Abstract
Protein degradation by the ubiquitin-proteasome system is critical for neuronal function. Neurons utilize microtubule-dependent molecular motors to allocate proteasomes to synapses, but how proteasomes are coupled to motors and how this is regulated to meet changing demand for protein breakdown remains largely unknown. We show that the conserved proteasome-binding protein PI31 serves as an adaptor to couple proteasomes with dynein light chain proteins (DYNLL1/2). Inactivation of PI31 inhibited proteasome motility in axons and disrupted synaptic proteostasis, structure and function. Moreover, phosphorylation of PI31 by p38 MAPK enhanced binding to DYNLL1/2 and promoted directional movement of proteasomes in axons, suggesting a mechanism to regulate loading of proteasomes onto motors. Inactivation of PI31 in mouse neurons attenuated proteasome movement in axons, indicating this process is conserved. Because mutations affecting PI31 activity are associated with human neurodegenerative diseases, impairment of PI31-mediated axonal transport of proteasomes may contribute to these disorders.
Keywords: proteasomes, ubiquitin-proteasome system, protein degradation, microtubule-dependent transport, axon, synapse, neurodegenerative diseases, Drosophila
eTOC Blurb
Proteasomes are actively transported between the soma and terminals of neurons. Liu et al. show that PI31 couples proteasomes to cellular motors for fast axonal transport and thereby brings proteasomes to sites where protein breakdown occurs. This mechanism is required for protein homeostasis and synaptic architecture.
Introduction
Protein homeostasis (proteostasis) is essential for cellular and organismal health (Balch et al., 2008; Collins and Goldberg, 2017; Glickman and Ciechanover, 2002; Goldberg, 2003; Labbadia and Morimoto, 2015; Varshavsky, 2005; Vilchez et al., 2014; Wolff et al., 2014). Eukaryotic cells have two major systems for clearing unwanted, damaged and potentially toxic proteins: the ubiquitin-proteasome system (UPS) and the autophagy-lysosome system(Bento et al., 2016; Collins and Goldberg, 2017; Dikic and Elazar, 2018; Glickman and Ciechanover, 2002; Goldberg, 2003; Levine and Kroemer, 2008; Murata et al., 2009; Nakatogawa et al., 2009; Schmidt and Finley, 2014). The UPS carries out the degradation of the vast majority of intracellular proteins, whereas autophagy is primarily responsible for the removal of protein aggregates and damaged organelles. In the UPS, proteins are tagged for destruction by the linkage of multiple copies of ubiquitin(Finley, 2009; Hershko and Ciechanover, 1998; Varshavsky, 2012). The actual degradation of proteins in the UPS is carried out by multi-subunit protease particles termed 26S proteasomes(Baumeister et al., 1998; Collins and Goldberg, 2017; Finley, 2009; Glickman and Ciechanover, 2002). The 26S proteasomes consist of 20S core particles, which contain the active proteases, and 19S regulatory subunits(Collins and Goldberg, 2017; Murata et al., 2009).
It has been suggested that proteasomes are typically present in excess and that their capacity is not fully used, at least in the absence of stress(Asano et al., 2015). On the other hand, some cell types appear to require special mechanisms to position proteasomes to appropriate sub-cellular compartments where protein breakdown occurs. This is especially important for neurons, since they are very large, structurally complex and highly compartmentalized cells that require transport mechanisms to allocate proteasomes to sites distant from the cell body (Bingol and Schuman, 2006; Bingol and Sheng, 2011; Tai and Schuman, 2008).
Proteasome function is required for growth and pruning of both axons and dendrites during development (DiAntonio et al., 2001; Erturk et al., 2014; Kuo et al., 2005; Wan et al., 2000; Watts et al., 2003). Moreover, in mature neurons UPS-mediated protein degradation at synapses is critical for activity-dependent plasticity, learning and memory(Bingol and Sheng, 2011; Campbell and Holt, 2001; Ding et al., 2007; Hegde et al., 2014; Lee et al., 2008; Pak and Sheng, 2003; Patrick, 2006; Yi and Ehlers, 2005). Recently, proteasomes were found to insert into the plasma membrane of neurons and acutely regulate synaptic activity and calcium signaling(Ramachandran et al., 2018; Ramachandran and Margolis, 2017). Due to the considerable distances from the soma, neurons employ microtubule-based transport mechanisms to deliver proteasomes to the periphery of neurons(Gorbea et al., 2010; Hsu et al., 2015; Kreko-Pierce and Eaton, 2017; Otero et al., 2014). However, despite the critical function of proteasomes at synapses, much remains to be learned about the underlying transport mechanisms. One of the molecular motors, the dynein complex, plays a key role in motility of proteasomes in axons(Hsu et al., 2015; Kreko-Pierce and Eaton, 2017). A recent study identified dynein light chain LC8-type proteins (DYNLL1/2) as components of microtubule-dependent proteasome transport in Drosophila neurons (Kreko-Pierce and Eaton, 2017). However, it remains unclear how proteasomes are coupled to microtubule-based motor complexes, and how proteasome transport is regulated to meet changing demands for protein breakdown.
Here we show that the conserved proteasome-binding protein PI31 (“proteasome inhibitor of 31kD”) mediates fast axonal transport of proteasomes in both Drosophila motor neurons and mouse hippocampal neurons by acting as an adapter between proteasomes and dynein light chain LC8-type proteins (DYNLL1/2). PI31 was originally identified based on its ability to inhibit 20S proteasome-mediated hydrolysis of peptides in vitro (Chu-Ping et al., 1992; McCutchen-Maloney et al., 2000; Zaiss et al., 1999). On the other hand, inactivation of PI31 impairs protein degradation during sperm maturation in Drosophila, and PI31 can stimulate the proteolytic capacity of cells by promoting the assembly of 19S and 20S particles into the fully active 26S proteasomes(Bader et al., 2011; Cho-Park and Steller, 2013). Similarly, the PI31 ortholog in yeast, Fub1, is required to alleviate proteotoxic stress, and the Arabidopsis ortholog, PTRE1, is a positive proteasome regulator that mediates auxin signaling (Yang et al., 2016; Yashiroda et al., 2015). However, studies in cultured HEK293 cells failed to reveal a clear effect of PI31 for proteasome activity, suggesting that PI31 function may be different in different cell types and systems(Li et al., 2014).
PI31 also binds directly and strongly to the F-box protein Ntc/FBXO7/PARK15, and this interaction is conserved from Drosophila to mammals(Bader et al., 2011; Kirk et al., 2008). Interestingly, mutations in FBXO7/PARK15 impair proteasome function and cause a juvenile form of Parkinson’s Disease (PD) in humans and PD-like symptoms in mice(Conedera et al., 2016; Di Fonzo et al., 2009; Paisan-Ruiz et al., 2010; Vingill et al., 2016). Since many age-related neurodegenerative diseases, including Alzheimer’s Disease (AD), PD, and amyotrophic lateral sclerosis (ALS), are characterized by the accumulation of protein aggregates, it is possible that mutations in FBXO7/PARK15 impair the clearance of pathological proteins (Ballatore et al., 2007; Irvine et al., 2008; Li and Li, 2011; Oddo, 2008; Ross and Poirier, 2004; Tai and Schuman, 2008). F-box proteins serve as substrate recognition modules in multi-component E3-ligase complexes for UPS-mediated protein degradation (Bai et al., 1996; Kipreos and Pagano, 2000; Skaar et al., 2013; Skowyra et al., 1997). However, very unexpectedly for an E3 ligase, binding of FBXO7 to PI31 does not result in PI31 degradation but rather serves to protect PI31 from proteolytic cleavage (Bader et al., 2011; Vingill et al., 2016). These observations are consistent with the idea that loss of FBXO7 leads to impaired proteasome function through inactivation of PI31, but direct evidence for this model is lacking due to insufficient knowledge about the precise function of PI31 in neurons. Moreover, PI31 was linked to AD by a genome-wide association study, and mutations in another PI31-interacting protein, Valosin-Containing Protein (VCP/P97), cause ALS (Clemen et al., 2015; Johnson et al., 2010; Sherva et al., 2011).
In order to address the function of PI31 in neurons, we investigated the consequences of PI31 inactivation in Drosophila motor neurons. We found a striking requirement of PI31 for synaptic structure and function. Loss of PI31 function altered the structure of presynaptic active zones and caused defects in protein homeostasis in the periphery of neurons. Significantly, we identified dynein light chain proteins (dDYNLL1/Ctp and dDYNLL2/Cdlc2) as direct binding partners of PI31, and PI31 was able to load proteasomes onto dDYNLL1/2 in vivo and in vitro. Importantly, inactivation of PI31 blocked proteasome motility in axons. We conclude that PI31 is a direct adaptor protein to load proteasomes onto dynein complexes and thereby mediates axonal transport of proteasomes. The interaction of PI31 and DYNLL1/2 is conserved from Drosophila to mammals, and inactivation of PI31 in mouse hippocampal neurons impairs proteasome movement in axons, indicating that the transport mechanism is conserved. Finally, we show that phosphorylation of PI31 by the stress-activated p38 MAP kinase modulates the formation of dDYNLL1/2-proteasome complexes and proteasome movement. These findings suggest that PI31 phosphorylation functions as a molecular switch to regulate the loading of proteasome onto motors. This work also reveals a molecular link between cellular stress pathways and proteasome transport. Because mutations affecting PI31 activity are associated with human neurodegenerative diseases, impairment of PI31-mediated axonal transport of proteasomes may contribute to these disorders.
Results
Requirement of PI31 for neuronal function and synaptic structure
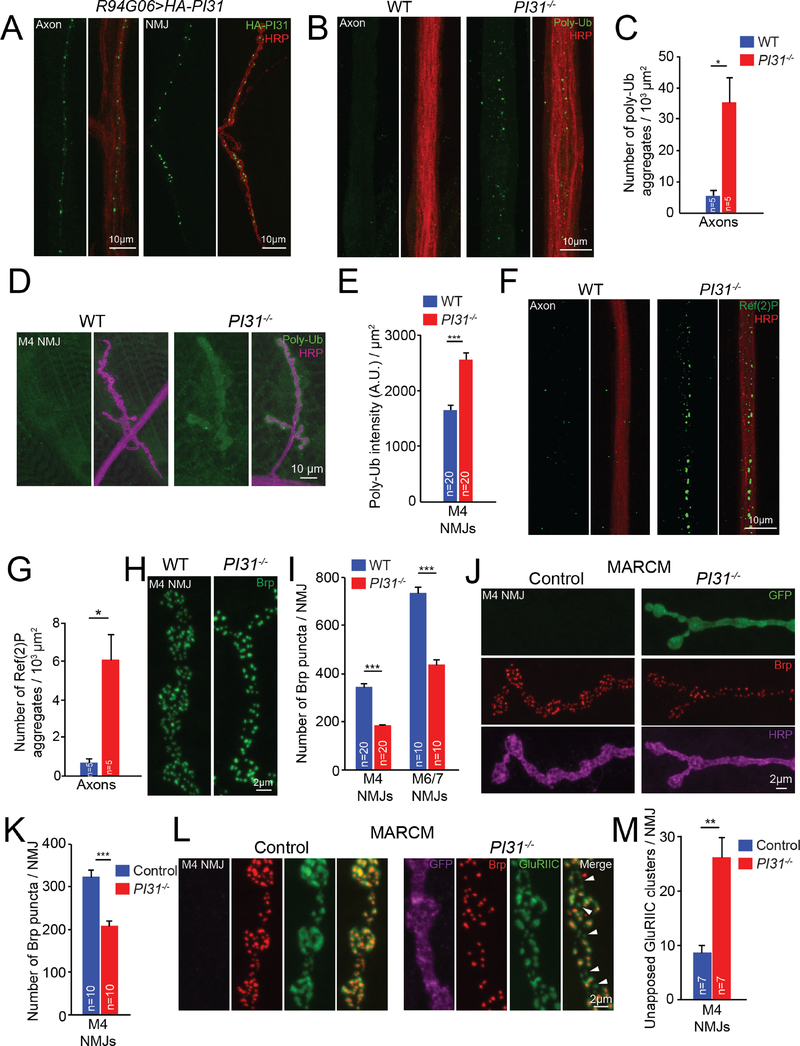
PI31 is a conserved proteasome-binding protein, and its expression is enriched in heads and testes in Drosophila (Bader et al., 2011; Chintapalli et al., 2007; Cho-Park and Steller, 2013; McCutchen-Maloney et al., 2000; Yashiroda et al., 2015). To study the role of PI31 in neurons, we used R94G06-GAL4 to drive expression of UAS-HA-PI31 and examined its localization in neurons. The R94G06-GAL4 driver allows for highly specific expression in a subset of motor neurons (MN1-IB) (Figure S1A). We found that PI31 forms puncta along axons and at neuromuscular junctions (NMJs), suggesting it may have a local function there (Figure 1A). Local activity of proteasomes is important for turnover of synaptic proteins and maintenance of protein homeostasis at neuronal terminals (Bingol and Schuman, 2006; Ehlers, 2003; Speese et al., 2003). To investigate a possible role of PI31 for protein homeostasis in axons and synapses, we conducted immunofluorescence studies to compare the amounts of poly-ubiquitinated proteins (poly-Ub), the substrates of proteasomes, in axons and at NMJs of wild-type and PI31−/− larvae. Quantification of these results demonstrates that PI31−/− larval axons contained significantly more poly-Ub aggregates compared to wild-type axons (Figure 1B and 1C). Likewise, the intensity of poly-Ub staining at NMJs was also elevated (Figure 1D and 1E). Since PI31 is a proteasome binding protein, this suggests that PI31 may play a role for protein degradation in axons and synapses. In order to explore this possibility further, we looked for other hallmarks of proteotoxic stress. Compromised proteasome function can induce compensatory mechanisms to remove unwanted proteins, such as clearance of aggregates by autophagy (Nezis et al., 2008; Pandey et al., 2007). For example, the Sequestosome 1 (SQSTM1/p62) protein delivers poly-Ub aggregates to the autophagic machinery for degradation in the lysosome, and formation of p62-positive aggregates is a hallmark for proteotoxic stress (Katsuragi et al., 2015; Moscat and Diaz-Meco, 2009). Loss of PI31 induced the formation of p62 aggregates in axons, consistent with the idea that PI31 is required for protein homeostasis at nerve terminals (Figure 1F and 1G).
Figure 1. PI31 is required for proteostasis and synaptic structure in motor neurons.
(A) PI31 forms puncta in axons and at neuromuscular junctions (NMJs) of Drosophila motor neurons. The expression of UAS-HA-PI31 was driven by R94G06-GAL4, a specific motor neuron driver (Figure S1A). HA antibody (green) labels HA-PI31, and HRP antibody (red) marks neuronal membranes.
(B and C) Representative images (B) and quantification(C) show that loss of PI31 leads to accumulation of poly-ubiquitinated (poly-Ub) protein aggregates in axons of Drosophila neurons. Poly-Ub proteins were detected with an Ub-conjugate antibody (clone FK2, green), and neuronal membranes were stained with anti-HRP (red) to visualize axons. Poly-Ub protein aggregates from three different images were counted and normalized for the axon area for each larva. n=5 larvae for each genotype. Mean+s.e.m., unpaired two-tailed t-test, * P<0.05.
(D and E) Representative images (D) and quantification (E) show that loss of PI31 leads to accumulation of poly-Ub proteins at Drosophila NMJs. Immunofluorescence intensities of poly-Ub proteins were measured and normalized for the NMJ area. n=20 M4 NMJs from four larvae for each genotype. Mean+s.e.m. Unpaired two-tailed t-test, *** P<0.001.
(F and G) Representative images (F) and quantification (G) show that loss of PI31 leads to accumulation of Ref(2)P aggregates (the Drosophila homologue of p62) in axons, indicative of proteotoxic stress. Ref(2)P aggregates from three different images were counted and normalized for the axon area for each larva. n=5 larvae for each genotype. Mean+s.e.m., unpaired two-tailed t-test, * P<0.05.
(H and I) High-magnification images (H) and quantification (I) of Brp puncta at NMJs indicate that loss of PI31 causes structural defects of synapses. n=20 NMJs from four larvae for M4, and n=10 NMJs from six larvae for M6/7 at A2 segments. Mean+s.e.m., unpaired two-tailed t-test, *** P<0.001. An overview of M4 NMJs is shown in Figure S1B, and overview and high-magnification images of M6/7 NMJs are presented in Figure S1C and S1D.
(J and K) Representative images (J) and quantification (K) of MARCM experiments show that loss of Brp puncta at PI31−/− NMJs is cell-autonomous and indicative of a requirement of PI31 in neurons. GFP+ is PI31−/−, and GFP− is PI31+/− from the same larva as an internal control. n=10 M4 NMJs from eight larvae for each genotype. Mean+s.e.m., paired two-tailed t-test, *** P<0.001. Shown here are high-magnification images, see Figure S1F for overview of MARCM M4 NMJs.
(L and M) High-magnification images (L) and quantification (M) of MARCM NMJs costained with anti-Brp (red) and anti-GluRIIC (green, a marker for postsynaptic densities) antibodies show that some synapses at PI31−/− NMJs lost Brp puncta but maintained postsynaptic densities (white arrowheads). The GFP-positive neuron (shown in a pseudo magenta color) is PI31−/−, whereas the GFP-negative neuron is PI31+/−. n=7 M4 NMJs from seven larvae for each genotype. Mean+s.e.m., paired two-tailed t-test, ** P<0.01. Quantification of GluRIIC intensity is provided in Figure S1G; no significant difference between control and PI31−/− MARCM NMJs were detected.
See also Figure S1.
Local activity of the UPS is important for the growth, activity and plasticity of synapses (Bingol and Sheng, 2011; DiAntonio et al., 2001; Ding et al., 2007; Djakovic et al., 2012; Erturk et al., 2014; Hegde et al., 2014; Kuo et al., 2005; Lee et al., 2008; Speese et al., 2003; Willeumier et al., 2006). To explore a role of PI31 for synaptic structure, we analyzed NMJ anatomy of wild-type and PI31−/− larvae. For this purpose, we conducted immunofluorescence analyses using anti-Horseradish peroxidase (HRP) and anti-Bruchpilot (Brp) antibodies, and examined Muscle 4 (M4) and Muscle 6/7 (M6/7) NMJs. The anti-HRP antibody binds to neuronal membranes and serves as a marker for neuronal morphology (Jan and Jan, 1982). Brp is a core scaffold protein for presynaptic active zone (AZ) and has been widely used as a marker for proper AZ assembly. It has homology to the human AZ protein ELKS/CAST/ERC and is crucial for AZ assembly, proper clustering of Ca2+ channels within AZs, and efficient neurotransmitter release(Kittel et al., 2006; Owald and Sigrist, 2009; Wagh et al., 2006). Although the overall morphology of NMJs was similar between wild-type and PI31−/− larvae, the number of Brp puncta was significantly reduced in PI31−/− larvae, suggesting a defect of AZ assembly (Figure 1H, 1I, and S1B–E). Our results indicate that PI31 is required for the proper development of synapses at NMJ.
