SUMMARY
During translation, an mRNA is typically occupied by multiple ribosomes sparsely distributed across the coding sequence. This distribution, mediated by slow rates of initiation relative to elongation, ensures that they rarely collide with each other, but given the stochastic nature of protein synthesis, collision events do occur. Recent work from our lab suggested that collisions signal for mRNA degradation through no-go decay (NGD). We have explored the impact of stalling on ribosome function when NGD is compromised and found it to result in +1 frameshifting. We used reporters that limit the number of ribosomes on a transcript to show that +1 frameshifting is induced through ribosome collision in yeast and bacteria. Furthermore, we observe a positive correlation between ribosome density and frameshifting efficiency. It is thus tempting to speculate that NGD, in addition to its role in mRNA quality control, evolved to cope with stochastic collision events to prevent deleterious frameshifting events.
In Brief
Ribosome collisions, resulting from stalling, activate quality control processes to degrade the aberrant mRNA and the incomplete peptide. mRNA degradation proceeds through an endonucleolytic cleavage between the stacked ribosomes, which resolves the collisions. Simms et al. show that, when cleavage is inhibited, colliding ribosomes move out of frame.
Graphical Abstract
INTRODUCTION
At any point during protein synthesis, a single mRNA is typically occupied by multiple ribosomes (Warner et al., 1963). Biochemical assays, as well as computational studies, have shown ribosomes to be thinly distributed across transcripts, rarely coming in close proximity to one another (Brandt et al., 2009, 2010; Kennell and Riezman, 1977; Kierzek et al., 2001; Siwiak and Zielenkiewicz, 2013). This sparse distribution is the result of at least two distinct mechanisms. The first involves vastly differential translation-initiation and -elongation rates. In E. coli, the rate of initiation has been estimated to be in the range of ~1 codon s−1 (Mitarai et al., 2008), whereas that of elongation has been measured to be ~20 codons s−1 (Sørensen and Pedersen, 1991). The second mechanism involves the apparent clustering of rare codons near the start codon (Eyre-Walker and Bulmer, 1993; Tuller et al., 2010), which has been hypothesized to slow down early elongating ribosomes, acting like a ramp and hence ensuring that ribosomes do not collide into each other downstream (Dobrzynski and Bruggeman, 2009). Despite these mechanisms, modeling studies suggest that ribosome collisions are inevitable given the stochastic nature of translation (Mitarai et al., 2008). Collisions are predicted to slow down the lagging ribosome significantly, which would, in turn, increase the likelihood of additional collisions and subsequent ribosome pileups. These effects can be minimized by decreasing the stability of the mRNA, but this comes at a cost of reduced protein output per mRNA. It has been estimated that, for a codon-optimized lacZ mRNA, which is translated an average of 30 times in E. coli, stochastic collisions lengthen the translation time by 5% compared to an mRNA that is translated once (Mitarai et al., 2008). These models suggest that not only do ribosome collisions occur frequently, even on codon-optimized mRNAs, but that they are costly, slowing down protein synthesis overall.
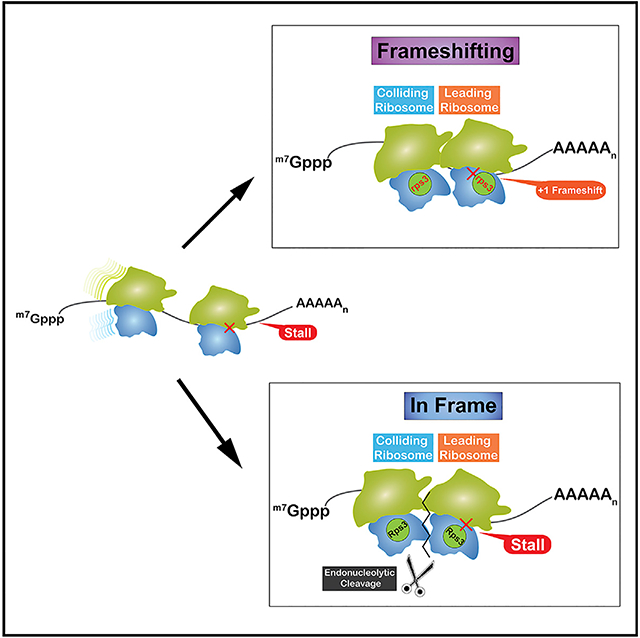
Ribosome collisions have been best studied in the context of defective mRNAs that stall ribosomes. More specifically, mRNAs harboring stable secondary structures, inhibitory codons, internal polyA sequences, damaged nucleobases, and ones that encode for peptides that interact with the exit tunnel of the ribosome are all well documented to dramatically inhibit translation (Brandman and Hegde, 2016; Simms et al., 2016). These mRNAs activate the processes of no-go decay (NGD) and ribosome-associated quality control (RQC) to rapidly degrade the defective mRNAs and the incomplete nascent peptide (Bengtson and Joazeiro, 2010; Doma and Parker, 2006). NGD is initiated through an endonucleolytic cleavage—by an unknown enzyme—upstream of the stall site, exposing the mRNA for the exonucleolytic action of Xrn1 and the exosome (Doma and Parker, 2006; Tsuboi et al., 2012). RQC requires the action of a number of factors that recognize a pep-tidyl-tRNA-bound large subunit (Brandman et al., 2012; Defenouillè re et al., 2013). Briefly, the E3 ligase Ltn1 (listerin in mammals) adds K48-linked ubiquitin chains to the peptide (Bengtson and Joazeiro, 2010). Following release by Vms1 (ANZF1 in mammals; Verma et al., 2018), the ubiquitinated peptide is recognized by the adaptor protein Cdc48 and presented to the proteasome for degradation (Defenouillè re et al., 2013; Verma et al., 2013). In addition to RQC and NGD, dissociation of the stalled ribosomes into their respective small and large subunits by Dom34/Hbs1/Rli1 ensures that they are rescued to be utilized for new bouts of translation. Ribosome rescue, NGD, and RQC are all dependent on the action of the E3 ligase Hel2 (Znf598 in mammals; Matsuo et al., 2017; Garzia et al., 2017; Juszkiewicz and Hegde, 2017; Sundaramoorthy et al., 2017). The factor ubiquitinates various ribosomal proteins through K63-linked chains, and deletion of the factor inhibits all of the processes associated with ribosome stalling.
Until recently, an important question for the field was: how are these stalls recognized? In other words, how does Hel2 distinguish stalled ribosomes from slow ones? Initial models suggested that stalled ribosomes adopt a distinct conformation that is recognized by the E3 ligase (Matsuo et al., 2017). Although this model was tempting, it lacked mechanistic insights into how this conformation could be attained. Instead, a more appealing model emerged recently that describes the factor recognizing collided ribosomes resulting from stalls. A major basis of this model, as put forward by our group, is the observation that ribosomal-protein ubiquitination by Hel2 is significantly increased when global ribosome collisions are induced (Simms et al., 2017). Recent studies from the Hegde, Ramakrishnan, Inada, and Beckman groups provided important structural support for this model; collided ribosomes form an interface on the small subunits that bring together the target ribosomal proteins for ubiquitination by Hel2 (Ikeuchi et al., 2019; Juszkiewicz et al., 2018). Further biochemical characterization of these reactions showed the factor to prefer higher order structures of ribosomes, i.e., polysomes. Analysis of Hel2 binding to RNA in vivo revealed that, although it binds most mRNA species, binding to the initial 150 nt of the coding sequence is significantly depleted, suggesting that it requires multiple ribosomes to initiate quality control (Winz et al., 2019). This is consistent with the observation that NGD is only robust when the stall sequence is >100 nt from the initiation codon (Simms et al., 2017). Therefore, it appears that eukaryotic cells have evolved a pathway to recognize and respond to ribosome collisions, suggesting that they are detrimental to cellular homeostasis if they go unresolved.
Analogous biochemical studies on replication and transcription have revealed that DNA and RNA polymerases often collide with each other, resulting in so-called “conflicts” (García-Muse and Aguilera, 2016). These “conflicts” are especially problematic for replisome integrity, thus necessitating mechanisms for their resolution (Azvolinsky et al., 2009; Merrikh et al., 2011). To this end, DNA damage response factors are able to sense these collision events and in most cases remove the RNA polymerase (Poli et al., 2016; Tehranchi et al., 2010; Tuduri et al., 2009). Failure to resolve collided polymerases has been shown to be associated with genomic instability (Prado and Aguilera, 2005; Helmrich et al., 2013). In contrast to replication, the consequences of unresolved collision events during translation are poorly understood. In particular, whether they could modify the decoding properties of the ribosome, and especially frame maintenance, is unknown.
This potential connection between mRNA quality control and frame maintenance is consistent with the observation that deletion of asc1 (RACK1 in mammals), which is a key factor in RQC (Kuroha et al., 2010; Brandman et al., 2012) and is predicted to act upstream of hel2 (Juszkiewicz and Hegde, 2017; Sundaramoorthy et al., 2017; Matsuo et al., 2017; Sitron et al., 2017), leads to +1 frameshifting in yeast (Wolf and Grayhack, 2015). However, a connection between frame maintenance and ribosome collision has not been explored. Equally important is the fact that stochastic ribosome collisions are predicted to be frequent and are likely to activate Hel2. Indeed, we have previously shown that increasing stochastic collisions by addition of sub-inhibitory concentrations of cycloheximide leads to robust ribosomal protein ubiquitination by Hel2 (Simms et al., 2017). These observations beg the question of whether ribosomebased quality control evolved to cope with stochastic collision events that would otherwise lead to frameshifting.
