Abstract
Objective
Acute myeloid leukemia (AML) is a malignant clonal disorder. Despite enormous progress in its diagnosis and treatment, the mortality rate of AML remains high. The aim of this study was to identify prognostic biomarkers by using the gene expression profile dataset from public database, and to improve the risk-stratification criteria of survival for patients with AML.
Materials and methods
The gene expression data and clinical parameter were acquired from the Therapeutically Applicable Research to Generate Effective Treatment (TARGET) database. A total of 856 differentially expressed genes (DEGs) were obtained from the childhood AML patients classified into first complete remission (CR1) group (n=791) and not CR group (n=249). We performed a series of bioinformatics analysis to screen key genes and pathways, further comprehending these DEGs through Gene Ontology (GO) function and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses.
Results
Six genes (SLC17A7, MSX2, CDC26, MSLN, CTSZ and DEFA3) identified by univariate, Kaplan-Meier survival and multivariate Cox regression analyses were used to develop the prognostic model. Further analysis showed that the survival estimations in the high-risk group had an increased risk of death compared with the low-risk group based on the model. The area under the curve of the receiver operator characteristic curve in the prognostic model for predicting the overall survival was 0.729, confirming good prognostic model. We also performed a nomogram to provide an individual patient with the overall probability, and internal validation in the TARGET cohort.
Conclusion
We identified a six-gene prognostic signature for risk-stratifying in patients with childhood AML. The risk classification model can be used to predict CR markers and may assist clinicians in providing realize the individualized treatment in this patient population.
Keywords: childhood acute myeloid leukemia, remission induction, gene expression profiling, prognosis, bioinformatics, survival analysis
Introduction
Acute myeloid leukemia (AML) is a malignant clonal disorder characterized by abnormal proliferation of immature myeloid cells at various stages of maturation.1 About 4% of AML cases occur in children and adolescents. The 5-year overall survival (OS) rate for patients under 19 is about 65%, but drops to 50%, 32%, and 6%, respectively, when the patients aged 20–49, 50–64, and 65 years and older.2 The cytogenetic karyotype and molecular abnormalities at diagnosis are considered the most significant prognostic factors and are highly predictive of complete remission (CR) rates, OS, risk of relapse and disease-free survival.3–5 During the last decades, accumulating evidence has proposed that many abnormal expressions and mutations of genes are involved in the progression and carcinogenesis of AML. Mutated genes with prognostic significance that have been reported include KIT, WT1, RUNX1, FLT3, KIT, CEBPA, NPM1, and MYC.6,7 However, only aberrations in NPM1, WT1, CEBPA and FLT3 are being widely utilized in clinical practice.8 Despite extensive research that has been carried out to identify find prognostic markers, the mortality rate of AML remains high. Therefore, prognostic risk stratification needs to be improved because it has the potential to develop effective diagnostic and therapeutic strategies.
With recent developments in microarray technology and bioinformatic analysis, the complex molecular architecture of AML has been widely used to inform disease classification, prognostic stratification and novel drug target discovery. Multiple studies have suggested that patients whose leukemic blasts contain the NPM1 mutation without FLT3-ITD have a favorable prognosis, whereas patients with TET2 or AXSL1 mutation have a poor prognosis.9,10 In addition, patients with CBF rearrangements or CEBPA mutations are assigned to the low-risk subgroup.11,12 Recently, Ng et al13 developed a risk-stratification model that generates a prognostic score based on 17-gene expression for rapid determination in patients with acute leukemia, Patel et al14 proposed a model of somatic mutations for risk stratification based on microarray technology of a set of 18 genes. These models were found to have prognostic value in their studies. However, even with these progresses, pediatric AML risk classification remains suboptimal as a large number of patients with AML have not achieved CR regardless of the known high-risk factors.
To improve the risk-stratification criteria for predicting prognosis in patients with childhood AML, our study analyzed the differentially expressed genes (DEGs) based on first CR15,16 using mRNA-seq datasets from the TARGET. We performed a systematic evaluation of mRNAs for the diagnosis of childhood AML by univariate analysis of gene expression and Cox regression analysis. We pooled the specificity and sensitivity of all genes in the files and constructed a time-dependent receiver operator characteristic (ROC) curve. We ranked and screened out the genes with high diagnostic accuracy based on area under the curve (AUC) values. The final risk-stratification model represents a potentially useful tool for predicting, CR but needs to be further evaluated in clinical practice.
Materials and methods
Data sources and processing
Gene mRNA expression data and clinical parameters associated with childhood AML patients up to April 29, 2019 were download from the NCI TARGET database (https://ocg.cancer.gov/). Series matrix files were extracted to assess mRNA expression, and mRNA-seq datasets preprocessed by quantile normalization or log2 transformation. According to the annotation platform file, we translated the mRNA IDs into symbol names. Then, we divided the patients into CR1 group (791 samples) and not CR group (249 samples) based on the sample annotation, see Table 1. The flow chart of the analysis procedure is shown in Figure 1. The gene mRNA expression data and clinical characteristics are publicly available and open to access, so this study did not need the approval from the ethics committee.
Table 1.
