Abstract
ADP-ribosylation—the addition of one or multiple ADP-ribose units onto proteins—is a therapeutically important post-translational modification implicated in cancer, neurodegeneration, and infectious diseases. The protein modification regulates a broad range of biological processes, including DNA repair, transcription, RNA metabolism, and the structural integrity of nonmembranous structures. The polymeric form of ADP-ribose, poly(ADP-ribose), was recently identified as a signal for triggering protein degradation through the ubiquitin-proteasome system. Using informatics analyses, we found that these ubiquitinated substrates tend to be low abundance proteins, which may serve as rate-limiting factors within signaling networks or metabolic processes. In this review, we summarize the current literature on poly(ADP-ribose)-dependent ubiquitination (PARdU) regarding its biological mechanisms, substrates, and relevance to diseases.
Keywords: ADP-ribosylation, Poly(ADP-ribose), Poly(ADP-ribose)-dependent ubiquitination, Ubiquitin- Proteasome System, Protein degradation, Rate-limiting factors, Tankyrase
Graphical Abstract
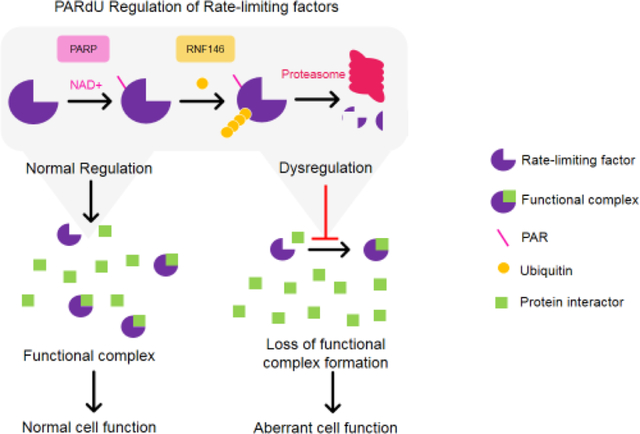
1. Introduction
Post-translational modification regulates protein function through the addition of polypeptides or chemical groups. Post-translational modification enables a relatively limited number of genes to generate a far greater number of proteoforms with differential functionalities, localizations and stabilities compared to the unmodified form [1]. Here we focus on a protein degradation process that requires two post-translational modifications: ADP-ribosylation and ubiquitination.
1.1. ADP-ribosylation
ADP-ribosylation refers to the covalent addition of one or more ADP-ribose (ADPr) onto proteins, whereby the ADPr units are transferred from NAD+ by ADP-ribosyltransferases, including a subset commonly known as Poly(ADP-ribose) Polymerases (PARPs) [2–7]. The modification exists as a monomeric [mono(ADP-ribosyl)ation or MARylation], or polymeric form [poly(ADP-ribosyl)ation or PARylation] (Figure 1). Amongst the 17 PARPs in humans, four synthesize poly(ADP-ribose) (PAR; PARPs 1, 2, 5a and 5b), two are catalytically dead (PARPs 9 and 13), and the rest synthesize mono(ADP-ribose) (MAR) based on automodification analyses [2]. While PARP5a only synthesizes linear PAR chains [3], the founding member PARP1 can also synthesize branched forms of poly(ADP-ribose) [4], further increasing the structural complexity of PARylation (Figure 1). PARylation has been implicated in a number of biological processes, including DNA damage-response [5], chromatin organization [6,7], liquid-liquid phase transition for the assembly of nonmembranous structures [8,9], as well as the focus of this review—protein degradation through ubiquitination.
Figure 1. ADP-ribosylation.
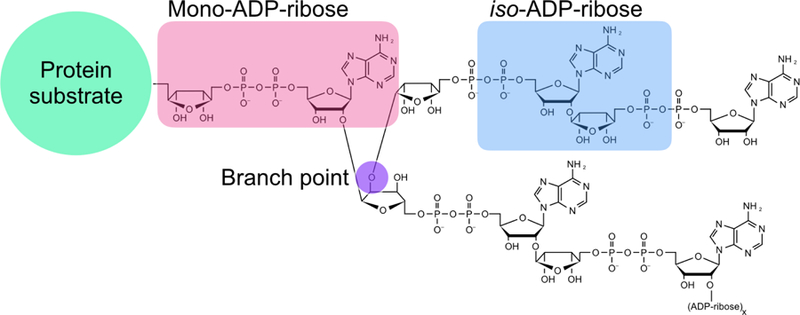
Proteins may be MARylated (pink) or PARylated. Branching of ADP- ribose can be synthesized by PARP1 and the branch point is shown in purple. The internal structure of poly(ADP-ribose) referred to as iso-ADP-ribose is highlighted in blue.
1.2. Ubiquitination
Ubiquitin is a small protein of 8.5 kDa that is covalently attached to substrates at lysine residues [10,11]. Ubiquitin itself can also be modified on any of its own seven lysines by another ubiquitin molecule, resulting in a variety of different ubiquitin chains with unique linkages and functions [11]. For example, poly-ubiquitin chains linked at the lysine-48 (K48) position are targeted for proteasomal degradation while K63-linked chains play signaling roles, for example, during DNA repair. The conjugation of ubiquitin to target proteins requires an enzymatic cascade of several proteins [12]. An ubiquitin is activated by the ubiquitin-activating enzyme E1 with ATP, then the activated ubiquitin is transferred to an ubiquitin-conjugating enzyme (E2) before finally conjugating to the protein targets through an E3 ubiquitin ligase, which confers substrate specificity [13]. There are three main types of E3 ligases, which differ in their domain architecture and the mechanism of ubiquitin transfer [14]. Both HECT (Homologous to the E6AP Carboxyl Terminus) and RBR (RING-between-RING) E3 ligases first transfer ubiquitin to a catalytic cysteine on the enzyme before transferring ubiquitin to its substrate [15, 16]. RING (Really Interesting New Gene) E3 ligases position the E2 and the target substrate for direct ubiquitin transfer [17,18]. This review focuses on a RING E3 ligase that possesses a domain that binds PAR to trigger ubiquitination, a process recently coined PAR-dependent ubiquitination (PARdU) [19]. Here we will take a historical perspective on its discovery, explore the underlying biological mechanisms, and highlight contemporary strategies for substrate identification. The review will conclude with a discussion of the open questions regarding the mechanism of PARdU, its disease relevance, and the potential therapeutic opportunities.