Here, we show, using various reporters in yeast as well as a well-defined in vitro bacterial translation system, that collisions result in +1 frameshifting. In yeast, inhibition of NGD was found to be accompanied by a significant increase in +1 frameshifting. We further show that efficient +1 frameshifting does not occur unless the stall sequence is placed beyond ~20 nt from the initiation codon, suggesting that it requires multiple ribosomes to load onto the transcript. Furthermore, we observe a positive correlation between ribosome density and frameshifting efficiency. No such correlation was observed for programmed frameshifting, suggesting that they exploit a distinct mechanism for recoding. In agreement with our model that ribosome collisions induce +1 frameshifting, we find that, in a well-defined bacterial system, frameshifting on a stall sequence within the downstream gene of a polycistronic mRNA depends on the translation of the upstream gene and, hence, collisions. In conclusion, our data strongly suggest that collisions are detrimental to frame maintenance and that NGD is likely activated frequently on regular mRNAs to resolve stochastic collision events.
RESULTS
Inhibition of NGD Leads to +1 Frameshifting
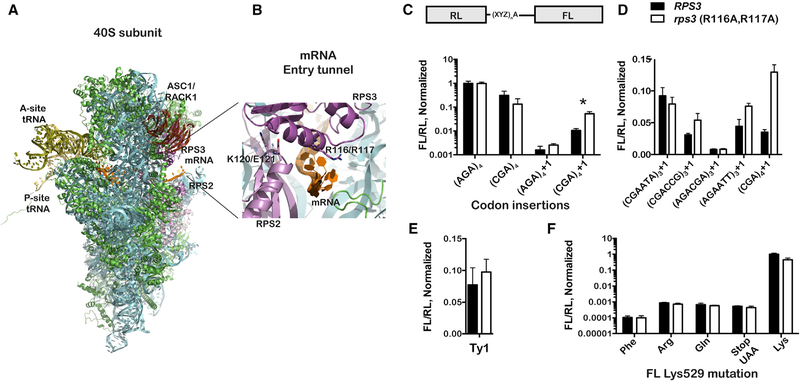
As collisions activate NGD and are accompanied by the rapid degradation of the mRNA (Simms et al., 2017), their effect on ribosome function could only be studied in the absence of pathway activity. We have recently reported that the entry tunnel residues of ribosomal protein Rps3 (Figures 1A and 1B) are important for the endonucleolytic cleavage reaction (Simms et al., 2018), thus providing us a means to examine frameshifting in the context of inhibited NGD. To probe frameshifting in these variant yeast strains, we utilized a dual luciferase reporter (Salas-Marco and Bedwell, 2005), for which the downstream firefly luciferase (FL) coding region is in different frames relative to the upstream Renilla luciferase (RL) coding region, either −1, in frame, or +1 (Figure 1C). Stalling on this reporter was promoted through the addition of inhibitory CGA codons between the two genes (Letzring et al., 2010). The ratio of FL luminescence to that of RL was used to measure the amount of frameshifting that occurred. We note that, because this reporter is a translational fusion, any effects of the mutations on the RNA stability, and hence its levels, should be minimized. Indeed, qRT-PCR analysis using primers specific to the FL region and the RL one showed that the mutations in RPS3 as well as in ASC1, known to affect NGD, have little to no effect on the RNA ratio of the two reporters (Figure S1).
Figure 1. Mutation of Rps3’s Entry Tunnel Residues Increases +1 Frameshifting on Stalling Sequences.
(A) Overview of the structure of the eukaryotic small ribosomal subunit complexed with mRNA and P-site tRNA (PDB: 5AJ0). The mRNA entry tunnel, encompassing residues from Rps3 and Rps2, is highlighted as well as the proximity of Rack1 (Asc1 in yeast) and its interaction with Rps3.
(B) A close-up view of the entry tunnel, where the ribosome makes the most intimate contacts with the mRNA (in orange). Rps3 and Rps2 are shown in dark and light magenta, respectively; other ribosomal proteins are shown in green; whereas the rRNA is depicted in cyan.
(C) The R116A; R117A mutations increase +1 frameshifting on CGA codons. The architecture of the dual-luciferase reporter is shown on top; the XYZ indicates the different nucleotides that were inserted in the linker region between the two coding sequences. An additional adenosine (A) was added for the +1 frameshifting reporters. Relative luminescence of firefly luciferase (FL) to that of Renilla luciferase (RL) from the indicated reporters in the wild-type and mutant Rps3 strains is plotted. The readings were normalized to those measured from the non-stalling (AGA)4 reporter. Asterisk denotes a significant difference between wild-type and mutant rps3 for (CGA)4+1 (p < 0.005 by unpaired t test). For comparison, p = 0.096 for the (AGA)4+1 reporter.
(D) The Rps3 mutations promote frameshifting on previously described stalling sequences. Bar graphs of the normalized luminescence ratio (to the original reporter with no insertion between the two reporters) for the indicated reporters in the wild-type and Rps3 mutant strains are shown.
(E) The Rps3 mutations have little to no effect on programmed frameshifting. Normalized ratios of FL luminescence to that of RL for a reporter harboring the Ty1 transposon frameshifting site in wild-type and rps3 strains are shown.
(F) The entry tunnel mutations do not affect missense and nonsense miscoding. Normalized luminescence ratios for reporters, which have the active-site FL Lys residue (K529) mutated to the indicated amino acid, are shown.
In all cases, the mean + SD of at least three biological repeats is plotted.
See also Figures S1, S2, and S3.
On a reporter containing AGA codons, which are synonymous for CGA codons but do not cause stalling, mutation of Rps3’s entry tunnel residues had no discernable effect on relative FL expression levels. In contrast and as expected, FL expression was significantly reduced (to 31% ± 15%) in wild-type cells carrying the reporter with in-frame CGA codons. Interestingly, in the presence of the Rps3 mutations, the relative FL expression was further reduced (to 13% ± 9.2%), presumably due to increased frameshifting when stalling occurs. Consistent with this idea, direct measurements using frameshifting reporters showed that, in the presence of CGA codons, +1 frameshifting is significantly higher relative to that seen in the presence of AGA codons. In particular, the relative FL expression was measured to be 0.16% ± 0.1% and 1.0% ± 0.2% for the (AGA)4+1 and (CGA)4+1 reporters, respectively. Including the Rps3 mutations increased frameshifting a further five-fold (to 5.3% ± 1.2%) on the (CGA)4+1 reporter (Figure 1C). To provide further support for this observed effect of the mutations on frameshifting, we also tested 4 more sequences that were previously identified by the Grayhack group to promote frameshifting in the absence of Asc1 (Gamble et al., 2016; Wang et al., 2018). On two of these sequences, namely (CGACCG)3+1 and (AGAATT)3+1, the mutations in the entry tunnel increased frameshifting, albeit to a lesser extent than that observed on the CGA codons (Figure 1D). This suggests that collision-induced +1 frameshifting occurs in the absence of functional NGD.
As expected, the mutations did not promote frameshifting on codons that do not induce stalling and also did not promote −1 frameshifting (Figure S1). Furthermore, the Rps3 mutations had no effect on programmed frameshifting efficiency, as assessed using a construct that had the frameshifting sequence from the Ty1 transposable element (Figure 1E). This particular programmed frameshifting construct was chosen due to its simple sequence requirement of a 7-nt motif and high frameshifting efficiency (Belcourt and Farabaugh, 1990). Western analysis corroborated these results and revealed that the mutations increase +1 frameshifting, specifically on the (CGA)4+1 reporter, but not on the Ty1 one (Figure S2). These findings suggest that, because programmed frameshifting likely occurs in a collision-independent manner, inhibition of NGD is inconsequential. Similarly, the mutations do not promote nonsense and missense types of miscoding as assessed by constructs that replace the active site Lys residue of the firefly luciferase (K529; Kramer and Farabaugh, 2007) by other amino acids or a stop codon (Figure 1E). In these constructs, firefly luminescence reports on the frequency of the ribosome misreading the mutated codon as a Lys. Collectively, our data support the notion that, when NGD does not occur, ribosomes undergo +1 frameshifting at a stall; it appears that other aspects of decoding are not affected
Entry Tunnel Residues of Rps2 Do Not Affect Frameshifting
Alongside Rps3, the mRNA entry tunnel of the ribosome encompasses residues of Rps2 (Figure 1B), namely K119, E120, Q94, and R95, suggesting that the protein may also play a role in frame maintenance. However, previous data from our group showed mutations in this region of Rps2 to have no observable effect on mRNA cleavage during NGD (Simms et al., 2018). Consistent with their apparent lack of effect on NGD, these mutations did not change frameshifting efficiency on the (CGA)4+1 reporter (Figure S3). Furthermore, the RPS2 mutations did not alter the increased levels of frameshifting observed in the rps3 (R116A; R117A) background. These observations suggest that Rps3 plays an especially important role in frame maintenance beyond its role as a constituent of the helicase domain of the ribosome. In agreement with this notion, genetic screens by the Culbertson and Grayhack groups identified a separate mutation in Rps3 (K108E), which promotes efficient +1 frameshifting (Hendrick et al., 2001; Juszkiewicz et al., 2018; Wang et al., 2018). This mutation also caused increased frameshifting on our (CGA)4+1 reporters (Figure S3), lending additional support to the idea that Rps3 plays a general role in preventing unwanted +1 frameshifting.