Next we used mosaic analysis with a repressible cell marker (MARCM) to inactivate PI31 in a limited number of cells (Wu and Luo, 2006). This analysis confirmed that PI31−/− neurons have reduced numbers of Brp puncta and revealed a cell-autonomous requirement of PI31 in neurons for proper synaptic morphology (Figure 1J, 1K, and S1F). Interestingly, using a GluRIIC antibody that labels postsynaptic densities to co-stain MARCM NMJs with Brp antibody, we found that some synapses in PI31−/− neurons lost Brp puncta but maintained postsynaptic densities (Figure 1L, 1M and S1G, indicated by white arrowheads). This is similar to the synaptic phenotypes caused by knockdown of proteasome subunits in motor neurons (Valakh et al., 2012).
We also inactivated PI31 with a neuron-specific driver, Nrv2-GAL4(Sun et al., 1999). Nrv2>PI31 RNAi homozygotes died as pharate adults that appeared overall well-developed. These mutants initiated but did not complete the eclosion process as they displayed severe leg motor defects that made it impossible for them to escape from pupal cases (Table S1). PI31 is also required for the function and long-term survival of many other neuronal cell types in Drosophila, including photoreceptor neurons and dopaminergic neurons (data not shown). Taken together, our results indicate that inactivation of PI31 causes proteotoxic stress in axons and synapses, which in turn leads to structural defects of synapses.
P31 binds to LC8-type dynein light chain proteins
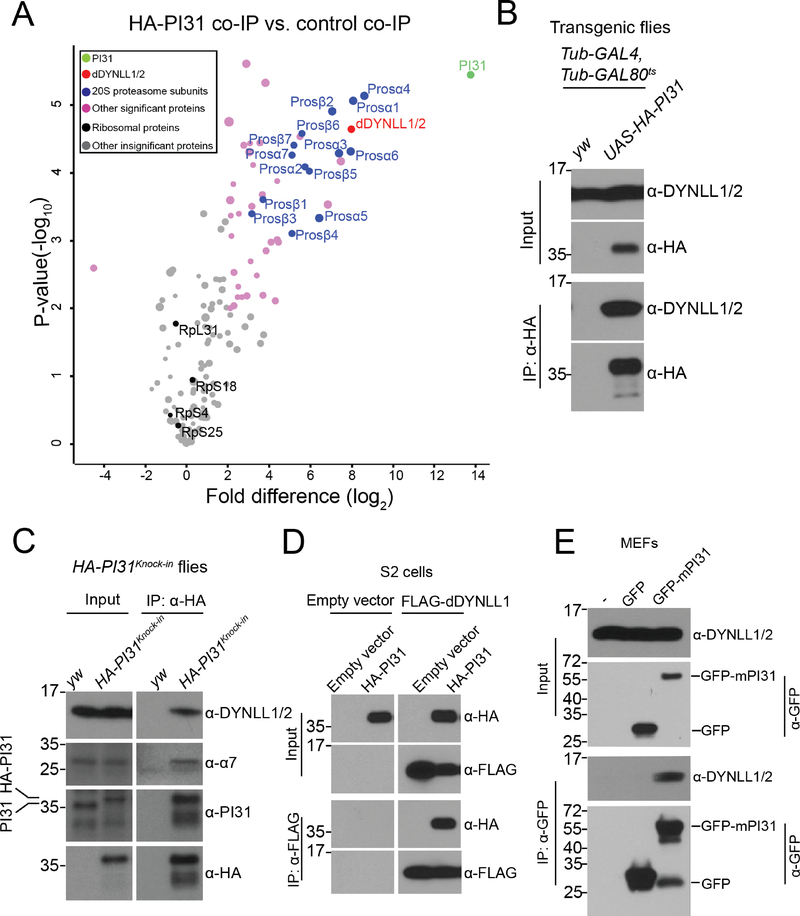
To understand the mechanism by which PI31 promotes proteostasis in the periphery of neurons, we looked for new binding partners of PI31. For this purpose, we expressed N-terminal HA-tagged PI31 (HA-PI31) in flies and performed anti-HA co-immunoprecipitation (co-IP) followed by label-free quantitative mass spectrometry (MS). Since HA-PI31 rescued the lethality of PI31 knockout flies, we conclude that the N-terminal tag did not interfere with PI31 functions (Bader et al., 2011). Consistent with the role of PI31 as a proteasome-binding protein, HA-PI31 pulled down all 20S proteasome subunits (Figure 2A and Table S2). Interestingly, two dynein light chain LC8-type proteins, Ctp (Drosophila homolog of dynein light chain LC8-type 1, dDYNLL1) and Cdlc2 (Drosophila homolog of dynein light chain LC8-type 2, dDYNLL2) were among the statistically most significant interactors (Figure 2A). Both proteins consist of 89 amino acids, share 98.9% identity, and differ in only one amino acid (Figure S2A). Since both unique peptides were identified by MS, this suggests that both proteins can bind to PI31 (Figure S2B). Dynein proteins are microtubule-dependent molecular motors that are responsible for fast intracellular transport of various cargos (Hirokawa et al., 2010; Reck-Peterson et al., 2018; Schliwa and Woehlke, 2003; Vale, 2003). Because dDYNLL1/Ctp is involved in axonal transport of proteasomes in Drosophila motor neurons, it is possible that PI31 mediates proteasome transport (Kreko-Pierce and Eaton, 2017).
Figure 2. DYNLL1/2 are binding partners of PI31 in Drosophila and mammalian cells.
(A) Volcano plot of quantitative MS data of anti-HA-PI31 co-IP (Tub-GAL4, Tub-GAL80ts >UAS-HA-PI31 flies) versus control co-IP (Tub-GAL4, Tub-GAL80ts flies). Biological triplicates were conducted and analyzed. The cutoff line for significance is P-value<0.01(−log10P >2) and fold difference>4 (log2fold difference >2 or <−2). PI31, dDYNLL1/2, 20S proteasome subunits and other significant proteins are labeled in green, red, blue and pink; whereas ribosomal proteins and other statistically insignificant proteins are labeled in black and grey. The size of dots roughly indicates the amount of a given protein detected by MS. dDYNLL1/2 are among the statistically most significant interactors of PI31, along with 20S proteasome subunits. See Table S2 for the raw data of MS analysis.
(B) The interaction between PI31 and dDYNLL1/2 was validated by co-IP-western blot assays. A human DYNLL1 antibody that cross-reacts with fly dDYNLL1/2 proteins was used for western blot analysis.
(C) Co-IP-western blot experiments using HA-PI31Knock-in flies demonstrate that PI31 interacts with dDYNLL1/2 at endogenous expression levels. A proteasome subunit α7 antibody that cross-reacts with the homologous fly protein was used to indicate that PI31 is also in a complex with proteasomes.
(D) Reciprocal co-IP using FLAG-dDYNLL1 as the bait validates the interaction between PI31 and dDYNLL1.
(E) The interaction between PI31 and DYNLL1/2 is conserved in mouse embryonic fibroblasts (MEFs).
To further study the interaction between PI31 and dDYNLL1/2, we conducted co-IP-western blot assays. HA-PI31 was able to pull down dDYNLL1/2 in both S2 cells and transgenic flies, indicating the existence of a PI31-dDYNLL1/2 complex in vivo (Figure 2B and S2C). To avoid possible overexpression artifacts and examine the interaction of PI31 and dDYNLL1 at endogenous expression levels, we generated a HA-PI31Knock-in fly strain through CRISPR-Homology Directed Repair (HDR)(Port et al., 2014). The expression level of HA-PI31Knock-in was comparable to PI31 levels in wild-type flies (Figure 2C). Using this system, we were able to show that PI31 can pull down dDYNLL1/2 and proteasomes at physiological expression levels (Figure 2C). We also performed the reciprocal co-IP using FLAG-dDYNLL1 as the bait and found it to successfully pull down PI31 (Figure 2D).
Dynein motor complexes are highly conserved in evolution (Hirokawa et al., 2010; Reck-Peterson et al., 2018; Schliwa and Woehlke, 2003; Vale, 2003). To investigate whether the interaction between PI31 and dynein light chain LC8-type proteins is conserved in mammals, we performed co-IP experiments in both MEFs and HEK293 cells and found that PI31 interacted with DYNLL1/2 in both cell types (Figure 2E and S2D). Taken together, these results indicate that PI31 interacts with DYNLL1/2 proteins, and that this interaction is conserved from Drosophila to mammals.
Our co-IP-MS experiments identified 20S proteasome subunits as binding partners of PI31 but did not detect 19S proteasome subunits (Figure 2A and Table S2). However, since we did not include ATP to preserve the stability of 26S particles, this does not mean that PI31 is only in a complex with 20S proteasomes (Liu et al., 2006). To investigate whether PI31 can form a complex with 26S proteasomes, we used HA-PI31Knock-in and conducted co-IP-native gel experiments in the presence of ATP. Our results indicate that PI31 interacts with both 20S and single-capped 26S proteasomes in vivo (Figure S2E).
PI31 phosphorylation stimulates direct binding to dDYNLL1/2
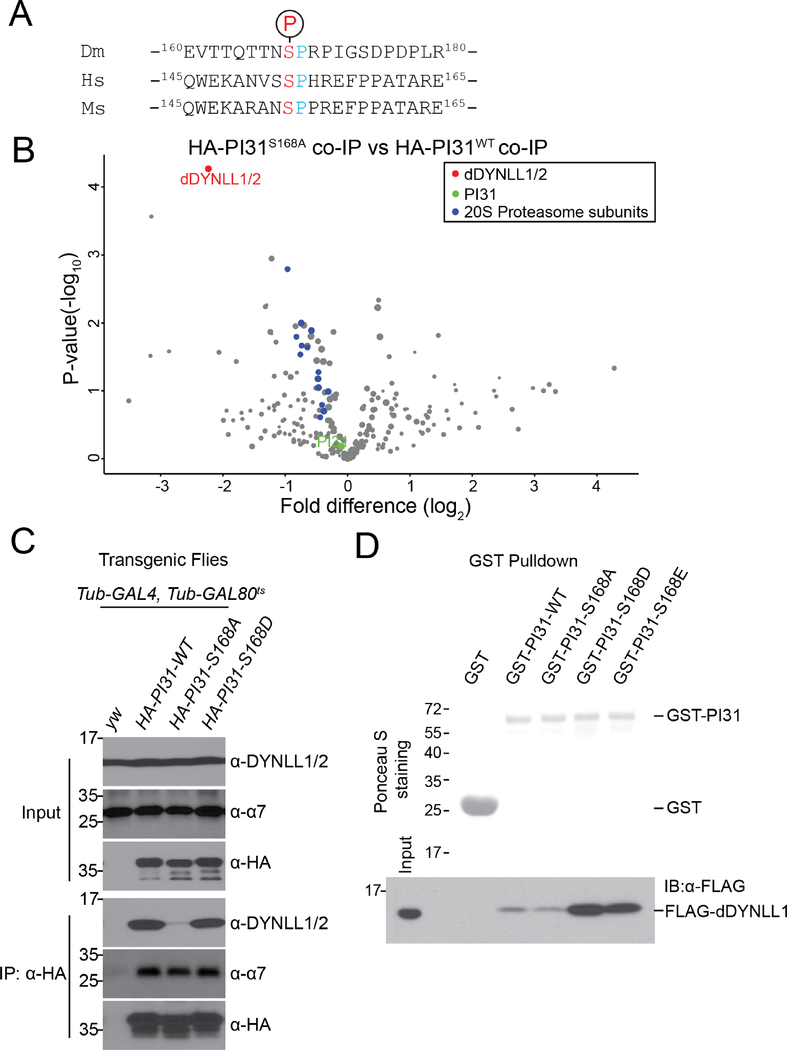
Co-IP-MS results revealed that Drosophila PI31 is phosphorylated at Serine 168 (Figure 3A and S3A). The corresponding site in human PI31 (Serine 153) is also phosphorylated, indicating that this modification is conserved from flies to human (Figure 3A and S3B). To study the function of this phosphorylation, we generated transgenic fly strains expressing a non-phosphorable mutant of Drosophila PI31 (Serine 168 to Alanine mutant, S168A) and compared its interactome with the interactome of wild-type PI31. Significantly, we identified dDYNLL1/2 as the most prominently affected binding partners of PI31 (Figure 3B and Table S3). This suggests that S168 phosphorylation stimulates the formation of a complex between PI31 and dDYNLL1/2.
Figure 3. PI31 phosphorylation enhances interaction with dDYNLL1/2.
(A) Alignment of the phosphorylation motifs of Drosophila, human and mouse PI31, indicates that PI31 is phosphorylated on a conserved Serine residue.
(B) Volcano plot of quantitative MS data of anti-HA-PI31S168A co-IP (Tub-GAL4, Tub-GAL80ts >UAS-HA-PI3S168A flies) versus anti-HA-PI31WT co-IP (Tub-GAL4, Tub-GAL80ts >UAS-HA-PI3wt flies). Biological triplicates were conducted and analyzed. PI31, dDYNLL1/2, and 20S proteasome subunits are labeled in green, red, and blue. The size of dots roughly indicates the amount of a given protein detected by MS. dDYNLL1/2 are the PI31-interacting proteins most affected by the non-phosphorable mutation, whereas binding of 20S proteasome subunits to PI31 was not significantly changed. See Table S3 for the raw data of MS analysis.
(C) Co-IP-western blot analysis shows that a non-phosphorable mutation of PI31 (S168A) significantly reduces interaction with dDYNLL1/2.
(D) GST-Pull down assays indicate that PI31 phosphorylation enhances direct binding to dDYNLL1. All recombinant proteins were produced in E. coli. Ponceau S that stains total proteins was used to assess the amount and purity of recombinant GST and GST-PI31 proteins. Anti-FLAG antibody was used to detect FLAG-dDYNLL1 pulled down by different forms of GST-PI31 proteins. The results show that purified GST-PI31, but not GST, can pull down recombinant FLAG-dDYNLL1. This demonstrates that PI31 and dDYNLL1 can directly bind to each other. Furthermore, this binding was strongly enhanced by two phospho-mimetic mutations of PI31 (GST-PI31-S168D and GST-PI31-S168E), suggesting that phosphorylation of PI31 at S168 stimulates complex formation. See also Figure S3 and Table S3.
Next we conducted co-IP-western blot assays. These results indicate that dDYNLL1/2 can efficiently interact with wild-type and phospho-mimetic PI31 (Serine 168 to Aspartic acid mutant, S168D) (Figure 3C and S3C, D). In contrast, this interaction was significantly reduced for the non-phosphorable mutant of PI31 (S168A) (Figure 3C and S3C, D).
We also investigated whether PI31 and dDYNLL1 directly interact with each other, and whether phosphorylation enhances interaction in vitro. For this purpose, we expressed recombinant FLAG-dDYNLL1 and GST-tagged wild-type (GST-PI31-WT), non-phosphorable mutant (GST-PI31-S168A), and two phospho-mimetic mutants (GST-PI31-S168D and GST-PI31-S168E) of PI31 in E. coli and conducted GST-pull down assays. The results show that GST-PI31-WT can interact with FLAG-dDYNLL1, whereas GST alone can not (Figure 3D). Importantly, the two phospho-mimetic mutants dramatically enhanced binding, whereas the non-phosphorable mutant had no activity (Figure 3D). We conclude that PI31 is a direct binding partner of dDYNLL1, and that phosphorylation of PI31 stimulates binding.
PI31 is required for the formation of dDYNLL1-proteasome complexes
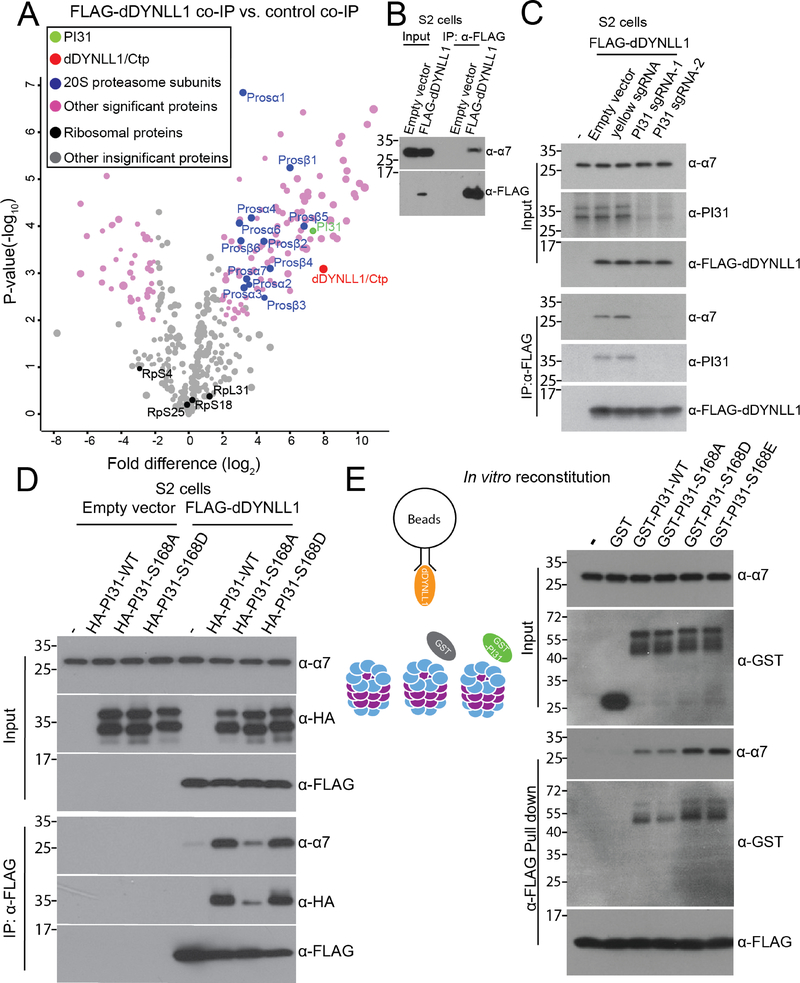
Since PI31 can directly bind to both dDYNLL1 and proteasomes, we explored the idea that this protein may serve as an adapter to mediate formation of a multi-protein complex containing proteasomes and dynein. We expressed FLAG-dDYNLL1 in S2 cells and conducted anti-FLAG co-IP experiments to see if proteasomes can be pulled down. Through MS analysis, we identified most of the 20S proteasome subunits (Figure 4A and Table S4). This supports the existence of a dDYNLL1-proteasome complex. Interestingly, PI31 was among the statistically most significant binding partners of dDYNLL1. We further validated the existence of this complex by co-IP-western blot assays (Figure 4B).
