Abstract
Carbapenem-resistant Enterobacteriaceae (CRE) are an emerging antimicrobial resistance threat for which few if any therapeutic options remain. Identification of new agents that either inhibit CRE or restore activity of existing antimicrobials is highly desirable. Therefore, a high throughput screen of 182,427 commercially available compounds was used to identify small molecules, which either enhanced activity of meropenem against a carbapenem-resistant Klebsiella pneumoniae ST258 screening strain and/or directly inhibited its growth. The primary screening methodology was a whole cell screen/counterscreen combination assay that tested for reduction of microbial growth in the presence or absence of meropenem, respectively. Screening hits demonstrating eukaryotic cell toxicity based on an orthogonal screening effort or identified as pan-assay interference (PAINS) compounds by computational methods were triaged. Primary screening hits were then clustered and ranked according to favorable physicochemical properties. Among remaining hits, we found ten compounds that enhanced activity of carbapenems against a subset of CRE. However, direct antimicrobials that passed toxicity and PAINS filters were not identified in this relatively large screening effort. It was previously shown that the same screening strategy was productive for identifying candidates for further development in screening known bioactive libraries inclusive of natural products. Our findings therefore further highlight liabilities of commercially available small molecule screening libraries in the Gram-negative antimicrobial space. In particular, there was especially low yield in identifying compelling activity against a representative, highly multidrug-resistant, carbapenemase-producing Klebsiella pneumoniae.
Keywords: High-throughput screening, whole-cell assay, medicinal chemistry, antibiotic resistance, carbapenem-resistant Enterobactericeae, CRE, KPC, Klebsiella pneumoniae
Introduction
Enterobacteriaceae are a common cause of bacterial bloodstream, urinary tract, and surgical site infections. Concerningly, these organisms also are commonly associated with resistance to clinically useful first and second-line antimicrobials including penicillins, cephalosporins, fluoroquinolones, and aminoglycosides.1 Carbapenems are the major last line of defense against multidrug-resistant Gram-negative pathogens. Unfortunately, carbapenem-resistant Enterobacteriaceae (CRE) emerged rapidly in the past two decades.2 They have now been detected worldwide3 with particularly high prevalence in Asia4 and Southern Europe.5 CRE are also isolated with increasing frequency in the United States.6 Few, if any treatments remain and those few often have dose-limiting toxicity.7 Recently, truly pandrug-resistant CRE have appeared,8 along with highly pathogenic hypermucoviscous strains that cause metastatic multi-organ infection in otherwise healthy adults,9, 10 highlighting the pressing need for new antimicrobials with activity against these pathogens.
Development of new antimicrobials active against multidrug-resistant Gram-negative pathogens has proven difficult due to relative impermeability of the Gram-negative cell membrane11 and ubiquitous expression of efflux pumps.12 However, carbapenems overcome both of these challenges and may retain detectable in vitro13 and in vivo14 activity even in strains expressing enzymes that degrade carbapenems (carbapenemases). Therefore, we hypothesized that this partial activity could be potentiated by small molecules through a variety of mechanisms to restore carbapenem efficacy against otherwise resistant CRE.
We therefore chose to use our previously validated screening/counterscreening approach to evaluate activity of a large collection of small molecules for their ability to either directly inhibit or potentiate activity of a representative carbapenem (meropenem) against a CRE screening strain.15 To rapidly triage compounds with non-specific activity, we used data from an orthogonal screening effort to eliminate those with eukaryotic cytotoxicity.16 Finally, we identified a series of compounds with optimal physicochemical properties, tested their spectrum of activity against representative CRE strains using commercially available compounds, and confirmed activity upon re-synthesis. Based on our observations, we believe the screening strategy will prove an efficient method for identifying direct and indirect antimicrobials, however, only in libraries optimized for the Gram-negative antimicrobial space.
Materials and Methods
Primary screening.
Our primary screening strain was Klebsiella pneumoniae BIDMC12A, a CRE strain of sequence type 258 (ST258), the most common sequence type of K. pneumoniae CRE strains circulating in the United States, which expresses the KPC-3 carbapenemase, and blaSHV-11, blaSHV-134, and blaTEM-1 β-lactamases.17 The screen was performed as a screening/counterscreening experiment as described in our previous work where only known bioactive compounds were examined.15 Briefly, prior to screening, 30 μl of cation-adjusted Mueller-Hinton broth (CAMHB, BD Diagnostics, Sparks, MD) containing 20 μg ml−1 meropenem (ArkPharm, Libertyville, IL) (screen) or no antibiotic (counterscreen) was added to clear, untreated polystyrene 384-well plates (Greiner Bio-One, Monroe, NC) using a MultiDrop Combi liquid handler (ThermoFisher Scientific, Waltham, MA). Compounds were added using pin-transfer robot calibrated to deliver 300 nL to each well and screened in duplicate in separate screening plates.
We screened commercially available libraries available at the Institute of Chemistry and Cell Biology (Harvard Medical School, Boston, MA), listed in Supplemental Material 2, which consist of small molecules without previously characterized activity. Compound concentrations varied by library. For libraries with concentrations expressed in μg mL−1, screening concentrations were 2.5, 10, 25, or 75 μg mL−1. For libraries with concentrations expressed as molarity, screening concentrations were 0.5, 5, 16.5, 44, or 50 μM. Immediately after compound transfer, 30 μl of K. pneumoniae BIDMC12A (1 × 106 colony forming units (CFU) ml−1) in CAMHB was added, bringing the final concentration of cells to approximately 5 × 10 5 CFU ml−1 per CLSI guidelines18 and meropenem (where applicable) to 10 μg mL−1 in a final assay volume of 60 μL.
Plates were incubated for 48 hours at 37 °C in 100% humidity. Bacterial growth was quantified by optical density at 600 nm (OD600) using an EnVision multimode plate reader (PerkinElmer, Waltham, MA). For each plate, Z’ was calculated based on positive (5 μg mL−1 colistin) and negative controls (CAMHB alone).19 Graphical representations of screening results was created using a custom Python script using the matplotlib library20 with point density calculated using the kernel density function as implemented in the scipy library.21
Hit Identification and Confirmation.
