig. 1). Within each sublevel, similar looking structures may be analogous (not sharing common structural origin) or morphologically homologous (sharing common structural origin).
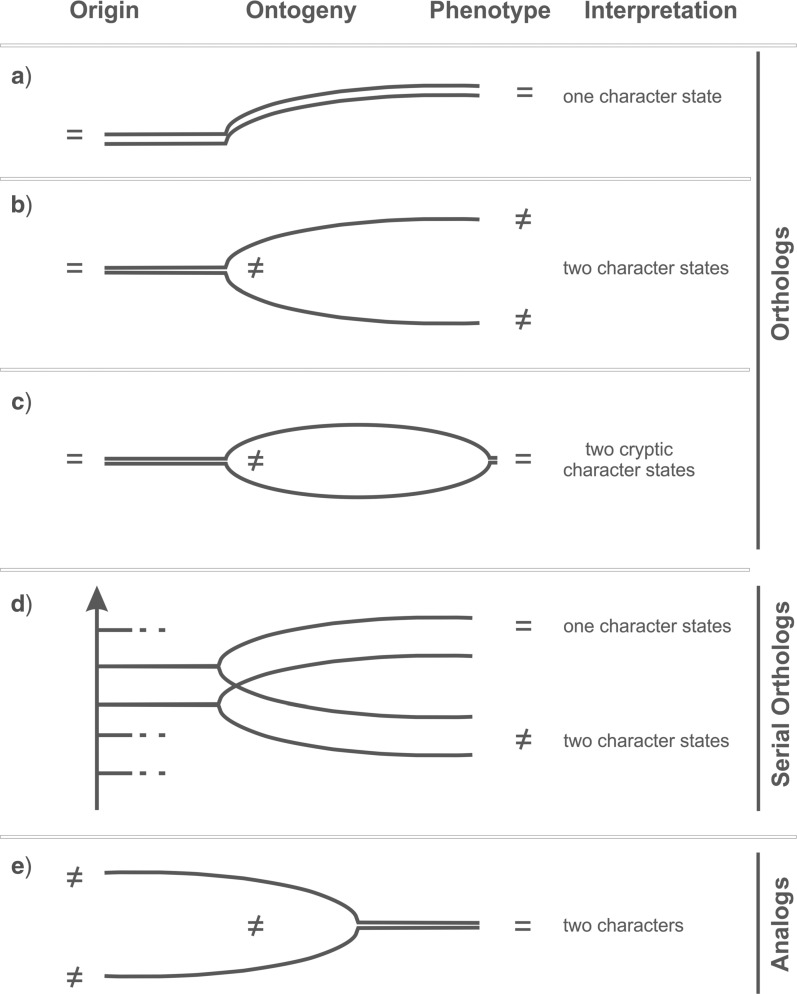
When in the before mentioned definition of orthology from Fitch (1970)—“the relationship among DNA sequences that diversify through speciation”—the words “DNA sequences” are replaced by “structures,” morphological homologs correspond to Fitch’s definition of orthologs. Consequently, we propose to extend the term “orthologs” to all kinds of morphological homologs (Fig. 1; in color: see the online version). Whether the structures compared are orthologs or analogs can be inferred from their ontogenetic origin and development (Fig. 2; see also Hunter 1964).
Figure 2.
Methodological procedure to identify orthology, serialogy, and analogy at the organism level. To identify orthologs, the common origin of the structures should be confirmed. Same ontogenies result in same phenotypes (one character state; a), different ontogenies in different phenotypes (two character states, b). Similar phenotypes can reflect the same (a) or two cryptic characters states (c). Duplication of orthologs results in serialogs irrespective of the phenotype (d). Analogs have different origins corresponding to different characters irrespective of their phenotypes (e). = indicates respectively same origin, same developmental pathways, similar phenotypes;  indicates respectively different origins, different developmental pathways, different phenotypes.
indicates respectively different origins, different developmental pathways, different phenotypes.
Similar looking structures are orthologs (morphological homologs) if they have the same ontogenetic origin and analogs if they have different ontogenetic origins (Figs. 1 and 2). In any case, what is meant by “ontogenetic origin of a structure” depends on the level/sublevel of the underlying question. It can be the same primordium (plants) or cell lineage (animals) at the organ level, the same relative position of the tissue at the histological level, the same precursor molecule at the molecule level, the same initial sequence at the protein or gene level, or the same position of an amino acid or mononucleotide within a sequence at the molecular level. Thus, having the same origin at a given level identifies structures as orthologs at that particular level. For example, floral structures assigned to the character “petal” are orthologs at the organ level because of the similar position of their primordia in a developing flower. In contrast, a petal and a petaloid bract have different origins, considering the respective meristems they originate from (Fig. 2e). They are analogous and should be assigned to different characters.
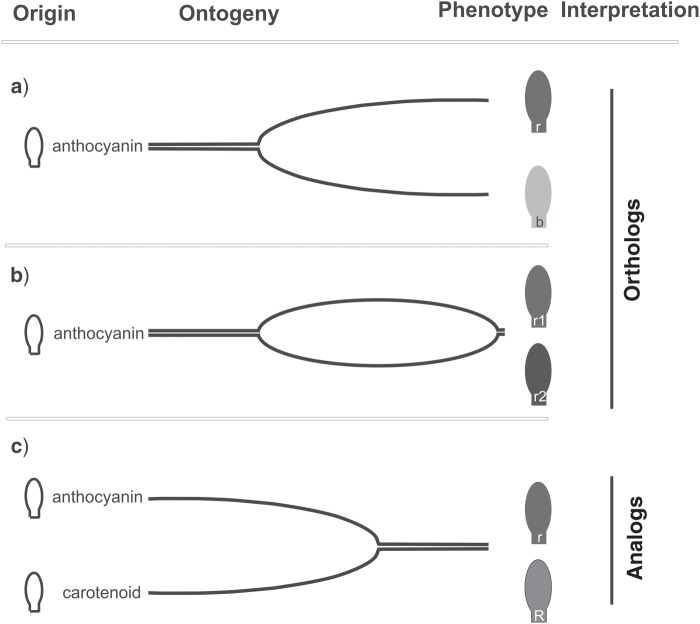
Often, different characters are combined into a single structure, which then is assigned to a composite character (e.g., colored petals). If the composing parts do not evolve in a parallel way, there is no concerted evolution and confusion in the homology assessment may result from it. For example, in “colored petals,” the petals are orthologous but the color can be caused by several analogs, such as a red of carotenoid origin and a red of betalain origin. Structures with composite characters may erroneously be interpreted as homologs at one hierarchical level and analogs at another level. In the above example, “colored petals” is a composite character consisting of “color” and “petals,” which should be restricted to “petals.” Indeed, if petals are under study, the origin of the pigments does not affect the nature of the petals. Each kind of colored petal is a character state of the character “petal.” In contrast, if pigments are under study (Fig. 3), the origin of the pigments matters to distinguish between orthologs (Fig. 3a), cryptic orthologs (Fig. 3b), or analogs (Fig. 3c).
