Abstract
Glutamate-gated AMPA receptors mediate the fast component of excitatory signal transduction at chemical synapses throughout all regions of the mammalian brain. AMPA receptors are tetrameric assemblies composed of four subunits, GluA1–4. Despite decades of study, the subunit composition, subunit arrangement and molecular structure of native AMPA receptors remain unknown. Here we elucidate the structures of 10 distinct native AMPA receptor complexes by single particle cryo-EM. We find that receptor subunits are arranged non stochastically, with the GluA2 subunit preferentially occupying the B and D positions of the tetramer and with triheteromeric assemblies comprising a major population of native AMPA receptors. Cryo-EM maps define the structure for S2-M4 linkers between the ligand binding and transmembrane domains, suggesting how neurotransmitter binding is coupled to ion channel gating.
One Sentence Summary
Cryo-EM illuminates the subunit composition and spatial configuration of native AMPA receptors in the mammalian brain.
AMPA receptors (AMPARs), which are expressed throughout the central and peripheral nervous systems are glutamate-activated, cation permeable ion channels that mediate the majority of fast excitatory signaling (1–3). AMPARs are essential to the genetically-encoded development of native circuitry in the nervous system, and they also underpin experience-dependent strengthening or weakening of signaling at synapses and, in turn, multiple forms of learning and memory (4, 5). Rapid, repetitive and synapse-specific AMPAR activation results in postsynaptic membrane depolarization, voltage-dependent block of NMDA receptors and ultimately entry of calcium and initiation of downstream signaling events (6). Perturbation of AMPAR signaling can disrupt region-specific signaling or wide spread neuronal homeostasis, and thus AMPARs are implicated in a range of nervous system disorders and diseases (7–11).
AMPARs are tetrameric receptor assemblies that display functional diversity arising from four distinct subunits, GluA1-A4, posttranscriptional splicing and editing, and multiple forms of posttranslational modification. Together these variations yield glutamate gated ion channels with a range of gating kinetics, ion channel permeation properties and pharmacological sensitivities (1, 12–15). AMPARs also assemble with many auxiliary subunits, including TARPs and cornichons, that further modulate receptor properties (16–20). The GluA2 subunit is central to AMPAR function, not only because it is widely expressed throughout the brain, but also because most GluA2 RNA transcripts undergo editing, in which a pore-lining ‘Q’ residue becomes a cationic ‘R’, resulting in calcium impermeable ion channels (20, 21). Immunoprecipitation (22) and single cell genetic (23) studies, suggest that diheteromeric GluA1/GluA2 and GluA2/GluA3 receptors are the most common assemblies in the mammalian brain. Despite the advances in AMPAR studies, however, the previous experiments largely involve averages over ensembles of receptor populations and thus the actual subunit stoichiometry, subunit arrangement and molecular structures of individual, native AMPARs (nAMPARs) remain unknown.
The first structural studies of an intact recombinant AMPAR showed that the 4 GluA2 subunits were arranged with overall 2-fold symmetry, with the extracellular amino terminal domains (ATDs) and the ligand binding domains (LBDs) organized as a dimer-of-dimers (24). In resting and non desensitized activated states, AMPARs adopt a capital ‘Y’ like shape, with broad and narrow ‘faces’ (25, 26). More recently, however, structures of a recombinant heteromeric GluA2/GluA3 receptor show an ‘O’ like shape, more reminiscent of NMDA receptor conformation, with GluA2 and GluA3 subunits in the A/C and B/D subunit positions, respectively (27). To faithfully define the subunit stoichiometry and arrangement of nAMPARs, and to map the positions of auxiliary subunits, we isolated native receptors, tagged GluA1-GluA3 subunits with specific antibody fragments, and elucidated reconstructions of 10 distinct nAMPARs by cryo-EM.
Characterization of antibody fragments against AMPAR subunits
To elucidate the architecture of nAMPARs, we employed subunit-specific antibody fragments to isolate receptor complexes and to label specific subunits. We generated three monoclonal antibodies (mAb), 11B8, 15F1 (28) and 5B2 against the GluA1, GluA2 or GluA3 subunits, respectively. To isolate the nAMPAR complex from rat brain, we employed the 11B8 single chain variable domain (scFv) and the 15F1 Fab, both coupled to carboxy-terminal affinity tags. The 5B2 Fab was produced by papain cleavage of purified mAb. We next carried out fluorescence-detection, size-exclusion chromatography experiments (FSEC) (29) using the scFv and Fabs, in combination with fluorescently labeled homomeric GluA1–4 receptors, to define antibody fragment subunit specificity and to estimate affinity. The antibody fragments form complexes with their cognate AMPAR subunits at nanomolar concentrations (fig. S1).
The nAMPAR complex is widely distributed in the CNS and consists of synaptic and extra-synaptic AMPARs (30, 31). Synaptic AMPARsare positioned in the postsynaptic density (PSD) by direct interactions between receptor auxiliary proteins and PSD-95, which is predominantly associated with PSD membranes (32). We first asked if digitonin- containing buffer, which allows for isolation of the recombinant AMPAR-TARP γ2 complex (25), releases synaptic AMPARs from the PSD. Following solubilization of membranes using digitonin, insoluble material was treated with solutions augmented by 1% Triton X-100 and 1% sodium deoxycholate (w/v), conditions that are routinely employed to solubilize the PSD (33, 34). Based on Western blot analysis, a substantial fraction of the PSD-95 from the membrane preparation was rendered soluble using digitonin (fig. S1H), suggesting that digitonin releases the synaptic AMPARs from PSD. We note that compared with the synaptic AMPARs, extrasynaptic AMPARs are not bound by synaptic scaffolding proteins and should be more readily extracted by mild detergents, such as digitonin. Thus we hypothesize that our purified sample is composed of synaptic and extra-synaptic AMPARs.