Figure 4. PI31 is required for the formation of dDYNLL1-proteasome complexes.
(A) Anti-FLAG-dDYNLL1 co-IP and MS analysis reveals that dDYNLL1 forms a protein complex with proteasomes. S2 cells expressing FLAG-dDYNLL1 were collected for anti-FLAG co-IP experiments. Volcano plot of label-free quantitative MS data is shown. Biological triplicates were conducted and analyzed. The cutoff line for significance is P-value<0.01(-log10P >2) and fold difference>4 (log2fold difference >2 or <−2). PI31, dDYNLL1/2, 20S proteasome subunits and other significant proteins are labeled in green, red, blue and pink; ribosomal proteins and other statistically insignificant proteins are labeled in black and grey. The size of dots roughly indicates the amount of a given protein detected by MS. See Table S4 for the raw data of MS analysis.
(B) Co-IP-western blot analysis confirms the existence of dDYNLL1-proteasome complexes. S2 cells expressing FLAG-dDYNLL1 were collected for anti-FLAG co-IP experiments.
(C) PI31 is required for the formation of dDYNLL1-proteasome complexes. PI31 knockout cell lines were generated by CRISPR using two independent sgRNAs. Control cell lines were generated either using empty vector for sgRNA cloning or using a sgRNA targeting the yellow gene. The results indicate that dDYNLL1 does not bind to proteasomes in PI31 knockout cells.
(D) PI31 phosphorylation stimulates the formation of dDYNLL1-proteasome complexes in S2 cells. FLAG-dDYNLL1 was expressed in S2 cells with or without different forms of HA-PI31, and then anti-FLAG co-IP-western blot analysis was conducted to assess proteasomes pulled down by dDYNLL1. The results indicate that PI31 can dramatically increase the amount of proteasomes bound to dDYNLL1, and this ability is diminished by the non-phosphorable mutation (S168A).
(E) PI31 phosphorylation stimulates the formation of dDYNLL1-proteasome complexes in vitro. Recombinant FLAG-dDYNLL1 was expressed in E. coli and bound to anti-FLAG magnetic beads. Next, purified bovine 20S proteasomes were added alone, or with GST or different forms of GST-PI31 proteins, which were expressed and purified from E.coli. GST-PI31, but not GST, was able to load 20S proteasomes onto dDYNLL1 in vitro, and this ability was further enhanced by two phospho-mimetic mutations (S168D and S168E). The left panel depicts a schematic model for the in vitro binding assay, and representative results of western blot analyses are shown in the right panel.
See also Table S4.
If PI31 is an adaptor protein mediating the interaction between dDYNLL1 and proteasomes, loss of PI31 should block the formation of these complexes. To test this possibility, we generated PI31-knockout S2 cell lines using two independent sgRNA targeting PI31 (Figure 4C) (Bassett et al., 2014). The interaction between dDYNLL1 and proteasomes was abolished in PI31 knockout cell lines (Figure 4C), indicating that PI31 is required for the formation of dDYNLL1-proteasome complexes. This suggests that PI31 functions as an adaptor to couple proteasomes with dyneins.
To examine the role of PI31 phosphorylation in the formation of dDYNLL1-proteasome complexes, we expressed FLAG-dDYNLL1 with different forms of HA-PI31 in S2 cells, performed anti-FLAG co-IP experiments, and compared the amount of proteasomes pulled down by FLAG-dDYNLL1. This demonstrates that elevated levels of PI31 can increase the amount of proteasomes bound to dDYNLL1 (Figure 4D). Interestingly, the non-phosphorable mutant of PI31 (S168A) was not significantly active (Figure 4D).
To test whether these effects are due to direct interactions mediated by PI31, we reconstituted the complex in vitro using purified 20S proteasomes and recombinant GST-PI31 and FLAG-dDYNLL1 expressed in E. coli. First, FLAG-dDYNLL1 was bound to anti-FLAG-magnetic beads. Then, 20S proteasomes were added alone, with GST, or with GST tagged wild-type or mutant PI31 proteins (Figure 4E). This approach showed that GST-PI31 was able to load 20S proteasomes onto dDYNLL1, whereas 20S proteasomes alone or with GST did not bind to dDYNLL1 (Figure 4E). Importantly, the two phospho-mimetic mutants (GST-PI31-S168D and GST-PI31-S168E) showed stronger ability than wild-type (GST-PI31-WT) or the non-phosphorable mutant (GST-PI31-S168A) to load proteasomes (Figure 4E). Taken together, these results show that PI31 is a direct adaptor protein that mediates the formation of a complex between proteasomes and dDYNLL1, and this activity is enhanced by phosphorylation of PI31.
PI31 is required for axonal transport of proteasomes in Drosophila motor neurons
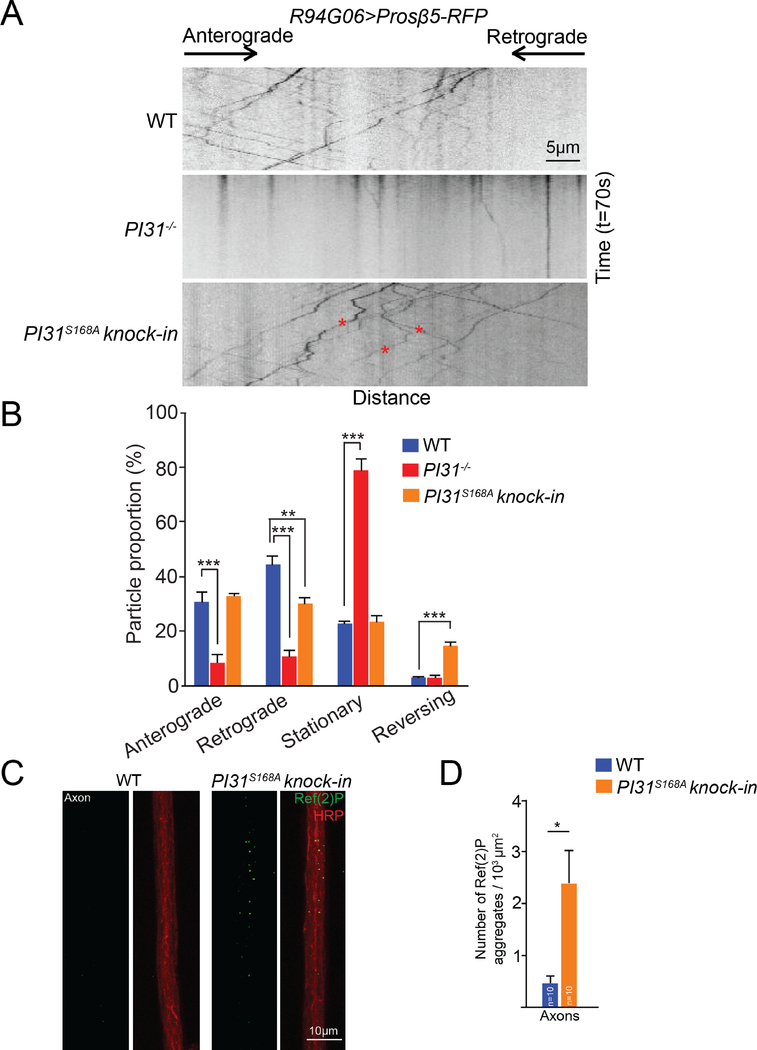
To examine a requirement of PI31 for axonal transport of proteasomes, we conducted live-imaging of proteasome movement in axons using the Prosβ5-RFP reporter, which is incorporated into functional proteasomes (Kreko-Pierce and Eaton, 2017). Expression was driven by R94G06-GAL4 to ensure specificity for motor neurons (Figure S1A). Inactivation of PI31 significantly reduced motility of proteasomes in axons (Figure 5A and 5B, and Movie S1 and S2). Both anterograde and retrograde movement of proteasomes was severely reduced, whereas the number of stationary proteasome particles was largely increased (Figure 5A and 5B, and Movie S1 and S2). These phenotypes are comparable to the defects described for dDYNLL1/ctp−/− motor neurons, supporting the idea that they function in the same protein complex (Kreko-Pierce and Eaton, 2017). Collectively, these results demonstrate that PI31 is required for proteasome transport along motor neuron axons.
Figure 5. PI31 and its phosphorylation are required for axonal transport of proteasomes in Drosophila motor neurons.
(A) Representative kymographs of Prosβ5-RFP motility in motor neurons of wild-type, PI31−/− and PI31S168A knock-in larvae. Prosβ5-RFP expression was driven by R94G06-GAL4, a motor neuron-specific driver (See Figure S1). The kymographs were generated from 70-second live imaging experiments (100 frames). The x-axis represents distance, while the y-axis represents time. The directions of anterograde and retrograde movement are indicated on top of the kymographs. Stationary particles appear as vertical lines, whereas motile particles appear as diagonal lines on kymographs. The red asterisks indicate the tracks of three proteasome particles in PI31S168A knock-in axons that changed their moving direction during live imaging. See Figure S4 for the generation of the PI31S168A knock-in strain.
(B) Quantification of proteasome movement. Shown are the percentages of Prosβ5-RFP particles that moved in anterograde direction, moved in retrograde direction, appeared stationary and reversely moved in axons of wild-type, PI31−/− and PI31S168A knock-in motor neurons. Mean + s.e.m. n=7 larvae for wild-type, n=12 larvae for PI31−/−, and n=7 larvae for PI31S168A knock-in. One-way Anova with Tukey’s honestly significant difference post hoc test. ***P<0.001, ** P<0.01. In axons of PI31−/− motor neurons, both anterograde and retrograde movements of proteasome particles were drastically reduced, while the portion of stationary proteasome particles increased accordingly; PI31S168A reduced retrograde transport of proteasomes and greatly increased the frequency of proteasomes that reversed the direction of their movement.
(C and D) Representative images (C) and quantification (D) show that PI31S168A knock-in flies accumulated Ref(2)P aggregates (the Drosophila homologue of P62) in axons, indicative of proteotoxic stress. Ref(2)P aggregates from three different images were counted for each larva. The numbers were added and normalized for the total axon area. n=10 larvae for each genotype. Mean+s.e.m., unpaired two-tailed t-test, * P<0.05.
Our biochemical data indicate that S168 phosphorylation of PI31 enhances loading of proteasomes onto dDYNLL1. To examine the physiological role of PI31-phosphorylation for proteasome transport, we generated PI31S168A knock-in mutants in which the wild-type PI31 gene was replaced with the non-phosphorable mutant (Figure S4). Live imaging of Prosβ5-RFP in homozygous PI31S168A knock-in larvae revealed that retrograde movement of proteasome particles was significantly reduced, and we also saw a dramatic increase in the number of particles that changed their direction of movement (Figure 5A, indicated by red asterisks, Figure 5B, and Movie S3). These results indicate that PI31 phosphorylation is important for directional transport of proteasomes.
Complete loss of PI31 disrupted proteasome transport and caused proteotoxic stress in axons (Figure 1B, 1C, 1F, 1G, 5A and 5B). Likewise, we detected accumulation of p62 aggregates in axons of the PI31S168A mutant, indicating that this mutation also causes proteotoxic stress (Figure 5C and 5D). However, since PI31S168A is hypomorphic, the effects of this mutant on both proteasome movement and p62 accumulation are less severe. We also found that PI31-S168A showed similar localization to the wild-type PI31 (Figure S4D). This suggests that S168 phosphorylation does not affect the overall localization of PI31 itself. This is expected because the mutant affects primarily the directionality of proteasome movement, but not binding of PI31 to proteasomes.
PI31 is required for axonal transport of proteasomes in mouse neurons
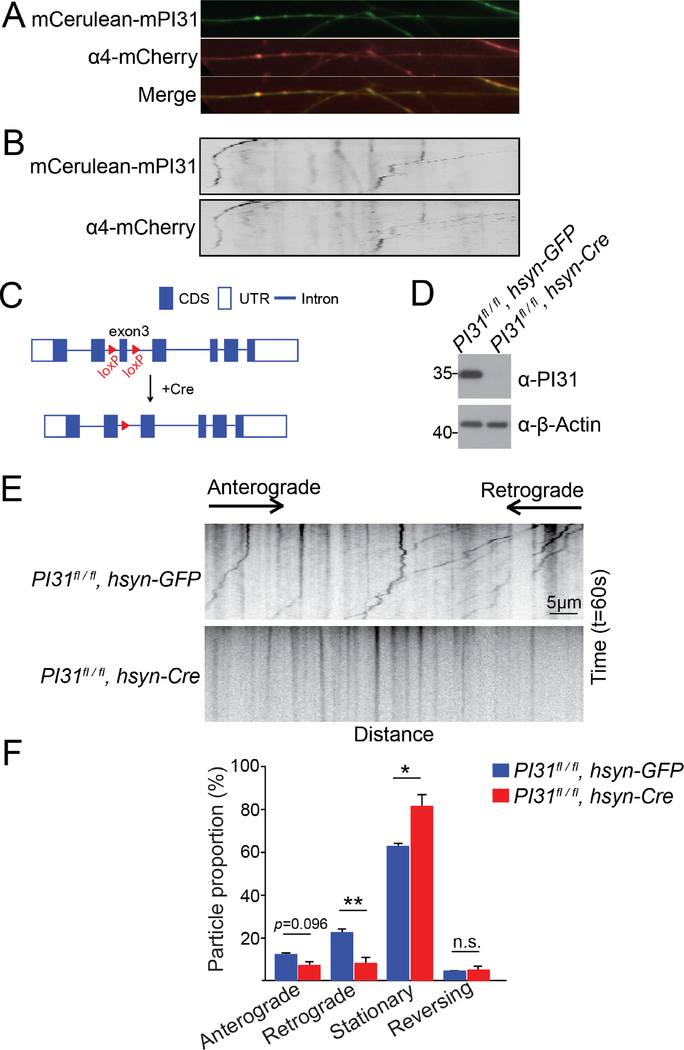
We also explored the possibility that the function of PI31 as an adapter to mediate axonal transport of proteasomes is conserved in evolution. First, we examined the subcellular localization of PI31 and proteasomes in axons of cultured mouse dorsal root ganglion (DRG) neurons. This showed that α4-mCherry and mCerulean-mPI31 co-localized and moved together in DRG axons (Figure 6A and 6B, and Movie S4).
Figure 6. PI31 is required for axonal transport of proteasomes in mouse neurons.
(A and B) Representative fluorescence images (A) and kymographs (B) of cultured mouse DRG neurons expressing mCerulean-mPI31 (green) and α4-mCherry (red) demonstrate that both proteins co-localized and moved together in axons. The kymographs were generated from a 140-second live imaging experiment (70 frames).
(C) Scheme for generating PI31-knockout neurons from PI31fl/fl mice. Two loxP sites were inserted flanking exon3 of PI31. Introduction of Cre recombinase into hippocampal neuron cultures with Adeno-associated virus (AAV) lead to the deletion of exon3 and inactivation of PI31.
(D) Western blot analysis was used to confirm that AAV-mediated introduction of Cre recombinase into hippocampal neurons succeeded in complete ablation of PI31 protein.
(E) Representative kymographs of α4-mScarlet-I motility in control (PI31fl/fl, hsyn-GFP) and PI31-knockout (PI31fl/fl, hsyn-Cre) hippocampal neurons. The α4-mScarlet-I expression cassette was delivered by lentiviral transduction. Kymographs were generated from 60-second live imaging experiments (100 frames). The x-axis represents distance, and the y-axis represents time. The directions of anterograde and retrograde movement are indicated on top of the kymographs.
(F) Quantification of proteasome movement. The percentages of α4-mScarlet-I particles that moved in anterograde direction, moved in retrograde direction, appeared stationary or reversed their direction were quantified in axons of control (PI31fl/fl, hsyn-GFP) and PI31 knockout (PI31fl/fl, hsyn-Cre) neurons. Mean + s.e.m. n=4 independent cultures for both genotypes. unpaired two-tailed t-test. **P<0.01, *P<0.05. Inactivation of PI31 had a modest effect on anterograde transport of proteasomes, whereas retrograde transport of proteasomes was strongly attenuated, and the fraction of stationary proteasomes was significantly increased as well.
To test the functional importance of PI31 for proteasome transport in mammals, we cultured hippocampal neurons derived from conditional PI31 knockout mice (PI31fl/fl) (Figure 6C and 6D). Proteasomes were visualized with a red fluorescent protein mScarlet-I-tagged proteasome subunit α4 reporter (α4-mScarlet-I) (Bindels et al., 2017). Live imaging revealed that the retrograde transport of proteasomes was strongly attenuated in PI31 knockout neurons (PI31fl/fl, hsyn-Cre), and we also saw a significant increase of stationary particles (Figure 6E and 6F). These results suggest that PI31 has a conserved function for axonal proteasome transport.
p38 MAPK can phosphorylate PI31
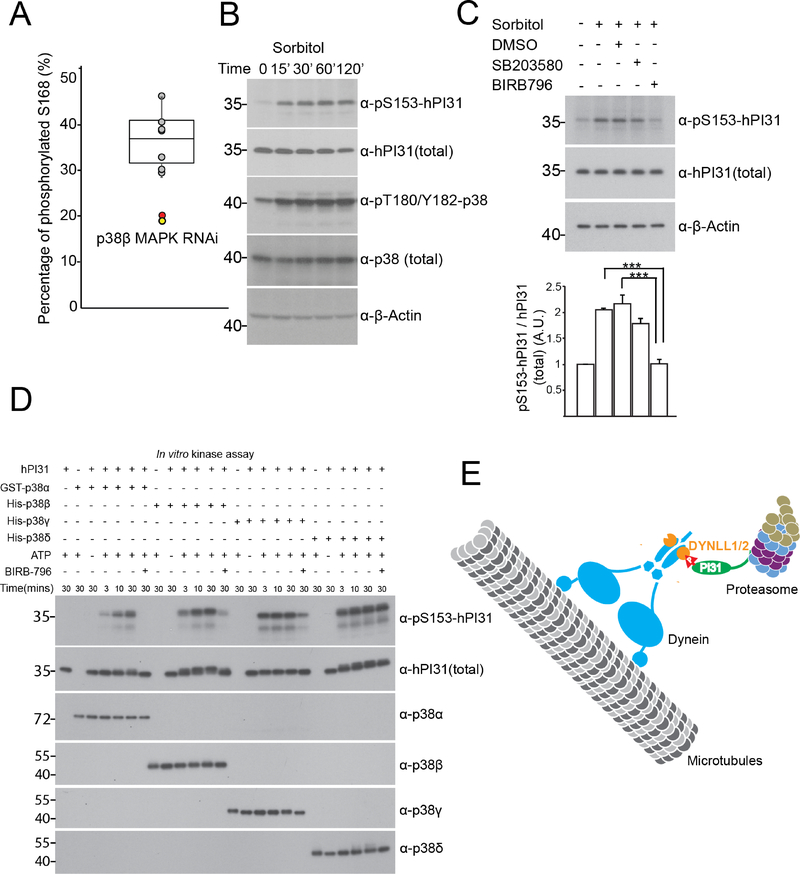
To identify the kinase responsible for PI31 phosphorylation, we conducted a candidate-based RNAi screen. Quantitative MS results indicated that knockdown of p38β MAPK reduced PI31 phosphorylation by almost 50%, suggesting it as a candidate for PI31 phosphorylation (Figure 7A).