Figure 3.
Example of an orthology assessment of “colored petals.” Petals are orthologs at the organ level. Diversification at the pigment level results in different character states (different red and blue petals, represented by petals in different grey shades from white to black) of the character “petal.” When considering the character “pigment,” if the pigments have the same origin, they constitute character states of the character “pigment” (a, b). If the pigments have different origins, they are analogs (c). b = blue; r, r1, r2 = red originating from anthocyanins; R = red originating from carotenoids. This figure appears in color in the online version of the article.
Characters and character states
Structures assigned to a same character implicitly have a common origin, thus being orthologs (Fig. 2a–c). While the definition of a character depends on origin, the definition of a character state is rather dependent on developmental processes or ontogenies. Different ontogenies in structures with a same origin result into a series of orthologous structures which constitute a character transformation series (Fig. 2b). The corresponding character states always result from diversification at subordinate levels. In our former example, petals with different colors in different species constitute character states of the character “petal,” regardless of the origin of the colors themselves. An illustration from literature of the application of the notions character and character states is the early attempt of Smets and Cresens (1988) to code characters and character states of nectaries at different hierarchical levels in nectaries.
In the course of evolution, new structures may be added (by duplication, becoming modular, or in plants by fragmentation of meristems as in dédoublement) or structures may be reduced (neoteny, pedogenesis, and ontogenetic abbreviation; Remane 1956; Schlichting and Pigliucci 1998), possibly leading to new characters when the new structures differentiate from the original ones (see further p. 5, where do characters come from?). Because character transformation is always part of an ontogenetic process, it was argued before that phylogeny is a chain of ontogenies (Zimmermann 1965), an idea that Kupiec (2009) further elaborated in his concept of ontophylogenesis.
Cryptic character states
If two structures with the same ontogenetic origin look similar, they either develop in the same way (Fig. 2a) or they obtain a similar phenotype through different developmental pathways (Fig. 2c). In the latter case, different developmental processes converge into a same phenotype, which can easily be mistaken as the same character state but actually consists of two different “hidden” or “cryptic” character states. In the “petal” example, we have cryptic character states of the character “petal” if we consider two petals of different organisms with indistinguishable red color. However, in the one petal the red is of carotenoid and in the other of betalain origin. An example from literature is the occurrence of tubular corollas in Rubiaceae. According to Vrijdaghs et al. 2015, rubiaceous tubular corollas result from three possible developmental processes which may act (or not) simultaneously. The proportion to which each of these processes contributes to the development of the corolla varies according to the species, but always results in similar, tubular corollas albeit with variable epipetaly with respect to the stamens.
Where do characters come from?
Since character states result from different processes in structures with a same origin, they can be easily confused with characters, resulting from structures with different origins. New characters originate by anagenesis, cladogenesis, or paralogy. Anagenesis is character transformation as a function of time within a lineage, cladogenesis is the diversification of characters through speciation, and paralogy is multiplication of a structure followed by diversification.
“Serial homology” or serialogy
Repeated structures in modular organisms are known as “serial homologs” (Carus 1828; Owen 1843; Boyden 1943), since the modules within the individual are considered to be structures with a same ontogenetic origin. However, in our framework, it is proposed to restrict the term “homology” to the species level, and therefore, we think that terms with combinations of the word “homology” such as “serial homology” are confusing. For the same reason, we avoid combinations with the word “orthology,” orthology being restricted to the organism level and to structures with a same ontogenetic origin in different individuals. Consequently, we recommend replacing the term “serial homology” by serialogy for structures with a same ontogenetic origin which are compared within a single individual. Within the individual, the repeated modules are then serialogs.
Using the same ontogenetic origin as major criterion to define orthology raises problems in modular organisms. Animals with a modular (or metameric) organization usually have a fixed number of segments. Therefore, the absolute position of a structure is a helpful criterion for homology assessment. For example, millipedes (Diplopods) have two leg pairs for each segment, with exception for the first three or four segments and have a fixed number of segments. Consequently, segments of different individuals can be “aligned” and compared. In contrast, plants have open growth caused by apical and axillary meristems which are active during all life stages. The number of modules is usually not fixed and identifying corresponding modules at the same absolute positions is difficult (Fig. 4; in color: see the online version). In plants and other organisms with an undetermined number of repeated modules, the relative position of structures fits better for homology assessments. For example, in plants, leaves always originate in a subapical exogenous position at the shoot apical meristem and are therefore orthologs irrespective of their absolute position within the plant or their development and adult appearance.
Figure 4.
