Abstract
Retinal pigment epithelial (RPE) cells are well known to play a central role in the progression of numerous retinal diseases. Changes in the structure and function of these cells thus may serve as sensitive biomarkers of disease onset. While in vivo studies have focused on structural changes, functional ones may better capture cell health owing to their more direct connection to cell physiology. In this study, we developed a method based on adaptive optics optical coherence tomography (AO-OCT) and speckle field dynamics for characterizing organelle motility in individual RPE cells. We quantified the dynamics in terms of an exponential decay time constant, the time required for the speckle field to decorrelate. Using seven normal subjects, we found the RPE speckle field to decorrelate in about 5 s. This result has two fundamental implications for future clinical use. First, it establishes a path for generating a normative baseline to which motility of diseased RPE cells can be compared. Second, it predicts an AO-OCT image acquisition time that is 36 times faster than used in our earlier report for individuating RPE cells, thus a major improvement in clinical efficacy.
1. Introduction
Retinal pigment epithelial (RPE) cells are composed of organelles that are under constant motion as they execute cellular and molecular tasks, which encompass essentially every aspect of RPE cell physiology. Thus, organelle motion is fundamental to RPE function [1]. Because of the key role RPE cells play in the interchange of nutrients, ions, water and waste between the choriocapillaris and the photoreceptors, this monolayer of cells is the initiation point of many retinal degenerative diseases, most notably age-related macular degeneration, but also Best’s disease, Stargardt’s disease, retinitis pigmentosa, and others [2]. Early manifestation of RPE degeneration occurs at the cellular level and so detection, monitoring, and treatment should be most effective when targeting processes at this level. Consistent with this, animal studies have shown that diseased RPE cells exhibit abnormal organelle motility [3–5], thus pointing to motility as a potentially sensitive indicator of cell health. However, how these findings translate to the living human eye remains unknown. The primary obstacles to making use of RPE organelle motility as a biomarker has been the limited resolution of in vivo imaging techniques necessary to delineate these cells and the strong waveguiding property of the overlying photoreceptors that mask the RPE. Adaptive optics optical coherence tomography (AO-OCT) and scanning laser ophthalmoscope (AO-SLO) approaches have overcome these obstacles to provide detailed mapping of the RPE cell mosaic [6–16]. AO-OCT delineates RPE cells by taking advantage of the cells’ intrinsic organelle motion to increase cell contrast [10,17], while AO-SLO uses the intrinsic fluorescence of lipofuscin [6,7,14] and melanin [12–14], extrinsic fluorescence of an ICG dye [11], or multiply-scattered light (dark-field detection) [8].
Despite these early demonstrations, measurements have been limited to morphological properties of RPE cells, such as cell density, area, and photoreceptor-to-RPE ratio. Measurement of cell function as expressed by organelle motion remains unexplored in living eyes. In a previous RPE imaging study, we found that AO-OCT volumes acquired at 3-minute intervals exhibited different speckle patterns in the RPE cells, sufficiently so that averaging of these volumes suppressed speckle noise and revealed the RPE cell mosaic [10]. Cancellation of speckle by averaging confirmed that the motility dynamics of these cells occurred on a time scale no greater than 3 minutes. In a subsequent AO-OCT pilot study, we characterized the temporal dynamics of RPE motility in two normal subjects and found them to be significantly shorter, being on the order of seconds rather than minutes. Averaging of images acquired at this shorter time interval also resulted in delineation of the RPE cell mosaic, thus substantiating the presence of faster dynamics [18].
In this study, we extend these first motility measurements: acquiring measurements on seven subjects, optimizing the sampling protocol for more complete characterization of the RPE motility dynamics, and confirming results on two separate AO-OCT imaging systems. We quantify dynamics in terms of an exponential decay time constant (τ), the time for motility to decorrelate the amplitude speckle field across individual RPE cells. This characterization has two fundamental uses. First, it provides a path for obtaining a motility baseline for normal, healthy RPE to which we can compare diseased RPE. Second, it predicts the extent to which our previously reported imaging experiments for individuating RPE cells can be reduced from its 90-minute length, a necessary requirement for clinical translation. We further demonstrate generalization of the clinical utility of RPE motility by obtaining results that are consistent on two AO-OCT platforms. These platforms, while different, share similar performance attributes, most critically the coherence volume of their imaging beam, which determines the number of organelles that contribute to each speckle.
2. Methods
2.1. Description of AO-OCT imaging systems
RPE motility measurements were acquired using two AO-OCT imaging systems, one at Indiana University and the other at the U.S. Food and Drug Administration (FDA). The two systems share similar optical designs (see [17,19,20] for description) and acquired RPE images of similar clarity [10,17]. Key system parameters are summarized in Table 1. Critical for this study, the imaging coherence (speckle) volumes of the two AO-OCT systems were essentially the same: 24.2 µm3 (Indiana) and 23.1 µm3 (FDA) in retinal tissue (see Table 1). Thus speckle formed by each system encapsulated the same total number of organelles and therefore should yield similar speckle dynamics when imaging in the same retinal tissue. The systems acquired images fast enough to track RPE speckle dynamics up to 2.75 Hz.
Table 1. AO-OCT system technical parameters for RPE motility imaging.
| Parameters | Indiana AO-OCT system | FDA AO-OCT system |
|---|---|---|
| Center wavelength (λc) | 785 nm | 830 nm |
| Bandwidth (Δλ) | 47 nm | 60 nm |
| Optical resolution (w × l × d) | 2.4 × 2.4 × 4.2 µm3 | 2.5 × 2.5 × 3.7 µm3 |
| Light power entering eye | 400 µW | 430 µW |
| Coherence volume | 24.2 µm3 | 23.1 µm3 |
2.2. Experimental design
Subject: Seven subjects, ranging in age from 21 to 49 yr (S1 = 21, S2 = 26, S3 = 47, S4 = 49, S5 = 27, S6 = 32 and S7 = 34 yr old) and free of ocular disease, were recruited for the study. Four subjects (S1-S4) were imaged with the Indiana AO-OCT system, and three different subjects (S5-S7) were imaged with the FDA AO-OCT system. All subjects had best corrected visual acuity of 20/20 or better. Eye lengths ranged from 23.56 mm to 25.40 mm as measured with IOLMaster (Zeiss, Oberkochen, Germany) and were used to correct for axial length differences in retinal image scale following Bennett et al. [21]. All procedures on the subjects adhered to the tenets of Helsinki declaration and approved by the Institutional Review Board of Indiana University and the FDA, respectively. Written informed consent was obtained after the nature and possible risks of the study were explained.
AO-OCT beam intensity was measured at the cornea (Table 1) and below the safe limits established by ANSI [22] for the retinal illumination pattern used (bidirectional point scan over a square area) and length of the experiment. The right eye was cyclopleged and dilated with one drop of Tropicamide 0.5% for imaging. The eye and head were aligned and stabilized using a bite bar (Indiana) or chin and head rest (FDA) attached to a XYZ translation stage.
Imaging protocol: AO-OCT volumes were acquired at 7° temporal to the fovea. Foveal scans were avoided to mitigate scanning beam distraction and additional eye motion by the subjects. For each subject, 30 to 62 AO-OCT videos were collected at 1 min or less interval and time stamped. As we did not know which video acquisition parameters would best capture the dynamics of RPE organelle motility, three experiments were conducted using different imaging parameters (Table 2). We used two main video durations: short videos (1.8 s for Experiment 1 and 1.3 s for Experiment 2) were acquired in all seven subjects, and long videos (8.6 s and 9 s video durations: Experiment 2 and Experiment 3) were acquired in four of the seven subjects (S2, S5-S7). Experiment 1 and 3 images were acquired at high speed (500 kHz A-line rate; 5.5 Hz volume rate), and Experiment 2 images were acquired at lower speed (210 kHz A-line rate; 2.3 Hz volume rate). These speeds enabled temporal dynamics of RPE cell motility to be captured up to 2.75 Hz and 1.2 Hz, respectively. During the long video acquisitions, subjects were instructed to blink once every 5 s to maintain good tear film quality while minimizing the number of volumes lost due to blinking. Time interval between consecutive videos ranged from 25 s to 80 s.
