Abstract
The International Committee on Taxonomy of Viruses (ICTV) is tasked with classifying viruses into taxa (phyla to species) and devising taxon names. Virus names and virus name abbreviations are currently not within the ICTV’s official remit and are not regulated by an official entity. Many scientists, medical/veterinary professionals, and regulatory agencies do not address evolutionary questions nor are they concerned with the hierarchical organization of the viral world, and therefore, have limited use for ICTV-devised taxa. Instead, these professionals look to the ICTV as an expert point source that provides the most current taxonomic affiliations of viruses of interests to facilitate document writing. These needs are currently unmet as an ICTV-supported, easily searchable database that includes all published virus names and abbreviations linked to their taxa is not available. In addition, in stark contrast to other biological taxonomic frameworks, virus taxonomy currently permits individual species to have several members. Consequently, confusion emerges among those who are not aware of the difference between taxa and viruses, and because certain well-known viruses cannot be located in ICTV publications or be linked to their species. In addition, the number of duplicate names and abbreviations has increased dramatically in the literature. To solve this conundrum, the ICTV could mandate listing all viruses of established species and all reported unclassified viruses in forthcoming online ICTV Reports and create a searchable webpage using this information. The International Union of Microbiology Societies could also consider changing the mandate of the ICTV to include the nomenclature of all viruses in addition to taxon considerations. With such a mandate expansion, official virus names and virus name abbreviations could be catalogued and virus nomenclature could be standardized. As a result, the ICTV would become an even more useful resource for all stakeholders in virology.
Keywords: Arbovirus, classification, International Committee on Taxonomy of Viruses, (ICTV), nomenclature, species, taxonomy, virus
Virus Classification
Virus classification is a branch of virology that defines categories of viruses on the basis of their shared physical and genetic characteristics. Virus taxonomy differs from other taxonomic frameworks in various ways. For instance, algal/fungal/plant, animal, and prokaryotic nomenclatural codes exclusively address taxon nomenclature but do not govern classification of organisms into taxa (International Committee on Systematic Bacteriology 1992; International Association for Plant Taxonomy 2011; International Commission on Zoological Nomenclature 2012). Virus taxonomy, on the other hand, governs both nomenclature and classification. Consequently, a taxonomic committee, rather than the scientific community, decides which taxa are official (International Committee on Taxonomy of Viruses 2018b). Second, in contrast to other biological taxonomic frameworks, virus taxonomy currently permits individual species to have several members. For instance, Dobrava virus (DOBV), Kurkino virus (KURV), Saaremaa virus (SAAV), and Sochi virus (SOCV) are currently considered distinct viruses all belonging to the same species, Dobrava-Belgrade orthohantavirus (Maes et al. 2019a). Third, as this species name exemplifies, species names do not yet follow the Linnaean naming format, and in fact they do not follow any proscribed format at all in virus taxonomy (Postler et al. 2017). Furthermore, in virus taxonomy, reminiscent of prokaryotic taxonomy but in contrast to algal/fungal/plant and animal taxonomies, all taxon names are italicized for easy recognition (International Committee on Taxonomy of Viruses 2018b). Last, but not least, the ICTV currently does not engage deeply in the discussion on the nature of “species” (Mayden et al. 1997; Hey 2001) but for practical purposes adopted a somewhat debated (Peterson 2014; Van Regenmortel 2018)simple species definition (“A species is the lowest taxonomic level in the hierarchy approved by the ICTV. A species is a monophyletic group of viruses whose properties can be distinguished from those of other species by multiple criteria” (International Committee on Taxonomy of Viruses 2018b).
In the current virus taxonomy framework, viruses are grouped together in defined units called taxa. If appropriate, taxa are included in other higher ranked taxa, resulting in hierarchies. In descending sequence, currently used ranks are phylum (composed of subphyla of related viruses), subphylum (composed of classes of related viruses), class (composed of orders of related viruses), order (composed of families of related viruses), family (composed of genera for related viruses), genus (composed of species for related viruses), and species (the fundamental unit of virus classification). Viruses are assigned to taxa (species) if they satisfy certain membership conditions. If a species is included in a genus and that genus is included in a family, then the virus belonging to this species also belongs to the respective genus and family. For instance, measles virus (abbreviated MeV) is a member of the species Measles morbillivirus, which is included in the genus Morbillivirus in the family Paramyxoviridae. Therefore, measles virus is a morbillivirus and a paramyxovirus.
In virus taxonomy, taxa are intangible, abstract, and hypothetical “averages” of groupings designed to unite the relationships of the viruses assigned to them (Van Regenmortel et al. 1991; Kuhn and Jahrling 2010; Calisher 2016). This categorization simplifies grasping the commonality of, or differences among, the properties possessed by the viruses that are under study. Viruses, in contrast to taxa, are not defined but are tangible entities that can be manipulated, visualized by electron microscopy, and otherwise studied to reveal their lineages, strains, genotypes, and phenotypes. Thus, taxa are categories associated with lists of viruses that are assigned to them much like shopping lists contain the names of real items to be obtained. Ideally, virus taxon hierarchies reflect evolutionary relationships of member viruses, although they often still reflect practical phenotypes that are easier to establish, such as virion characteristics or features of virus-induced disease. Categorization of viruses into taxa informs communities about which viruses are or are not related and, if related, how closely they are related. As opposed to classification, nomenclature is the selection of names for both categories (here: taxa) and physical objects (here: viruses).
International Committee on Taxonomy of Viruses (ICTV)
Virus taxonomy is the task of the ICTV, a member body of the Virology Division of the International Union of Microbiology Societies (IUMS). Established in 1966 as the International Committee on Nomenclature of Viruses (emphasis added), the original intent was to classify viruses into hierarchical taxa based upon experimental virus characterization (Wildy 1971). Today, the ICTV is still responsible for classification of viruses into taxa and the nomenclature of these taxa but is not responsible for the nomenclature of viruses (Adams et al. 2017; International Committee on Taxonomy of Viruses 2018d). Furthermore, viruses are increasingly classified into taxa based on genome sequence comparisons, phylogenies, and sequence-inferred phenotypes rather than through direct study in culture. Indeed, the ICTV has begun a very promising initiative to establish taxa for novel viruses known only from complete or coding-complete genome sequences in an attempt to accommodate the exponential increase in virus discovery made possible by new sequencing technologies (Adams et al. 2017). Following its Statutes and Code (International Committee on Taxonomy of Viruses 2018a, 2018d), the ICTV appropriately focuses on listing taxa in ICTV publications that serve both taxonomists and evolutionary biologists.