Theoretical outline of a paralogy assessment. The left column shows how new characters arise through paralogy. The right column gives a survey of the interpretations of structures resulting from multiplication and diversification, in function of the different hierarchical levels. Multiplication of a given structure (in casu duplication of a leaf bearing node) at the organism level (a) results in serialogs (a1) which might diversify into thorns or tendrils at the population level (b). Within a single individual, the repeated structures are serialogs, irrespective of their phenotype (b1, framed). Comparing serialogs with different phenotypes among different individuals results in two possible statements: referring to the same absolute position, structures are genealogs irrespective of their phenotype (b2, framed), referring to different absolute positions, they are paralogs (b3, framed). At the species level, different character states of “leaf” might have been fixed in different individuals (c). When considering the relative position, leaves, tendrils, and thorns are characters states (c1), when considering their absolute positions, they are two characters (c2, each frame contains a character). Symbols:  tendril;
tendril;  thorn;
thorn;  leaf.
leaf.
Gene duplication (commonly known as paralogy) creates modularity at the molecular level. Duplication, segmentation, meristem fragmentation, and dédoublement are all phenomena resulting in a repetition of sequences, segments, modules, or structures. Because such repetitions, irrespective of the sublevel to which they belong or what caused them, result in identical “modules” sharing a same origin, they can be considered as serialogs. Therefore, we propose to extend the term “serialogs” to the molecular level for repeated, nonmodified sequences. Furthermore, we propose to use the term paralogs for diversified serialogs on all sublevels (from molecules to organs).
Population Level
Paralogy
If serialogs from different individuals are morphologically identical, they can be treated as orthologs. However, if a serialog diversifies, it becomes a paralog and forms a new structure, which may be considered as a new character. The interpretation of serialogs as orthologous (same character) or paralogous (different characters) depends on the kind of comparison made: serialogs within the same individual are orthologous, irrespective of their phenotype (e.g., leaves and tendrils within a single individual; Fig. 4b1). Structures belonging to modules with the same absolute position in different individuals are also orthologous, irrespective of their phenotype (e.g., leaves or tendrils or thorns at the same node in different plants, see Fig. 4b1,b2). However, in plants, determining the same absolute position (or same node) is difficult, if not impossible. Therefore, in homology assessments of plant structures, the relative position of these structures is considered. The relative position of a structure is determined by its origin, compared to its immediate surroundings in the plant, irrespectively of its absolute position. When in plants the relative position of phenotypically different structures in different individuals appears to be the same, the different phenotypes may be interpreted as character states (e.g., tendrils and leaves; Fig. 4c1). In contrast, if the exact position of divergent structures can be determined, and if they are located on different modules (animals) or different nodes (plants), these structures unambiguously are paralogs. In that case, they should be assigned to different characters. This is illustrated in our example of leaves, tendrils, and thorns (Fig. 4b3).
Paralogs may easily be confused with orthologs if their serialogous nature remains unknown. Indeed, when duplicates with different phenotypes are not recognized as diversified serialogs, hence paralogs, they could erroneously be considered as character states.
Genealogs and xenologs
Testing the hypothesis of common origin at the level of population assumes that the structures under consideration are orthologs at the organism level (Fig. 1). We define orthologs that are transferred from one generation to the following within one lineage (same species, without mixing) as genealogs (Fig. 1). In contrast, when orthologs are transferred from one individual to another belonging to a different species by hybridization or horizontal gene transfer, the lineages are mixed, and we adopt the term xenologs (Gray and Fitch 1983) for the resulting structures (Fig. 1). Only genealogs should be subject to further homology assessment. Indeed, xenologs do not have a common origin at the level of population and therefore should be excluded from further homology (=similarity due to common ancestry) assessment.
Species Level
“Phylogenetic homologs” or homologs and homoplasts
Since Darwin (1859), “origin” has two meanings: the structural origin of an organ and its ancestral (phylogenetic) origin. If a structure appears in an ancestor and is passed on its descendants, it should have in all descendants the same structural origin as in the ancestor (same character), although the structure may be modified in shape and function (character states) due to the selection pressure each of the descendants undergoes. As a consequence, accepting common origin at the level of species assumes that the structures under consideration are genealogous implying that they are also orthologous (Fig. 1). If horizontal gene transfer is excluded, a set of orthologs assigned to a same character should have a single phylogenetic origin (Nixon and Carpenter 2012). Consequently, these orthologs and genealogs could also be phylogenetic homologs and only then, they are ’homologs’ in the here newly defined sense (Fig. 1).
When orthologs have a different phylogenetic origin, they are considered to be homoplastic. Common ancestry of homologous structures does, however, not imply that they are per definition synapomorphic or symplesiomorphic (see Fig. 6a; Nixon and Carpenter 2012). Consequently, for homology assessment, polarity of characters (ancestral vs. derived) does not need to be determined a priori, and synapomorphy should not be equated to homology (Nixon and Carpenter 2012; see further p. 9). A plesiomorphic character with several character states might be a synapomorphy at another hierarchical level at another node in the phylogenetic tree (when such a synapomorphy is equated to homology at that given level). However, it is only the derived character state that is explicitly tested and corroborated as a homolog, while all the other character states of the synapomorphic character should also be tested.
Possible causes of homoplasy
The hypothesis of common origin or homology can only be fully corroborated when the common origin of each ortholog is verified at the levels of population and species (Fig. 1). When structures were only assumed, but not tested to be orthologs and/or genealogs, errors in the interpretation of homology versus homoplasy may occur. Structures interpreted as homoplastic, assuming (but not having assessed) orthology and genealogy, may, in fact, be analogous, xenologous, or paralogous.