Mapping the GluA1 or GluA2 subunits in the nAMPAR complex
To define the locations of the GluA1 or GluA2 subunits in nAMPARs, the solubilized membranes were separately incubated with an excess of the StrepII tagged 11B8 scFv (anti-GluA1) or 15F1 Fab (anti-GluA2), respectively, without additional scFv/Fab labeling. GluA1 or GluA2-containing nAMPARs were isolated by affinity chromatography and further purified by size exclusion chromatography (SEC) for cryo-EM studies (fig. S2, A and J). For the GluA1-containing, 11B8 scFv single-tagged nAMPAR-11B8 complex, we observed densities protruding from two sides of the ATD layer in some of the 2D classes, due to bound 11B8 scFv (fig. S2, B and C). Three-dimensional (3D) classification yielded three classes featuring two 11B8 scFvs at the A and C positions, one 11B8 scFv at either the A or C position and the final class with a splayed ATD layer, with the 11B8 scFv not well defined (fig. S2, D, G and I). The other non 11B8 scFv labeled subunits could be GluA2, GluA3 or GluA4. The doubly-bound 11B8 scFv class accounted for 57% of total particles (table S1). Subsequent LBD-TMD focused classification resolved two different classes, with or without two auxiliary proteins associated with the receptor TMD at the B’-D’ positions underneath the B-D LBDs, but with density for the M2 helices weak or absent in both maps (fig. S2, E and F). This result is consistent with previous studies showing that fully occupied auxiliary subunits are essential to stabilize M2 helices (25). We also carried out ATD-focused refinement for the singly-bound 11B8 scFv class and obtained a map at ~8.0 Å resolution. To explore the composition of the non scFv bound A/C subunit of this complex, the structures of all four GluA1–4 ATDs were separately docked into the density at the non scFv bound A/C position of the complex. Upon inspecting the fit of the structures to the maps, we found that the GluA3 or GluA4 ATDs are more compatible with map features, especially at predicted glycosylation sites involving Asn35 and Asn352 in GluA3 (or Asn35 and Asn350 in GluA4; fig. S2, G–H). We thus conclude that the GluA1 subunit tends to occupy the A or C sites in the receptor and that, in some of the nAMPARs composed of a single GluA1 subunit, there is a GluA3 or a GluA4 subunit at the remaining A/C position.
In the GluA2-containing, 15F1 Fab single tagged nAMPAR-15F1 Fab complex, particles on micrographs and the 2D classes show 15F1 Fab densities protruding from the middle of the ATD layer and perpendicular to the membrane, suggesting that the GluA2 subunit occupies different positions in the nAMPARs in comparison to the GluA1 subunit (fig. S2, K–L). Further 3D classification generated three classes. Class I is the largest population, with two 15F1 Fabs at the B and D positions and GluA1, GluA3 or GluA4 at the A-C positions (fig. S2M and table S1). Here, the ATD layer is organized as a dimer-of-dimers and focused reconstruction gives rise to a 6.7 Å map. The secondary structural elements are well defined, especially for the GluA2 subunits at B and D positions. The ATD from a homomeric GluA2 crystal structure was docked into the density and all of the secondary structural elements agree well with the density map (fig. S2N) (24), demonstrating that the GluA2 ATD dimer-dimer interface is conserved between homomeric receptors and the native heteromeric receptor. We also observed two small populations featuring 15F1 Fab labeling at the A, B and D or A and D positions (fig. S2, O and P, table S1).
Architecture and symmetry of GluA2-containing nAMPARs
To unambiguously explore subunit composition and spatial arrangement of the GluA2 containing 11B8/15F1/5B2 tagged nAMPARs, nAMPARs were isolated using the 15F1 Fab from digitonin-solubilized material, and then incubated with an excess of 11B8 scFv and 5B2 Fab, followed by SEC (Fig. 1, A and B) and cryo-EM grid preparation. Western blots confirmed the presence of the GluA1–4 subunits, as well as PSD-95, in the final sample (Fig. 1C). Radiolabel binding studies using 3H AMPA shows that the purified nAMPAR harbors active agonist binding sites (Fig. 1D).
Fig. 1. nAMPAR purification and biochemical analysis.
(A) Flow chart for nAMPAR isolation. (B) Representative SEC profile of the nAMPAR complex. Inset shows a SDS-PAGE of a nAMPAR sample used for cryo-EM grid preparation and of antibody fragments, visualized by Coomassie blue staining. (C) Western blot analysis of isolated nAMPAR blotted using antibodies against GluA1, GluA2, GluA3, GluA4 or PSD-95. (D) Single point SPA radio-ligand binding assay of 3H-AMPA binding to the nAMPAR complex. Non specific binding was estimated by the addition of 10 mM cold glutamate (right column). Data are shown as means ± SEM (n=3). (E) nAMPAR subunit composition determination by LC-SRM. Each point represents the peptide-level quantitation shown as percent composition of the original sample. Analysis shows both the median with standard deviation (box plot) and the mean (diamond plot) for each protein subunit after protein-level roll-up.
To independently estimate the relative abundance of each of the AMPAR subunits, we carried out liquid chromatography-selected reaction monitoring (LC-SRM) mass spectrometry experiments, using three surrogate peptides per subunit and heavy isotope-labeled peptide internal standards (fig. S3). Our results indicated that in all preparations the GluA2 subunit was most abundant. This was followed by GluA1 with a roughly 2:3 ratio compared to GluA2, with the GluA3 and GluA4 subunits in overall lower, yet approximately equivalent, abundance (Fig. 1E).
We next measured a relatively large single particle data set on the GluA2-containing 11B8/15F1/5B2 tagged nAMPAR complex, purified using the 15F1 Fab (anti-GluA2) and incubated with the antagonist MPQX, the 11B8 scFv (anti-GluA1) and the 5B2 Fab (anti-GluA3). 2D classification revealed 11B8 scFv and 15F1 Fab features (fig. S4), consistent with observations derived from the nAMPAR-11B8 and nAMPAR-15F1 data sets (fig. S2, C and L). Additional 2D classes were resolved that featured a ‘crown-like’ ATD layer due to bound 5B2 Fab (fig. S4). All of the particles showing typical receptor features were subjected to 3D reconstruction followed by ATD-focused refinement and multiple rounds of 3D classification focused on the antibody fragments, ultimately resulting in ten distinct nAMPAR complexes (fig. S4). These 10 complexes are representative of nAMPARs across the whole brain and thus deconvolution of subunit composition and spatial arrangement of nAMPAR in specific brain regions will require further study.
