Abstract
Primary cancers disseminate both single circulating tumor cells (CTCs) and CTC “clusters,” the latter of which have been shown to demonstrate greater metastatic propensity and adverse impact on prognosis. Many devices developed to isolate single CTCs also capture CTC clusters, but there is translational potential for a platform specifically designed to isolate CTC clusters. Herein, we introduce our microfluidic device for isolating CTC clusters (“Microfluidic Isolation of CTC Clusters” or MICC), which is equipped with ∼10 000 trap chambers that isolate tumor cell clusters based on their large sizes and dynamic force balance against a pillar obstacle in the trap chamber. Whole blood is injected, followed by a wash step to remove blood cells and a final backflush to release intact clusters for downstream analysis. Using clusters from tumor cell-line and confocal microscopy, we verified the ability of the MICC platform to specifically capture tumor cell clusters in the trap chambers. Our flow rate optimization experiments identified 25 μl/min for blood injection, 100 μl/min as wash flow rate, and 300 μl/min as the release flow rate – indicating that 1 ml of whole blood can be processed in less than an hour. Under these optimal flow conditions, we assessed the MICC platform’s capture and release performance using blood samples spiked with different concentrations of clusters, revealing a capture efficiency of 66%–87% and release efficiency of 76%–90%. The results from our study suggest that the MICC platform has the potential to isolate CTC clusters from cancer patient blood, enabling it for clinical applications in cancer management.
INTRODUCTION
To metastasize, primary tumors must shed cells that enter the circulation, lodge in secondary organs, and proliferate.1 These circulating tumor cells (CTCs) can be solitary or be released as groups of cells, known as a cluster.2,3 CTC clusters originate not from the proliferation of a single tumor cell progeny in the vasculature or the intravascular aggregation of single CTCs, but from cell clumps released by the primary tumor into the bloodstream.4 First reported in the 1950s,5,6 CTC clusters consist of tumor cells as well as blood, immune, and/or stromal cells.6,7 Studies in mice have shown that CTC clusters have higher metastatic potential than single CTCs.5 Moreover, the presence of CTC clusters in patients with breast,4,8 prostate,4 colorectal,9,10 or lung cancer11 has been associated with lower survival rates and worse prognosis. Thus, CTC clusters have significant potential to not only offer insights into cancer metastasis but also serve as a clinical marker for cancer management in patients.
One of the main obstacles to realizing the full clinical potential of CTC clusters is that their isolation from patient blood samples is technically challenging since clusters are rare and moreover can be prone to breakage under shear forces.7 Nevertheless, several microfluidic devices developed to specifically isolate single CTCs12–14 have also reported the presence of CTC clusters. Since these devices are not designed to specifically isolate CTC clusters, they are less reliable in capturing them or preserving their integrity.
To date, only a few dedicated devices have been developed to isolate CTC clusters.9,15 Microfilters have been used to capture clusters,9,16,17 where the cluster can be arrested at a pore in the filter due to the significant larger size of clusters than single cells. However, these methods are prone to loss of some clusters due to their deformability and subsequent transit through the pores.18 In contrast to this method, Sarioglu et al.15 developed the Cluster-Chip, which contains a parallelized array of pillars with each pillar having the shape of an equilateral triangle. The authors showed that a cluster can be wedged within a three-pillar unit and arrested due to a dynamic force balance between cell-cell adhesion strength, fluid drag forces, wall friction, and reactionary forces from the pillars. A potential drawback of the device design could be that the sharp edge of the triangular pillar in combination with the opportunity for the clusters to interact with multiple three-pillar units in the device may lead to cluster damage and inefficiency in capture.
In this study, we build on the previously-mentioned devices and developed a new microfluidic device for isolating CTC clusters (“Microfluidic Isolation of CTC Clusters” or MICC platform) from unprocessed blood samples. The basic geometry of the capture unit involves a trap chamber with a pillar that bifurcates flow. Similar to Sarioglu et al.,15 this capture unit uses the dynamic force balance between cells, fluid and channel walls to trap tumor cell clusters. The capture units are embedded within a unique fluidic network, where fluid is distributed uniformly and interaction of clusters with the bifurcating-flow pillars is minimized. The MICC platform offers a new method for the reliable isolation of intact CTC clusters, which can be analyzed for prognostication and to better understand mechanisms of metastasis.