2. A brief history of PARdU
Current understandings of the mechanism of PARdU have been assembled from several independent studies (Figure 2). PARdU was first discovered through identifying the target of the Wnt signaling inhibitor XAV939, which inhibits PARP5a/b (commonly referred to as Tankyrase1/2) [20]. Inhibition of PARP5a/b increases the protein level of the rate-limiting factors within the Wnt signaling pathway—Axin1/2 [20]. Subsequent studies revealed that degradation of Axin1/2 was mediated by the E3 ligase RING finger protein 146 (RNF146), which is activated by binding of PAR [21–23]. The substrates of PARP5a/b, such as Axin1/2, possess an amino acid motif defined as the “Tankyrase binding motif” (TBM), which was later shown to be critical for PARdU [24–26]. Additional PARdU substrates, such as 3BP2 and PTEN, were then defined and characterized, with hundreds of candidate substrates identified in recent proteomics studies [21,26–29]. These studies provide the foundation to elucidate the mechanism of how PARylation leads to ubiquitination and protein degradation.
Figure 2.
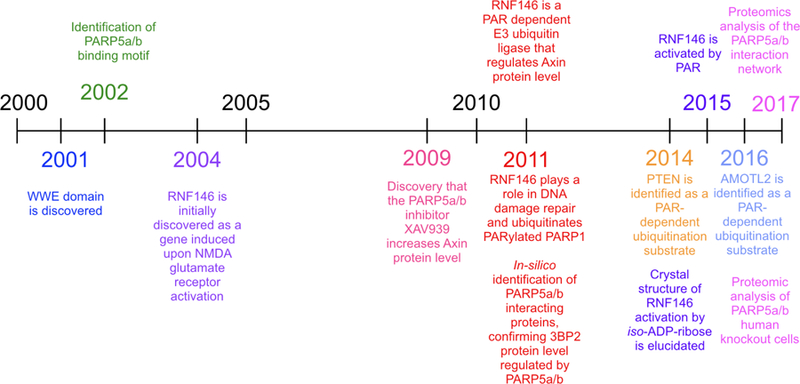
A timeline of PARdU discoveries.
2.1. Identification of the WWE domain and PARP5a/b binding motif
In 2001, a PSI-BLAST search to investigate an 80 amino acid-stretch in the notch-signaling protein Deltex revealed a novel domain called WWE, named after its most conserved residues [30]. The WWE domain is found among several RING and HECT E3 ubiquitin ligases as well as five PARPs, leading to the authors’ prediction that both enzyme classes share similar targets [30]. Subsequent biochemical studies revealed that the E3 ligase RNF146 WWE domain binds to iso-ADP-ribose (iso-ADPr)—the shortest internal unit of PAR—with ~370 nM affinity (Figure 1; Figure 3A). The binding to PAR by RNF146 via the WWE domain later proved to be critical to activate the E3 ubiquitin ligase activity.
Figure 3. Structures of the RNF146-iso-ADPr complex and ankyrin repeat clusters (ARCs) of PARP5a binding to Tankyrase binding motif (TBM) of RNF146.
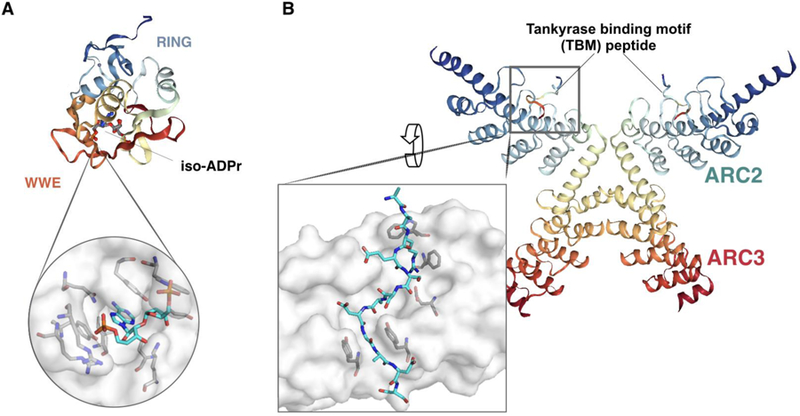
(A) Cartoon model of the RNF146- iso-ADPr complex highlighting the RING finger and WWE domain (PDB ID: 4QPL), with a close-up of the RNF146-iso-ADPr binding interface. (B) Cartoon model of the complex between RNF146 TBM and PARP5a (PDB ID: 6CF6), with a close-up of the TBM binding interface at an ARC of PARP5a. Structures were generated via Protein Data Bank and PyMOL.
Around the same time in the early 2000s, an investigation of PARP5a/b binding to the protein IRAP revealed that IRAP binds an ankyrin repeat cluster (ARC), a conserved domain within PARP5a/b, through the specific hexapeptide motif: RQSPDG [24,25]. Mutagenesis studies of the known IRAP interacting motif revealed that the Q and S residues are less important for binding and that the RXXPDG motif is sufficient for a functional TBM. This notion was further confirmed in two other PARP5a/b binding proteins, TRF-1 and TAB182 [25]. Using a peptide library screen, Guettler and colleagues further defined an 8-residue consensus R-X-X-[small hydrophobic or G]-[D/E]-[G]-[no P]-[D/E] that binds to ARCs, resulting in a ranked list of thousands of proteins that match to this peptide motif [26]. In combination with structural studies of the PARP5a/b ARCs binding to peptides [31–33] (e.g., Figure 3B), models to predict PARP5a/b interactors were further improved [34], enabling the prediction of additional PARdU candidates.
