Abstract
Viburnum erubescens Wall., Rhododendron arboretum Sm., Eurya japonica Thumb., Symplocos lucida (Thunb.) Siebold & Zucc, and Symplocos pyrifolia Wall. ex G. Don are extensively used by the native and ethnic populations of the South-Eastern Himalayan region for several dermatological conditions, yet their phytochemical composition remained largely unknown. Therefore, the aim of the study was to explore the therapeutically relevant volatile phytochemical compositions and study the molecular interactions against intracellular cytoregulatory transcription factors. Leaves of the five plants were subjected to Gas chromatography-Mass spectrometry (GCMS) post silylation derivation. The results were further analyzed using multivariate statistical methods such as Principal Component Analysis (PCA) and Hierarchical Cluster Analysis (HCA). A total of 115 compounds were identified in the five plants. Multivariate analysis revealed optimum metabolomic correlation between S. pyrifolia and S. lucida (0.876), whereas lowest correlation was found between E. japonica and V. erubescens (−0.242). Arbutin, β-amyrin, betulin, β-sitosterol and stigmasterol demonstrated highest interaction with the molecular targets. Collectively, the present study revealed the bioactive volatile phytochemicals responsible the therapeutic uses against diverse skin conditions.
Keywords: Gas chromatography-Mass spectrometry, Molecular docking, Multivariate statistics, Silylation, Volatiles
Graphical abstract
1. Introduction
The South-Eastern Himalayan biodiversity hub is constantly being explored to identify and characterize new floral and faunal species. According to the World Wide Fund for Nature report of 2015, more than 130 new plant species have been identified only in last few years.1 Essential oils, volatiles and organic extracts of many of these plants are extensively used by the native people for therapeutic purposes, yet their phytochemical profiles remain largely unknown. Ethnopharmacological survey reveals that herbal remedies in the Eastern Himalayan region are primarily focused to cure dermatological problems, pain, and respiratory disorders.2
The present study has documented the phytometabolomic fingerprinting of the volatiles in five major high altitudinal ethnomedicinal plants (supplementary Table 1) of Kurseong hills of the South-Eastern Himalayas, and explored their possible interaction with intracellular transcription factors. The plants were selected based on their relevance in therapeutic use. Apart from their diverse ethnopharmacological activities, the individual aqueous/oil extracts of these plants or polyherbal formulations containing one or multiple of these plants are topically applied for the treatment of diverse skin related conditions such as microbial infections, cuts, wounds, cancer and acute inflammatory swellings by the local people of the South-Eastern Himalaya. In high altitudinal Himalaya of neighboring country Nepal, R. arboreum and S. pyrifolia are used by the traditional healers against wounds and skin diseases, respectively.3 E. japonica is externally used in Chinese traditional medicine for the treatment of rheumatoid arthritis and hemostasis of external injuries.4 V. erubescens along with some other plants in a polyherbal formulation has been claimed to lower skin pigmentation by inhibiting melanin synthesis.5 Moreover, a glycoside arbutin which lowers melanin formation by inhibiting tyrosinase activity and also used in various skin-care products, was isolated from the root extract of V. erubescens.6 The tribals of the Mizoram hills at Eastern Indian province use E. japonica poultice on skin swellings and its anti-microbial effect against various gram + ve and –ve bacteria has already been evaluated.7 R. arboretum is externally used to relieve headache in the Garhwal Himalaya, India.8
However, despite tremendous use in traditional medicine, these plants lack proper pharmacognostic evaluation or standardized phytometabolomic fingerprinting, and their volatile constituents has not been profiled comprehensively. Moreover, the active volatile constituents in the plants has never before correlated with their ethnomedicinal uses. Therefore, the present study was undertaken to explore the phytochemical status of the leaves of five selected plants of the South-Eastern Himalayan region using GC-MS method and to correlate the phytochemical fingerprints using multivariate statistical methods.