Asc1/RACK1 Promotes Frame Maintenance Independently of Rps3
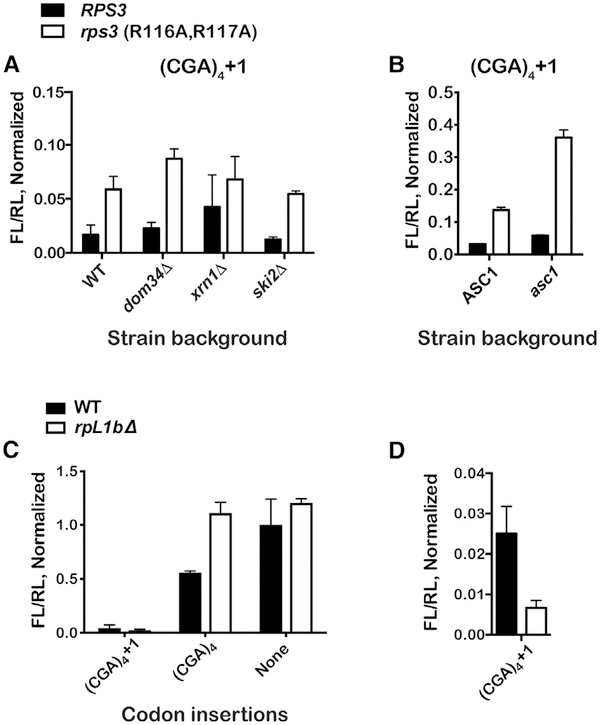
To examine the potential contribution of other NGD components, we introduced the Rps3 mutations into yeast backgrounds that affect the pathway, but none were found to alter the observed increased levels of frameshifting seen in the Rps3 mutant alone (Figure 2A). Furthermore, in addition to Rps3, the ribosome-associated factor Asc1 (human RACK1) has been documented to affect multiple facets of NGD and ribosome quality control (RQC) of nascent peptides during stalling (Ikeuchi and Inada, 2016; Letzring et al., 2013; Sitron et al., 2017; Wolf and Grayhack, 2015). Most relevant to our studies is the observation that, in yeast, its deletion results in increased frameshifting on CGA codons (Wolf and Grayhack, 2015); furthermore, the protein interacts with the C-terminal end of Rps3 on the ribosome (Ben-Shem et al., 2011). Hence, it is possible that the increased frame-shifting we observe in the presence of the Rps3 mutations is due to Asc1-specific defects. We addressed this by introducing mutations to Asc1 (R38D and K40E) known to greatly reduce its association with the ribosome (Coyle et al., 2009). As has been reported earlier for the asc1 deletion (Wolf and Grayhack, 2015), these mutations enhanced frameshifting on our (CGA)4+1 construct to 5.8% ± 0.18%. In the presence of the Rps3 mutations alone, the observed occurrence of frameshifting was 13.7% ± 0.9%, and surprisingly, when combined with the asc1 mutations, we observe a compounding effect, resulting in 36.0% ± 2.4% frameshifting, suggesting that the factors may, at least in part, contribute to NGD independently (Figure 2B). This is consistent with structural and biochemical studies showing that Asc1 interacts with Rps3 away from the entry tunnel (Ben-Shem et al., 2011) and that mutations of Rps3’s entry-tunnel residues do not affect the interaction between Asc1 and the ribosome (Simms et al., 2018). The same mutations appear to have little to no effect on +1 frameshifting on polyA sequences, presumably because frameshifting on these sequences results in the same sequence in the A site and does not alleviate stalling (Figure S4). Similarly, frameshifting on reporters harboring other stretches of positively charged amino acids was not affected (Figure S4).
Figure 2. +1 Frameshifting Does Not Depend on NGD Components but Is Correlated with Ribosome Density.
(A) Deletion of NGD factors does not alter the effect of the Rps3 mutations on frameshifting. Normalized ratios of FL luminescence to that of RL for the (CGA)4+1 reporter in the indicated strains are shown.
(B) The effects of Rps3 mutation on frameshifting are additive with mutations in Asc1. The bar graph shows the normalized luminescence ratios for the (CGA)4+1 reporter in the indicated strains. In the double mutant (rps3; asc1), frameshifting efficiency is nearly 40%.
(C) Bar graph of normalized FL/RL luminescence ratios for the indicated reporters in wild-type and rpl1bΔ strains.
(D) Similar to first dataset in (C). Deletion of rpL1b results in decreased +1 frameshifting.
In all cases, the mean + SD is plotted from at least three biological replicates, normalized to values for the (AGA)4 reporter.
See also Figure S4.
Ribosome Density Is Correlated with +1 Frameshifting
Our analysis suggests that there is a correlation between NGD inhibition and increased +1 frameshifting, but the underlying mechanism is unclear. We suggest that, in the absence of efficient cleavage of the aberrant mRNA, an increase in collision events, which go unresolved, results in a steric clash between ribosomes. Consequently, this somehow causes the leading ribosome to move out of frame, which in turn alleviates stalling (the P-site codon changes from the inhibitory CGA one to GAC). A prediction of this model is that frameshifting should be correlated to ribosome density. Earlier work by our laboratory has shown that reducing cellular ribosome concentrations by depleting ribosomal proteins strongly inhibits NGD (Simms et al., 2017). This in turn results in readthrough of CGA codons. It is important to note that the mechanism by which reduction of ribosome density inhibits NGD—decreased collision events—is different from the one induced by the rps3 mutations—increased collision events; hence, their effects on frameshifting should also be divergent. In complete agreement with our model, deletion of one of the two genes that encode ribosomal protein RpL1 (rpl1b) decreased frameshifting by four-fold (from 2.8% ± 0.17% to 0.7% ± 0.15%; Figures 2C and 2D). Deletion of rpl1b has a dramatic effect on ribosome concentrations, but not its composition, that is apparent in polysome profiles of the deletion strain; a marked decrease in the amount of polysomes indicates a reduction in the density of ribosomes on mRNAs (Simms et al., 2017). Interestingly, this relationship between ribosome density and “unwanted” +1 frameshifting is in direct contrast to what we know about programmed frameshifting, for which slowed translation is a major determinant for increased recoding (Kawakami et al., 1993).
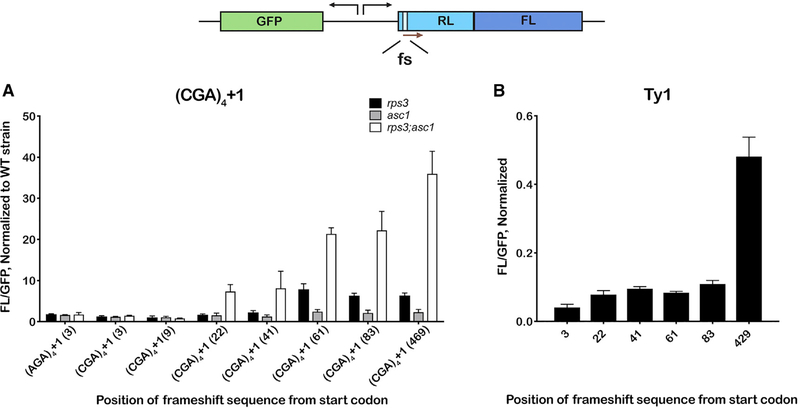
+1 Frameshifting Increases at Greater Distances from the Initiation Codon
To further probe the mechanism for collision-induced frameshifting, we placed the frameshifting site (the inhibitory CGA codons) at various distances from the initiation codon. The rationale behind these experiments is that, at distances close to the initiation codon, the stalling sequence would prevent multiple ribosomes from loading onto the transcript, precluding collisions. Indeed, we observe no increase in frameshifting relative to the wild-type strain in the rps3 and asc1 mutants, unless the stalling sequence is placed at least 22 nt from the initiation codon (Figure 3A). The efficiency of frameshifting increases as the stalling sequence moves farther from the initiation codon, consistent with the idea that loading multiple ribosomes behind the primary stalled ribosome increases frameshifting. This pattern was by and large independent of the rps3 mutations; we observe a very similar dependence on stalling-sequence placement in the K108E mutant (Figure S5), which is also in agreement with data reported by Wolf and Grayhack (2015). Although programmed frameshifting also appears to be dependent on the frameshifting site, there was no obvious correlation between distance from start site and frameshifting efficiency. For Ty1, we observe equivalent efficiencies for constructs harboring the site between 3 and 83 nt from the start codon, but frameshifting significantly increases when it was placed at position 429 (Figure 3B). For the programmed frameshift sequence EST3, frameshifting efficiency was higher for position 3 relative to positions 22 and 83 but significantly increased for position 429 (Figure S5). These observations, again, highlight the important distinctions between programmed and unwanted frameshifting.
Figure 3. Frameshifting on CGA Inhibitory Codons Depends Critically on the Position of the Stalling Sequence and Is Likely to Require Multiple Ribosomes to Load onto the mRNA.
A schematic of the reporter indicates the dual-luciferase-fusion reporter together with a GFP reporter under a constitutive promoter that is oriented opposite to the luciferase reporter. The frameshifting site (fs) was placed at different positions throughout the Renilla coding sequence.
(A) Frameshifting only occurs on reporters that have the fs site placed at least 22 nt from the initiation codon. Ratios of FL luminescence to GFP fluorescence for the indicated reporters, normalized to values obtained using the wild-type strain, obtained in the presence of the rps3 (R116A; R117A), asc1 (R38D; K40E) and the double mutation are plotted.
(B) Similar to (A), but the (CGA)4+1 fs sequence was substituted with the (programmed fs) Ty1 transposon sequence.
In all cases, the mean + SD of at least three biological repeats is plotted.
See also Figure S5.