In our cryo-EM analysis, GluA1/GluA2 or GluA2/GluA3 heteromers are predominant populations. For the GluA1/GluA2 complex, the major population has two GluA1 subunits at the A and C positions and two GluA2 subunits at the B and D positions. We thus define this complex as A1A2A1A2, where the subunits occupy the ABCD positions of the receptor, respectively (Fig. 2A) (24). A minor population is composed of a 1:3 heteromer of GluA1:GluA2, with GluA1 at the C position and GluA2 at the A-B-D positions, yielding a A2A2A1A2 complex (Fig. 2B). The GluA2/GluA3 complex was also determined with stoichiometries of either 2:2 or 3:1 that are the major A3A2A3A2 assembly and the minor A2A2A3A2 complex (Fig. 2, C and D).
Fig. 2. Cryo EM analysis of the nAMPAR complex.
(A-J) Cryo EM maps of the 10 resolved complexes, viewed parallel to the membrane. Three rules were applied to define subunit position: a) When GluA2 is present at the A/C position, we prioritize A; b) GluA1 is preferentially assigned to the A position; c) GluA3 is preferentially assigned to the C position. GluA1, GluA2, GluA3 and unidentified subunits are blue, red, orange and grey, respectively. The 11B8 scFv, 15F1 Fab and 5B2 Fab are light blue, pink and light cyan, respectively. A schematic cartoon in each panel shows the subunit arrangement of the ATD layer and of antibody fragment binding. (K) Complex distribution illustrated as a pie chart. (L) A pie chart, organized like the ATD layer, showing how the GluA1, GluA2, GluA3 and non-Fab/scFv bound subunit populate the A, B, C, or D positions. Assignment of each subunit position is derived from the view of nAMPAR shown in panel A-J.
In addition to diheteromeric nAMPARs, analysis of the nAMPAR-11B8–15F1–5B2 data set unambiguously revealed a previously unknown triheteromeric A1A2A3A2 complex, featuring an 11B8 scFv on one side and a 5B2 Fab on the other side of the ATD layer, in accord with class II in the nAMPAR-11B8 data set showing nonequivalent A and C subunits (fig. S2, G–H). Strikingly, this complex comprises the largest population of particles (25%; table S1, fig. S4). Interestingly, we also discovered a native homomeric GluA2 complex (Fig. 2F), accounting for 1.1% of total particles (table S1, fig. S4). We did not observe this complex in the nAMPAR-15F1 data set, likely due to the smaller size of the data set.
We also discovered four complexes that have a single non tagged subunit and that we define as A1A2AxA2, AxA2A3A2, A2AxA1A2 and A2AxA3A2, where ‘Ax’ represents a subunit not bound by an scFv/Fab domain (Fig. 2, G to J). These untagged subunits could be the result of a scFv/Fab domain dissociating from the complex or they could represent a GluA4 subunit. These complexes account for a substantial portion of total particles (~29%; Fig. 2K). Because FSEC experiments show that the scFv/Fab-receptor complex is stable, on the time-scale associated with receptor isolation (fig. S1, E to G), we suggest that a substantial fraction of the complexes harbor the GluA4 subunit. Thus, GluA4 might form four different triheteromeric nAMPARs with GluA2 and with GluA1 or GluA3. We note that GluA4 can also access the B position and interact with the GluA2 ATD to bridge the ATD dimer of dimers, unlike the GluA1 or GluA3 subunits, which appear to not substantially populate the B or D positions (Fig. 2L and table S2). Moreover, the AMPAR subunit distribution determined by cryo-EM is consistent with the results of mass spectrometry experiments (fig. S3N).
We next carried out focused classification and refinement on the three major complexes –A1A2A1A2, A3A2A3A2 and A1A2A3A2 – dissecting the structures into the scFv/Fab-ATD, ATD-LBD and LBD-TMD layers, and achieved reasonable maps (figs. S5, S6 and table S3). All of the glycosylation sites found in the crystal structures are compatible with the cryo-EM maps and thus the subunit specific glycosylation further bolsters subunit identification (fig. S7, A to C and G). Moreover, these scFv/Fab-ATD density maps unambiguously define how the antibody fragments bind to their cognate subunit. The residues involved in the ATD and scFv/Fab interfaces are spread throughout elements of secondary structure and are not conserved between subunits, consistent with the conclusion that the antibody fragments bind to conformational and subunit specific epitopes (fig. S7, D to G).
Our cryo-EM study requires that the scFv/Fab fragments access the requisite subunits at the A-B-C-D positions without steric hindrance. The 15F1 Fab (GluA2) can access all four positions because we observed the homomeric GluA2 structure with four bound Fabs (Fig. 2F). We next superimposed the 11B8 scFv – GluA1 ATD structure on the homomeric GluA2 ATD layer and found that one 11B8 scFv can access the B or D positions without steric clash and that there are only a few overlaps when two 11B8 scFvs occupy both the B and D positions (fig. S8, A and B). Notably, we did not observe any 11B8 labeling at the B or D positions. The superimposition of 5B2 Fab – GluA3 ATD on the homomeric GluA2 ATD layer demonstrates that the 5B2 Fab can access all four positions simultaneously (fig. S8C). We thus conclude that the locations of the GluA1 and GluA3 subunits at the A and C positions are not biased by sterically restricted 11B8 scFv or 5B2 Fab labeling (Fig. 2L). Interestingly, we do not find evidence, in the cryo-EM reconstructions, for extracellular auxiliary proteins, such as N-cadherin or pentraxin (35, 36), presumably because these proteins bind relatively weakly to the receptors and unbind during complex isolation.