For each well, z-scores were calculated based on average and standard deviation of all experimental wells from the same assay plate. Direct antimicrobial hits were defined as strong (z < −6), moderate (−3 > z > −6), or weak (−1.5 > z > −3) based on the least significant z-score between replicates. Compounds were defined as potential adjunctives when the z-score for the screen was >3-fold that of the counterscreen. Based on previous work,15 we selected hits with >50% inhibition in the screen as candidates for follow-up testing.
Hits with eukaryotic cell cytotoxicity were identified based on results from a separate orthogonal high throughput screening effort using the same compound libraries.16 Briefly, the cytotoxicity assay consisted of application of compounds to J774A.1 macrophages incubated in the presence of 125 nM SYTOX Green, a membrane impermeant nucleic acid binding dye. In this assay, cytotoxicity results in increased eukaryotic cell membrane permeability and associated increase in SYTOX Green fluorescence, which is measured relative to controls. The assay was described previously as part of a combined screen for intracellular bacterial growth and eukaryotic cell death.16 Cytotoxic compounds were defined conservatively as those with cytotoxicity z-scores > 1.5.
Hits were cherry picked for confirmatory testing from library plates using a Tecan EVO75 liquid handler (Tecan, Morrisville, NC). We then used an HP D300 digital dispenser (HP Inc., Palo Alto, CA) to add 300 nL of compound to CAMHB or CAMHB containing 10 μg mL−1 meropenem to replicate conditions of the screen and counterscreen. K. pneumoniae BIDMC12A was added to a concentration of 5 × 105 CFU mL−1 with a final assay volume of 60 μL. Plates were incubated at 37 °C in 100% humidity for 48 hours and growth quantitated as described in the primary screen. Adjunctive activity was considered confirmed if it resulted in >25% growth inhibition in the presence of meropenem, but <25% growth inhibition in CAMHB alone, while direct activity was consider confirmed if inhibition were >25% in the absence of meropenem.
Secondary Analysis using Commercially Available Compounds.
Select compounds were ordered as powder from ChemDiv (San Diego, CA), ChemBridge (San Diego, CA), Enamine (Monmouth Jct., NJ), or Asinex (Winston-Salem, NC). Compounds were dissolved in 100% DMSO (Sigma-Aldrich, St. Louis, MO) to a concentration of 5 mg mL−1 and stored at −80 °C. For each compound, we performed two-dimensional synergy assays in combination with meropenem using a previously validated protocol.22, 23 Briefly, we used an HP D300 digital dispenser to prepare combinatorial two-fold orthogonal dilution series of meropenem and compounds of interest. Minimal inhibitory concentrations were defined as the lowest concentration of antimicrobial resulting in complete growth inhibition (OD600 < 0.08), as previously validated by our laboratory.24
In these experiments, we only tested meropenem potentiators with no detectable MIC value on their own. Therefore, synergy was assessed solely based on the greatest fold reduction of the meropenem MIC in the presence of compound, i.e., the MIC of meropenem in the presence of compound divided by the MIC of meropenem alone, which is expressed as the fractional inhibitory concentration ratio or FIC. FIC values ≤ 0.5, consistently observable in biological replicates, were considered to indicate synergy.23, 25
In-House Synthesis of Confirmed Hits.
Selected compounds were re-synthesized in-house for follow-up activity confirmation experiments. Materials, instrumentation used, and experimental details can be found in Supplemental Material 1 (Materials and Instrumentation) and Supplemental Material 6 (Synthesis).
Cheminformatics.
Pan-assay interference compound (PAINS)26 filtering was performed through an available PAINS filter27 (Eli Lilly, Cambridge MA). Next, using Scaffold Hunter, hit compounds were arranged into clusters based on Tanimoto distance measurements of fingerprints generated for each molecule.28 Physicochemical properties of compounds were predicted with Microsoft Excel (Microsoft, Redmond WA) using add-ins from ChemDraw (PerkinElmer, Walthan MA) and ChemAxon (Cambridge MA). Compounds were scored through two multi-parameter optimization (MPO) tools using these predicted properties.
First, compounds were ranked by a previously reported multi-parameter optimization (MPO) algorithm for calculating optimal physicochemical properties of drug molecules with good bioavailability29. Furthermore, an “in-house” MPO algorithm was designed to predict the ability of a compound to penetrate into a bacterial cell and avoid efflux, both characteristics of effective Gram-negative antimicrobial compounds. For this reason, we refer to our “in-house” MPO algorithm as PEMPO (Permeation and Efflux Multiparameter Optimization).
PEMPO scoring focused on assessing optimal ranges (shown in parentheses) for targeted physicochemical properties of Gram-negative antimicrobials including the isoelectric point (6.1–8.7), the total polar surface area (100–200 Å2), the number of hydrogen bond donors (2–6), the number of hydrogen bond acceptors (6–11), the partition coefficient clogP (≤3), and the distribution coefficient clogD7.4 (≤0.2). Optimal ranges were defined by analysis of average physicochemical properties of 100 known Gram-negative active antimicrobials from a study by Moser et al.30. Compounds were then scored based on how similar each physicochemical property related to the optimal value (Supplemental Material 3c). Thus, a high scoring compound suggested a high probability for bacterial cell permeation and a low probability for efflux.
Two antibacterial classes were excluded from development of the PEMPO model: macrocycles (such as macrolides or cyclic peptides such as colistin) and aminoglycosides. Both compound classes exhibit a significantly higher molecular weight than most “drug-like” compounds found within screening libraries and as a result would disproportionately influence the scoring of compounds based on extreme characteristics compared with other classes. Aminoglycosides contain on average 30 HBD/HBA whereas the other 6 classes of antibacterials (penicillins, cephems, carbapenems, sulfa drugs, fluoroquinolones, and tetracyclines) contain on average 13 HBD/HBA30 Macrolides display many more lipophilic residues, contributing to a higher average cLogD7.4 of 2.6, whereas an average cLogD7.4 of −2.77 is observed among the other 6 antimicrobial classes.30 Molecular weight was not used to calculate PEMPO scores as molecular weight of known Gram-negative compounds can vary widely based on compound class. The formula for calculating the PEMPO score is described in Supplemental Material 3a.
