Abstract
Short (“seed”) or extended base-pairing between microRNAs (miRNAs) and their target RNAs enable post-transcriptional silencing in many organisms. These interactions allow the computational prediction of potential targets. In model organisms, predicted targets are frequently validated experimentally, hence meaningful miRNA-regulated processes are reported. However, in non-models, these reports mostly rely on computational prediction alone. Many times, further bioinformatic analyses such as gene ontology (GO) enrichment are based on these in silico projections. Here we review such approaches, highlight their caveats and demonstrate the ease of picking false targets from predicted lists. We discuss discoveries that shed new light on how miRNAs evolved to regulate targets in various phyletic groups, and discuss the pitfalls of target identification in non-model organisms. Our goal is to prevent the misuse of bioinformatic tools, as they cannot bypass the biological understanding of miRNA-target regulation.
Introduction
microRNAs (miRNAs) are small non-coding RNAs of 20-24 nucleotides (nt) that regulate messenger RNA (mRNA) levels in many eukaryotic lineages [1–3]. In bilaterian animals (the vast majority of extant animals) and land plants they play important roles in disease and most aspects of physiology because they are major components in canalization of development, fine tuning of expression and genetic switching [3–5].
Their biogenesis begins from accurate cleavage of longer precursors by the RNAse III enzymes Drosha and Dicer in animals, or just Dicer in plants (reviewed in [3, 6]). The product of this process is a duplex of small RNAs (sRNA), of which one strand is preferentially chosen to load into an Argonaute (AGO) protein, the core of the RNA Induced Silencing Complex (RISC). The chosen strand then guides the RISC via base-pairing to suppress its targets [7]. In theory, the relatively constant regularity of miRNA biogenesis should allow their efficient bioinformatic identification and annotation. However, even this initial stage in the study of miRNA biology raises various challenges that were extensively discussed elsewhere [8–13].
Different lineages display various strategies by which miRNAs together with RISC execute the target downregulation. In plants the dominating mechanism is a high miRNA-target complementarity recognition that is required to mediate the direct cleavage of the target (slicing) and/or its translational inhibition and destabilization [14–20]. The majority of animals, however, utilize partial complementarity as a main mode of target recognition. This weak interaction, mediates de-adenylation and translational inhibition of the targets (reviewed in [21]).
The different modes of target recognition and repression by miRNAs in plants and bilaterians might influence the number of targets each miRNA would have, and the magnitude of their downregulation (reviewed in [22]). The fact that each miRNA in bilaterian animals can bind to numerous transcripts together with the fact that transcription factors are among the targets, led to the suggestion that whole genetic networks are regulated by miRNAs [4]. The translational inhibition and transcript destabilization, that is carried out by miRNAs in many animals is on one hand very subtle [23], but on the other can enable reversibility [4, 24], hence contributing to complex regulatory networks. Mild reduction of dosage sensitive genes such as transcription factors or RNA-binding proteins might be sufficient to generate significant downstream effects as each of these have numerous targets of its own. In plants, slicing is a prominent mechanism that has a stronger and non-reversible effect on targets [14, 17, 19]. However, the co-existence of milder translational inhibition [15, 16, 18, 20, 25] and the tendency of miRNAs to target transcription factors also in plants [26–28] imply that complex target regulation is likely to occur also in this kingdom.
Discoveries of miRNA regulated processes rely on identification of genes regulated by specific miRNAs. For this purpose, target identification methods were developed [29–32]. These programs rely on principles of miRNA-target interaction in plants and animals and were discussed in contexts of ways of improving predictability of real targets, and in contexts of comparing their accuracies [32, 33]. These software and the discussions regarding their limitations were always oriented to the knowledge we had from a handful of model organisms. However, the recent expansion of miRNA studies, of functional miRNA-target interactions in previously uncharacterized organisms, shed new light on alternative modes of target regulation [34–36]. For example, cnidarians (sea anemones, corals, hydroids and jellyfish) are non-bilaterian animals whose miRNAs to targets with a high complementarity, similarly to plants [34, 35]. Therefore, it is time to revise the previous concepts of “animal-like” or “plant-like” target regulation as these might not precisely reflect the real biology of miRNAs. Moreover, miRNAs are being discovered in neither plant nor animal lineages of eukaryotes [37–39], where targets are being predicted without experimental validation despite the lack of knowledge of their mode of target recognition.
Using recent experimental data about miRNA-target interactions from model and non-model organisms we re-evaluate here the levels of false positives among predicted targets. We further show that the reported conclusions about miRNA regulated biological processes when based on prediction alone in non-model organisms are most likely false. We postulate that evolutionary understanding of how the miRNA pathway evolved is essential for understanding the limitations in miRNA-target identification methods and we highlight previously undiscussed considerations that should be taken into account in non-model organisms to help distinguishing real miRNA targets from false.
The majority of predicted targets is false
Why do researchers trust target prediction software?
Some regulatory non-coding RNAs such as long non-coding RNAs (lncRNAs) do not share features that enable an accurate prediction of their function, however, the mechanisms by which miRNAs regulate gene expression are well understood. By base-pairing, miRNAs guide the RISC to reduce the levels of the targeted mRNA (reviewed in [2, 3]). As it was experimentally shown that the miRNA system is involved in crucial biological processes such as development and disease in plants and animals [2, 3, 40, 41], many studies aim to identify miRNAs, their targets and the biological process that they regulate. Base-pairing enables to predict targets of miRNAs; the fly in the ointment is that the binding sites are often as short as 7 nucleotides and can be easily found by chance in large datasets of sequences. It has been more than a decade since it was shown that the exclusion of non-conserved binding sites from the lists of predicted targets considerably reduces such noise [42]. Since then, many prediction software were designed, some that take evolutionary conservation of target sites into account (for example: TargetScan [32]) and some that do not (for example: miRanda [30] or PITA [31]). The latter are more frequently used when genomic data from closely related species is unavailable, but not always. It seems that some researchers tend to overestimate the prediction accuracy of such software, to a degree that experimentally non-validated miRNA targets are speculated to control various biological processes [37–39, 43–51]. This is probably due to the misconception that the parameters that these software take into account (e.g., computation of miRNA-mRNA favorable thermodynamics and secondary structure of mRNA) enable to efficiently eliminate most false positives.
We believe that probable or at least reasonable, speculations and suggestions about putative processes promote science. However, how probable is a computationally predicted miRNA target to be real? Now that high-throughput techniques for miRNA target identification are available [52–55], we decided to make this estimation.
What are the actual noise levels of target prediction?