Figure 7. p38 MAPK can phosphorylate PI31.
(A) A candidate-based kinase RNAi screen identified p38β as the kinase that phosphorylates PI31 at S168. Tub-GAL4, Tub-GAL80ts>UAS-HA-PI31 flies were crossed with RNAi transgenic fly strains targeting each kinase of MAPK or CDK family. HA-PI31 protein was immunoprecipitated and digested. The peptides containing the phosphorylated and non-phosphorylated S168 (EVTTQTTN168SPRPIGSDPDPLR) were quantified by MS analysis. The ratio of phospho-peptide signal versus total signal was used to estimate the percentage of S168-phosphorylated PI31. The distribution of S168 phosphorylation percentages of the screen is presented as a box plot with the following percentiles: 10% and 90% (bottom and top whiskers), 25% and 75% (bottom and top of box) and 50% (middle line). The percentages of S168 phosphorylation for control RNAi strains are shown as grey dots, and for replicates of p38β RNAi as red and yellow dots. This analysis indicates a ~50% decrease of PI31-phosphorylation upon RNAi-mediated down-regulation of p38β See Table S5 for the summary of MS analysis.
(B) S153 phosphorylation of human PI31 (pS153-hPI31) increased in response to osmotic stress, which activates p38. HEK293 cells were treated with 0.3M sorbitol for the indicated times. See Figure S5 for the generation and validation of the pS153-hPI31 antibody.
(C) The increase of pS153-hPI31 under osmotic stress was suppressed by p38 inhibitors. HEK293 cells were pretreated with indicated compounds (SB203580 and BIRB 796) at 10 μM for 2 hours, followed by treatment with 0.3M sorbitol for 1 hour. The upper panel shows representative western blot results, and the bottom panel is their quantification. Mean+ s.e.m. n=3. One-way Anova with Tukey’s honestly significant difference post hoc test. ***P<0.001. The results indicate that SB203580, which inhibits p38α and p38β, only mildly suppressed pS153-hPI31. In contrast, BIRB 796, which targets all four isoforms of p38, dramatically reduced pS153-hPI31. This indicated that PI31 phosphorylation in response to osmotic stress requires p38 activity.
(D) In vitro kinase assays using recombinant proteins generated in E. coli. All four isoforms of p38 MAPK can directly phosphorylate hPI31 at S153. Reactions without kinases, without PI31, without ATP or with BIRB 796 served as controls to validate specificity. The effects of BIRB 796 on p38δ were subtle under these conditions, consistent with the observation that p38δ had the highest IC50 for in vitro assays. Samples were also collected at three time points (3, 10 and 30 mins) to show kinetics of the reactions.
(E) Model for PI31-mediated transport of proteasomes in axons. Proteasomes can move rapidly along microtubules to fulfill dynamic local demands for protein degradation in different cellular compartments(DiAntonio et al., 2001; Hegde et al., 2014; Hsu et al., 2015; Kreko-Pierce and Eaton, 2017; Watts et al., 2003; Zhao et al., 2003). PI31 binds directly to both proteasomes and dynein light chain LC8-type proteins (DYNLL1/2). Inactivation of PI31 severely reduces proteasome movement and leads to proteotoxic stress in the periphery of neurons. This suggests that PI31 functions as an adaptor to directly couple proteasomes to microtubule-based motors. PI31 phosphorylation by the stress-activated p38 MAP kinase regulates the PI31-dDYNLL1 interaction and thereby modulates proteasome transport in axons. This may serve to dynamically adjust the spatio-temporal distribution of proteasomes in response to stress to match changing demands for protein breakdown. If this mechanism is impaired, mis-regulation of proteasome distribution occurs and leads to a failure of localized protein degradation. See also Figure S5 and Table S5.
p38 MAPKs can be activated by a wide range of cellular stresses as well as inflammatory cytokines(Cuenda and Rousseau, 2007; Zarubin and Han, 2005). One classical stress to activate p38 is sorbitol-induced osmotic stress(Cuenda and Rousseau, 2007; Han et al., 1994; Zarubin and Han, 2005). Therefore, we asked whether PI31 phosphorylation is elevated in response to osmotic stress. First, we generated a phosphorylation-specific antibody for Serine 153 of human PI31 (hPI31) and verified its specificity (Figure S5). Next, we treated HEK293 cells with sorbitol and assessed S153 phosphorylation (pS153) of endogenous hPI31. This showed that pS153 levels were indeed elevated after treatment, suggesting that stress promotes phosphorylation of PI31 at S153 (Figure 7B). To test whether this stress-induced phosphorylation depends on p38, we applied distinct p38 inhibitors. Inhibition of p38α and p38β by SB203580 had only minor effects, but BIRB 796, which targets all four isoforms of p38, dramatically reduced S153 phosphorylation of PI31 (Figure 7C). This suggests that multiple isoforms of p38 can phosphorylate PI31. Finally, we used purified recombinant p38 and hPI31 to conduct in vitro kinase assays and found that all four isoforms of p38 can directly phosphorylate PI31 (Figure 7D). We conclude that p38 can phosphorylate PI31 in response to stress.
Discussion
The degradation of unwanted and potentially toxic proteins by the ubiquitin-proteasome system (UPS) is of critical importance for protein homeostasis(Balch et al., 2008; Collins and Goldberg, 2017; Glickman and Ciechanover, 2002; Goldberg, 2003). In addition, UPS-mediated protein degradation also sculpts cell shape during development and modifies tissues throughout life(Bader et al., 2011; Bingol and Sheng, 2011; Campbell and Holt, 2001; Hegde et al., 2014; Lecker et al., 1999; Lee et al., 2008; Pak and Sheng, 2003; Qian et al., 2013; Sandri, 2013; Watts et al., 2003; Yi and Ehlers, 2005; Zhong and Belote, 2007). Neurons pose a particular challenge for regulated protein degradation due to their large size and highly compartmentalized structure. Localized regulation of UPS activity is crucial for synaptic growth, function and plasticity, and remodeling of axons and dendrites (Bingol and Schuman, 2006; Bingol and Sheng, 2011; DiAntonio et al., 2001; Djakovic et al., 2012; Ehlers, 2003; Erturk et al., 2014; Hamilton and Zito, 2013; Kuo et al., 2005; Pak and Sheng, 2003; Ramachandran and Margolis, 2017; Speese et al., 2003; Wan et al., 2000; Watts et al., 2003; Zhao et al., 2003). Nerve cells employ microtubule-based transport mechanisms to quickly distribute proteasomes between soma and neurites (Bingol and Schuman, 2006; Hsu et al., 2015; Kreko-Pierce and Eaton, 2017; Otero et al., 2014). However, central unresolved questions are how proteasomes are coupled to microtubule-based motor complexes, and how this process is regulated to meet dynamic demands of localized protein degradation. Advances in this area are important since they provide the foundation for specific manipulation of proteasome transport, including to potentially boost clearance of toxic proteins in the periphery of neurons.
One candidate protein as a transport adaptor for proteasomes is Ecm29(Hsu et al., 2015). This study used a fluorescent reporter, MV151, to visualize proteasome movement in axons of cultured hippocampal neurons. One concern with the use of this compound is that MV151 binds to catalytic β-subunits and inhibits proteasome activity(Verdoes et al., 2006). Significantly, Ecm29 is a proteasome quality control factor that is recruited to aberrant proteasomes (De La Mota-Peynado et al., 2013; Finley et al., 2016; Lee et al., 2011; Leggett et al., 2002; Lehmann et al., 2010; Park et al., 2011; Wang et al., 2010). Therefore, an alternative interpretation is that Ecm29-modulated proteasome movement may represent a quality control mechanism in response to proteasome inhibition by MV-151. Moreover, for Ecm29 to qualify as an “adapter” it remains to be demonstrated that Ecm29 can bind directly to motor proteins. Therefore, the precise role of Ecm29 in the formation of proteasome-motor complexes and the physiological function of this protein for neuronal development and function in vivo deserves further examination.
Here we identified dynein light chain LC8-type proteins (DYNLL1/2) as direct binding partners of the conserved proteasome regulator PI31 (“Proteasome Inhibitor of 31kD”). This raised the possibility that PI31 serves as an adapter to couple proteasomes via dynein light chain proteins to microtubule-based motors. In support of this idea, PI31 forms a complex with proteasomes and DYNLL1/2 in vivo, and we were able to reconstitute this complex with purified components in vitro. Moreover, we found that inactivation of PI31 disrupted axonal proteasome movement in both Drosophila and mouse neurons. Furthermore, loss of PI31 function led to the accumulation of poly-Ub proteins and p62 aggregates at nerve terminals and in axons. This indicates that insufficient amounts of proteasomes were available in the periphery of neurons to maintain protein homeostasis, which in turn caused structural defects of synapses. Collectively, our results suggest that PI31 is a conserved adaptor protein for microtubule-dependent transport of proteasomes and thereby regulates allocation of proteasomes between soma and synapses (Figure 7E).
Our work also revealed a role of p38 MAPK-mediated phosphorylation of PI31 in the formation of proteasome-DYNLL1/2 complexes. Phosphorylation of a conserved site in PI31 enhanced binding to DYNLL1/2 and stimulated the formation of proteasome-DYNLL1/2 complexes. Importantly, mutating the phospho-acceptor site of PI31 (PI31S168A knock-in) impaired proteasome transport and protein homeostasis in axons. This suggests that PI31 phosphorylation serves as a molecular switch to regulate proteasome movement (Figure 7E). p38 MAPKs can be activated by a wide range of cellular stresses, including proteotoxic stress(Cuenda and Rousseau, 2007; Zarubin and Han, 2005). Our findings are consistent with the idea that p38 MAPKs stimulate proteasome movement in response to stress by promoting the formation of proteasome-DYNLL1/2 complexes. Although both PI31−/− and PI31S168A knock-in mutants showed impairment of proteasome transport in axons, their phenotypes were not identical. Whereas complete inactivation of PI31 disrupted both anterograde and retrograde proteasome movement, the major defects in the PI31S168A mutant were reduced retrograde movement and loss of directionality (i.e., the number of proteasomes changing direction was strikingly increased compared to wild type). The most likely explanation for this difference is that PI31S168A is a hypomorphic mutant, which reduces but does not completely abolish the interaction between PI31 and dDYNLL1/2. Consistent with this possibility, we found that PI31S168A caused accumulation of p62 aggregates in axons that, while significant, was milder compared to PI31 full knockout. However, we cannot rule out that PI31 interacts with other motor proteins through phosphorylation-independent mechanisms, and/or that other post-translational modifications affect this process. We also attempted to quantify the steady-state levels of proteasomes at synapses, but unfortunately current techniques do not readily permit quantification of proteasomes at synapses in vivo because of their close proximity to surrounding cells.
This study may help reconcile somewhat conflicting reports in the literature. PI31 was initially identified as a proteasome-interacting protein, and binding of PI31 to 20S particles can inhibit peptide proteolysis in vitro(Bader et al., 2011; Chu-Ping et al., 1992; McCutchen-Maloney et al., 2000; Zaiss et al., 1999). On the other hand, genetic studies in Drosophila, yeast and Arabidopsis indicate that the physiological function of PI31 is to promote protein breakdown(Bader et al., 2011; Yang et al., 2016; Yashiroda et al., 2015). In contrast, no overt requirements for PI31 were detected in cultured HEK293 and HeLa cells, and PI31-inactivation produces only modest biochemical changes at the level of whole-cell extracts(Cho-Park and Steller, 2013; Li et al., 2014; Vingill et al., 2016). The current study provides an explanation for these seemingly conflicting reports as it reveals a role of PI31 for localized regulation of proteasomes. Such a localized function is not readily detectable by standard methodologies for assessing proteasome activity in whole-cell extracts. It is also noteworthy that large cells with highly compartmentalized cytoarchitecture, such as neurons and spermatids, are particularly sensitive to the loss of PI31. The terminal differentiation of both neurons and spermatids involves the combined action of caspases and proteasome-mediated protein degradation, and in both systems proteasomes move over considerable distances (Arama et al., 2003; Bader et al., 2011; Bingol and Sheng, 2011; Schoenmann et al., 2010; Simon et al., 2012; Watts et al., 2003; Zhong and Belote, 2007). Therefore, it appears that proper spatial allocation of proteasomes is particularly important in these contexts and relies on PI31 function.
The results presented here may also have important implications for human neurological disorders. Many age-related neurodegenerative diseases, including Alzheimer’s Disease (AD), Parkinson’s Disease (PD), and amyotrophic lateral sclerosis (ALS) are characterized by the accumulation of protein aggregates(Balch et al., 2008; Ballatore et al., 2007; Blokhuis et al., 2013; Irvine et al., 2008; Li and Li, 2011; Oddo, 2008; Ross and Poirier, 2004; Tai and Schuman, 2008). The proteins in these pathognomonic aggregates are typically ubiquitin-conjugated (Bilen and Bonini, 2005; de Vrij et al., 2004; Mori et al., 1987; Morimoto, 2008; Morishima-Kawashima et al., 1993; Perry et al., 1987; Rubinsztein, 2006; Zoghbi and Orr, 2000). This suggests that the affected neurons attempted to remove abnormal proteins by tagging them with ubiquitin, but subsequently failed to degrade them. One possible explanation for this is that proteasomes were not available in sufficient numbers to degrade all ubiquitin-conjugates. Because the first signs of impaired proteostasis and accumulation of pathological proteins are generally seen in the periphery of neurons, it has been suggested that defects in proteasome transport may be responsible for abnormal protein clearance in neurodegenerative diseases (Coleman and Perry, 2002; Gendron and Petrucelli, 2009; Hoover et al., 2010; Ittner et al., 2010; Otero et al., 2014; Yoshiyama et al., 2007; Yu and Lu, 2012). PI31 is implicated in this process since human mutations affecting PI31 activity are associated with neurodegenerative diseases, including AD, PD and ALS (Conedera et al., 2016; Di Fonzo et al., 2009; Johnson et al., 2010; Koppers et al., 2012; Paisan-Ruiz et al., 2010; Sherva et al., 2011). In particular, mutations in FBXO7/PARK15 lead to an early-onset, familiar form of PD in humans, and inactivation of FBXO7 in mice causes neuronal degeneration and PD-like phenotypes (Conedera et al., 2016; Di Fonzo et al., 2009; Paisan-Ruiz et al., 2010; Vingill et al., 2016). FBXO7 is a strong and direct binding partner of PI31 in both insects and mammals, and loss of FBXO7 function results in proteolytic cleavage and reduced levels of PI31(Bader et al., 2011; Kirk et al., 2008; Vingill et al., 2016). Strikingly, FBXO7-null mice were found to have impaired proteasome activity and significantly reduced PI31 levels, but a critical function of PI31 was dismissed based on analysis of whole-cell extracts(Vingill et al., 2016). Since a role of PI31 in proteasome transport would not have been detected by this approach, we suggest that this topic deserves further investigation.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Hermann Steller (steller@rockefeller.edu).
EXPERIMENTAL MODELS AND SUBJECT DETAILS
Drosophila melanogaster
All Drosophila melanogaster strains were raised on standard molasses formulation food at 25°C (except flies carrying GAL80ts, see below) with a 12-hour light/12-hour dark cycle. The yw strain was used as wild-type controls. For immunofluorescence and live imaging assays, wandering 3rd-instar larvae (96–120 hours after egg laying) were used. Both males and females were tested and showed similar phenotypes. Three-day old male HA-PI31Knock-in flies were used for co-IP experiments; for the co-IP experiments using flies that carried GAL80ts to achieve the inducible expression, 0–3-day-old male flies were collected from 25°C and moved to 29°C for 6 days. Male flies were used to avoid potential interference from egg-laying and the development of cultures.
Primary neuron cultures
All mouse neuron culture experiments were approved by the Animal Care and Use Committee (IACUC) of The Rockefeller University (protocol number 17006). Dorsal root ganglion (DRG) neurons were cultured from E13 mouse embryos (CD-1 IGS pregnant mouse, Charles River, #022). Hippocampal neurons were cultured from E18 embryos of PI31fl/fl mice, in which the third exon of PI31 is flanked by two loxP sites (Minis, Rodriguez, Levin & Steller, manuscript in preparation). All primary neurons were cultured in a 37 °C, 5% CO2 incubator.
METHOD DETAILS
Fly husbandry and strains
All flies were raised on standard molasses formulation food at 25°C with a 12-hour light/12-hour dark cycle. The following fly strains were used: R94G06-GAL4 (Janelia GAL4 collection, Bloomington Drosophila Stock Center, BDSC#40701) (Pfeiffer et al., 2008), UAS-mCD8-mCherry (BDSC #27392), UAS-HA-PI31 (2nd chromosome)(Bader et al., 2011), PI31−/−(Bader et al., 2011), hsp70-FLP1 FRT42D tubP-GAL80 (BDSC #9917), D42-GAL4 (BDSC # 8816), UAS-IVS-myr::GFP (BDSC #32197) (Pfeiffer et al., 2010), tubP-GAL4 (BDSC # 5138), tubP-GAL80ts (BDSC #7018), Nrv2-GAL4 UAS-GFP (BDSC #6794), UAS- PI31 RNAiKK105476 (Vienna Drosophila Resource Center, #105476) and UAS-Prosβ5-RFP(Kreko-Pierce and Eaton, 2017) (kindly provided by Dr. Benjamin A. Eaton, University of Texas). UAS-HA-PI31-WT attP_ ZH–68E, UAS-HA-PI31-S168A attP_ ZH–68E, UAS-HA-PI31-S168D attP_ZH–68E, HA-PI31Knock-in and PI31S168A knock-in (this study, see below).
Fly genotypes for each figure, table and movie are listed below:
Figure 1A. w; UAS-HA-PI31/+; R94G06-GAL4/+.