MATERIALS AND METHODS
MICC platform design and fabrication
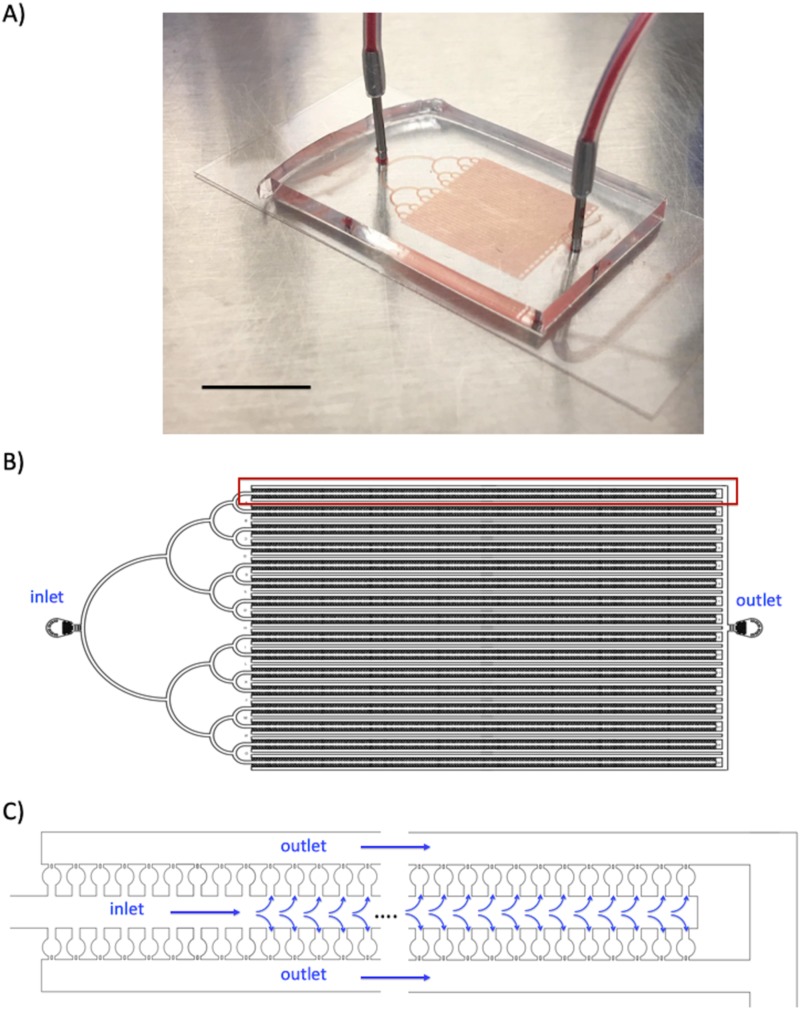
We designed the MICC platform to capture spiked tumor cell clusters from unprocessed blood samples [Fig. 1(a)]. The fluidic network in the device [Fig. 1(b)] comprises of three sections: (1) a treelike entrance section that evenly distributes the flow inside the device; (2) a capture section with thousands of individual trap chambers connected in a series of parallel units, in which isolation of the clusters occurs; and (3) an outlet section that collects the outlet flow from each row of chambers. As shown in Fig. 1(c), the basic unit in the capture section contains a long central inlet channel lined with entrances to 312 trap chambers and 2 peripheral outlet channels. The inlet and outlet channels are 203.3 ± 2.6 μm wide. Because the inlet channel has a dead-end, the fluid must flow through the trap chambers to the outlet channels. Each unit has its own outlet channel, which prevents cells in the outlet channels of one unit from flowing into the inlet channel of another unit. The capture section has 32 units × 312 traps per unit, amounting to 9984 traps (∼10 000) for isolation of clusters. The entire network has a uniform height of 23.2 ± 1.9 μm.
FIG. 1.
MICC device design and flow network. (a) Photograph of the PDMS-based MICC device. The scale bar represents 10 mm. (b) AutoCAD design of the device, which has 3 sections: (1) an inlet section on the left for distributing flow, (2) a capture section in the middle for trapping tumor cell clusters with blood flowing through, and (3) an outlet section on the right. (c) Schematic of one row of trap chambers in the MICC device.