2.2. Discovery of PARdU while characterizing the effect of PARP5a/b inhibitor in Wnt signaling
Fundamental to the discovery of PARdU was the identification of the PARP5a/b inhibitor XAV939 through a high-throughput screen of inhibitors of the Wnt/β-catenin signaling pathway [20]. XAV939 treatment increased the protein levels of the β-catenin destruction complex component Axin1/2 in cells. Using an immobilized XAV939 analog, an affinity-capture technique was used to identify molecular targets that bind to this small molecule from cell extracts. Several PARPs bound to XAV939, although with varying affinities: PARP1 (Kd = 1.2 μM), PARP5a (Kd = 0.099 μM) and PARP5b (Kd = 0.093 μM). Additionally, knockdown of PARP5a/b, but not PARP1, led to an increase in Axin1/2 protein level similar to XAV939 treatment. The use of IWR-1—a previously published inhibitor of the Wnt signaling pathway shown to increase Axin1/2 levels—also inhibited PARP5a/b activity, further supporting that the catalytic activity of PARP5a/b is critical for regulating Axin protein level [22,35]. Using Axin1 as an example, Huang and colleagues further showed that Axin1 interacted with PARP5a/b through a conserved TBM [20], and its disruption by mutagenesis resulted in high Axin1 protein levels. These data suggest that Axin1/2 regulation by PARP5a/b requires both PARylation activity and physical interactions. In addition, Axin1/2 regulation by PARP5a/b was identified to be mediated via the ubiquitin-proteasome system; treatment with the proteasome inhibitor MG132 increased levels of polyubiquitinated Axin1/2 [20,22]. The authors of these studies concluded that XAV939 may stabilize Axin1/2 by preventing its ubiquitination, consistent with later findings that PARylation of Axin1/2 leads to its ubiquitination and subsequent degradation [20–23]. However, it would take two years before the link between PAR and ubiquitination was uncovered through the discovery of the first known PAR-dependent E3 ubiquitin ligase, RNF146 [17–19,32].
2.3. RNF146 is the missing link of PARdU regulation of Axin1/2
RNF146 (also known as Iduna) was initially discovered as a gene induced in response to the N-methyl-D-aspartate (NMDA) glutamate receptor [36]. Later work revealed that RNF146 possesses two highly conserved domains: the WWE and RING finger domains [21,37–39]. Overexpression of RNF146 in neurons protected against NMDA-induced cell death caused by PARP1 activation and subsequent PAR synthesis [40–42]. In 2011, several groups independently identified RNF146 as the E3 ubiquitin ligase that ubiquitinates Axin1/2 [21–23]. Specifically, the binding of RNF146 to Axin1/2 and subsequent regulation of Axin1/2 protein levels was dependent upon the WWE domain of RNF146 [21]. Importantly, the interaction between RNF146 and Axin1/2 is abolished upon PARP5a/b inhibitor treatment and Axin1/2 PARylation in cells is reduced upon XAV939 treatment [21,22]. These findings suggest that (1) RNF146 regulation of Axin1/2 is mediated through PAR-binding ability of WWE, and (2) Axin1/2 PARylation is PARP5a/b-dependent, thereby linking PARP5a/b activity and PAR-binding ability of RNF146 in this novel PARdU pathway. A yeast two-hybrid screen for other PARP5a/b substrates in one of these pioneering studies further revealed two TBM-containing proteins (BLZF1 and MLN51) as potential PARdU candidates [21]. The protein levels of GFP-tagged BLZF1 and MLN51 proteins were increased upon PARP5a/b inhibitor treatment as well as knockdown of PARP5a/b or RNF146. Both GFP- tagged BLZ1 and MLN51 require a functional TBM for an increase in protein level to be observed upon these chemical or genetic perturbations, further suggesting that these proteins are likely PARdU substrates. Yet, it is unclear whether these two proteins are endogenous substrates of PARdU.
The PAR-binding ability of RNF146 and its roles in ubiquitination have also been explored in the context of DNA damage. RNF146 binds to many DNA damage-associated proteins, such as PARP1, XRCC6, DNA Polymerase III and KU70 in a PAR-dependent manner [38]. An in vitro ubiquitination assay showed that PAR-binding to RNF146 activates the ubiquitin ligase activity, and this PAR-dependent ligase activity protects cells from NMDA-induced cell death and DNA damage [38].
2.4. Structural Insights into PARdU
After the discovery was made that RNF146 requires PAR for its E3 ubiquitin ligase activity [21,23,38], efforts were focused on determining which structures of PAR were important for RNF146 activation. Biochemical analysis revealed that RNF146 does not bind to the ADPr monomer, but instead binds avidly to the smallest internal subunit of PAR, iso-ADPr (Figure 1) [37]. In particular, iso-ADPr binding is sufficient to activate E3 ubiquitin ligase activity [19]. X-ray crystallography studies showed that the adenine ring of iso-ADPr fits into the half-β-barrel pocket of the WWE domain structure, and the phosphate-ribose moieties on either side of the iso-ADPr structure make specific contacts with WWE domain residues [37], providing insights into why RNF146 binds iso-ADPr but not ADPr (Figure 3A). Without iso-ADPr or PAR, RNF146 is unable to transfer E2-conjugated ubiquitin to its lysine substrate [19]. RNF146 protein that contains a RING domain and a WWE domain binds iso-ADPr with ten-times higher affinity compared to the RNF146 WWE domain alone, suggesting a plausible role of the RING domain in iso-ADPr binding [19]. Comparisons between the apo-structure and iso-ADPr bound RNF146 revealed that the RING domain adopts a conformation with a more available E2 binding surface upon ligand binding [19].
Additional binding domains of RNF146 and PARP5a/b play a role in PARdU. The Sterile Alpha Motif (SAM) domain of PARP5a/b enables PARP5a/b polymerization and facilitates its localization to cytoplasmic signaling complexes enriched with Axin1/2 [43,44]. Disruption of PARP5a/b polymerization via mutation of the SAM domain prevents PARP5a/b regulation of canonical Wnt/β-catenin signaling, suggesting that PARP5a/b polymerization promotes its catalytic activity and PARdU [43,44]. Further structural studies of the PARP5a/b ARC domain indicate that certain ARCs function together to aid in substrate binding [31]. ARCs vary in rigidity and flexibility, serving as a versatile binding platform for diverse PARP5a/b interactors [31]. RNF146 possesses one canonical (section 2.1) and four non-canonical TBMs [45]. The non-canonical TBMs of RNF146 are “extended” with additional amino acids between the conserved Arg and residue in position 4 (small hydrophobic or Gly residue) [19] (Figure 3B). The strongest binding non-canonical motif (Motif I), which is the most phylogenetically conserved, binds to PARP5a at 5.8 μM similar to canonical TBMs, which are in the range of 0.3–20 μM [15,22,41]. Individual RNF146 TBMs bind weakly to PARP5a but together facilitate strong multivalent binding [19,45]. Thus, in addition to the individual enzymatic activities, PARdU is mediated through multiple binding interactions of PARP5a/b and RNF146, as well as their interactions with the substrates.