2. Materials and methods
2.1. Sample processing and silylation
Healthy and disease-free leaves of the five plants were collected from Dowhill Arboreum at the Eco Park under Kurseong division, West Bengal, India. The leaves were thrice washed with water to remove any dirt or foreign matter, and then cut into pieces and shade dried at laboratory temperature (24 °C) for three weeks. The leaves were then grinded to powder using pestle and mortar. The resultants (250 mg) were mixed with 5 mL n-hexane in reacti-vials (Thermo Scientific) and incubated at 4 °C with occasional shaking.9 After 48-h, N,O-Bis(trimethylsilyl)trifluoroacetamide + Trimethylchlorosilane (BSTFA + TMCS; 99:1 v/v) was added separately to the previous individual mixtures (1:10 v/v; BSTFA + TMCS: mixture) and incubated at 4 °C for 60-min under constant shaking (60 rpm). The mixtures were then centrifuged twice (15,000 rpm at 4 °C for 20-min) and the clear supernatant was then passed through Whatman filter paper no. 1 (11 μm) and the resultant was used for GC-MS analysis.
2.2. GC-MS analysis
In brief, samples were run in Thermo Scientific Trace 1300 gas chromatography instrument attached with Thermo Scientific ISQ QD single quadrupole mass spectrophotometer and analyzed using Thermo Xcalibur™ 2.2 software. The GC was equipped with TG-5MS column (30 m × 0.25 mm × 0.25 μm). The inlet temperature was maintained at 250 °C. The initial temperature was set at 60 °C (solvent delay 5-min) with a hold of 4-min, followed by a ramp of 5 °C–290 °C with a hold of 4-min (54-min program). Samples were (1 μL) injected in a splitless mode (split flow 50 mL/min) with splitless time of 0.80-min, using a Thermo Scientific AI-1310 auto-sampler. The carrier gas was helium (99.99%), with a constant flow of 1 mL/min. MS transfer line temperature was set at 290 °C with an ion source temperature of 230 °C (electron ionization). The individual samples were analyzed at electron energy 70 eV (vacuum pressure- 2.21e-0.5 Torr). The mass analyzer range was set at 50–650 amu (scan duration 0.2-sec) with positive ion polarity. All samples were analyzed thrice for confirmation. Solvent wash (2 μL) was performed after each sample run with a 17-min program of 60 °C–300 °C (20 °C/min ramp) temperature and 5-min hold at 300 °C, resulting in baseline normalization.
2.3. Data analysis
MS data analysis was performed by Automated Mass Spectral Deconvolution and Identification System (AMDIS) version 2.70. The major and essential compounds were identified by mass fragmentation patterns (m/z) of the reference of parent compound (mol. peak and base peak *) using MS interpreter Version 2.0 and by matching with the reference database of National Institute Standard and Technology (NIST) with a MS library version 2011.
2.4. Multivariate statistics
Data were statistically analyzed using IBM SPSS statistics version 20.0 software package for Windows. The inter relationship between the metabolomic profiles of the five different plants were analyzed by Principal Component Analysis (PCA) based on the correlation matrix. Under descriptive statistics, KMO and Bartley's test was performed to study the underlying dimensions of the five variables. Two factors were extracted under varimax method. Data were further analyzed by multivariate method by employing a hierarchical cluster analysis (HCA). The cluster method employed was between group linkage with interval of square Euclidian distance. Transform values of variables (average zero and S.D. 1) called Z scores was carried out as a pre-treatment of the data. Proximity heat map was generated using Microsoft Excel 2010.
2.5. Target proteins for the docking study
High resolution X-ray diffraction structure of activator protein-1 (AP1) clathrin adaptor core (1W63), human FOXO3 (2UZK), nuclear factor kappa-β (1NFK) and Keap1 Kelch domain (5FNQ) were retrieved from Protein Data Bank (PDB). Information about the quality of these structures were retrieved from the PDB database and their resolutions were found to be 4.1, 2.7, 2.3 and 1.9, respectively. These proteins were fed into CASTp server (http://sts.bioe.uic.edu/castp/) for predicting the active sites or pockets. Autodock Tool 1.5.6 was used for preparing these proteins ready to dock. Water molecules in the structures were replaced by polar hydrogen. After calculation of the Gasteiger charge of these aforementioned proteins they were saved in .pdbqt format.