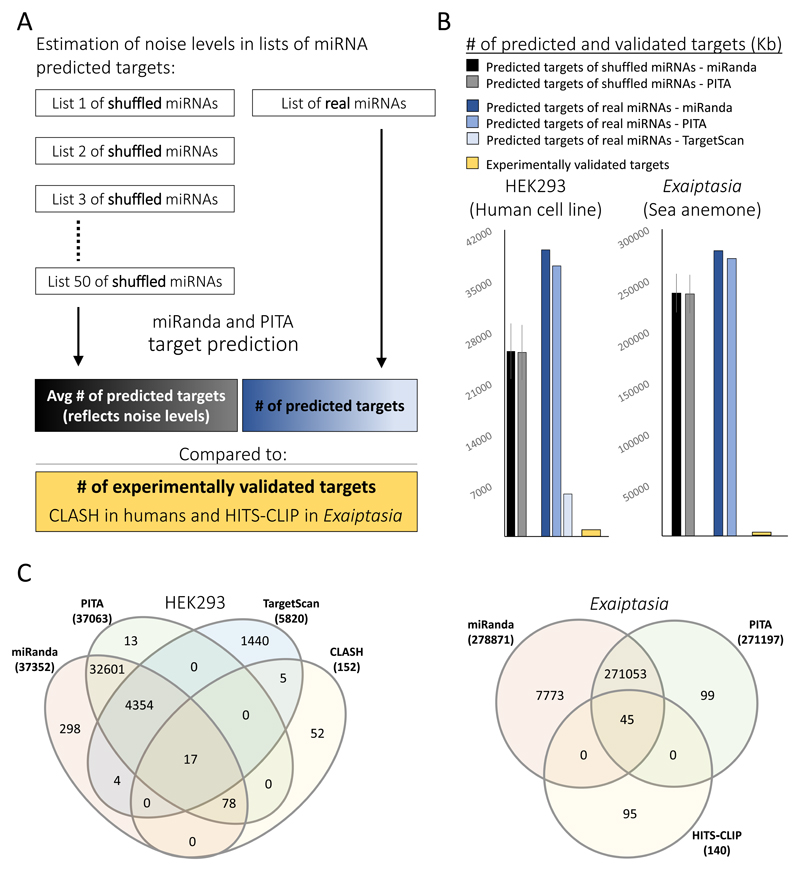
We compared the number of predicted targets to the number of targets that were experimentally validated by Cross-linking, Ligation And Sequencing of Hybrids (CLASH) and High-Throughput Sequencing of RNA isolated by Cross-Linking and Immunoprecipitation (HITS-CLIP) in a HEK293 cell line [55] and in the sea anemone Exaiptasia pallida [35] (previously called Aiptasia pallida), respectively (Fig. 1A). The first example (HEK293), was chosen as a well-studied reference cell line to highlight the consequences of using prediction tools alone. The second example (Exaiptasia) was chosen to highlight the consequences of applying these tools on a lesser studied organism, where such prediction approaches are used in spite of not knowing [37–39] or ignoring [48] the miRNA-mRNA interaction mode. To maximize the accuracy of the analysis we chose 20 abundant HEK293-specific miRNAs [56], and predicted their targets in a dataset that includes 11,741 transcripts that are expressed in this cell line. This was done to minimize the non-relevant biological noise and to provide the best conditions for the software to generate its optimal output. In Exaiptasia, there are no available transcriptomes for specific cell-types, therefore we predicted targets for all 46 known miRNAs [35] in the entire transcriptome [57], both derived from whole animals. We compared these results to the average level of noise, which is represented as the mean number of targets for 50 lists of shuffled miRNAs (Fig. 1B). The fact that the peaks of predicted targets of real miRNAs are higher than the peak of the shuffled list might result from some enrichment for real targets. Even if this explanation is correct, then ~65% and ~85% of the predicted targets in HEK293 and Exaiptasia, respectively, are noise (calculated by dividing the noise levels -- represented by the black and gray bars -- to the number of real-miRNA predicted targets represented by the blue bars). This analysis strongly suggests that these prediction tools cannot confidently discern real targets from false ones.
Figure 1. Predicted lists of miRNA seed matching targets consist mainly of false positives.
Target prediction using miRanda and PITA strict conditions (allowing no mismatches in the seed) was performed on the 3′ UTRs of HEK293 specific transcripts obtained from The Human Protein Atlas (https://www.proteinatlas.org), and on transcripts of the sea anemone Exaiptasia pallida obtained from [57]. TargetScan results were obtained from http://www.targetscan.org [32] and filtered to exclude non-HEK293 transcripts. HEK293 targets were predicted for 20 HEK293 miRNAs from [56], and targets of Exaiptasia were predicted for all the reported miRNAs from [35]. For each organism the noise levels was calculated as follows (see also panel A for graphic description): The miRNAs in the list were shuffled 50 times, and the number of targets was predicted for each of the lists and the mean number of targets is presented by the black and gray bars in panel B. The blue bars represent the number of targets predicted for the real miRNAs. The yellow bars represent the number of experimentally validated targets from CLASH experiments in HEK293 [55], and HITS-CLIP experiments in Exaiptasia [35]. Because the prediction in HEK293 was performed on the 3′ UTRs, only experimentally-validated targets in 3′ UTRs were counted in the CLASH data. The data used for these analyses are available in https://figshare.com/s/f858ae4266c680d162be (C) Overlapping targets from the outputs of prediction software with experimentally validated targets that contain a canonical seed match. Diagram was generated using http://www.interactivenn.net [58].