3. Characterized PARdU substrates
Three methods have been commonly used to characterize PARdU substrates (Table 1): (1) PARP5a/b inhibitors, such as XAV939 and IWR-1, have been used to show that PARP5a/b catalytic activity regulates the protein level of substrates; (2) the PARdU substrate interacts with PARP5a/b and/or RNF146; (3) the PARdU substrate increases following RNAi-mediated knockdown of PARP5a/b and/or RNF146. Conversely, PARP5a/b or RNF146 overexpression may decrease substrate levels. Although mass spectrometry methods for identifying ADP-ribosylated sites on PARdU substrates are available (reviewed in [46]), only one study (PTEN) so far has identified the site of ADP-ribosylation necessary for ubiquitination and subsequent degradation [47]. Additionally, an in vitro ubiquitination protocol by RNF146 has been established [38] and could potentially be used to determine which site of ADP-ribosylation is necessary for binding and ubiquitination by RNF146. Below we will highlight several characterized PARdU substrates and their biological roles:
Table 1.
A summary of methods to characterize PARdU substrates.
| Axin1 | Axin2 | 3BP2 | PTEN | AMOTL2 | PARP1 | PARP5a | RNF146 | |
|---|---|---|---|---|---|---|---|---|
| PARP5a/b inhibition | ● | ● | ● | ● | ● | ● | ||
| PARP5a/b knockdown/ overexpression | ● | ● | ● | ● | ||||
| RNF146 knockdown/ overexpression | ● | ● | ● | ● | ● | ● | ● | |
| In vitro ubiquitination assay | ● | ● | ● | ● | ||||
| Ubiquitination sites identified | ● | ● | ● | |||||
| ADP-ribosylation sites identified to be critical for PARdU | ● |
Axin1/2:
Axin1/2, the first well-defined substrate that was pivotal in uncovering PARdU [21–23,35], is integral to the β-catenin destruction complex and negatively regulates canonical Wnt signaling [48–50]. Wnt signaling plays a critical role in development by regulating cell proliferation, differentiation and survival [48–50]. Axin1/2 degradation by PARdU may prevent the formation of the β-catenin destruction complex, resulting in the constitutive activation of Wnt signaling and leading to aberrant cell proliferation observed in cancer [48–50]. Consistently, PARP5a/b and β-catenin are overexpressed in lung adenocarcinoma A549 cells, and PARP5a/b inhibition with inhibitor XAV939 reduces cell proliferation of the cell line [51]. In addition, the ubiquitin-specific protease USP25 antagonizes Wnt signaling by promoting deubiquitination and stabilization of PARP5a/b, and disruption of PARP5a interaction with USP25 destabilizes PARP5a/b, leading to Axin1/2 stabilization and subsequent attenuation of Wnt signaling [52]. However, PARdU regulation of Axin1/2 and Wnt signaling play diverse roles beyond cell proliferation. RNF146-mediated degradation of Axin1/2 is also required for proper embryonic development in Xenopus embryos, where depletion of RNF146 leads to downregulation of critical pattern-organization proteins [53].
3BP2:
Before 3BP2 was identified as possessing a TBM, it was known that mutations in the gene encoding for 3BP2 were the cause of the bone disease cherubism [54–56]. Further study found that the mutations in the 3BP2 gene are often in the region encoding the TBM [27]. When PARdU-regulated degradation of 3BP2 is disrupted, elevated 3BP2 protein levels persist, and osteoclast formation and function are promoted in vitro, consistent with the pathogenesis of cherubism [27]. 3BP2 has been further identified as an adaptor protein that plays a role in several signaling pathways important for osteoclast and immune-cell function, including the SRC, SYC, and VAV signaling pathways [27].
PTEN:
PTEN is a tumor suppressor, and knockdown of PARP5a/b has been found to stabilize PTEN, and slow cell proliferation [47]. Recent work by Li and colleagues have identified PTEN as a novel PARdU substrate that binds to both PARP5a (via a non-canonical RYQEDGFD motif) and RNF146, as shown through biochemical methods (Table 1) [47]. Importantly, the PARdU regulation was inhibited upon mutating ADP-ribosylated sites identified from recent proteomics studies, suggesting specific sites of ADP-ribosylation may signal for ubiquitination and subsequent degradation.
AMOT-family proteins:
After the initial observation that PARP5a/b inhibition or RNF146 knockdown perturbed the localization of proteins important for cellular tight junction integrity, a screen for interactors for both RNF146 and PARP5a/b identified the AMOT-family proteins, e.g., AMOTL2 [57]. AMOT proteins play a role in maintaining tight junctions and can affect the localization of other tight junction proteins if over-expressed. Therefore, controlling AMOT protein levels through PARdU is critical for preserving epithelial integrity [57]. Additionally, PARP5a/b inhibition suppresses the nuclear functions of the Hippo pathway effector and oncoprotein YAP through stabilization of AMOT-family proteins, suggesting PARdU of AMOT-family proteins may play a role in regulation of Hippo signaling [58].
PARP1:
PARP1, the founding PARP family member, was also identified as a PARdU substrate in one of the pioneering studies identifying RNF146 as a PAR-dependent E3 ubiquitin ligase [38]. PARP1 PARylation is required for RNF146-mediated ubiquitination of PARP1, and PAR binding ability of RNF146 is required to decrease PARP1 protein levels [38]. PARP1 plays a crucial role in DNA damage repair and participates in the PAR-dependent cell death pathway, parthanatos [40,59]. PARP1 is auto-modified following recognition of DNA damage [60], resulting in recruitment of DNA damage repair proteins to the sites of damage [61,62]. Thus, PARdU may control the right amount of PARP1 for effective DNA repair or remove PARP1 from DNA damage sites.
RNF146 and PARP5a/b:
Importantly, RNF146 and PARP5a/b are substrates of one another [21,38]. Therefore, chemical inhibition or genetic knockdown of one leads to the increase in protein levels of the other, which may have other downstream effects that should be considered.