As physicochemical property calculators vary between platforms, we evaluated PEMPO scores calculated from properties generated by Pipeline Pilot (Accelrys, San Diego, California) and ACD/Labs (Toronto, Ontario, Canada) compared to scores generated from Chemdraw and ChemAxon. Known Gram-negative antimicrobials had an average PEMPO score of 4.97 and 5.08 out of 6.0, respective to property prediction platform. Results listed by compound can be found in Supplemental Material 3b. Therefore, we observed an average increased PEMPO score of +0.11 using the latter compared to the former property generation tools, but considered this difference to be negligible.
Spectrum of Activity Testing.
Follow-up activity spectrum studies were performed for selected compounds. We tested commercially available or re-synthesized compound in combination with meropenem as described above using thirty de-identified CRE isolates collected at our institution including Escherichia coli (n = 8), K. pneumoniae (n = 20), Serratia marcescens (n = 1) and Enterobacter cloacae (n = 1). The genome sequences of all strains are available.17
Construction of Carbapenemase-Expressing E. coli Strains.
KPC-2, KPC-3, and NDM-1 carbapenemases were PCR amplified with Q5 DNA polymerase (New England Biolabs, Ipswich, MA, annealing temperature = 60 °C) using primers listed in Supplemental Material 4. PCR products were introduced into the pUC19 vector using the NEBuilder HiFi DNA Assembly Kit (New England Biolabs) according to manufacturer’s instructions. Vectors alone or vector containing carbapenemases were transformed into electrocompetent DH5α (New England Biolabs); tolC mutant strain, JW5503–1 (E. coli Genetic Resources Stock Center, Yale University, New Haven, CT); or lptD mutant strain, RFM795 (E. coli Genetic Resources Stock Center), and selected with 100 μg mL−1 ampicillin (ThermoFisher Scientific, Waltham, MA). Cloning fidelity was confirmed by DNA sequencing (Genewiz, Boston, MA). Carbapenemase expression was confirmed phenotypically by determination of meropenem MICs.24 Select re-synthesized compounds were tested for synergy with meropenem in all constructed strains as outlined in the secondary analysis section.
Results
Primary Screening.
Gram-negative bacteria are intrinsically resistant to a variety of antibiotics owing to the relative impermeability of the cell envelope. Additionally, multidrug-resistant organisms have an extensive system of efflux pumps with broad and unpredictable specificities, which also prevent molecules from reaching the cytoplasm.12 Therefore, we chose to perform a whole cell bacterial growth inhibition screen so that screening hits would have already passed these two significant hurdles. Furthermore, our screening strain was a representative ST258, multidrug-resistant clinical K. pneumoniae isolate, representative of the most common CRE strains circulating in the United States, and resistant to a variety of antimicrobial agents including penicillins, cephalosporins, carbapenems, fluoroquinolones, nitrofurantoin, trimethoprim/sulfamethoxazole, tobramycin, and amikacin.15 Thereby, a high bar for activity was set, which is appropriate for identifying efficacy against an emerging multidrug-resistant pathogen target.
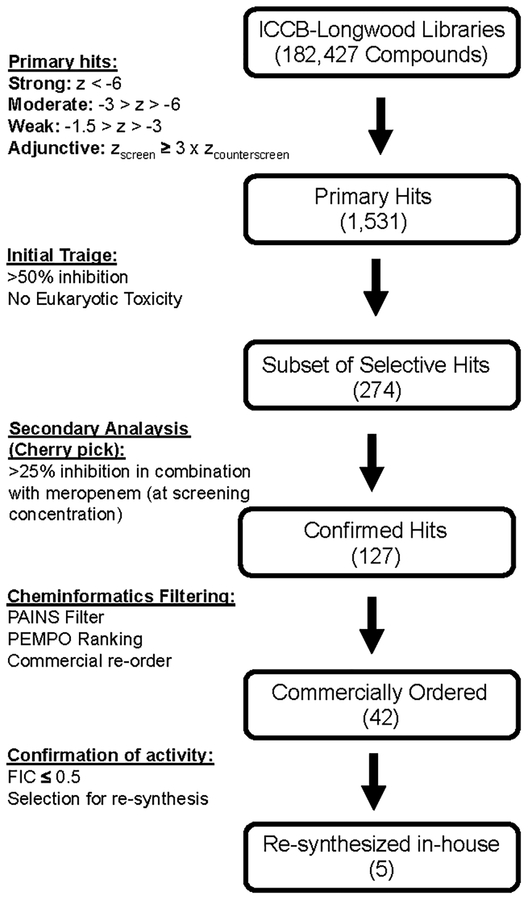
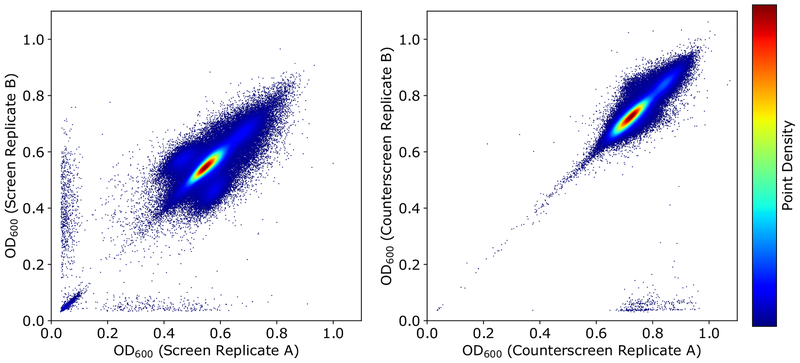
Our high throughput screening assay was designed as a screen/counterscreen. Screening wells contained meropenem at a subinhibitory concentration of 10 μg mL−1, and the counterscreen contained no antimicrobial. Therefore, compounds that potentiated meropenem would demonstrate activity in the screen (growth inhibition in the presence of meropenem), but not in the counterscreen (without meropenem). Direct antimicrobials would exhibit inhibitory effects independent of meropenem and therefore demonstrate activity in both the screen and counterscreen. In total, we screened 182,427 compounds without previously characterized biological activity in duplicate using this two-tiered assay. An overview of the screening effort and secondary analysis is summarized in Fig. 1. During the screening effort, we found good reproducibility between replicates for both the screen and counterscreen experiments (Fig. 2). Average cumulative Z’ was 0.61 for the screen and 0.67 for the counterscreen based on positive and negative screening wells from screening plates.