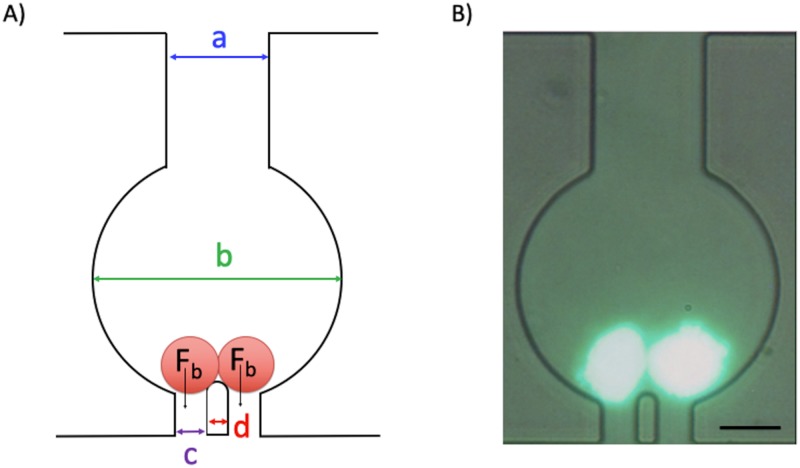
Each trap chamber has a narrow inlet channel that feeds into a circular region and is connected to an exit channel [Fig. 2(a)]. The chamber’s narrow exit contains two gaps separated by a pillar obstacle—allowing single cells and red blood cells to pass through while retaining cell clusters. We chose a gap size of 15.7 ± 1.1 μm because the average size of HCT116 tumor cells is 16.2 ± 3.7 μm (also, many other cell lines such as the ones originated from breast and prostate cancers are in the same size, mean ∼18 μm19,20). Given that a cluster would comprise at least two or more cells, this gap size is small enough to retain clusters but large enough to allow single cells to pass. Also, the pillar obstacle between the gaps acts as a fulcrum that provides balanced forces Fb to clusters encountering the pillar and if the fluid forces are not too strong, clusters are retained behind the obstacle and do not squeeze through one of the gaps [Figs. 2(a) and 2(b)]. In addition, once a cluster enters the trap chamber, it encounters the pillar obstacle only once, minimizing cluster breakup (unlike the design of Sarioglu et al.,15 where the cluster can interact with several three-pillar units as it is traversing through the network).
FIG. 2.
Design of the trap chamber for capturing tumor cell clusters. (a) Schematic of one trap chamber. The 2 red circles represent cells in a cluster. Fb is the balance force acting when a binary cluster encounters a pillar that bifurcates the flow. The geometric dimensions highlighted in the chamber are a = 51.6 ± 1.3 μm, b = 122.4 ± 1.6 μm, c = 15.7 ± 1.1 μm and d = 11.4 ± 0.9 μm. (b) Fluorescent image of a cluster with two cells captured inside a chamber. Celltracker™ green CMFDA dye has been used for the fluorescent staining of the cells. The scale bar represents 50 μm.
To fabricate the MICC platform, we used a standard soft lithography method described previously.21 Briefly, molds were made by spin-coating SU8-2035 negative photoresist (MicroChem, Westborough, MA) on a silicon wafer with a 7.63-cm diameter. The thickness of the spin-coated layer corresponded to the height of the inlet and outlet channels. Polydimethylsiloxane (PDMS), a transparent biocompatible polymer, was poured into the molds. It was then degassed in desiccators and incubated in oven for 2 h at 80 °C. The solid PDMS was cut and peeled from the molds. The inlet and outlet holes were punched using Harris Uni-Core sampling tools (1-mm outer diameter). The MICC devices were then bonded to glass cover slides using the air plasma (Plasma Cleaner, Harrick Plasma, Ithaca, NY), filled with phosphate-buffered saline (PBS) to keep them wet and refrigerated until use.
Flow simulations
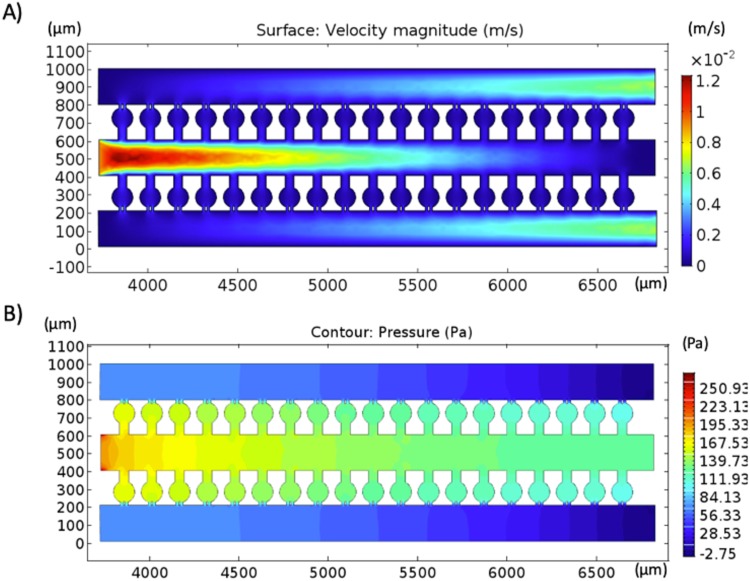
To estimate the flow velocity and pressure within the MICC platform, we used COMSOL Multiphysics modeling software (version 5.2; Comsol, Inc., Burlington, MA) to simulate 2D flow through a 20-trap chamber section of the MICC device using a fixed flow rate of ∼1 μl/min (calculated from 30 μl/min in device inlet divided by 32 units) in the inlet channel [Figs. 3(a) and 3(b)]. The simulation showed that the flow velocity inside the trap chambers is <2 mm/s [Fig. 3(a)], which is within the limit of flow velocities in blood capillaries in vivo (<2 mm/s)22 and far below that of most other microfluidic- or filtration-based methods of CTC isolation.23,24 This velocity estimate from the 2D simulations should be considered the upper limit since the flow velocity in the actual 3D chamber would be lower, and also the inlet flow rate (of 1 μl/min) is splitting into 312 trap chambers in the MICC device rather than the 20-trap chambers we simulated. Simulations also show that the pressure inside each chamber remains within 80–170 Pa, which is below the capillary pressure (1300–4500 Pa25), leaving the clusters intact [Fig. 3(b)]. Also, the pressure drop across each trap chamber is <200 Pa, imposing low stress on the clusters.