Limitations of the Current ICTV Framework
We propose that the ICTV could and should strive to increase its usefulness for the virology community, including medical and veterinary professionals and regulatory agencies. Most of these communities are primarily concerned with viruses, rather than virus taxa, and therefore, perceive limited use for ICTV-devised concepts and categories (taxa). Yet, to place their research or other activities into proper context, these professionals need to be able to state the correct and most up-to-date taxonomic affiliation of the viruses they study in their publications and oral proceedings (Calisher 2016). Currently, a considerable effort needs to be undertaken to provide taxonomic affiliations. For instance, via the ICTV webpage (International Committee on Taxonomy of Viruses 2018a), one can indeed find that the species Measles morbillivirus is included in the genus Morbillivirus, which is included in the family Paramyxoviridae, which is included in the order Mononegavirales (https://talk.ictvonline.org/taxonomy/, under “Virus Taxonomy: 2018 Release,” expanding “Phylum Negarnaviricota”  “Subphylum Haploviricotina”
“Subphylum Haploviricotina”  “Class Monjiviricetes”
“Class Monjiviricetes”  “Order Mononegavirales”
“Order Mononegavirales”  “Family Paramyxoviridae”
“Family Paramyxoviridae”  “Genus Morbillivirus”). A direct search for
“Genus Morbillivirus”). A direct search for  measles
measles yields the result
yields the result
“Negarnaviricota › Haploviricotina › Monjiviricetes › Mononegavirales › Paramyxoviridae › Morbillivirus › Measles morbillivirus.”
However, the site does not list the member of the species Measles morbillivirus, namely measles virus, nor does the ICTV webpage provide information on the abbreviation(s) for virus names (here: MeV). Indeed, a direct search for  measles virus
measles virus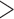 or
or 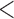 MeV
MeV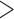 does not yield results. Infrequently, virus information can be found in linked historical ICTV documents, but finding this information is cumbersome. Browsing for this information is not obvious to nontaxonomists. As a result, an increasing number of scientists wrongfully use species names, rather than virus names, in their communications (e.g., scientists may wrongfully state that they are working with “Measles morbillivirus” rather than with the correct “measles virus”).
does not yield results. Infrequently, virus information can be found in linked historical ICTV documents, but finding this information is cumbersome. Browsing for this information is not obvious to nontaxonomists. As a result, an increasing number of scientists wrongfully use species names, rather than virus names, in their communications (e.g., scientists may wrongfully state that they are working with “Measles morbillivirus” rather than with the correct “measles virus”).
This problem is exacerbated by the fact that many ICTV-approved species currently have several virus members with different names, which most often cannot even be found in historical ICTV documents. For instance, the species California encephalitis orthobunyavirus is listed on https://talk.ictvonline.org/taxonomy/ as one of the 49 species currently recognized in the genus Orthobunyavirus (Bunyavirales: Peribunyaviridae). The 9th ICTV Report indicates that this species has only a single member, La Crosse virus (LACV) (Plyusnin et al. 2011) (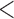 La Crosse virus
La Crosse virus cannot be found using the ICTV search engine). However, the community of bunyavirologists recognizes at least 18 distinct members in this species, each of which has its own name (Table 1). These viruses differ widely with respect to recognized pathogenicity for humans, geographic distribution, and arthropod vector(s) (Blitvich et al. 2018). A search for most virus names or abbreviations linked to California encephalitis orthobunyavirus does not yield results on the ICTV webpage, despite the fact that these virus names and their abbreviations are in common usage not only in early as well as current publications but also in major reference works, such as the modernized and updated International Catalog of Arboviruses and Certain Other Viruses on the US Centers for Disease Control and Prevention (CDC) website (US Department of Health and Human Services et al. 2009; US Centers for Disease Control and Prevention 2018). The length of Table 1, which only lists all currently accepted species in the bunyaviral families Hantaviridae and Peribunyaviridae, emphasizes the magnitude of this problem. This problem is not only restricted to bunyaviruses but also extends to the order Mononegavirales (in particular the family Bornaviridae) (Maes et al. 2019b) and numerous other taxa.
cannot be found using the ICTV search engine). However, the community of bunyavirologists recognizes at least 18 distinct members in this species, each of which has its own name (Table 1). These viruses differ widely with respect to recognized pathogenicity for humans, geographic distribution, and arthropod vector(s) (Blitvich et al. 2018). A search for most virus names or abbreviations linked to California encephalitis orthobunyavirus does not yield results on the ICTV webpage, despite the fact that these virus names and their abbreviations are in common usage not only in early as well as current publications but also in major reference works, such as the modernized and updated International Catalog of Arboviruses and Certain Other Viruses on the US Centers for Disease Control and Prevention (CDC) website (US Department of Health and Human Services et al. 2009; US Centers for Disease Control and Prevention 2018). The length of Table 1, which only lists all currently accepted species in the bunyaviral families Hantaviridae and Peribunyaviridae, emphasizes the magnitude of this problem. This problem is not only restricted to bunyaviruses but also extends to the order Mononegavirales (in particular the family Bornaviridae) (Maes et al. 2019b) and numerous other taxa.