Figure 1B–I and S1B–E. WT: yw; +/+; +/+.
PI31−/−: w; PI3IΔ/PI3IΔ;+/+.
Figure 1J–M, S1F and S1G. yw, hsp70-FLP1; FRT42D, tubP-GAL80/FRT42D, PI31Δ; UAS- IVS-myr:: GFP/D42-GAL4.
Figure 2A and 2B, and Table S2.
Control: yw; +/+; tubP-GAL4, tubP-GAL80ts /+.
HA-PI31: w; +/+; tubP-GAL4, tubP-GAL80ts/UAS-HA-PI31-WTattP_ZH–68E.
Figure 2C and S2E. Control: yw; +/+; +/+.
HA-PI31Knock-in: w; HA-PI31Knock-in/HA-PI31Knock-in; +/+.
HA-PI31-WT: w; +/+; tubP-GAL4, tubP-GAL80ts/UAS-HA-PI31-WTattP_ZH–68E.
HA-PI31-S168A: w; +/+; tubP-GAL4, tubP-GAL80ts/UAS-HA-PI31-S168A attP_ZH–68E.
yw: w; +/+; tubP-GAL4, tubP-GAL80ts/+.
HA-PI31-WT: w; +/+; tubP-GAL4, tubP-GAL80ts/UAS-HA-PI31-WT attP_ZH–68E.
HA-PI31-S168A: w; +/+; tubP-GAL4, tubP-GAL80ts/UAS-HA-PI31-S168A attP_ZH–68E.
HA-PI31-S168D: w; +/+; tubP-GAL4, tubP-GAL80ts/UAS-HA-PI31-S168D attP_ZH–68E.
Figure 5A and 5B, and Movies S1–3.
WT: w; +/+; R94G06-GAL4, UAS-Prosβ5-RFP/R94G06-GAL4, UAS-Prosβ5-RFP.
PI31−/−: w; PI31Δ/PI31Δ; R94G06-GAL4, UAS-Prosβ5-RFP/R94G06-GAL4, UAS-Prosβ5-RFP.
PI31S168A knock-in: w; PI31S168A/PI31S168A; R94G06-GAL4, UAS-Prosβ5-RFP/R94G06-GAL4, UAS-Prosβ5-RFP.
WT: yw; +/+; +/+. PI31S168A knock-in: w; PI31S168A/PI31S168A; +/+.
Figure 7A and Table S5. F1 non-Tb flies from the cross of w; UAS-HA-PI31/UAS-HA-PI31; tubP-GAL4, tubP-GAL80ts/TM6B, Tb with kinase RNAi transgenic lines.
Figure S1A. w; +/+; R94G06-GAL4/UAS-mCD8-mCherry.
Figure S3A. w; +/+; tubP-GAL4, tubP-GAL80ts/UAS-HA-PI31-WTattP_ZH–68E.
Figure S4C. w; PI31S168A/+;+/+.
Figure S4D. w; +/+; R94G06-GAL4/UAS-HA-PI31-S168A attP_ ZH–68E.
Nrv2-GAL4: w;+/+; Nrv2-GAL4, UAS-GFP/Nrv2-GAL4, UAS-GFP.
PI31 RNAi: w; PI31 RNAiKK105476/PI31 RNAiKK105476.
Nrv2>PI31 RNAi: w; PI31 RNAiKK105476/PI31 RNAiKK105476; Nrv2-GAL4, UAS-GFP/Nrv2-GAL4, UAS-GFP.
Constructs and virus production
The gRNA constructs for making HA-PI31Knock-in and PI31S168A knock-in fly strains or making PI31 knockout S2 cell lines were generated by annealing of two ssDNA oligonucleotides (ssODN) together and inserting the annealed products into the BbsI sites of pCFD3-dU6:3gRNA vector (Port et al., 2014) (a gift from Dr. Simon Bullock, Addgene#49410) or BspQI sites of pAC-sgRNA-Cas9 vector (Bassett et al., 2014) (a gift from Dr. Ji-Long Liu, Addgene#49330). Listed are the ssODN sequences:
HA-PI31Knock-in_F: GTCGAGTCGACCCACTTTCCATTG;
HA-PI31Knock-in_R: AAACCAATGGAAAGTGGGTCGACT;
PI31S168A knock-in_F: GTCGTCCAATGGGGCGTGGTGAGT;
PI31S168A knock-in_R: AAACACTCACCACGCCCCATTGGA;
S2 cells_PI31_knockout_1_F: TTCGCGGTGGCCGCAATGGAAAGT;
S2 cells_ PI31_knockout_1_R: AACACTTTCCATTGCGGCCACCGC;
S2 cells_ PI31_knockout_2_F: TTCGAGTCGACCCACTTTCCATTG;
S2 cells_ PI31_knockout_2_R: AACCAATGGAAAGTGGGTCGACTC.
To generate S2 cell vectors to express HA-PI31 and FLAG-dDYNLL1, the HA-PI31 fragment was cut from pcDNA3-HA-PI31(Bader et al., 2011) using EcoRI and XhoI (New England Biolabs), and the FLAG-dDYNLL1 was cloned from a yw cDNA library by PCR and the FLAG-tag sequence was added on the forward primer in frame with the dDYNLL1 coding sequence (CDS). HA-PI31 (EcoRI/XhoI) and FLAG-dDYNLL1 (EcoRI/XbaI) fragments were inserted into the S2-cell expression vector pAc5.1/V5-His-A (Thermo Fisher Scientific, V411020).
To generate the S168 point mutation transgenic fly strains (UAS-HA-PI31-WT attP_ ZH–68E, UAS-HA-PI31-S168A attP_ ZH–68E, UAS-HA-PI31-S168D attP_ ZH–68E ), we first mutated the site on pcDNA3-HA-PI31(Bader et al., 2011) through PCR-based site directed mutagenesis. The PCR primers we used are:
S168A_F: CCACGCAGACGACCAACGCCCCACGCCCCATTGGATC;
S168A_R: GATCCAATGGGGCGTGGGGCGTTGGTCGTCTGCGTGG;
S168D_F: CCACGCAGACGACCAACGATCCACGCCCCATTGGATCG;
S168D_R: CGATCCAATGGGGCGTGGATCGTTGGTCGTCTGCGTGG.
After PCR, the template DNA was removed by DpnI treatment. The mutated clones were isolated by Sanger sequencing, and sub-cloned into pUAST-attB vector with EcoRI and XhoI (New England Biolabs)(Bischof et al., 2007).
To generate S2 cell vectors to express HA-PI31 S168 point mutants, the above mutants are sub-cloned into pAc5.1/V5-His-A (Thermo Fisher Scientific, V411020) with EcoRI and XhoI (New England Biolabs).
To generate E coli. vectors to recombinantly express different forms of GST-HA-PI31 and FLAG-dDYNLL1, the HA-PI31-WT, -S168A, -S168D and -S168E fragments were cut from pcDNA3-HA-PI31 vectors using EcoRI and XhoI and inserted into the pGEX-4T1 vector (GE Healthcare, #28954549) to generate pGEX-4T1-PI31 clones; the FLAG-dDYNLL1 fragment was produced by PCR using pAc5.1A-FLAG-dDYNLL1 as the template, digested with NcoI and EcoRI (New England Biolabs), and inserted into pET28a vector (Novagen, #69864–3) to generate the pET28a-FLAG-dDYNLL1 clone.
Mouse PI31 (mPI31) CDS was cloned from a cDNA library of mouse small intestine, which was generated by Superscript III First Strand Synthesis Kit (Thermo Fisher Scientific, #18080051) using oligo(dT) amplification, and inserted into the BstI/XmaI sites of pLVX-EF1a-AcGFP-C1 vector (Clontech, #631984). The lentivirus was packaged by transfection of pLVX-EF1a-AcGFP-C1-mPI31, pMD2.G (a gift from Dr. Didier Trono, Addgene #12259) and psPAX2 (a gift from Dr. Didier Trono, Addgene #12260) into HEK293 cells, and collected 48 hours after transfection. Human PI31 (hPI31) CDS was cloned from a cDNA library of HEK293 cells, and inserted into pcDNA3.2/V5/GW/D-TOPO vector (Thermo Fisher Scientific, K244020) through TOPO cloning method (kindly provided by Dr. Park Cho-Park, University of Pennsylvania).
Mouse proteasome subunit α4 CDS was cloned from a mouse cDNA library, and mCherry CDS was subcloned from the mCherry-Mito-7 vector (Olenych et al., 2007) (a gift from Dr. Michael Davidson, Addgene #55102). The α4-mCherry cassette was inserted between the EcoRI/NotI sites of pCAGGS/ES vector. mCerulean CDS was amplified by PCR using mCerulean-N1 vector as the template (Koushik et al., 2006)(a gift from Dr. Steven Vogel, Addgene #27795), and mPI31 CDS was cloned from a mouse cDNA library. The mCerulean-mPI31 cassette was inserted between the EcoRI/NheI sites of pCAGGS/ES vector. The α4-mScarlet-I expression cassette was synthesized by IDT Inc., and inserted into the AgeI and EcoRI sites of pLenti-hSynapsin- Cre-WPRE to replace the Cre CDS(Sakurai et al., 2016) (a gift from Dr. Fan Wang, Addgene #86641). Lentivirus was packaged by transfection of pLenti-hSynapsin-α4-mScarlet-I-WPRE, pMD2.G (a gift from Dr. Didier Trono, Addgene #12259) and psPAX2 (a gift from Dr. Didier Trono, Addgene #12260) into HEK293 cells, and collected 48 hours after transfection. Lentivirus was concentrated with Lenti-X concentrator (Takara Inc., #631231), and resuspended in Neurobasal medium.
All constructs were validated by Sanger sequencing (Genewiz).
Antibody production
To generate a polyclonal antibody against Drosophila PI31, GST-PI31 was expressed in BL21 Star (DE3) E.coli cells(Thermo Fisher Scientific, #C601003), and purified using Glutathione Sepharose 4B beads (GE Healthcare, #17–0756-01). The purified protein was injected into rabbits and the antisera were collected (Cocalico).
To generate the phosphorylation-specific antibody towards S153 phosphorylation of human PI31, a phospho-peptide was synthesized (Figure S5A), conjugated to carrier proteins and injected into rabbits (YenZym Antibodies, LLC). The elicited antiserum was first affinity-purified against the phospho-peptide. Bound antibodies were eluted and further affinity-absorbed against the non-phosphorylated peptide counterpart to remove antibodies recognizing the other parts of the peptide instead of the phosphorylation site.
Generation of MARCM clones
PI31−/− mutant clones were made using the mosaic analysis with a repressible cell marker (MARCM) system(Wu and Luo, 2006). The fly genotype is: yw, hsp70-FLP1; FRT42D, tubP-GAL80/FRT42D, PI31Δ; UAS- IVS-myr::GFP/D42-GAL4. The eggs were laid on fly food at 25°C. The 0–6-hour embryos were heat-shocked at 37°C for 30mins followed by recovery at 25 °C for 30mins, and then heat-shocked again at 37°C for 45mins. The wandering 3rd -instar larvae with the appropriate genotype were dissected and proceeded for immunostaining.
Generation of knock-in fly strains
CRISPR-Cas9 technology was used to generate the HA-PI31Knock-in and PI31S168A knock-in fly strains. The off-target effect of gRNAs were assessed by CRISPR target finder (http://tools.flycrispr.molbio.wisc.edu/targetFinder/) (Gratz et al., 2014) and E-CRISPR (www.e-crisp.org/E-CRISP/)(Heigwer et al., 2014). The selected sequences (HA-PI31Knock-in:AGTCGACCCACTTTCCATTG, on the “-” strand; PI31S168A knock-in: TCCAATGGGGCGTGGTGAGT, on the “-” strand) were cloned into pCFD3-dU6:3gRNA vector as described above(Port et al., 2014). ssODNs, which serve as donor templates for HDR, were designed with ~50nt homology arm on each side, and synthesized (IDT). Their sequences are listed below:
HA-PI31Knock-in:
CTTGTACAGCAGATCCCAACCGTAAAAGAAATCGCCCGTCTTGGCAGTCGACCCACTTTCGCCACCGCCGCTAGCATAGTCGGGCACATCATATGGGTACATTGCGGCCACCGATAATCCTTTACAAAATCTGGGTGGGAGTAAACAGAACGAATGGAAC;
PI31S168A knock-in:
GTTCTTACCCGCCACGCCTTGGCTCGCCGATGCGCAAGGGATCTGGATCCGAT CCAATGGGGCGTGGTGCGTTCGTCGTCTGCGTGGTAACTTCGCGCGAGTTTCCCGTGAATACAGGGTCTAGAAGCTC.
The pCFD3-dU6:3-PI31 gRNA constructs (final concentration 150ng/μl) and the donor template ssODNs (final concentration 200ng/μl) were mixed together and injected into they, sc, v; +/+; nos-Cas9attP2 fly (Ren et al., 2013). Knock-in flies were initially identified by PCR and then confirmed by Sanger sequencing (Genewiz).
Generation of PI31-knockout S2 cell lines
S2 cells cultured in 10cm-plates were transfected with 12μg pAC-sgRNA-Cas9 (empty vector, a gift from Dr. Ji-Long Liu, Addgene#49330)(Bassett et al., 2014), pAC-y1_sgRNA-Cas9 (a gift from Dr. Ji-Long Liu, Addgene#49331)(Bassett et al., 2014), pAC-PI31_1_sgRNA-Cas9, or pAC-PI31_2_sgRNA-Cas9 with TransIT-Insect transfection reagent (Mirus, #MIR6100). After three days the cells were treated with 5μg/ml puromycin for seven days. Knockout efficiency was examined by western blot analysis.
Immunofluorescence
Wandering 3rd-instar larvae were pinned on a silicone petri dish, and dissected in HL3 saline. The fillet preparations were fixed in 4% paraformaldehyde/PBS for 20mins. After washing with PBS/0.1% Triton X-100 (PBST), the fillets were blocked in 5% NGS/0.2% BSA/PBST, followed by incubation with primary antibodies overnight at 4°C. After washing for 3 times with 0.2%BSA/PBST, the secondary antibodies were applied at room temperature for 3 hours. The fillets were washed 4 times and proceeded for mounting and examination on a confocal microscope (LSM780, Zeiss). The following antibodies were used: chicken anti-mCherry (1:500, Novus, #NBP2–25158), rat anti-HA (1:100, clone 3F10, Sigma-Aldrich, #11867423001), mouse anti-ubiquitin conjugates (1:500, clone FK2, Enzo, #PW8810), rabbit anti-Ref(2)P (Nezis et al., 2008) (1:5000, kindly provided by Dr. Tor Erik Rusten, University of Oslo), mouse anti-Brp (1:300, clone nc82, Developmental Studies Hybridoma Bank, #nc82), chicken anti-GFP (1:1000, Aves Labs, #GFP-1020), rabbit anti-GluRIIC(Marrus et al., 2004) (1:1000, kindly provided by Dr. Aaron DiAntonio, Washington University in St. Louis), goat anti-rat-Alexa 488(1:500, Jackson ImmunoResearch, #112–545-167), goat anti-mouse-Alexa 488 plus (1:500, Thermo Fisher Scientific, #A32723), goat anti-chicken-Alexa 488(1:500, Jackson ImmunoResearch, #103–545-155), goat anti-chicken-Cy3(1:500, Jackson ImmunoResearch, #103–165-155), goat anti-mouse-Alexa 568(1:500, Thermo Fisher Scientific, #A11031), goat anti-HRP-Cy3(1:3000, Jackson ImmunoResearch, #123–165-021), goat anti-rabbit-Alexa 633(1:500, Thermo Fisher Scientific, #A21071), and goat anti-HRP-Alexa 647(1:300, Jackson ImmunoResearch, #123–605-021). Quantification of puncta number and intensity of poly-Ub proteins was done with Fiji software(Schindelin et al., 2012), and Fiji and Imaris (Bitplane) softwares were used for quantification of numbers and volume of Brp puncta.
Co-Immunoprecipitation (Co-IP)
For the co-IP experiments shown in Figure 2A, 2B, 3B, and 3C, tubP-GAL4, tubP-GAL80ts flies were crossed with indicated forms of UAS-HA-PI31 or yw (as control) flies and raised at 25°C. The 0–3-day-old F1 progenies were collected and moved to 29°C for 6 days. Three hundred male flies were collected and homogenized in 1.5ml buffer A [50mM Tris-HCl (pH7.5), 137mM NaCl, 0.25% NP-40, protease inhibitor cocktail (Roche, #11873580001) and phosphatase inhibitor cocktail (Roche, #04906837001)]. After centrifuged at 20000g for 15mins and filtered through a low-protein binding 0.45μm PVDF membrane syringe filter (Millipore, #SLHVX13NL), the extracts were incubated with 30μl mouse anti-HA agarose beads (clone HA-7, Sigma-Aldrich, #A2095) at 4°C for 3 hours. After six washes in buffer A and two additional washes in buffer A without NP-40 and protease and phosphatase inhibitors, the beads were eluted in 30μl 0.1M glycine (pH2.5) at room temperature for 10mins. The eluted proteins were taken either for mass spectrometry (MS) analysis (Figure 2A and 3B) or for western blot analysis (Figure 2B and 3C).
For the co-IP experiment shown in Figure 2C, 150 3-day old male HA-PI31Knock-in flies and yw flies (as control) were collected separately and homogenized in 1ml buffer A with 50U/ml DNase I (Thermo Fisher Scientific, #18047019). After centrifuged at 20000g for 15mins, sonicated briefly to break genomic DNAs, and filtered through a 0.45μm syringe filter, the extracts were incubated with 30μl mouse anti-HA magnetic beads (Thermo Fisher Scientific, #88836) at 4°C for 3 hours. After five washes in buffer A and two additional washes in buffer A without NP-40 and protease and phosphatase inhibitors, the beads were eluted in 30μl 500μg/ml HA peptide (Thermo Fisher Scientific, #26184) at 37°C for 30mins. The eluted proteins were analyzed by western blotting.
For the IP or co-IP experiments shown in Figure 2D, 2E, 4B, 4C, 4D, S2C, S2D, S3C and S3D, 10cm-plate cultures of indicated cell lines were transfected/infected with indicated constructs using TransIT-Insect transfection reagent (Mirus, #MIR6100) for S2 cells, Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific, #11668–019) for HEK293 cells and lentivirus for primary MEFs, respectively. After 48 hours for the transfected cells or 7 days for the infected MEFs, the cell cultures were rinsed in cold PBS, collected, and resuspended in 0.6ml buffer A. Lysis was facilitated by freeze/thaw for three times. After centrifugation at 20000g for 15mins and filtration through a 0.45μm syringe filter, the extracts were incubated with 30μl mouse anti-HA agarose beads (clone HA-7, Sigma-Aldrich, #A2095), or 30μl mouse anti-FLAG magnetic beads (clone M2, Sigma-Aldrich, #M8823), or 30μl GFP-Trap agarose beads (Chromotek, gta-20), which were pre-blocked with 2% BSA, at 4°C for 3 hours. The beads were washed in buffer A for six times and then were eluted in 30μl 1.5XSDS-PAGE sample buffer by boiling at 95°C for 5mins.