FIG. 3.
Two-dimensional simulation of the flow in the MICC device. (a) Velocity field and (b) pressure field.
Cell lines and blood samples
For the experiments, we used with HCT116 colorectal cancer cells (kindly provided by E. Vilar, MD Anderson Cancer Center) spiked in the blood of healthy donors. The blood from healthy donors were obtained from MD Anderson Cancer Center through informed written consent involving two protocols approved by the Institutional Review Board (IRB) (PA14-0867 and PA11-0670). HCT116 was grown in McCoy’s 5A media (1×, with L-glutamine) with 5% fetal bovine serum (FBS) and 1% Penicillin-streptomycin. To harvest the cells, after removing the media and washing with PBS, 5 ml trypsin/EDTA solution was added to the cells in 75 cm2 cell-culture flasks, incubated for 3–4 min and neutralized by 550–600 μl FBS. The clusters were generated by gentle pipetting of cells in the culture flasks during trypsinization process, in order to prevent the breakup of cell clumps by high shear stress.
MICC optimization and performance assays
We used a syringe pump (kdScientific, Holliston, MA) to inject 1-ml samples of healthy donor blood spiked with HCT116 colorectal cancer cells into the MICC device at fixed flow rates of 10, 25, 50, or 100 μl/min. We tested the device performance at various concentrations of spiked clusters ∼100, 500, 1000, or 5000 clusters in 1 ml of normal blood. The clusters were counted manually from microscopy images and their concentration accordingly adjusted. In order to recapitulate the range of clusters in the clinical settings, the concentration of clusters was kept below 5000 and as a result we did not observe fouling to be a main concern in our microfluidic device. In addition, coatings such as bovine serum albumin could cause slippery obstacles and less capture efficiency of the clusters.19 Thus, no coating was used on the device.
Moreover, to maintain an even distribution of clusters in the flow and prevent blood clotting inside the device, a magnetic stir bar (3.2-mm diameter, 12-mm length) was inserted in the syringe (BD 1-ml TB syringe, diameter, 4.1 mm) and the blood sample was mixed periodically. After blood injection was complete, we injected PBS into the MICC device at 100 μl/min to flush the red blood cells and other single cancer cells. To release the captured clusters, we reversed the flow by injecting PBS into the device outlet. This PBS backflush was done at 300 μl/min and the tumor cells clusters were collected.
To count the number of cells in each captured cluster, we stained their nuclei with NucBlue Live Cell Stain (Invitrogen, Carlsbad, CA) and used confocal microscopy to visually observe and manually count the captured clusters within the device. To visualize the cluster morphology, we fluorescently labeled the epithelial cell adhesion molecule (EpCAM) (Alexa Flour 488 conjugate, Cell Signaling Technology, Danvers, MA) and nucleus (NucBlue) (Thermofisher Scientific, Waltham, MA). Celltracker™ green CMFDA dye was used to stain the whole cell. The devices were imaged with an LSM 880 Confocal Laser Scanning Microscope (Zeiss, Oberkochen, Germany), and fluorescent images were captured by oil immersion optics (63× magnification). Also, alamarBlue cell viability reagent (ThermoFisher Scientific, Waltham, MA) and Spectramax M2 plate reader (Molecular devices, San Jose, CA) were used to monitor the viability and proliferation rate of captured and released clusters. To measure the capture efficiency of the MICC device, for each sample, we compared the number of clusters before the injection with the number of clusters captured inside the device; dividing the latter number by the former number yielded the device’s capture efficiency. Moreover, to measure the device’s release efficiency, we divided the number of clusters released from the device by the number of clusters captured inside the device. All experiments were done at room temperature. We used the nonparametric Kolmogorov-Smirnov test to analyze the differences between each data set and considered p-values <0.05 to be statistically significant.