Notably many of these characterized PARdU substrates are rate-limiting factors of signaling pathways. Rate-limiting factors are proteins whose intracellular abundances must be maintained at particular levels to ensure proper cellular function. For example, Axin1/2 is critical to canonical Wnt signaling and formation of the β-catenin destruction complex [63]. In another example, genetic mutation of the TBM stabilizes the 3BP2 protein, and it is no longer regulated by PARdU. Elevated and stabilized 3BP2 contributes to downstream hyperactivation of SRC, SYC, and VAV signaling pathways, resulting in the disease pathogenesis of cherubism [27]. In these examples, PARdU regulation seems to be necessary for maintaining the abundance of these proteins at a critical level. To test whether this pattern holds true for all characterized PARdU substrates, we examined four datasets that quantitated the copy number of individual proteins in human cell lines [64–67]. Intriguingly, most characterized PARdU substrates are at least 10-fold lower in abundance than the median copy number of all proteins and ≥100-fold lower than the mean copy number across the four different studies (Figure 4). Similarly, two previously identified PARdU candidates BLZF1 and MLN51 are also low in abundance [21]. One possibility is that the observed low abundance could be because the protein level is suppressed by PARdU. However, given that these characterized substrates do not increase by 10–100 fold upon knockdown of PARP5a/b or RNF146, other factors must be at play in selecting which cellular proteins are PARdU substrates. We postulate here that PARdU may regulate low-abundance proteins that are rate- or concentration-limiting factors in their respective biological processes. Notably, the level of PARP5a protein in the cell is also similar to the protein levels of other characterized PARdU substrates. Therefore, PARP5a/b may be a rate-limiting factor in PARdU, which, in turn, influences the abundance of many substrate proteins.
Figure 4. PARdU substrates are often low abundance proteins.
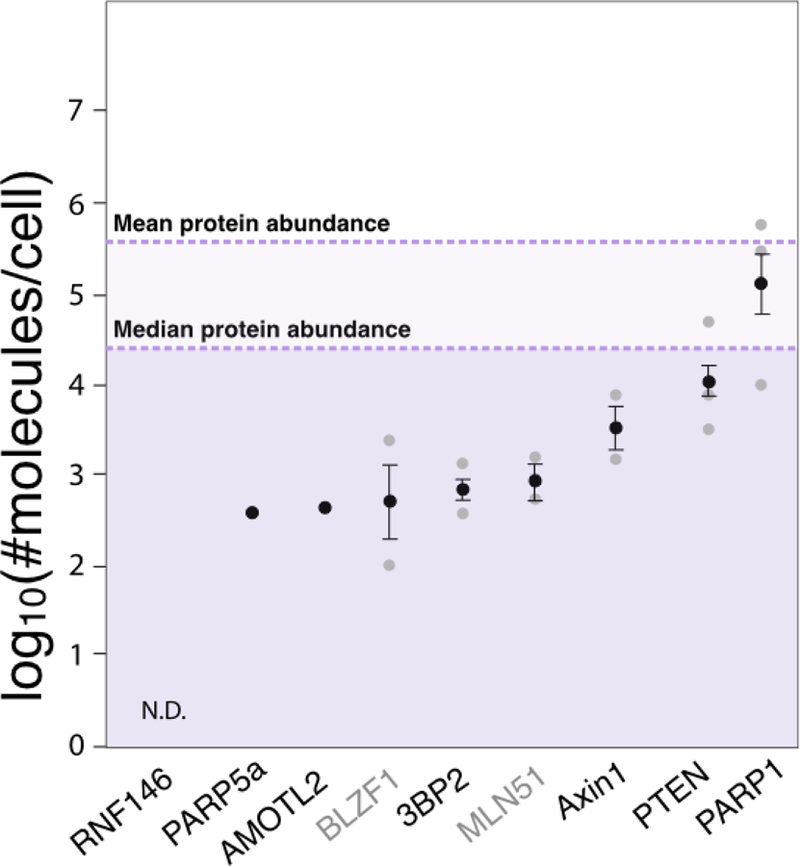
Protein levels measured in log10(#molecules/cell) from four human cell line studies are plotted as mean±standard error (in black) overlaid over individual data points (grey). All characterized PARdU substrates from Table 1 with protein copy number measured are plotted along with two potential PARdU candidates BLZF1 and MLN51. The mean and median protein abundance across all studies are noted. N.D., not detected by mass spectrometry across all studies.
4. Global analyses of the PARP5a/b interactome and potential PARdU substrates
Given that all characterized PARdU substrates interact with PARP5a/b, recent efforts have been focusing on identifying novel substrates through identifying these binding interactors. While in silico studies from Guettler and colleagues have generated a number of PARdU candidate substrates by identifying proteins possessing one or more TBM [26], two recent proteomics studies identified PARP5a/b interactors using immunoprecipitation [28] and determined which proteins change in abundance upon PARP5a/b knockout [29].
To identify PARP5a/b interacting proteins, Li and colleagues have generated 293T cells that stably express tagged PARP5a/b. After treatment of these cells with either DMSO (control) or PARP5a/b inhibitor XAV939, they immunoprecipitated these tagged constructs and performed mass spectrometry analysis to identify the PARP5a/b binding partners under these conditions [28]. All but one previously identified PARdU substrate (3BP2) was identified as a high confidence interactor. Significant overlap was observed between the PARP5a/b interactors identified in DMSO compared to the PARP5a/b inhibitor-treated condition, suggesting that the enzymatic activity of PARP5a/b is not critical for maintaining PARP5a/b interactions. Gene ontology revealed that PARP5a/b interactors participate in diverse signaling pathways, from Wnt and Hippo signaling, to hypoxia, autophagy, and Rho GTPase signaling. Consistent with the diverse roles in signaling pathways, PARP5a/b interactors localize throughout the cell, from the mitochondria to the Golgi and outwards to cellular membranes and the cytoskeleton. These observations paint a picture of PARP5a/b as a critical node linking diverse cellular processes through physical associations regardless of its enzymatic activity.
An additional proteomics screen by Bhardwaj and colleagues was performed to identify proteins whose abundance is altered in the absence of PARP5a/b [29]. 287 proteins increased in abundance following PARP5a/b double knockout, of which 74 possess TBMs. Amongst these 74 candidates, some of them have also been demonstrated to increase in abundance following PARP5a/b inhibition, suggesting these proteins are likely PARdU substrates. These proteins participate in diverse biological processes including Wnt signaling, microRNA processing, Notch signaling, and glucose transport. Li et al. and Bhardwaj et al. have each identified enrichment in Hippo signaling, suggesting a potential role of PARP5a/b in this pathway [28,29].
Upon comparison of these three studies, 13 proteins were identified to overlap in all studies (Figure 5A). They have three characteristics common for PARdU substrates: possession of a PARP5a/b binding motif, interaction with PARP5a/b, and increased abundance upon PARP5a/b knockout. Two established substrates (AMOTL2 and RNF146) appear in this set of 13 proteins, suggesting that this list may posit candidate PARdU substrates [21], that warrant future investigation. Intriguingly, we also found that most of these PARdU candidates are of low abundance, including multiple ones that cannot be detected by mass spectrometry in all studies analyzed (Figure 5B). Given that many of these substrates were curated in ADPriboDB with site information [68], it will be of interest to test whether any specific sites of ADP-ribosylation plays a role in PARdU.