Table 2. Image acquisition parameters.
| Parameters | Exp. 1 (S1-S4) | Exp. 2 (S5-S7) | Exp. 3 (S2) | |
|---|---|---|---|---|
| A-line acquisition rate | 500 kHz | 210 kHz | 500 kHz | |
| A-line lateral sampling | 1 µm/px | 1.5 µm/px | 1 µm/px | |
| # of A-lines per B-scan | 300 | 300 | 300 | |
| # of B-scans per volume | 300 | 300 | 300 | |
| Volume rate | 5.5 Hz | 2.3 Hz | 5.5 Hz | |
| AO-OCT image FOV | 1° × 1° | 1.5° × 1.5° | 1° × 1° | |
| Volume time interval (TI) | 0.18 s | 0.46 s | 0.18 s | |
| # of volumes per video | 10 | 3 | 20 | 50 |
| Video duration | 1.8 s | 1.3 s | 8.6 s | 9 s |
| Video time interval | ≤1 min | ≤1 min | ≤1 min | ≤1 min |
| # of videos / location | 30-62 | 30-40 | 2-3 | 2 |
Prior to collection of the AO-OCT volumes, system focus was adjusted to optimize cone image quality, determined by visual inspection of cones in en face images that were projected axially through the portion of the AO-OCT volume that contained the cone inner/outer segment junction (IS/OS) and cone outer segment tip (COST) reflectance bands [23].
2.3. 3-D image registration and data analysis
3-D registration was applied to the AO-OCT volumes, followed by layer segmentation of the cone IS/OS, COST, and RPE layers. The B-scan based 3-D registration algorithm [24] was processed on a Graphical Processing Unit (GPU) via the Compute Unified Device Architecture (CUDA; NVIDIA, Santa Clara CA) parallel programming platform. The segmentation and data analysis steps were processed with custom algorithms developed in MATLAB (Mathworks; Nattick, MA, U.S.A.). Registration and segmentation were based on further advancements of previously published algorithms [24,25], and data analysis algorithms were new for this study.
Volumes with excessive motion artifacts were removed. Remaining volumes were combined into a single, time-stamped video and then registered in all three dimensions with subcellular accuracy to correct for motion artifacts. Registration to this level was necessary to prevent masking of organelle motility within individual RPE cells, which themselves were manually identified in the averaged, registered RPE en face image. Two additional layers were selected as motility controls (see Fig. 1): reflections from the cone layer (IS/OS + COST) and outer nuclear layer (ONL). Without visible light stimulation, the cone reflectance (IS/OS + COST) is known to be largely stable over minutes and exceedingly so over several seconds [26], the temporal bandwidth of the RPE motility dynamics we measured in this study. Reflectance changes associated with disc shedding are infrequent and occur over much longer periods [27–31]. Therefore, we expected the cone reflections to have a relatively long decay time constant (τ), which we used for normalizing the RPE signal. In contrast, the ONL reflection is considerably weaker, so much so in our images that white noise dominates and results in a delta-like τ.
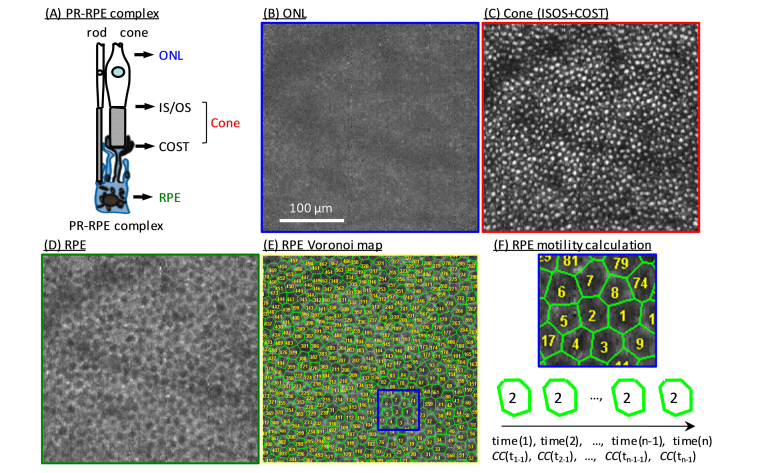
Fig. 1.
Assessing motility dynamics in RPE and cone cells in a 26-year-old subject (S2) at 7° temporal retina using AO-OCT imaging. (A) Schematic depicts the three layers analyzed: (1) ONL, (2) cone (IS/OS + COST), and (3) RPE. En face images were extracted from the same registered and averaged AO-OCT volume at depths of (B) ONL, (C) cone (projection of cone IS/OS and COST), and (D) RPE. Average was over 45 registered AO-OCT volumes. Each RPE cell in (D) is represented by a Voronoi cell in (E). (F) Shown is a magnified view of Voronoi cells in the blue box superimposed on the RPE map in (E). A motility function (CF) was calculated for each Voronoi cell—an example for one RPE cell is diagrammed at bottom of (F)—and then averaged across cells to increase SNR.
To quantify the temporal dynamics of RPE motility, a correlation function (CF) was calculated from the dynamic speckle pattern of individual RPE cells. Pixels within each RPE cell, defined by a Voronoi region [10] (see Fig. 1(E and F)), were used to compute the correlation coefficient (CC) between a reference (at time t1) and each consecutive image (at time t1, t2, t3, …) in the image series of that RPE cell:
| (1) |
where is the intensity of the rth row and cth column pixel in the reference image, is the mean intensity of all pixels in the cell of the reference image, is the intensity of the rth row and cth column pixel of the nth image in the image series at time point tn-1, and is the mean intensity of all pixels in the cell of the nth image at time point tn-1. ∑r∑c is summation over all pixels within the RPE cell as defined by its Voronoi region. CC equals one if no changes occur between the two images (i.e., absence of motility). CC was computed for each RPE cell (see Fig. 1(F)) then averaged across all of the identified RPE cells at the same time point and plotted against time as CF in the local area. The same analysis was also applied to the corresponding regions of the two control layers: cone and ONL for comparison and normalization purposes. On four of the subjects (S2, S5-S7), additional CF values were computed at different depths across the RPE-Bruch’s membrane (BM) complex, thus testing for motility differences that might occur as organelle types and concentrations change across the complex depth, e.g., melanosomes, phagosome, and lipofuscin [32]. The five tested depths were apical, middle and basal areas of RPE, rod outer segment tip (ROST), and BM layer. The CF is predicted to follow an exponential decay model [33], which is described as
| (2) |
The correlation function is defined by two independent terms. The first term, , captures residual motion contributions from the eye and system, e.g., uncorrected scanning artifacts, which are reduced in magnitude to the sub-cellular level after image registration is applied. The second term, , captures organelle motility. is the de-correlation time of the speckle pattern and the metric we used to quantify RPE motility. Unfortunately, separating the two terms is problematic as both are unknown. Here we took advantage of the fact that all pixels of a given A-scan are simultaneously acquired, and therefore A(t) must contain the same system and uncorrected eye motion artifacts regardless of depth in the AO-OCT volume. Our strategy was therefore to remove the first term in our measurements by dividing the measured CF of the RPE by the measured CF of the cone signal. As the cone signal is stable, its CF can be approximated as A(t), i.e., τ >>1. We then fit the normalized CF of the RPE (CFnorm) to an exponential function:
| (3) |
where A and B are fitting parameters.