RESULTS AND DISCUSSION
MICC platform efficiently captures tumor cell clusters
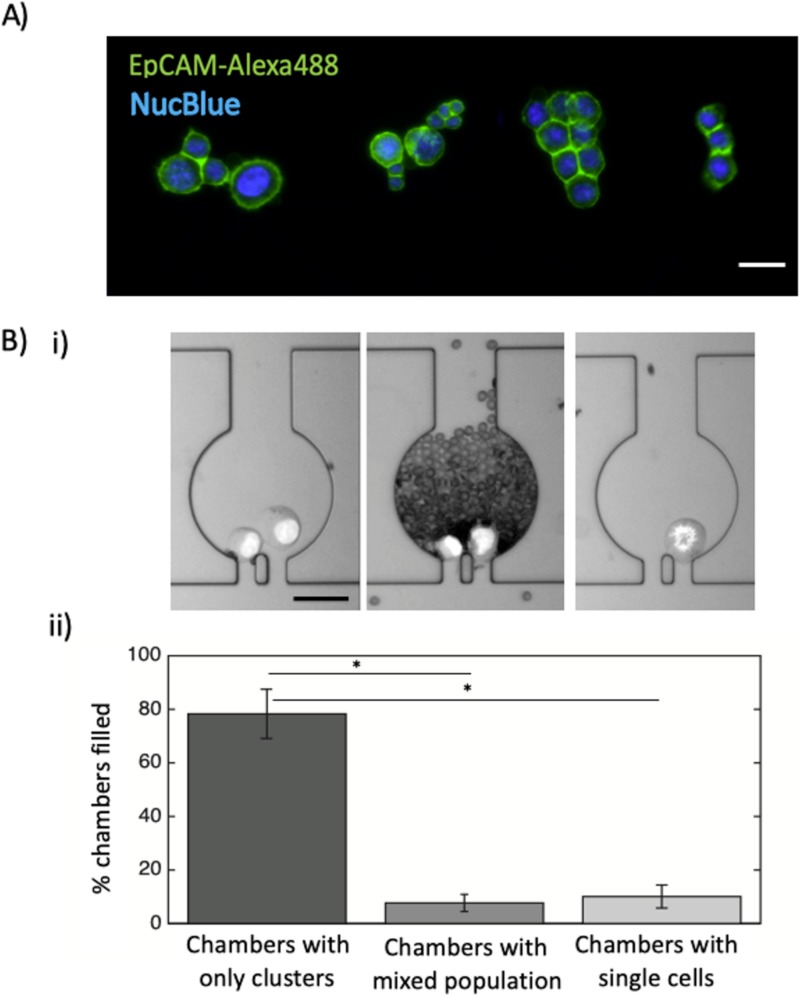
Direct visualization with confocal microscopy confirmed that the MICC platform captures cell clusters [Fig. 4(a)]. Confocal microscopy of EpCAM- and NucBlue-labeled HCT116 cells (N = 3 independent replicate experiments) revealed that the device could capture clusters of 2–13 cells with different morphologies from unprocessed blood [Fig. 4(a)]. This demonstrates that the network design and the fluid stresses are suitable for capturing spike cell clusters with a large range in size.
FIG. 4.
Imaging and enumeration of clusters captured in the MICC device. (a) Fluorescent confocal microscopy images of captured clusters inside the MICC device. Each cluster is within one chamber. The scale bar represents 50 μm. (b-i) Bright field snapshots of different chambers containing tumor cell clusters (left), red blood cells and clusters (middle), and single cells (right). (b-ii) Mean percentages of chambers containing only clusters or mixed-cell populations or single cells (N = 3 devices). The whiskers show standard deviation. *p-value < 0.01.
We next tested how many of the chambers in the MICC platform would contain pure clusters, single cells and mixed-cell populations after injection of blood sample spiked with ∼1000 clusters/ml. We found that total number of chambers only filled with clusters (78.3 ± 9.2%, N = 3) [Fig. 4(b)] is much larger than the chambers filled with only single cells (10.1 ± 4.2%, N = 3, p-value < 0.01) or mixed-cell populations (<10%, N = 3, p-value < 0.01). The mixed-cell population chambers were mostly filled with red blood cells because clusters blocked the chamber exits. The relatively low percentage of chambers filled with single cells or mixed-cell populations suggests that the MICC platform is highly efficient in specifically capturing clusters of tumor cells.