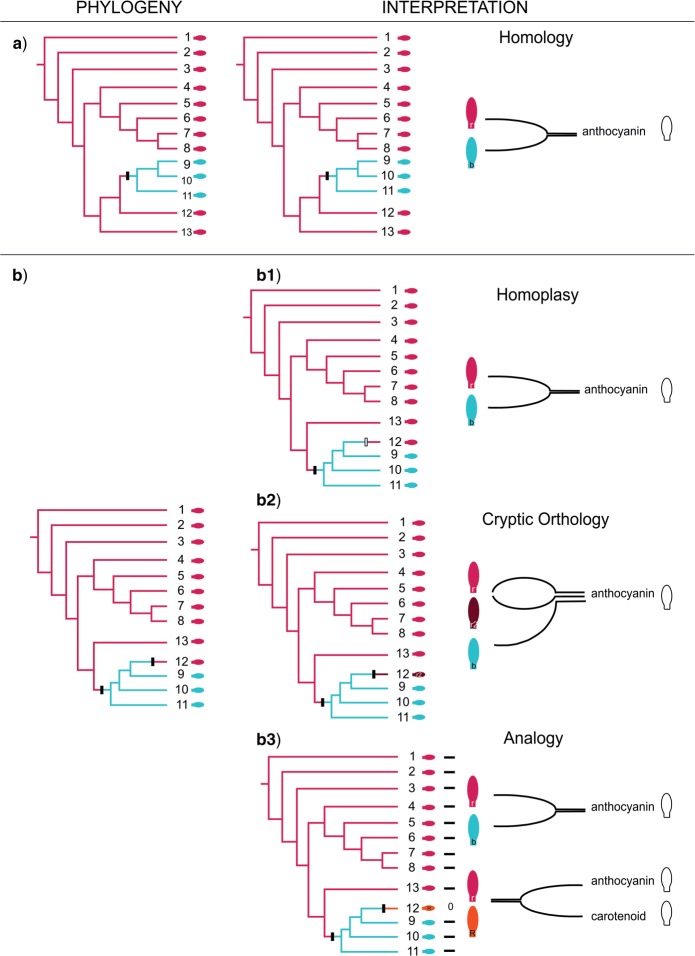
During the assessment of orthology (morphological homology), two major types of errors may occur: 1) Cryptic character states are erroneously coded as the same character state due to their similar phenotype. 2) Analogs are erroneously considered to be orthologs, which results in the interpretation of different characters as character states. To illustrate these possible errors (Fig. 5; in color: see the online version), we start from two a priori defined character states “red (light grey)” and “blue (white)” of the character “pigment” and a phylogenetic tree corroborating a single origin for blue (white) and red (light grey) (both are homologs), only blue (white) being synapomorphic (Fig. 5a). In Figure 5b, another case is presented where blue (white) was corroborated as a homolog, but not red (light grey). In that case, there are three possible explanations for the assumed homoplasy of red (light grey): 1) it is a “true” homoplasy [e.g., a reversal of red (light grey) that switched the color off and on; Fig. 5b1] or, an error occurred 2) either in the phylogenetic hypothesis, or 3) in the character coding (orthology assessment). In the case that “red (light grey, dark grey)” comprises cryptic character states, each cryptic character state of “red (light grey, dark grey)” on the cladogram should correspond to a different character state (r1, r2); if two “reds (light grey, black)” originated from different precursors (r, R), they are analogs and should have been coded as two characters (Fig. 5b3). In practice, such cases can be detected by iterative phylogenetic analyses (e.g., Franz 2014), although these tests are limited to the species level.
Figure 5.
Theoretical outline of a homology assessment. The left column shows two hypothetical cladograms (a, b) which are used for homology assessments of a specific structure, in casu petal pigments. The middle column shows the homology assessments in function of each of the cladograms. The right column shows the interpretations of the observations of petal pigments, reflected in the character coding (as a single character with two or three character states; or as two characters). Excluding horizontal gene transfer, if two as orthologous confirmed character states blue (white) and red (light grey) have the same origin in the phylogenetic tree, they are corroborated as homologs (a). Apparent two independent origins for a single character state (red-light grey, b) can be explained in three different ways: homoplasy (b1), cryptic orthology (b2), or analogy (b3). Homoplasy implies independent origins for the same ortholog, for example a reversal. Cryptic orthologs and analogs imply error in the character coding: if in b2, the red (light and dark grey) petals are erroneously interpreted to be a single character state, apparent homoplasy results. If in contrast the cryptic orthologs are recognized as such and coded as two different character states (r1 and r2), the apparent homoplasy is explained by the existence of three (blue-white and two reds-light and dark grey) character states. If in b3 the red pigments (light grey and black) are interpreted as a single character state (r) next to blue (white) (b), both of anthocyanin origin, apparent homoplasy results. If in contrast the caretenoid origin of one of the reds (R) is recognized as such, this red (black) is assigned to another, analogous character and the apparent homoplasy is explained. Symbols: black hash mark = synapomorphy; gray hash mark = homoplasy. This figure appears in color in the online version of the article.
From the first case in the above example, it is clear that in contrast to Nixon and Carpenter (2012), we do accept that homoplasy can be the result of a natural process, in the case that orthologous and genealogous nature has been corroborated, but appear in different lineages at the species level. In our opinion, “true homoplasy” can be explained by reversals caused by so-called internal constraints (Gould 2002) or “deep homology” (Shubin et al. 2009; see also below, p. 19) and therefore, we consider it useful to maintain and acknowledge the concept of homoplasy. An example from literature is the one-armed staminal lever in Salvia (Lamiaceae), which evolved at least four times independently within Salvia s.l. by parallel reduction of the common posterior lever arm (Claßen-Bockhoff 2017). Not calling the one-armed staminal lever in Salvia homoplastic because the character state appears in independent lineages, in our opinion, becomes a rather semantic issue.
Justification of terminological propositions
In our framework, we initially considered to use only existing terminology in its traditional meaning, namely “morphological homology” and “phylogenetic homology.” To continue using these traditional terms would maintain the confusion about the term “homology” given that each of the traditional terms we propose to replace, consists of an adjective, which consequently changes context and meaning of homology, and the term “homology.” Our idea, on the other hand, is to define homology in an unambiguous way. Furthermore, for duplicated and subsequently modified morphological structures, apparently no particular term exists. In contrast, when looking at the traditional definitions of orthology and paralogy, which are both originally defined in a molecular context, it is easy to see that a gene can be considered as a unit or module. We believe that the meanings of the definitions of orthology and paralogy are not affected whether the unit considered is indeed a gene, or a DNA sequence, an amino acid sequence, or an anatomical/morphological unit. For this reason and for the sake of clarity, we propose to limit the meaning of homology to the species level and to extend the meanings of orthology and paralogy to “morphology.” Moreover, we consider it necessary to distinguish between duplication (currently called paralogy) and duplication followed by modification (our definition of paralogy), since duplication without subsequent modification does not affect the homology assessment. We thus prevent to use the term “homology” in different contexts and meanings and propose a solution for the absence of a specific term for a duplicated and subsequently modified morphological structure.