Clinical characteristics of patients with CR1 and not CR
| ID | CR1 (n=791) | Not CR (n=249) | X2 | P-value |
|---|---|---|---|---|
| Age | 2.969 | 0.085 | ||
| <14 | 549 (69.4%) | 187 (75.1%) | ||
| ≥14 | 242 (30.6%) | 62 (24.9%) | ||
| Gender | 0.609 | 0.435 | ||
| Male | 416 (52.6%) | 138 (55.4%) | ||
| Female | 375 (47.4%) | 111 (44.6%) | ||
| White blood cell | 4.567 | 0.033 | ||
| <150 | 720 (91%) | 215 (86.3%) | ||
| ≥150 | 71 (9%) | 34 (13.7%) | ||
| Bone marrow leukemic blast | 0.386 | 0.534 | ||
| <90% | 603 (76.2%) | 185 (74.3%) | ||
| ≥90% | 188 (23.8%) | 64 (25.7%) | ||
| Peripheral blasts | 1.657 | 0.198 | ||
| <90% | 715 (90.4%) | 218 (87.6%) | ||
| ≥90% | 76 (9.6%) | 31 (12.4%) | ||
| CNS disease | 2.560 | 0.110 | ||
| Yes | 47 (5.9%) | 22 (8.8%) | ||
| No | 744 (94.1%) | 227 (91.2%) | ||
| Chloroma | 4.110 | 0.043 | ||
| Yes | 86 (10.9%) | 39 (15.7%) | ||
| No | 705 (89.1%) | 210 (84.3%) | ||
| FAB category | 24.741 | 0.001 | ||
| MO | 16 (2%) | 15 (6%) | ||
| M1 | 86 (10.9%) | 33 (13.3%) | ||
| M2 | 178 (22.5%) | 52 (20.9%) | ||
| M3 | 2 (0.3%) | 0 (0%) | ||
| M4 | 192 (24.3%) | 33 (13.3%) | ||
| M5 | 148 (18.7%) | 46 (18.5%) | ||
| M6 | 13 (1.6%) | 4 (1.6%) | ||
| M7 | 31 (3.9%) | 15 (6%) | ||
| Unknown | 125 (15.8%) | 51 (20.5%) | ||
| Primary cytogenetic code | 25.819 | <0.001 | ||
| inv (16) | 115 (14.5%) | 12 (4.8%) | ||
| MLL | 146 (18.5%) | 44 (17.7%) | ||
| t (8;21) | 123 (15.5%) | 29 (11.6%) | ||
| Other | 189 (23.9%) | 85 (34.1%) | ||
| Normal | 180 (22.8%) | 68 (27.3%) | ||
| Unknown | 38 (4.8%) | 11 (4.4%) | ||
| FLT3/ITD positive | 7.974 | 0.005 | ||
| Yes | 133 (16.8%) | 62(24.9%) | ||
| No | 655 (82.8%) | 187(75.1%) | ||
| Unknown | 3 (0.4%) | 0 (0%) | ||
| FLT3 PM | 0.057 | 0.811 | ||
| Yes | 54 (6.8%) | 16 (6.4%) | ||
| No | 733 (92.7%) | 233 (93.6%) | ||
| Unknown | 4 (0.5%) | 0 (0%) | ||
| NPM mutation | 5.225 | 0.022 | ||
| Yes | 77 (9.7%) | 13 (5.2%) | ||
| No | 698 (88.2%) | 236 (94.8%) | ||
| Unknown | 16 (2%) | 0 (0%) | ||
| CEBPA mutation | 5.156 | 0.023 | ||
| Yes | 52 (6.6%) | 7 (2.8%) | ||
| No | 727 (91.9%) | 241 (96.8%) | ||
| Unknown | 12 (1.5%) | 1 (0.4%) | ||
| WT1 mutation | 12.147 | <0.001 | ||
| Yes | 45 (5.7%) | 31(12.4%) | ||
| No | 731 (92.4%) | 218(87.6%) | ||
| Unknown | 15 (1.9%) | 0 (0%) | ||
| c-Kit mutation exon 8 | 0.457 | 0.499 | ||
| Yes | 36 (4.6%) | 5 (2%) | ||
| No | 189 (23.9%) | 37 (14.9%) | ||
| Not done | 566 (71.6%) | 207 (83.1%) | ||
| c-Kit mutation exon 17 | 2.321 | 0.128 | ||
| Yes | 24 (3%) | 8 (3.2%) | ||
| No | 200 (25.3%) | 34(13.7%) | ||
| Not done | 567 (71.7%) | 207 (83.1%) | ||
| MRD at end of course 1 | 161.121 | <0.001 | ||
| Yes | 126 (15.9%) | 143 (57.4%) | ||
| No | 492 (62.2%) | 68 (27.3%) | ||
| Unknown | 173 (21.9%) | 38 (15.3%) | ||
Abbreviations: CR, complete remission; FAB, French-American-British; MRD, minimal residual disease.
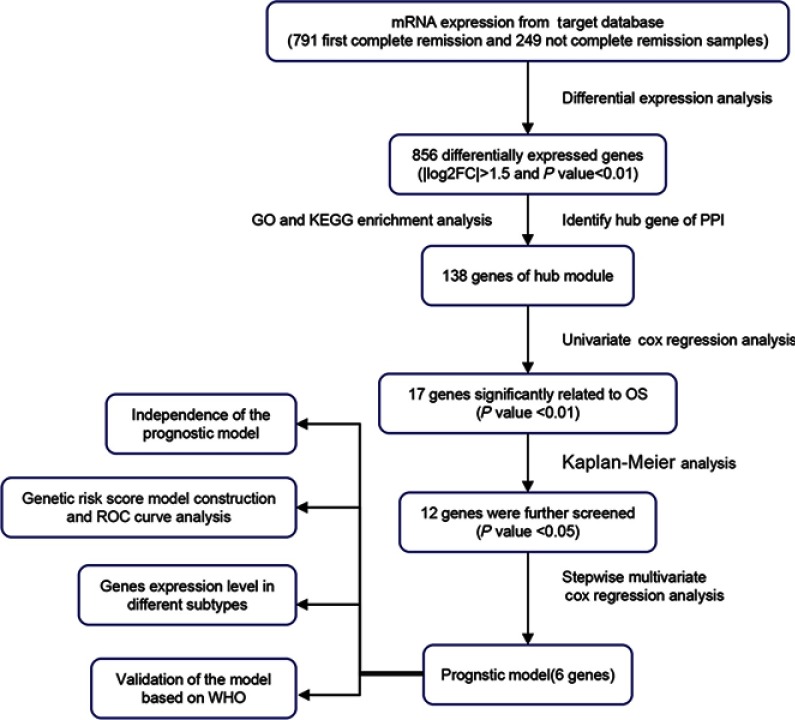
Figure 1.
Flow diagram of the analysis procedure.
Abbreviations: OS, overall survival; ROC, receiver operator characteristic; WHO, World Health Organization.
Identifying genes of differential expression
All data were analyzed with the R 3.5.2 software (https://www.r-project.org/). The differential expression of mRNA in childhood AML (260 CR1 and 93 not CR samples with full survival information along with mRNA-seq datasets) was calculated by using R/Bioconductor package of edgeR.17 We defined the cut-off criteria DEGs as |log2 fold-change(log2FC)|>1.5 and adjusted P-value (adj.P) <0.01. Finally, hierarchical cluster analysis was used to show the heat map and volcano plot of two groups by using gplots package in R platform.
Functional and pathway enrichment analysis
To explore the biological effects and pathways of the identified DEGs. The top 10 of Gene Ontoloy (GO) Biological Process analyses were conducted by using the R/Bioconductor package of Clusteprofiler.18 The significant results of biological process (BP), cellular component (CC), and molecular function (MF) were based on the threshold of P<0.05. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis19 was performed for the selected genes using The Database for Annotation, Visualization and Integrated Discovery (DAVID; https://david.ncifcrf.gov/). A P<0.05 was considered statistically significant.
Integration of the protein–protein interaction (PPI) network
The Search Tool for the Retrieval of Interacting Genes version 11.0 (STRING; https://string-db.org/) was used for the exploration of potential DEGs interactions at the protein level.20 In the present study, the parameter of interactions was set as interaction score >0.55 could be considered statistically significant, hiding disconnected nodes in the network. Then, the Cytoscape software (version 3.7.1; https://cytoscape.org/) was used for constructing and visualizing a PPI network of common DEGs.21 The plug-in Molecular Complex Detection (MCODE) (version 1.5.1) of Cytoscape was used to Cluster a given network based on topology to find densely connected regions. The criteria for selection were as follows: degree cut-off=2, node score cut-off=0.2, k-core=2, and max depth=100.