Architecture of the GluA2/GluA3 complex
The major population of the nAMPAR GluA2/GluA3 complex has a capital ‘Y’ shape and consists of three layers – the ATD, LBD and TMD (Fig. 3A). The GluA3 subunits occupy the A-C positions and GluA2 subunits reside in the B-D positions, suggesting that the GluA2 subunit may play a greater role in gating (37). Like GluA2 homomers (24), the A3A2A3A2 complex displays a dimer-of-dimers organization of the ATD and LBD layers, with domain swapping from the ATD layer to the LBD layer. Within the ATD layer, the GluA3 subunits at the A and C positions form local ATD heterodimers with GluA2 subunits from the B and D positions, respectively. By contrast, in the LBD layer, GluA3 subunits in the A and C positions form local LBD heterodimers with GluA2 subunits from the D and B positions, respectively. Distinct from the 2-fold related ATD and LBD layers, the TMD ion channel adopts pseudo 4-fold symmetry, thus resulting in symmetry mismatch between the LBD and the TMD layers and generating two distinct subunit pairs, the proximal A-C and the distal B-D pairs (Fig. 3B). Like the A3A2A3A2 complex, all of the other complexes have a similar domain organization (Fig. 2, A to J).
Fig. 3. Architecture of the heteromeric GluA2/GluA3 AMPA receptor complex.
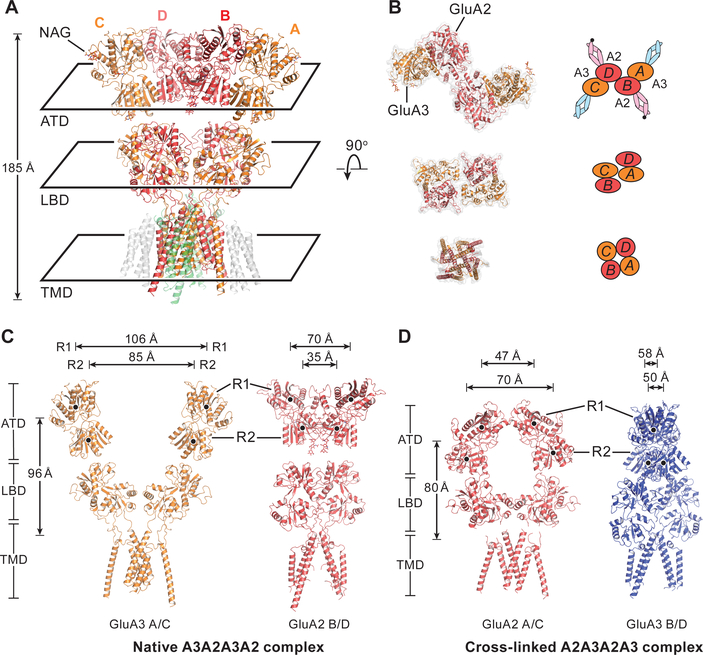
(A) Structure of the nAMPAR A3A2A3A2 complex. N-linked carbohydrates are shown as ‘sticks’. (B) ATD, LBD and TMD layers viewed from the ‘top’ of the receptor, along the overall 2-fold axis of symmetry, showing the ATD, LBD and TMD layers. (C-D) Cartoon representations of the A-C and B-D subunits of the nAMPAR A3A2A3A2 complex (C) or the cross-linked A2A3A2A3 complex (27) (D), viewed parallel to the membrane. COMs of R1 and R2 lobes are indicated by black dots. The separations of the R1 or R2 lobes from opposing subunits are labeled at the top of panels. The COM distances between T625 and the entire ATD layer is indicated on the left of the panel. GluA2 and GluA3 subunits of the recombinant cross-linked receptor are red and blue, respectively.
The structure of the nAMPAR GluA2/GluA3 complex provides an opportunity to compare it with the previously determined recombinant GluA2/GluA3 structure (rGluA2/GluA3) (27). Surprisingly, the rGluA2/GluA3 structure has the GluA2 subunit at the A-C positions and the GluA3 subunit at the B-D positions (Fig. 3D). Moreover, the conformation of the ATD layer is more compact (O shape), which brings the ATD closer to the membrane by ~16 Å, relative to the nAMPAR A3A2A3A2 complex, resulting in a NMDA receptor-like architecture (Fig. 3, C and D). While the authors hypothesized that the GluA2/GluA3 receptor transits between the O shape and the N shape of the nAMPARs, we did not observe any ‘O’-like class or any with a compacted ATD layer in our nAMPAR reconstructions, even though the 15F1 and 5B2 Fabs can label the compacted ATD layer (fig. S8D). Thus, we propose that in the rGluA2/GluA3 study, heterologous expression and cross-linking lead to a non native subunit arrangement and an aberrant receptor conformation.
Organization and conformation of the ATD and LBD layers
In all of the ten nAMPAR complexes, the ATD layer forms a dimer-of-dimers assembly bridged by subunits at the B and D positions (Fig. 2, A to J). In the three major populations, the assembly of the ATD layer is nearly identical to the homomeric GluA2 complex (Fig. 4, A to D) (24). Within the B-D interface, a histidine in GluA2 (His208GluA2), close to the overall pseudo 2-fold axis, is positioned near the negative dipole end of the α7 helix of an opposing subunit (24), perhaps forming a favorable charge-helix dipole interaction (Fig. 4E). Sequence alignment reveals that the His208GluA2 is conserved in GluA3 and GluA4, but is not present in GluA1 (fig. S7G). Superimposition of the crystal structures of the GluA3 or GluA4 ATDs (27, 38) to B-D positions of the homomeric GluA2 ATD structure (24) indicates the structure around the dimer-dimer interface is nearly identical in GluA2, GluA3 and GluA4 (fig. S8, E and F), consistent with the GluA2 and GluA4 subunits forming stable heteromeric ATD dimer-dimer interface (Fig. 2, G to J). When the GluA1 ATD (39) is similarly examined, we found clashes between the two GluA1 subunits or between the GluA1 and GluA2 subunits (fig. S8G). Considering that His208GluA2 is not conserved in GluA1, we speculate that the ATD dimer-of-dimers assembly is disfavored or perhaps even disrupted if GluA1 occupies one or both of the B and D positions.In the nAMPAR-11B8 data set, we blurred ATDs in some 2D classes and splayed opened ATD layers in 3D classification (fig. S2, C and I). Thus, we suggest that when GluA1 accesses the B/D positions there is a tendency for the receptor to adopt an ATD ‘splayed opened’ complex. The splayed opened ATD was only resolved in the 11B8 scFv isolated nAMPAR complex, while not found in the 15F1 Fab isolated nAMPAR complex (figs. S2I, S4), indicating that the former complex could be a GluA2-lacking, calcium permeable GluA1 homomer, GluA1/GluA3 heteromer, or GluA1/GluA4 heteromer, all of which are present in the brain and are essential for synaptic plasticity and learning (22, 23, 40, 41).