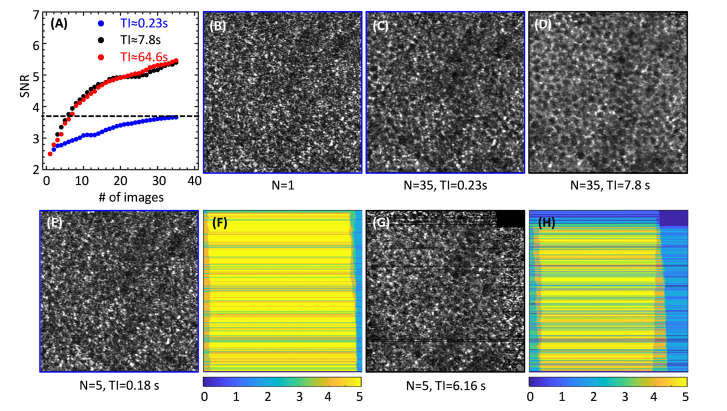
We hypothesize that in Eq. (3) is the smallest time interval that should be used between acquired AO-OCT images for the purpose of improving RPE cell contrast by averaging. To test, we computed the signal-to-noise ratio (SNR) of the RPE mosaic for different time intervals and number of images averaged. The SNR of the RPE was defined as the ratio between the RPE peak signal (cusp of concentrated energy) and the average of the noise floor in the 2-D power spectra.
3. Results
3.1 AO-OCT imaging of cones and RPE cells
We successfully observed the RPE cell mosaic and quantified its organelle motility in all seven subjects. Figure 1 shows a representative AO-OCT result on subject S2 and illustrates the contrasting appearances of the ONL, cone (IS/OS + COST), and RPE layers at the cellular level. Depth locations are marked in Fig. 1(A) that correspond to the averaged, registered en face images shown in Fig. 1(B-D). As expected, the image of ONL in Fig. 1(B) shows no clear cell structure owing to the weak reflectance of the ONL and dominating effect of noise. Conversely, bright regularly spaced cells are observed in the IS/OS + COST and RPE images in Fig. 1(C, D) with densities (cone: 11,406 cells/mm2; RPE: 4,834 cells/mm2) consistent with previously reported histology [34] and in vivo imaging [10]. The RPE Voronoi patches in Fig. 1(E & F) were used to calculate CC of each RPE cell using Eq. (1).
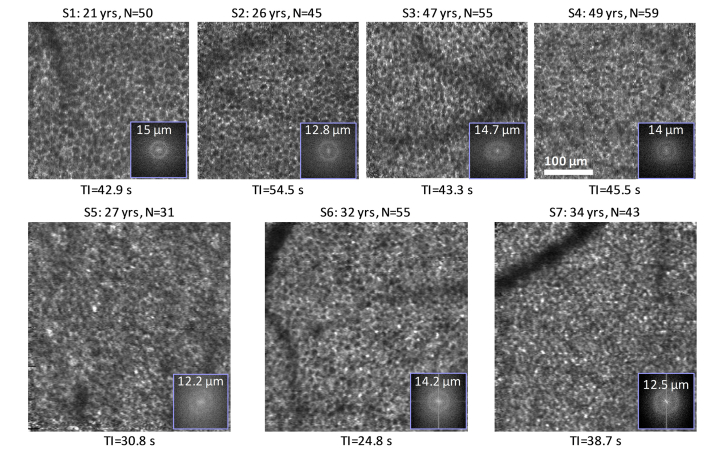
As shown in Fig. 2, the RPE cell mosaic is visually evident in all seven subjects using AO-OCT volumes that were separated by an average time interval of less than 1 min. This interval was realized by using one volume from each of the 30-62 videos acquired per subject. The fact that the RPE cells in these images are of comparable clarity (visual comparison) as those in our earlier study in which we used a 3-min imaging interval [10] suggests that organelle motility occurs on a time scale no larger than 1 min (specifically 24.8 s to 54.5 s). Also shown in Fig. 2 is the corresponding 2-D power spectra of the RPE images, which show rings of concentrated energy. Average ring radius equates to a row-to-row cell spacing of 13.6 ± 1.1 μm (7°) for the seven subjects, which agrees with measured RPE cell spacing in the literature [10,34].
Fig. 2.
Averaged and registered RPE images for the seven subjects imaged at 7° temporal to the fovea. N is the total number of images averaged. TI denotes mean time intervals between RPE images, which were selected one per AO-OCT video. 2-D power spectra are superimposed at bottom right of each en face image.
3.2 Characterization of RPE cell motility
We quantified RPE organelle motility using the correlation coefficient (Eq. (1). A representative example is given by the green data points in Fig. 3(A) that were averaged across 475 RPE cells. Motility of the two control layers (IS/OS + COST and ONL) are also plotted (red and blue). As expected, correlation of the RPE layer decreases at a rate faster than the cone layer and slower than the ONL. Cone signal is relatively stable, confirming our original expectations (see Visualization 1 (183.1MB, avi) for the intensity comparison between cone and RPE). Likewise, ONL shows a delta-like correlation with near zero correlation measured for the first sample point at 0.2 s, again confirming expectation. Similar behavior of the cone and ONL signals were found in the other six subjects.
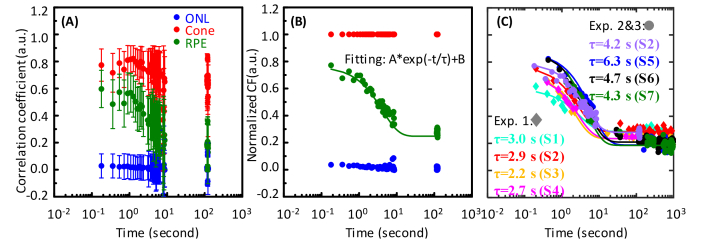
Fig. 3.
Motility dynamics measured at three retinal layers in seven subjects. (A) Representative raw motility function from S2 (Experiment 3) before normalization to the cone layer. Error bars represent standard deviation across 475 RPE cells. Colors represent measurements taken at three retinal layers: ONL (blue), Cone (IS/OS + COST) (red), and RPE (green). Colors in (A) also apply in (B). (B) Motility function is normalized to cone layer to remove residual eye motion and system errors. (C) Normalized RPE motility measurements are shown for the seven subjects. Scatterplots with ♦ symbols were from Experiment 1 with short video durations of 1.8 s, and scatterplots with ● symbols were from Experiments 2 and 3 with long video durations of ~9 s (see Table 2 for acquisition parameters). The gap between ~10 s and ~100 s for this data set is due to the time interval between two consecutive videos. (see Section 2.2). Fitted time constants are given in the key for each subject in each experiment.
To determine τ for RPE, system and residual motion errors were removed by normalizing to the cone reflectance as shown in the motility functions plotted in Fig. 3(B). The plot shows that CC is high at the beginning of the image series, indicating an initially stable speckle pattern. But with the lapse of time, CC decreases monotonically until after a few seconds it reaches a plateau determined by features common to all images in the series. Figure 3(C) shows the normalized CC measurements and corresponding CF fit (Eq. (3) for the seven subjects. From Experiment 1, the CF fit gives an average τ of 2.7 s for RPEs based on 363 to 475 RPE cells per subject. However, the Experiment 1 video duration of 1.8 s was too short to sample across the drop of the motility curve, including near the time constant value (2.7 s). To assess whether this undersampling may have caused an error in our CF fit, Experiment 2 and 3 used longer video durations (~9 s) in order to more completely capture the RPE motility exponential decay. These experiments yielded an average τ = 4.9 s based on 475 to 651 cells (see Fig. 3(c)). Note that the three subjects in Experiment 2 were imaged with the FDA system, and the averaged time constant (τ = 5.1 s) from those three subjects was almost twice long as the averaged time constant (τ = 2.7 s) from Experiment 1, but close to the time constant (τ = 4.2 s) from Experiment 3 obtained with the Indiana system.