For the co-IP experiment shown in Figure 4A, 10-cm plate cultures of S2 cells were transected with 15 μg pAc5.1A-FLAG-dDYNLL1 or pAc5.1A empty vector (as control) constructs using TransIT-Insect transfection reagent. After 48 hours, the cells were rinsed, collected and resuspended in 600μl buffer A, and lysis was facilitated by freeze/thaw for three times. After centrifugation at 20000g for 15mins and filtration through a 0.45μm syringe filter, the extracts were incubated with 30μl mouse anti-FLAG magnetic beads (clone M2, Sigma-Aldrich, #M8823) at 4°C for 3 hours. After six washes in buffer A and two additional washes in buffer A without NP-40 and protease and phosphatase inhibitors, the beads were eluted in 30μl 300 μg/ml 3ΧFLAG peptide (Sigma-Aldrich, #F4799) at 4°C for 45mins. The eluted proteins were run on a SDS-PAGE gel for ~1cm to remove the 3ΧFLAG peptide. Finally, the gel was cut for MS analysis. Biological triplicates of the co-IP were used for quantification.
For the co-IP experiment shown in Figure S2E, eight hundreds of 3-day-old male HA-PI31Knock-in flies and yw flies (as control) were collected separately and homogenized in 5ml buffer B [25mM HEPES (pH7.5), 100mM NaCl, 5mM MgCl2, 2mM ATP, 10% glycerol, 0.25% NP-40, protease inhibitor cocktail and phosphatase inhibitor cocktail] with 50U/ml DNase I (Thermo Fisher Scientific, #18047019). After centrifugation at 20000g for 15mins, lysates were sonicated briefly to break genomic DNA and filtered through a 0.45μm syringe filter, and then the extracts were incubated with 200μl mouse anti-HA magnetic beads (Thermo Fisher Scientific, #88836) at 4°C for 3 hours. After five washes in buffer B and two additional washes in buffer C [25mM HEPES (pH7.5), 150mM NaCl, 5mM MgCl2, 2mM ATP, 10% glycerol], the beads were eluted by 200μl 500μg/ml HA peptide in buffer C at 37°C for 30mins. The eluted proteins were concentrated 5X with 10KDa-cutoff Amicon centrifugal filter unit (Millipore, #UFC501024), and analyzed by native gels and western blot assays.
Mass spectrometry analysis
Glycine (0.1M, pH2.5) eluted samples (Figure 2A, 3B, 7A, S3A and S3B) were dried in Speedvac concentrator (Thermo Fisher Scientific) and dissolved in 8 M urea, 0.1 M ammonium bicarbonate, and 10 mM DTT. After reduction, cysteines were alkylated in 30mM iodoacetamide. The proteins were digested in less than 4 M urea by endoproteinase LysC (Wako Chemicals) followed by digestion with Trypsin Gold (Promega) in less than 2 M urea. FLAG-peptide eluted samples (Figure 4A) were cleaned up using SDS-PAGE, reduced (10 mM DTT) and alkylated (30 mM iodoacetamide), followed by digestion with a cocktail of LysC and Trypsin Gold. Digestions were halted by adding trifluoroacetic acid (TFA) and digests were desalted and analyzed by reversed phase nano-LC-MS/MS using either a Fusion Lumos or a Q-Exactive Plus (Thermo Fisher Scientific) both operated in high/high mode(Rappsilber et al., 2007).
Data were searched and quantified against Uniprot’s Drosophila and human proteome databases using ProteomeDiscoverer v. 1.4.0.288 (Thermo Scientific) combined with Mascot v. 2.5.1 (Matrix Science) and/or MaxQuant v. 1.6.0.13. (Cox et al., 2014). Oxidation of methionine and protein N-terminal acetylation were allowed as variable modifications and all cysteines were treated as being carbamidomethylated. Peptide matches were filtered using a Percolator-calculated false discovery rate (FDR) of 1%(Kall et al., 2007). For the quantitative MaxQuant analysis, FDR thresholds for peptides and proteins were set at 2% and 1%, respectively. For assignment of phosphorylated residues, we used PhosRS3.0 combined with Mascot(Taus et al., 2011).
Label Free Quantitation (LFQ) or intensity-based absolute quantification (iBAQ) were used for the identification and quantitation of binding partners(Cox et al., 2014; Schwanhausser et al., 2011). Data were processed using Perseus v1.6.0.7(Tyanova et al., 2016). In short, reverse matches and common contaminations were disregarded, and protein signals in minimum 2 out of 3 replicates for at least one condition was required. Data were validated by scatter plots, LFQ or iBAQ based principal component analysis (PCA) and distributions of LFQ or iBAQ values, and found to be comparable. T-tests were performed and proteins were considered as interesting hits if showing a difference of more than 4-linear folds and a p-value of less than 0.01.
GST pull-down assays
BL21 Star (DE3) competent E.coli cells were transformed with pGEX-4T1-PI31 vectors (-WT, -S168A, -S168D, and -S168E), pGEX-4T1 empty vector or pET28a-FLAG-dDYNLL1 vector. Single colonies were picked and cultured in 5ml LB, followed by an enlarged culture in 100ml LB. When OD600≈0.4, IPTG was added to a final concentration of 0.3mM. After 4 hours, the bacteria were collected by centrifugation and rinsed by cold PBS. The bacteria were resuspended in 5ml cold PBS with protease inhibitor cocktail, lysed by sonication and centrifuged at 20000g for 15mins.
1ml pGEX-4T1 empty vector lysate or 1ml pGEX-4T1-PI31 lysate were incubated with 30μl Glutathione Sepharose 4B beads at 4°C for 3 hours. Then, the beads were washed with PBS for three times, followed by incubation with 1ml pET28a-FLAG-dDYNLL1 lysate at 4°C for 3 hours. The beads were washed in buffer A for five times and in 50 mM Tris-HCl (pH 8.0) once. Elution was done by incubation of the beads in 30μl GST elution buffer [50 mM Tris-HCl (pH 8.0) and 20mM reduced glutathione (Sigma-Aldrich, #G4251)] at room temperature for 20mins. The eluted proteins were analyzed by SDS-PAGE and western blot assays. Ponceau S (Sigma-Aldrich, #P7170) staining was used to assess purity and amount of GST and different forms of GST-PI31 proteins. Anti-FLAG-HRP (1:1000, Sigma-Aldrich, #A8592) was used to evaluate amount of FLAG-dDYNLL1 bound to GST or different forms of GST-PI31 proteins.
In vitro reconstitution
To reconstitute the dDYNLL1-PI31-proteasome complex in vitro, six-milliliter lysate of BL21 Star (DE3) expressing pET28a-FLAG-dDYNLL1 was incubated with 200μl mouse anti-FLAG magnetic beads (clone M2, Sigma-Aldrich, #M8823) at 4°C for 3 hours. After washed with buffer A for four times, the beads were split equally into six tubes. Then, 5μg purified bovine 20S proteasomes (UBPbio, #A1401), or 5μg GST+5μg purified bovine 20S proteasomes, or 5μg indicated forms of GST-PI31 proteins+5μg purified bovine 20S proteasomes were added to the six tubes, respectively, and incubated at 4°C for 3 hours. After four washes in buffer A, the beads were eluted in 35μl 300μg/ml 3ΧFLAG peptide at 4°C for 45mins.
Kinase screen
The conserved PI31 phosphorylation site (pSerine-Proline) pointed to Proline-directed kinases, such as cyclin-dependent kinases (CDKs) and mitogen-activated protein kinases (MAPKs) (Wagih et al., 2016; Zhu et al., 2005). Therefore, we screened all available CDK and MAPK RNAi fly strains from the Transgenic RNAi Project (TRiP) collection. Tub-GAL4, Tub-GAL80ts>UAS-HA-PI31 flies were crossed with RNAi transgenic fly strains. HA-PI31 protein was immunoprecipitated and digested. The peptides containing the phosphorylated and non-phosphorylated S168 (EVTTQTTN168SPRPIGSDPDPLR) were targeted and measured in parallel reaction monitoring (PRM) experiments. Phospho-peptide signal versus total signal was used to estimate the phosphorylation occupancy percentage.
In vitro kinase assay
Purified recombinant human p38α (MAPK14, GST-tagged, Thermo Fisher Scientific, #PV3304), p38β (MAPK11, His-tagged, Thermo Fisher Scientific, #PV3679), p38γ (MAPK12, His-tagged Thermo Fisher Scientific, #PV3654), p38δ (MAPK13, His-tagged, Thermo Fisher Scientific, #PV3656) and hPI31 (UBPBio, #A3901) were used for in vitro kinase assay.
One microgram of hPI31 was incubated with 200ng indicated kinases in 30μl kinase assay buffer [20 mM HEPES (pH 7.5), 2 mM DTT, 10 mM MgCl2, 200μM ATP, and phosphatase inhibitors] at 30°C for indicated time (3, 10 or 30 mins). Reactions without kinases, without PI31 or without ATP were conducted as control to validate that the PI31 phosphorylation signal was specific. p38 MAPK inhibitor BIRB 796 was pre-incubated with kinases on ice for 10mins in indicated reactions to validate the dependence of PI31 phosphorylation on p38. The reaction was terminated by adding 15μl 3XSDS-PAGE loading buffer and boiling at 95°C for 5mins.
In vitro phosphatase assay
To validate the specificity of p153-hPI31 antibody, HA-hPI31 was expressed in HEK293 cells and IP with anti-HA agarose beads. After four washes, the beads were split into three tubes. One was incubated with 50μl phosphatase assay buffer (50mM HEPES, 100mM NaCl, 2mM DTT, 0.01% Brij-35) as a control, one was treated with 400U Lambda protein phosphatase (Lambda PP, New England Biolabs, #P0753S) in 50μl phosphatase assay buffer, and the other one was treated with Lambda PP in the presence of phosphatase inhibitor cocktail (Roche, #04906837001). After incubated at 30°C for 30mins, the beads were precipitated and hPI31 proteins were eluted in 1XSDS-PAGE loading buffer and boiled at 95°C for 5mins.
Native gel assays
Protein complexes captured by anti-HA co-IP using extracts of yw (as control) and HA-PI31Knock-in flies were eluted in buffer C with HA peptide under the native condition (see the “Co-IP” section for more details). The eluted proteins, as well as 15μg total lysates of yw and HA-PI31Knock-in flies (as inputs for the co-IP experiment), were mixed with 2X native sample buffer (Bio-rad, #1610738), and loaded onto 3–8% Tris-acetate gels (Bio-rad, #3450131). Purified bovine 19S (UBPbio, #A1301), 20S (UBPbio, #A1401) and 26S (UBPbio, #A1201) proteasomes were also loaded onto the gels as standards to mark positions of different proteasome complexes. Electrophoresis was started with 50V for 1 hour in running buffer (90mM Tris, 90mM Boric Acid, 1mM EDTA, 2.5 mM MgCl2, 1mM ATP and 0.5mM DTT). After samples fully entered the gel, voltage was increased to 120V and ran 5 hours at 4°C. Then, proteins in the gels were transferred to PVDF membrane for western blot.
Western blot analysis
Proteins in SDS-PAGE gels or native gels were transferred onto 0.45μm Immobilon-P PVDF membranes (Millipore. #IPVH00010). The membranes were blocked with 5% non-fat milk in TBST [20mM Tris (pH7.5), 300mM NaCl, 0.1% Tween-20] at room temperature for 1 hour, followed by incubation with primary antibodies in 5% BSA/TBST at 4°C overnight. After washed for three times with TBST, the membranes were incubated with HRP-conjugated secondary antibodies at room temperature for 1.5 hours. The membranes were washed for four times with TBST, and detection was performed with ECL reagent (GE Healthcare, #RPN2134) or SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific, # 34096). The following antibodies were used for western blot: rabbit anti-DYNLL1/2(1:3000, clone EP1660Y, Abcam, #ab51603), mouse anti-α7 (1:3000, clone MCP72, Enzo, # PW8110), mouse anti-Rpt3 (1:3000, clone TBP7–27, Enzo, # PW8765), rabbit anti-dPI31(1:5000), goat anti-hPI31(1:3000, Thermo Fisher Scientific, #PA5–18133), rabbit anti-hPI31(1:3000, Abcam, #ab140497), rabbit anti-phospho-p38 MAPK (Thr180/Tyr182) (1:3000, clone 3D7, Cell Signaling Technology, #9215), rabbit anti-p38 MAPK (total) (1:3000, Cell Signaling Technology, #9212), rabbit anti-p38α (1:2000, Cell Signaling Technology, #9218), rabbit anti-p38β (1:2000, Cell Signaling Technology, #2339), rabbit anti-p38γ (1:2000, Cell Signaling Technology, #2307), rabbit anti-p38δ (1:2000, Cell Signaling Technology, #2308), rat anti-HA-HRP(1:3000, clone 3F10, Roche, #12013819001), mouse anti-FLAG-HRP(1:1000, clone M2, Sigma-Aldrich, #A8592), mouse anti-GFP-HRP(1:1000, clone B2, Santa Cruz Biotechnology, #sc-9996-HRP), rabbit anti-GST-HRP(1:1000, Abcam, #ab3416), rabbit anti-β-Actin-HRP (1:10000, clone 13E5, Cell Signaling Technology, #5125), mouse anti-β-Actin-HRP (1:10000, clone AC-15, Sigma-Aldrich, #A3854), donkey anti-rabbit-HRP(1:5000, Jackson ImmunoResearch, #711–035-152), donkey anti-mouse-HRP(1:5000, Jackson ImmunoResearch, #715–035-150), goat anti-mouse Fc-HRP(1:3000, Thermo Fisher Scientific, #31437).
Live imaging
For analysis of proteasome transport in axons of Drosophila motor neurons, wandering 3rd-instar larvae of R94G06-GAL4>Prosβ5-RFP (as control), PI31−/−, R94G06-GAL4>Prosβ5-RFP and PI31S168A, R94G06-GAL4>Prosβ5-RFP were dissected in HL3 saline supplemented with 7mM L-glutamic acid, and flipped the inside out to expose motor neuron axons. The larval preparations were mounted on a microscope slide with coverslip bridges to avoid crash of tissues. Live imaging was done on a 60X silicone oil objective lens (NA1.3, Olympus) of inverted Olympus IX-70 microscope equipped with Evolve-512 EMCCD camera (Photometrics). Images were captured using a 500-ms exposure at 1.4 Hz, and each recording consisted of 100 frames in 70 seconds. Kymographs were generated using ImageJ with the “Multiple Kymograph” plugin. All particles were picked and tracked by the software.
The mouse neuron culture experiments were approved by the Animal Care and Use Committee (IACUC) of The Rockefeller University (protocol number 17006). For live imaging of α4-mCherry and mCerulean–PI31 in mouse DRG neuron cultures, DRGs were dissected from E13 mouse embryos (CD-1 IGS pregnant mouse, Charles River, #022) and dissociated to single cell suspension using Trypsin. Dissociated sensory neurons were transfected with pCAGGS/ES-α4-mCherry and pCAGGS/ES-mCerulean–mPI31 plasmids using the Neon transfection system (Thermo Fisher Scientific, MPK5000) at 1400 V, 20 ms, and 1 pulse. After transfection, 5X105 neurons were centrifuged, resuspended in 9μl growth medium [Neurobasal medium (Thermo Fisher Scientific, #21103049), B-27 (Thermo Fisher Scientific, #17504044), penicillin-streptomycin (Thermo Fisher Scientific, #15140122) with 12.5ng/ml mouse growth factor 2.5S (NGF, Alomone Labs, #N-100)] and transferred to 15μl growth medium as a hanging drop overnight to form cell aggregates. Aggregates were cut into four pieces, plated on Poly-D-Lysine (Sigma-Aldrich, #P6407)/Laminin (Sigma-Aldrich, #L2020)-coated 8-well μ-Slide (ibidi, # 80826) and cultured for additional 24–48 hours. Before imaging, culture medium was changed to Hibernate E low fluorescence media (BrainBits, #HELF) supplemented with B27 and NGF, in order to maintain cells in ambient CO2 levels. Images were taken at 0.5Hz for 1–3 minutes on a 60X silicone oil objective lens (NA1.3, Olympus) of inverted Olympus IX-70 microscope equipped with Evolve-512 EMCCD camera (Photometrics) and with a heating chamber at 37°C.
For live imaging experiments of α4-mScarlet-I in mouse hippocampal neuron cultures, hippocampi were dissected from E18 PI31fl/fl mouse embryos and incubated with papain-DNase I mix (Worthington Biochemical, #LK003160) at 37°C for 10mins. After washed and treated with Ovomucoid protease inhibitor, hippocampi were dissociated into single cell suspension by gentle pipetting. Cells were collected by centrifugation at 300g for 10mins, and resuspended in phenol red-free Neurobasal medium (Thermo Fisher Scientific, #12348017) with B-27 supplement (Thermo Fisher Scientific, #17504044), GlutaMAX supplement (Thermo Fisher Scientific, 35050061) and penicillin-streptomycin (Thermo Fisher Scientific, #15140122). After counted using a hemocytometer, 6X104 cells were plated into each well of 8-well μ-Slide (ibidi, # 80826) coated with Poly-D-Lysine (Sigma-Aldrich, #P6407), and cultured in a 37 °C, 5% CO2 incubator. After 48 hours, the cultures were treated with 10μM cytosine arabinoside (AraC) to remove any glial cell contamination. Medium was completely changed after 24 hours, and Adeno-associated virus 8 (AAV8)-hSyn-GFP-Cre or AAV8-hSyn-GFP (control) (University of North Carolina, Vector Core) was added to cultures to induce recombination between loxP sites and inactivation of PI31. After 24 hours, lentivirus carrying the expression cassette of α4-mScarlet-I was added to cultures. Half the medium was changed every three days. Live imaging experiments were conducted 12 days after AAV infection. Images were taken at 1.6 Hz for 1min on a 60X silicone oil objective lens (NA1.3, Olympus) of inverted Olympus IX-70 microscope equipped with Evolve-512 EMCCD camera (Photometrics) and with a heating chamber at 37°C.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis was performed using Prism 7 (Graphpad Software Inc.) and Excel (Microsoft). Significance was determined by two-tailed Student’s t-test for comparison of two groups or by one-way Anova with Tukey’s honestly significant difference post hoc test for multiple comparisons. All detailed information of statistics has been indicated in figure legends.