2.6. Ligand preparation for the docking study
Phytochemicals identified in GC-MS analysis were used as ligand molecules for the docking study. The structures of these molecules (in .sdf format) were obtained from NCBI PubChem database. SMILES server was utilized for converting the .sdf format to PDB. These structures were then fed into Autodock Tool 1.5.6 and saved in .pdbqt format. Information regarding the ADME and QSAR property of considered phytochemicals were obtained from PubChem database.
2.7. Docking algorithm
We have used a blind a blind docking method in this study. The whole protein surface of all considered proteins was subjected to dock with each ligand. Grid was selected in such a way that the whole protein is covered. All the steps including the final run for docking was done in Autodock Tool 1.5.6. The docked structures were viewed through PyMol version 1.7.4.
2.8. Root mean square validation of docked structures
Autodock predicted 9 different docking sites for each studied ligands. All of them had different RMSD values. However, we have considered only those sites which gave RMSD value 0 for the betterment of our understanding.
3. Result and discussion
The GC-MS analysis revealed the presence of several volatile compounds and derivatized phenolics, many of which are well established for their potent pharmacognostic properties (Fig. 1 and supplementary table 2-6). Stigmasterol (m/z 483ˉ, 83*) was found in V. erubescens, S. lucida and S. pyrifolia by m/z in its TMS (m/z 72) form, identified by m/z 394 (C29H46) and 255 (C19H27) (Fig. 2). Stigmasterol, known as wulzen anti-stiffness factor, is used to treat arthritic pains and its chemopreventive effects against skin carcinoma by its topical application has been established.10 