Table 1.
Current composition of the bunyaviral families Hantaviridae and Peribunyaviridae
| Genus | Species | Virus (abbreviation) |
|---|---|---|
| Family Hantaviridae | ||
| Orthohantavirus | Amga orthohantavirus | Amga virus (MGAV) |
| Andes orthohantavirus | Andes virus (ANDV) | |
| Castelo dos Sonhos virus (CASV) | ||
| Lechiguanas virus (LECV = LECHV) | ||
| Orán virus (ORNV) | ||
| Asama orthohantavirus | Asama virus (ASAV) | |
| Asikkala orthohantavirus | Asikkala virus (ASIV) | |
| Bayou orthohantavirus | bayou virus (BAYV) | |
| Catacamas virus (CATV) | ||
| Black Creek Canal orthohantavirus | Black Creek Canal virus (BCCV) | |
| Bowe orthohantavirus | Bowé virus (BOWV) | |
| Bruges orthohantavirus | Bruges virus (BRGV) | |
| Cano Delgadito orthohantavirus | Caño Delgadito virus (CADV) | |
| Cao Bang orthohantavirus | Cao B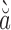 ng virus (CBNV) ng virus (CBNV) |
|
| Liánghé virus (LHEV) | ||
| Choclo orthohantavirus | Choclo virus (CHOV) | |
| Dabieshan orthohantavirus | Dàbiésh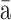 n virus (DBSV) n virus (DBSV) |
|
| Dobrava-Belgrade orthohantavirus | Dobrava virus (DOBV) | |
| Kurkino virus (KURV) | ||
| Saaremaa virus (SAAV) | ||
| Sochi virus (SOCV) | ||
| El Moro Canyon orthohantavirus | Carrizal virus (CARV) | |
| El Moro Canyon virus (ELMCV) | ||
| Huitzilac virus (HUIV) | ||
| Fugong orthohantavirus | Fúgòng virus (FUGV) | |
| Fusong orthohantavirus | Fŭsōng virus (FUSV) | |
Hantaan orthohantavirus
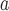
|
Amur virus (AMRV) | |
| Hantaan virus (HTNV) | ||
| Soochong virus (SOOV) | ||
| Jeju orthohantavirus | Jeju virus (JJUV) | |
| Kenkeme orthohantavirus | Kenkeme virus (KKMV) | |
| Khabarovsk orthohantavirus | Khabarovsk virus (KHAV) | |
| Topografov virus (TOPV) | ||
| Laguna Negra orthohantavirus | Laguna Negra virus (LANV) | |
| Maripa virus (MARV) | ||
| Río Mamoré virus (RIOMV) | ||
| Luxi orthohantavirus | Lúxī virus (LUXV) | |
| Maporal orthohantavirus | Maporal virus (MAPV) | |
| Montano orthohantavirus | Montaño virus (MTNV) | |
| Necocli orthohantavirus | Necoclí virus (NECV) | |
| Oxbow orthohantavirus | Oxbow virus (OXBV) | |
| Prospect Hill orthohantavirus | Prospect Hill virus (PHV) | |
| Puumala orthohantavirus | Hokkaido virus (HOKV) | |
| Muju virus (MUJV) | ||
| Puumala virus (PUUV) | ||
| Rockport orthohantavirus | Rockport virus (RKPV) | |
| Sangassou orthohantavirus | Sangassou virus (SANGV) | |
| Seoul orthohantavirus | g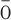 u virus (GOUV) u virus (GOUV) |
|
| Seoul virus (SEOV) | ||
| Sin Nombre orthohantavirus | New York virus (NYV) | |
| Sin Nombre virus (SNV) | ||
| Thailand orthohantavirus | Anjozorobe virus (ANJZV) | |
| Serang virus (SERV) | ||
| Thailand virus (THAIV) | ||
| Tula orthohantavirus | Adler virus (ADLV) | |
| Tula virus (TULV) | ||
| Yakeshi orthohantavirus | Yákèshí virus (YKSV) | |
| Loanvirus |
Longquan loanvirus
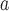
|
Lóngquán virus (LQUV) |
| Mobatvirus | Laibin mobatvirus | Láib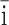 n virus (LAIV) n virus (LAIV) |
Nova mobatvirus
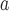
|
Nova virus (NVAV) | |
| Quezon mobatvirus | Quezon virus (QZNV) | |
| Thottimvirus | Imjin thottimvirus | Imjin virus (MJNV) |
Thottapalayam thottimvirus
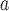
|
Thottapalayam virus (TPMV) | |
| Family Peribunyaviridae | ||
| Herbevirus |
Herbert herbevirus
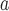
|
Herbert virus (HEBV) |
| Kibale herbevirus | Kibale virus (KIBV) | |
| Tai herbevirus | Taï virus (TAIV) | |
| Orthobunyavirus | Acara orthobunyavirus | Acará virus (ACAV) |
| Moriche virus (MORV) | ||
| Akabane orthobunyavirus | Akabane virus (AKAV) | |
| Sabo virus (SABOV) | ||
| Tinaroo virus (TINV) | ||
| Yaba-7 virus (Y7V) | ||
| Alajuela orthobunyavirus | Alajuela virus (ALJV) | |
| Brus Laguna virus (BLAV) | ||
| San Juan virus (SJV) | ||
| Anopheles A orthobunyavirus | Anopheles A virus (ANAV) | |
| Arumateua virus (ARTV = ARMTV) | ||
| Caraipé virus (CPEV = CRPV) | ||
| Las Maloyas virus (LMV) | ||
| Lukuni virus (LUKV) | ||
| Trombetas virus (TRMV) | ||
| Tucuruí virus (TUCV = TUCRV) | ||
| Anopheles B orthobunyavirus | Anopheles B virus (ANBV) | |
| Boracéia virus (BORV) | ||
| Bakau orthobunyavirus | Bakau virus (BAKV) | |
| Ketapang virus (KETV) | ||
| Nola virus (NOLAV) | ||
| Tanjong Rabok virus (TRV) | ||
| Telok Forest virus (TFV) | ||
| Batama orthobunyavirus | Batama virus (BMAV) | |
| Benevides orthobunyavirus | Benevides virus (BVSV = BENV) | |
| Bertioga orthobunyavirus | Bertioga virus (BERV) | |
| Cananéia virus (CNAV) | ||
| Guaratuba virus (GTBV) | ||
| Itimirim virus (ITIV) | ||
| Mirim virus (MIRV) | ||
| Bimiti orthobunyavirus | bimiti virus (BIMV) | |
| Botambi orthobunyavirus | Botambi virus (BOTV) | |
Bunyamwera orthobunyavirus
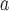
|
Anadyr virus (ANADV) | |
| Batai virus (BATV) | ||
| Birao virus (BIRV) | ||
| Bozo virus (BOZOV) | ||
| Bunyamwera virus (BUNV) | ||
| Cache Valley virus (CVV) | ||
| Fort Sherman virus (FSV) | ||
| Germiston virus (GERV) | ||
| Ilesha virus (ILEV) | ||
| Lokern virus (LOKV) | ||
| Maguari virus (MAGV) | ||
| Mboké virus (MBOV) | ||
| Ngari virus (NRIV) | ||
| Northway virus (NORV) | ||
| Playas virus (PLAV) | ||
| Potosi virus (POTV) | ||
| Santa Rosa virus (SARV) | ||
| Shokwe virus (SHOV) | ||
| Stanfield virus (STAV) | ||
| Tensaw virus (TENV) | ||
| Tlacotalpan virus (TLAV) | ||
| Xingu virus (XINV) | ||
| Bushbush orthobunyavirus | Benfica virus (BENV = BNFV) | |
| Bushbush virus (BSBV) | ||
| Juan Díaz virus (JDV) | ||
| Bwamba orthobunyavirus | Bwamba virus (BWAV) | |
| Pongola virus (PGAV) | ||
| California encephalitis orthobunyavirus | Achiote virus (ACHOV) | |
| California encephalitis virus (CEV) | ||
| infirmatus virus (INFV) | ||
| Inkoo virus (INKV) | ||
| Jamestown Canyon virus (JCV) | ||
| Jerry Slough virus (JSV) | ||
| Keystone virus (KEYV) | ||
| Khatanga virus (KHATV) | ||