Figure 1.
Overview of high throughput screening hit analysis.
Figure 2. Correlation between in high throughput screen and counterscreen.
Assays were performed in duplicate. Screening wells contained 10 μg/ml of meropenem, counterscreening wells did not contain antibiotic. Readout was the OD600 of microwells after a 48 h incubation. The values for each pair of duplicate measurements were plotted on X and Y axes for the screen (A) and counterscreen (B), respectively. Higher relative data point density is represented by warmer colors as indicated in legend. Inclusive of control wells, r2 = 0.82 for the screen and 0.92 for the counterscreen, indicating excellent correlation between replicate wells.
Hit Identification.
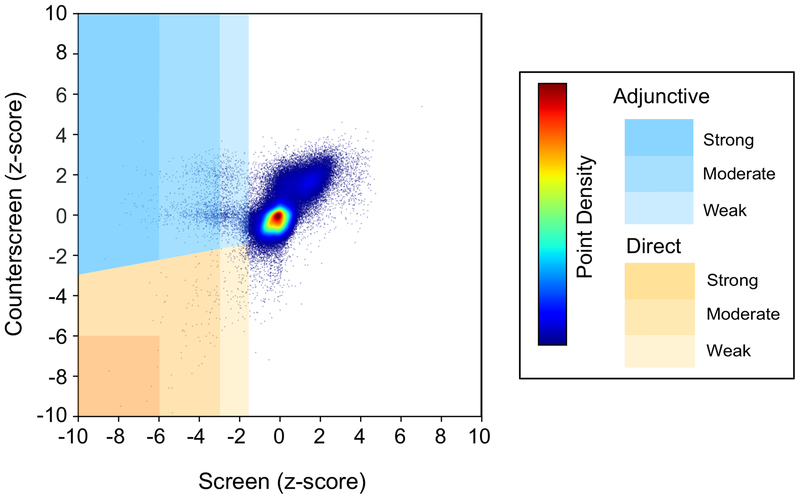
We initially identified 1,531 (0.84% of total compounds screened) total adjunctive and direct antimicrobial screening hits. Of the adjunctive hits, 605 (0.332%), 599 (0.328%) and 43 (0.02%) were weak, medium, and strong, respectively. Of the direct antimicrobial hits 205 (0.11%), 71 (0.04%), and 8 (0.004%) were weak, medium, and strong, respectively. The z-score distribution for the screen/counterscreen is graphically summarized in Fig. 3.
Figure 3. Plot of least significant z-scores for duplicate compound testing in the screen and counterscreen.
Z-criteria hit ranking (strong, medium, weak) are represented in shades of yellow (direct antimicrobials) or blue (adjunctive antimicrobials). Higher relative data point density is represented by warmer colors as indicated in legend.
We previously established that our screening assay yielded a high false positive rate based on z-score criterion alone and that those hits demonstrating <50% inhibition (compared to control wells) were unlikely to confirm in secondary analysis.15 Accordingly, we applied a potency requirement of >50% inhibition for at least one of the duplicate measurements. After applying this filter, 439 (72.6%) weak, 598 (99%) medium, and all strong adjunctive hits and 20 (9.8%) weak, 52 (73.2%) moderate, and all strong direct hits were retained.
Additionally, we filtered out compounds that showed cytotoxicity to eukaryotic cells, a marker for non-specific activity, or a target shared by both prokaryotes and eukaryotes, which as a consequence would not be druggable. Eukaryotic cytotoxicity data were from a previously described screening assay using the same compound libraries16. After applying this filter, 252 (57.4%) weak, 375 (62.7%) moderate, and 31 (72.1%) strong adjunctive hits were retained; 9 (45%) weak, 31 (59.6%) moderate, and no strong direct antimicrobial hits were retained.
Hit Confirmation.
We selected 274 filtered adjunctive and direct antimicrobial hits based on primary screening potency for confirmatory testing using cherry picks from commercial library plates in a manner identical to the primary screening assay. Here, we set a less stringent 25% inhibition cutoff in recognition that hits may not recapitulate activity exactly upon secondary analysis. In total, 127 (44.2%) adjunctive hits and no direct antimicrobial hits confirmed on retesting.
Cheminformatics Triage.
Cheminformatics filtering was then performed to remove nonspecific, pan-assay interference compounds (PAINS) with features of covalent modifiers (for example electrophiles such as aldehydes, ketones, or boronic acids) or metal binders (for example hydroxamic acids or phosphonates). Even though many antibacterial drugs contain such reactive structural features (approximately 58% of known antibiotics from our testing set fail the PAINS filter), PAINS are considered to be problematic for hit-to-lead optimization and drug development.26, 31 Therefore, of the 127 confirmed, adjunctive hits, 20 compounds were identified as PAINS and excluded from further analysis. The remaining 107 were clustered based on common substructure, which resulted in identification of 15 clusters and 17 singletons. Further prioritization within clusters was performed based on a compound activity profile and scoring of physicochemical properties characteristic of known antibacterials, using a cheminformatic pipeline called PEMPO (Permeation and Efflux Multiparameter Optimization) described in the materials and methods section. Select singletons were removed after visual inspection because of limited synthetic tractability or the presence of unfavorable functional groups known to possibly pose bioavailability limitations, narrowing our future analysis to 42 compounds representing 15 clusters and 6 singletons. PEMPO and MPO scoring for these compounds is shown in Supplemental Material 3c. Representative structures of top scoring clusters and singletons are shown in Fig. 4.