Efficiency of cluster capture for various cluster concentrations and injection flow rates
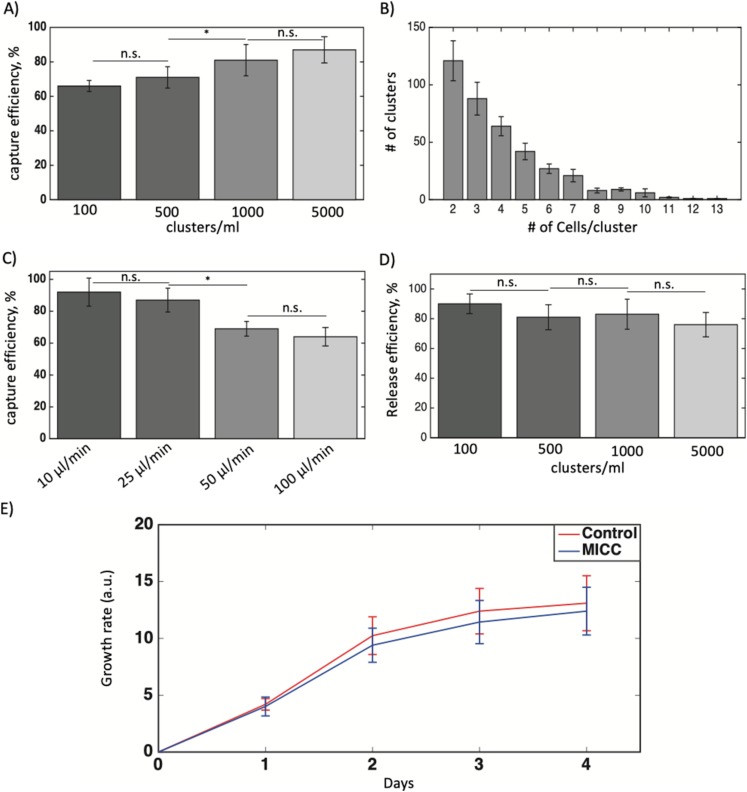
To assess our device’s capability to capture tumor clusters, we injected blood samples spiked with different concentrations of HCT116 clusters (∼100, 500, 1000, or 5000 clusters/ml) into the device at flow rate of 50 μl/min. The mean efficiencies of capture for samples with ∼100 or 500 clusters/ml (66–71%, N = 3) were significantly lower than those for samples with ∼1000 or 5000 clusters/ml [81–87%, N = 3; Fig. 5(a)] (p-value < 0.05 between ∼500 and 1000 clusters/ml). This observation suggests that the capture efficiency of MICC device for clinical samples, which tend to have lower concentration of clusters (<500 clusters/ml7,9), is still high. We also determined the cluster-size distribution (in the device) for the blood sample spiked with 500 clusters/ml [Fig. 5(b)] and found that the clusters are mostly smaller in sizes (2–4 cells/cluster). Even though the clusters were generated manually during the incubation with trypsin and spiked in the blood of healthy donors, the size distribution is similar to what was observed for clusters in cancer patients.15
FIG. 5.
Performance and optimization experiments for determining cluster capture and release efficiency. (a) The MICC device’s capture efficiencies (%) for blood samples with different concentrations of tumor cell clusters. The flow rate was fixed at 50 μl/min. (b) Distribution of clusters according to the number of cells per cluster in a blood sample. Cluster concentration is ∼500 clusters/ml. (c) The capture efficiencies (%) for blood samples with a cluster concentration of ∼500 clusters/ml at different flow rates. (d) The cluster release efficiencies (%) for blood samples with different cluster concentrations. Three devices were used in each experiment. (e) Growth rate of captured and released clusters from the MICC device (N = 6) vs control clusters (N = 6) over the course of 4 days. The whiskers show standard deviation in all figures. *p < 0.05. n.s., not significant.
The high injection flow rate (50 μl/min) used in earlier experiments may have contributed to our device’s low capture efficiency for blood samples spiked with lower cluster concentrations. To identify the optimal injection flow rate, we introduced samples spiked with ∼500 clusters/ml into the device at flow rates of 10, 25, 50, and 100 μl/min and found that the capture efficiencies using flow rates of 10 and 25 μl/min were significantly higher than those using rates of 50 and 100 μl/min (p-value < 0.05) [Fig. 5(c)]. This higher capture efficiency at lower flow rates (velocity) is due to less shear stress imposed on clusters leading to less chance of cluster escape through the gaps at low flow rates. Considering the total time of operation, we chose 25 μl/min as the optimal flow rate for the MICC platform. At this flow rate, injecting 1 ml of whole blood takes 40 min; given the additional time needed to perform the backflush step (injecting 0.5 ml of PBS at a flow rate of 0.3 ml/min), the total time for CTC cluster isolation and recovery is less than 1 h.