Hub genes selection and analysis
To identify DEGs predictive of clinical factors and survival outcomes, the information of 138 hub genes in the training dataset was utilized to perform univariate Cox regression analysis using the Survival package of R software (Version 2.44-1.1). The HR with 95% CI were calculated and log-rank test (P<0.01) was conducted to further select the most significant candidate genes. The OS analyses of candidate genes were performed using Kaplan–Meier plots by using Bioconductor R package. Gene expression value was labeled as high or low using a dichotomy method, with P<0.05 being considered significantly different. Multivariate Cox proportional hazards regression model was used to calculate the risk score (RS) based on the 12 potentially relevant genes in the preliminary screening, and the impact of OS information. The RS of each sample was calculated using the formula of RS=β1Exp1+β2Exp2+…+βxExpx (βi: the coefficient value, Expx: the gene expression level). The childhood AML patients were classified into low-risk and high-risk groups according to the median RS survival analysis and log-rank test were performed to evaluate the differences between the two groups. The ROC analyses were performed by using SurvivalROC package of R (Version 1.0.3) based the prognostic model that incorporates genes expression factors to predict the probability of 3- and 5-year OS. Then, identifying prognostic genes between CR1 and not CR samples, according to this research, we used performed the nomogram-based model to predict the survival probability by using the R package “rms” (Version 5.1-3.1).22 We divided the patients into eight groups by the French-American-British (FAB) category from database to analyze the six candidate genes expression level in different subtypes of childhood AML. The statistical analysis this study is performed by using the GraphPad Prism (Version 8.0.2; GraphPad Software, Inc., La Jolla, CA, USA).
Results
Identification of differential molecules in childhood AML
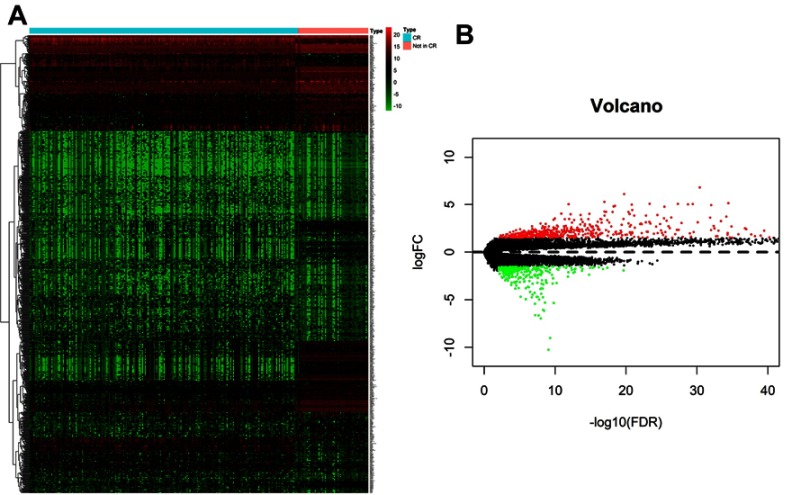
A gene expression database generated by RNA-Seq was downloaded from TARGET. The database included the expression levels detected in childhood AML samples with clinical information on whether the patient achieved first CR or not. A total of 856 differential genes met the criteria of |log2FC|>1.5 and adj-P<0.01, including 543 up-regulated genes and 313 down-regulated genes in childhood AML compared with CR1 group. The heat map and volcano plots that demonstrated significant differential distribution among each data set are shown in Figure 2A and B.
Figure 2.
(A) Heat map for potential mRNAs based on the expression profles of signifcantly diferentially expressed genes. (B) Volcano plot of genes detected in childhood AML, red dots represent upregulated and green dots represent downregulated.
DEGs functional and pathway enrichment analysis
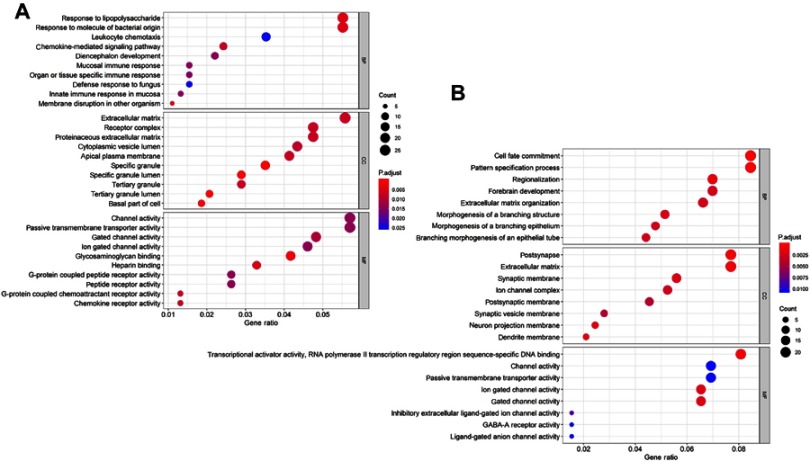
To explore the biological functional implication of DEGs, the top 10 GO enrichment analysis of up-regulated and down-regulated DEGs was performed, see Figure 3A and B. The up-regulated genes were mostly associated with the BP terms response to lipopolysaccharide, molecule of bacterial origin, leukocyte chemotaxis, and chemokine-mediated signaling pathway, while the down-regulated genes were mostly enriched in cell fate commitment, pattern specification process, regionalization, and morphogenesis of a branching structure. In addition, CC analysis showed that the up-regulation genes were related to extracellular matrix, receptor complex, proteinaceous extracellular matrix, and apical plasma membrane, and the down-regulated genes were mostly found in the postsynapse, extracellular matrix, neuron projection membrane, and dendrite membrane. Additionally, for MF terms, up-regulated genes were enriched in channel activity, passive transmembrane transporter activity, and G-protein coupled peptide receptor activity, while the down-regulated genes were relevant to transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific DNA binding, and passive transmembrane transporter activity.
Figure 3.
GO enrichment analysis of aberrantly diferentially expressed genes with no complete remission. The top 10 up-regulated (A) and down-regulated (B) genes GO analysis (The size of each dot represents the count of genes, the color represents the adj-P).
KEGG pathway enrichment analysis was performed using DAVID. Table 2 shows the most significant KEGG pathway of the up-regulated and down-regulated DEGs, including cytokine-cytokine receptor interaction, neuroactive ligand-receptor interaction, cell adhesion molecules (CAMs), hematopoietic cell lineage, and signaling pathways regulating pluripotency of stem cells, etc.
Table 2.