Corroboration tests
The common ancestry hypothesis can be tested independently at the organism, population, and species level. A proper set of more or less rigorous tests at those levels exists, each restricting the number of initial homology hypotheses (Fig. 6; in color: see online version). At first glance, when designing a study, there are probably many possible characters and character states that can be hypothesized as homologous. However, as tests are applied, the number of features being corroborated as characters or character states is usually reduced. In Figure 6, the area of each triangle depicts the number of putative characters or character states before the homology tests and the colored area depicts the number of the confirmed characters and character states after the tests. At the organism level (Fig. 6a), testing orthology can be done by establishing the position or precursors of structures based on organogenetic studies or secondarily by considering “qualities,” which are clearly connected with only one single structure (sensu Remane 1956). For example, the position within a whorled flower can be used to identify structures as petals, and this identification can be supported by “qualities,” such as color. At the population level (Fig. 6b), genealogs can be identified by studying features and properties such as chromosome numbers or phenotypes, haplotype networks, (phylo)geography (indicating the potential for horizontal gene flow), intermediate morphologies, gene trees, and common garden experiments. For example, a pink petal color can be considered as intermediate between red and white, suggesting hybridization. This can be simply tested by plotting the colors on a distribution map, and if pink only occurs in geographical contact areas between red and white, pink can be hypothesized as a xenolog; a more rigorous test would be to try to artificially hybridize both populations. At the species level (Fig. 6c), possible tests are character compatibility, parsimony, Bayesian or likelihood analyses using morphological and/or molecular data. The hypothesis becomes more probable if corroborated at two levels (Fig. 6d–f). The hypothesis is fully corroborated when all three levels pass the tests when the structures compared are orthologs, vertically transferred to the next generation, and belong to different species with a common ancestor (Fig. 6g). Note that assessment of homology does not imply directionality; tests can be started at any level. As long as there is no conflict in the initial homology hypothesis, there is no need to apply tests at all hierarchical levels.
Implications for organ identity
Orthology determines organ identity in the sense of morphological homology. However, the term organ identity as used in evo-devo studies (e.g., Theißen 2001; Rutishauser and Moline 2005; Hirayama et al. 2007) refers to a developmental process and not to the same origin (Fig. 2). We elucidate the difference between our point of view and the evo-devo approach with an example in Arabidopsis. Bowman et al. (1991) described homeotic mutants of Arabidopsis where carpel-like structures are formed at sepal position. From a developmental point of view, these carpel-like structures are carpels at sepal positions because they have the same development (same “organ identity”) as carpels in the wild type. However, considering its position, the carpel-like structure is a sepal that develops like a carpel; it differs from wild type sepals by a diverging development. Both views are important and interesting, but differ in their aim. To assess homology (=determining common origin), using “origin” instead of “development” is essential. Moreover, developmental pathways may diverge through time without affecting the phenotype of the structures considered. This phenomenon is known as developmental system drift (True and Haag 2001). We believe that developmental system drift shows that, in homology assessments, structural origin and developmental process should be decoupled. Consequently, the carpel-like sepal of the above example is, in our view, a character state of the character “sepal” and not a carpel placed on a wrong position. In our framework, character states result from different developmental processes in structures with a same origin. Hence, recognizing the origin and subsequent development of structures enables the identification of characters and character states (Fig. 2e) and reduces conflicts at the phylogenetic level (indeed, analogs lead to apparent homoplasy; Fig. 4b3). Ontogenetic studies are very useful to show possible ways of diversification in the course of evolution (e.g., Gravendeel et al. 2017) and may explain the cause of homoplasy.
Homeosis
In molecular investigations the exclusion of paralogs in phylogenetic analyses is common practice. An obvious case of positionally fixed, divergent serialogs (thus paralogs) are the whorls in a flower. In a flower, the expression of a structure of a given whorl in another whorl (such as carpel-like structures at the position of sepals), is referred to as “homeosis” (Sattler 1988). In zoology, this term was introduced for organs appearing at unusual positions (Bateson 1894). Since developmental programs are independent of the absolute position in plants (due to the open growth pattern), the term homeosis is misleading in botany (see e.g., Kirchoff 1991).
“Hybrid Organ” and “Partial Homology”
In our framework, “mixed homologies” (Baum and Donoghue 2002) or “hybrid organs” cannot occur as each structure has a unique origin that defines the character. The idea of hybrid organs results from using two reference systems at the same time, such as origin and developmental process (see Claßen-Bockhoff 2001, 2005). For example, a phylloclade was considered to be a hybrid organ between a shoot (position and origin) and a leaf (development; Sattler 1994), but, according to our framework, a phylloclade is a character state of “shoot” and analogous to a “leaf.” A consequence of combining two reference frameworks is that the same structure (phylloclade) has two different identities, which results in apparent “partial homology” (Sattler and Rutishauser 1992). However, if a structure is accepted to be partially homologous, this implies that: 1) the structure can have more than one origin and 2) development and origin are considered at an equal level. Both implications inevitably result in confusion for character determination. Instead, in our conceptual framework, each structure has only one origin and the development of a structure can only be consecutive to its origin, imposing us to regard origin and development in a sequential order. Therefore, partial homology is discarded in our framework: a structure is either orthologous (morphologically homologous) or analogous. This is the major difference between the hybrid organ concept and our conceptual framework.