β-Sitosterol was abundant in V. erubescens and R. arboreum, identified by m/z 485ˉ, 396 (C25H52OSi), 255 (C15H31OSi), 145 (C7H16OSi) and 129* (C6H13OSi). Stigmasterol and β-Sitosterol rich extracts of another ethnomedicinal plant Bowdichia virgilioides demonstrated potent wound healing and antimicrobial activities.11 The triterpene betulin was identified in both the Symplocos sp. with highest abundance in S. lucida. The identifying mass fractions (m/z) were 441ˉ, 411 (C28H43O2), 207 (C14H24O), 189* (C14H21) and 135 (C10H15). Apart from its extensive use in skincare cosmetics as skin toner, restorer and scar remover,12 betulin also restricts A431 skin epidermoid carcinoma cell proliferation and ear inflammation upon external treatment.13 Highest abundance of azelaic acid was found in R. arboretum in its bis-TMS (m/z 144) form and identified in all the plants primarily by m/z 201 (C10H21O2Si), and 129 (C5H9O2Si). Azelaic acid is pharmacologically known for its therapeutic use in the treatment of skin disorders such as papulopustular rosacea and melasma.14,15 Moreover, it has been successfully evaluated against acne causing bacteria Propionibacterium acnes.16
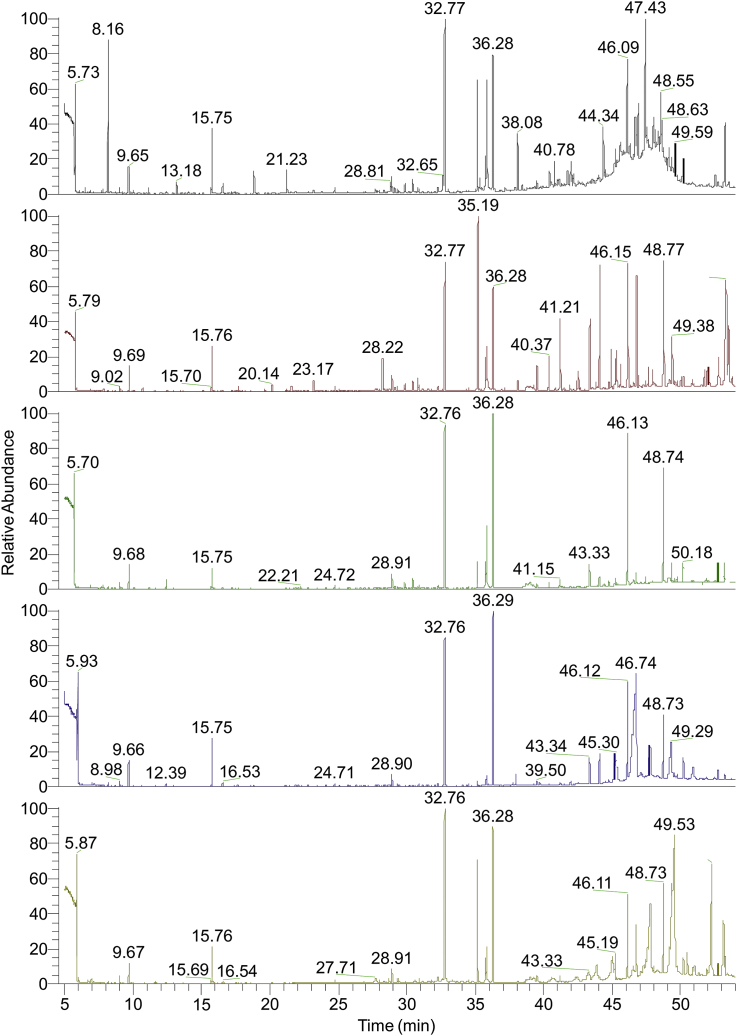
Fig. 1.
GC chromatogram of (A) V. erubescens; (B) R. arboreum; (C) E. japonica; (D) S. lucida; (E) S. pyrifolia. The GC-MS analysis revealed quite distinct yet overlapping phytochemical profiles comprising of several discrete volatile constituents. A total of 38, 48, 38, 39 and 49 compounds were identified in V. erubescens, R. arboreum, E. japonica, S. lucida and S. pyrifolia, respectively (Supplementary data). However, a total of 115 individual compounds were identified in the five plants.
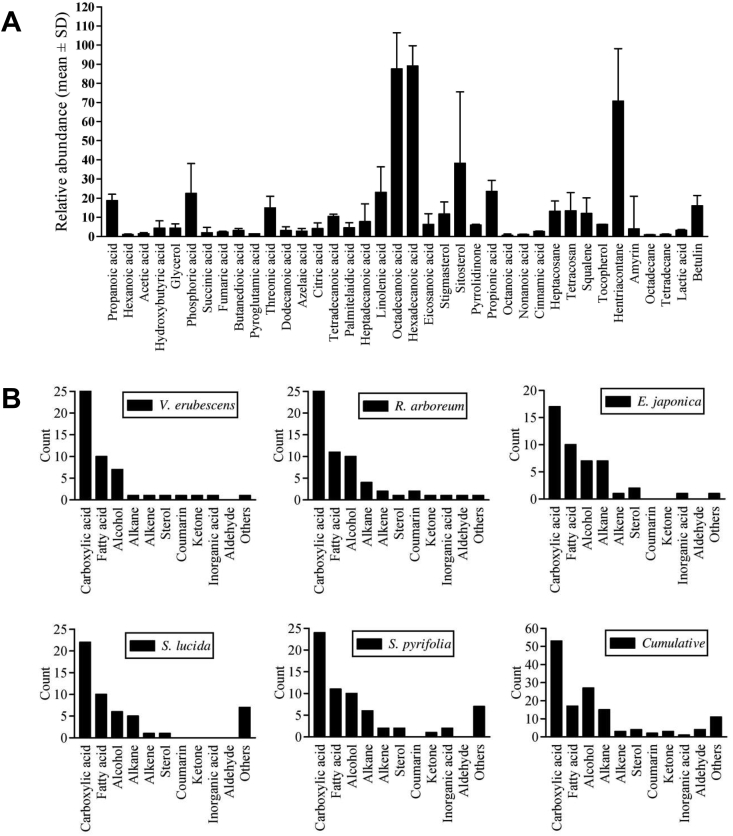
Fig. 2.