Fig. 4. ATD layer and ATD-LBD interface.
(A-C) Capital ‘Y’ view of di-heteromeric A1A2A1A2 (A), A3A2A3A2 (B) or tri-heteromeric A1A2A3A2 complexes (C). The COM distances between the ATD and LBD layers, and between the LBD and TMD layers, are shown. The spaces between the ATD and LBD layers are indicated by a square bracket. A’-C’ and B’-D’ auxiliary proteins are grey and green. (D) ATD layer analysis of the A1A2A1A2 and A3A2A3A2 complexes. The ATD model of the A1A2A1A2 complex is in the left panel, in which the COMs of the R1 and R2 lobes are indicated by black dots. The distances and angles of vectors defined by COMs of the A1A2A1A2 (upper right) or the A3A2A3A2 (lower right) complex are shown in the right two panels. (E) Interactions that may stabilize the ATD dimer-dimer interface. ‘Top’ view of the ATD layer of the A3A2A3A2 complex is in the left panel. Boxed regions in the right panels highlight dimer-dimer contacts, viewed parallel or perpendicular to the overall 2-fold axis. The α7 helix is colored from blue at the N-terminus to red at the C-terminus. His 208 and N-linked carbohydrates are in ‘sticks’. (F-G) ATD-LBD interface comparison between GluA1 and GluA3 subunits from A1A2A1A2 and A3A2A3A2 complexes or from the A1A2A3A2 complex. Structures were superimposed using the LBD layer. COMs of the D1 lobe, GluA1 R1/R2 lobes and GluA3 R1/R2 lobes are indicated by black, blue and orange dots. Distances are in ängstroms (Å).
To address the question of why specific subunits are found in particular positions of the ABCD receptor complex, we note that the ATDs at the A and C positions are more accessible for protein binding than ATDs at the B and D positions because the proximal B-D subunit interactions across the ATD dimer-dimer interface occlude ATD surface area (fig. S8A). Thus, we speculate that the GluA1/GluA3 subunits at the A/C positions and are more accessible for interactions with synaptic or trafficking proteins (Fig. 2L). This is consistent with a study showing that the GluA1 ATD, rather than the GluA2 ATD, is critical for synaptic trafficking of AMPARs by possible interactions with proteins in the synaptic cleft (42).
The distance between ATD-LBD layers is shorter in the A3A2A3A2 complex than in the A1A2A1A2 structure (Fig. 4, A and B). Thus, the GluA1 ATD is ~10 Å farther away from the LBD layer in comparison with the GluA3 subunit, ablating nearly all ATD-LBD interaction (Fig. 4, F and G). To explore if different ATD-LBD interfaces could influence LBD arrangements, we compared LBD layers from A1A2A1A2 and A3A2A3A2 complexes using one of the LBD dimers as a reference. Although there is no clear difference detected between the heteromeric LBD dimers of GluA1/GluA2 and GluA2/GluA3, the gating ring formed by two LBD dimers is less expanded in the A3A2A3A2 complex (fig. S9, A to C). The difference is subtle in the current antagonist bound state, but we speculate that the ATD layer might act as a ‘clamp’ to restrict the LBD layer rearrangement during activation, desensitization or recovery from desensitization through ATD-LBD interactions at the A-C positions, consistent with the ATD affecting desensitization kinetics (43).
S2-M4 linker is positioned to modulate gating
RNA splicing sites are located at helix J/K of the LBD layer and within the S2-M4 linker connecting the LBD and TMD layers (fig. S9A and Fig. 5A), yielding two variants, flip and flop, that differ in gating kinetics (12, 44, 45).The structure of the S2-M4 has not been resolved previously, but there is interpretable density for the S2-M4 linker in all of three major nAMPAR complexes, allowing us to fit the main chain (Fig. 5, A to C). The flip/flop splicing site adopts a similar conformation in different subunits, forming a ‘hook’ shaped structure in combination with helix K (Fig. 5, A–C). Because the nAMPAR sample is composed of flip and flop isoforms, the S2-M4 linker density is weaker than the other LBD-TMD linkers, consistent with chemical heterogeneity and possible additional conformational flexibility. To understand if the ‘hook’ shape of the S2-M4 linker in nAMPAR complexes is present in previously determined structures, we inspected cryo-EM maps of antagonist-bound MPQX GluA2flop-TARP γ2 and GluA2flip-TARP γ2 complexes (25, 46), finding a similar feature in the latter, suggesting that the densities of the splice regions in the nAMPAR complexes arise from the flip isoform (fig. S9D). In agreement with this hypothesis, there is no density for a flop splicing site in the GluA2flop-TARP γ2 maps (fig. S9E), likely because the sequence is glycine rich (SGGGD) and conformationally heterogeneous.
Fig. 5. S2/M4 linkers interact with gating machinery.