Figure 4. Representative structures of clusters and selected singletons identified by filtering and PEMPO analysis.
For clusters, representative structures shown are the highest PEMPO scoring compounds within each cluster. Compounds highlighted in gray demonstrated a synergistic adjunctive activity against representative CRE strains after repurchase from commercial suppliers.
Secondary Analysis Using Commercially Synthesized Compounds.
We ordered these 42 compounds from commercial suppliers and performed synergy assays in combination with meropenem using our primary screening strain. In total, 23.8% (n = 10) had evidence of synergistic activity with meropenem (FIC ≤ 0.5).
All compounds with an FIC ≤ 0.5 were tested for activity spectrum against a panel of CRE strains consisting of E. coli and K. pneumoniae containing either KPC-2 or KPC-3 carbapenemases. All selected compounds had activity against ≥50% of strains tested (Table 1).
Table 1.
Spectrum of Activity of Commercially Synthesized and Re-synthesized Compounds.
| Average FICa | % CRE Activityb | |||
|---|---|---|---|---|
| Compound | Commercialc | Re-synthesizedd | Commercial | Re-synthesized |
| KP40 | 0.31 | 0.42 | 75 | 23 |
| KP14 | 0.5 | -e | 70 | - |
| KP17 | 0.5 | - | 70 | - |
| KP5 | 0.38 | - | 60 | - |
| KP13 | 0.5 | - | 60 | - |
| KP11 | 0.38 | 0.5 | 50 | 27 |
| KP8 | 0.38 | - | 50 | - |
| KP9 | 0.38 | >1 | 50 | 33 |
| KP19 | 0.5 | 0.5 | 50 | 17 |
| KP56 | 0.19 | 0.75 | - | 17 |
Calculated from quadtruplicate testing of K. pneumoniae BIDMC 12A.
Percent of CRE strains with FIC ≤ 0.5 on combinatorial testing with meropenem. Calculated using at least 10 representative CRE strains for commercial compounds and 30 CRE strains for re-synthesized compounds.
Compounds purchased from commercial suppliers
Compounds synthesized in our laboratory
Not determined
Analysis of Re-synthesized Compounds.
To this point, we had been using compounds available in limited quantities from the commercial suppliers of our screening libraries. We added an additional layer of confirmation by resynthesizing hit compounds and confirming structural identity and purity by liquid chromatography with diode array detection, mass spectrometry and NMR (see Supplemental Material 6 and 7, respectively for details). We used potency (based on FIC) and spectrum of activity as primary criteria, and predicted physicochemical property data as secondary criteria to select KP40 and KP11 for re-synthesis. Other compounds chosen for re-synthesis were KP9 and KP19, which displayed excellent PEMPO scores (4.4 and 4.8 respectively), along with KP56, which demonstrated good activity in primary screening. Following synthesis, these compounds were tested again using our screening strain to confirm activity using our standard synergy assay.
Of the re-synthesized compounds, KP11, KP40, and KP19 demonstrated synergy with meropenem against our screening strain (Table 1). However, two compounds, KP9 and KP56, did not. We then tested re-synthesized compounds against our 30 strain CRE panel. Re-synthesized compounds showed synergy against 17 to 33% of CRE strains. Interestingly, KP9 and KP56, while not showing synergy against the screening strain, demonstrated synergy against a subset of clinical CRE strains.
Synergy testing in a non-CRE background.
Confirmed adjunctive hits might interfere with carbapenemase activity or alternatively affect the physiology of specific bacterial strains to enhance potency of meropenem by other mechanisms. To distinguish phenotypically between these possibilities, we first constructed isogenic E. coli strains expressing the serine carbapenemases (KPC-2 or KPC-3) or metallo-carbapenemase (NDM-1). The strain background used was DH5α, a laboratory-adapted E. coli K-12 strain with no intrinsic resistance to β-lactams including meropenem. However, we found no synergy of compounds with meropenem, suggesting that effects might be strain specific, and as a result not effective on E. coli K-12.
We therefore considered whether the lack of activity in the E. coli K-12 background might relate to either lack of permeation and/or efflux. To distinguish between these two possibilities, we introduced our KPC-2, KPC-3, and NDM-1 containing plasmids into E. coli strains with defects in the outer membrane permeability barrier (lptD) or efflux activity (tolC). The lptD mutant expresses a truncated form of LptD, a protein critical in transporting lipopolysaccharide (LPS) to the outer membrane.32 The resulting deficit in LPS in the outer membrane leads to increased permeability. The tolC mutant inactivates a critical shared component of several efflux pumps.33 Interestingly, we did not observe synergy with meropenem in either strain. However, we did observe direct antimicrobial activity of KP40 in the tolC mutant. MICs were independent of carbapenemase production and ranged from 16–128 μM during replicate testing, an unusual degree of biological variability not typical of established antimicrobials with specific mechanisms of action (Supplemental Material 5a–c).
Cheminformatic characterization of high throughput screening libraries.
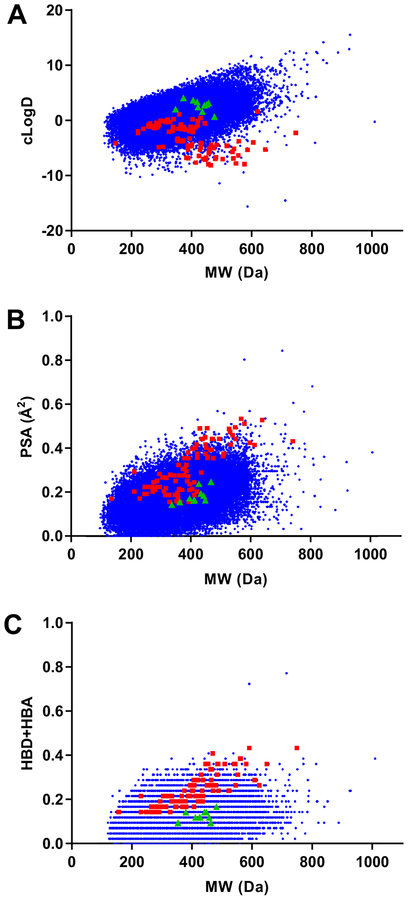
The physicochemical properties of screening libraries used in this effort were characterized and compared to the chemical space occupied by 100 known Gram-negative antimicrobials. Molecular weight, polar surface area, cLogD7.4, and the summation of hydrogen bond donor/acceptors of screening compounds and known antimicrobials were calculated. Plots of physicochemical properties versus molecular weight are shown in Fig. 5.