(A) Demonstrated the variation of relative abundance of different phytochemicals commonly found in the selected plants. Phytochemicals were selected based on presence in two or more plants. Data represented as mean ± SD of relative abundance in 2–5 plants. Apart from this, as summarized in Supplementary table 2-6, several phytochemicals were identified in a single plant. For example: Valeric acid, butenoic acid, hexenoic acid, phenylethanol, glyceric acid, levulinic acid and scopoletin were only detected in V. erubescens; farnesol, coumaric acid, linoleic acid, heneicosane, docosanol, methyl lignocerate and lupenone in R. arboretum; lauric acid, hexadecanethiol and cholesterol in E. japonica; ethyloctane, dicyclopentadiene, isocaproic acid, benzoic acid derivative, pimaric acid and carotene in S. lucida; oxalic acid, malonic acid, mevalonic acid, lanosterol, urosoloc acid and matairesinol in S. pyrifolia were not identified in other plants. (B) Summarizes the absolute count of different phytochemicals of the major organochemical species. The carboxylic acid group also includes fatty acid compounds. The ‘other’ group includes identified minor chemical species. Cumulatively, carboxylic acid species (53 nos.) followed by alcohol (27 nos.), fatty acids (17 nos.) and alkane hydrocarbons (15 nos.) were the most abundant chemical species identified. Three different aldehydes (nonanal, decanal and benzaldehyde) were identified only in R. arboretum. Levulinic acid (V. erubescens), lupenone (R. arboreum) and 2′,6′-dihydroxyacetophenone (S. pyrifolia) were the three ketone compounds identified.
Multivariate analysis (Supplementary figure 1) of the complete phytochemical fingerprints of the five plants demonstrated greater metabolomic similarity between S. pyrifolia and S. lucida (0.876), whereas E. japonica and V. erubescens (−0.242) possessed least similarities. These data were further supported by the proximity scores resulting in least proximity score between S. pyrifolia and S. lucida (9.207) and highest between E. japonica and V. erubescens (91.897). Details of the variance scores, component matrix, agglomeration schedule plot and proximity matrix details are provided in Supplementary data. Based on the phytochemical profiles, the loading plot of PCA indicated partial convergence due to similar primary and secondary metabolic pathways, giving rise to commonly identified phytochemicals. For instance, the component plot (Supplementary Figure 1A) shows a nearness of S. lucida and S. pyrifolia representing common phytochemical profiles in these two plants belonging to the same genus Symplocos. The loading of first and second principal component cumulatively accounted for 81.86% and 50.39% variance, respectively. Cluster analysis (CA) performed using HCA identified the similar phytochemical profiles, and also validated the component analysis. Among the five plants, the proximity score of S. lucida vs S. pyrifolia were lowest (9.207), representing greatest similarity of the phytochemical profiles, whereas V. erubescens vs E. japonica demonstrated highest proximity score (91.897), representing lowest similarity (Supplementary Figure 1C).
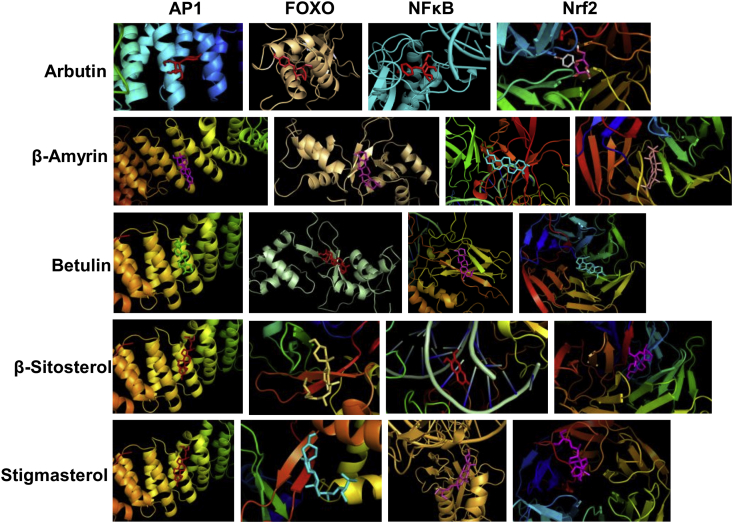