Figure 5. Comparison of three PARP5a/b interactome studies.
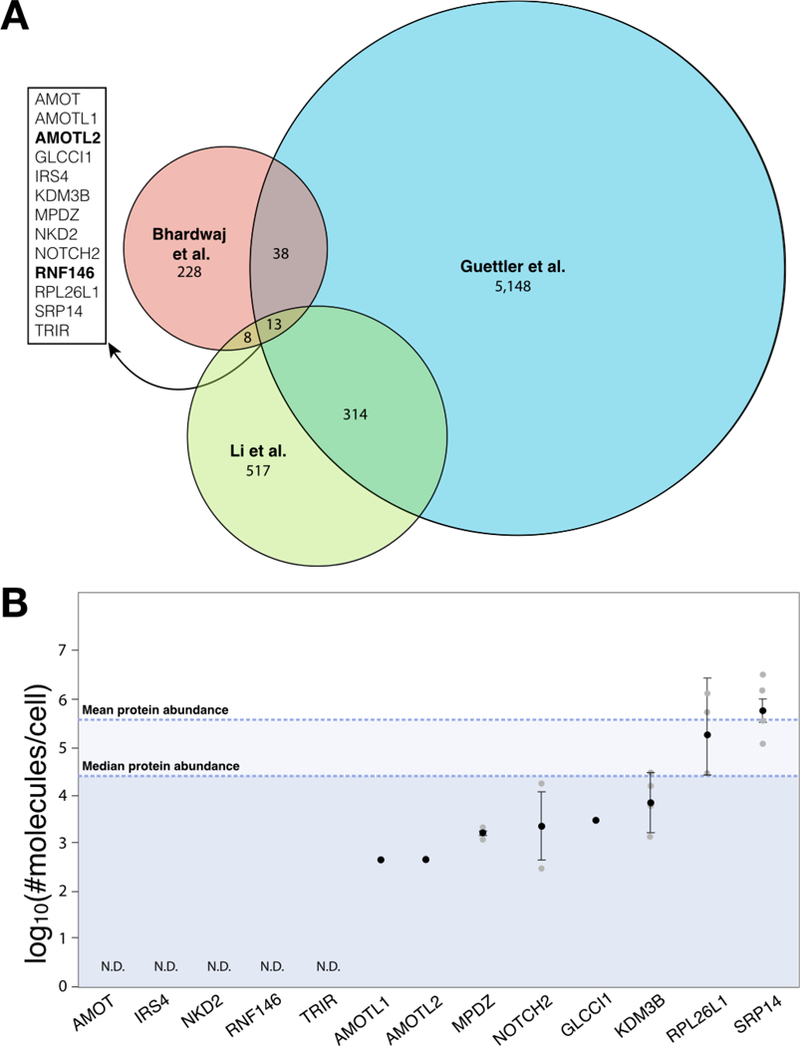
A. Venn diagram displaying the overlap of protein identifications among an in silico study identifying proteins with at least one Tankyrase- binding motif (5,513 proteins; Guettler), global analyses of proteins that increase in abundance upon PARP5a/b knockout (287 proteins; Bhardwaj), and proteomics analyses of PARP5a/b interactors (852 proteins; Li). For the Guettler study, only proteins with IUPred disorder score ≥ 0.45 are included for the analyses, which increases the stringency of the in silico prediction based on ref. [26]. Proteins identified in all three studies are listed on the left, with established PARdU substrates in bold. B. As in Figure 4, protein levels measured in log10(#molecules/cell) from four human cell line studies are plotted as mean±standard error (in black) overlaid over individual data points (grey). The mean and median protein abundance across all studies are noted. N.D., not detected by mass spectrometry across all studies.
5. Open questions
While it has been established that RNF146 binds PAR through its WWE domain and that binding leads to a conformational change that activates its E3 ligase activity [21,37,38], several questions remain regarding PARdU. Could other E3 ubiquitin ligases be activated upon PAR binding? Could other PARPs besides PARP5a/b synthesize the modification necessary for E3 ubiquitin ligase binding, activation and ubiquitination?
5.1. Other PARPs?
Binding of RNF146 to iso-ADPr requires linear PAR, which can be synthesized by PARP5a/b, and also by PARPs 1 and 2 [2,69,70]. Indeed, RNF146 is known to bind PARP1 and PARP2 [38,71,72], suggesting the possibility of the involvement of these PARPs in PARdU. Notably, a recent report suggests that PARP1 mediates PARdU of the E3 ubiquitin ligase Siah1, which in turn regulates the level of protein factors that are critical for HIV-1 transactivation [73]. The diversity of PARP localization presents a possibility that different pools of substrates can be targeted for PARdU by different PARPs (e.g., PARP1/2 in the nucleus versus PARP5a/b in the cytoplasm and nucleus [74]), adding an additional layer of specificity to the PARdU mechanism. In addition, it is possible that PARylation can be mediated through sequential actions of different PARPs, where one PARP adds MAR to its substrate, and then PARP5a/b or other PAR-synthesizing PARPs add additional ADPr units. The resultant iso-ADPr structure or PAR may then activate RNF146 for subsequent ubiquitination of the substrate. A recent study of auto-PARylation of PARP-1 suggests this possibility, where the initial ADPr was conjugated at a lysine residue by a non-PARP enzyme SIRT6 [75]. Notably, proteasomal degradation of Zika virus nonstructural proteins ns1 and ns3 depends on the catalytic activity of PARP12, which can only add single ADPr units [76]. It is unclear whether MARylation is sufficient for such degradation or an endogenous PARP synthesizes PAR on the substrates for PARdU.