KEGG pathway enrichment analysis of aberrantly differentially expressed genes in childhood AML with no complete remission
| Pathway ID | Description | P-value | Gene count | Genes |
|---|---|---|---|---|
| hsa04060 | Cytokine-cytokine receptor interaction | 3.231E-05 | 25 | CSF3, CXCL1, IL1R2, CSF2, CXCL5, IL7, TNFSF15, CXCR1, CXCR2, IL24, PF4V1, CCL7, ACVR2A, CCR8, TNFRSF10C, CCR7, TNFRSF11B, CCR6, CXCL14, CCL20, PRLR, IFNB1, IL12B, BMP7, PRL |
| hsa04080 | Neuroactive ligand-receptor interaction | 3.293E-03 | 22 | OPRM1, F2RL2, GABRG1, CGA, GABRA2, GLRB, GRIK1, GABRA4, GRIN3B, BDKRB2, GRM4, GABRR1, PRLR, ADRA2A, AVPR1A, CHRND, UTS2R, PRL, ADRA1D, GLP1R, CHRNG, GRID1 |
| hsa04514 | CAMs | 1.083E-02 | 13 | PTPRM, CD8B, CD276, LRRC4B, CLDN22, CLDN10, CLDN11, CDH5, NCAM2, SIGLEC1, CDH15, CLDN2, CNTNAP2 |
| hsa05033 | Nicotine addiction | 2.033E-02 | 6 | SLC17A7, GABRG1, GABRA2, GABRR1, GABRA4, GRIN3B |
| hsa04640 | Hematopoietic cell lineage | 2.203E-02 | 9 | CSF3, CSF2, IL1R2, CD3G, CD3D, DNTT, IL7, CD8B, ITGA1 |
| hsa04530 | Tight junction | 2.203E-02 | 9 | PARD6B, MPDZ, CLDN22, CRB3, CLDN2, ACTN2, CLDN10, MYH14, CLDN11 |
| hsa04978 | Mineral absorption | 2.953E-02 | 6 | TF, MT1M, HMOX1, SLC26A9, MT1H, MT1G |
| hsa04550 | Signaling pathways regulating pluripotency of stem cells | 0.052215 | 11 | WNT5A, ACVR2A, HNF1A, OTX1, DLX5, PAX6, NEUROG1, IGF1, WNT6, WNT8A, KLF4 |
| hsa04950 | Maturity onset diabetes of the young | 0.082815 | 4 | HNF1A, MNX1, PAX6, NEUROG3 |
| hsa05410 | HCM | 0.089722 | 7 | ACE, CACNG8, CACNG6, ITGA1, CACNB2, IGF1, TTN |
Abbreviations: KEGG, Kyoto Encyclopedia of Genes and Genomes; AML, acute myeloid leukemia; CAMs, cell adhesion molecules; HCM, hypertrophic cardiomyopathy.
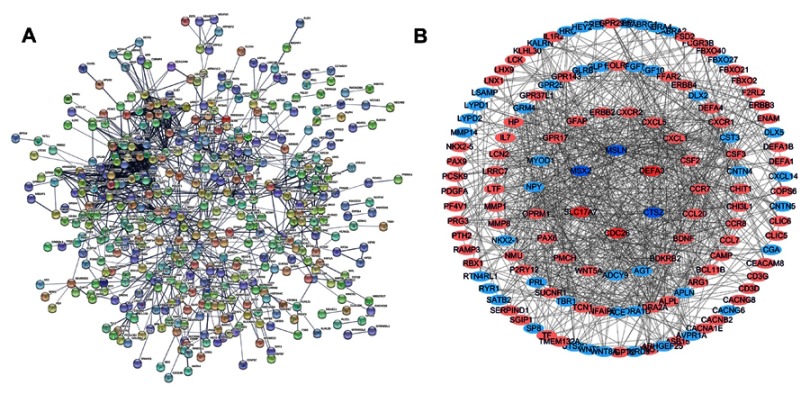
PPI network construction and module analysis
The STRING was used to construct PPI networks of DEGs, see Figure 4A. The plug‐in MCODE of the Cytoscape software was used to identify the most significant module. Ultimately, 138 nodes and 885 edges were established from the most significant genes with differential expression, including 88 up-regulated genes and 50 down-regulated genes, see Figure 4B, which possibly play an important role in childhood AML progression and can be used as a predictor of CR.
Figure 4.
(A) PPI network of signifcantly diferentially expressed genes. (B) The most significant module was established from PPI network with 138 nodes and 885 edges, up-regulated genes are marked with light red; down-regulated genes are marked with light blue.
Abbreviation: PPI, protein–protein interaction.
Prognostic gene marker screening
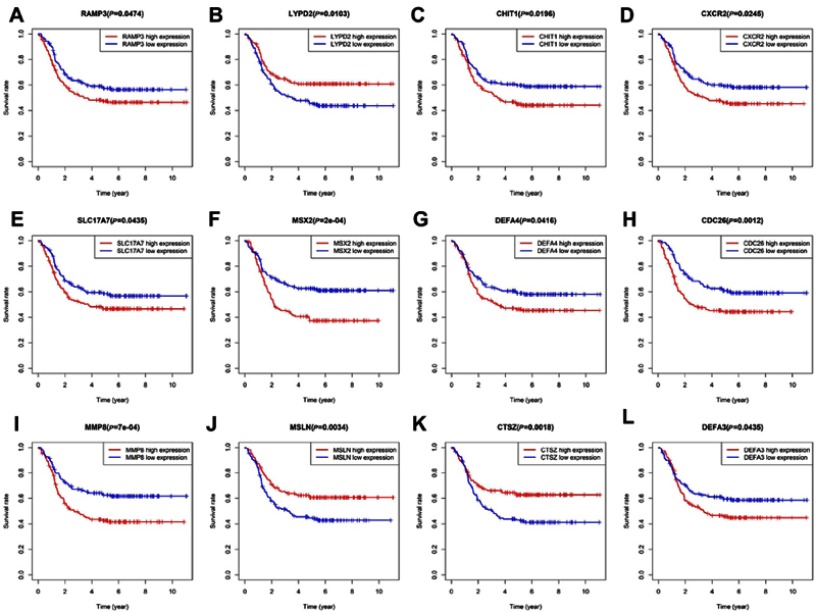
To assess the prognostic value of the most significant module form 138 genes, we performed Cox regression analysis, OS analysis and ROC curve analyses along with calculations of the area under the curve (AUC). The results of log-rank test showed that 17 genes were significantly associated with OS evidenced by positive coefficients in the Cox regression, suggesting that they may have a low risk of recurrence, see Table 3. Secondly, we analyzed the association between these candidate gene expression of patients with childhood AML by Kaplan-Meier analysis. The results showed that 12 genes expression (RAMP3, LYPD2, CHIT1, CXCR2, SLC17A7, MSX2, DEFA4, CDC26, MMP8, MSLN, CTSZ, DEFA3) was associated with OS for childhood AML, see Figure 5.
Table 3.
Univariate Cox regression analysis for the candidate genes in the training dataset
| Gene | HR | Lower 95% CI | Upper 95% CI | z | P-value |
|---|---|---|---|---|---|
| RAMP3 | 1.107431 | 1.035415 | 1.184456 | 2.974399 | 0.002936 |
| LYPD2 | 0.896117 | 0.834845 | 0.961886 | −3.035332 | 0.002403 |
| FBXO2 | 1.127005 | 1.032227 | 1.230485 | 2.667657 | 0.007638 |
| CHIT1 | 1.100148 | 1.032955 | 1.171711 | 2.968346 | 0.002994 |
| CXCL1 | 1.114886 | 1.043231 | 1.191462 | 3.208677 | 0.001333 |
| FBXO21 | 1.198487 | 1.062074 | 1.352422 | 2.936787 | 0.003316 |
| CXCR2 | 1.153750 | 1.069802 | 1.244286 | 3.710558 | 0.000207 |
| SLC17A7 | 1.094191 | 1.024202 | 1.168963 | 2.669037 | 0.007607 |
| FFAR2 | 1.099628 | 1.023025 | 1.181966 | 2.577853 | 0.009942 |
| MSX2 | 1.071317 | 1.020138 | 1.125063 | 2.758274 | 0.005811 |
| DEFA4 | 1.055876 | 1.013570 | 1.099949 | 2.605988 | 0.009161 |
| CDC26 | 1.390649 | 1.168735 | 1.654700 | 3.717825 | 0.000201 |
| DEFA1B | 1.082348 | 1.028912 | 1.138560 | 3.063311 | 0.002189 |
| MMP8 | 1.071428 | 1.026945 | 1.117837 | 3.188925 | 0.001428 |
| MSLN | 0.940093 | 0.908923 | 0.972332 | −3.590855 | 0.000330 |
| CTSZ | 0.908571 | 0.846955 | 0.974670 | −2.676011 | 0.007450 |
| DEFA3 | 1.053126 | 1.015175 | 1.092494 | 2.764295 | 0.005705 |
Figure 5.