The mode of application of these plants in traditional medicine is primarily topical and therefore, selected identified phytochemicals were evaluated in silico for their cell permeability, absorptivity and potentials for pharmacological leads. As summarized in Table 1, the physiochemical properties of the selected ligands indicated smooth absorption, distribution and enhanced metabolic clearance, supporting potentials as bioactive leads. Moreover, the absorbance of the bioactive phytochemicals on skin, post topical application is in part likely justified by the favorable score as per Lipinski's rule. The selected compounds were further subjected to in silico molecular docking studies (Table 1 and Fig. 3) to predict their physical interactions with cytoregulatory transcription factors (activator protein-1; forkhead box class O; nuclear factor kappa-light-chain-enhancer of activated B cells; nuclear factor (erythroid-derived 2)-like 2), known to have central regulatory role in dermatological complications. For instance AP-1 and the ubiquitously expressed FOXO group of transcription factors regulates several aspects of skin physiology and pathology like keratinocyte proliferation, photoaging, epidermal morphogenesis, dermal wound healing, and is also associated with conditions like psoriasis, skin papilloma and acne.17, 18, 19 NFκB is a key mediator of psoriasis due to its integral role in inflammation and apoptosis.20 Nrf2 is associated with cytoprotective response against oxidative damages to melanocytes, keratinocytes and fibroblasts.21 Arbutin, β-amyrin, betulin, β-sitosterol and stigmasterol were found to be interacting effectively with all the selected targets. All the ligands demonstrated considerably strong binding affinity with the Nrf2 protein 5FNQ. Despite these interactions, the bioactivities of these plants whether mediated by the additive properties of the phytocompounds or through individual interactions with intracellular targets, needs to be explored by evidence-based pharmacological evaluations. Although beyond the scope of the study, these underexplore phytotherapeutics should be systematically fractionated to isolate and purify possible novel bioactive phytochemicals. Moreover, whether these molecular interactions result in antagonist or agonist activities, needs to be explored by bioassays. Nevertheless, the present phytometabolomic screening coupled with in silico approach provides a foundational basis for the exploration of ethnomedicinal flora in the relatively underexplored South-Eastern Himalayan region.
Table 1.
Binding energies and physico-chemical properties (as per Lipinski rule) of selected ligands.
| Ligand | AP1(1W63)a | FOXO (2UZK)a | NFκB (1NFK)a | Nrf2 (5FNQ)a | MW (g/mol) | Hydrogen Bond Donor | Hydrogen Bond Acceptor | Cell permeability | XLogP3-AA |
|---|---|---|---|---|---|---|---|---|---|
| α-Tocopherol | −4.8 | −4.8 | −4.7 | −7.2 | 430.717 | 1 | 2 | + | 10.7 |
| Arbutin | −5.2 | −5.1 | −6.5 | −7.8 | 272.253 | 5 | 7 | + | −0.7 |
| Azelaic acid | −4.2 | −4.5 | −4.1 | −5.7 | 188.223 | 2 | 4 | + | 1.6 |
| β-Amyrin | −7.9 | −7.4 | −7.3 | −9.4 | 426.729 | 1 | 1 | + | 9.2 |
| Betulin | −7.3 | −7.5 | −7.2 | −8.9 | 442.728 | 2 | 2 | + | 8.3 |
| Cinnamic acid | −4.7 | −5.2 | −4.1 | −5.9 | 148.161 | 1 | 2 | + | 2.1 |
| Eugenol | −4.3 | −4.6 | −4.5 | −6.1 | 164.204 | 1 | 2 | + | 2 |
| Ferulic acid | −4.7 | −4.9 | −4.5 | −4.5 | 194.186 | 2 | 4 | + | 1.5 |
| Gallic acid | −4.7 | −5.4 | −6.2 | −7.0 | 170.12 | 4 | 5 | + | 0.7 |
| Hexadecanoic acid | −4.4 | −2.9 | −2.8 | −3.5 | 256.43 | 1 | 2 | + | 6.4 |
| Levulinic acid | −3.9 | −3.6 | −4.1 | −4.4 | 116.116 | 1 | 3 | + | −0.5 |
| Linolenic acid | −3.2 | −2.7 | −4.1 | −3.5 | 278.436 | 1 | 2 | + | 5.9 |
| O-coumaric acid | −4.7 | −5.0 | −5.9 | −6.5 | 164.16 | 2 | 3 | + | 1.5 |
| Scopoletin | −4.6 | −4.8 | −5.8 | −5.6 | 192.17 | 1 | 4 | + | 1.5 |
| β-sitosterol | −5.2 | −5.9 | −5.2 | −6.7 | 414.718 | 1 | 1 | + | 9.3 |
| Squalene | −3.5 | −3.0 | −4.0 | −2.0 | 410.73 | 0 | 0 | NA | 11.6 |
| Stigmasterol | −6.0 | −5.7 | −6.3 | −5.9 | 412.702 | 1 | 1 | + | 8.6 |
Abbreviations: AP1: activator protein-1; FOXO: forkhead box class O; NFκB: nuclear factor kappa-light-chain-enhancer of activated B cells; Nrf2: nuclear factor (erythroid-derived 2)-like 2; MW: molecular weight
Binding energy calculated in kcal/mol.