5.2. Other E3 Ubiquitin Ligases?
Besides RNF146, multiple groups have noted that DTX1–4 (deltex homologs 1–4), HUWE1 (HECT, UBA, and WWE domain containing 1), and TRIP12 (thyroid hormone receptor interactor 12) are WWE-containing E3 ubiquitin ligases, which may regulate protein turnover in a PARylation-dependent manner [30,37]. Supporting this possibility, Callow and colleagues reported Axin1/2 can be stabilized upon silencing of PARP5a/b, but not RNF146, in a colorectal cancer cell line [22]. Notably, two classes of E3 ubiquitin ligases—HECT and RING E3 ubiquitin ligases—possess WWE domains, and they employ different catalytic mechanisms [77]. While RING E3 ligases (e.g., RNF146, DTX1–4) aid in the direct transfer of ubiquitin from an E2 ubiquitin carrier protein to protein substrates, HECT E3 ubiquitin ligases (e.g., HUWE1, TRIP12) form a covalent intermediate with ubiquitin before transferring the ubiquitin moiety to substrates (Section 1.2). If HECT E3 ligases are found to have functional WWE domains that bind iso-ADPr, it is worth questioning whether PAR binding is necessary for HECT E3 ligase activation? Or is binding to PAR enough to bring the E3 into proximity with its substrate? However, not all WWE domains bind iso-ADPr to the same extent, despite the fact that the domain is highly conserved [37,78]. For example, DDHD2 possesses a WWE domain that fails to bind PAR [37]. On the other hand, E3 ligases may possess non-WWE PAR-binding domains or motifs [79,80]. For example, RING-type E3 ligase CHFR possesses a PAR-binding zinc finger motif [81,82]. Future studies should determine whether other E3 ligases are PAR dependent, and if so, which pool of substrates these new PARdU players target.
5.3. Other PARdU Mechanisms?
While most current data support the model that RNF146 binds to and ubiquitinates a PARylated substrate, an alternative model is possible based on existing data (Figure 6). Kang and colleagues observed that RNF146 interacts with PARP1 only when PARP1 is PARylated, suggesting that the interaction between RNF146 and PAR, and not that between RNF146 and PARP1, determines whether PARP1 undergoes PARdU [38]. These data suggest that the activation of the E3 ligase does not require binding to PARylated substrates and is instead mediated by PAR. Perhaps, RNF146 binding to a PAR molecule is sufficient for the activation of its E3 ligase activity and ubiquitination of substrates, especially to those PAR binding proteins that are proximal to the ligase. Furthermore, autoubiquitination of RNF146 increases in a dose-dependent manner with the addition of free PAR polymers [38]. Taken together, these data suggest that free PAR polymer, if it exists in cells, or PAR conjugated to protein may play a role in activating RNF146 activity.
Figure 6. Working models of PARdU.
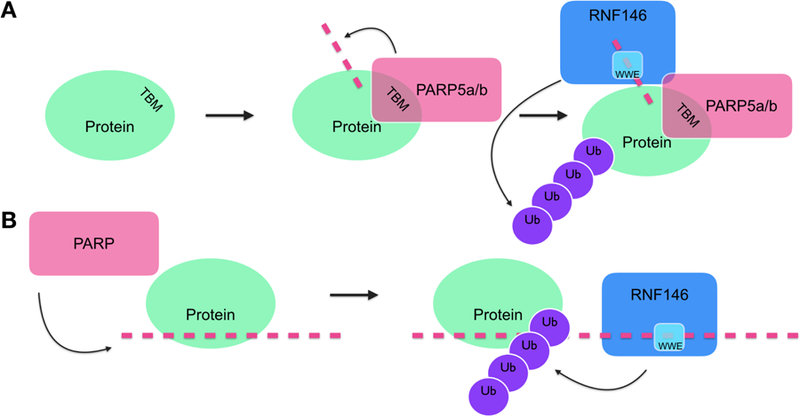
(A) PARP5a/b binds its substrate via Tankyrase Binding Motif (TBM) before RNF146 binds the iso-ADPr within PAR (pink dashed line) through its WWE domain and becomes catalytically active and poly-ubiquitinates the substrate to tag it for proteasomal degradation. (B) PARPs synthesize PAR, which is then bound by PAR-binding proteins. RNF146 binds PAR, becomes catalytically active, and ubiquitinates proximal PAR-binding proteins.
6. Considerations on exploiting PARdU to treat diseases
Several PARdU substrates, such as Axin1/2, 3BP2, and PTEN, are relevant to specific disease contexts, and dysregulation of PARdU of these factors leads to potentially deleterious effects (Section 3 and Table 2). Thus, the enzymes regulating PARdU may be attractive therapeutic targets. Inhibition of PARP5a/b has shown promising results in downregulating aberrant Wnt/β-catenin signaling and reducing growth in colorectal cancer cell lines and may, therefore, be used in the clinic to treat colorectal cancer, particularly those with overactive Wnt signaling [48,83]. Yet, given that PARP5a/b plays a role in a multitude of different biological functions, from mitosis and telomere maintenance to glucose metabolism [84], selective inhibition of PARP5a/b may lead to unwanted effects. Side effects such as intestinal toxicity have also been observed with various PARP5a/b inhibitors [85,86]. Instead, it may be more favorable to target the E3 ligase RNF146. Targeting RNF146 may, however, prove difficult since RING and WWE domains are highly conserved [19]. That said, several selective kinase inhibitors target highly-conserved ATP-binding pockets [87], which lends hope to prospective therapies targeting these conserved domains of RNF146. Finally, as discussed in Section 3, RNF146 and PARP5a/b regulate PARdU and degradation of one another [21,22]; therefore, perturbing the stability or activity of one enzyme may inevitably affect the stability and intracellular levels of the other.
Table 2.
A summary of PARdU substrates, their associated biological processes and possible pathological outcomes upon PARdU dysregulation.
| PARdU substrate | Biological Pathway | Pathology |
|---|---|---|
| Axin1/2 | Wnt signaling | Cancer |
| 3BP2 | SRC, SYK, VAV signaling | Cherubism |
| PTEN | AKT signaling; tumor suppressor | Cancer |
| AMOTL2 | Tight junctions | Cancer |
| PARP1 | DNA damage, stress response | Cancer |
Careful consideration must also be given to the context of PARdU regulation. Is the PARdU substrate a rate- or concentration-limiting factor? Is the protein an activator or suppressor in its biological pathway? For example, Axin1/2 is a rate-limiting factor and a negative regulator of the Wnt signaling pathway. PARdU of Axin1/2 limits the formation of the β-catenin destruction complex, affecting Wnt signaling and sensitizing cells to Wnt factors. In contrast, excessive PARdU of PTEN results in aberrant cell proliferation and potential tumorigenesis. Therefore, PARdU can either drive disease progression or serve as a safeguard against disease initiation. Thus, while PARdU is an attractive therapeutic target, inhibiting or activating this system in a targeted manner may be challenging, and several questions must be addressed prior to therapeutic considerations: in which contexts does PARdU dysregulation lead to disease? What factors dictate the specificity and selection of PARdU targets? Does PARdU target selection vary among various cell types?