Prognostic value of twelve key genes (A) RAMP3 (B) LYPD2 (C) CHIT1 (D) CXCR2 (E) SLC17A7 (F) MSX2 (G) DEFA4 (H) CDC26 (I) MMP8 (J) MSLN (K) CTSZ (L) DEFA3 in childhood AML from TARGET database.
Genetic risk score model construction and ROC curve analysis
Among the 12 prognostic genes identified for which multiple stepwise Cox regression was performed to explore the effect of these genes on the survival time and the patient’s outcome, six gene markers were found to be independent predictors in childhood AML patients, see Table 4. As a result, six genes were finally selected to build a predictive model. Patient RSs were determined using the formula below.
Table 4.
A six-gene signature identified by multivariate Cox regression analysis
| id | coef | exp (coef) | se (coef) | z | Pr(>|z|) |
|---|---|---|---|---|---|
| SLC17A7 | 0.108993 | 1.115154 | 0.034211 | 3.185898 | 0.001443 |
| MSX2 | 0.110729 | 1.117092 | 0.028377 | 3.902084 | 0.000095 |
| CDC26 | 0.319005 | 1.375758 | 0.085776 | 3.719048 | 0.000200 |
| MSLN | −0.048660 | 0.952505 | 0.019963 | −2.437433 | 0.014792 |
| CTSZ | −0.068123 | 0.934146 | 0.039113 | −1.741674 | 0.081566 |
| DEFA3 | 0.045649 | 1.046707 | 0.020474 | 2.229620 | 0.025773 |
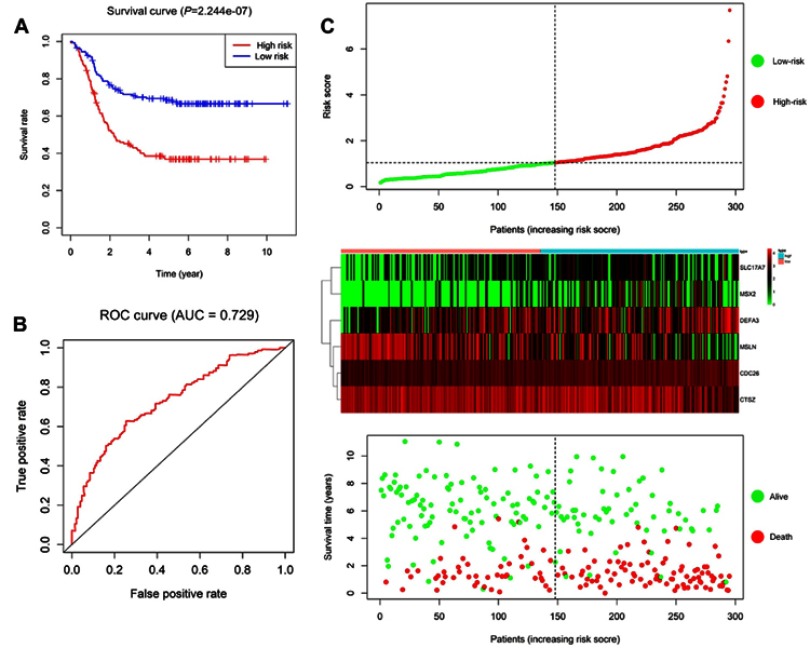
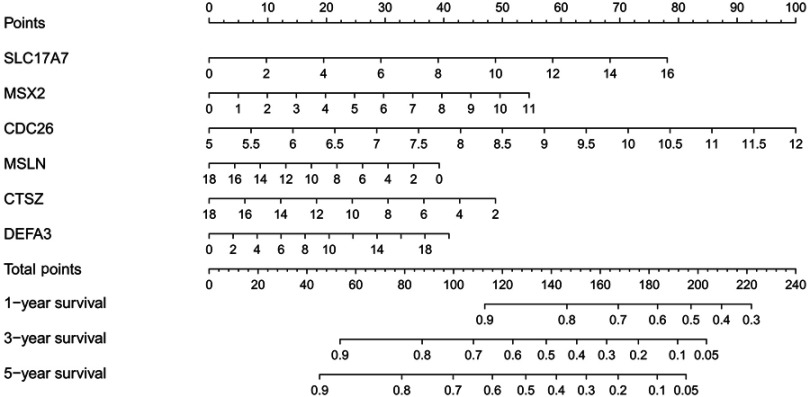
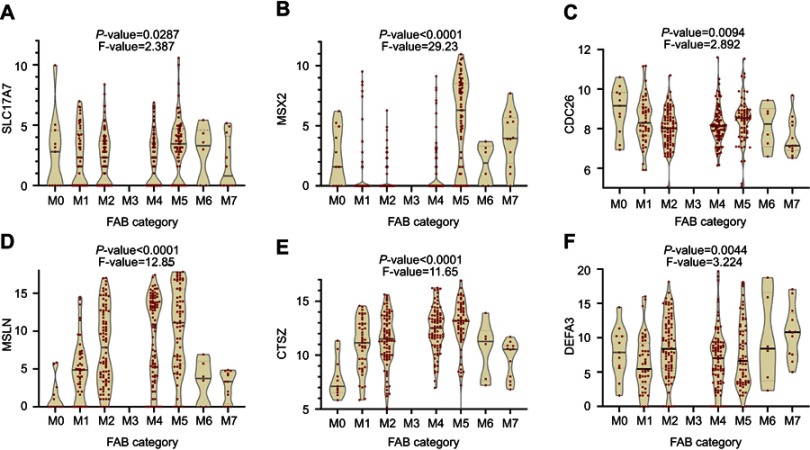
A total of 295 patients were classified into a high-risk group and a low-risk group by using the median of the RSs as a cut-off point. The survival estimated for childhood AML patients in the high-risk group and those in the low-risk group were significantly different, with an increased risk of death in the high-risk group. The results show that the 3- and 5-year survival rate were significantly different between the high-risk group and low-risk group, see Figure 6A. The prognostic capacity of the six-gene signature was evaluated by using the AUC of a time-dependent ROC curve. The AUC of genes biomarker prognostic model was 0.729, see Figure 6B. The RS, expression heat map, and patients’ survival status distribution of the 6 prognostic genes in two groups are shown in Figure 6C, indicating that the predictive model had a high sensitivity and specificity. We developed a nomogram to predict the probability of the 1‐, 3‐ and 5‐year OS. The predictors of the nomogram included six independent prognostic factors including SLC17A7, MSX2, CDC26, MSLN, CTSZ and DEFA3, see Figure 7. We analyzed the six candidate genes expression level in different subtypes of childhood AML based on the FAB category, but M3 data are scarce, see Figure 8.