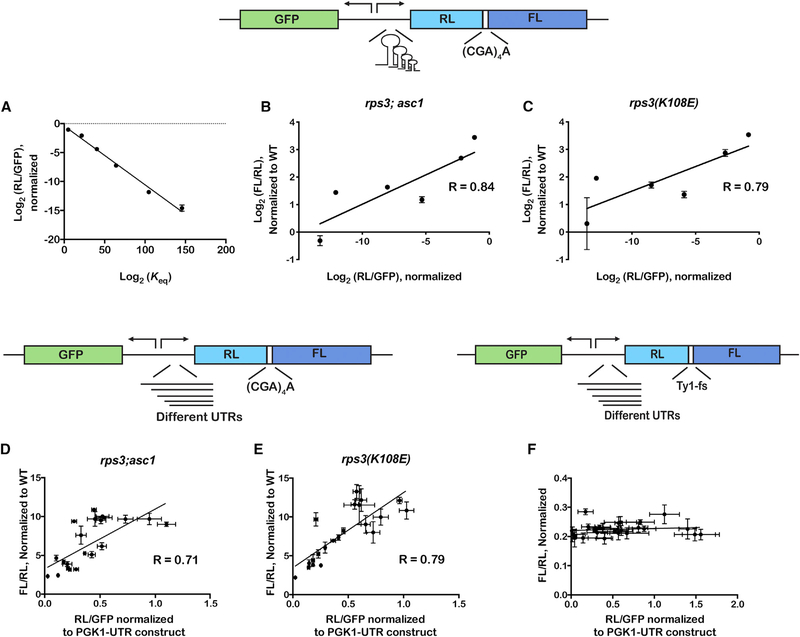
+1 Frameshifting Correlates with the Efficiency of Gene Expression
Next, we sought to change ribosome density specifically on our frameshifting reporters without affecting overall ribosome homeostasis. We reasoned that, by modifying the 5′ UTR sequence, we should be able to change initiation efficiency and hence ribosome density. In the first approach, we introduced stem loops of various lengths in the 5′ UTR of the reporters; these should slow or inhibit scanning by the small sub-unit during initiation (Hinnebusch et al., 2016). These reporter plasmids, in addition to having the dual luciferase gene to measure frameshifting, harbor a gene that encodes superfolder-green fluorescent protein (sf-GFP) (Pédelacq et al., 2006). Both genes are under constitutive promoters. As a result, the ratio of Renilla luminescence to GFP fluorescence reports on translation efficiency of the reporters with the different stem loops, and that of firefly luminescence to the Renilla one reports on frameshifting. As expected, the introduction of the stem loops reduced the expression of Renilla luciferase by as much as four orders of magnitude and this reduction correlated well with the equilibrium constant of unwinding for the stem loops (Figure 4A). Importantly, the level of frameshifting correlated well with gene expression, and presumably ribosome density, regardless of the mutation. For the rps3; asc1 mutant, we measured an R value of 0.84; for the rps3(K108E), we measured an R value of 0.79 (Figures 4B, 4C, and S6). Therefore, initiation efficiency, and likely ribosome density, is a good predictor for frameshifting efficiency.
Figure 4. Efficiency of Stalling-Induced Frameshifting Correlates with Gene Expression.
A schematic of the reporter, which harbors the fs site between RL and FL coding sequence, is shown on top. Stem loops of various lengths were introduced in the UTR of the dual-luciferase gene.
(A) Plot of the ratio of RL luminescence relative to GFP fluorescence versus the equilibrium constant of unwinding of the stem loops. The equilibrium constants were calculated from ΔG values determined by mfold (Zuker, 2003).
(B) A plot of luminescence ratios (RL/FL), which reports on fs efficiency, against that of RL luminescence to GFP fluorescence, which reports on gene expression, in the rps3; asc1 double mutant.
(C) Similar to (B) but in the rps3 K108E mutant.
(D) A schematic of the reporter is shown on top. It is similar to the one shown in (A), but instead of stem loops, various native yeast UTRs were introduced upstream of the dual-luciferase gene. The plot is similar to (B) but uses the UTR reporter set.
(E) Similar to (C) with UTR reporter set.
(F) A schematic of the reporter used to assess the effect of gene-expression efficiency on programmed fs is shown. The reporter is similar to that used in (D) but harbors the Ty1 fs sequence between RL and FL instead of the stalling (CGA)4+1 sequence. The plot shows FL/RL luminescence ratios versus RL luminescence divided by GFP fluorescence. Note the lack of correlation between the two.
In all cases, the mean + SD of at least three biological repeats is plotted.
See also Figure S6.
As a second approach for this collision model, we introduced different natural yeast 5′ UTRs into our frameshifting reporter. In total, we generated 22 reporters with 5′ UTRs that span a broad length (30–829 nt), which we expected to have drastic effects on initiation efficiency and hence gene expression (Hinnebusch et al., 2016). We measured ratios of Renilla luminescence to GFP fluorescence (our measure for translation efficiency) that were more than an order of magnitude different among the different UTRs. Likewise, frameshifting efficiency varied by more than five-fold among the different reporters (Figures 4D, 4E, and S6). Most pertinent to our model, however, is the observation that, similar to our stem-loop reporters, the amount of frameshifting we observed was found to correlate tightly with gene expression, with R values of 0.71 and 0.79 for the rps3; asc1 and rps3(K108E) mutants, respectively. Similar to what we documented for the stem-loop reporters, no correlation between translation efficiency and frameshifting was observed for programmed Ty1 frameshifting (Figure 4F). The results for asc1 may highlight differences in its role with respect to frameshifting as it correlates with gene expression in the stem loop experiments, but not in the UTR experiments (Figure S6). It may also be due to our choice of strain—we observe lower frameshifting efficiencies with the R38DK40E mutations than are reported for the asc1 deletion strain (Wolf and Grayhack, 2015). Collectively, our use of reporters with disparate initiation rates suggests that translation efficiency, and hence ribosome density, contributes significantly to unwanted frameshifting.
Collision Is Required for +1 Frameshifting-Dependent Gene Expression In Vitro
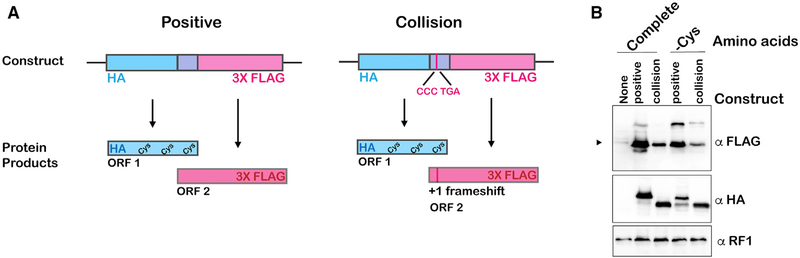
To address the collision model more directly, we chose to use the bacterial reconstituted PURE system (New England Biolabs) because of the polycistronic organization of genes in bacteria. This allowed us to generate mRNA reporters consisting of two genes with overlapping coding sequences. In particular, the initiation codon of the downstream gene resides within the coding region of the upstream one, and translation of its full-length product depends on an early frameshifting event (poor stop-codon context with CCC UGA coding for Pro-STOP at codon 7 from the start codon; Gurvich et al., 2003). In other words, translation of this gene in isolation would allow for a maximum of one ribosome to load before the frameshift site is encountered. As a result, if frameshifting is dependent on ribosome collision, then the translation of the downstream gene should be dependent on the translation of the upstream gene. As a positive control, we generated a reporter for which translation of the downstream gene does not depend on frameshifting (Figure 5A; Table S1). In both cases, the upstream gene contains sequences that encode cysteine, whereas the downstream one does not and, as expected, translation of the upstream gene, which contains an N-terminal hemagglutinin (HA) tag, was dependent on cysteine for both reporters. In the absence of cysteine, the amount of protein products as assessed by western blotting decreased by ~two-fold (Figure 5B). This incomplete inhibition of synthesis of the upstream product is likely due to the presence of residual amounts of Cys-tRNA in the tRNA preparation used in the translation reaction. Regardless, synthesis of the downstream protein products, as assessed by western blotting against the C-terminal FLAG tag, differed between the two reporters. For the positive-control reporter, depletion of cysteine had little to no effect on its translation (Figure 5B). This was expected as the downstream gene does not encode cysteine residues. In contrast and in agreement with our prediction, for the frameshifting reporter, we observed a ~two-fold reduction of protein produced for the downstream gene in the absence of cysteine. This suggests that its synthesis depends on translation of the upstream gene, because the downstream protein does not contain any cysteine residues, and therefore it is likely that collisions from ribosomes translating the upstream gene are required to induce frameshifting on the downstream gene. Not only do these observations add support for ribosome collisions playing an important role in promoting frameshifting, but they also suggest that the mechanism is conserved, occurring in bacteria as well as in eukaryotes.
Figure 5. Collisions Result in Frameshifting in a Reconstituted Bacterial System.
(A) A schematic of the reporters used to assess the effect of collisions on frameshifting. The reporters are bicistronic, with the initiation codon of the downstream gene residing within the upstream gene. The first open reading frame (ORF) codes for an N-terminal HA tag as well as cysteine residues throughout. ORF 2 codes for a C-terminal FLAG tag and no cysteine residues; it is in the +1 frame relative to the upstream gene. For the collision reporter, the downstream gene has a stop codon in a poor context (CCC TGA) and requires a frameshift to translate the C-terminal FLAG.
(B) Western blot analysis of in vitro translation reactions with the indicated reporters in the presence or absence of cysteine. Synthesis of the upstream protein, assessed by probing with HA, is more robust in the presence of cysteine. Synthesis of the downstream protein from the collision reporter is twice as abundant in the presence of cysteine and hence the synthesis of the upstream protein. Arrowhead indicates the band for the downstream product.
See also Table S1.
DISCUSSION
The ability of the ribosome to precisely and repetitively move 3 nt at a time during the elongation phase of protein synthesis is an indispensable feature of the translation machinery. This maintenance of reading frame ensures that the genetic code is faithfully deciphered. To this end, frameshifting errors are among the most deleterious ones, especially when compared to missense errors, as they result in peptide products that bear no resemblance to the intended protein. Precise translocation, and hence the prevention of frameshifting, is not an easy task, given the multiple coordinated movements of the mRNA, tRNA, and the ribosomal subunits that must occur (Noller et al., 2017). After peptidyl transfer, the peptidyl-tRNA residing in the A site must move to the adjacent P site. Initially, the acceptor stem of the tRNA moves from the A site to the P site of the large subunit, and the anticodon stem remains in the A site of the small subunit, forming the so-called “hybrid” state (Moazed and Noller, 1989). Following EF-G (eEF2) binding, guanosine triphosphate (GTP) hydrolysis, and Pi release, full translocation of the tRNA to the P site of the ribosome is completed (Brilot et al., 2013; Pulk and Cate, 2013; Ramrath et al., 2013; Tourigny et al., 2013; Voorhees and Ramakrishnan, 2013; Zhou et al., 2013, 2014; Belardinelli et al., 2016; Noller et al., 2017). During this process, multiple interactions between the tRNA-mRNA complex and the ribosome must be disrupted and new ones established. Multiple elements of the ribosome, translation factors, and the tRNA contribute to these interactions and hence the precision of the translocation process. Indeed, mutations in the tRNA, mRNA, and the ribosome that alter some of these interactions have been shown to modulate translocation mechanics and accuracy (Atkins and Björk, 2009).