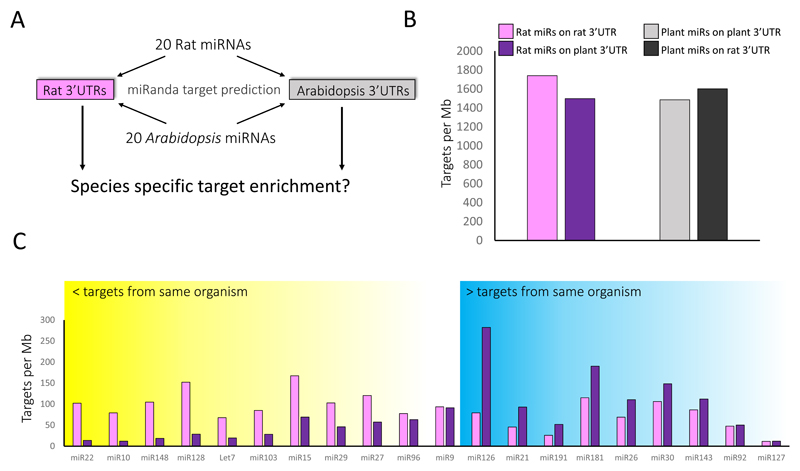
Another way to estimate the false positive rate is to compare the number of targets from the experimental data to the number of targets in the predicted lists. This comparison implies that the actual frequency of false positives might even be higher. In HEK293, only 95 out of ~37,000 PITA and miRanda, and only 22 out of 5,820 TargetScan predicted targets overlapped with the canonical targets from the experimental data obtained with CLASH (Fig. 1C, left panel). In Exaiptasia only 45 out of ~270,000 PITA and miRanda predicted targets overlapped with the experimental data from HITS-CLIP (Fig. 1C, right panel). The tradeoff between specificity and sensitivity is well known, and high-throughput experimental methods, such as CLIP and CLASH are missing real targets because of their reliance on efficiency of enzymatic reactions, and stringent purification conditions during which real targets are getting lost. However, can we confidently treat the extra ~99% of the predicted targets that are not experimentally supported, as a pool that is considerably enriched with real targets? If true, the majority of miRNAs are expected to display an enrichment in targets, when searched against the mRNA transcripts from the same organism, compared to when searched against a set of transcripts without any relevance to them, like for example transcripts of a far-related organism. Although cross-species interacting miRNAs (known as “xenomiRs”) have been previously suggested, it is now known the majority of these are false [59, 60]. Even when real [61], it is only a subset of miRNAs that act in a cross-species manner, while the vast majority are host miRNAs that regulate host transcripts. We chose two evolutionary very distant organisms that are not known to interact with one another, do not share common miRNAs, but share a similar number of genes: Arabidopsis thaliana (thale cress) and Rattus norvegicus (brown rat). For each of these organisms we chose 20 highly confident miRNAs [62–64], which were used for target prediction in a reciprocal manner (Self miRNA vs. self transcripts, and self miRNAs vs. non-self transcripts) (Fig. 2A). The number of predicted targets of each miRNA was normalized to the number of nucleotides in the transcriptome of each dataset. Whether self on self, or self on non-self, the normalized number of predicted targets per a list of miRNAs was high and very similar (fig. 2B). As for specific miRNAs, there was no tendency to exhibit a species specific enrichment in the number of targets (Fig. 2C). In some cases (9 of 20 miRNAs) miRNA had more predicted targets when checked self on self (yellow shaded), in other cases more targets were predicted for self on non-self (7 of 20 miRNA, blue shaded), and the rest were indistinguishable (white). Altogether, these results (Fig. 2) demonstrate that picking a random target from a predicted list is highly probable to be false unless the target was validated experimentally.
Figure 2. Prediction of miRNA targets in the species that express the miRNAs, is not increased compared to cross-species prediction.
(A) 20 miRNAs from brown rat (Rattus norvegicus) or Thale cress (Arabidopsis thaliana) were used to predict the number of targets that each set generates with the 3′ UTRs of each of the two organisms. (B) The sum of the predicted targets for each combination. (C) the number of putative targets for each individual rat miRNA in rat 3′ UTRs (pink) compared to the number in A. thaliana. To compare the number of targets between these two species number of targets was normalized to the total length of each 3′ UTR dataset. The number of targets are presented as targets per mega base (targets per Mb). Rat miRNAs with high confidence were chosen from MirGeneDB2.0 [64]. Arabidopsis highly confident miRNAs were chosen according to [62, 63]. The data used for these analyses is available in https://figshare.com/s/7db0263cf3e3085443b6.
Resist the urge to tell stories based on prediction alone
It can be difficult to resist the temptation not to report the putative role of a predicted target, when the putative binding site falls inside a gene that fits well with the story we desire to tell. However, our analysis shows that even a careful usage of a prediction tool (using cell-type specific transcriptome with cell-type specific miRNA list as input) (Fig. 1B, left panel) when lacking experimental support, can yield predicted lists consisting mainly of false positives (Figs. 1 and 2). It seems that speculations about miRNA-target regulation, based on prediction alone are less frequent in highly studied model organisms as experimental support can be obtained relatively easily. However, when it comes to non-model organisms, recent publications continue with bold speculations regarding the putative role of miRNAs that regulate targets that were chosen from a predicted list without any experimental support [37–39, 43–51]. A disturbing fact is that in non-model organisms the search is often made by assuming miRNA-target recognition rules that might not reflect the biology of the studied organisms [38, 39, 48]. Thus, in the next section we will focus on what biological information needs to be taken into account regarding miRNA-target identification in non-model organisms.
Target prediction in non-model organisms
The miRNA pathway originated either once or multiple times from an ancient RNA interference (RNAi) system [22, 65]. Therefore, the primary miRNA pathways, similarly to the siRNA pathways they evolved from, must have recognized targets through high complementarity. This feature enables the sRNA carriers, the Argonaute proteins (AGOs), to directly cleave target RNA (reviewed in [66]). Since its appearance, the miRNA pathway had a long time to evolve and hence the mode of action and target recognition significantly vary between different organisms.