Hybrid organs have been postulated when considering the ever on-going structural change in the course of evolution, often resulting in a morphological continuum. However, the terminology “hybrid organ” is misleading as the origin of a structure is not a product of hybridization. Instead, it results from the simultaneous activities of different developmental programs in a structure with one single, previously fixed origin, causing its phenotype to converge to one or another organ. For example, based on its origin, a phylloclade is assigned to the character “shoot.” Its leaf properties represent a particular character state. Another example of how hybridization cannot result in homology is the following commonly known situation: a particular phenotype (e.g., pink petal) might appear in a population after mixing genetic information from two individuals (e.g., one with white and one with red petals). At the organism level, petals have a same origin and consequently, pink petals are orthologous with respect to the parental white and red petals. Pink, white, and red petals are character states of the character “petal.” However, at the population level, the pink color of the petals results from horizontal gene transfer. Consequently, pink petals are xenologs and should be excluded from further homology assessment (e.g., they cannot be considered to support a synapomorphy). At the species level, again, a genealog is either homologous or homoplastic, but never a hybrid.
Homoplasy
Phenotypically identical structures that occur in different lineages (clades) are homoplastic genealogs in the case of true homoplasy, or analogs or xenologs. It has been argued that homoplasy cannot naturally exist, and that it is always the result of an error, either in the phylogenetic hypothesis or in the character coding. However, we consider it useful to retain the term “homoplasy” for structures that occur in different clades but nevertheless passed the orthology and genealogy tests. Several such cases were explained in evo-devo studies by shared regulatory systems (see e.g., Wake et al. 2011; and “factorial homology” in Minelli and Fusco 2013). Moreover, we think that “deep homology” (Shubin et al. 2009) may offer an explanation of some cases of true homoplasy. According to Shubin et al. (2009, p. 818), ancient genetic systems involved in complex regulatory processes are inherited from a very early common ancestor and maintained in the course of evolution, causing “cryptic homology,” such as in the photoreceptors of various extant zoological clades, which “on morphological or phylogenetic base would not be recognized as homologous without the observation of the common underlying genetic cassettes.” We think that “deep homology” sensu Shubin et al. (2009) concurs with the “internal constraints” of Gould (2002).
Accepting the possibility of “true” homoplasy does not preclude the justification of parsimony as a method for postulating phylogenetic hypotheses (in contrast to Nixon and Carpenter 2012). Indeed, parsimony minimizes error in evaluating identified orthology and allows the objective identification of phylogenetically recurrent orthologs, or the recognition of cryptic character states.
Conclusions
In contrast to previous approaches of homology (e.g., Hall 1994; Sanderson and Hufford 1996; Scotland Pennington editors 2000; Scotland 2010; Richter 2017; Vogt 2017), we restrict the term homology to common ancestry of compared structures at the species level. This implies that the presumed homologous structures also have a common origin at the organism and population levels.
The origin of structures can be determined using morphological studies on the organism level resulting in the identification of orthologs (“morphological homologs”). We expect that orthologs, corresponding to character states of a same character, define a clade when mapped on a species tree. In case the orthologs are phylogenetically recurrent and consequently fail to define a clade, we deal with homoplasy and need to re-evaluate the origin of the structures (Fig. 3g) or the phylogenetic hypothesis; note that we exclude in this case horizontal gene transfer. Consequently, putatively homoplastic characters prompt to re-evaluate the orthology, serialogy, and genealogy of the structures compared. At the species level, structures can appear to be homoplastic if analogs are erroneously considered to be orthologs, if cryptic character states are mistaken as a single character state, if the character is introduced in the lineage through horizontal gene transfer or lineage fusion (xenologs), or if paralogs are taken as genealogs.
Evo-devo studies also focus on the level of organism. They provide a basis to disentangle the origin of orthologs, serialogs, and analogs. This kind of studies is important to address the overwhelming appearance of apparent homoplasy in the tree of life because they could unravel why and how the same developmental pathway can be achieved in different lineages.
Most often the population level is overlooked in studies on homology. Genealogy (gene transfer from generation to generation) is usually assumed and xenology (orthologs which undergo hybridization or horizontal gene transfer) is neglected as a source for erroneous conclusions. Only when there is conflict among different gene trees xenology is assumed, but rarely tested.
By better defining the terms and concepts used in homology assessments and restricting them to specific hierarchical levels, we hope to reduce conflict and confusion related to the use of homology in biology. We repeat the most important advantages of our conceptual framework:
The terms homology (previously called “phylogenetic homology”) and homoplasy are restricted to the species level; the terms genealogy, paralogy, and xenology to the population level; and the terms orthology (previously called “morphological homology”), serialogy (previously called “serial homology”), and analogy to the organism level (including all sublevels). All these levels allow for the postulation of falsifiable hypotheses that can be tested separately. To fully corroborate homology, tests must support common origin at each level.
Only mapping morphological characters onto molecular phylogenetic trees (assuming that the phylogenetic hypothesis is correct) is not sufficient to test the homologous nature of the mapped characters. Indeed, for full corroboration of homology, the presence of analogs, paralogs, or xenologs needs to be excluded. As a consequence, mapping only assesses correlations between characters and phylogenetic lineages. Absence of such correlations (apparent homoplasy) implies that further assessment of common origin at the subordinated levels is needed (or that the phylogenetic hypothesis should be revised). The question is if the assumed homoplastic characters really represent orthologs and genealogs, or whether they merely share regulatory systems that explain the apparent homoplasy?
The identity of a subject under study is crucial in all disciplines regardless of its hierarchical level or the goal of the study. Therefore, unraveling to be or not to be the same is fundamental in all biological sciences, preventing inconsistencies, wrong conclusions and miscommunication. We hope to invite people with different backgrounds to use and test our proposed framework in the hope of further refinement or improvement.
Acknowledgments
Dr. Steve Donovan, Prof. David Bryant, and an anonymous reviewer are acknowledged for their valuable comments to the manuscript.
APPENDIX 1
Definitions:
Analogs are structures with different origins at the organism level irrespective of their phenotype or function.
Characters are qualities assigned to structures differing from each other by either another origin (analogy) or by deep divergence in serialogy (paralogy).