To determine whether these differences in time constant were due to the use of different imaging systems or to an undersampling of CC, we used the data from Experiment 3 and refitted the exponential decay after removing the data points between 1.8 s and 9 s to mimic the short video scenario. The model yielded a similar time constant (τ = 2.8 s) as the one (τ = 2.9 s) achieved from the short video in Experiment 1. A shorter time constant (average τ = 2.7 s) was also produced when the same strategy (remove data points between 1.8 s and 9 s) was applied to the Experiment 2 data, thus confirming that undersampling was the root cause of the underestimation in Experiment 1. Regardless, results from any of the three experiments point to a motility time constant that is orders of magnitude less than the 3-minute interval we used in our first RPE imaging study [10].
A repeated measurement on RPE motility, performed using two long videos from the same subject (S6) on the same patch of retina, resulted in time constants of 4.74 s and 4.61 s. The small difference (<3%) indicates strong consistency of the motility measurement despite possible variability between RPE cells imaged in the two data sets.
3.3 Optimizing RPE image contrast with predicted cell motility
In this study, we first reduced the image acquisition interval from our previously reported 3 minutes to less than 1 minute and observed that RPE cell mosaic clarity was maintained as assessed visually (see Fig. 2). This factor of three reduction results in a total imaging session duration of 30 minutes for collecting ~30 volumes, which while improved still precludes clinical use. This raises the question as to how short the imaging session can be while still retaining clarity of the RPE cell mosaic. Analysis of our motility time constant measurements of the previous section provides a direct prediction (Fig. 3). Specifically, our motility measurements indicate a minimum time interval of ~5 s between consecutively acquired RPE images. This additional factor of 12 reduction (5 s instead of 1 minute) results in a total imaging session duration of just 2.5 minutes for collecting the ~30 volumes.
Motivated by this prediction, we further optimized our imaging protocol to test the optimal imaging time for RPE cell contrast enhancement. To do so required increasing the speed of our AO-OCT image saving routines. This proved non-trivial given the large size of the 4-D data streams to be saved. Three different imaging protocols provided average time intervals of 0.23 s, 7.8 s, and 64.6 s with the shortest two straddling the measured motility time constant (4.9 s). The same number of AO-OCT volumes were averaged for the three protocols, acquired on the approximate same patch of retina (within 0.5° of each other) in the same eye (S2), and followed the same processing steps established in Section 2.3. Note that while we would have liked to image the exact same patch of RPE cells for the three protocols, we know of no reason why the slightly different locations that we did image would influence our results. The structural shape, dimension, and composition of the RPE mosaic is well known to vary slowly across the retina, differences that would be inconsequential over the different locations imaged here. Consistent with this, our measurements of RPE cell spacing for the patches differed by only 3.6%. In addition, neither location contained major blood vessels, thus avoiding possible vasculature effects. Results are presented in Fig. 4.
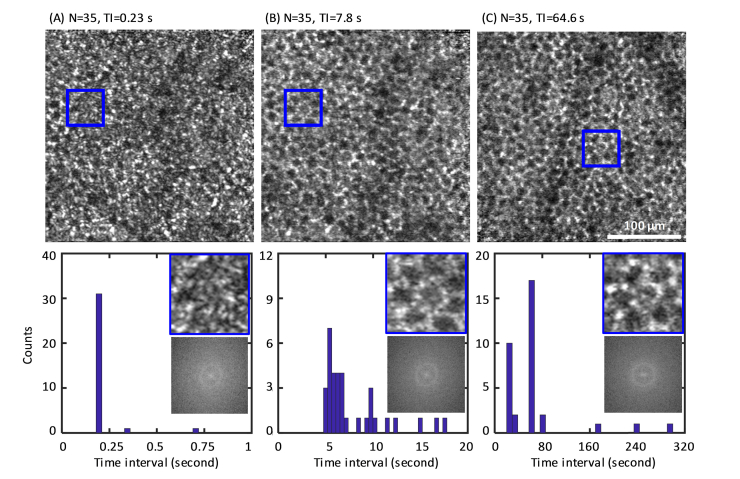
Fig. 4.
Clarity of the RPE cell mosaic depends on the time interval between acquired AO-OCT images. En face images are averages of 35 volumes with an average time interval of (A) 0.23 s, (B) 7.8 s, and (C) 64.6 s in subject S2. The corresponding histograms of time interval (TI), 2-D power spectra and magnified sub-images (50 μm × 50 μm) indicated by the blue box in (A-C) are shown below each image. Note: Images of (A) and (B) are from the same patch of the retina; (C) is from a slightly different patch.
With an average TI of 0.23 s (TI <), individual RPE cells are difficult to identify visually in the en face and magnified images of Fig. 4(A). In contrast, the average time interval of 7.8 s (TI >) yielded an improved image (Fig. 4(B)) with individual RPE cells more clearly visible and retaining cell clarity similar to that with the longer ~1-min TI (Fig. 4(C). Improved RPE clarity with TI > is also evident in the 2-D power spectra, which show better defined power at the RPE fundamental frequency and decreased noise floor compared to the one in Fig. 4(A). The similar RPE quality between Fig. 4(B) (7.8 s) and Fig. 4(C) (64.6 s) as assessed visually suggests that cell clarity might be preserved at even shorter TIs. Consistent with this, almost two thirds of the AO-OCT volumes (22 of 35) in Fig. 4(B) were collected with an average time interval of 5.9 s (see corresponding histogram), thus suggesting shorter TIs are likely possible and supporting our hypothesis that the motility time constant (~5 s as measured in this study) plays a fundamental role in determining RPE cell clarity.
To further explore the relative contributions of RPE motility (speckle pattern variation) and averaging on RPE en face image contrast, we calculated the SNR ratio as described in Section 2.3 for the three imaging protocols (with TI = 0.23 s, 7.8 s and 64.6 s, respectively). Figure 5(A) shows SNR as function of number of images averaged for the three different TIs, which were directly calculated from the OCT file timestamps. SNR increases with number of images averaged, but at different rates depending on TI. SNR increase is 2.2 × higher for the two longest TIs compared to the shortest one. The additional SNR improvement with TI further confirms the results observed in Fig. 4. The slight SNR increase with TI< is partially from the RPE speckle pattern variation, given the total image duration is 8 s (0.23 (TI) × 35 (# of images)), about twice longer than the predicted time constant ( s on S2). However, the SNR improvement is primarily attributed to averaging, which reduces the variation of OCT image noise (mainly photon noise) that sets the noise floor in the 2-D power spectra of our images. We observed the speckle patterns change little between Fig. 5(B) (N = 1) and Fig. 5(E) (N = 5), and somewhat alter their appearance in Fig. 5(C) (N = 35) in which the RPE clarity is clearly inferior to the en face image in Fig. 5(D) (N = 35). The comparison between Fig. 5(C) and Fig. 5(D) suggests the speckles remain largely correlated for TI = 0.23 s, and longer TI is required to enable speckle decorrelation to further improve RPE clarity. This is further illustrated by comparison of the en face images in Fig. 5(C) and 5(G), which appear qualitatively similar even though one is produced with 35 images but TI< and the other is produced with 5 images but TI, confirming the relative averages that produce equivalent SNR (horizontal crossing point of dash line in Fig. 5(A)). Similar results were obtained for the same analysis in other subjects as well. Note, eye motion may result in black pixels and/or lines in single en face images, which results in zero contribution to those pixels and/or lines in the image average. Thus variation in the effective number of images that are averaged at any pixel is expected. This is demonstrated by the false-colored images of Figs. 5(F) and 5(H) that show the number of volumes averaged varies from 0 to 5 depending on pixel location in the image.
Fig. 5.