| La Crosse virus (LACV) | ||
| Lumbo virus (LUMV) | ||
| Melao virus (MELV) | ||
| Morro Bay virus (MBV) | ||
| San Angelo virus (SAV) | ||
| Serra do Navio virus (SDNV) | ||
| snowshoe hare virus (SSHV) | ||
| South River virus (SORV) | ||
| Ťahyňa virus (TAHV) | ||
| trivittatus virus (TVTV) | ||
| Capim orthobunyavirus | Capim virus (CAPV) | |
| Caraparu orthobunyavirus | Apeú virus (APEUV) | |
| Bruconha virus (BRUV) | ||
| Caraparú virus (CARV) | ||
| El Huayo virus (EHUV) | ||
| Itaya virus (ITYV) | ||
| Ossa virus (OSSAV) | ||
| Vinces virus (VINV) | ||
| Catu orthobunyavirus | Catú virus (CATUV) | |
| Estero Real orthobunyavirus | Estero Real virus (ERV) | |
| Gamboa orthobunyavirus | Calchaquí virus (CQIV) | |
| Gamboa virus (GAMV) | ||
| Pueblo Viejo virus (PVV) | ||
| Soberanía virus (SOBV) | ||
| Guajara orthobunyavirus | Guajará virus (GJAV) | |
| Guama orthobunyavirus | Ananindeua virus (ANUV) | |
| Guamá virus (GMAV) | ||
| Mahogany Hammock virus (MHV) | ||
| Moju virus (MOJUV) | ||
| Guaroa orthobunyavirus | Guaroa virus (GROV) | |
| Kaeng Khoi orthobunyavirus | Kaeng Khoi virus (KKV) | |
| Kairi orthobunyavirus | Kairi virus (KRIV) | |
| Koongol orthobunyavirus | koongol virus (KOOV) | |
| wongal virus (WONV) | ||
| Madrid orthobunyavirus | Madrid virus (MADV) | |
| Main Drain orthobunyavirus | Main Drain virus (MDV) | |
| Manzanilla orthobunyavirus | Buttonwillow virus (BUTV) | |
Cát Qu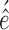 virus (CQV) virus (CQV) |
||
| Ingwavuma virus (INGV) | ||
| Inini virus (INIV) | ||
| Manzanilla virus (MANV) | ||
| Mermet virus (MERV) | ||
| Marituba orthobunyavirus | Gumbo Limbo virus (GLV) | |
| Marituba virus (MTBV) | ||
| Murutucú virus (MURV) | ||
| Nepuyo virus (NEPV) | ||
| Restan virus (RESV) | ||
| Zungarococha virus (ZUNV) | ||
| Minatitlan orthobunyavirus | Minatitlán virus (MNTV) | |
| Palestina virus (PLSV) | ||
| MPoko orthobunyavirus | M’Poko virus (MPOV) | |
| Yaba-1 virus (Y1V) |
| Nyando orthobunyavirus | Nyando virus (NDV) | |
| Eretmapodites virus (ERETV) | ||
| Olifantsvlei orthobunyavirus | Bobia virus (BIAV) | |
| Dabakala virus (DABV) | ||
| Olifantsvlei virus (OLIV) | ||
| Oubi virus (OUBIV) | ||
| Oriboca orthobunyavirus | Itaquí virus (ITQV) | |
| Oriboca virus (ORIV) | ||
| Oropouche orthobunyavirus | Facey’s paddock virus (FPV) | |
Iquitos virus (IQTV)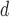
|
||
| Madre de Dios virus (MDDV) | ||
| Oropouche virus (OROV) | ||
| Perdões virus (PDEV) | ||
| Pintupo virus (PINTV) | ||
| Utinga virus (UTIV) | ||
| Utivé virus (UVV = UTVEV) | ||
| Patois orthobunyavirus | Abras virus (ABRV) | |
| Babahoya virus (BABV) | ||
| Pahayokee virus (PAHV) | ||
| Patois virus (PATV) | ||
| Shark River virus (SRV) | ||
| Sathuperi orthobunyavirus | Douglas virus (DOUV) | |
| Sathuperi virus (SATV) | ||
| Shamonda orthobunyavirus | Peaton virus (PEAV) | |
| Sango virus (SANV) | ||
| Shamonda virus (SHAV) | ||
| Shuni orthobunyavirus | Aino virus (AINOV) | |
| Kaikalur virus (KAIV) | ||
| Shuni virus (SHUV) | ||
| Simbu orthobunyavirus | Simbu virus (SIMV) | |
| Oya virus (OYAV) | ||
| Tacaiuma orthobunyavirus | Tacaiuma virus (TCMV) | |
| CoAr 1071 virus (CA1071V) | ||
| CoAr 3627 virus (CA3626V) | ||
| Virgin River virus (VRV) | ||
| Tete orthobunyavirus | Bahig virus (BAHV) | |
| Matruh virus (MTRV) | ||
| Tete virus (TETEV) | ||
| Tsuruse virus (TSUV) | ||
| Weldona virus (WELV) | ||
| Thimiri orthobunyavirus | Thimiri virus (THIV) | |
| Timboteua orthobunyavirus | Timboteua virus (TBTV) | |
| Turlock orthobunyavirus | Lednice virus (LEDV) | |
| Turlock virus (TURV) | ||
| Umbre virus (UMBV) | ||
| Wolkberg orthobunyavirus | Wolkberg virus (WBV) | |
| Wyeomyia orthobunyavirus | Anhembi virus (AMBV) | |
| BeAr 328208 virus (BAV) | ||
| Cachoeira Porteira virus (CPOV) | ||
| Iaco virus (IACOV) | ||
| Macauã virus (MCAV) | ||
| Rio Pracupi virus | ||
| Sororoca virus (SORV) | ||
| Taiassui virus (TAIAV) | ||
| Tucunduba virus (TUCV) | ||
| Wyeomyia virus (WYOV) | ||
| Zegla orthobunyavirus | Zegla virus (ZEGV) | |
| Shangavirus | Insect shangavirus | Shu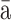 ngào insect virus 1 (SgIV-1) ngào insect virus 1 (SgIV-1) |
| Tospovirus | Groundnut bud necrosis tospovirus | groundnut bud necrosis virus (GBNV) |
| Groundnut ringspot tospovirus | groundnut ringspot virus (GRSV) | |
| Groundnut yellow spot tospovirus | groundnut yellow spot virus (GYSV) | |
| Impatiens necrotic spot tospovirus | impatiens necrotic spot virus (INSV) | |
| Iris yellow spot tospovirus | iris yellow spot virus (IYSV) | |
| Polygonum ringspot tospovirus | Polygonum ringspot virus (PolRSV) | |
| Tomato chlorotic spot tospovirus | tomato chlorotic spot virus (TCSV) | |
Tomato spotted wilt tospovirus
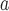
|
tomato spotted wilt virus (TSWV) | |
| Watermelon bud necrosis tospovirus | watermelon bud necrosis virus (WBNV) | |
| Watermelon silver mottle tospovirus | watermelon silver mottle virus (WSMoV) | |
| Zucchini lethal chlorosis tospovirus | zucchini lethal chlorosis virus (ZLCV) |
Modified from Maes et al. (2019a).
 Type species.
Type species.
Potential Consequences of Current Shortcomings in the ICTV Framework
Mixing up species and viruses, distinct viruses within a species, viruses and subtypes, and subtypes and isolates leads to confusion as to what is actually being studied (even the ICTV webpage sometimes confuses “isolate” and “strain” or “exemplar” when mentioning what should be called “virus”—isolates and strains are representatives of viruses). Importantly, such taxonomic confusion resulting in the mislabeling of a virus may lead to clinically inappropriate treatment of a patient or, in case of some plant viruses, inadequate control measures. Also, the misperception regarding the virus’s geographic distribution, vector and vertebrate hosts, pathogenicity, biosafety classification, or import/export requirements, may create biosafety, biosecurity, and permit challenges and may hinder appropriate outbreak control responses (Blitvich et al. 2018).