Figure 5. Cheminformatic analyses of screening libraries.
(A) Partition coefficient (cLogD7.4) (B) polar surface area (PSA), and (C) the summation of hydrogen bond donors (HBD) and hydrogen bond acceptors (HBA) versus molecular weight for screening library compounds (blue), known Gram-negative antimicrobials (red), and confirmed hit compounds (green). In contrast to Gram-negative antimicrobials, library compounds demonstrated increasing lipophilicity (cLogD7.4) with increasing molecular weight. Opposite trends were observed for polar surface area and summations of hydrogen bond donors/acceptors.
From data plots, it is apparent that the screening library consists of compounds with a greater degree of lipophilic substituents. More specifically, partition coefficients (cLogD7.4) for library compounds demonstrate increasing lipophilicity with increasing molecular weight (Fig. 5A). In contrast, partition coefficients of 100 known Gram-negative active antimicrobials show the opposite trend. Similar but inverted trends for the screening libraries and known antimicrobials were observed in plots of polar surface area (Fig. 5B) as well as summations of hydrogen bond donors/acceptors (Fig 5C).
Discussion
A screen of commercially available small molecule libraries was performed to identify carbapenem potentiators and direct antimicrobial inhibitors of a representative Klebsiella pneumoniae carbapenem-resistant clinical isolate. The goal was to identify hits that could be further improved upon using medicinal chemistry approaches. Furthermore, the whole cell screening approach was agnostic as to potential mechanism of action that could be further delineated at a later time for promising scaffolds.
Hits underwent initial triage based on combined use of cheminformatics approaches and data from an orthogonal screen to eliminate eukaryotic cell toxic compounds. However, based on this initial stringent, but likely appropriate down selection, ultimately only a few hits with potentiating activity and no hits with direct activity remained. Many, but not all, of these adjunctive hits retained activity on re-synthesis. The lack of complete reproducibility on re-synthesis is a well-known finding in commercial library screening efforts and may result from contaminants such as heavy metal catalysts, which may confer antimicrobial activity unrelated to the compound under study. Additionally, we observed reduction in activity spectrum of several of the compounds. Reasons for this are not immediately obvious, but may relate to borderline adjunctive activity, which did not reach a threshold for phenotypic detection with the resynthesized compound. It may also represent contributions of both compound and contaminants in the original commercial preparations that differed from resynthesized compounds.
We hypothesized that a subset of potentiators would represent hits that either directly or indirectly targeted carbapenemase activity. However, tests in isogenic E. coli strains expressing several types of carbapenemases failed to detect synergy with meropenem suggesting effects were specific to only a subset of clinical strains being tested based on shared regulatory and/or biophysical characteristics, potentially a reflection of the diversity of the CRE strain set.
To address target access, we tested previously well-characterized E. coli K-12 strains with known defects in either permeability barrier (lptD) or in a major class of efflux pumps (tolC). Neither strain allowed observation of carbapenem potentiation in the E. coli K-12 background, suggesting that differences other than efflux or outer membrane permeability barrier accounted for the observed activity spectrum. Interestingly, one compound, KP40, was noted to have direct, but highly variable, antimicrobial activity against the tolC mutant; this high biological variability suggests non-specific interference with bacterial growth, i.e., hitting multiple targets with total assay variability reflecting the sum of the variability of multiple events.
Our goal was to identify compounds that had already passed the high bar for activity against a multidrug-resistant pathogen. In that way there would be a stringent biological triage with hopes of later improving initial activity using medicinal chemistry approaches. Our clinical screening strain is known to encode multiple antimicrobial resistance elements17 and has a very high baseline carbapenem MIC (50 μg mL−1) which is 16 to 32-fold higher than in a laboratory E. coli strain expressing the same carbapenemase gene (data not shown). Therefore, the carbapenem resistance phenotype observed in this and other clinical isolates is likely complex and polygenic with contributions from efflux pumps, altered porins, membrane and cell wall characteristics, and/or β-lactamases with low-level ability to hydrolyze carbapenems.
Unfortunately, compounds with compelling direct or adjunctive antimicrobial activity were not identified through these efforts. One potential explanation for this is that the biological triage was too stringent. For example, we did not follow up on compounds that demonstrated statistically significant cytotoxicity for J774A.1 macrophages observed in a separate screening effort. Therefore, it remains possible that some compounds identified as eukaryotic cell toxic may have had some degree of selectivity for bacteria that could have been improved upon during structure-activity relationship studies.
Another possibility is that the commercial screening libraries available did not contain sufficiently diverse compounds with physicochemical properties conducive to Gram-negative antimicrobial activity. For example, a prior screening effort of 500,000 compounds at GlaxoSmithKline against a efflux competent strain of E. coli yielded no confirmed hits.34 This finding was attributed to lack of chemical diversity. Although the chemical space occupied by the libraries examined was not reported, it is well known that commercial and pharmaceutical libraries historically have been optimized for “drug-like” molecules based on metrics such as Lipinski’s rule of five.35
However, antimicrobials in general and Gram-negative agents in particular rarely satisfy these rules30. Upon analyses of the physicochemical properties of our screening libraries and representative hits, we observed trends suggesting that compounds with characteristics of Gram-negative antimicrobials were underrepresented. Gram-negative antimicrobials typically possess zwitterionic or polar moieties, which facilitate passage of compounds through water-filled transmembrane porins and entry into the periplasm. However, our libraries and screening hits had a paucity of such compounds. Instead, they were enriched for compounds with lipophilic substituents (high cLogD), decreased number of hydrogen bond donors and acceptors, and decreased polar surface area, which face an increased enthalpic barrier for entry into the Gram-negative cell. Compounds with these attributes are generally less challenging to synthesize and purify and therefore not unexpectedly are overrepresented in screening libraries.