(A) Signal-to-noise ratio (SNR) of RPE mosaic fundamental frequency as a function of AO-OCT images averaged from subject S2. The RPE mosaic fundamental frequency and noise were determined from the power spectrum of the registered, averaged images. Images were acquired at averaged TI of 0.23 s (blue), 7.8 s (black) and 64.6 s (red). RPE en face images of: (B) single volume (N = 1), (C) average of 35 volumes with averaged TI = 0.23 s, (D) average of 35 volumes with TI = 7.8 s, and average of 5 volumes with averaged (E) TI = 0.18 s, and (G) TI = 6.16 s. The number of effective volumes that generate each image pixel in (E) and (G) varies between 0 and 5 depending on eye motion as shown in (F) and (H), respectively. The fast B-scan direction of the AO-OCT system is horizontal.
4. Discussion
We developed a novel AO-OCT-based method for measuring organelle motility in individual RPE cells. Our method detects the time-dependent fluctuations in scattered light intensity (speckle) from the cells and relates these fluctuations to physical properties of the same cells (organelle motility in this study). This dynamic speckle technique has been successfully used in prior studies for capturing intra-cellular motion [35], enhancing subcellular details [36,37] and characterizing motility of other retinal tissue layers [38,39]. Using motion-evoked speckle changes here, we successfully delineated RPE cells and measured the RPE motility decay time constant (τ) in seven healthy subjects. Because our technique relies on the 3-D resolution provided by AO-OCT and the speckle decorrelation dynamics inherent in RPE organelle motility, we expect the same approach can be applied at any retinal location, including the fovea. In support of this, the RPE mosaic has previously been observed with AO-OCT at numerous locations across the macula with different densities of cones and rods [10,40].
4.1 Important parameters of the AO-OCT design
Our measurements of τ are consistent between the two imaging systems that were used in this study (Fig. 3(C)), a finding perhaps not unexpected given their similar designs. Consistency provides evidence of generalizability and repeatability of our method, but also raises questions about the role that differences between the two systems might play, as for example our systems differed in physical size and design strategies for the sample arm, and used different wavefront correctors, wavefront sensors, linescan detectors, light sources, and acquisition software. In AO-OCT and OCT images, speckle arises from the coherent interaction of the imaging beam with localized random scatterers in the tissue (in our case RPE organelles) that result in constructive and destructive interference. Because speckle is the fundamental physical phenomena that underlies our RPE motility measurements, the optical properties that control speckle size and shape (i.e., coherence volume) must drive system design and be fundamental for achieving consistency across AO-OCT platforms. Theoretically, these properties reduce to imaging wavelength and numerical aperture (for beam entering the human eye). For our two systems (Table 1), we used similar imaging wavelengths (785 nm and 830 nm) and same beam diameters at the eye (6.7 mm), resulting in almost identical coherence volumes (24.2 μm3 and 23.1 μm3) in tissue.
4.2 Possible organelle sources of the motility signal
The RPE cell interior is in constant motion with intracellular transport of organelles along three-dimensional scaffolding of actin filaments and microtubules. These protein-enabled thoroughfares allow organelles to traverse the cell in a second or so. Surprisingly, the literature on organelle content in human RPE is limited [32], and thus we can only speculate as to which types may contribute to the motility signal in our AO-OCT images. Prominent organelles in the RPE include melanosomes, phagosomes, and lipofuscin. The first two are well known for their motility behavior [1]. Both have high refractive indices (n = 1.7 of melanin [41] and 1.43 of photoreceptor outer segment remnants [42]) relative to their cytoplasmic surround (n = 1.37 [41]) and thus are likely sources of reflected light. Consistent with this, several studies point to melanin, which is synthesized and compartmentalized in melanosome organelles [43], as the primary source of the OCT signal in the RPE band [10,44,45]. Animal model studies show that the melanosome movement in RPE cells can be faster than 1 µm/s, enabling these organelles to quickly traverse the coherence volume of our AO-OCT systems (~2.5 × 2.5 × 4 μm3). If a primary contributing source in our images, melanin motility will have direct clinical significance as interruption of its transport across the cell has been shown to cause blindness in humans [4].
Phagosomes also exhibit motility behavior. They form from the membranous discs that are regularly pruned from the tips of photoreceptor outer segments. These pruned tips descend into the RPE cell where they are engulfed and phagocytized. Regulating phagosome movement is thought to involve the same molecular machinery that regulates melanosome movement [1,3,5]. We know the initial pruning event (disc shedding) causes a pronounced, transient disturbance in the reflection at the photoreceptor/RPE interface, as has been recently observed using AO-OCT [30,31]. Thus it is reasonable to expect the downstream phagocytosis process to generate similar reflectance disturbances within the RPE cell as the engulfed contents are digested, but this awaits testing.
Finally, lipofuscin is the undigested byproduct of the phagocytosis event, composed of a heterogeneous mixture of lipids and proteins that accumulate in the RPE cell with age. The optical properties of this byproduct must vary with mixture concentrations, which makes characterizing its reflectance contributions difficult. Nevertheless, if it were to strongly contribute, we would expect RPE image clarity to also vary with age (either increase or decrease). Our imaging results to date have not demonstrated an age dependence, but the number of subjects we have imaged is limited and the trend could be masked by other factors, as for example inter-subject variation and age-dependent properties of other organelles.
4.3 Depth-dependent motility in RPE layer
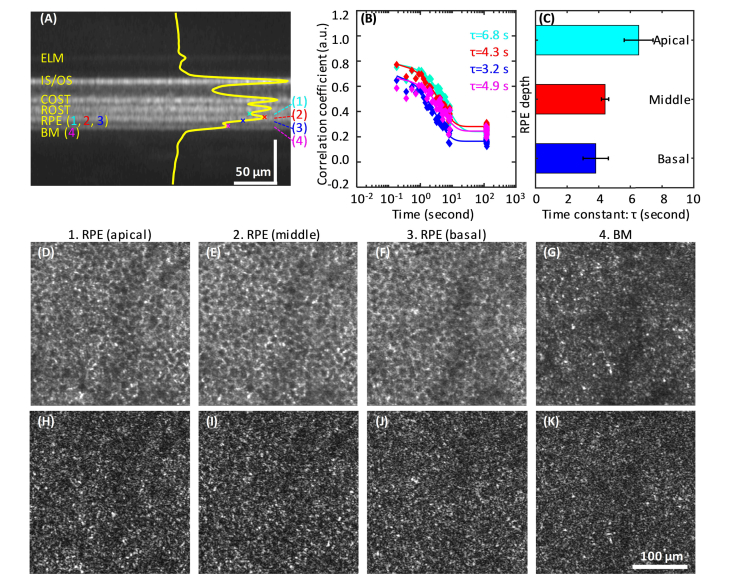
Some of the primary RPE organelles (e.g., melanosomes and lipofuscin) have been reported to stratify to different depths in the RPE [46]. Given that their different physical size and physiological role may lead to different motility dynamics, we hypothesized a possible variation in CF with depth in the RPE. To test, we calculated CF at three depths inside the RPE layer, a fourth one at BM, and a fifth at ROST. The three RPE depths (marked as (1-3) in Fig. 6(A)) were selected at the apical (expected high melanosome density [46]), middle (the strongest RPE reflectance in our AO-OCT images), and basal (expected high lipofuscin density [46]) portions of the RPE layer. The apical and basal locations were chosen to be ~4 µm above and below the peak intensity, a separation large enough to generate different speckle patterns in the corresponding en face images (Fig. 6(D-F)), but small enough to avoid signal contamination from the adjacent ROST and BM layers. Although the averaged en face views at the three RPE depths are similar, the single frames (e.g., in Fig. 6(H-J)) demonstrate different speckle patterns at the three depths indicating the sampling of different organelles.
Fig. 6.
Motion-evoked speckle changes with depth in the RPE-BM complex. (A) Averaged AO-OCT B-scan of S2 shows distinct hyper- and hypo-reflective bands in the outer retina. Four depths are marked on the superimposed averaged A-line profile that represent apical, middle, and basal sub-layers of the RPE and the BM layer. The color-coded CFs at the four corresponding depths are shown in (B). The average time constants of four subjects (S2, S5-S7) are shown in (C). Error bars denote standard deviation. Also shown are the corresponding en face images of the averaged volume (D-G) and single frame from the reference volume (H-K) of S2.