The P-site tRNA, through its interaction with the mRNA codon, appears to play an important role in reading frame maintenance (Baranov et al., 2004). More specifically, the likelihood of the tRNA anticodon to transiently dissociate and establish new out-of-frame base pairing interactions is governed by the energetics of binding to the P-site codon. In addition to tRNA-mRNA interactions, stalling plays a major role in promoting frameshifting. This is mainly dictated by the nature and availability of the A-site ligand (Farabaugh et al., 2006). For example, during RF2-programmed frameshifting in bacteria, the efficiency of frameshifting at the poorly decoded RF2-specific UGA stop codon is significantly impacted by the concentration of cellular RF2 (Craigen and Caskey, 1986). Finally, frameshifting is heavily influenced by structural impediments upstream or downstream of the ribosome. Using RF2-programmed frameshifting as an example again, this impediment is provided by a Shine-Dalgarno sequence upstream that has been hypothesized to base pair with the rRNA, pulling the mRNA out of frame (Márquez et al., 2004; Devaraj and Fredrick, 2010).
Here, we provide compelling evidence that, if left unresolved, ribosomes colliding behind the primary stalled one provide the necessary structural element for efficient frameshifting. To look at unintended frameshifting, we chose the CGA codon and yeast as a model system. The wobble adenosine nucleotide, as decoded by inosine on the anticodon, provides the necessary weak interaction in the P site, which presumably allows for dissociation and reassociation of the tRNA. The presence of an adjacent slowly decoded CGA codon in the A site lends the additional requirement of stalling for efficient frameshifting (Letzring et al., 2010). Our data suggest that these two requirements are not sufficient for frameshifting to occur. Ribosome queuing is needed to complete the requirements, similar to those noted for programmed frameshifting.
Previous data from our group as well as others have shown ribosome collisions to activate the process of NGD (Simms et al., 2017a; Juszkiewicz et al., 2018; Ikeuchi et al., 2019), which results in the endonucleolytic cleavage of the mRNA and hence resolving collisions. Consistent with our collision-induced frameshifting model, inhibition of the cleavage reaction by introducing mutations to the ribosomal protein Rps3 in the entry tunnel increases frameshifting significantly. This agrees with data from the Grayhack group that showed deletion of the NGD/RQC factor Asc1 also results in significant frameshifting (Wolf and Grayhack, 2015). Here, we too show that mutations of Asc1 that disrupt its interaction with the ribosome promote frameshifting. Ribosome-associated Asc1 appears to be important for the recruitment of the E3 ligase Hel2 (Juszkiewicz and Hegde, 2017; Sundaramoorthy et al., 2017; Matsuo et al., 2017; Sitron et al., 2017), which adds K63-linked ubiquitin chains on collided ribosomes (Ikeuchi et al., 2019). Deletion and mutation of Asc1 have been documented to inhibit NGD-associated cleavage of the mRNA and downstream ribosome rescue by Dom34/Hbs1/Rli1 (Ikeuchi and Inada, 2016). Asc1 likely senses collided ribosomes and somehow communicates that information to Hel2. In its absence, ribosomes pile up behind the primary stalled one and cause it to move out of frame. Interestingly, previous work also from the Grayhack group has suggested that the ribosome-associated protein Mbf1 is recruited to stalled ribosomes where, together with Rps3, they may act as bumpers, preventing adjacent ribosomes from colliding (Wang et al., 2018).
Although there are previous reports of a relationship between frameshifting and NGD, the connection between frameshifting and ribosome collision and the underlying mechanics have not been explored prior to the work presented here. We provide various pieces of evidence to support a direct influence of ribosome queuing on frameshifting efficiency. For instance, reducing the cellular concentration of ribosomes, which decreases their density on mRNA and hence their likelihood of collision, lowered frameshifting by more than three-fold (Figure 2D). Furthermore, when the stalling sequence is placed near the initiation codon so that a maximum of one ribosome can occupy the mRNA before encountering the stall site, we observe little to no increase in frameshifting as a result of NGD inhibition. The correlation between ribosome density and frameshifting efficiency is bolstered by our observation that alteration of ribosome loading onto the mRNA by modulating initiation efficiency impacted frameshifting by almost an order of magnitude (Figure 3). We document a significantly positive correlation between translation efficiency, and hence ribosome loading, and frameshifting (Figure 3). It should be noted that, in these experiments, ribosome density was only altered on our reporters without globally affecting translation. Finally, perhaps the strongest piece of supporting evidence comes from our experiments in a reconstituted bacterial translation system. Here, we observe that frameshifting on the downstream gene of a polycistronic mRNA depends critically on the ability of ribosomes to translate the upstream gene and collide with the ribosomes stalled on the downstream one (Figure 5). Collectively, through the use of various reporters and genetic manipulations in yeast as well as a high-resolution in vitro bacterial translation system, we provide data that strongly suggest that ribosome collisions result in +1 frameshifting.
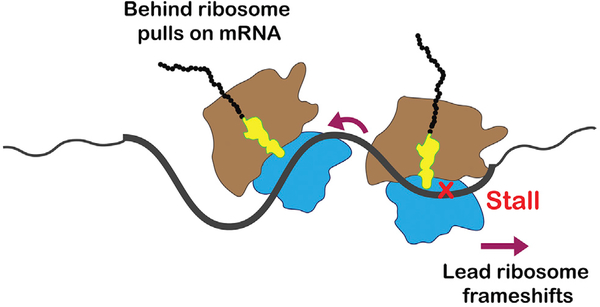
Given that ribosome collisions are likely to happen during translation under normal conditions, it should not be surprising that a mechanism for reducing their impact on cellular fitness may have evolved. In conclusion, we propose a mechanism for collision-induced frameshifting (Figure 6), whereby the behind ribosome translocates along the mRNA until it reaches the stalled ribosome, at which point the mRNA is stretched completely. Under normal conditions, this triggers cleavage and downstream events of NGD. Under conditions where cleavage is inhibited, the behind ribosome is free to pull on the mRNA—because its A-site codon is competent—exerting force on the fully stretched RNA, resulting in slippage by the stalled ribosome, causing it to frameshift.
Figure 6. A Model for Collision-Induced Frameshifting.
In this model, the colliding, behind ribosome pulls on the mRNA, resulting in its slippage from the A site of the lead, stalled ribosome.
Not only does this study add critical insights into frame maintenance and its relationship to mRNA quality control processes, but it also suggests that ribosomes are able to change the decoding properties of their neighboring ones. Although there has been a wealth of studies that have looked at the role of RNA structure and sequence on frameshifting (Farabaugh et al., 2006), this study may be the first to look at the ribosome itself acting in trans to affect the decoding properties of another ribosome. Beyond adding new mechanistic details into the role of trans factors in promoting frameshifting, our findings suggest that this is a widespread phenomenon that cells need to deal with, especially given that ribosome collisions are predicted to occur frequently. As a result, it is tempting to speculate that NGD, in addition to its primary role in mRNA quality control (Simms et al., 2016), evolved to cope with stochastic collisions events that are bound to take place occasionally and, if left unresolved, result in deleterious frameshifting events. At a minimum, our data reveal a profound connection between an mRNA surveillance mechanism and maintaining the integrity of the decoding process.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Hani Zaher (hzaher@wustl.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Yeast Strains and Growth Conditions
Yeast strains were cultured in YPD or defined media (when expressing reporter plasmids) at 30°C. Strains carrying mutations were constructed using standard PCR-based recombination techniques in the background BY4741 (MATa (his3Δ1 leu2Δ0 met15Δ0 ura3Δ0). Strains used in this study are included as Table S2.
The RPS2 mutant strains were generated by cloning a PCR fragment encoding RPS2-LEU2-rpS2 3′UTR into the BamHI/XbaI sites in pPROEX-Htb. Point mutations in RPS2 were introduced by site directed mutagenesis, the entire region PCR amplified and the resulting fragment used for transformation of target yeast strains. RPS3 (K108E) strains were made in the same manner, except a HIS3 gene was used to tag RPS3 and the PCR product was cloned into pPROEX-Htb using BamHI and XhoI sites. The HIS3 and LEU2 coding regions were amplified from plasmids pFA6a-6xGLY-FLAG-HIS3 and pAG415 respectively (Funakoshi and Hochstrasser, 2009; Alberti et al., 2007). pAG415-GPD-ccdB was a gift from Susan Lindquist (Addgene plasmid # 14146; http://addgene.org/14146).
METHOD DETAILS
Plasmids
The Renilla-Firefly dual luciferase reporter was constructed from plasmid pDB688 (Salas-Marco and Bedwell, 2005) using site-directed mutagenesis to insert various sequences between the two coding regions. These included the CGA and AGA codon repeats (with an extra A in the +1 constructs), and the Ty1 and EST3 fs sequences. The Lys 529 constructs were built similarly, with mutations introduced into the firefly luciferase gene.
The GFP-dual luciferase reporter was assembled from overlapping PCR products; the GPD promoter sequence from pAG415 driving GFP (Wolf and Grayhack, 2015) in one orientation, with a dual luciferase frameshifting cassette positioned in the opposite direction and driven by a PGK1 promoter. In the first set of constructs, frameshifting sequences ((CGA)4+1, Ty1, or EST3) were inserted into the renilla luciferase gene by PCR amplifying the entire plasmid and re-ligating using AgeI sites that were included in the primer oligos. For experiments with constructs containing various stem loop or 5′ UTR sequences, PCR fragments were cloned into BglII sites downstream of the PGK1 promoter or AscI/SbfI sites, respectively.