In highly studied model animals, target slicing occurs on a very small minority of targets (often mediating the degradation of the miRNA itself [67]), and the vast majority of targets are recognized through a short seed match, and repressed through a mild translational inhibition and destabilization carried out by components of RISC [68]. On the contrary, in land plants slicing is a prominent mechanism [14, 17, 19]. Translational inhibition also occurs in plants, however, it still requires high complementarity throughout the length of the miRNA, and unlike in bilaterian animals an interaction restricted to the seed sequence would not promote any noticeable target downregulation [17, 69].
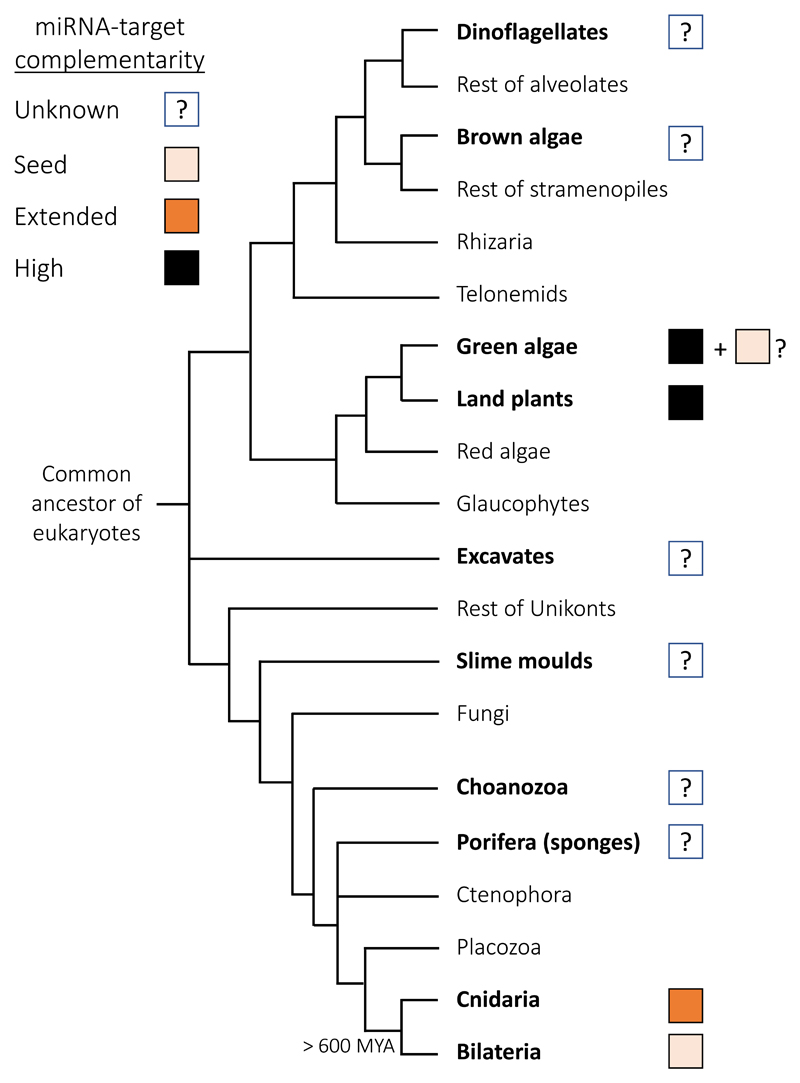
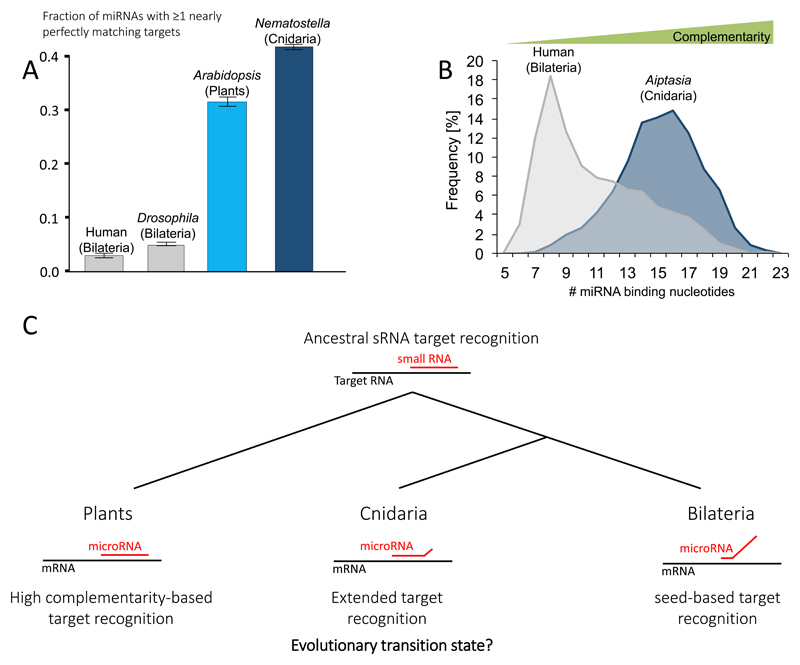
Such observations from highly studied plants and animals cause many to classify target recognition to be “plant-like” or “animal-like”, a statement that is factually false. Nematodes, flies, frogs, fish, mice and other highly studied model animals are all bilaterians. Cnidaria (sea anemones, corals, hydroids and jellyfish), is the sister group that separated from Bilateria more than 600 million years ago (MYA) (Fig. 3). It turned out that cnidarian miRNAs bind their targets with a high degree of complementarity and frequently mediate their cleavage [34]. The fraction of miRNAs with nearly perfectly matching targets in Cnidaria is even higher than in plants (Fig. 4A) [34]. The recent AGO-CLIP in Exaiptasia revealed that seed-restricted recognition does not occur in this sea anemone [35] (Fig. 4B) and combined with the previous work on other cnidarians [34] strongly suggest that matches restricted to the seed cannot mediate silencing in this animal phylum.
Figure 3. Phylogenetic topology of the major eukaryotic groups.
Groups known to have miRNAs appear in bold based on the following publications: Cnidaria and Porifera [70], Choanozoa [71], Slime molds [72], Excavates [50], Green algae [73, 74], Brown algae [75], Dinoflagellates [37], Plants [76], Bilateria [77, 78]. The current knowledge of the required complementarity levels between the miRNA to their targets in each group is indicated in colors. The tree topology is based on a previously published phylogeny [79].
Figure 4. Different modes of target recognition among bilaterians, cnidarians and plants.
(A) Fraction of miRNAs with at least one nearly perfect target in Cnidaria, Bilateria and plants. Nearly perfect matches are defined as 2.5 mismatches or less (G:U wobble is counted as 0.5 mismatches and mismatches at positions 2-7 of the miRNA are double penalized). Adapted from [34]. (B) AGO-CLIP summary of the miRNA-target complementarity levels that occur in a bilaterian and a cnidarian. Adapted from [35]. (C) Evolutionary scenario suggesting how miRNAs evolved to regulate targets, from ancestral high complementarity to seed-based target recognition in bilaterians.
Such findings demonstrate that we cannot automatically apply “animal-like” rules [48, 50], as various binding strategies exist in animals. The fact that in cnidarians miRNAs bind to their targets most frequently by 17 nt (Fig. 4B) [35], and not by full complementarity suggests that these basally branching animals possibly exhibit an evolutionary transition state, where the miRNAs evolved to not perfectly bind their targets, but still not reaching the extremely partial recognition that evolved in bilaterians (Fig. 4C). Thus, studies aiming to identify targets in non-bilaterian animals should take into account that it is highly probable that their system could not exert any regulatory effects through a seed match alone.
The existence of alternative binding strategies within a lineage is not restricted to Metazoa. A study on miRNAs in the green alga Chlamydomonas suggests that a match restricted to the seed is sufficient to mediate translational repression of a synthetic reporter [36]. The coexistence of such a system with a slicing mechanism in this species [74, 80] suggests that variation in target binding strategies might exist within some members of the Viridiplantae (plants and green algae).
Broadly, there are many eukaryotic lineages where the mechanism by which miRNAs regulate their targets is unknown (Fig. 3). Some of them, like dinoflagellates, being extremely distant from both plants and animals, leave us clueless about their miRNA mode of action. Yet studies of members of these species arbitrarily choose to apply “animal-like” or “plant-like” rules for target identification without justifying their choice. For example, two recent studies predicted targets in different dinoflagellate species (Symbiodinium kawagutii and Prorocentrum donghaiense) applying opposing prediction approaches [38, 39].
Notably, faulty target prediction is not restricted only to non-bilaterian animals and lineages where the mechanism is unknown as some studies in bilaterian non-model organisms are suffering from the same problems [43, 44, 46, 47, 49, 81]. These studies searched for seed-matching targets, which is indeed the relevant mechanism for bilaterians; however, their conclusions about the importance of miRNA related targets are most likely wrong because of the reasons discussed in the previous chapter. To summarize, we identify three sources that may cause erroneous conclusions regarding target regulation by miRNAs: (a) biological knowledge regarding the mode of action is present, but there is no awareness to the high degree of noise in a list of predicted targets, (b) biological knowledge of the miRNA-target regulation mechanism is absent and a prediction strategy is arbitrarily chosen, (c) lack of biological understanding combined with no awareness of the high degree of noise in the predicted lists.