DATA AND CODE AVAILABILITY
Raw data of all mass spectrometry analyses have been shown in Supplementary Tables 2–5.
Supplementary Material
Table S2. Raw data for the MS analysis of the anti-HA-PI31 co-IP experiments. Related to Figure 2.
Table S3. Raw data for the MS analysis of the anti-HA-PI31S168A co-IP versus anti-HA-PI31wt co-IP experiments. Related to Figure 3.
Table S4. Raw data for the MS analysis of the anti-FLAG-dDYNLL1 co-IP experiments. Related to Figure 4.
Table S5. Summary for the MS analysis of the kinase screen experiments. Related to Figure 7.
Movie S1. Representative video of Prosβ5-RFP particle movement in an axon of wild-type larva (w; +/+; R94G06-GAL4, UAS-Prosβ5-RFP/R94G06-GAL4, UAS-Prosβ5-RFP). Related to Figure 5. The video was acquired at 1.4Hz rate for 70 seconds (100 frames in total), and is shown in 10X of the original speed.
Movie S2. Representative video of Prosβ5-RFP particle movement in an axon of PI31−/− larva (w; PI31Δ/PI31Δ; R94G06-GAL4, UAS-Prosβ5-RFP/R94G06-GAL4, UAS-Prosβ5-RFP). Related to Figure 5. The video was acquired at 1.4Hz rate for 70 seconds (100 frames in total), and is shown in 10X of the original speed.
Movie S3. Representative video of Prosβ5-RFP particle movement in an axon of PI31S168Aknock-in larva (w; PI31S168A / PI31S168A; R94G06-GAL4, UAS-Prosβ5-RFP/R94G06-GAL4, UAS-Prosβ5-RFP). Related to Figure 5. The video was acquired at 1.4Hz rate for 70 seconds (100 frames in total), and is shown in 10X of the original speed.
Movie S4. Representative video of mCerulean-mPI31 (green) and α4-mCherry (red) movement in axons of cultured mouse DRG neurons. Related to Figure 5. The video was acquired at 0.5Hz rate for 140 seconds (70 frames in total), and is shown in 10X of the original speed.
Movie S5. Representative video of α4-mScarlet-I particle movement in an axon of control hippocampal neuron (PI31fl/fl, hsyn-GFP). Related to Figure 6. The video was acquired at 1.6Hz rate for 60 seconds (100 frames in total), and is shown in 10X of the original speed.
Movie S6. Representative video of α4-mScarlet-I particle movement in an axon of PI31 knockout hippocampal neuron (PI31fl/fl, hsyn-Cre). Related to Figure 6. The video was acquired at 1.6Hz rate for 60 seconds (100 frames in total), and is shown in 10X of the original speed.
Highlights.
PI31 directly mediates the formation of dynein light chain-proteasome complexes.
PI31 is required for axonal transport of proteasomes in Drosophila and mice.
Stress regulates PI31 activity through p38 MAPK-mediated phosphorylation.
This mechanism is required for protein homeostasis and structure of synapses.
Acknowledgement
We thank Drs. Aaron DiAntonio and Tor Erik Rusten for the GluRIIC and Ref(2)P antibodies; Dr. Benjamin Eaton, the Bloomington Drosophila Stock Center, Vienna Drosophila Resource Center, FlyLight team of Janelia Research Campus, and Flybase for Drosophila strains and relevant information. We are also grateful to Drs. Park Cho-Park, Simon Bullock, Michael Davidson, Ji-Long Liu, Didier Trono, Steven Vogel, and Fan Wang, and Addgene for plasmids. We thank Yetis Gultekin for generating the pLVX-EF1a-AcGFP-C1-mPI31 plasmid and anti-PI31 antibody, and Drs. Alison North, Christina Pyrgaki and Tao Tong for their help on live-imaging analysis. We also thank Drs. Michael Young, Brian Chait, Sarit Larisch, Park Cho-Park, Xiaochun Li, Mia Horowitz, Avi Levin, Sigi Benjamin-Hong, Dolors Ferres-Marco, Junko Shimazu, Thomas Hsiao, Wanhe Li, Erica Jacobs, and Alana Persaud for valuable discussions. This work was supported by NIH grant RO1GM60124 and a generous gift from the Loewenberg Foundation to H.S.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arama E, Agapite J, and Steller H (2003). Caspase activity and a specific cytochrome C are required for sperm differentiation in Drosophila. Developmental cell 4, 687–697. [DOI] [PubMed] [Google Scholar]
- Asano S, Fukuda Y, Beck F, Aufderheide A, Forster F, Danev R, and Baumeister W (2015). Proteasomes. A molecular census of 26S proteasomes in intact neurons. Science 347, 439–442. [DOI] [PubMed] [Google Scholar]
- Bader M, Benjamin S, Wapinski OL, Smith DM, Goldberg AL, and Steller H (2011). A conserved F box regulatory complex controls proteasome activity in Drosophila. Cell 145, 371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, and Elledge SJ (1996). SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86, 263–274. [DOI] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, Dillin A, and Kelly JW (2008). Adapting proteostasis for disease intervention. Science 319, 916–919. [DOI] [PubMed] [Google Scholar]
- Ballatore C, Lee VM, and Trojanowski JQ (2007). Tau-mediated neurodegeneration in Alzheimer’s disease and related disorders. Nat Rev Neurosci 8, 663–672. [DOI] [PubMed] [Google Scholar]
- Bassett AR, Tibbit C, Ponting CP, and Liu JL (2014). Mutagenesis and homologous recombination in Drosophila cell lines using CRISPR/Cas9. Biology open 3, 42–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumeister W, Walz J, Zuhl F, and Seemuller E (1998). The proteasome: paradigm of a self-compartmentalizing protease. Cell 92, 367–380. [DOI] [PubMed] [Google Scholar]
- Bento CF, Renna M, Ghislat G, Puri C, Ashkenazi A, Vicinanza M, Menzies FM, and Rubinsztein DC (2016). Mammalian Autophagy: How Does It Work? Annual review of biochemistry 85, 685–713. [DOI] [PubMed] [Google Scholar]
- Bilen J, and Bonini NM (2005). Drosophila as a model for human neurodegenerative disease. Annual review of genetics 39, 153–171. [DOI] [PubMed] [Google Scholar]
- Bindels DS, Haarbosch L, van Weeren L, Postma M, Wiese KE, Mastop M, Aumonier S, Gotthard G, Royant A, Hink MA, et al. (2017). mScarlet: a bright monomeric red fluorescent protein for cellular imaging. Nature methods 14, 53–56. [DOI] [PubMed] [Google Scholar]
- Bingol B, and Schuman EM (2006). Activity-dependent dynamics and sequestration of proteasomes in dendritic spines. Nature 441, 1144–1148. [DOI] [PubMed] [Google Scholar]
- Bingol B, and Sheng M (2011). Deconstruction for reconstruction: the role of proteolysis in neural plasticity and disease. Neuron 69, 22–32. [DOI] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, and Basler K (2007). An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proceedings of the National Academy of Sciences of the United States of America 104, 3312–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokhuis AM, Groen EJ, Koppers M, van den Berg LH, and Pasterkamp RJ (2013). Protein aggregation in amyotrophic lateral sclerosis. Acta neuropathologica 125, 777–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell DS, and Holt CE (2001). Chemotropic responses of retinal growth cones mediated by rapid local protein synthesis and degradation. Neuron 32, 1013–1026. [DOI] [PubMed] [Google Scholar]
- Chintapalli VR, Wang J, and Dow JA (2007). Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nature genetics 39, 715–720. [DOI] [PubMed] [Google Scholar]
- Cho-Park PF, and Steller H (2013). Proteasome regulation by ADP-ribosylation. Cell 153, 614–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu-Ping M, Slaughter CA, and DeMartino GN (1992). Purification and characterization of a protein inhibitor of the 20S proteasome (macropain). Biochimica et biophysica acta 1119, 303–311. [DOI] [PubMed] [Google Scholar]
- Clemen CS, Marko M, Strucksberg KH, Behrens J, Wittig I, Gartner L, Winter L, Chevessier F, Matthias J, Turk M, et al. (2015). VCP and PSMF1: Antagonistic regulators of proteasome activity. Biochemical and biophysical research communications 463, 1210–1217. [DOI] [PubMed] [Google Scholar]
- Coleman MP, and Perry VH (2002). Axon pathology in neurological disease: a neglected therapeutic target. Trends in neurosciences 25, 532–537. [DOI] [PubMed] [Google Scholar]
- Collins GA, and Goldberg AL (2017). The Logic of the 26S Proteasome. Cell 169, 792–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conedera S, Apaydin H, Li Y, Yoshino H, Ikeda A, Matsushima T, Funayama M, Nishioka K, and Hattori N (2016). FBXO7 mutations in Parkinson’s disease and multiple system atrophy. Neurobiology of aging 40, 192 e191–192 e195. [DOI] [PubMed] [Google Scholar]
- Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, and Mann M (2014). Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Molecular & cellular proteomics : MCP 13, 2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuenda A, and Rousseau S (2007). p38 MAP-kinases pathway regulation, function and role in human diseases. Biochimica et biophysica acta 1773, 1358–1375. [DOI] [PubMed] [Google Scholar]
- De La Mota-Peynado A, Lee SY, Pierce BM, Wani P, Singh CR, and Roelofs J (2013). The proteasome-associated protein Ecm29 inhibits proteasomal ATPase activity and in vivo protein degradation by the proteasome. The Journal of biological chemistry 288, 29467–29481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vrij FM, Fischer DF, van Leeuwen FW, and Hol EM (2004). Protein quality control in Alzheimer’s disease by the ubiquitin proteasome system. Prog Neurobiol 74, 249–270. [DOI] [PubMed] [Google Scholar]
- Di Fonzo A, Dekker MC, Montagna P, Baruzzi A, Yonova EH, Correia Guedes L, Szczerbinska A, Zhao T, Dubbel-Hulsman LO, Wouters CH, et al. (2009). FBXO7 mutations cause autosomal recessive, early-onset parkinsonian-pyramidal syndrome. Neurology 72, 240–245. [DOI] [PubMed] [Google Scholar]
- DiAntonio A, Haghighi AP, Portman SL, Lee JD, Amaranto AM, and Goodman S (2001). Ubiquitination-dependent mechanisms regulate synaptic growth and function. Nature 412, 449–452. [DOI] [PubMed] [Google Scholar]
- Dikic I, and Elazar Z (2018). Mechanism and medical implications of mammalian autophagy. Nature reviews Molecular cell biology 19, 349–364. [DOI] [PubMed] [Google Scholar]
- Ding M, Chao D, Wang G, and Shen K (2007). Spatial regulation of an E3 ubiquitin ligase directs selective synapse elimination. Science 317, 947–951. [DOI] [PubMed] [Google Scholar]
- Djakovic SN, Marquez-Lona EM, Jakawich SK, Wright R, Chu C, Sutton MA, and Patrick GN (2012). Phosphorylation of Rpt6 regulates synaptic strength in hippocampal neurons. The Journal of neuroscience : the official journal of the Society for Neuroscience 32, 5126–5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers MD (2003). Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nature neuroscience 6, 231–242. [DOI] [PubMed] [Google Scholar]
- Erturk A, Wang Y, and Sheng M (2014). Local pruning of dendrites and spines by caspase-3-dependent and proteasome-limited mechanisms. The Journal of neuroscience : the official journal of the Society for Neuroscience 34, 1672–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D (2009). Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annual review of biochemistry 78, 477–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D, Chen X, and Walters KJ (2016). Gates, Channels, and Switches: Elements of the Proteasome Machine. Trends in biochemical sciences 41, 77–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron TF, and Petrucelli L (2009). The role of tau in neurodegeneration. Molecular neurodegeneration 4, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman MH, and Ciechanover A (2002). The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiological reviews 82, 373–428. [DOI] [PubMed] [Google Scholar]
- Goldberg AL (2003). Protein degradation and protection against misfolded or damaged proteins. Nature 426, 895–899. [DOI] [PubMed] [Google Scholar]
- Gorbea C, Pratt G, Ustrell V, Bell R, Sahasrabudhe S, Hughes RE, and Rechsteiner M (2010). A protein interaction network for Ecm29 links the 26 S proteasome to molecular motors and endosomal components. The Journal of biological chemistry 285, 31616–31633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM, and O’Connor-Giles KM (2014). Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196, 961–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AM, and Zito K (2013). Breaking it down: the ubiquitin proteasome system in neuronal morphogenesis. Neural Plast 2013, 196848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee JD, Bibbs L, and Ulevitch RJ (1994). A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cells. Science 265, 808–811. [DOI] [PubMed] [Google Scholar]
- Hegde AN, Haynes KA, Bach SV, and Beckelman BC (2014). Local ubiquitin-proteasome-mediated proteolysis and long-term synaptic plasticity. Frontiers in molecular neuroscience 7, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heigwer F, Kerr G, and Boutros M (2014). E-CRISP: fast CRISPR target site identification. Nature methods 11, 122–123. [DOI] [PubMed] [Google Scholar]
- Hershko A, and Ciechanover A (1998). The ubiquitin system. Annual review of biochemistry 67, 425–479. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Niwa S, and Tanaka Y (2010). Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron 68, 610–638. [DOI] [PubMed] [Google Scholar]
- Hoover BR, Reed MN, Su J, Penrod RD, Kotilinek LA, Grant MK, Pitstick R, Carlson GA, Lanier LM, Yuan LL, et al. (2010). Tau mislocalization to dendritic spines mediates synaptic dysfunction independently of neurodegeneration. Neuron 68, 1067–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu MT, Guo CL, Liou AY, Chang TY, Ng MC, Florea BI, Overkleeft HS, Wu YL, Liao JC, and Cheng PL (2015). Stage-Dependent Axon Transport of Proteasomes Contributes to Axon Development. Developmental cell 35, 418–431. [DOI] [PubMed] [Google Scholar]
- Irvine GB, El-Agnaf OM, Shankar GM, and Walsh DM (2008). Protein aggregation in the brain: the molecular basis for Alzheimer’s and Parkinson’s diseases. Mol Med 14, 451–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ittner LM, Ke YD, Delerue F, Bi M, Gladbach A, van Eersel J, Wolfing H, Chieng BC, Christie MJ, Napier IA., et al. (2010). Dendritic function of tau mediates amyloid-beta toxicity in Alzheimer’s disease mouse models. Cell 142, 387–397. [DOI] [PubMed] [Google Scholar]
- Jan LY, and Jan YN (1982). Antibodies to horseradish peroxidase as specific neuronal markers in Drosophila and in grasshopper embryos. Proceedings of the National Academy of Sciences of the United States of America 79, 2700–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JO, Mandrioli J, Benatar M, Abramzon Y, Van Deerlin VM, Trojanowski JQ, Gibbs JR, Brunetti M, Gronka S, Wuu J, et al. (2010). Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron 68, 857–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kall L, Canterbury JD, Weston J, Noble WS, and MacCoss MJ (2007). Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nature methods 4, 923–925. [DOI] [PubMed] [Google Scholar]
- Katsuragi Y, Ichimura Y, and Komatsu M (2015). p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. The FEBS journal 282, 4672–4678. [DOI] [PubMed] [Google Scholar]
- Kipreos ET, and Pagano M (2000). The F-box protein family. Genome biology 1, REVIEWS3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk R, Laman H, Knowles PP, Murray-Rust J, Lomonosov M, Meziane el K, and McDonald NQ (2008). Structure of a conserved dimerization domain within the F-box protein Fbxo7 and the PI31 proteasome inhibitor. The Journal of biological chemistry 283, 22325–22335. [DOI] [PubMed] [Google Scholar]
- Kittel RJ, Wichmann C, Rasse TM, Fouquet W, Schmidt M, Schmid A, Wagh DA, Pawlu C, Kellner RR, Willig KI, et al. (2006). Bruchpilot promotes active zone assembly, Ca2+ channel clustering, and vesicle release. Science 312, 1051–1054. [DOI] [PubMed] [Google Scholar]
- Koppers M, van Blitterswijk MM, Vlam L, Rowicka PA, van Vught PW, Groen EJ, Spliet WG, Engelen-Lee J, Schelhaas HJ, de Visser M , et al. (2012). VCP mutations in familial and sporadic amyotrophic lateral sclerosis. Neurobiology of aging 33, 837 e837–813. [DOI] [PubMed] [Google Scholar]
- Koushik SV, Chen H, Thaler C, Puhl HL 3rd, and Vogel SS (2006). Cerulean, Venus, and VenusY67C FRET reference standards. Biophysical journal 91, L99–L101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreko-Pierce T, and Eaton BA (2017). The Drosophila LC8 homolog cut up specifies the axonal transport of proteasomes. Journal of cell science 130, 3388–3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CT, Jan LY, and Jan YN (2005). Dendrite-specific remodeling of Drosophila sensory neurons requires matrix metalloproteases, ubiquitin-proteasome, and ecdysone signaling. Proceedings of the National Academy of Sciences of the United States of America 102, 15230–15235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbadia J, and Morimoto RI (2015). The biology of proteostasis in aging and disease. Annual review of biochemistry 84, 435–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecker SH, Solomon V, Mitch WE, and Goldberg AL (1999). Muscle protein breakdown and the critical role of the ubiquitin-proteasome pathway in normal and disease states. The Journal of nutrition 129, 227S–237S. [DOI] [PubMed] [Google Scholar]
- Lee SH, Choi JH, Lee N, Lee HR, Kim JI, Yu NK, Choi SL, Lee SH, Kim H, and Kaang BK (2008). Synaptic protein degradation underlies destabilization of retrieved fear memory. Science 319, 1253–1256. [DOI] [PubMed] [Google Scholar]
- Lee SY, De la Mota-Peynado A, and Roelofs J (2011). Loss of Rpt5 protein interactions with the core particle and Nas2 protein causes the formation of faulty proteasomes that are inhibited by Ecm29 protein. The Journal of biological chemistry 286, 36641–36651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, Walz T, Ploegh H, and Finley D (2002). Multiple associated proteins regulate proteasome structure and function. Molecular cell 10, 495–507. [DOI] [PubMed] [Google Scholar]
- Lehmann A, Niewienda A, Jechow K, Janek K, and Enenkel C (2010). Ecm29 fulfils quality control functions in proteasome assembly. Molecular cell 38, 879–888. [DOI] [PubMed] [Google Scholar]