The in vitro fs constructs were designed with two overlapping coding regions, derived from sequences for E. coli InfA and DHFR, and the dsDNA templates were obtained from IDT.
All plasmid constructs and DNA oligos used in this study are included as Table S3.
Luminescence Assays - Dual Luciferase Reporters
5–10 mL of exponentially growing culture (defined media –Ura) was collected. The cell pellet was washed with TE and resuspended in 100–200 μL of passive lysis buffer (Promega). Lysis was accomplished by adding glass beads (~50 μL) to the sample and vortexing 5 × for a minute each time at high speed, with incubation on ice in between each interval. The lysate was clarified by centrifugation and diluted 30 to 50-fold. Luminescence was measured using the Dual-Luciferase Reporter Assay System (Promega) on a Tecan plate reader equipped with an automated injection system.
Luminescence Assays - GFP + Dual Luciferase Reporters
Overnight cultures were grown to saturation in 0.5 mL media in 96-well deep well plates, diluted 1:100 into 1 mL media and grown to OD ~0.5–0.8. Cells were then pelleted, washed once with sterile water and pelleted again. 25 μL of 1 mg/mL zymolase in buffer [50 mM Tris-Cl (pH 7.5); 10 mM MgCl2; 1 M sorbitol; 30 mM DTT] was added per well, shaken briefly to mix, and incubated at 37C for 30 minutes. Cells were lysed by adding 250 μL 1X Passive Lysis Buffer (Promega) and shaking vigorously for 10 minutes. After brief centrifugation, 50 μL of lysate was added to each well of a 96-well plate. GFP fluorescence was measured using a Tecan plate reader (485 nM excitation/530 nM emission) immediately followed by luminescence measurement using the Dual Luciferase Reporter Assay system (Promega).
For all luciferase reporter assays, experiments were run in triplicate and the data was analyzed and plotted using Graphpad Prism.
Quantitative RT-PCR
Total RNA from different yeast cells was isolated following the hot phenol method. cDNA was then generated with M-MuLV reverse transcriptase (Promega) from 2ug of total RNA that was treated with DNase 1 (Themo Fisher Scientific). Quantitative RT-PCR was conducted by using iTaq Universal SYBR Green Supermix (BIO-RAD) with ~50ng of cDNA. Relative fold change was obtained by following the ΔΔCt method between the RL and FL PCR products.
In vitro translation assays
Assays were performed using the PUREexpress in vitro protein translation kit (NEB), according to the manufacturer’s protocol.
Western blotting
Proteins were isolated using NaOH/TCA and were resuspended in HU buffer (8 M Urea, 5% SDS, 200 mM Tris pH 6.8, 100 mM DTT). Proteins were resolved on 12% SDS-PAGE gels and transferred to PVDF membrane using a semi-dry transfer apparatus (BioRad). Membranes were blocked in 5% milk/PBST for ~30 minutes at room temperature followed by incubation with primary antibody overnight at 4°C. After washing with PBST, the membrane was incubated with the appropriate HRP-conjugated secondary antibody for ~1hr at room temperature before washing 3–4 × with PBST. Detection was carried out on a GE ImageQuant LAS 4000 using Pierce SuperSignal West Pico Chemiluminescent Substrate. The following antibodies were used: mouse anti-FLAG [M2] from Sigma-Aldrich; HA-probe antibody, Y-11 (Santa Cruz, sc-805), Anti-Renilla Luciferase Antibody, clone 1D5.2 (milipore, MAB4410), Anti-Firefly Luciferase antibody (Abcam, ab21176) rabbit anti-eRF1 was a gift from R. Green (Eyler et al., 2013); goat anti-mouse IgG HRP (31430) and goat anti-rabbit IgG HRP (31460) from Thermo Scientific.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data for luciferase reporter experiments was analyzed and plotted using Graphpad Prism. In all figures the mean ± SD is shown with n representing the number of biological replicates that were performed. Additional details and definition of significance, where calculated, can be found in the figure legends. For Figures 4D–4F, SD for x values were calculated in Excel before plotting in Prism.
DATA AND CODE AVAILABILITY
Raw data has been deposited to Mendeley Data and is available at https://doi.org/10.17632/vbv4c9jwnj.2.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-FLAG [M2] | Sigma | Cat#: F3165; AB_259529 |
| Rabbit HA probe [Y-11] | Santa Cruz | Cat#: sc-805; AB_631618 |
| Mouse anti-Renilla luciferase 1D5.2 | Millipore Sigma | Cat#: MAB4410; AB_95102 |
| Rabbit anti-Firefly luciferase | Abcam | Cat#: ab21176; AB_446076 |
| Goat anti-mouse IgG(H+L) HRP | Thermo Fisher Scientific | Cat#: 31430; AB_228307 |
| Goat anti-rabbit IgG(H+L) HRP | Thermo Fisher Scientific | Cat#: 31460; AB_228341 |
| Rabbit anti-eRF1 | Gift from R.Green (Eyler et al., 2013) | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| BglII | NEB | Cat#: R0144 |
| AscI | NEB | Cat#: R0558 |
| SbfI | NEB | Cat#: R0642 |
| Phusion High Fidelity DNA Polymerase | NEB | Cat#: M0530 |
| DNase I | Thermo Scientific | Cat#: 89836 |
| M-MuLV reverse transcriptase | Promega | Cat#: M1701 |
| BamHI | NEB | Cat#: R0136 |
| iTaq Universal SYBR Green Supermix | Bio-rad | Cat#: 1725121 |
| Random hexamer | Invitrogen | Cat#: SO142 |
| Supersignal West Pico Chemiluminescent Substrate | Thermo Scientific | Cat#: 30480 |
| Passive Lysis Buffer | Promega | Cat#: E194A |
| XbaI | NEB | Cat#: R0145 |
| XhoI | NEB | Cat#: R0146 |
| AgeI | NEB | Cat#: R0552 |
| Critical Commercial Assays | ||
| PURExpress in vitro protein translation kit | NEB | Cat#: E6840 |
| Dual Luciferase Reporter Assay system | Promega | Cat#: E1960 |
| Deposited Data | ||
| Mendeley data: Raw data and blot files | This work | Mendeley data:https://doi.org/10.17632/vbv4c9jwnj.2 |
| Experimental Models: Organisms/Strains | ||
| Yeast Strains | See Table S2 | n/a |
| Oligonucleotides | ||
| DNA oligos | See Table S3 | n/a |
| Recombinant DNA | ||
| pDB688 | Salas-Marco and Bedwell, 2005 | n/a |
| pRL-(AGA)4-FL | This work | n/a |
| pRL-(CGA)4-FL | This work | n/a |
| pRL-(AGA)4+1-FL | This work | n/a |
| pRL-(CGA)4+1-FL | This work | n/a |
| pRL-(CGAATA)3+1-FL | This work | n/a |
| pRL-(CGACCG)3+1-FL | This work | n/a |
| pRL-(AGAATT)3+1-FL | This work | n/a |
| pRL-Ty1-FL | This work | n/a |
| pRL-FL-K529F | This work | n/a |
| pRL-FL-K529R | This work | n/a |
| pRL-FL-K529Q | This work | n/a |
| pRL-FL-K529X | This work | n/a |
| pGPD-GFP-PGK-RL-(AGA)4+1_3-FL | This work | n/a |
| pGPD-GFP-PGK-RL-(CGA)4+1_3-FL | This work | n/a |
| pGPD-GFP-PGK-RL-(CGA)4+1_9-FL | This work | n/a |
| pGPD-GFP-PGK-RL-(CGA)4+1_22-FL | This work | n/a |
| pGPD-GFP-PGK-RL-(CGA)4+1_41-FL | This work | n/a |
| pGPD-GFP-PGK-RL-(CGA)4+1_61-FL | This work | n/a |
| pGPD-GFP-PGK-RL-(CGA)4+1_83-FL | This work | n/a |
| pGPD-GFP-PGK-RL-(CGA)4+1_469-FL | This work | n/a |
| pGPD-GFP-PGK-RL-Ty1_3-FL | This work | n/a |
| pGPD-GFP-PGK-RL-Ty1_22-FL | This work | n/a |
| pGPD-GFP-PGK-RL-Ty1_41-FL | This work | n/a |
| pGPD-GFP-PGK-RL-Ty1_61-FL | This work | n/a |
| pGPD-GFP-PGK-RL-Ty1_83-FL | This work | n/a |
| pGPD-GFP-PGK-RL-Ty1_429-FL | This work | n/a |
| pGPD-GFP-PGK-SL1-RL-(CGA)4+1-FL | This work | n/a |
| pGPD-GFP-PGK-SL2-RL-(CGA)4+1-FL | This work | n/a |
| pGPD-GFP-PGK-SL3-RL-(CGA)4+1-FL | This work | n/a |
| pGPD-GFP-PGK-SL4-RL-(CGA)4+1-FL | This work | n/a |
| pGPD-GFP-PGK-SL5-RL-(CGA)4+1-FL | This work | n/a |
| pGPD-GFP-PGK-RL-(CGA)4+1-FL | This work | n/a |
| pGPD-GFP-PDC1-RL-(CGA)4+1-FL | This work | n/a |
| pGPD-GFP-YGL010W-RL-(CGA)4+1-FL | This work | n/a |
| pGPD-GFP-YLR327C-RL-(CGA)4+1 -FL | This work | n/a |
| pGPD-GFP-YFR053C-RL-(CGA)4+1-FL | This work | n/a |
| pGPD-GFP-ADY2-RL-(CGA)4+1-FL | This work | n/a |
| pGPD-GFP-YDL055C-RL-(CGA)4+1-FL | This work | n/a |
| pGPD-GFP-SSP2-RL-(CGA)4+1-FL | This work | n/a |
| pGPD-GFP-SEC61-RL-(CGA)4+1-FL | This work | n/a |
| pGPD-GFP-YPL154C-RL-(CGA)4+1-FL | This work | n/a |
| pGPD-GFP-YRO2-RL-(CGA)4+1-FL | This work | n/a |
| pGPD-GFP-YDL224C-RL-(CGA)4+1-FL | This work | n/a |
| pGPD-GFP-RSP5-RL-(CGA)4+1-FL | This work | n/a |
| pGPD-GFP-YGL215W-RL-(CGA)4+1-FL | This work | n/a |
| pGPD-GFP-SCH9-RL-(CGA)4+1-FL | This work | n/a |
| pGPD-GFP-ATG1 -RL-(CGA)4+1-FL | This work | n/a |
| pGPD-GFP-RPO21 -RL-(CGA)4+1-FL | This work | n/a |
| pGPD-GFP-YDR096W-RL-(CGA)4+1 -FL | This work | n/a |
| pGPD-GFP-CLB2-RL-(CGA)4+1-FL | This work | n/a |
| pGPD-GFP-YPL184C-RL-(CGA)4+1-FL | This work | n/a |
| pGPD-GFP-YNR051C-RL-(CGA)4+1-FL | This work | n/a |
| pGPD-GFP-YHR082C-RL-(CGA)4+1-FL | This work | n/a |
| pPROEX-Htb | Thermo Scientific | n/a |
| pGPD-GFP-PGK-RL-Ty1-FL | This work | n/a |
| pGPD-GFP-PDC1-RL-Ty1-FL | This work | n/a |
| pGPD-GFP-YGL010W-RL-Ty1-FL | This work | n/a |
| pGPD-GFP-YLR327C-RL-Ty1-FL | This work | n/a |
| pGPD-GFP-YFR053C-RL-Ty1-FL | This work | n/a |
| pGPD-GFP-ADY2-RL-Ty1-FL | This work | n/a |
| pGPD-GFP-YDL055C-RL-Ty1-FL | This work | n/a |
| pGPD-GFP-SSP2-RL-Ty1-FL | This work | n/a |
| pGPD-GFP-SEC61-RL-Ty1-FL | This work | n/a |