Figure 6.
Prognostic risk score model analysis of six prognostic genes. (A) The Kaplan–Meier curves for low-risk and high-risk groups. (B) The ROC curves for predicting OS by the risk score. (C) The distribution of risk score, expression heat map, and survival status.
Abbreviations: AUC, area under the curve; ROC, receiver operator characteristic; OS, overall survival.
Figure 7.
Nomogram for predicting 1-, 3-, and 5-year survival rate in childhood AML patients. By adding up the points identified on the point scale for each variable, the total score on the bottom scale shows the probability of survival.
Figure 8.
The French-American-British (FAB) category from database to analyze the six each candidate genes expression level in different subtypes of childhood AML. (A) SLC17A7 (B) MSX2 (C) CDC26 (D) MSLN (E) CTSZ (F) DEFA3.
In order to validate the prognostic model, we incorporate WHO risk-stratification criteria such as cytogenetics and genetics.23 A total of 295 patients was analyzed with favorable or adverse factors for internal validation, see Table 5. The results show that the CR rate was lower in children in the high-risk group (68.7%) than in those in low-risk group (87.8%) (P<0.01). In addition, the distribution of some favorable or adverse factors8 such as RUNX1-RUNX1T1, CBFB-MYH11, CEBPA mutation, cytogenetic complexity, and FLT3-ITD combined with WT1 mutation was consistent with the results of previous studies.
Table 5.
Evaluate the prognostic model by WHO classification (cytogenetics or genetics)
| High risk (n=147) | Low risk (n=148) | P-value | |
|---|---|---|---|
| CR status at end of course 1 | 101 (68.7%) | 130 (87.8%) | 6.7E-05* |
| Favorable factors | |||
| t(8;21)(q22;q22)/RUNX1-RUNX1T1 | 5 (3.4%) | 41 (27.7%) | 1.5E-08* |
| inv(16)(p13.1q22)/CBFB-MYH11 | 0 (0%) | 42 (28.4%) | 5.8E-12* |
| CEBPA mutation | 4 (2.7%) | 16 (10.8) | 5.7E-03* |
| NPM mutation | 14 (9.5%) | 14 (9.4%) | 9.8E-01 |
| Adverse factors | |||
| Cytogenetic complexity (3 or more) | 31 (21.1%) | 18 (12.1%) | 3.9E-02* |
| t(10;11)(p12;q23)/MLLT10-MLL | 5 (3.4%) | 2 (1.3%) | 2.2E-01 |
| t(6;9)(p23;q34)/DEK-NUP214 | 2 (1.4%) | 1 (0.6%) | 5.3E-01 |
| FLT3-ITD/combined with WT1 mutation | 12/21 (57.1%) | 2/14 (14.2%) | 8.6E-03* |
Note: *Difference between the two groups was significant (P<0.05).
Discussion
AML is one of the most common malignancies, with multiple types of molecular and cellular heterogeneity in childhood.3 Hematopoietic stem cell transplantation combined with chemotherapy are the basic means to treat AML, but the prognosis of childhood AML remains suboptimal due to high recurrence and high mortality.24,25 In particular, refractory acute leukemia has poor response to treatment, a short survival period and low-induced relieving rate in the second CR2 after relapse.26 Recently, many studies had reported that the prognosis of childhood AML is partly driven by genetic factors, and the expressions of multiple genes maybe beneficial to predicting prognosis and select treatment regimens.8,27,28 The clinical implementation of an improved child AML risk classification model is likely to provide more relevant information for clinical decisions and improve the prognosis of child AML patients by refining patient’s risk stratification.29,30 Therefore, understanding the etiological factors and molecular mechanisms of childhood AML progression is essential for the diagnosis and treatment of this disease. Microarray technology has been widely applied to identify potential therapeutic targets. Previously, Luo et al31 analyzed the GSE8970 dataset and revealed that ubiquitin-conjugating enzyme E2E1 (UBE2E1) as a prognostic factor may be involved in AML. Zhang et al32 analyzed the GSE12417 dataset and suggested that the long non-coding RNA H19 may serve roles in AML. Niu et al33 analyzed the TCGA dataset and constructed a risk prediction model based on relapse information, with the limitations that the number of AMLs cohorts was small and more specimens should be included to validate the ability of model. On the other hand, the TARGET database has the advantage of having large AML samples and complete clinical information for children. To reduce mortality and improve the risk-stratification criteria, there is an urgent need for the molecular screening of biomarkers of childhood AML.
In this study, we identified significant DEGs between the childhood AML into first CR and not CR samples from the TARGET database. Furthermore, we performed a series of bioinformatics analyses to screen key genes and pathways. As a result, a total of 856 DEGs were identified, consisting of 543 up-regulated genes and 313 down-regulated genes. GO function and KEGG pathway analyses were performed to acquire an in-depth understanding of these DEGs. The functional enrichment analyses demonstrated that the up-regulated genes were enriched in some BPs such as leukocyte chemotaxis, chemokine-mediated signaling pathway, receptor complex, apical plasma membrane, G-protein coupled peptide receptor activity, and channel activity. In addition, the down-regulated genes were mostly enriched in cell fate commitment, morphogenesis of a branching structure, projection membrane, transcriptional activator activity, and RNA polymerase II transcription regulatory region sequence-specific DNA binding. The results are consistent with previous knowledge proved that gain or loss of these functions plays an important role in AML tumorigenesis and progression. The KEGG pathway analysis revealed that the DEGs were significantly associated with cytokine-cytokine receptor interaction, neuroactive ligand–receptor interaction, hematopoietic cell lineage, and signaling pathways regulating pluripotency of stem cells. Our study results suggested that these DEGs may be involved in the onset and progression of childhood AML.
Based on these findings, the hub genes were screened, and univariate, multivariate Cox analyses were conducted to build a risk model to predict childhood AML prognosis. We identified six genes: SLC17A7, MSX2, CDC26, MSLN, CTSZ and DEFA3. High expression levels of SLC17A7, MSX2, CDC26 and DEFA3 were relevant to a poor prognosis in childhood AML patients, but MSLN and CTSZ were associated with a good prognosis. The AUC of the ROC curve for the prognostic model for predicting the OS was 0.729, indicating that the six-gene signature had a good performance for survival prediction. With the gene expression risk scoring prognostic model, the patients with childhood AML were divided into a high-risk group and a low-risk group. According to the results predicted by the model, the clinician can change the treatment plan and provide individualized treatment for childhood AML patients. There is a need for developing strategies to improve CR in the high-risk group. Patients in the high-risk group should be followed more frequently, and bone marrow aspiration and biopsy should be performed regularly to facilitate early detection of disease recurrence. Our prognostic mode is independent of other factors in childhood AML, and may have implication in guiding hematopoietic stem cell transplantation. Similarly, nomogram is a kind of statistical tools that provides an individual patient with the overall probability of a particular outcome. Whether this model is applicable to adult AML,34 warrants further investigation.