The resulting time constants on S2 were 6.8 s, 4.3 s, and 3.2 s at the apical, middle, and basal depths, respectively (Fig. 6(B)). Similar results were obtained from all four subjects as summarized in Fig. 6(C), the time constants were 6.5 ± 0.9 s (mean ± std) at apical, 4.4 ± 0.2 s at middle, and 3.8 ± 0.8 s at basal RPE. The change in time constant is small, but significantly different for the three RPE depths measured (p = 0.001, One-way ANOVA analysis). This nearly two-fold difference in τ supports our hypothesis that different types of organelles exhibit different motility dynamics. Assuming melanosomes and lipofuscin stratify to the apical and basal RPE [46], respectively, the observed decrease in τ with depth suggests that melanosomes are either less densely distributed, more slowly moving, or both compared that of the underlying protein- and lipid-rich lipofuscin organelles. This of course assumes both contribute to the RPE reflectance. Interestingly, the band with the largest time constant was ROST (τ = 7.8 s, not show here in figure), the site where rod end tips embed directly into the apical RPE surface. Figure 6(A)) shows ROST is clearly separate from the three RPE bands, indicating minimal influence on the underlying τ measurements. The BM band is similarly separated, albeit on the basal side. Its time constant (averaged and single en face images shown in Fig. 6(G,K)) of 4.9 s is closest to that of the middle RPE band (τ = 4.3 s). These measurements demonstrate a clear path for establishing motility baselines across the RPE complex in normals and with which comparisons can be made against pathologic retinas.
4.4 Influence of motion on speckle decorrelation
We removed motion contributions from the eye and imaging system by registering the acquired images with subcellular accuracy (confirmed by the resolution of photoreceptor and RPE cells in the averaged images) and normalizing the correlation function with the cone signal (Eq. (2)). While this approach appeared effective, we were unable to directly determine what residual motion artifacts might have remained and what their influence, if any, might have been on the observed speckle decorrelation, i.e., τ ~5 s for RPE cells. Unable to quantitatively assess, we resorted to a qualitative examination of the possible contributions of motion. To do so, we took advantage of the notable difference in RPE clarity in images acquired with TI = 0.23 s and 7.8 s (see Fig. 5). We examined whether this clarity difference could have been caused by angle compounding (a pupil effect caused by eye rotation, thus a consequence of eye motion) [47] or minute levels of image blur induced by residual eye motion.
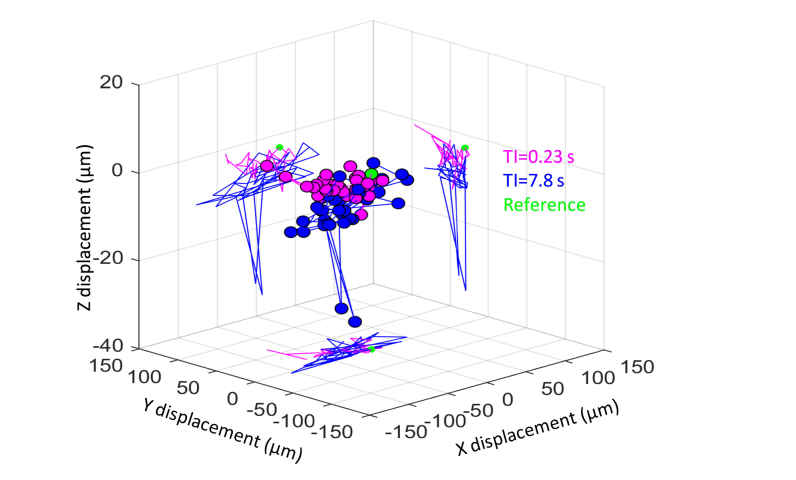
To start, we extracted the three-dimensional eye motion from a representative OCT data set and plotted as displacements in the x, y, and z directions in Fig. 7. The displacements were computed from the output of the 3-D registration. B-scans with registration coefficients larger than the selected coefficient threshold were used for this calculation (the mean displacement was calculated as the displacement of the volume, which is plotted in the figure. As evident from the figure, considerable eye motion occurred during both video acquisitions with motion traversals larger than the resolution element of the two imaging systems (2.4 × 2.4 × 4.2 µm3 and 2.5 × 2.5 × 3.7 µm3). The overall 3-D displacements were comparable, 29.2 ± 15.5 μm and 43.3 ± 19.8 μm (mean ± std) for TI of 0.23 s and 7.8 s, respectively. Similar results were obtained for the same analysis in other subjects as well. Comparable retinal motion implies comparable range of angular illumination of the retina and therefore comparable levels of contrast enhancement of the RPE.
Fig. 7.
Representative eye motion measured in two series of 35 AO-OCT images in three dimensions (3-D) from S2 with different time intervals (magenta: TI = 0.23 s, and blue: TI = 7.8 s). To better visualize the individual components, the 3-D motion is projected into the x-y, x-z, and y-z planes. Green point denotes the reference image coordinate: (x, y, z) = (0, 0, 0).
Next, we examined the possibility of motion-induced image blur. Based on the algorithmic design of our image registration method, we expect registration accuracy to worsen with the magnitude of the eye motion, both for motion parallel to and perpendicular to the fast B-scan direction. This trend coupled with the comparable motion displacements in the two imaging scenarios (TI = 0.23 s and 7.8 s) points to comparable registration accuracy for both scenarios and thus comparable motion-induced image blur and contrast improvement. The fact that we observed notably different RPE contrast suggests residual eye motion is an unlikely contributor. In summary, we have found no evidence to link our measured time constant and RPE contrast improvement to sources other than the physiological dynamics internal to the cells imaged. Consistent with this conclusion, the RPE time constants we have measured (~5 s) fall well within the range reported for other biological tissues (0.4-34 s) that were measured under highly controlled and stable environments [36,38,48,49], albeit this range is relatively large.
4.5 Clinical implications and prospects for measuring RPE motility
Our method has two important clinical implications. First, speckle decorrelation (as defined by its time constant, τ) provides a measure of cell function by quantifying the dynamic interactions of the cell’s organelles with the imaging beam. While interpretation of this complex light-tissue interaction remains incomplete, establishment of a normative baseline for τ will allow us to compare RPE motility dynamics in retinal diseases that afflict the RPE. Inhibited transport of ingested discs [3], abnormal phagosome movement during phagocytosis [5], and interrupted melanosomes movement [4] in mutant RPE cells have been suggested as causes for RPE motility changes that directly link to RPE function deficits and degeneration of photoreceptors found in many retinal diseases. We hypothesize that τ may be a sensitive functional metric of RPE health and likely increases (motility slows) with retinopathies that afflict RPE.
The second clinical implication relates to translation of AO-based methodologies such as the one described here for RPE cell imaging. Our results reveal that the 90-minute imaging protocol we used to observe individual RPE cells in our first study can be substantially decreased to just 2.5 minutes (36 × shorter) without loss in cell clarity. While we have not fully confirmed this prediction as the image acquisition intervals we tested (TI = 0.23 s and 7.8 s) only straddled the τ ~5 s prediction, our results and prediction show strong consistency. More relevant for improving clinical utility will be methodological advancements that reduce τ (and therefore TI), as for example by increasing organelle motion of these cells through stimulation. This remains future work.