(A-C) LBD-TMD structure from opposing subunit pairs, A-C (A) and B-D (B), and adjacent subunits A-D (C), viewed perpendicular to the ‘central’ 2-fold axis (vertical dashed lines). The densities are shown for helices J-K, the S2-M4 linkers and M4 helices. Cαs of the flip/flop splicing sites on the S2-M4 linkers are displayed as black spheres. (D-E) Densities with atomic models for helix K, the S2-M4 linkers from the A/C (D) or the B/D (E) subunits and helix E, the M3-S2 linker from an adjacent subunit, viewed parallel to the membrane. The putative interactions are highlighted by dotted line circles. (F) Density and model of helix K and S2-M4 are shown in the bottom left panel, with the side chains of Y768 and K776. (G) Sequence alignment of GluA1–3 flip and flop isoforms at helix K and the S2-M4 linker. Y768 and G/K776 are highlighted by red and blue boxes, respectively. (H-I) Bar plot showing the effect of Y768A/K776G mutants on desensitization rate and Iss/Ipeak ratio, respectively. An asterisk indicates a statistically significantly difference, p<0.05.
The S2-M4 linker is positioned near the receptor’s central axis through the ‘hook’ shaped splicing site (Fig. 5, A and B). Contacts between the S2-M4 splice sites and the gating-critical M3-S2 linkers from adjacent subunits were identified in all four subunits. Moreover, the splicing sites of the B/D subunits also interact with helices E from adjacent subunits (Fig. 5, D and E). We thus tested the role of the S2-M4 linker region in ion channel gating, noting that there is a possible cation - π interaction between Y768 and K776, within the ‘hook’ shaped conformation of the A3A2A3A2 complex. Both side chains were well fit in the density map (Fig. 5F), with the conserved Y768 located at the C terminus of helix K and K776 within the flip/flop cassette (Fig. 5G). To investigate the functional role of the flip/flop splicing sites on the S2-M4 linker, we designed a double mutation Y768A/K776G in the context of the full length wild-type flip GluA2 construct and performed outside-out patch recording experiments (fig. S9F). Whereas the desensitization rate of the double mutant is 7.6±0.34 ms (n=8), comparable to 8.5±0.36 ms (n=9) of the wild-type flip isoform and ~3-fold slower than 2.1±0.09 ms (n=10) of the flop isoform (Fig. 5H), the double mutant has significantly reduced Iss/Ipeak ratio, from 1.88±0.31% (n=10) of the wild-type flip isoform to 0.75±0.11% (n=9), similar to the flop variant (Fig. 5I). While further studies are required to fully elaborate the roles of the S2-M4 linker on ion channel gating, these experiments are consistent with the structural observation that the S2-M4 linker is positioned close to regions important for ion channel gating.
Assembly of auxiliary proteins in the nAMPAR complex
To explore the function of the nAMPAR preparation we carried out two-electrode, voltage-clamp experiments using oocytes injected with proteoliposomes reconstituted with the complex. These experiments show that kainate, a partial agonist, elicits ~50% of the current of glutamate, a full agonist, showing that auxiliary proteins, such as TARPs, are retained in the nAMPAR complex (Fig. 6A) (47). Western blot analysis bolsters this hypothesis and indicates the presence of TARP γ2, as well as lower quantities of TARP γ8 and cornichon 2/3 (Fig. 6B). Cryo-EM maps of the three major complexes reveal density for four auxiliary proteins surrounding the receptor TMD, each with four TM helices (Fig. 6, C and D). Because there is high sequence identity within the LBD and the TMD layers between the GluA1-A4 subunits, the LBD-TMD density map was improved by 3D refinement using all particles from the three major complexes. The resulting map (LBD-TMDmerged map) clearly defines the positions of the TM helices with resolved helix grooves and protrusions for bulky side chains (Fig. 6C). Phosphorylation and palmitoylation of the nAMPAR complex impacts receptor trafficking and pore properties and occurs at the C- terminus of receptor subunits (13–15). However, these regions of the receptor are not resolved in the density maps, likely because of conformational heterogeneity.
Fig. 6. Auxiliary proteins associated with nAMPAR.
(A) TEVC recordings from oocytes injected with proteoliposomes containing the isolated nAMPAR complex. A representative pair of currents recorded using the same oocyte is shown in the left panel. The right panel shows that the ratio of steady-state currents evoked by kainate (KA) and glutamate (Glu) is 0.50±4 (mean ± standard deviation, n=4). (B) Western blot of nAMPAR used for cryo-EM study with antibodies against TARP γ2, TARP γ8 and cornichon2/3. (C) The LBD-TMDmerged map illustrates architecture of the nAMPAR complex with auxiliary proteins. Auxiliary proteins at the A’/C’ and B’/D’ positions are in grey and green, respectively. (D) Cross section of cryo-EM maps at the ‘height’ indicated in panel C shows the auxiliary protein density features in the A1A2A1A2, A3A2A3A2 and A1A2A3A2 complexes. The auxiliary density features are separated from the receptor TMD by dotted lines. (E) Density with atomic model for B’/D’ auxiliary proteins, viewed parallel to the membrane. (F) Superimposition of auxiliary protein structures from the A’/C’ and B’/D’ positions using related receptor TMDs, viewed from extracellular (upper) or intracellular sides (lower). The relative rotation angles of TM1, TM2 and TM4 are indicated.
Based on the LBD-TMDmerged map and the Western blot data, we tentatively assigned the density at the B’/D’ positions as TARP γ2 (Fig. 6E). We observed side chain density features at the B’/D’ positions that include Y96, Y156, W178, F187 and F201, residues that are conserved across TARP subunits (fig. S9, G and H). However, the auxiliary subunit density at the A’/C’ positions has fewer features and the density for the TM helices is weaker than that at the B’/D’ positions. Thus, we conclude that different auxiliary proteins may access the A’/C’ and the B’/D’ positions and that there is a preference for a TARP, such as γ2, at the B’/D’ positions (Fig. 6, E and F). We hypothesize that TARP subunits at the B’-D’ positions play an important role in modulating channel activation and desensitization by interactions between the LBD ‘KGK’ motifs and the TARP γ2 acidic loops (26). Occupancy of A’-C’ positions by auxiliary subunits may be more permissive, by contrast, with multiple distinct auxiliary subunits participating in additional functional activities.