Orthobunyaviruses in general exemplify this dilemma perfectly. These viruses are historically considered a group of groups of similar individuals or genetic variants of viruses capable of exchanging genomic segments (reassortment). Viruses belonging to the group formerly known as “the California serogroup” (Casals and Whitman 1960) can reassort (Briese et al. 2013), so it is not illogical to consider them as representing a single species (reassortment could be considered a curious variety of “interbreeding,” a characteristic fundamental to Ernst Mayr’s definition of “species” (Mayr 1942)). Yet, reassortment can change virus properties dramatically—just because distinct viruses are assigned to the same species does not make them less distinct. For instance, the Garissa isolate of Ngari virus (NRIV) caused a severe viral hemorrhagic fever epidemic among humans in Kenya and Somalia from 1997 to 1998. NRIV was eventually shown to be a reassortant orthobunyavirus with a genome containing two segments from Bunyamwera virus (BUNV) and a third segment stemming from an unknown virus (Bowen et al. 2001; Gerrard et al. 2004; Briese et al. 2006). Both BUNV and NRIV have been classified in the same species, Bunyamwera orthobunyavirus, together with numerous other viruses (Table 1). If species names and virus names, such as “Bunyamwera orthobunyavirus” and “Bunyamwera virus,” are confused, or if NRIV is not listed as a member of the species Bunyamwera orthobunyavirus, one can imagine how public health responses may be hindered. Indeed, BUNV has never been associated with viral hemorrhagic fever in humans. Similarly, distinguishing viruses for biosafety and regulatory purposes and providing a risk assessment and justification for the shipment of viruses for import or export is of enormous practical importance. Some members of the species Bunyamwera orthobunyavirus have never been associated with human diseases (e.g., Tensaw virus [TENV]), whereas others, such as NRIV, may cause human deaths. Logically, transport, storage, and work permissions may differ for these viruses and are not necessarily (and probably should not be) generalized to all members of a species (Blitvich et al. 2018).
In short, all stakeholders in virology should have access to a frequently updated database that can be used to identify the current classification of the virus being studied or addressed. In the absence of such a database, scientist–administrators (e.g., at the US Centers for Disease Control and Prevention, the US National Institutes of Health, the World Health Organization, the World Organization for Animal Health, the Pan-American Health Organization, and Plant Quarantine Services), universities, state and local laboratories, commercial vector control groups, journal and textbook editors, internet reporting systems (such as ProMED, the Program for Monitoring Emerging Diseases), blogs and other forms of social media, human and veterinary clinicians, and historians will continue to be sorely confused by the seeming discordance of taxon lists in ICTV publications and incomplete virus lists maintained elsewhere. Unless a proper and complete listing of viruses is made available in an openly accessible database, funding proposals for crucial projects may be inexact, possibly incorrect, and ultimately unclear to those who must approve them.
Possible Solutions
The ICTV, an internationally recognized body with more than 50 years of tradition (Adams et al. 2017), is the logical choice to address the outlined problem. The ICTV is already in charge of taxon classification and nomenclature, whereas currently no regulatory body is in charge of virus nomenclature. The only other repository that maintains (incomplete) lists of viruses, the US National Center for Biotechnology Information (NCBI), is dependent on single user-provided information and is building its taxonomic framework on that of the ICTV as much as is feasible. Unfortunately, the ICTV is currently an underfunded organization that functions almost exclusively due to the tireless efforts of volunteers. Consequently, any new ICTV task may further strain the meager funding pool and/or increase the workload of the volunteers. With these constraints in mind, we suggest that lists of virus names and abbreviations be made available to the ICTV via the ICTV Study Groups after consultation of expert groups with mutual interests, such as the Centers for Disease Control and Prevention (CDC) or the Subcommittee on InterRelations of Catalogued Arboviruses (SIRACA) of the American Committee on Arthropod-borne Viruses (ACAV). ICTV Study Groups consist of a Chair (appointed by an ICTV Subcommittee Chair) and a theoretically unlimited number of members appointed by the ICTV Study Group Chair. The task of the ICTV Study Groups is to evaluate, facilitate, or create taxonomic proposals (“TaxoProps”) for virus taxa at any rank, including species. Each of these proposals already is required to list at least one member (“exemplar virus”) belonging to a species. In addition, the ICTV Study Groups are required to write chapters for the ICTV Report (International Committee on Taxonomy of Viruses 2018c) on the composition of the taxa they govern (typically a family).
The ICTV Study Groups are already explicitly advised to list all member viruses for each species in these report chapters. Finally, the ICTV Study Groups are continuously called upon by the ICTV Executive Committee to populate the ICTV Virus Metadata Repository (VMR) Excel sheet (International Committee on Taxonomy of Viruses 2018b) and are specifically asked to add all species members into the sheet. Consequently, ICTV Study Groups already carefully curate the list of viruses that belong to species as part of their routine taxon review and proposal work flow and they already establish criteria for species inclusion, and, outside of official ICTV purview, for valid and ideally unique virus names and abbreviations. The ICTV Study Groups would simply be asked to systematically evaluate whether distinct virus names refer to actually distinct viruses or whether these names are synonyms for distinct isolates of the same virus. In the latter case, one virus name and abbreviation could be retired, and its use could be officially discouraged. In the former case, the ICTV Study Group could consider creating a novel species for the distinct virus by changing species demarcation criteria to reduce the number of species members. We argue that these activities would not require any additional ICTV funding and that none of these activities would increase the workload of Study Groups significantly. In any case, since the ICTV Study Group membership is theoretically unlimited, any ICTV Study Group Chair could increase the group by appointing additional members to distribute increasing workloads.
The final list of virus names and abbreviations could then be forwarded to the ICTV Executive Committee, which could compare the list to, and if appropriate incorporate into, the already rapidly developing VMR and ICTV Report. We appreciate the current ICTV Executive Committee’s effort and strongly encourage further development of the VMR that could then be used as the source material to extend the existing ICTV taxon database by one level to include viruses as the lowest level beneath species. A direct search at https://talk.ictvonline.org/taxonomy/ for  measles virus
measles virus or
or  MeV
MeV would then yield the result
would then yield the result
“Negarnaviricota › Haploviricotina › Monjiviricetes › Mononegavirales › Paramyxoviridae › Morbillivirus › Measles morbillivirus › measles virus (MeV).”
Likewise, searches for  California encephalitis virus
California encephalitis virus and
and  La Crosse virus
La Crosse virus would then yield the results
would then yield the results
“Negarnaviricota › Polyploviricotina › Ellioviricetes › Bunyavirales › Peribunyaviridae › Orthobunyavirus › California encephalitis orthobunyavirus › California encephalitis virus (CEV),” and
“Negarnaviricota › Polyploviricotina › Ellioviricetes › Bunyavirales › Peribunyaviridae › Orthobunyavirus › California encephalitis orthobunyavirus › La Crosse virus (LACV),” respectively.
This final product, reminiscent of Wilson and Reeder’s database for mammalian species and mammal vernacular names (Wilson and Reeder 2005), could then serve as source material for the NCBI and ultimately be used to achieve complete harmony between the ICTV and other public databases (e.g., NCBI, EBI). This last step is critical and would allow established virus names to be communicated among both sequence data providers and users, improving acceptance and usage of these names. Both the NCBI Virus and Taxonomy groups are already digesting the ICTV VMR and updating taxonomy, exemplar isolates, and reference genome records (RefSeqs) accordingly (Brister et al. 2015). Extending this workflow to virus names and abbreviations would be a relatively trivial endeavor.