The adoption of RPE motility as a clinically useful metric of degenerative disease (e.g., AMD, etc.) requires several conditions and demonstrations. First, the technique must be accessible to the clinical population. As we have seen, RPE motility depends on the coherence volume, which for a research AO-OCT device is smaller than the RPE cell size. For heterogeneous retinal tissue, further expansion of the coherence volume would lead to distortion of the metric as the motility of other organelles become part of the measurement. And as we have shown, there is even some variability across the RPE cell that may be due to stratification of the different organelles and their densities (see Fig. 6). Thus repeatable coherence volume measurements matched to the target tissue is a pre-requisite, although other methods may be available to partially achieve this condition (e.g., ultrahigh bandwidth OCT sources to separate the RPE layer or other means of RPE cell contrast), and AO seems to be the most straight-forward technique to achieve the required lateral resolution. What we have demonstrated in comparable measurements on two different AO-OCT systems is that once the necessary coherence volume is achieved, other factors in system performance appear less important. This increases the potential of validation as the measurement is not tied to a highly specialized technique. The second condition is proper validation in normal, healthy eyes. Repeatability and reproducibility measurements must be performed on a sizeable cohort of subjects (perhaps hundreds) to establish a normative baseline. The relatively recent development of OCT-measured NFL thickness as a surrogate for GCL loss and glaucoma disease stage points to a similar validation pathway for RPE motility in degenerative diseases. Finally, motility differences in RPE-related diseases must be measured, along with studies that definitively determine the organelle source of motility speeds (and optical decorrelation). The path is arduous, while the reward in terms of AO clinical translation may be significant.
Acknowledgments
We thank Furu Zhang, Ayoub Lassoued and Tim Turner at Indiana University for technical assistance.
Funding
NIH (EY018339); FDA Critical Path Initiative (CPI).
Disclosures
The authors declare that there are no conflicts of interest related to this article.
Disclaimer: The mention of commercial products, their sources, or their use in connection with material reported herein is not to be construed as either an actual or implied endorsement of such products by the US Department of Health and Human Services.
References
- 1.Futter C. E., “The molecular regulation of organelle transport in mammalian retinal pigment epithelial cells,” Pigment Cell Res. 19(2), 104–111 (2006). 10.1111/j.1600-0749.2006.00303.x [DOI] [PubMed] [Google Scholar]
- 2.Strauss O., “The retinal pigment epithelium in visual function,” Physiol. Rev. 85(3), 845–881 (2005). 10.1152/physrev.00021.2004 [DOI] [PubMed] [Google Scholar]
- 3.Gibbs D., Kitamoto J., Williams D. S., “Abnormal phagocytosis by retinal pigmented epithelium that lacks myosin VIIa, the Usher syndrome 1B protein,” Proc. Natl. Acad. Sci. U.S.A. 100(11), 6481–6486 (2003). 10.1073/pnas.1130432100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gibbs D., Azarian S. M., Lillo C., Kitamoto J., Klomp A. E., Steel K. P., Libby R. T., Williams D. S., “Role of myosin VIIa and Rab27a in the motility and localization of RPE melanosomes,” J. Cell Sci. 117(26), 6473–6483 (2004). 10.1242/jcs.01580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang M., Esteve-Rudd J., Lopes V. S., Diemer T., Lillo C., Rump A., Williams D. S., “Microtubule motors transport phagosomes in the RPE, and lack of KLC1 leads to AMD-like pathogenesis,” J. Cell Biol. 210(4), 595–611 (2015). 10.1083/jcb.201410112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morgan J. I., Dubra A., Wolfe R., Merigan W. H., Williams D. R., “In vivo autofluorescence imaging of the human and macaque retinal pigment epithelial cell mosaic,” Invest. Ophthalmol. Vis. Sci. 50(3), 1350–1359 (2009). 10.1167/iovs.08-2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rossi E. A., Rangel-Fonseca P., Parkins K., Fischer W., Latchney L. R., Folwell M. A., Williams D. R., Dubra A., Chung M. M., “In vivo imaging of retinal pigment epithelium cells in age related macular degeneration,” Biomed. Opt. Express 4(11), 2527–2539 (2013). 10.1364/BOE.4.002527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scoles D., Sulai Y. N., Dubra A., “In vivo dark-field imaging of the retinal pigment epithelium cell mosaic,” Biomed. Opt. Express 4(9), 1710–1723 (2013). 10.1364/BOE.4.001710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felberer F., Kroisamer J. S., Baumann B., Zotter S., Schmidt-Erfurth U., Hitzenberger C. K., Pircher M., “Adaptive optics SLO/OCT for 3D imaging of human photoreceptors in vivo,” Biomed. Opt. Express 5(2), 439–456 (2014). 10.1364/BOE.5.000439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Z., Kocaoglu O. P., Miller D. T., “3D imaging of retinal pigment epithelial cells in the living human retina,” Invest. Ophthalmol. Vis. Sci. 57(9), OCT533 (2016). 10.1167/iovs.16-19106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tam J., Liu J., Dubra A., Fariss R., “In vivo imaging of the human retinal pigment epithelial mosaic using adaptive optics enhanced indocyanine green ophthalmoscopy,” Invest. Ophthalmol. Vis. Sci. 57(10), 4376–4384 (2016). 10.1167/iovs.16-19503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu T., Jung H., Liu J., Droettboom M., Tam J., “Noninvasive near infrared autofluorescence imaging of retinal pigment epithelial cells in the human retina using adaptive optics,” Biomed. Opt. Express 8(10), 4348–4360 (2017). 10.1364/BOE.8.004348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grieve K., Gofas-Salas E., Ferguson R. D., Sahel J. A., Paques M., Rossi E. A., “In vivo near-infrared autofluorescence imaging of retinal pigment epithelial cells with 757 nm excitation,” Biomed. Opt. Express 9(12), 5946–5961 (2018). 10.1364/BOE.9.005946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Granger C. E., Yang Q., Song H., Saito K., Nozato K., Latchney L. R., Leonard B. T., Chung M. M., Williams D. R., Rossi E. A., “Human retinal pigment epithelium: In vivo cell morphometry, multispectral autofluorescence, and relationship to cone mosaic,” Invest. Ophthalmol. Vis. Sci. 59(15), 5705–5716 (2018). 10.1167/iovs.18-24677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Azimipour M., Zawadzki R. J., Gorczynska I., Migacz J., Werner J. S., Jonnal R. S., “Intraframe motion correction for raster-scanned adaptive optics images using strip-based cross-correlation lag biases,” PLoS One 13(10), e0206052 (2018). 10.1371/journal.pone.0206052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burns S. A., Elsner A. E., Sapoznik K. A., Warner R. L., Gast T. J., “Adaptive optics imaging of the human retina,” Prog. Retin. Eye Res. 68, 1–30 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Z., Tam J., Saeedi O., Hammer D. X., “Trans-retinal cellular imaging with multimodal adaptive optics,” Biomed. Opt. Express 9(9), 4246–4262 (2018). 10.1364/BOE.9.004246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Z., Kurokawa K., Zhang F., Miller D. T., “Characterizing motility dynamics in human RPE cells,” Ophthalmic Technologies XXVII 10045, 1004515 (2017). 10.1117/12.2256144 [DOI] [Google Scholar]