The protein encoded by SLC17A7 is a vesicle-bound, sodium-dependent phosphate transporter that is particularly expressed in neuron-rich regions of the brain. Wan et al35 identified SLC17A7 as the potential diagnostic and prognostic biomarkers of uveal melanoma by Co-expression modules. Homeobox-containing (HOX) genes encode transcription factors, which play an important regulatory role in signal transduction pathways such as cell development, migration, and differentiation, and are frequently found to be aberrantly expressed in cancer.36 Up-regulation of muscle segment homeobox genes 2 (MSX2), a member of the homeobox gene family, was found in pancreatic cancer and prostate cancer patients. Many clinical studies showed MSX2 was involved in the occurrence and development of tumors.37,38 Zhai et al39 have discovered that MSX2 is a direct downstream target of WNT signaling and correlated with the invasiveness of endometrioid adenocarcinoma. Moreover, MSX2 has been identified as a physiological NKL in hematopoietic cells. It is involved in NOTCH3-signaling,and this pathway interacts between the physiological and oncogenic homeobox signaling in T-ALL.40 Cell division control protein 26 (CDC26) is part of the protein modification and involved in the pathway protein ubiquitination. It catalyzes the formation of protein-ubiquitin conjugates that are subsequently degraded by the proteasome.41 Mesothelin (MSLN) is a glycosylphosphatidylinositol-anchored cell-surface protein and may be a CAM. Steinbach et al42 prospectively evaluated the prognostic value of monitoring treatment response in AML by measuring the expression of 7 leukemia-related genes. Among them, MSLN is regarded as the important prognostic indicator. Cathepsin Z (CTSZ), a lysosomal cysteine protease and a member of the peptidase C1 family is widely expressed in tumor cell lines and primary tumors. Like other members of the family, it may be involved in the occurrence of tumors.43 Defensin alpha 3 (DEFA3) is present in the bactericidal granules of neutrophils and may play a role in phagocyte-mediated host defense. The proliferation rate was affected by the stimulation of defensin in tumor cell lines.44
The six-gene prognostic model may facilitate the development of new prognostic predictors for childhood AMLs. In addition, our solution significantly reduces the cost of sequencing, which makes the application of gene-specific targeted sequencing more cost-effective and routine. In future, we plan to use single-cell transcriptome sequencing in bone marrow to detect the expression of these six genes in patients who are poor candidates for transplantation. The prognostic assessment is crucial in selecting the suitable treatment. Since patients with the same subtype and stage can have different clinical outcomes, we developed this predictive model for risk stratification in childhood AML, and the model may become routinely used in the future.
Our study has several limitations. First, our results were derived from data in TARGET dataset and generated by bioinformatic analysis. The TARGET database does not provide information about specific treatments received by each patient. Thus, the results of our study need to be validated in other databases. Further investigations are needed to validate our results based on childhood AML samples and clinical data. Second, the number of samples without CR was smaller than those with CR in childhood AML. Therefore, more specimens need to be included to validate the predictive model capability we developed.
In conclusion, our study results indicate that the six-gene prognostic model is a reliable tool for predicting the OS of childhood AML, and a nomogram comprising a prognostic model can serve as a predictor for CR and may assist clinicians in providing individualized treatment in this patient population. This discovery has the potential to provide new therapeutic targets for childhood AML.
Acknowledgment
This study was supported by the Natural Science Foundation Project of CQ (grant no. cstc2017jcyjAX0239).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Lim SH, Dubielecka PM, Raghunathan VM. Molecular targeting in acute myeloid leukemia. J Transl Med. 2017;15(1):183. doi: 10.1186/s12967-017-1281-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66:271–289. doi: 10.3322/caac.21349 [DOI] [PubMed] [Google Scholar]
- 3.Prada-Arismendy J, Arroyave JC, Röthlisberger S. Molecular biomarkers in acute myeloid leukemia. Blood Rev. 2017;31:63–76. doi: 10.1016/j.blre.2016.08.005 [DOI] [PubMed] [Google Scholar]
- 4.Hourigan CS, Gale RP, Gormley NJ, Ossenkoppele GJ, Walter RB. Measurable residual disease testing in acute myeloid leukaemia. Leukemia. 2017;31:1482–1490. doi: 10.1038/leu.2017.113 [DOI] [PubMed] [Google Scholar]
- 5.Tarlock K, Meshinchi S. Pediatric acute myeloid leukemia: biology and therapeutic implications of genomic variants. Pediatr Clin North Am. 2015;62:75–93. doi: 10.1016/j.pcl.2014.09.007 [DOI] [PubMed] [Google Scholar]
- 6.Komanduri KV, Levine RL. Diagnosis and therapy of acute myeloid leukemia in the era of molecular risk stratification. Annu Rev Med. 2016;67:59–72. doi: 10.1146/annurev-med-051914-021329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marcucci G, Haferlach T, Döhner H. Molecular genetics of adult acute myeloid leukemia: prognostic and therapeutic implications. J Clin Oncol. 2011;29:475–486. doi: 10.1200/JCO.2010.30.2554 [DOI] [PubMed] [Google Scholar]
- 8.Creutzig U, van Den Heuvel-Eibrink MM, Gibson B, et al. Diagnosis and management of acute myeloid leukemia in children and adolescents: recommendations from an international expert panel. Blood. 2012;120(16):3187–3205. doi: 10.1182/blood-2012-03-362608 [DOI] [PubMed] [Google Scholar]
- 9.Staffas A, Kanduri M, Hovland R, et al.; Nordic Society of Pediatric Hematology and Oncology (NOPHO). Presence of FLT3-ITD and high BAALC expression are independent prognostic markers in childhood acute myeloid leukemia. Blood. 2011;118:5905–5913. doi: 10.1182/blood-2011-05-353185 [DOI] [PubMed] [Google Scholar]
- 10.Wu X, Feng X, Zhao X, et al. Prognostic significance of FLT3-ITD in pediatric acute myeloid leukemia: a meta-analysis of cohort studies. Mol Cell Biochem. 2016;420:121–128. doi: 10.1007/s11010-016-2775-1 [DOI] [PubMed] [Google Scholar]
- 11.Heidrich K, Thiede C, Schäfer-Eckart K, Study Alliance Leukemia (SAL), et al. Allogeneic hematopoietic cell transplantation in intermediate risk acute myeloid leukemia negative for FLT3-ITD, NPM1- or biallelic CEBPA mutations. Ann Oncol. 28;2017:2793–2798. doi: 10.1093/annonc/mdx500 [DOI] [PubMed] [Google Scholar]
- 12.Mrózek K, Marcucci G, Paschka P, Bloomfield CD. Advances in molecular genetics and treatment of core-binding factor acute myeloid leukemia. Curr Opin Oncol. 2008;20:711–718. doi: 10.1097/CCO.0b013e32831369df [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ng SW, Mitchell A, Kennedy JA, et al. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature. 2016;540:433–437. doi: 10.1038/nature20598 [DOI] [PubMed] [Google Scholar]