Efficiency of cluster release for various cluster concentrations and injection flow rates
Release of clusters enables downstream molecular analysis. We, therefore, tested the mean release efficiency across all samples with different cluster concentrations and found the efficiency (76%–90%) to not vary significantly [Fig. 5(d)]. The loss of a few clusters during the release process may be due to the handling of the device during changing of syringes before performing the backflush step. The device is fully clean and devoid of clusters after the backflush, thus omitting the chance of clusters remaining inside the device. For all samples with different cluster concentrations, we used a backflush flow rate of 300 μl/min. To determine whether the backflush flow rate influences release efficiency, we also used a flow rate of 150 μl/min for a set of samples with cluster concentrations of ∼500 clusters/ml and found that the release efficiency using this flow rate (84 ± 7.2%, N = 3) was not significantly different from 300 μl/min (88.3 ± 5.7%, N = 3) (data not shown).
Our results show that cluster release in the MICC device is independent of cluster concentration or backflush flow rate for the tested rates because in our device geometry, the cluster interacts with only one trap chamber. This is a potential advantage over the Cluster-Chip15 whose release efficiency depends on flow rate probably because the clusters interact with the parallel arrays of triangular pillars during release. In addition, these authors released the clusters at 4 °C to improve the recovery rate, which is not the case here where experiments were conducted at room temperature.
Viability and proliferation rate of captured and released clusters
Finally, to find whether the captured and released clusters by MICC device remain viable and proliferative, we used alamarBlueTM cell viability reagent and monitored their viability and growth rate (N = 6 devices) over the course of 4 days [Fig. 5(e)]. We found the released clusters have statistically similar growth rate as control clusters, which were not exposed to MICC device. This suggests that MICC device keeps the captured and released clusters intact and do not impose a traumatic condition.
Conclusions
In this study, we developed a microfluidic device, MICC, to isolate CTC clusters from unprocessed blood. The MICC platform is equipped with ∼10 000 chambers that can physically (label-free) capture and release intact tumor cell clusters based on entrapment in a trap chamber where the dynamic forces holding a cluster are stronger than shear forces. The MICC platform demonstrated high capture and release efficiencies [on average 66%–87% (N = 3) and 76%–90% (N = 3), respectively] for 1 ml of blood at different concentrations of clusters. As future work, this pilot study must be validated using samples from cancer patients. These assessments will enable us to fine-tune our MICC device to ensure that its future clinical use reliably yields relevant data for the detection and treatment of various cancers. Also, to further validate MICC device performance and better comparison, it is necessary to test more cell lines and clinical samples from cancer patients.
ACKNOWLEDGMENTS
This research was supported in part by the Cancer Prevention and Research Institute of Texas (CPRIT) (No. RP160517) and the National Cancer Institute (NCI) (No. CA218230) grants for which A.M. is a co-principal investigator. N.K. was supported by the CPRIT Research Training Program (No. RP170067). V.B. was supported by the CPRIT Research Training Program (Nos. RP140106 and RP170067) and NCI (Nos. T32CA217789-03 and U54CA096297). J.J.L. was supported by the National Institutes of Health (NIH) (No. T32CA009599). S.A.V. acknowledges support from the CPRIT (No. RP140298). The manuscript was edited by Department of Scientific Publications at University of Texas MD Anderson Cancer Center. A.S. was supported by the German Research Foundation (DFG Grant) (No. SE-2616/2-1). A.M. discloses receiving royalties from Hangzhou Guangkeande (Cosmos) Biotechnology Company LTD for being a coinventor on a license related to pancreatic cancer early detection, which is managed by the MD Anderson Conflict of Interest Committee. There are no other conflicts of interests to declare.
Note: This paper is part of the special issue on Microfluidics, Circulating Biomarkers and Cancer.