- 19.Liu Z., Kocaoglu O. P., Miller D. T., “In-the-plane design of an off-axis ophthalmic adaptive optics system using toroidal mirrors,” Biomed. Opt. Express 4(12), 3007–3029 (2013). 10.1364/BOE.4.003007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kocaoglu O. P., Turner T. L., Liu Z., Miller D. T., “Adaptive optics optical coherence tomography at 1 MHz,” Biomed. Opt. Express 5(12), 4186–4200 (2014). 10.1364/BOE.5.004186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bennett A. G., Rudnicka A. R., Edgar D. F., “Improvements on Littmann’s method of determining the size of retinal features by fundus photography,” Graefes Arch. Clin. Exp. Ophthalmol. 232(6), 361–367 (1994). 10.1007/BF00175988 [DOI] [PubMed] [Google Scholar]
- 22.A.N.S.I. Z136, 1, “Safe use of lasers,” (Laser Institute of America, 2014). [Google Scholar]
- 23.Jonnal R. S., Kocaoglu O. P., Zawadzki R. J., Lee S. H., Werner J. S., Miller D. T., “The cellular origins of the outer retinal bands in optical coherence tomography images,” Invest. Ophthalmol. Vis. Sci. 55(12), 7904–7918 (2014). 10.1167/iovs.14-14907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.N. H. Do, “Parallel processing for adaptive optics optical coherence tomography (ao-oct) image registration using gpu.,” Master’s thesis (Indiana University–Purdue University Indianapolis, Indianapolis, IN). (2016). [Google Scholar]
- 25.Kocaoglu O. P., Ferguson R. D., Jonnal R. S., Liu Z., Wang Q., Hammer D. X., Miller D. T., “Adaptive optics optical coherence tomography with dynamic retinal tracking,” Biomed. Opt. Express 5(7), 2262–2284 (2014). 10.1364/BOE.5.002262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jonnal R. S., Rha J., Zhang Y., Cense B., Gao W., Miller D. T., “In vivo functional imaging of human cone photoreceptors,” Opt. Express 15(24), 16141–16160 (2007). 10.1364/OE.15.016141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pallikaris A., Williams D. R., Hofer H., “The reflectance of single cones in the living human eye,” Invest. Ophthalmol. Vis. Sci. 44(10), 4580–4592 (2003). 10.1167/iovs.03-0094 [DOI] [PubMed] [Google Scholar]
- 28.Pircher M., Kroisamer J. S., Felberer F., Sattmann H., Götzinger E., Hitzenberger C. K., “Temporal changes of human cone photoreceptors observed in vivo with SLO/OCT,” Biomed. Opt. Express 2(1), 100–112 (2011). 10.1364/BOE.2.000100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cooper R. F., Dubis A. M., Pavaskar A., Rha J., Dubra A., Carroll J., “Spatial and temporal variation of rod photoreceptor reflectance in the human retina,” Biomed. Opt. Express 2(9), 2577–2589 (2011). 10.1364/BOE.2.002577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kocaoglu O. P., Liu Z., Zhang F., Kurokawa K., Jonnal R. S., Miller D. T., “Photoreceptor disc shedding in the living human eye,” Biomed. Opt. Express 7(11), 4554–4568 (2016). 10.1364/BOE.7.004554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.F. Zhang, Z. Liu, K. Kurokawa, and D. T. Miller, “Tracking dynamics of photoreceptor disc shedding with adaptive optics-optical coherence tomography.” SPIE proceeding 10045, (2017). [Google Scholar]
- 32.Pollreisz A., Messinger J. D., Sloan K. R., Mittermueller T. J., Weinhandl A. S., Benson E. K., Kidd G. J., Schmidt-Erfurth U., Curcio C. A., “Visualizing melanosomes, lipofuscin, and melanolipofuscin in human retinal pigment epithelium using serial block face scanning electron microscopy,” Exp. Eye Res. 166, 131–139 (2018). 10.1016/j.exer.2017.10.018 [DOI] [PubMed] [Google Scholar]
- 33.Kurokawa K., Makita S., Hong Y. J., Yasuno Y., “Two-dimensional micro-displacement measurement for laser coagulation using optical coherence tomography,” Biomed. Opt. Express 6(1), 170–190 (2015). 10.1364/BOE.6.000170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ach T., Huisingh C., McGwin G., Jr., Messinger J. D., Zhang T., Bentley M. J., Gutierrez D. B., Ablonczy Z., Smith R. T., Sloan K. R., Curcio C. A., “Quantitative autofluorescence and cell density maps of the human retinal pigment epithelium,” Invest. Ophthalmol. Vis. Sci. 55(8), 4832–4841 (2014). 10.1167/iovs.14-14802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeong K., Turek J. J., Nolte D. D., “Speckle fluctuation spectroscopy of intracellular motion in living tissue using coherence-domain digital holography,” J. Biomed. Opt. 15(3), 030514 (2010). 10.1117/1.3456369 [DOI] [PubMed] [Google Scholar]
- 36.Pan Y. T., Wu Z. L., Yuan Z. J., Wang Z. G., Du C. W., “Subcellular imaging of epithelium with time-lapse optical coherence tomography,” J. Biomed. Opt. 12(5), 050504 (2007). 10.1117/1.2800007 [DOI] [PubMed] [Google Scholar]
- 37.Liu Z., Kurokawa K., Zhang F., Lee J. J., Miller D. T., “Imaging and quantifying ganglion cells and other transparent neurons in the living human retina,” Proc. Natl. Acad. Sci. U.S.A. 114(48), 12803–12808 (2017). 10.1073/pnas.1711734114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang X. R., Knighton R. W., Zhou Y., Zhao X. P., “Reflectance speckle of retinal nerve fiber layer reveals axonal activity,” Invest. Ophthalmol. Vis. Sci. 54(4), 2616–2623 (2013). 10.1167/iovs.12-11347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.K. Kurokawa, Z. Liu, J. Crowell, F. Zhang, and D. T. Miller, “Method to investigate temporal dynamics of ganglion and other retinal cells in the living human eye.” Proc. SPIE Ophthalmic Technologies XXVIII 10474, 104740W (2018). [Google Scholar]
- 40.Zhang F., Kurokawa K., Lassoued A., Crowell J. A., Miller D. T., “Cone photoreceptor classification in the living human eye from photostimulation-induced phase dynamics,” Proc. Natl. Acad. Sci. U.S.A. 116(16), 7951–7956 (2019). 10.1073/pnas.1816360116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dunn A. K., Smithpeter C. L., Welch A. J., Richards-Kortum R. R., “Finite-difference time-domain simulation of light scattering from single cells,” J. Biomed. Opt. 2(3), 262–266 (1997). 10.1117/12.275219 [DOI] [PubMed] [Google Scholar]
- 42.Snyder A. W., Pask C., “The stiles-crawford effect-explanation and consequences,” Vision Res. 13(6), 1115–1137 (1973). 10.1016/0042-6989(73)90148-X [DOI] [PubMed] [Google Scholar]
- 43.Barral D. C., Seabra M. C., “The melanosome as a model to study organelle motility in mammals,” Pigment Cell Res. 17(2), 111–118 (2004). 10.1111/j.1600-0749.2004.00138.x [DOI] [PubMed] [Google Scholar]
- 44.Zhang Q. X., Lu R. W., Messinger J. D., Curcio C. A., Guarcello V., Yao X. C., “In vivo optical coherence tomography of light-driven melanosome translocation in retinal pigment epithelium,” Sci. Rep. 3(1), 2644 (2013). 10.1038/srep02644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilk M. A., Huckenpahler A. L., Collery R. F., Link B. A., Carroll J., “The effect of retinal melanin on optical coherence tomography images,” Transl. Vis. Sci. Technol. 6(2), 8 (2017). 10.1167/tvst.6.2.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weiter J. J., Delori F. C., Wing G. L., Fitch K. A., “Retinal pigment epithelial lipofuscin and melanin and choroidal melanin in human eyes,” Invest. Ophthalmol. Vis. Sci. 27(2), 145–152 (1986). [PubMed] [Google Scholar]
- 47.Bashkansky M., Reintjes J., “Statistics and reduction of speckle in optical coherence tomography,” Opt. Lett. 25(8), 545–547 (2000). 10.1364/OL.25.000545 [DOI] [PubMed] [Google Scholar]
- 48.Yuan Z., Chen B., Ren H., Pan Y., “On the possibility of time-lapse ultrahigh-resolution optical coherence tomography for bladder cancer grading,” J. Biomed. Opt. 14(5), 050502 (2009). 10.1117/1.3223246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bruder J. M., Pfeiffer Z. A., Ciriello J. M., Horrigan D. M., Wicks N. L., Flaherty B., Oancea E., “Melanosomal dynamics assessed with a live-cell fluorescent melanosomal marker,” PLoS One 7(8), e43465 (2012). 10.1371/journal.pone.0043465 [DOI] [PMC free article] [PubMed] [Google Scholar]