| pGPD-GFP-YPL154C-RL-Ty1-FL | This work | n/a |
| pGPD-GFP-YRO2-RL-Ty1-FL | This work | n/a |
| pGPD-GFP-YDL224C-RL-Ty1-FL | This work | n/a |
| pGPD-GFP-RSP5-RL-Ty1-FL | This work | n/a |
| pGPD-GFP-YGL215W-RL-Ty1-FL | This work | n/a |
| pGPD-GFP-SCH9-RL-Ty1-FL | This work | n/a |
| pGPD-GFP-ATG1-RL-Ty1-FL | This work | n/a |
| pGPD-GFP-RPO21-RL-Ty1-FL | This work | n/a |
| pGPD-GFP-YDR096W-RL-Ty1-FL | This work | n/a |
| pGPD-GFP-CLB2-RL-Ty1-FL | This work | n/a |
| pGPD-GFP-YPL184C-RL-Ty+1-FL | This work | n/a |
| pGPD-GFP-YNR051C-RL-Ty1-FL | This work | n/a |
| pGPD-GFP-YHR082C-RL-Ty1-FL | This work | n/a |
| pAG415-GPD-ccdB | Addgene; Alberti et al., 2007 | Cat#: 14146 |
| pEKD1024 | Wolf and Grayhack, 2015 | n/a |
| p-FA6a-6XGLY-FLAG-HIS3 | Addgene; Funakoshi and Hochstrasser, 2009 | Cat#: 20750 |
Highlights.
Mutations that inhibit NGD promote +1 frameshifting on stalling sequences
+1 frameshifting efficiency depends critically on the stallsequence location
Higher translation initiation rates lead to increased +1 frameshifting
Ribosome bumping is required to promote +1 frameshifting in a reconstituted system
ACKNOWLEDGMENTS
The authors wish to thank Allen Buskirk, Joe Jez, Doug Chalker, Nima Mosammaparast, Bob Kranz, and members of the Zaher laboratory for comments on earlier versions of the manuscript. This work was supported by the NIH (NIH R01GM112641 to H.S.Z.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.07.046.
REFERENCES
- Alberti S, Gitler AD, and Lindquist S (2007). A suite of Gateway cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. Yeast 24, 913–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins JF, and Björk GR (2009). A gripping tale of ribosomal frameshifting: extragenic suppressors of frameshift mutations spotlight P-site realignment. Microbiol. Mol. Biol. Rev 73, 178–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azvolinsky A, Giresi PG, Lieb JD, and Zakian VA (2009). Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol. Cell 34, 722–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranov PV, Gesteland RF, and Atkins JF (2004). P-site tRNA is a crucial initiator of ribosomal frameshifting. RNA 10, 221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belardinelli R, Sharma H, Caliskan N, Cunha CE, Peske F, Wintermeyer W, and Rodnina MV (2016). Choreography of molecular movements during ribosome progression along mRNA. Nat. Struct. Mol. Biol. 23, 342–348. [DOI] [PubMed] [Google Scholar]
- Belcourt MF, and Farabaugh PJ (1990). Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7 nucleotide minimal site. Cell 62, 339–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, and Yusupov M (2011). The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 334, 1524–1529. [DOI] [PubMed] [Google Scholar]
- Bengtson MH, and Joazeiro CAP (2010). Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature 467, 470–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O, and Hegde RS (2016). Ribosome-associated protein quality control. Nat. Struct. Mol. Biol 23, 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O, Stewart-Ornstein J, Wong D, Larson A, Williams CC, Li G-W, Zhou S, King D, Shen PS, Weibezahn J, et al. (2012). A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell 151, 1042–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt F, Etchells SA, Ortiz JO, Elcock AH, Hartl FU, and Baumeister W (2009). The native 3D organization of bacterial polysomes. Cell 136, 261–271. [DOI] [PubMed] [Google Scholar]
- Brandt F, Carlson LA, Hartl FU, Baumeister W, and Grünewald K (2010). The three-dimensional organization of polyribosomes in intact human cells. Mol. Cell 39, 560–569. [DOI] [PubMed] [Google Scholar]
- Brilot AF, Korostelev AA, Ermolenko DN, and Grigorieff N (2013). Structure of the ribosome with elongation factor G trapped in the pretranslocation state. Proc. Natl. Acad. Sci. USA 110, 20994–20999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyle SM, Gilbert WV, and Doudna JA (2009). Direct link between RACK1 function and localization at the ribosome in vivo. Mol. Cell. Biol. 29, 1626–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigen WJ, and Caskey CT (1986). Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature 322, 273–275. [DOI] [PubMed] [Google Scholar]
- Defenouille re, Q., Yao Y, Mouaikel J, Namane A, Galopier A, Decourty L, Doyen A, Malabat C, Saveanu C, Jacquier A, and Fromont-Racine M. (2013). Cdc48-associated complex bound to 60S particles is required for the clearance of aberrant translation products. Proc. Natl. Acad. Sci. USA 110, 5046–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaraj A, and Fredrick K (2010). Short spacing between the Shine-Dalgarno sequence and P codon destabilizes codon-anticodon pairing in the P site to promote +1 programmed frameshifting. Mol. Microbiol 78, 1500–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrzynski M, and Bruggeman FJ (2009). Elongation dynamics shape bursty transcription and translation. Proc. Natl. Acad. Sci. USA 106, 2583–2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma MK, and Parker R (2006). Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 440, 561–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyler DE, Wehner KA, and Green R (2013). Eukaryotic release factor 3 is required for multiple turnovers of peptide release catalysis by eukaryotic release factor 1. J. Biol. Chem 288, 29530–29538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A, and Bulmer M (1993). Reduced synonymous substitution rate at the start of enterobacterial genes. Nucleic Acids Res. 21, 4599–4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh PJ, Kramer E, Vallabhaneni H, and Raman A (2006). Evolution of +1 programmed frameshifting signals and frameshift-regulating tRNAs in the order Saccharomycetales. J. Mol. Evol 63, 545–561. [DOI] [PubMed] [Google Scholar]
- Funakoshi M, and Hochstrasser M (2009). Small epitope-linker modules for PCR-based C-terminal tagging in Saccharomyces cerevisiae. Yeast 26, 185–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble CE, Brule CE, Dean KM, Fields S, and Grayhack EJ (2016). Adjacent codons act in concert to modulate translation efficiency in yeast. Cell 166, 679–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Muse T, and Aguilera A (2016). Transcription-replication conflicts: how they occur and how they are resolved. Nat. Rev. Mol. Cell Biol. 17, 553–563. [DOI] [PubMed] [Google Scholar]
- Garzia A, Jafarnejad SM, Meyer C, Chapat C, Gogakos T, Morozov P, Amiri M, Shapiro M, Molina H, Tuschl T, and Sonenberg N (2017). The E3 ubiquitin ligase and RNA-binding protein ZNF598 orchestrates ribosome quality control of premature polyadenylated mRNAs. Nat. Commun. 8, 16056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurvich OL, Baranov PV, Zhou J, Hammer AW, Gesteland RF, and Atkins JF (2003). Sequences that direct significant levels of frameshifting are frequent in coding regions of Escherichia coli. EMBO J. 22, 5941–5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmrich A, Ballarino M, Nudler E, and Tora L (2013). Transcription-replication encounters, consequences and genomic instability. Nat. Struct. Mol. Biol 20, 412–418. [DOI] [PubMed] [Google Scholar]
- Hendrick JL, Wilson PG, Edelman II, Sandbaken MG, Ursic D, and Culbertson MR (2001). Yeast frameshift suppressor mutations in the genes coding for transcription factor Mbf1p and ribosomal protein S3: evidence for autoregulation of S3 synthesis. Genetics 157, 1141–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG, Ivanov IP, and Sonenberg N (2016). Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science 352, 1413–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi K, and Inada T (2016). Ribosome-associated Asc1/RACK1 is required for endonucleolytic cleavage induced by stalled ribosome at the 3′ end of nonstop mRNA. Sci. Rep 6, 28234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi K, Tesina P, Matsuo Y, Sugiyama T, Cheng J, Saeki Y, Tanaka K, Becker T, Beckmann R, and Inada T (2019). Collided ribosomes form a unique structural interface to induce Hel2-driven quality control pathways. EMBO J 38, e100276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juszkiewicz S, and Hegde RS (2017). Initiation of quality control during poly(A) translation requires site-specific ribosome ubiquitination. Mol. Cell 65, 743–750.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juszkiewicz S, Chandrasekaran V, Lin Z, Kraatz S, Ramakrishnan V, and Hegde RS (2018). ZNF598 is a quality control sensor of collided ribosomes. Mol. Cell 72, 469–481.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K, Pande S, Faiola B, Moore DP, Boeke JD, Farabaugh PJ, Strathern JN, Nakamura Y, and Garfinkel DJ (1993). A rare tRNA-Arg(CCU) that regulates Ty1 element ribosomal frameshifting is essential for Ty1 retrotransposition in Saccharomyces cerevisiae. Genetics 135, 309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennell D, and Riezman H (1977). Transcription and translation initiation frequencies of the Escherichia coli lac operon. J. Mol. Biol 114, 1–21. [DOI] [PubMed] [Google Scholar]