Here we elucidated the molecular structures of ten nAMPAR complexes, illuminating the spectrum of subunit composition, stoichiometry and arrangement and demonstrating the existence of a substantial population of triheteromeric nAMPARs composed of GluA1, GluA2 and GluA3 subunits. We show how subunit arrangement is non stochastic, with the GluA2 subunit preferentially occupying the B and D positions, and the GluA1 and GluA3 subunits predominantly accessing the A and C positions. In the three major complexes, we consistently observed a more extensive ATD-LBD interface in the GluA3 subunit than in the GluA1 subunit, leading to a subtle rearrangement of LBD dimers, which suggests, in turn, how the ATD layer is poised to modulate receptor function. The S2-M4 linkers were clearly resolved and were ‘plugged’ into the gating machinery by the ‘hook’ shaped splicing site, interacting with M3-S2 linkers from adjacent subunits and thus poised to modulate receptor gating. The nAMPAR complex involves an ensemble of auxiliary proteins (19, 20) and our studies show how the A’/C’ and B’/D’ positions have distinct density features, with the B’/D’ positions described by occupancy of TARP subunits and the A’/C’ sites, with less well resolved density features, consistent with occupancy by a broader spectrum of auxiliary subunits.
Supplementary Material
Acknowledgements
We thank D. Cawley for generating mAbs, the Multiscale Microscopy Core (OHSU) for support with microscopy, the Advanced Computing Center (OHSU) for computational support, L. Vaskalis for assistance with figures, H. Owen for help with manuscript preparation and Gouaux laboratory members for helpful discussions.
Funding: Mass spectrometry experiments were supported by the NIH Grant P41 GM103493 and performed in the DOE Environmental Molecular Sciences Laboratory at Pacific Northwest National Laboratory, operated by Battelle Memorial Institute under Contract DE-AC05-76RL0 1830. This work was supported by a NINDS grant of the National Institutes of Health under award number R01NS038631 to E.G., who is also an investigator of the Howard Hughes Medical Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of National Institutes of Health.
Footnotes
Competing interests: The authors declare no competing interests.
Data and materials availability: The cryo-EM maps, including maps for overall, ATD, LBD-TMD and LBD-TMD layers and coordinates of the A1A2A1A2, A3A2A3A2 and A1A2A3A2 complexes have been deposited in the Electron Microscopy Data Bank (EMDB) under accession numbers EMD-9387, EMD-9388 and EMD-9389 and in the Protein Data Bank under accession codes 6NJL, 6NJM and 6NJN, respectively. The cryo-EM maps of the A2A2A3A2, A2A2A1A2, A2A2A2A2, A1A2AxA2, AxA2A3A2, A2AxA1A2 and A2AxA3A2 complexes have been deposited in the Electron Microscopy Data Bank (EMDB) under accession numbers EMD-0426, EMD-0427, EMD-0428, EMD-0429, EMD-0430, EMD-0431 and EMD-0432, respectively.
Supplementary Materials:
Materials and Methods
Figures S1-S9
Tables S1-S3
References (50–64)
References
- 1.Keinänen K et al. , A family of AMPA-selective glutamate receptors. Science 249, 556–560 (1990). [DOI] [PubMed] [Google Scholar]
- 2.Mosbacher J et al. , A molecular determinant for submillisecond desensitization in glutamate receptors. Science 266, 1059–1062 (1994). [DOI] [PubMed] [Google Scholar]
- 3.Traynelis SF et al. , Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev 62, 405–496 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kessels HW, Malinow R, Synaptic AMPA receptor plasticity and behavior. Neuron 61, 340–350 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diering GH, Huganir RL, The AMPA receptor code of synaptic plasticity. Neuron 100, 314–329 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mayer ML, Westbrook GL, Guthrie PB, Voltage-dependent block by Mg2+ of NMDA responses in spinal cord neurones. Nature 309, 261–263 (1984). [DOI] [PubMed] [Google Scholar]
- 7.Alt A, Nisenbaum ES, Bleakman D, Witkin JM, A role for AMPA receptors in mood disorders. Biochem Pharmacol 71, 1273–1288 (2006). [DOI] [PubMed] [Google Scholar]
- 8.O’Neill MJ, Witkin JM, AMPA receptor potentiators: application for depression and Parkinson’s disease. Curr Drug Targets 8, 603–620 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Dirnagl U, Iadecola C, Moskowitz MA, Pathobiology of ischaemic stroke: an integrated view. Trends Neurosci 22, 391–397 (1999). [DOI] [PubMed] [Google Scholar]
- 10.Francis PT, Glutamatergic systems in Alzheimer’s disease. Int J Geriatr Psychiatry 18, S15–21 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Henley JM, Wilkinson KA, Synaptic AMPA receptor composition in development, plasticity and disease. Nat Rev Neurosci 17, 337–350 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Sommer B et al. , Flip and flop: A cell-specific functional switch in glutamate-operated channels of the CNS. Science 249, 1580–1585 (1990). [DOI] [PubMed] [Google Scholar]
- 13.Roche KW, O’Brien RJ, Mammen AL, Bernhardt J, Huganir RL, Characterization of multiple phosphorylation sites on the AMPA receptor GluR1 subunit. Neuron 16, 1179–1188 (1996). [DOI] [PubMed] [Google Scholar]
- 14.Hayashi T, Rumbaugh G, Huganir RL, Differential regulation of AMPA receptor subunit trafficking by palmitoylation of two distinct sites. Neuron 47, 709–723 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Lee HK et al. , Phosphorylation of the AMPA receptor GluR1 subunit is required for synaptic plasticity and retention of spatial memory. Cell 112, 631–643 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Chen L et al. , Stargazin regulates synaptic targeting of AMPA receptors by two distinct mechanisms. Nature 408, 936–943 (2000). [DOI] [PubMed] [Google Scholar]
- 17.Jackson AC, Nicoll RA, The expanding social network of ionotropic glutamate receptors: TARPs and other transmembrane auxiliary subunits. Neuron 70, 178–199 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gill MB, Kato AS, Wang H, Bredt DS, AMPA receptor modulation by cornichon-2 dictated by transmembrane AMPA receptor regulatory protein isoform. Eur J Neurosci 35, 182–194 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Greger IH, Watson JF, Cull-Candy SG, Structural and functional architecture of AMPA-type glutamate receptors and their auxiliary proteins. Neuron 94, 713–730 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Schwenk J et al. , Regional diversity and developmental dynamics of the AMPA-receptor proteome in the mammalian brain. Neuron 84, 41–54 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Sommer B, Kohler M, Sprengel R, Seeburg PH, RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell 67, 11–19 (1991). [DOI] [PubMed] [Google Scholar]