Problems of functional annotation and enrichment of miRNA targets
Studies of miRNAs often do not stop at lists of predicted targets. Many perform functional annotation on the predicted targets, followed by gene ontology (GO) enrichment analysis for finding biological pathways regulated by certain miRNAs [38, 39, 43–46, 82]. When significant enrichment for biological processes is detected, the level of confidence in the “newly discovered” miRNA-regulated pathway is increased. There are several problems with this approach: The first concerns the functional annotation step. The most prevalent way by which annotation algorithms annotate an uncharacterized protein is by BLAST or PSI-BLAST search of homologs with a known function, assuming that the function is conserved [83]. This assumption might be a source for biases as conserved sequences between very distant organisms would get the annotation of the better studied organism, despite the fact that their function might have considerably changed in the distant relative.
The next two problems are related to two assumptions made by the researchers regarding enrichment analysis of predicted targets:
-
(1)
Predicted targets from the list are affected by the miRNA to a degree that is sufficient to drive changes in biological processes.
-
(2)
The predicted list is enriched with real targets.
The problem with the first assumption is that a seed-based interaction in animals reduces the target expression level by up to two-fold [84, 85], while the majority of the genes are not dosage sensitive [86]. Therefore, it was shown that such mild reduction in target levels, which often does not exceed the intrinsic variability of their expression levels, might have only negligible biological consequences [87].
The second assumption was revoked in the first chapter, and ideally, under such circumstances we would expect the analysis not to detect any significant enrichments in predicted lists consisting mainly of noise. Previously it was shown in a cell line that enrichment analysis produced different results when applied on predicted targets or validated targets [52]. However, one might think that such differences might result from miRNA targets that were missed by the experimental approach and are present in the predicted list. If true, it is reasonable to expect that enrichment for biological processes would be detected almost exclusively for predicted targets of real miRNAs.
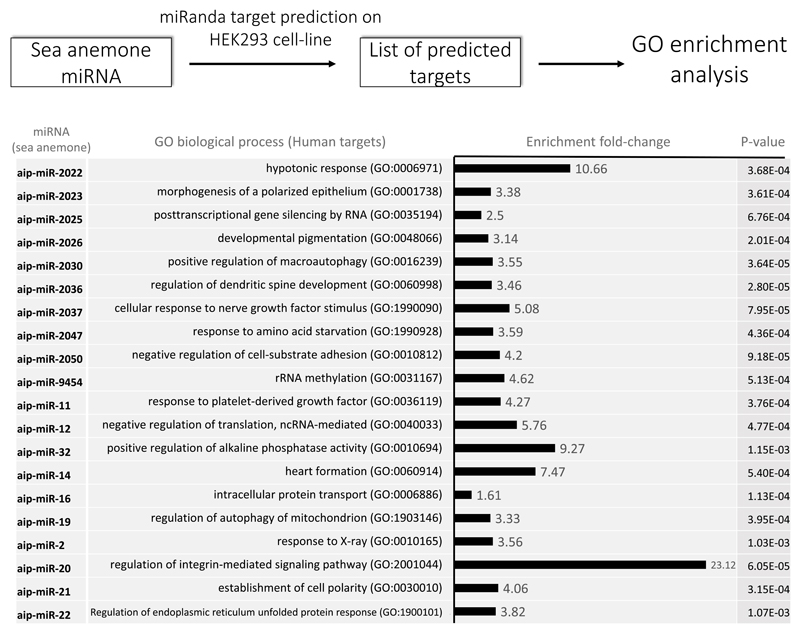
To test this idea we selected 20 sea anemone miRNAs from Exaiptasia that are not conserved with humans, thus predicting their targets in a human transcriptome would reflect noise. Targets of these miRNAs were predicted in a transcriptome of HEK293 cell-line using miRanda, and an enrichment analysis for the sets of predicted genes of each miRNA was performed (Fig. 5). Strikingly, significantly-enriched biological processes were found in the predicted lists for all of the sea anemone miRNAs (Fig. 5). A non-plausible interpretation of this absurd example is that sea anemones evolved to regulate human transcriptomes. The plausible interpretation from this analysis is that enrichment for predicted targets should not be assumed to be real.
Figure 5. Gene Ontology (GO) enrichment analysis on HEK293 predicted targets of sea anemone (Exaiptasia) miRNAs.
Target prediction was performed on the 3′ UTR of HEK293 transcripts using miRanda with strict conditions allowing no mismatches in the seed. Enrichment analyses were performed on predicted target lists of each of the individual miRNAs. The most highly enriched GO biological process for each miRNA is presented. GOs were obtained from the Gene Ontology Consortium [88, 89] and enrichment analysis was performed using PANTHER [90]. The data used for these analyses is available in https://figshare.com/s/f50b76a2ffd0153c813f.
In summary, we discussed major problems related to computational predictions of miRNA targets and their regulatory pathways. The high level of noise in lists of predicted targets (Figs 1 and 2), the failure to consider the type of the relevant miRNA-target interaction [38, 39, 48], and the pitfalls of using GO enrichment analyses on predicted targets, altogether strongly suggest that miRNA-regulated pathways being reported without experimental support [37–39, 43–51] are false.
What can be done?
In the study of the miRNAs of model organisms many of the problems we presented throughout this paper rarely occur because of the available experimental approaches to study targets. Hence, we would like to focus on what we believe should be done in order to get meaningful insights about miRNA-regulated processes in non-model organisms where the experimental possibilities are much more limited.
Taking into account the evolutionary history of the miRNA pathway
We need to start by asking ourselves whether we have an indication about the target repression mechanism of miRNAs in the examined organism (Fig. 3). Phylogenetic relationships might assist at this step. For example, because in all studied members of Bilateria such as insects, nematodes and vertebrates target repression is mediated through a short seed-match, it is very likely that non-model bilaterians exhibit the same mode of regulation. In more basally branching animals, such as cnidarians a seed recognition is most probably insufficient to mediate repression as it was experimentally shown that high-complementarity based target cleavage occur in both sea anemones and hydroids [34]. As these are distantly related cnidarians, it is most probable that all cnidarians regulate their targets in a similar manner. This notion is also supported by AGO-CLIP experiments from a sea anemone [35] (Fig. 4B).
High and low throughput methods
Whether dealing with a seed or high complementarity-based target regulation, high throughput methods such as CLIP variants or CLASH can provide a list of highly confident targets [52–55]. Success in CLIP and CLASH depends on available antibodies capable to efficiently precipitate the AGOs. If the non-model organism is close enough to a model organism the antibodies of the latter might be suitable. Otherwise, the procedure depends on a successful generation of custom antibodies, which is far from guaranteed to succeed. In organisms with a high complementarity-based mechanism such as cnidarians and plants, degradome sequencing is an additional high throughput method that enables to detect mRNAs that are cleaved at the position where they are bound by the miRNA [34, 91–94]. However, such high-throughput techniques are challenging, as they are expensive, require time-consuming experimental calibrations and adjustments and require non-trivial analysis skills. Nevertheless, their big advantage is that they can be used to study organisms lacking genetic manipulation tools.