- 14.Patel JP, Gönen M, Figueroa ME, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366:1079–1089. doi: 10.1056/NEJMoa1112304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fasslrinner F, Schetelig J, Burchert A, et al. Long-term efficacy of reduced-intensity versus myeloablative conditioning before allogeneic haemopoietic cell transplantation in patients with acute myeloid leukaemia in first complete remission: retrospective follow-up of an open-label, randomised phase 3 trial. Lancet Haematol. 2018;5(4):e161–161e169. [DOI] [PubMed] [Google Scholar]
- 16.Tallman MS, Dewald GW, Gandham S, et al. Impact of cytogenetics on outcome of matched unrelated donor hematopoietic stem cell transplantation for acute myeloid leukemia in first or second complete remission. Blood. 2007;110(1):409–417. doi: 10.1182/blood-2006-10-043299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–353D361. doi: 10.1093/nar/gkw1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–607D613. doi: 10.1093/nar/gky1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doncheva NT, Morris JH, Gorodkin J, Jensen LJ. Cytoscape StringApp: network analysis and visualization of proteomics data. J Proteome Res. 2019;18:623–632. doi: 10.1021/acs.jproteome.8b00702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han Y, Yang J, Liu P, et al. Prognostic nomogram for overall survival in patients with diffuse large B-cell lymphoma. Oncologist. 2019. doi: 10.1634/theoncologist.2018-0361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swerdlow SH, Campo E, Pileri SA, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127(20):2375–2390. doi: 10.1182/blood-2016-01-643569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van der Linden MH, Creemers S, Pieters R. Diagnosis and management of neonatal leukaemia. Semin Fetal Neonatal Med. 2012;17:192–195. doi: 10.1016/j.siny.2012.03.003 [DOI] [PubMed] [Google Scholar]
- 25.Mo XD, Lv M, Huang XJ. Preventing relapse after haematopoietic stem cell transplantation for acute leukaemia: the role of post-transplantation minimal residual disease (MRD) monitoring and MRD-directed intervention. Br J Haematol. 2017;179:184–197. doi: 10.1111/bjh.14778 [DOI] [PubMed] [Google Scholar]
- 26.Gorman MF, Ji L, Ko RH, et al. Outcome for children treated for relapsed or refractory acute myelogenous leukemia (rAML): a Therapeutic Advances in Childhood Leukemia (TACL) consortium study. Pediatr Blood Cancer. 2010;55(3):421–429. doi: 10.1002/pbc.22612 [DOI] [PubMed] [Google Scholar]
- 27.Radhi M, Meshinchi S, Gamis A. Prognostic factors in pediatric acute myeloid leukemia. Curr Hematol Malig Rep. 2010;5:200–206. doi: 10.1007/s11899-010-0060-z [DOI] [PubMed] [Google Scholar]
- 28.Aziz H, Ping CY, Alias H, Ab Mutalib NS, Jamal R. Gene mutations as emerging biomarkers and therapeutic targets for relapsed acute myeloid leukemia. Front Pharmacol. 2017;8:897. doi: 10.3389/fphar.2017.00897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang M, Lindberg J, Klevebring D, et al. Validation of risk stratification models in acute myeloid leukemia using sequencing-based molecular profiling. Leukemia. 2017;31:2029–2036. doi: 10.1038/leu.2017.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanchez M, Levine RL, Rampal R. Integrating genomics into prognostic models for AML. Semin Hematol. 2014;51:298–305. doi: 10.1053/j.seminhematol.2014.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo H, Qin Y, Reu F, et al. Microarray-based analysis and clinical validation identify ubiquitin-conjugating enzyme E2E1 (UBE2E1) as a prognostic factor in acute myeloid leukemia. J Hematol Oncol. 2016;9:125. doi: 10.1186/s13045-016-0356-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang TJ, Zhou JD, Zhang W, et al. H19 overexpression promotes leukemogenesis and predicts unfavorable prognosis in acute myeloid leukemia. Clin Epigenetics. 2018;10:47. doi: 10.1186/s13148-018-0486-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niu P, Yao B, Wei L, Zhu H, Fang C, Zhao Y. Construction of prognostic risk prediction model based on high-throughput sequencing expression profile data in childhood acute myeloid leukemia. Blood Cells Mol Dis. 2019;77:43–50. doi: 10.1016/j.bcmd.2019.03.008 [DOI] [PubMed] [Google Scholar]
- 34.Sandahl JD, Kjeldsen E, Abrahamsson J, et al. The applicability of the WHO classification in paediatric AML. A NOPHO-AML study. Br J Haematol. 2015;169(6):859–867. doi: 10.1111/bjh.13366 [DOI] [PubMed] [Google Scholar]
- 35.Wan Q, Tang J, Han Y, Wang D. Co-expression modules construction by WGCNA and identify potential prognostic markers of uveal melanoma. Exp Eye Res. 2018;166:13–20. doi: 10.1016/j.exer.2017.10.007 [DOI] [PubMed] [Google Scholar]
- 36.Samuel S, Naora H. Homeobox gene expression in cancer: insights from developmental regulation and deregulation. Eur J Cancer. 2005;41:2428–2437. doi: 10.1016/j.ejca.2005.08.014 [DOI] [PubMed] [Google Scholar]
- 37.Chua CW, Chiu YT, Yuen HF, et al. Differential expression of MSX2 in nodular hyperplasia, high-grade prostatic intraepithelial neoplasia and prostate adenocarcinoma. APMIS. 2010;118:918–926. doi: 10.1111/j.1600-0463.2010.02626.x [DOI] [PubMed] [Google Scholar]
- 38.Satoh K, Hamada S, Shimosegawa T. MSX2 in pancreatic tumor development and its clinical application for the diagnosis of pancreatic ductal adenocarcinoma. Front Physiol. 2012;3:430. doi: 10.3389/fphys.2012.00430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhai Y, Iura A, Yeasmin S, et al. MSX2 is an oncogenic downstream target of activated WNT signaling in ovarian endometrioid adenocarcinoma. Oncogene. 2011;30:4152–4162. doi: 10.1038/onc.2011.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nagel S, Venturini L, Przybylski GK, et al. NK-like homeodomain proteins activate NOTCH3-signaling in leukemic T-cells. BMC Cancer. 2009;9:371. doi: 10.1186/1471-2407-9-371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang J, Dye BT, Rajashankar KR, Kurinov I, Schulman BA. Insights into anaphase promoting complex TPR subdomain assembly from a CDC26-APC6 structure. Nat Struct Mol Biol. 2009;16:987–989. doi: 10.1038/nsmb.1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steinbach D, Bader P, Willasch A, et al. Prospective validation of a new method of monitoring minimal residual disease in childhood acute myelogenous leukemia. Clin Cancer Res. 2015;21:1353–1359. doi: 10.1158/1078-0432.CCR-14-1999 [DOI] [PubMed] [Google Scholar]
- 43.Akkari L, Gocheva V, Quick ML, et al. Combined deletion of cathepsin protease family members reveals compensatory mechanisms in cancer. Genes Dev. 2016;30:220–232. doi: 10.1101/gad.270439.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winter J, Kraus D, Reckenbeil J, Probstmeier R. Oncogenic relevant defensins: expression pattern and proliferation characteristics of human tumor cell lines. Tumour Biol. 2016;37:7959–7966. doi: 10.1007/s13277-015-4701-7 [DOI] [PubMed] [Google Scholar]