An alternative screening approach using a screening strain with a lower barrier for activity may have been more productive in identifying lead candidates. For example, a screen of 150,000 small molecules using fully antimicrobial susceptible E. coli and Pseudomonas aeruginosa strains identified several confirmed hits with weak activity.36 However, further development of novel compounds from this screen has not been described to the best of our knowledge. Additional perturbation, such as use of a tolC or lptD mutants may further lower the bar for Gram-negative inhibitor detection37 but later require additional chemistry efforts to address efflux and permeability effects that may or not prove productive.
Taken together, our results support previous observations that Gram-negative antimicrobial lead candidates may be largely absent from commercially available screening libraries. Certainly, this appeared to be the case for compounds with intrinsic activity against a highly multidrug- and carbapenem-resistant Klebsiella pneumoniae clinical strain. Although we were able to find detectable activity for several compounds, the overall potency was low. Therefore, we provide further data that whole cell screening efforts for Gram-negative antimicrobials should be conducted using libraries with diverse scaffolds and substituents outside the Lipinski space, for example, including compounds with greater hydrophilicity.
Further supporting this view, our prior efforts using the same screening strategy to examine known bioactive libraries with higher diversity inclusive of natural products was highly productive. This led to identification of apramycin and several nucleoside analogues as lead direct antimicrobial candidates for development against highly drug-resistant CRE and for the former against MDR Acinetobacter baumannii, where apramycin is now recognized more generally as a candidate for pre-clinical development38–40. In this earlier screening effort, we also identified potent meropenem adjunctive activity of triclosan confirming the underlying ability of the screening strategy to detect both direct and adjunctive antimicrobials15. Therefore, the fundamental ability of the whole cell, high throughput screening assay to detect antimicrobials with activity against CRE, and by extension other MDR Gram-negative pathogens of concern, offers promise as libraries with appropriate physicochemical properties become available.
Supplementary Material
Acknowledgements
K.P.S. and J.E.K. were supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award numbers F32AI124590, and R21AI119114 and R33AI119114, respectively. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The screening work was performed at ICCB-Longwood (Boston, MA). We would like to thank Jennifer Smith, Jennifer Nale, David Wrobel, Stewart Rudnicki, Rachel Warden, and Richard Siu (ICCB-Longwood) for their assistance. The HP D300 digital dispenser was provided for our use by Tecan (Morrisville, NC). Tecan had no role in study design, data collection/interpretation, manuscript preparation, or decision to publish.
References
- 1.Weiner LM; Webb AK; Limbago B; et al. Antimicrobial-Resistant Pathogens Associated With Healthcare-Associated Infections: Summary of Data Reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011–2014. Infect Control Hosp Epidemiol 2016, 37, 1288–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradford PA; Bratu S; Urban C; et al. Emergence Of Carbapenem-Resistant Klebsiella Species Possessing The Class A Carbapenem-Hydrolyzing KPC-2 And Inhibitor-Resistant TEM-30 Beta-Lactamases In New York City. Clin Infect Dis 2004, 39, 55–60. [DOI] [PubMed] [Google Scholar]
- 3.Nordmann P; Naas T; Poirel L Global spread of Carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 2011, 17, 1791–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsu LY; Apisarnthanarak A; Khan E; et al. Carbapenem-Resistant Acinetobacter baumannii and Enterobacteriaceae in South and Southeast Asia. Clin Microbiol Rev 2017, 30, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Albiger B; Glasner C; Struelens MJ; et al. Carbapenemase-producing Enterobacteriaceae in Europe: Assessment By National Experts From 38 Countries, May 2015. Euro Surveill 2015, 20, 1–18. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease, C.; Prevention. Vital Signs: Carbapenem-Resistant Enterobacteriaceae. MMWR Morb Mortal Wkly Rep 2013, 62, 165–70. [PMC free article] [PubMed] [Google Scholar]
- 7.Thaden JT; Pogue JM; Kaye KS Role Of Newer And Re-Emerging Older Agents In The Treatment Of Infections Caused By Carbapenem-Resistant Enterobacteriaceae. Virulence 2017, 8, 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen L; Todd R; Kiehlbauch J; et al. Notes from the Field: Pan-Resistant New Delhi Metallo-Beta-Lactamase-Producing Klebsiella pneumoniae - Washoe County, Nevada, 2016. MMWR Morb Mortal Wkly Rep 2017, 66, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu L; Tang L; Wang S; et al. Co-location of the blaKPC-2, blaCTX-M-65, rmtB And Virulence Relevant Factors In An IncFII Plasmid From A Hypermucoviscous Klebsiella pneumoniae isolate. Microb Pathog 2018, 124, 301–304. [DOI] [PubMed] [Google Scholar]
- 10.Liu Z; Gu Y; Li X; et al. Identification and Characterization of NDM-1-producing Hypervirulent (Hypermucoviscous) Klebsiella pneumoniae in China. Ann Lab Med 2019, 39, 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Masi M; Refregiers M; Pos KM; et al. Mechanisms Of Envelope Permeability And Antibiotic Influx And Efflux In Gram-Negative Bacteria. Nat Microbiol 2017, 2, 17001. [DOI] [PubMed] [Google Scholar]