When high-throughput techniques are not available because of limitations (resources, analysis skills, technical aspects etc.) there are some low throughput options. This will limit the researchers to test a smaller number of target candidates and some of these methods are only possible if genetic manipulation techniques are available for the organism. For high complementarity based interactions, RLM-RACE can be used to detect sliced targets. This method enables to assay if the putative target transcript is cleaved at the conserved position of AGO-slicing within the miRNA binding site [26, 28, 34, 91]. This method is also a good supplement when performing degradome sequencing, as it is highly sensitive and can help testing targets missed by the degradome [34]. In both high complementarity and seed restricted interactions, if genetic manipulations and/or transfections are available, individual targets can be validated using reporters with miRNA binding sites [36, 95]. Alternatively, one can knock-down, knock-out or mis-express individual miRNAs and then measure changes in target levels [96]. When such genetic manipulations are not possible, one can test the ability to repress by less direct methods such as co-transfection of a miRNA and a reporter construct harboring a miRNA binding site into a cell line [97, 98]. It is important to keep in mind that some of the described methods are made in artificial contexts that might not reflect the real biological conditions, and caveats of such approaches were previously discussed [99].
There is a very important point to consider regarding the interpretation of the above described methods that is equally relevant to model as well as to non-model organisms: These methods can provide reliable information about miRNA-mRNA interactions and levels of target down-regulation. However, they do not guarantee that the consequence of each of these interactions is biologically meaningful [87]. Therefore, we encourage researchers to test the biological relevance of a miRNA target by genetic tests, whenever possible.
A computational approach for finding high-complementary targets
Sometimes evolutionary relations cannot provide any meaningful clues regarding how targets are regulated by miRNAs because of the large distance of the studied organism from relatives with a known mechanism of target recognition (e.g. dinoflagelates, see Fig. 3). In such cases, we advise taking a computational approach that was previously described to provide an estimation if high matches are prevalent in the studied organisms [34]. By counting the fraction of miRNAs with at least one nearly perfect target, enrichment for highly complement target sites can be considered reliable. This is because unlike seed-restricted matches, the long nearly-perfect matches are far less prone to be noise [34]. Such analyses can provide a good indication of whether it is worth proceeding with experimental approaches to identify targets. However, a negative result of this approach is not a proof that binding via seed-restricted matches is a functional mechanism in the tested organism.
In the case of seed-restricted matches, programs such as miRanda, Targetscan, PITA, psRNATarget [29] and others are useful for getting the initial lists of predicted targets, from which it is easy to identify which transcripts had potential binding site to which miRNAs. The predicted lists generated by different programs might differ from each other and such comparisons of their performances have been previously discussed [32, 33]. In cases when studying an organism whose genome and the genomes of its closely-related species were sequenced, prediction programs that take evolutionary conservation into account would probably provide lists of more reliable targets. Yet, because of the high noise levels generated by seed-restricted target prediction algorithms (Figs. 1 and 2), discussions regarding their biological relevance should preferably be limited to cases where additional experimental support is available. Some studies try to bypass the experimental validation step by discussing only targets that were predicted by several different algorithms. We show here that this approach would probably not increase the credibility of these predicted targets: out of the 4,371 targets that overlapped between PITA, miRanda and TargetScan, only 17 were shared with the experimentally validated targets in HEK293 (Fig. 1C).
Enrichment analysis on predicted targets requires caution
When performing enrichment analysis we highly advise to include only experimentally validated targets. It was previously shown that the results of enrichment analysis on experimentally validated targets vary from the enrichments obtained with predicted ones [52]. In any case, as a result of the ease by which significant but irrelevant enrichments can be obtained (Fig. 5) we advise to be cautious about the interpretation of such results and treat them as not more than a hint for processes that are regulated by miRNAs. To further increase the chances for getting meaningful enrichments, the genes included in such analyses should be carefully annotated. However the last suggestion depends on the community that studies the given non-model organism to constantly update the terms of newly studied genes in the relevant databases.
Conclusions and outlook
miRNA-regulated processes are frequently reported in the literature. While in model organisms such reports are many times reliable because of the capabilities of experimental confirmation, in non-model organisms putative miRNA-based regulation of processes relies mostly on target prediction alone. In the first section, we exploited the recent high throughput experimental information available from model and non-model organisms and provided analyses demonstrating that the signal to noise ratios are insufficient to reliably identify miRNA targets only by computational means. Based on findings that shed new light on how the miRNA pathway evolved in animals; in the second section we highlighted the considerations that have to be taken into account to enable the identification of real targets. Moreover, we discussed the additional problems of target prediction in non-model organisms for which we lack the understanding of how targets are regulated by miRNAs. We showed that the gene ontology enrichment analyses, which are often performed on experimentally non-validated targets, are most likely meaningless, and should not be coupled to target prediction. To the best of our knowledge this work is the first to discuss caveats and pitfalls of target prediction in non-model organisms where the conclusions about miRNA regulated processes are frequently reported without experimental validation of the targets. In addition to raising these glaring issues, we provide advice on how to avoid the common pitfalls, and analyze miRNAs and their interaction with their targets even in little-studied species in a biologically meaningful way.