- Levine B, and Kroemer G (2008). Autophagy in the pathogenesis of disease. Cell 132, 27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Thompson D, Kumar B, and DeMartino GN (2014). Molecular and cellular roles of PI31 (PSMF1) protein in regulation of proteasome function. The Journal of biological chemistry 289, 17392–17405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XJ, and Li S (2011). Proteasomal dysfunction in aging and Huntington disease. Neurobiology of disease 43, 4–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CW, Li X, Thompson D, Wooding K, Chang TL, Tang Z, Yu H, Thomas PJ, and DeMartino GN (2006). ATP binding and ATP hydrolysis play distinct roles in the function of 26S proteasome. Molecular cell 24, 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrus SB, Portman SL, Allen MJ, Moffat KG, and DiAntonio A (2004). Differential localization of glutamate receptor subunits at the Drosophila neuromuscular junction. The Journal of neuroscience : the official journal of the Society for Neuroscience 24, 1406–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutchen-Maloney SL, Matsuda K, Shimbara N, Binns DD, Tanaka K, Slaughter CA, and DeMartino GN (2000). cDNA cloning, expression, and functional characterization of PI31, a proline-rich inhibitor of the proteasome. The Journal of biological chemistry 275, 18557–18565. [DOI] [PubMed] [Google Scholar]
- Mori H, Kondo J, and Ihara Y (1987). Ubiquitin is a component of paired helical filaments in Alzheimer’s disease. Science 235, 1641–1644. [DOI] [PubMed] [Google Scholar]
- Morimoto RI (2008). Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes & development 22, 1427–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishima-Kawashima M, Hasegawa M, Takio K, Suzuki M, Titani K, and Ihara Y (1993). Ubiquitin is conjugated with amino-terminally processed tau in paired helical filaments. Neuron 10, 1151–1160. [DOI] [PubMed] [Google Scholar]
- Moscat J, and Diaz-Meco MT (2009). p62 at the crossroads of autophagy, apoptosis, and cancer. Cell 137, 1001–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata S, Yashiroda H, and Tanaka K (2009). Molecular mechanisms of proteasome assembly. Nature reviews Molecular cell biology 10, 104–115. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Suzuki K, Kamada Y, and Ohsumi Y (2009). Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nature reviews Molecular cell biology 10, 458–467. [DOI] [PubMed] [Google Scholar]
- Nezis IP, Simonsen A, Sagona AP, Finley K, Gaumer S, Contamine D, Rusten TE, Stenmark H, and Brech A (2008). Ref(2)P, the Drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain. The Journal of cell biology 180, 1065–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oddo S (2008). The ubiquitin-proteasome system in Alzheimer’s disease. J Cell Mol Med 12, 363–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olenych SG, Claxton NS, Ottenberg GK, and Davidson MW (2007). The fluorescent protein color palette. Current protocols in cell biology Chapter 21, Unit 21 25. [DOI] [PubMed] [Google Scholar]
- Otero MG, Alloatti M, Cromberg LE, Almenar-Queralt A, Encalada SE, Pozo Devoto VM, Bruno L, Goldstein LS, and Falzone TL (2014). Fast axonal transport of the proteasome complex depends on membrane interaction and molecular motor function. Journal of cell science 127, 1537–1549. [DOI] [PubMed] [Google Scholar]
- Owald D, and Sigrist SJ (2009). Assembling the presynaptic active zone. Current opinion in neurobiology 19, 311–318. [DOI] [PubMed] [Google Scholar]
- Paisan-Ruiz C, Guevara R, Federoff M, Hanagasi H, Sina F, Elahi E, Schneider SA, Schwingenschuh P, Bajaj N, Emre M, et al. (2010). Early-onset L-dopa-responsive parkinsonism with pyramidal signs due to ATP13A2, PLA2G6, FBXO7 and spatacsin mutations. Movement disorders : official journal of the Movement Disorder Society 25, 1791–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak DT, and Sheng M (2003). Targeted protein degradation and synapse remodeling by an inducible protein kinase. Science 302, 1368–1373. [DOI] [PubMed] [Google Scholar]
- Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, Schwartz SL, DiProspero NA, Knight MA, Schuldiner O, et al. (2007). HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature 447, 859–863. [DOI] [PubMed] [Google Scholar]
- Park S, Kim W, Tian G, Gygi SP, and Finley D (2011). Structural defects in the regulatory particle-core particle interface of the proteasome induce a novel proteasome stress response. The Journal of biological chemistry 286, 36652–36666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick GN (2006). Synapse formation and plasticity: recent insights from the perspective of the ubiquitin proteasome system. Current opinion in neurobiology 16, 90–94. [DOI] [PubMed] [Google Scholar]
- Perry G, Friedman R, Shaw G, and Chau V (1987). Ubiquitin is detected in neurofibrillary tangles and senile plaque neurites of Alzheimer disease brains. Proceedings of the National Academy of Sciences of the United States of America 84, 3033–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BD, Jenett A, Hammonds AS, Ngo TT, Misra S, Murphy C, Scully A, Carlson JW, Wan KH, Laverty TR, et al. (2008). Tools for neuroanatomy and neurogenetics in Drosophila. Proceedings of the National Academy of Sciences of the United States of America 105, 9715–9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BD, Ngo TT, Hibbard KL, Murphy C, Jenett A, Truman JW, and Rubin GM (2010). Refinement of tools for targeted gene expression in Drosophila. Genetics 186, 735–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port F, Chen HM, Lee T, and Bullock SL (2014). Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proceedings of the National Academy of Sciences of the United States of America 111, E2967–2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian MX, Pang Y, Liu CH, Haratake K, Du BY, Ji DY, Wang GF, Zhu QQ, Song W, Yu Y, et al. (2013). Acetylation-mediated proteasomal degradation of core histones during DNA repair and spermatogenesis. Cell 153, 1012–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran KV, Fu JM, Schaffer TB, Na CH, Delannoy M, and Margolis SS (2018). Activity-Dependent Degradation of the Nascentome by the Neuronal Membrane Proteasome. Molecular cell 71, 169–177 e166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran KV, and Margolis SS (2017). A mammalian nervous-system-specific plasma membrane proteasome complex that modulates neuronal function. Nature structural & molecular biology 24, 419–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Mann M, and Ishihama Y (2007). Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nature protocols 2, 1896–1906. [DOI] [PubMed] [Google Scholar]
- Reck-Peterson SL, Redwine WB, Vale RD, and Carter AP (2018). The cytoplasmic dynein transport machinery and its many cargoes. Nature reviews Molecular cell biology 19, 382–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Sun J, Housden BE, Hu Y, Roesel C, Lin S, Liu LP, Yang Z, Mao D, Sun L, et al. (2013). Optimized gene editing technology for Drosophila melanogaster using germ line-specific Cas9. Proceedings of the National Academy of Sciences of the United States of America 110, 19012–19017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CA, and Poirier MA (2004). Protein aggregation and neurodegenerative disease. Nature medicine 10 Suppl, S10–17. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC (2006). The roles of intracellular protein-degradation pathways in neurodegeneration. Nature 443, 780–786. [DOI] [PubMed] [Google Scholar]
- Sakurai K, Zhao S, Takatoh J, Rodriguez E, Lu J, Leavitt AD, Fu M, Han BX, and Wang F (2016). Capturing and Manipulating Activated Neuronal Ensembles with CANE Delineates a Hypothalamic Social-Fear Circuit. Neuron 92, 739–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandri M (2013). Protein breakdown in muscle wasting: role of autophagy-lysosome and ubiquitin-proteasome. The international journal of biochemistry & cell biology 45, 2121–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nature methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliwa M, and Woehlke G (2003). Molecular motors. Nature 422, 759–765. [DOI] [PubMed] [Google Scholar]
- Schmidt M, and Finley D (2014). Regulation of proteasome activity in health and disease. Biochimica et biophysica acta 1843, 13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenmann Z, Assa-Kunik E, Tiomny S, Minis A, Haklai-Topper L, Arama E, and Yaron A (2010). Axonal degeneration is regulated by the apoptotic machinery or a NAD+-sensitive pathway in insects and mammals. The Journal of neuroscience : the official journal of the Society for Neuroscience 30, 6375–6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, and Selbach M (2011). Global quantification of mammalian gene expression control. Nature 473, 337–342. [DOI] [PubMed] [Google Scholar]
- Sherva R, Baldwin CT, Inzelberg R, Vardarajan B, Cupples LA, Lunetta K, Bowirrat A, Naj A, Pericak-Vance M, Friedland RP, et al. (2011). Identification of novel candidate genes for Alzheimer’s disease by autozygosity mapping using genome wide SNP data. Journal of Alzheimer’s disease : JAD 23, 349–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon DJ, Weimer RM, McLaughlin T, Kallop D, Stanger K, Yang J, O’Leary DD, Hannoush RN, and Tessier-Lavigne M (2012). A caspase cascade regulating developmental axon degeneration. The Journal of neuroscience : the official journal of the Society for Neuroscience 32, 17540–17553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaar JR, Pagan JK, and Pagano M (2013). Mechanisms and function of substrate recruitment by F-box proteins. Nat Rev Mol Cell Biol 14, 369–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra D, Craig KL, Tyers M, Elledge SJ, and Harper JW (1997). F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91, 209–219. [DOI] [PubMed] [Google Scholar]
- Speese SD, Trotta N, Rodesch CK, Aravamudan B, and Broadie K (2003). The ubiquitin proteasome system acutely regulates presynaptic protein turnover and synaptic efficacy. Current biology : CB 13, 899–910. [DOI] [PubMed] [Google Scholar]
- Sun B, Xu P, and Salvaterra PM (1999). Dynamic visualization of nervous system in live Drosophila. Proceedings of the National Academy of Sciences of the United States of America 96, 10438–10443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai HC, and Schuman EM (2008). Ubiquitin, the proteasome and protein degradation in neuronal function and dysfunction. Nature reviews Neuroscience 9, 826–838. [DOI] [PubMed] [Google Scholar]
- Taus T, Kocher T, Pichler P, Paschke C, Schmidt A, Henrich C, and Mechtler K (2011). Universal and confident phosphorylation site localization using phosphoRS. Journal of proteome research 10, 5354–5362. [DOI] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, and Cox J (2016). The Perseus computational platform for comprehensive analysis of (prote)omics data. Nature methods 13, 731–740. [DOI] [PubMed] [Google Scholar]
- Valakh V, Naylor SA, Berns DS, and DiAntonio A (2012). A large-scale RNAi screen identifies functional classes of genes shaping synaptic development and maintenance. Developmental biology 366, 163–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD (2003). The molecular motor toolbox for intracellular transport. Cell 112, 467–480. [DOI] [PubMed] [Google Scholar]
- Varshavsky A (2005). Regulated protein degradation. Trends in biochemical sciences 30, 283–286. [DOI] [PubMed] [Google Scholar]
- Varshavsky A (2012). The ubiquitin system, an immense realm. Annual review of biochemistry 81, 167–176. [DOI] [PubMed] [Google Scholar]
- Verdoes M, Florea BI, Menendez-Benito V, Maynard CJ, Witte MD, van der Linden WA, van den Nieuwendijk AM, Hofmann T, Berkers CR, van Leeuwen FW, et al. (2006). A fluorescent broad-spectrum proteasome inhibitor for labeling proteasomes in vitro and in vivo. Chemistry & biology 13, 1217–1226. [DOI] [PubMed] [Google Scholar]
- Vilchez D, Saez I, and Dillin A (2014). The role of protein clearance mechanisms in organismal ageing and age-related diseases. Nature communications 5, 5659. [DOI] [PubMed] [Google Scholar]
- Vingill S, Brockelt D, Lancelin C, Tatenhorst L, Dontcheva G, Preisinger C, Schwedhelm-Domeyer N, Joseph S, Mitkovski M, Goebbels S, et al. (2016). Loss of FBXO7 (PARK15) results in reduced proteasome activity and models a parkinsonism-like phenotype in mice. The EMBO journal 35, 2008–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagh DA, Rasse TM, Asan E, Hofbauer A, Schwenkert I, Durrbeck H, Buchner S, Dabauvalle MC, Schmidt M, Qin G, et al. (2006). Bruchpilot, a protein with homology to ELKS/CAST, is required for structural integrity and function of synaptic active zones in Drosophila. Neuron 49, 833–844. [DOI] [PubMed] [Google Scholar]
- Wagih O, Sugiyama N, Ishihama Y, and Beltrao P (2016). Uncovering Phosphorylation-Based Specificities through Functional Interaction Networks. Molecular & cellular proteomics : MCP 15, 236–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan HI, DiAntonio A, Fetter RD, Bergstrom K, Strauss R, and Goodman CS (2000). Highwire regulates synaptic growth in Drosophila. Neuron 26, 313–329. [DOI] [PubMed] [Google Scholar]
- Wang X, Yen J, Kaiser P, and Huang L (2010). Regulation of the 26S proteasome complex during oxidative stress. Science signaling 3, ra88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts RJ, Hoopfer ED, and Luo L (2003). Axon pruning during Drosophila metamorphosis: evidence for local degeneration and requirement of the ubiquitin-proteasome system. Neuron 38, 871–885. [DOI] [PubMed] [Google Scholar]
- Willeumier K, Pulst SM, and Schweizer FE (2006). Proteasome inhibition triggers activity-dependent increase in the size of the recycling vesicle pool in cultured hippocampal neurons. The Journal of neuroscience : the official journal of the Society for Neuroscience 26, 11333–11341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff S, Weissman JS, and Dillin A (2014). Differential scales of protein quality control. Cell 157, 52–64. [DOI] [PubMed] [Google Scholar]
- Wu JS, and Luo L (2006). A protocol for mosaic analysis with a repressible cell marker (MARCM) in Drosophila. Nature protocols 1, 2583–2589. [DOI] [PubMed] [Google Scholar]
- Yang BJ, Han XX, Yin LL, Xing MQ, Xu ZH, and Xue HW (2016). Arabidopsis PROTEASOME REGULATOR1 is required for auxin-mediated suppression of proteasome activity and regulates auxin signalling. Nat Commun 7, 11388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashiroda H, Toda Y, Otsu S, Takagi K, Mizushima T, and Murata S (2015). N-terminal alpha7 deletion of the proteasome 20S core particle substitutes for yeast PI31 function. Molecular and cellular biology 35, 141–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi JJ, and Ehlers MD (2005). Ubiquitin and protein turnover in synapse function. Neuron 47, 629–632. [DOI] [PubMed] [Google Scholar]
- Yoshiyama Y, Higuchi M, Zhang B, Huang SM, Iwata N, Saido TC, Maeda J, Suhara T, Trojanowski JQ, and Lee VM (2007). Synapse loss and microglial activation precede tangles in a P301S tauopathy mouse model. Neuron 53, 337–351. [DOI] [PubMed] [Google Scholar]
- Yu W, and Lu B (2012). Synapses and dendritic spines as pathogenic targets in Alzheimer’s disease. Neural Plast 2012, 247150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaiss DM, Standera S, Holzhutter H, Kloetzel P, and Sijts AJ (1999). The proteasome inhibitor PI31 competes with PA28 for binding to 20S proteasomes. FEBS letters 457, 333–338. [DOI] [PubMed] [Google Scholar]
- Zarubin T, and Han J (2005). Activation and signaling of the p38 MAP kinase pathway. Cell research 15, 11–18. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Hegde AN, and Martin KC (2003). The ubiquitin proteasome system functions as an inhibitory constraint on synaptic strengthening. Current biology : CB 13, 887–898. [DOI] [PubMed] [Google Scholar]
- Zhong L, and Belote JM (2007). The testis-specific proteasome subunit Prosalpha6T of D. melanogaster is required for individualization and nuclear maturation during spermatogenesis. Development 134, 3517–3525. [DOI] [PubMed] [Google Scholar]
- Zhu G, Fujii K, Belkina N, Liu Y, James M, Herrero J, and Shaw S (2005). Exceptional disfavor for proline at the P + 1 position among AGC and CAMK kinases establishes reciprocal specificity between them and the proline-directed kinases. The Journal of biological chemistry 280, 10743–10748. [DOI] [PubMed] [Google Scholar]
- Zoghbi HY, and Orr HT (2000). Glutamine repeats and neurodegeneration. Annual review of neuroscience 23, 217–247. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S2. Raw data for the MS analysis of the anti-HA-PI31 co-IP experiments. Related to Figure 2.
Table S3. Raw data for the MS analysis of the anti-HA-PI31S168A co-IP versus anti-HA-PI31wt co-IP experiments. Related to Figure 3.
Table S4. Raw data for the MS analysis of the anti-FLAG-dDYNLL1 co-IP experiments. Related to Figure 4.
Table S5. Summary for the MS analysis of the kinase screen experiments. Related to Figure 7.
Movie S1. Representative video of Prosβ5-RFP particle movement in an axon of wild-type larva (w; +/+; R94G06-GAL4, UAS-Prosβ5-RFP/R94G06-GAL4, UAS-Prosβ5-RFP). Related to Figure 5. The video was acquired at 1.4Hz rate for 70 seconds (100 frames in total), and is shown in 10X of the original speed.
Movie S2. Representative video of Prosβ5-RFP particle movement in an axon of PI31−/− larva (w; PI31Δ/PI31Δ; R94G06-GAL4, UAS-Prosβ5-RFP/R94G06-GAL4, UAS-Prosβ5-RFP). Related to Figure 5. The video was acquired at 1.4Hz rate for 70 seconds (100 frames in total), and is shown in 10X of the original speed.
Movie S3. Representative video of Prosβ5-RFP particle movement in an axon of PI31S168Aknock-in larva (w; PI31S168A / PI31S168A; R94G06-GAL4, UAS-Prosβ5-RFP/R94G06-GAL4, UAS-Prosβ5-RFP). Related to Figure 5. The video was acquired at 1.4Hz rate for 70 seconds (100 frames in total), and is shown in 10X of the original speed.
Movie S4. Representative video of mCerulean-mPI31 (green) and α4-mCherry (red) movement in axons of cultured mouse DRG neurons. Related to Figure 5. The video was acquired at 0.5Hz rate for 140 seconds (70 frames in total), and is shown in 10X of the original speed.
Movie S5. Representative video of α4-mScarlet-I particle movement in an axon of control hippocampal neuron (PI31fl/fl, hsyn-GFP). Related to Figure 6. The video was acquired at 1.6Hz rate for 60 seconds (100 frames in total), and is shown in 10X of the original speed.
Movie S6. Representative video of α4-mScarlet-I particle movement in an axon of PI31 knockout hippocampal neuron (PI31fl/fl, hsyn-Cre). Related to Figure 6. The video was acquired at 1.6Hz rate for 60 seconds (100 frames in total), and is shown in 10X of the original speed.
Data Availability Statement
Raw data of all mass spectrometry analyses have been shown in Supplementary Tables 2–5.