- 12.Li XZ; Plesiat P; Nikaido H The Challenge Of Efflux-Mediated Antibiotic Resistance In Gram-Negative Bacteria. Clin Microbiol Rev 2015, 28, 337–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fattouh R; Tijet N; McGeer A; et al. What Is the Appropriate Meropenem MIC for Screening of Carbapenemase-Producing Enterobacteriaceae in Low-Prevalence Settings? Antimicrob Agents Chemother 2015, 60, 1556–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daikos GL; Tsaousi S; Tzouvelekis LS; et al. Carbapenemase-Producing Klebsiella pneumoniae Bloodstream Infections: Lowering Mortality By Antibiotic Combination Schemes And The Role Of Carbapenems. Antimicrob Agents Chemother 2014, 58, 2322–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith KP; Kirby JE Validation of a High-Throughput Screening Assay for Identification of Adjunctive and Directly Acting Antimicrobials Targeting Carbapenem-Resistant Enterobacteriaceae. Assay Drug Dev Technol 2016, 14, 194–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiaraviglio L; Kirby JE High-Throughput Intracellular Antimicrobial Susceptibility Testing of Legionella pneumophila. Antimicrob Agents Chemother 2015, 59, 7517–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cerqueira GC; Earl AM; Ernst CM; et al. Multi-Institute Analysis Of Carbapenem Resistance Reveals Remarkable Diversity, Unexplained Mechanisms, And Limited Clonal Outbreaks. Proc Natl Acad Sci U S A 2017, 114, 1135–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Suceptibility Tests for Bacteria that Grow Aerobically: Tenth Edition M07-A10. CLSI: Wayne, PA, USA, 2015. [Google Scholar]
- 19.Zhang JH; Chung TD; Oldenburg KR A Simple Statistical Parameter For Use In Evaluation And Validation Of High Throughput Screening Assays. J Biomol Screen 1999, 4, 67–73. [DOI] [PubMed] [Google Scholar]
- 20.Hunter JD Matplotlib: A 2D Graphics Environment. Computing in Science and Engineering 2007, 9, 90–95. [Google Scholar]
- 21.Jones E; Oliphant E; Peterson P SciPy: Open Source Scientific Tools for Python. Accessed Sept. 12, 2017 http://www.scipy.org/
- 22.Brennan-Krohn T; Pironti A; Kirby JE Synergistic Activity of Colistin-Containing Combinations against Colistin-Resistant Enterobacteriaceae. Antimicrob Agents Chemother 2018, 62, e00873–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brennan-Krohn T; Truelson KA; Smith KP; et al. Screening For Synergistic Activity Of Antimicrobial Combinations Against Carbapenem-Resistant Enterobacteriaceae Using Inkjet Printer-Based Technology. J Antimicrob Chemother 2017, 72, 2775–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith KP; Kirby JE Verification of an Automated, Digital Dispensing Platform for At-Will Broth Microdilution-Based Antimicrobial Susceptibility Testing. J Clin Microbiol 2016, 54, 2288–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Odds FC Synergy, Antagonism, And What The Chequerboard Puts Between Them. J Antimicrob Chemother 2003, 52, 1. [DOI] [PubMed] [Google Scholar]
- 26.Bruns RF; Watson IA Rules For Identifying Potentially Reactive Or Promiscuous Compounds. J Med Chem 2012, 55, 9763–72. [DOI] [PubMed] [Google Scholar]
- 27.Baell J; Walters MA Chemistry: Chemical Con Artists Foil Drug Discovery. Nature 2014, 513, 481–3. [DOI] [PubMed] [Google Scholar]
- 28.Schafer T; Kriege N; Humbeck L; et al. Scaffold Hunter: A Comprehensive Visual Analytics Framework For Drug Discovery. J Cheminform 2017, 9, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wager TT; Hou X; Verhoest PR; et al. Moving beyond rules: the development of a central nervous system multiparameter optimization (CNS MPO) approach to enable alignment of druglike properties. ACS Chem Neurosci 2010, 1, 435–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Shea R; Moser HE Physicochemical Properties Of Antibacterial Compounds: Implications For Drug Discovery. J Med Chem 2008, 51, 2871–8. [DOI] [PubMed] [Google Scholar]
- 31.Baell JB; Holloway GA New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J Med Chem 2010, 53, 2719–40. [DOI] [PubMed] [Google Scholar]
- 32.Okuda S; Sherman DJ; Silhavy TJ; et al. Lipopolysaccharide transport and assembly at the outer membrane: the PEZ model. Nat Rev Microbiol 2016, 14, 337–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nikaido H Structure and Mechanism of RND-type Multidrug Efflux Pumps. Adv Enzymol Relat Areas Mol Biol 2011, 77, 1–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Payne DJ; Gwynn MN; Holmes DJ; et al. Drugs For Bad Bugs: Confronting The Challenges Of Antibacterial Discovery. Nat Rev Drug Discov 2007, 6, 29–40. [DOI] [PubMed] [Google Scholar]
- 35.Lipinski CA; Lombardo F; Dominy BW; et al. Experimental And Computational Approaches To Estimate Solubility And Permeability In Drug Discovery And Development Settings. Adv Drug Deliv Rev 2001, 46, 3–26. [DOI] [PubMed] [Google Scholar]
- 36.De La Fuente R; Sonawane ND; Arumainayagam D; et al. Small molecules with Antimicrobial Activity Against E. coli and P. aeruginosa Identified By High-Throughput Screening. Br J Pharmacol 2006, 149, 551–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fan J; de Jonge BL; MacCormack K; et al. A Novel High-Throughput Cell-Based Assay Aimed At Identifying Inhibitors Of DNA Metabolism In Bacteria. Antimicrob Agents Chemother 2014, 58, 7264–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kang AD; Smith KP; Berg AH; et al. Efficacy of Apramycin against Multidrug-Resistant Acinetobacter baumannii in the Murine Neutropenic Thigh Model. Antimicrob Agents Chemother 2018, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Truelson KA; Brennan-Krohn T; Smith KP; et al. Evaluation Of Apramycin Activity Against Methicillin-Resistant, Methicillin-Sensitive, And Vancomycin-Intermediate Staphylococcus aureus Clinical Isolates. Diagn Microbiol Infect Dis 2018, 92, 168–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Riedel S; Vijayakumar D; Berg G; et al. Evaluation Of Apramycin Against Spectinomycin-Resistant And -Susceptible Strains Of Neisseria gonorrhoeae. J Antimicrob Chemother 2019, 1311–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.