Acknowledgements
This research was supported by a European Research Council Starting Grant (CNIDARIAMICRORNA, 637456) to YM.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- [1].Ameres SL, Zamore PD. Nat Rev Mol Cell Biol. 2013;14:475. doi: 10.1038/nrm3611. [DOI] [PubMed] [Google Scholar]
- [2].Bartel DP. Cell. 2018;173:20. doi: 10.1016/j.cell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Voinnet O. Cell. 2009;136:669. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- [4].Flynt AS, Lai EC. Nature reviews Genetics. 2008;9:831. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hornstein E, Shomron N. Nature genetics. 2006;38(Suppl):S20. doi: 10.1038/ng1803. [DOI] [PubMed] [Google Scholar]
- [6].Kim VN, Han J, Siomi MC. Nature reviews Molecular cell biology. 2009;10:126. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- [7].Czech B, Hannon GJ. Nature reviews Genetics. 2011;12:19. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N. Nucleic acids research. 2012;40:37. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Shahid S, Axtell MJ. Methods. 2014;67:20. doi: 10.1016/j.ymeth.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Axtell MJ. RNA. 2013;19:740. doi: 10.1261/rna.035279.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Taylor RS, Tarver JE, Foroozani A, Donoghue PC. Bioessays. 2017;39 doi: 10.1002/bies.201600113. [DOI] [PubMed] [Google Scholar]
- [12].Axtell MJ, Meyers BC. Plant Cell. 2018;30:272. doi: 10.1105/tpc.17.00851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Fromm B, Billipp T, Peck LE, Johansen M, Tarver JE, King BL, Newcomb JM, Sempere LF, Flatmark K, Hovig E, Peterson KJ. Annu Rev Genet. 2015;49:213. doi: 10.1146/annurev-genet-120213-092023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Baumberger N, Baulcombe DC. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:11928. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O. Science. 2008;320:1185. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- [16].Li S, Liu L, Zhuang X, Yu Y, Liu X, Cui X, Ji L, Pan Z, Cao X, Mo B, Zhang F, et al. Cell. 2013;153:562. doi: 10.1016/j.cell.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Liu Q, Wang F, Axtell MJ. Plant Cell. 2014;26:741. doi: 10.1105/tpc.113.120972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Yang L, Wu G, Poethig RS. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:315. doi: 10.1073/pnas.1114673109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Qi Y, Denli AM, Hannon GJ. Molecular cell. 2005;19:421. doi: 10.1016/j.molcel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- [20].Gandikota M, Birkenbihl RP, Hohmann S, Cardon GH, Saedler H, Huijser P. Plant J. 2007;49:683. doi: 10.1111/j.1365-313X.2006.02983.x. [DOI] [PubMed] [Google Scholar]
- [21].Fabian MR, Sonenberg N. Nature structural & molecular biology. 2012;19:586. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- [22].Moran Y, Agron M, Praher D, Technau U. Nat Ecol Evol. 2017;1:27. doi: 10.1038/s41559-016-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Nature. 2008;455:58. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- [24].Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Cell. 2006;125:1111. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- [25].Yu B, Bi L, Zhai J, Agarwal M, Li S, Wu Q, Ding SW, Meyers BC, Vaucheret H, Chen X. Nucleic acids research. 2010;38:5844. doi: 10.1093/nar/gkq348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Llave C, Xie Z, Kasschau KD, Carrington JC. Science. 2002;297:2053. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- [27].Nodine MD, Bartel DP. Genes Dev. 2010;24:2678. doi: 10.1101/gad.1986710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Palatnik JF, Wollmann H, Schommer C, Schwab R, Boisbouvier J, Rodriguez R, Warthmann N, Allen E, Dezulian T, Huson D, Carrington JC, et al. Developmental cell. 2007;13:115. doi: 10.1016/j.devcel.2007.04.012. [DOI] [PubMed] [Google Scholar]
- [29].Dai X, Zhuang Z, Zhao PX. Nucleic acids research. 2018;46:W49. doi: 10.1093/nar/gky316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. Genome biology. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. Nat Genet. 2007;39:1278. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- [32].Agarwal V, Bell GW, Nam JW, Bartel DP. Elife. 2015;4 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Peterson SM, Thompson JA, Ufkin ML, Sathyanarayana P, Liaw L, Congdon CB. Front Genet. 2014;5:23. doi: 10.3389/fgene.2014.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Moran Y, Fredman D, Praher D, Li XZ, Wee LM, Rentzsch F, Zamore PD, Technau U, Seitz H. Genome Res. 2014;24:651. doi: 10.1101/gr.162503.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Baumgarten S, Cziesielski MJ, Thomas L, Michell CT, Esherick LY, Pringle JR, Aranda M, Voolstra CR. Mol Ecol. 2018;27:403. doi: 10.1111/mec.14452. [DOI] [PubMed] [Google Scholar]
- [36].Yamasaki T, Voshall A, Kim EJ, Moriyama E, Cerutti H, Ohama T. Plant J. 2013;76:1045. doi: 10.1111/tpj.12354. [DOI] [PubMed] [Google Scholar]
- [37].Baumgarten S, Bayer T, Aranda M, Liew YJ, Carr A, Micklem G, Voolstra CR. BMC Genomics. 2013;14:704. doi: 10.1186/1471-2164-14-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lin S, Cheng S, Song B, Zhong X, Lin X, Li W, Li L, Zhang Y, Zhang H, Ji Z, Cai M, et al. Science. 2015;350:691. doi: 10.1126/science.aad0408. [DOI] [PubMed] [Google Scholar]
- [39].Shi X, Lin X, Li L, Li M, Palenik B, Lin S. ISME J. 2017;11:2209. doi: 10.1038/ismej.2017.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. Science. 2005;308:833. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- [41].Chen X. Annual review of cell and developmental biology. 2009;25:21. doi: 10.1146/annurev.cellbio.042308.113417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Cell. 2003;115:787. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- [43].Jebessa E, Ouyang H, Abdalla BA, Li Z, Abdullahi AY, Liu Q, Nie Q, Zhang X. Oncotarget. 2018;9:17309. doi: 10.18632/oncotarget.22457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Feng X, Zhou X, Zhou S, Wang J, Hu W. Parasites & Vectors. 2018;11:172. doi: 10.1186/s13071-018-2734-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Liu M, Bruni GO, Taylor CM, Zhang Z, Wang P. Sci Rep. 2018;8:5243. doi: 10.1038/s41598-018-23611-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wang L, Zhu W, Dong Z, Song F, Dong J, Fu J. International journal of molecular sciences. 2018:19. doi: 10.3390/ijms19041209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Caballero-Perez J, Espinal-Centeno A, Falcon F, Garcia-Ortega LF, Curiel-Quesada E, Cruz-Hernandez A, Bako L, Chen X, Martinez O, Alberto Arteaga-Vazquez M, Herrera-Estrella L, et al. Dev Biol. 2018;433:227. doi: 10.1016/j.ydbio.2017.08.022. [DOI] [PubMed] [Google Scholar]
- [48].Gajigan AP, Conaco C. Mol Ecol. 2017;26:3472. doi: 10.1111/mec.14130. [DOI] [PubMed] [Google Scholar]
- [49].Durban J, Sanz L, Trevisan-Silva D, Neri-Castro E, Alagon A, Calvete JJ. J Proteome Res. 2017;16:3370. doi: 10.1021/acs.jproteome.7b00414. [DOI] [PubMed] [Google Scholar]