Fig. 3.
Molecular interaction modelling of GC-MS identified compounds with cytoregulatory transcription factors. The binding energies (kcal/mol) has been summarized in Table 1. Abbreviation: AP1: activator protein-1; FOXO: forkhead box class O; NFκB: nuclear factor kappa-light-chain-enhancer of activated B cells; Nrf2: nuclear factor (erythroid-derived 2)-like 2.
Author contributions
PD performed the GCMS analysis and drafted the manuscript. IS and MRS performed the molecular interaction studies. SD collected the plant material and assisted in drafting of manuscript. TKC conceived the idea, supervised the study and finalized the manuscript. All authors have read and approved the final version of the manuscript.
Acknowledgements
The authors are thankful to central instrumentation facility of department of Zoology, University of North Bengal for the GCMS facility. Prof. Abhaya Prasad Das, department of Botany, University of North Bengal is acknowledged for authenticating the plants. The study was not funded by any external funding agencies. The authors declare that they have no conflict of interest.
Footnotes
Peer review under responsibility of The Center for Food and Biomolecules, National Taiwan University.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.jtcme.2018.08.001.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Nature WWFF . 2015. Hidden Himalayas: Asia's Wonderland. [Google Scholar]
- 2.Tangjang S., Namsa N.D., Aran C., Litin A. An ethnobotanical survey of medicinal plants in the Eastern Himalayan zone of Arunachal Pradesh, India. J Ethnopharmacol. 2011;134(1):18–25. doi: 10.1016/j.jep.2010.11.053. [DOI] [PubMed] [Google Scholar]
- 3.Poudel P. 2008. Medicinal Plants of Change VDC of Taplejung, Eastern Nepal. [Google Scholar]
- 4.Rosalind T., Dutta B.K., Paul S. 2014. Anti-diabetic Activity of Leaf Methanolicextract of Eurya Japonica Thunb. Instreptozotocin Induced Diabetes in Mice and Isolation of Fraction (EJ-1) from the Extract. [Google Scholar]
- 5.Kenichi Umishio K.M., Kobayashi Koji. 2006. Inventor. External Preparation for Skin and Skin-whitening Agent. 10.07.2008. [Google Scholar]
- 6.Prabhu K., Karar P., Hemalatha S., Ponnudurai K. Isolation and spectral identification of arbutin from the roots of viburnum erubescens. Int J Res Ayurveda Pharm. 2011;2(3):889–892. [Google Scholar]
- 7.Rosalind T., Dutta B., Paul S. Evaluation of in vitro antioxidant activity, estimation of total phenolic and flavonoid content of leaf extract of Eurya japonica Thunb. Asian J Pharmaceut Clin Res. 2013;6(1):152–155. [Google Scholar]
- 8.Bhat J.A., Kumar M., Bussmann R.W. Ecological status and traditional knowledge of medicinal plants in Kedarnath Wildlife Sanctuary of Garhwal Himalaya, India. J Ethnobiol Ethnomed. 2013;9(1):1. doi: 10.1186/1746-4269-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dutta S., Dey P., Saha M.R. Differential interaction with O2 and N2 free-radicals, phytochemical fingerprinting and molecular docking reveals potent antioxidant activities of three major recreational foods of the Indian subcontinent. Journal of Functional Foods. 2017;39:112–122. [Google Scholar]