Some impediments are certain to occur during establishment of a robust, community-accepted list of virus names and unique virus name abbreviations. The potential scope is large. Four thousand nine hundred and fifty-eight ICTV viral species are currently approved, which means that the number of currently classified viruses is higher than that. Moreover, duplications or multiplications of virus name abbreviations are already rampant. However, we have good reason to believe that an ICTV-curated virus name list can be achieved. Removing or at least clearly identifying virus name synonyms and creation of unique virus name abbreviations would result in clear advantages for virologists globally because literature database searches and communication overall would be simplified. Ongoing community efforts outside of the official ICTV framework, but often with the support of ICTV Study Groups, have already demonstrated that similar standards can be achieved, disseminated, and accepted. For instance, naming standards have been established for human adenovirus genotypes (Seto et al. 2011), papillomavirus types (Bzhalava et al. 2015; Mühr et al. 2018), rotaviruses (Matthijnssens et al. 2011), and filoviruses (Kuhn et al. 2013a, 2013b, 2014a, 2014b), and the results of these efforts have been integrated into GenBank submissions standards and NCBI taxonomy. In addition, individual ICTV Study Group members have created specific databases for their taxa (The Pirbright Institute 2017), thereby demonstrating that a curated virus name and abbreviation database can indeed be achieved with the help of community members.
As outlined, resolving the current confusion requires some investment from all interested and/or affected parties. The virology community needs to better understand the ins and outs of virus taxonomy and the process involved with establishing novel taxonomy, whereas the ICTV needs to better understand the needs of the virology community. Subtle changes in the ICTV framework like those outlined above would be extremely beneficial for both the virology community and the ICTV. Finally, we call on the IUMS to consider extending the mandate of the ICTV to administer the nomenclature of viruses to achieve name harmonization and standardization in virology. Virus discoverers would still retain the right to name new viruses which, along with abbreviations, could become an official part of ICTV-guided taxonomic proposals. Upon submission, the ICTV would cross-check the submitted name and abbreviation against its database to ensure the name and abbreviation are original, sufficiently distinct, and do not violate the ICTV Code. If there are no obvious problems, the name and abbreviation would be added to the official ICTV database, and thereby benefit all interested parties.
Acknowledgments
The authors thank Laura Bollinger, Integrated Research Facility, for critically editing the manuscript. The views and conclusions contained in this document are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of the US Department of Defense or the US Army, of the US Department of Health and Human Services, the Department of Homeland Security (DHS) Science and Technology Directorate (S&T), of the International Committee on Taxonomy of Viruses, or of the institutions and companies affiliated with the authors. In no event shall any of these entities have any responsibility or liability for any use, misuse, inability to use, or reliance upon the information contained herein. The US departments do not endorse any products or commercial services mentioned in this publication.
Funding
This work was supported by Battelle Memorial Institute’s prime contract with the US National Institute of Allergy and Infectious Diseases (NIAID) under Contract No. HHSN272200700016I (J.H.K.), by US National Institutes of Health (NIH) grant R24 AI120942 (S.C.W and N.V.), and by the Intramural Research Program of the NIH, National Library of Medicine (J.R.B.). S.S. acknowledges support from Mississippi State University (MSU)—Mississippi Agricultural and Forestry Experiment Station (MAFES) Strategic Research Initiative grants. This work was also funded in part under ContractNo. HSHQDC-15-C-00064 awarded by DHS S&T for the management and operation of the National Biodefense Analysis and Countermeasures Center (NBACC), a federally funded research and development center (V.W.).
References
- Adams M.J., Lefkowitz E.J., King A.M.Q., Harrach B., Harrison R.L., Knowles N.J., Kropinski A.M., Krupovic M., Kuhn J.H., Mushegian A.R., Nibert M.L., Sabanadzovic S., Sanfaçon H., Siddell S.G., Simmonds P., Varsani A., Zerbini F.M., Orton R.J., Smith D.B., Gorbalenya A.E., Davison A.J.. 2017. 50 years of the International Committee on Taxonomy of Viruses: progress and prospects. Arch. Virol. 162:1441–1446. [DOI] [PubMed] [Google Scholar]
- Blitvich B.J., Beaty B.J., Blair C.D., Brault A.C., Dobler G., Drebot M.A., Haddow A.D., Kramer L.D., LaBeaud A.D., Monath T.P., Mossel E.C., Plante K., Powers A.M., Tesh R.B., Turell M.J., Vasilakis N., Weaver S.C.. 2018. Bunyavirus taxonomy: limitations and misconceptions associated with the current ICTV criteria used for species demarcation. Am. J. Trop. Med. Hyg. 99:11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen M.D., Trappier S.G., Sanchez A.J., Meyer R.F., Goldsmith C.S., Zaki S.R., Dunster L.M., Peters C.J., Ksiazek T.G., Nichol S.T.; RVF Task Force . 2001. A reassortant bunyavirus isolated from acute hemorrhagic fever cases in Kenya and Somalia. Virology. 291:185–190. [DOI] [PubMed] [Google Scholar]
- Briese T., Bird B., Kapoor V., Nichol S.T., Lipkin W.I.. 2006. Batai and Ngari viruses: M segment reassortment and association with severe febrile disease outbreaks in East Africa. J. Virol. 80:5627–5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briese T., Calisher C.H., Higgs S.. 2013. Viruses of the family Bunyaviridae: are all available isolates reassortants? Virology. 446:207–216. [DOI] [PubMed] [Google Scholar]
- Brister J.R., Ako-adjei D., Bao Y., Blinkova O.. 2015. NCBI viral genomes resource. Nucleic Acids Res. 43:D571–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bzhalava D., Eklund C., Dillner J.. 2015. International standardization and classification of human papillomavirus types. Virology. 476:341–344. [DOI] [PubMed] [Google Scholar]
- Calisher C.H. 2016. The taxonomy of viruses should include viruses. Arch. Virol. 161:1419–1422. [DOI] [PubMed] [Google Scholar]
- Casals J., Whitman L.. 1960. A new antigenic group of arthropod-borne viruses: the Bunyamwera group. Am. J. Trop. Med. Hyg. 9:73–77. [DOI] [PubMed] [Google Scholar]
- Gerrard S.R., Li L., Barrett A.D., Nichol S.T.. 2004. Ngari virus is a Bunyamwera virus reassortant that can be associated with large outbreaks of hemorrhagic fever in Africa. J. Virol. 78:8922–8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J. 2001. The mind of the species problem. Trends Ecol. Evol. 16:326–329. [DOI] [PubMed] [Google Scholar]
- International Association for Plant Taxonomy. 2011. International Code of Nomenclature for algae, fungi, and plants (Melbourne Code). Regnum Vegetabile 154 Oberreifenberg (Germany): Koeltz Scientific Books; Available from: URL http://www.iapt-taxon.org/nomen/main.php. [Google Scholar]
- International Commission on Zoological Nomenclature. 2012. International Code of Zoological Nomenclature. 4th ed.London (UK): The International Trust for Zoological Nomenclature; 1999. Available from: URL http://www.iczn.org/iczn/index.jsp#. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Committee on Systematic Bacteriology. 1992. International Code of Nomenclature of Bacteria: Bacteriological Code, 1990 Revision. Washington (DC): ASM Press. [PubMed] [Google Scholar]
- International Committee on Taxonomy of Viruses (ICTV). 2018a. Complete ICTV statutes. Available from: URL https://talk.ictvonline.org/information/w/ictv-information/383/ictv-code.