- 22.Wenthold RJ, Petralia RS, Blahos JI, Niedzielski AS, Evidence for multiple AMPA receptor complexes in hippocampal CA1/CA2 neurons. J. Neurosci. 16, 1982–1989 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu W et al. , Subunit composition of synaptic AMPA receptors revealed by a single-cell genetic approach. Neuron 62, 254–268 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sobolevsky AI, Rosconi MP, Gouaux E, X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature 462, 745–756 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao Y, Chen S, Yoshioka C, Baconguis I, Gouaux E, Architecture of fully occupied GluA2 AMPA receptor-TARP complex elucidated by cryo-EM. Nature 536, 108–111 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen S et al. , Activation and desensitization mechanism of AMPA receptor-TARP complex by cryo-EM. Cell 170, 1234–1246 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herguedas B et al. , Structure and organization of heteromeric AMPA-type glutamate receptors. Science 352, aad3873 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Penn AC et al. , Hippocampal LTP and contextual learning require surface diffusion of AMPA receptors. Nature 549, 384–388 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawate T, Gouaux E, Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure 14, 673–681 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Pickard L, Noel J, Henley JM, Collingridge GL, Molnar E, Developmental changes in synaptic AMPA and NMDA receptor distribution and AMPA receptor subunit composition in living hippocampal neurons. J Neurosci 20, 7922–7931 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee SH et al. , Super-resolution imaging of synaptic and extra-synaptic AMPA receptors with different-sized fluorescent probes. Elife 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schnell E et al. , Direct interactions between PSD-95 and stargazin control synaptic AMPA receptor number. Proc Natl Acad Sci U S A 99, 13902–13907 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baucum AJ 2nd et al. , Identification and validation of novel spinophilin-associated proteins in rodent striatum using an enhanced ex vivo shotgun proteomics approach. Mol Cell Proteomics 9, 1243–1259 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baucum AJ 2nd, Shonesy BC, Rose KL, Colbran RJ, Quantitative proteomics analysis of CaMKII phosphorylation and the CaMKII interactome in the mouse forebrain. ACS Chem Neurosci 6, 615–631 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saglietti L et al. , Extracellular interactions between GluR2 and N-cadherin in spine regulation. Neuron 54, 461–477 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Pelkey KA et al. , Pentraxins coordinate excitatory synapse maturation and circuit integration of parvalbumin interneurons. Neuron 85, 1257–1272 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kazi R, Dai J, Sweeney C, Zhou HX, Wollmuth LP, Mechanical coupling maintains the fidelity of NMDA receptor-mediated currents. Nat Neurosci 17, 914–922 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sukumaran M et al. , Dynamics and allosteric potential of the AMPA receptor N-terminal domain. EMBO J 30, 972–982 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yao G et al. , Crystal structure of the glutamate receptor GluA1 N-terminal domain. Biochem J 438, 255–263 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saab AS et al. , Bergmann glial AMPA receptors are required for fine motor coordination. Science 337, 749–753 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Liu SQ, Cull-Candy SG, Synaptic activity at calcium-permeable AMPA receptors induces a switch in receptor subtype. Nature 405, 454–458 (2000). [DOI] [PubMed] [Google Scholar]
- 42.Diaz-Alonso J et al. , Subunit-specific role for the amino-terminal domain of AMPA receptors in synaptic targeting. Proc Natl Acad Sci U S A 114, 7136–7141 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moykkynen T, Coleman SK, Semenov A, Keinanen K, The N-terminal domain modulates alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor desensitization. J Biol Chem 289, 13197–13205 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Partin KM, Fleck MW, Mayer ML, AMPA receptor flip/flop mutants affecting deactivation, desensitization, and modulation by cyclothiazide, aniracetam, and thiocyanate. J. Neurosci. 16, 6634–6647 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seeburg PH, The role of RNA editing in controlling glutamate receptor channel properties. J Neurochem 66, 1–5 (1996). [DOI] [PubMed] [Google Scholar]
- 46.Twomey EC, Yelshanskaya MV, Grassucci RA, Frank J, Sobolevsky AI, Elucidation of AMPA receptor-stargazin complexes by cryo-electron microscopy. Science 353, 83–86 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Turetsky D, Garringer E, Patneau DK, Stargazin modulates native AMPA receptor functional properties by two distinct mechanisms. J Neurosci 25, 7438–7448 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coleman JA, Green EM, Gouaux E, X-ray structures and mechanism of the human serotonin transporter. Nature 532, 334–339 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Turski L et al. , ZK200775: a phosphonate quinoxalinedione AMPA antagonist for neuroprotection in stroke and trauma. Proc Natl Acad Sci U S A 95, 10960–10965 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yi L et al. , Targeted quantification of phosphorylation dynamics in the context of EGFR-MAPK pathway. Anal Chem 90, 5256–5263 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.MacLean B et al. , Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng SQ et al. , MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang K, Gctf: Real-time CTF determination and correction. J Struct Biol 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scheres SH, RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kucukelbir A, Sigworth FJ, Tagare HD, Quantifying the local resolution of cryo-EM density maps. Nat Methods 11, 63–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pettersen EF et al. , UCSF Chimera – a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Biasini M et al. , SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42, W252–258 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Afonine PV et al. , Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Karakas E, Furukawa H, Crystal structure of a heteromeric NMDA receptor ion channel. Science 344, 992–997 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen VB et al. , MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.DeLano WL (DeLano Scientific, San Carlos, CA, USA, 2002). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.