Character states are observable traits of a given character (to which orthologs are assigned), caused by developmental divergence of structures with a same origin.
Genealogs are orthologs transferred without mixing at the population level (vertical gene transfer).
Homologs are genealogs that also share common origin at the species level (same phylogenetic origin).
Homology means similarity due to common origin at all levels (organism, population and species).
Homoplasts are genealogs that do not share common origin at the species level.
Homoplasy means similarity due to common origin at the organism and population levels, but without common origin at the species level.
Ontogeny is the development of an organism including different life stages and all developmental processes at all sublevels such as organogenesis and biosynthetic pathways.
Orthologs are structures with the same origin at the organism level.
Paralogs are diversified serialogs at the population level (they do not undergo concerted evolution).
Serialogs are copies of orthologs at the organism level.
Xenologs are orthologs transferred with mixing at the population level (horizontal gene transfer or hybridization).
Helga Ochoterena, Alexander Vrijdaghs, Erik Smets, and Regine Claßen-Bockhoff contributed equally to this article.
Funding
This work was supported by the Research Foundation - Flanders (WO.005.05N).
References
- Assis L.C.S. 2015. Homology assessment in parsimony and model-based analyses: two sides of the same coin. Cladistics 31:315–320. [DOI] [PubMed] [Google Scholar]
- Baum D.A., Donoghue M.J.. 2002. Transference of function, heterotopy and the evolution of plant development. In: Cronk Q., Bateman R., Hawkins J., editors. Developmental genetics and plant evolution London, UK: Taylor and Francis; p. 52–69. [Google Scholar]
- Bateson W. 1894. Materials for the study of variation. London: Macmillan. [Google Scholar]
- Belon P. 1555. L’histoire de la Nature des Oyseaux. Paris: Guillaume Cavellet. [Google Scholar]
- Bowman J.L., Smyth D.R., Meyerowitz E.M.. 1991. Genetic interactions among floral homeotic genes of Arabidopsis. Development 112:1–20. [DOI] [PubMed] [Google Scholar]
- Boyden A. 1943. Homology and analogy: a century after the definitions of “homologue” and “analogue” of Richard Owen. Q. Rev. Biol. 18:2288241. [Google Scholar]
- Brigandt I. 2003. Homology in comparative, molecular, and evolutionary developmental biology: the radiation of a concept. J. Exp. Zool. 299B:9–17. [DOI] [PubMed] [Google Scholar]
- Carus C.G. 1828. Grundzüge der vergleichende Anatomie und Physiologie. Dresden: Hilschersche Buchhandlung; 81 p. [Google Scholar]
- Charlesworth D. 2010. Don’t forget the ancestral polymorphisms. Heredity 105:509–510. [DOI] [PubMed] [Google Scholar]
- Claßen-Bockhoff R. 2001. Vom Umgang mit der Vielfalt—eine kurze Geschichte der Pflanzenmorphologie. Wulfenia 8:125–144. [Google Scholar]
- Claßen-Bockhoff R. 2005. Aspekte, Typifikationsverfahren und Aussagen der Pflanzenmorphologie. In: Harlan V., editor. Wert und Grenzen des Typus in der botanischen Morphologie. Nürnbrecht, Germany: Galunder; p. 31–52. [Google Scholar]
- Claßen-Bockhoff R. 2017. Stamen construction, development and evolution in Salvia s.l. Nat. Volatiles Essent. Oils 4:28–48. [Google Scholar]
- Darwin C. 1859. The origin of species. Ed. Beer G.1996. Great Britain: Oxford University Press. [Google Scholar]
- Fitch W.M. 1970. Distinguishing homologous from analogous proteins. Syst. Zool. 19:99–113. [PubMed] [Google Scholar]
- Franz N.M. 2014. Anatomy of a cladistic analysis. Cladistics 30:294–321. [DOI] [PubMed] [Google Scholar]
- Fusco G., Minelli A.. 2010. Phenotypic plasticity in development and evolution: facts and concepts. Philos. Trans. R. Soc. B365:547–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould S.J. 2002. The structure of evolutionary theory. Cambridge, Massachusetts, USA: The Belknap Press of Harvard University Press; 1433 p. [Google Scholar]
- Gravendeel B., Butôt R., van Schaik P., Wijnands J.W., van den Berg R., Krol L., Doebar S., van Kooperen K., de Boer H., Kramer E., Smets E., Vos R., Vrijdaghs A., Dirks-Mulder A.. 2017. Exploring the evolutionary origin of floral organs of Erycina pusilla, an emerging orchid model system. BMC Evol. Biol. 17:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray G.S., Fitch W.M.. 1983. Evolution of antibiotic resistance genes: the DNA sequence of a kanamycin resistance gene from Staphylococcus aureus. Mol. Biol. Evol. 1:57–66. [DOI] [PubMed] [Google Scholar]
- Haeckel E. 1866. Generelle Morphologie der Organismen, Bd 1-2. Berlin: Reimer. [Google Scholar]
- Hall B.K. (Ed.) 1994. Homology, the hierarchical basis of comparative homology. San Diego, California, USA: Academic Press Inc; 475 p. [Google Scholar]
- Hennig W. 1950. Grundzüge einer Theorie der Phylogenetischen Systematik. Berlin: Deutcher Zentralverlag. [Google Scholar]
- Hennig W. 1966. Phylogenetic systematics. Urbana: University of Illinois Press. [Google Scholar]
- Hirayama Y.,Yamada T., Oya Y., Ito M., Kato M., Imaichi R.. 2007. Expression patterns of class I KNOX and YABBY genes in Ruscus aculeatus (Asparagaceae) with implications for cladode homology. Dev. Genes Evol. 217:363–372. [DOI] [PubMed] [Google Scholar]