- International Committee on Taxonomy of Viruses (ICTV). 2018b. ICTV Code. Available from: URL https://talk.ictvonline.org/information/w/ictv-information/383/ictv-code.
- International Committee on Taxonomy of Viruses (ICTV). 2018c. The ICTV Report Virus Taxonomy: The Classification and Nomenclature of Viruses. Available from: URL https://talk.ictvonline.org/ictv-reports/ictv_online_report/.
- International Committee on Taxonomy of Viruses (ICTV). 2018d. VMR: virus metadata resource. Available from: URL https://talk.ictvonline.org/taxonomy/vmr/.
- Kuhn J.H., Andersen K.G., Bào Y., Bavari S., Becker S., Bennett R.S., Bergman N.H., Blinkova O., Bradfute S., Brister J.R., Bukreyev A., Chandran K., Chepurnov A.A., Davey R.A., Dietzgen R.G., Doggett N.A., Dolnik O., Dye J.M., Enterlein S., Fenimore P.W., Formenty P., Freiberg A.N., Garry R.F., Garza N.L., Gire S.K., Gonzalez J.-P., Griffiths A., Happi C.T., Hensley L.E., Herbert A.S., Hevey M.C., Hoenen T., Honko A.N., Ignatyev G.M., Jahrling P.B., Johnson J.C., Johnson K.M., Kindrachuk J., Klenk H.-D., Kobinger G., Kochel T.J., Lackemeyer M.G., Lackner D.F., Leroy E.M., Lever M.S., Mühlberger E., Netesov S.V., Olinger G.G., Omilabu S.A., Palacios G., Panchal R.G., Park D.J., Patterson J.L., Paweska J.T., Peters C.J., Pettitt J., Pitt L., Radoshitzky S.R., Ryabchikova E.I., Saphire E.O., Sabeti P.C., Sealfon R., Shestopalov A.M., Smither S.J., Sullivan N.J., Swanepoel R., Takada A., Towner J.S., van der Groen G., Volchkov V.E., Volchkova V.A., Wahl-Jensen V., Warren T.K., Warfield K.L., Weidmann M., Nichol S.T.. 2014a. Filovirus RefSeq entries: evaluation and selection of filovirus type variants, type sequences, and names. Viruses. 6:3663–3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn J.H., Bào Y., Bavari S., Becker S., Bradfute S., Brauburger K., Rodney Brister J., Bukreyev A.A., Caì Y., Chandran K., Davey R.A., Dolnik O., Dye J.M., Enterlein S., Gonzalez J.-P., Formenty P., Freiberg A.N., Hensley L.E., Hoenen T., Honko A.N., Ignatyev G.M., Jahrling P.B., Johnson K.M., Klenk H.-D., Kobinger G., Lackemeyer M.G., Leroy E.M., Lever M.S., Mühlberger E., Netesov S.V., Olinger G.G., Palacios G., Patterson J.L., Paweska J.T., Pitt L., Radoshitzky S.R., Ryabchikova E.I., Saphire E.O., Shestopalov A.M., Smither S.J., Sullivan N.J., Swanepoel R., Takada A., Towner J.S., van der Groen G., Volchkov V.E., Volchkova V.A., Wahl-Jensen V., Warren T.K., Warfield K.L., Weidmann M., Nichol S.T.. 2014b. Virus nomenclature below the species level: a standardized nomenclature for filovirus strains and variants rescued from cDNA. Arch. Virol. 159:1229–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn J.H., Bao Y., Bavari S., Becker S., Bradfute S., Brister J.R., Bukreyev A.A., Caì Y., Chandran K., Davey R.A., Dolnik O., Dye J.M., Enterlein S., Gonzalez J.-P., Formenty P., Freiberg A.N., Hensley L.E., Honko A.N., Ignatyev G.M., Jahrling P.B., Johnson K.M., Klenk H.-D., Kobinger G., Lackemeyer M.G., Leroy E.M., Lever M.S., Lofts L.L., Mühlberger E., Netesov S.V., Olinger G.G., Palacios G., Patterson J.L., Paweska J.T., Pitt L., Radoshitzky S.R., Ryabchikova E.I., Saphire E.O., Shestopalov A.M., Smither S.J., Sullivan N.J., Swanepoel R., Takada A., Towner J.S., van der Groen G., Volchkov V.E., Wahl-Jensen V., Warren T.K., Warfield K.L., Weidmann M., Nichol S.T.. 2013a. Virus nomenclature below the species level: a standardized nomenclature for laboratory animal-adapted strains and variants of viruses assigned to the family Filoviridae. Arch. Virol. 158:1425–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn J.H., Bao Y., Bavari S., Becker S., Bradfute S., Brister J.R., Bukreyev A.A., Chandran K., Davey R.A., Dolnik O., Dye J.M., Enterlein S., Hensley L.E., Honko A.N., Jahrling P.B., Johnson K.M., Kobinger G., Leroy E.M., Lever M.S., Mühlberger E., Netesov S.V., Olinger G.G., Palacios G., Patterson J.L., Paweska J.T., Pitt L., Radoshitzky S.R., Saphire E.O., Smither S.J., Swanepoel R., Towner J.S., van der Groen G., Volchkov V.E., Wahl-Jensen V., Warren T.K., Weidmann M., Nichol S.T.. 2013b. Virus nomenclature below the species level: a standardized nomenclature for natural variants of viruses assigned to the family Filoviridae. Arch. Virol. 158:301–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn J.H., Jahrling P.B.. 2010. Clarification and guidance on the proper usage of virus and virus species names. Arch. Virol. 155:445–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes P., Adkins S., Alkhovsky S.V., Avšič-Županc T., Ballinger M.J., Bente D.A., Beer M., Bergeron É., Blair C.D., Briese T., Buchmeier M.J., Burt F.J., Calisher C.H., Charrel1 R.N., Choi I.R., Clegg C.S., de la Torre J. C, Lamballerie X.D., DeRisi J.L., Digiaro M., Drebot M., Ebihara H., Elbeaino T., Ergünay K., Fulhorst C.F., Garrison A.R., Gāo G.F., Gonzalez J.-P.J., Groschup M.H., Günther S., Haenni A.-L., Hall R.A., Hewson R., Hughes H.R., Jain R.K., Jonson M.G., Junglen S., Klempa B., Klingström J., Kormelink R., Lambert A.J., Langevin S.A., Lukashevich I.S., Marklewitz M., Martelli G.P., Mielke-Ehret N., Mirazimi A., Mühlbach H.-P., Naidu R., Nunes M.R.T., Palacios G., Papa A., Pawęska J.T., Peters C.J., Plyusnin A., Radoshitzky S.R., Resende R.O., Romanowski V., Sall A.A., Salvato M.S., Sasaya T., Schmaljohn C., Shí X., Shirako Y., Simmonds P., Sironi M., Song J.-W., Spengler J.R., Stenglein M.D., Tesh R.B., Turina M., Wèi T., Whitfield A.E., Yeh S.