- [50].Liew YJ, Aranda M, Carr A, Baumgarten S, Zoccola D, Tambutte S, Allemand D, Micklem G, Voolstra CR. PLoS One. 2014;9:e91101. doi: 10.1371/journal.pone.0091101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Durban J, Perez A, Sanz L, Gomez A, Bonilla F, Rodriguez S, Chacon D, Sasa M, Angulo Y, Gutierrez JM, Calvete JJ. BMC Genomics. 2013;14:234. doi: 10.1186/1471-2164-14-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Chi SW, Zang JB, Mele A, Darnell RB. Nature. 2009;460:479. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp AC, Munschauer M, Ulrich A, et al. Cell. 2010;141:129. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Broughton JP, Pasquinelli AE. Methods. 2013;63:119. doi: 10.1016/j.ymeth.2013.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Helwak A, Kudla G, Dudnakova T, Tollervey D. Cell. 2013;153:654. doi: 10.1016/j.cell.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Panwar B, Omenn GS, Guan Y. Bioinformatics. 2017;33:1554. doi: 10.1093/bioinformatics/btx019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Baumgarten S, Simakov O, Esherick LY, Liew YJ, Lehnert EM, Michell CT, Li Y, Hambleton EA, Guse A, Oates ME, Gough J, et al. Proc Natl Acad Sci U S A. 2015;112:11893. doi: 10.1073/pnas.1513318112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Heberle H, Meirelles GV, da Silva FR, Telles GP, Minghim R. BMC Bioinformatics. 2015;16:169. doi: 10.1186/s12859-015-0611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Kang W, Bang-Berthelsen CH, Holm A, Houben AJ, Muller AH, Thymann T, Pociot F, Estivill X, Friedlander MR. RNA. 2017;23:433. doi: 10.1261/rna.059725.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Fromm B, Tosar JP, Lu Y, Halushka MK, Witwer KW. J Nutr. 2018;148:1506. doi: 10.1093/jn/nxy144. [DOI] [PubMed] [Google Scholar]
- [61].Shahid S, Kim G, Johnson NR, Wafula E, Wang F, Coruh C, Bernal-Galeano V, Phifer T, dePamphilis CW, Westwood JH, Axtell MJ. Nature. 2018;553:82. doi: 10.1038/nature25027. [DOI] [PubMed] [Google Scholar]
- [62].Axtell MJ, Bowman JL. Trends in plant science. 2008;13:343. doi: 10.1016/j.tplants.2008.03.009. [DOI] [PubMed] [Google Scholar]
- [63].Arribas-Hernandez L, Kielpinski LJ, Brodersen P. Plant physiology. 2016;171:2620. doi: 10.1104/pp.16.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Fromm B, Domanska D, Hackenberg M, Mathelier A, Hoye E, Johansen M, Hovig E, Flatmark K, Peterson KJ. bioRxiv. 2018 258749. [Google Scholar]
- [65].Shabalina SA, Koonin EV. Trends Ecol Evol. 2008;23:578. doi: 10.1016/j.tree.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Huntzinger E, Izaurralde E. Nat Rev Genet. 2011;12:99. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- [67].Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, Zamore PD. Science. 2010;328:1534. doi: 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Huntzinger E, Izaurralde E. Nature reviews. Genetics. 2011;12:99. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- [69].Iwakawa HO, Tomari Y. Molecular cell. 2013;52:591. doi: 10.1016/j.molcel.2013.10.033. [DOI] [PubMed] [Google Scholar]
- [70].Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, Degnan BM, Rokhsar DS, Bartel DP. Nature. 2008;455:1193. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Brate J, Neumann RS, Fromm B, Haraldsen AAB, Tarver JE, Suga H, Donoghue PCJ, Peterson KJ, Ruiz-Trillo I, Grini PE, Shalchian-Tabrizi K. Curr Biol. 2018;28:3288. doi: 10.1016/j.cub.2018.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Avesson L, Reimegard J, Wagner EG, Soderbom F. Rna. 2012;18:1771. doi: 10.1261/rna.033175.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Molnar A, Schwach F, Studholme DJ, Thuenemann EC, Baulcombe DC. Nature. 2007;447:1126. doi: 10.1038/nature05903. [DOI] [PubMed] [Google Scholar]
- [74].Zhao T, Li G, Mi S, Li S, Hannon GJ, Wang XJ, Qi Y. Genes & development. 2007;21:1190. doi: 10.1101/gad.1543507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Tarver JE, Cormier A, Pinzon N, Taylor RS, Carre W, Strittmatter M, Seitz H, Coelho SM, Cock JM. Nucleic Acids Res. 2015;43:6384. doi: 10.1093/nar/gkv578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. Genes Dev. 2002;16:1616. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Lee RC, Feinbaum RL, Ambros V. Cell. 1993;75:843. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- [78].Wightman B, Ha I, Ruvkun G. Cell. 1993;75:855. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- [79].Burki F, Okamoto N, Pombert JF, Keeling PJ. Proc Biol Sci. 2012;279:2246. doi: 10.1098/rspb.2011.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Zhao T, Li G, Mi S, Li S, Hannon GJ, Wang XJ, Qi Y. Genes Dev. 2007;21:1190. doi: 10.1101/gad.1543507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Durban J, Perez A, Sanz L, Gomez A, Bonilla F, Rodriguez S, Chacon D, Sasa M, Angulo Y, Gutierrez J, Calvete J. BMC Genomics. 2013;14:234. doi: 10.1186/1471-2164-14-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Chai J, Chen L, Luo Z, Zhang T, Lou P, Sun W, Long X, Lan J, Wang J, Pu H, Qiu J, et al. Bioscience, biotechnology, and biochemistry. 2018;82:1081. doi: 10.1080/09168451.2018.1459175. [DOI] [PubMed] [Google Scholar]
- [83].Soding J, Remmert M. Curr Opin Struct Biol. 2011;21:404. doi: 10.1016/j.sbi.2011.03.005. [DOI] [PubMed] [Google Scholar]
- [84].Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Nature. 2008;455:58. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- [85].Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. Nature. 2008;455:64. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Seitz H. Current biology : CB. 2009;19:870. doi: 10.1016/j.cub.2009.03.059. [DOI] [PubMed] [Google Scholar]
- [87].Pinzon N, Li B, Martinez L, Sergeeva A, Presumey J, Apparailly F, Seitz H. Genome Res. 2017;27:234. doi: 10.1101/gr.205146.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Nucleic acids research. 2017;45:D331. doi: 10.1093/nar/gkw1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, et al. Nat Genet. 2000;25:25. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, Thomas PD. Nucleic acids research. 2017;45:D183. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Karginov FV, Cheloufi S, Chong MM, Stark A, Smith AD, Hannon GJ. Molecular cell. 2010;38:781. doi: 10.1016/j.molcel.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ. Current biology : CB. 2008;18:758. doi: 10.1016/j.cub.2008.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].German MA, Pillay M, Jeong DH, Hetawal A, Luo S, Janardhanan P, Kannan V, Rymarquis LA, Nobuta K, German R, De Paoli E, et al. Nature biotechnology. 2008;26:941. doi: 10.1038/nbt1417. [DOI] [PubMed] [Google Scholar]
- [94].Shin C, Nam JW, Farh KK, Chiang HR, Shkumatava A, Bartel DP. Molecular cell. 2010;38:789. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Brennecke J, Stark A, Russell RB, Cohen SM. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Kluiver J, Slezak-Prochazka I, Smigielska-Czepiel K, Halsema N, Kroesen B-J, van den Berg A. Methods. 2012;58:113. doi: 10.1016/j.ymeth.2012.07.019. [DOI] [PubMed] [Google Scholar]
- [97].Clement T, Salone V, Rederstorff M. Methods Mol Biol. 2015;1296:187. doi: 10.1007/978-1-4939-2547-6_17. [DOI] [PubMed] [Google Scholar]
- [98].Jin Y, Chen Z, Liu X, Zhou X. Methods Mol Biol. 2013;936:117. doi: 10.1007/978-1-62703-083-0_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Thomson DW, Bracken CP, Goodall GJ. Nucleic acids research. 2011;39:6845. doi: 10.1093/nar/gkr330. [DOI] [PMC free article] [PubMed] [Google Scholar]