- Hunter I.J. 1964. Paralogy, a concept complementary to homology and analogy. Nature 204:604. [DOI] [PubMed] [Google Scholar]
- Kaplan D.R. 1984. The concept of homology and its central role in the elucidation of plant systematic relationships. In: T. Duncan T., Stuessy T.F., editors. Cladistics: perspectives on the reconstruction of evolutionary history. New York, NY, USA: Columbia University Press; p. 51–70. [Google Scholar]
- Kirchoff B.K. 1991. Homeosis in the flowers of Zingiberales. Am. J. Bot. 78:833. [Google Scholar]
- Kupiec J.-J. 2009. The origin of individuals: a Darwinian approach to developmental biology. Singapore: World Scientific Publishing; 273 p. [Google Scholar]
- Lankester E.R. 1870. On the use of the term homology in modern zoology, and the distinction between homogenetic and homoplastic agreements. Ann. Mag. Nat. Hist. (4th Series) 6:34–43. [Google Scholar]
- Minelli A., Fusco G.. 2013. Homology. In: Kampourakis K. editor. The philosophy of biology: a companion for educators, history, philosophy and theory of the life Sciences. Dordrecht: Springer; p. 289–322. [Google Scholar]
- Nixon K.C., Carpenter J.M.. 2012. On homology. Cladistics 28:160–169. [DOI] [PubMed] [Google Scholar]
- Owen R. 1843. Lectures on the comparative anatomy and physiology of the invertebrate animals. London: Longman, Brown, Green & Longmans. [Google Scholar]
- Owen R. 1848. On the archetype and homologies of the vertebrate skeleton. London: Van Voorst. [Google Scholar]
- Patterson A.C. 1982. Problems of phylogenetic reconstruction. In: Joysey K.A., Friday A.E., editors. Systematics association special, vol. 21 New York: Academic Press; p. 21–74. [Google Scholar]
- Prashant P.S., Clouse R.M., Wheeler W.C.. 2017. Hennig’s semaphoront concept and the use of ontogenetic stages in phylogenetic reconstruction. Cladistics 33:93–108. [DOI] [PubMed] [Google Scholar]
- Remane A. 1956. Grundlagen des natürlichen Systems, der vergleichenden Anatomie und der Phylogenetik. Leipzig: Geest & Portig. [Google Scholar]
- Richter S. 2017. Homology and synapomorphy-symplesiomorphy—neither synonymous nor equivalent but different perspectives on the same phenomenon. Cladistics 33:540–544. [DOI] [PubMed] [Google Scholar]
- Rutishauser R., Moline P.. 2005. Evo-devo and the search for homology (“sameness”) in biological systems. Theor. Biosci. 125:213–241. [DOI] [PubMed] [Google Scholar]
- Sanderson M.J., Hufford L., editors. 1996. Homoplasy: recurrence of similarity in evolution. San Diego, CA: Academic Press. [Google Scholar]
- Sattler R. 1966. Towards a more adequate approach to comparative morphology. Phytomorphology 16:417–429. [Google Scholar]
- Sattler R. 1988. Homeosis in plants. Am. J. Bot. 75:1606-1617. [Google Scholar]
- Sattler R. 1994. In: B. K. Hall B.K., Homology: the hierarchical basis of comparative biology. New York: Academic Press; p. 423–475. [Google Scholar]
- Sattler R., Rutishauser R.. 1992. Partial homology of pinnate leaves and shoots. Orientation of leaflet inception. Bot. Jahrb. Syst. 114: 61–79. [Google Scholar]
- Sanderson M. J., Hufford L., editors. 1996. Homoplasy: recurrence of similarity in evolution. San Diego: Academic Press. [Google Scholar]
- Scotland R.W. 2010. Deep homology: a view from systematics. BioEssays 32:438–449. [DOI] [PubMed] [Google Scholar]
- Scotland R.W., Pennington R.T. editors. 2000. Homology and systematics. Systematic Association Special Volume Series 58, London, UK: Taylor & Francis; 217 p. [Google Scholar]
- Schlichting C.D., Pigliucci M.. 1998. Phenotypic evolution. A reaction norm perspective. Sunderland: Sinauer. [Google Scholar]
- Sharma P., Clouse R.M., Wheeler W.C.. 2017. Hennig’s semaphoront concept and the use of ontogenetic stages in phylogenetic reconstruction. Cladistics 33:97–108. [DOI] [PubMed] [Google Scholar]
- Shubin N., Tabin C., Caroll S.. 2009. Deep homology and the origins of evolutionary novelty. Nature 457:818–823. [DOI] [PubMed] [Google Scholar]
- Smets E., Cresens E.. 1988. Types of floral nectaries and the concepts ‘character’ and ‘character-state’—a reconsideration. Acta Bot. Neerl. 37:121–128. [Google Scholar]
- Theißen G. 2001. Development of floral organ identity: stories from the MADS house. Curr. Opin. Plant Biol. 4: 75–85. [DOI] [PubMed] [Google Scholar]
- True J. R., Haag E. S.. 2001. Developmental system drift and flexibility in evolutionary trajectories. Evol. Dev. 3:109–119. [DOI] [PubMed] [Google Scholar]
- Vogt L. (2017), Assessing similarity: on homology, characters and the need for a semantic approach to non-evolutionary comparative homology. Cladistics 33:513–539. [DOI] [PubMed] [Google Scholar]
- Vrijdaghs A., De Block P., Verstraete B., Groeninckx I., Smets E., Dessein S.. 2015. A developmental model for the corolla in Rubiaceae. Cryptic character states in corollas of the Spermacoceae alliance. Plecevo 148:237–255. [Google Scholar]
- Wagner G.P. 1996. Homologues, natural kinds and the evolution of modularity. Am. Zool. 36:36–43. [Google Scholar]
- Wake D.B., Wake M.H., Specht C.D.. 2011. Homoplasy: from detecting pattern to determining process and mechanism of evolution. Science 331:1032. [DOI] [PubMed] [Google Scholar]
- Zimmermann W. 1965. Die Telomtheorie (Fortschritte der Evolutionsforschung, Band I). Stuttgart: Fischer. [Google Scholar]