-D., Zerbini F.M., Zhang Y.-Z., Zhou X., Kuhn J.H.. 2019a. Taxonomy of the order Bunyavirales: second update 2018. Arch. Virol. DOI: 10.1007/s00705-018-04127-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes P., Amarasinghe G.K., Ayllón M.A., Basler C.F., Bavari S., Blasdell K.R., Briese T., Brown P.A., Bukreyev A., Balkema-Buschmann A., Buchholz U.J., Chandran K., Crozier I., Swart R.L.D., Dietzgen R.G., Dolnik O., Domier L.L., Drexler J.F., Dürrwald R., Dundon W.G., Duprex W.P., Dye J.M., Easton A.J., Fooks A.R., Formenty P.B.H., Fouchier R.A.M., Freitas-Astúa J., Ghedin E., Griffiths A., Hewson R., Horie M., Hurwitz J.L., Hyndman T.H., Jiāng D., Kobinger G.P., Kondō H., Kurath G., Kuzmin I.V., Lamb R.A., Lee B., Leroy E.M., Lĭ J., Marzano S.-Y.L., Mühlberger E., Netesov S.V., Nowotny N., Palacios G., Pályi B., Pawęska J.T., Payne S.L., Rima B.K., Rota P., Rubbenstroth D., Simmonds P., Smither S.J., Song Q., Song T., Span K., Stenglein M.D., Stone D.M., Takada A., Tesh R.B., Tomonaga K., Tordo N., Towner J.S., Hoogen B.V.D., Vasilakis N., Wahl V., Walker P.J., Wang D., Wang L.-F., Whitfield A.E., Williams J.V., Yè G., Zerbini F.M., Zhang Y.-Z., Kuhn J.H.. 2019b. Taxonomy of the order Mononegavirales: second update 2018. Arch. Virol. DOI: 10.1007/s00705-018-04126-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens J., Ciarlet M., McDonald S.M., Attoui H., Bányai K., Brister J.R., Buesa J., Esona M.D., Estes M.K., Gentsch J.R., Iturriza-Gómara M., Johne R., Kirkwood C.D., Martella V., Mertens P.P.C., Nakagomi O., Parreño V., Rahman M., Ruggeri F.M., Saif L.J., Santos N., Steyer A., Taniguchi K., Patton J.T., Desselberger U., Van Ranst M.. 2011. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG). Arch. Virol. 156:1397–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayden R.L. 1997. A hierarchy of species concepts: the denouement in the saga of the species problem. In: Claridge MF, Dawah AH, Wilson MR, editors. Species: the units of biodiversity. London (UK): Chapman & Hall; p. 381–424. [Google Scholar]
- Mayr E. 1942. Systematics and the origin of species. New York (NY): Columbia University Press. [Google Scholar]
- Mühr L.S.A., Eklund C., Dillner J.. 2018. Towards quality and order in human papillomavirus research. Virology. 519:74–76. [DOI] [PubMed] [Google Scholar]
- Peterson A.T. 2014. Defining viral species: making taxonomy useful. Virol. J. 11:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plyusnin Å., Beaty B.J., Elliott R.M., Goldbach R., Kormelink R., Lundvist A., Schmaljohn C.S., Tesh R.B.. 2011. Family Bunyaviridae. London (UK): Academic Press; Available from: URL https://talk.ictvonline.org/ictv-reports/ictv_9th_report/negative-sense-rna-viruses-2011/w/negrna_viruses/205/bunyaviridae. [Google Scholar]
- Postler T.S., Clawson A.N., Amarasinghe G.K., Basler C.F., Bavari S., Benkő M., Blasdell K.R., Briese T., Buchmeier M.J., Bukreyev A., Calisher C.H., Chandran K., Charrel R., Clegg C.S., Collins P.L., de la Torre J.C., DeRisi J.L., Dietzgen R.G., Dolnik O., Dürrwald R., Dye J.M., Easton A.J., Emonet S., Formenty P., Fouchier R.A.M., Ghedin E., Gonzalez J.-P., Harrach B., Hewson R., Horie M., Jiāng D., Kobinger G., Kondo H., Kropinski A.M., Krupovic M., Kurath G., Lamb R.A., Leroy E.M., Lukashevich I.S., Maisner A., Mushegian A.R., Netesov S.V., Nowotny N., Patterson J.L., Payne S.L., Paweska J.T., Peters C.J., Radoshitzky S.R., Rima B.K., Romanowski V., Rubbenstroth D., Sabanadzovic S., Sanfaçon H., Salvato M.S., Schwemmle M., Smither S.J., Stenglein M.D., Stone D.M., Takada A., Tesh R.B., Tomonaga K., Tordo N., Towner J.S., Vasilakis N., Volchkov V.E., Wahl-Jensen V., Walker P.J., Wang L.-F., Varsani A., Whitfield A.E., Zerbini F.M., Kuhn J.H.. 2017. Possibility and challenges of conversion of current virus species names to Linnaean binomials. Syst. Biol. 66:463–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto D., Chodosh J., Brister J.R., Jones M.S.; Members of the Adenovirus Research Community. 2011. Using the whole-genome sequence to characterize and name human adenoviruses. J. Virol. 85:5701–5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Pirbright Institute. 2017. Picornavirales SG. Available from: URL http://www.picornavirales.org/study_group/picornavirales_sg.htm.
- US Centers for Disease Control and Prevention. 2018. Arbovirus catalog. Available from: URL https://wwwn.cdc.gov/arbocat/.
- US Department of Health and Human Services, Centers for Disease Control and Prevention, National Institutes of Health. 2009. Biosafety in microbiological and biomedical laboratories (BMBL). HHS Publication No. (CDC) 93–8395. 5 ed.Washington (DC): US Government Printing Office; Available from: URL https://www.cdc.gov/biosafety/publications/bmbl5/. [Google Scholar]
- Van Regenmortel M.H.V. 2018. The species problem in virology. Adv. Virus Res. 100:1–18. [DOI] [PubMed] [Google Scholar]
- Van Regenmortel M.H.V., Maniloff J., Calisher C.. 1991. The concept of virus species. Arch. Virol. 120:313–314. [DOI] [PubMed] [Google Scholar]
- Wildy P. 1971. Classification and nomenclature of viruses—First Report of the International Committee on Nomenclature of Viruses. Basel (Switzerland): S. Karger. [Google Scholar]
- Wilson D.E., Reeder D.M.. 2005. Mammal species of the world. A taxonomic and geographic reference. 3rd ed Baltimore (MD): Johns Hopkins University Press; Available from: URL http://www.departments.bucknell.edu/biology/resources/msw3/. [Google Scholar]