REFERENCES
- 1.O’Flaherty J. D. et al. , “Circulating tumour cells, their role in metastasis and their clinical utility in lung cancer,” Lung Cancer 76, 19–25 (2012). 10.1016/j.lungcan.2011.10.018 [DOI] [PubMed] [Google Scholar]
- 2.Plaks V., Koopman C. D., and Werb Z., “Cancer. Circulating tumor cells,” Science 341, 1186–1188 (2013). 10.1126/science.1235226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aceto N., Toner M., Maheswaran S., and Haber D. A., “En route to metastasis: Circulating tumor cell clusters and epithelial-to-mesenchymal transition,” Trends Cancer 1, 44–52 (2015). 10.1016/j.trecan.2015.07.006 [DOI] [PubMed] [Google Scholar]
- 4.Aceto N. et al. , “Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis,” Cell 158, 1110–1122 (2014). 10.1016/j.cell.2014.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fidler I. J., “The relationship of embolic homogeneity, number, size and viability to the incidence of experimental metastasis,” Eur. J. Cancer 9, 223–227 (1973). 10.1016/S0014-2964(73)80022-2 [DOI] [PubMed] [Google Scholar]
- 6.Duda D. G. et al. , “Malignant cells facilitate lung metastasis by bringing their own soil,” Proc. Natl. Acad. Sci. U.S.A. 107, 21677–21682 (2010). 10.1073/pnas.1016234107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fabisiewicz A. and Grzybowska E., “CTC clusters in cancer progression and metastasis,” Med. Oncol. 34, 12 (2017). 10.1007/s12032-016-0875-0 [DOI] [PubMed] [Google Scholar]
- 8.Paoletti C. et al. , “Significance of circulating tumor cells in metastatic triple-negative breast cancer patients within a randomized, phase II trial: TBCRC 019,” Clin. Cancer Res. 21, 2771–2779 (2015). 10.1158/1078-0432.CCR-14-2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cima I. et al. , “Tumor-derived circulating endothelial cell clusters in colorectal cancer,” Sci. Transl. Med. 8, 345ra389 (2016). 10.1126/scitranslmed.aad7369 [DOI] [PubMed] [Google Scholar]
- 10.Molnar B., Ladanyi A., Tanko L., Sreter L., and Tulassay Z., “Circulating tumor cell clusters in the peripheral blood of colorectal cancer patients,” Clin. Cancer Res. 7, 4080–4085 (2001). [PubMed] [Google Scholar]
- 11.Hou J. M. et al. , “Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small-cell lung cancer,” J. Clin. Oncol. 30, 525–532 (2012). 10.1200/JCO.2010.33.3716 [DOI] [PubMed] [Google Scholar]
- 12.Stott S. L. et al. , “Isolation of circulating tumor cells using a microvortex-generating herringbone-chip,” Proc. Natl. Acad. Sci. U.S.A. 107, 18392–18397 (2010). 10.1073/pnas.1012539107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sheng W. et al. , “Capture, release and culture of circulating tumor cells from pancreatic cancer patients using an enhanced mixing chip,” Lab Chip 14, 89–98 (2014). 10.1039/C3LC51017D [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagrath S. et al. , “Isolation of rare circulating tumour cells in cancer patients by microchip technology,” Nature 450, 1235–1239 (2007). 10.1038/nature06385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarioglu A. F. et al. , “A microfluidic device for label-free, physical capture of circulating tumor cell clusters,” Nat. Methods 12, 685–691 (2015). 10.1038/nmeth.3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang Y. T., Doh I., Byun J., Chang H. J., and Cho Y. H., “Label-free rapid viable enrichment of circulating tumor cell by photosensitive polymer-based microfilter device,” Theranostics 7, 3179–3191 (2017). 10.7150/thno.19686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adams D. L. et al. , “The systematic study of circulating tumor cell isolation using lithographic microfilters,” RSC Adv. 9, 4334–4342 (2014). 10.1039/C3RA46839A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Au S. H. et al. , “Clusters of circulating tumor cells traverse capillary-sized vessels,” Proc. Natl. Acad. Sci. U.S.A. 113, 4947–4952 (2016). 10.1073/pnas.1524448113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamyabi N., Khan Z. S., and Vanapalli S. A., “Flow-induced transport of tumor cells in a microfluidic capillary network: Role of friction and repeated deformation,” Cell. Mol. Bioeng. 10, 563–576 (2017). 10.1007/s12195-017-0499-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamyabi N. and Vanapalli S. A., “Microfluidic cell fragmentation for mechanical phenotyping of cancer cells,” Biomicrofluidics 10, 021102 (2016). 10.1063/1.4944057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia Y. and Whitesides G. M., “Soft lithography,” Annu. Rev. Mater. Sci. 28, 153–184 (1998). 10.1146/annurev.matsci.28.1.153 [DOI] [Google Scholar]
- 22.Bollinger A., Butti P., Barras J. P., Trachsler H., and Siegenthaler W., “Red blood cell velocity in nailfold capillaries of man measured by a television microscopy technique,” Microvasc. Res. 7, 61–72 (1974). 10.1016/0026-2862(74)90037-5 [DOI] [PubMed] [Google Scholar]
- 23.Coumans F. A., van Dalum G., Beck M., and Terstappen L. W., “Filtration parameters influencing circulating tumor cell enrichment from whole blood,” PLoS One 8, e61774 (2013). 10.1371/journal.pone.0061774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vona G. et al. , “Isolation by size of epithelial tumor cells: A new method for the immunomorphological and molecular characterization of circulating tumor cells,” Am. J. Pathol. 156, 57–63 (2000). 10.1016/S0002-9440(10)64706-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guyton A. C. and Hall J. E., Textbook of Medical Physiology, 11th ed. (Elsevier Saunders, 2006), pp. 161–194. [Google Scholar]