- Kierzek AM, Zaim J, and Zielenkiewicz P (2001). The effect of transcription and translation initiation frequencies on the stochastic fluctuations in prokaryotic gene expression. J. Biol. Chem. 276, 8165–8172. [DOI] [PubMed] [Google Scholar]
- Kramer EB, and Farabaugh PJ (2007). The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA 13, 87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroha K, Akamatsu M, Dimitrova L, Ito T, Kato Y, Shirahige K, and Inada T (2010). Receptor for activated C kinase 1 stimulates nascent polypeptide-dependent translation arrest. EMBO Rep. 11, 956–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letzring DP, Dean KM, and Grayhack EJ (2010). Control of translation efficiency in yeast by codon-anticodon interactions. RNA 16, 2516–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letzring DP, Wolf AS, Brule CE, and Grayhack EJ (2013). Translation of CGA codon repeats in yeast involves quality control components and ribosomal protein L1. RNA 19, 1208–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Márquez V, Wilson DN, Tate WP, Triana-Alonso F, and Nierhaus KH (2004). Maintaining the ribosomal reading frame: the influence of the E site during translational regulation of release factor 2. Cell 118, 45–55. [DOI] [PubMed] [Google Scholar]
- Matsuo Y, Ikeuchi K, Saeki Y, Iwasaki S, Schmidt C, Udagawa T, Sato F, Tsuchiya H, Becker T, Tanaka K, et al. (2017). Ubiquitination of stalled ribosome triggers ribosome-associated quality control. Nat. Commun 8, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrikh H, Machón C, Grainger WH, Grossman AD, and Soultanas P (2011). Co-directional replication-transcription conflicts lead to replication restart. Nature 470, 554–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitarai N, Sneppen K, and Pedersen S (2008). Ribosome collisions and translation efficiency: optimization by codon usage and mRNA destabilization. J. Mol. Biol 382, 236–245. [DOI] [PubMed] [Google Scholar]
- Moazed D, and Noller HF (1989). Intermediate states in the movement of transfer RNA in the ribosome. Nature 342, 142–148. [DOI] [PubMed] [Google Scholar]
- Noller HF, Lancaster L, Zhou J, and Mohan S (2017). The ribosome moves: RNA mechanics and translocation. Nat. Struct. Mol. Biol 24, 1021–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pédelacq JD, Cabantous S, Tran T, Terwilliger TC, and Waldo GS (2006). Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol 24, 79–88. [DOI] [PubMed] [Google Scholar]
- Poli J, Gerhold CB, Tosi A, Hustedt N, Seeber A, Sack R, Herzog F, Pasero P, Shimada K, Hopfner KP, and Gasser SM (2016). Mec1, INO80, and the PAF1 complex cooperate to limit transcription replication conflicts through RNAPII removal during replication stress. Genes Dev. 30, 337–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado F, and Aguilera A (2005). Impairment of replication fork progression mediates RNA polII transcription-associated recombination. EMBO J. 24, 1267–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulk A, and Cate JHD (2013). Control of ribosomal subunit rotation by elongation factor G. Science 340, 1235970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramrath DJF, Lancaster L, Sprink T, Mielke T, Loerke J, Noller HF, and Spahn CMT (2013). Visualization of two transfer RNAs trapped in transit during elongation factor G-mediated translocation. Proc. Natl. Acad. Sci. USA 110, 20964–20969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas-Marco J, and Bedwell DM (2005). Discrimination between defects in elongation fidelity and termination efficiency provides mechanistic insights into translational readthrough. J. Mol. Biol. 348, 801–815. [DOI] [PubMed] [Google Scholar]
- Simms CL, and Zaher HS (2016). Quality control of chemically damaged RNA. Cell Mol. Life Sci. 73, 3639–3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms CL, Thomas EN, and Zaher HS (2017). Ribosome-based quality control of mRNA and nascent peptides. Wiley Interdiscip. Rev. RNA 8, 10.1002/wrna.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms CL, Yan LL, and Zaher HS (2017a). Ribosome collision is critical for quality control during no-go decay. Mol. Cell 68, 361–373.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms CL, Kim KQ, Yan LL, Qiu J, and Zaher HS (2018). Interactions between the mRNA and Rps3/uS3 at the entry tunnel of the ribosomal small subunit are important for no-go decay. PLoS Genet. 14, e1007818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitron CS, Park JH, and Brandman O (2017). Asc1, Hel2, and Slh1 couple translation arrest to nascent chain degradation. RNA 23, 798–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siwiak M, and Zielenkiewicz P (2013). Transimulation - protein biosynthesis web service. PLoS ONE 8, e73943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen MA, and Pedersen S (1991). Absolute in vivo translation rates of individual codons in Escherichia coli. The two glutamic acid codons GAA and GAG are translated with a threefold difference in rate. J. Mol. Biol. 222, 265–280. [DOI] [PubMed] [Google Scholar]
- Sundaramoorthy E, Leonard M, Mak R, Liao J, Fulzele A, and Bennett EJ (2017). ZNF598 and RACK1 regulate mammalian ribosome-associated quality control function by mediating regulatory 40S ribosomal ubiquitylation. Mol. Cell 65, 751–760.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tehranchi AK, Blankschien MD, Zhang Y, Halliday JA, Srivatsan A, Peng J, Herman C, and Wang JD (2010). The transcription factor DksA prevents conflicts between DNA replication and transcription machinery. Cell 141, 595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourigny DS, Fernández IS, Kelley AC, and Ramakrishnan V (2013). Elongation factor G bound to the ribosome in an intermediate state of translocation. Science 340, 1235490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi T, Kuroha K, Kudo K, Makino S, Inoue E, Kashima I, and Inada T (2012). Dom34:hbs1 plays a general role in quality-control systems by dissociation of a stalled ribosome at the 30 end of aberrant mRNA. Mol. Cell 46, 518–529. [DOI] [PubMed] [Google Scholar]
- Tuduri S, Crabbé L, Conti C, Tourrière H, Holtgreve-Grez H, Jauch A, Pantesco V, De Vos J, Thomas A, Theillet C, et al. (2009). Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat. Cell Biol. 11, 1315–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller T, Carmi A, Vestsigian K, Navon S, Dorfan Y, Zaborske J, Pan T, Dahan O, Furman I, and Pilpel Y (2010). An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell 141, 344–354. [DOI] [PubMed] [Google Scholar]
- Verma R, Oania RS, Kolawa NJ, and Deshaies RJ (2013). Cdc48/p97 promotes degradation of aberrant nascent polypeptides bound to the ribosome. eLife 2, e00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Reichermeier KM, Burroughs AM, Oania RS, Reitsma JM, Aravind L, and Deshaies RJ (2018). Vms1 and ANKZF1 peptidyl-tRNA hydrolases release nascent chains from stalled ribosomes. Nature 557, 446–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhees RM, and Ramakrishnan V (2013). Structural basis of the translational elongation cycle. Annu. Rev. Biochem. 82, 203–236. [DOI] [PubMed] [Google Scholar]
- Wang J, Zhou J, Yang Q, and Grayhack EJ (2018). Multi-protein bridging factor 1(Mbf1), Rps3 and Asc1 prevent stalled ribosomes from frameshifting. eLife 7, e39637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR, Knopf PM, and Rich A (1963). A multiple ribosomal structure in protein synthesis. Proc. Natl. Acad. Sci. USA 49, 122–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winz ML, Peil L, Turowski TW, Rappsilber J, and Tollervey D (2019). Molecular interactions between Hel2 and RNA supporting ribosome-associated quality control. Nat. Commun. 10, 563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf AS, and Grayhack EJ (2015). Asc1, homolog of human RACK1, prevents frameshifting in yeast by ribosomes stalled at CGA codon repeats. RNA 21, 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Lancaster L, Donohue JP, and Noller HF (2013). Crystal structures of EF-G-ribosome complexes trapped in intermediate states of translocation. Science 340, 1236086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Lancaster L, Donohue JP, and Noller HF (2014). How the ribosome hands the A-site tRNA to the P site during EF-G-catalyzed translocation. Science 345, 1188–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data has been deposited to Mendeley Data and is available at https://doi.org/10.17632/vbv4c9jwnj.2.