- 10.Ali H., Dixit S., Ali D., Alqahtani S.M., Alkahtani S., Alarifi S. Isolation and evaluation of anticancer efficacy of stigmasterol in a mouse model of DMBA-induced skin carcinoma. Drug Des Dev Ther. 2014;9:2793–2800. doi: 10.2147/DDDT.S83514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Agra I.K., Pires L.L., Carvalho P.S., Silva-Filho E.A., Smaniotto S., Barreto E. Evaluation of wound healing and antimicrobial properties of aqueous extract from Bowdichia virgilioides stem barks in mice. An Acad Bras Ciências. 2013;85(3):945–954. doi: 10.1590/S0001-37652013005000049. [DOI] [PubMed] [Google Scholar]
- 12.Metelmann H.-R., Podmelle F., Waite P.D., Müller-Debus C.F., Hammes S., Funk W. Conditioning in laser skin resurfacing–Betulin emulsion and skin recovery. J Cranio-Maxillofacial Surg. 2013;41(3):249–253. doi: 10.1016/j.jcms.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 13.Dehelean C.A., Şoica C., Ledeţi I. Study of the betulin enriched birch bark extracts effects on human carcinoma cells and ear inflammation. Chem Cent J. 2012;6(1):1. doi: 10.1186/1752-153X-6-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu R.H., Smith M.K., Basta S.A., Farmer E.R. Azelaic acid in the treatment of papulopustular rosacea: a systematic review of randomized controlled trials. Arch Dermatol. 2006;142(8):1047–1052. doi: 10.1001/archderm.142.8.1047. [DOI] [PubMed] [Google Scholar]
- 15.Sarkar R., Bhalla M., Kanwar A.J. A comparative study of 20% azelaic acid cream monotherapy versus a sequential therapy in the treatment of melasma in dark-skinned patients. Dermatology. 2002;205(3):249–254. doi: 10.1159/000065851. [DOI] [PubMed] [Google Scholar]
- 16.Bojar R.A., Holland K.T., Cunliffe W.J. The in-vitro antimicrobial effects of azelaic acid upon Propionibacterium acnes strain P37. J Antimicrob Chemother. 1991;28(6):843–853. doi: 10.1093/jac/28.6.843. [DOI] [PubMed] [Google Scholar]
- 17.Angel P., Szabowski A., Schorpp-Kistner M. Function and regulation of AP-1 subunits in skin physiology and pathology. Oncogene. 2001;20(19):2413–2423. doi: 10.1038/sj.onc.1204380. [DOI] [PubMed] [Google Scholar]
- 18.Mori R., Tanaka K., de Kerckhove M. Reduced FOXO1 expression accelerates skin wound healing and attenuates scarring. Am J Pathol. 2014;184(9):2465–2479. doi: 10.1016/j.ajpath.2014.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsitsipatis D., Klotz L.O., Steinbrenner H. Multifaceted functions of the forkhead box transcription factors FoxO1 and FoxO3 in skin. Biochim Biophys Acta. 2017;1861(5 Pt A):1057–1064. doi: 10.1016/j.bbagen.2017.02.027. [DOI] [PubMed] [Google Scholar]
- 20.Goldminz A.M., Au S.C., Kim N., Gottlieb A.B., Lizzul P.F. NF-kappaB: an essential transcription factor in psoriasis. J Dermatol Sci. 2013;69(2):89–94. doi: 10.1016/j.jdermsci.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 21.Gęgotek A., Skrzydlewska E. The role of transcription factor Nrf2 in skin cells metabolism. Arch Dermatol Res. 2015;307(5):385–396. doi: 10.1007/s00403-015-1554-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.