7. Concluding remarks
ADP-ribosylation is a functionally diverse post-translational modification with the ability to affect protein function, stability and activity. As a signal for ubiquitination and subsequent degradation, ADP- ribosylation seems to regulate the stability of proteins that are of low abundance. Perhaps, possession of two post-translational modifications adds an additional layer of specificity that facilitates tighter regulation—more than one enzymatic reaction is necessary for these low abundance substrates to be destined for proteasome degradation. Knowing that other post-translational modifications, like phosphorylation [88] and SUMOylation [89] can also target a protein for ubiquitin-mediated proteasomal degradation, one may wonder if these dual-modification degradation mechanisms also target proteins of low abundance, or if they target a specific pool of proteins that all share another common trait. The crosstalk between ADP-ribosylation and ubiquitination is not limited to PARdU. For example, ubiquitin itself is ADP-ribosylated, which has been shown to block subsequent attachment of ubiquitin to target proteins [90,91]. On the other hand, PARP5a/b ADP-ribosylates the proteasome regulator PI31, promoting the assembly of 26S proteasome [92]. Given the precedent set forth by PARdU, it is possible that ADP-ribosylation may also serve as a signal for regulating the synthesis of other post-translational modifications. It will, therefore, be of interest to see how many other modifying enzymes (such as kinases or SUMO E3 ligases) possess PAR-binding motifs.
With the availability of proteomics-based tools to globally identify the sites of ADP-ribosylation and ubiquitination, we may define better rules for PARdU substrates at the amino acid level, including its relationships with PARP-binding sites, such as the TBM, or specific motifs surrounding PTM sites. Through mutagenesis, we can rigorously test whether MARylation by a specific PARP (e.g., PARP12 [76]) is sufficient as a degradation signal for the ubiquitin-proteasome system. As novel methods to identify and characterize PARylation substrates are developed [93,94], we may identify additional degradation signals within PAR, such as its length and structure.
Acknowledgments
We thank Mr. Lyle McPherson for making the inserts of Figure 3 and the members of the Leung laboratory for critical inputs of this manuscript. This work was partly supported by R01GM104135 (A.K.L.L.), U54GM103520 (A.K.L.L.) and T32CA009110 (C.A.V) from National Institutes of Health, and the American Cancer Society Research Scholar Award RSG-16–062-01-RMC (A.K.L.L.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interest: none
References
- [1].Prabakaran S, Lippens G, Steen H, Gunawardena J, Post-translational modification: nature’s escape from genetic imprisonment and the basis for dynamic information encoding, Wiley Interdiscip Rev Syst Biol Med 4 (2012) 565–583. doi: 10.1002/wsbm.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Vyas S, Matic I, Uchima L, Rood JE, Zaja R, Hay RT, et al. , Family-wide analysis of poly(ADP-ribose) polymerase activity, Nature Communications 5 (2014) 4426. doi: 10.1038/ncomms5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Rippmann JF, Damm K, Schnapp A , Functional characterization of the poly(ADP-ribose) polymerase activity of tankyrase 1, a potential regulator of telomere length, J Mol Biol 323 (2002) 217–224. [DOI] [PubMed] [Google Scholar]
- [4].Alvarez-Gonzalez R, Jacobson MK, Characterization of polymers of adenosine diphosphate ribose generated in vitro and in vivo, Biochemistry 26 (1987) 3218–3224. [DOI] [PubMed] [Google Scholar]
- [5].Gupte R, Liu Z, Kraus WL, PARPs and ADP-ribosylation: recent advances linking molecular functions to biological outcomes, Genes Dev 31 (2017) 101–126. doi: 10.1101/gad.291518.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Chaudhuri A. Ray, Nussenzweig A, The multifaceted roles of PARP1 in DNA repair and chromatin remodelling, Nat Rev Mol Cell Biol (2017). doi: 10.1038/nrm.2017.53. [DOI] [PMC free article] [PubMed]
- [7].Smith S, Giriat I, Schmitt A, de Lange T, Tankyrase, a poly(ADP-ribose) polymerase at human telomeres, Science 282 (1998) 1484–1487. [DOI] [PubMed] [Google Scholar]
- [8].Leung AKL, Poly(ADP-ribose): An organizer of cellular architecture, 205 (2014) 613–619. doi: 10.1083/jcb.201402114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Altmeyer M, Neelsen KJ, Teloni F, Pozdnyakova I, Pellegrino S, Grøfte M, et al. , Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose), Nature Communications 6 (2015) 8088. doi: 10.1038/ncomms9088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ozkaynak E, Finley D, Varshavsky A, The yeast ubiquitin gene: head-to-tail repeats encoding a polyubiquitin precursor protein, Nature 312 (1984) 663–666. [DOI] [PubMed] [Google Scholar]
- [11].Komander D, Rape M, The ubiquitin code, Annu Rev Biochem 81 (2012) 203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- [12].Scheffner M, Nuber U, Huibregtse JM, Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade, Nature 373 (1995) 81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- [13].Hershko A, Heller H, Elias S, Ciechanover A, Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown, J Biol Chem 258 (1983) 8206–8214. [PubMed] [Google Scholar]
- [14].Ardley HC, Robinson PA, E3 ubiquitin ligases, Essays Biochem 41 (2005) 15–30. doi: 10.1042/EB0410015. [DOI] [PubMed] [Google Scholar]
- [15].Spratt DE, Walden H, Shaw GS, RBR E3 ubiquitin ligases: new structures, new insights, new questions, Biochem. J 458 (2014) 421–437. doi: 10.1042/BJ20140006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kee Y, Huibregtse JM, Regulation of catalytic activities of HECT ubiquitin ligases, Biochem Biophys Res Commun 354 (2007) 329–333. doi: 10.1016/j.bbrc.2007.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Deshaies RJ, Joazeiro CAP, RING domain E3 ubiquitin ligases, Annu Rev Biochem 78 (2009) 399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- [18].Metzger MB, Pruneda JN, Klevit RE, Weissman AM, RING-type E3 ligases: master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination, Biochim Biophys Acta 1843 (2014) 47–60. doi: 10.1016/j.bbamcr.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].DaRosa PA, Wang Z, Jiang X, Pruneda JN, Cong F, Klevit RE, et al. , Allosteric activation of the RNF146 ubiquitin ligase by a poly(ADP-ribosyl)ation signal, Nature (2014). doi: 10.1038/nature13826. [DOI] [PMC free article] [PubMed]
- [20].Huang S-MA, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, et al. , Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling, Nature 461 (2009) 614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- [21].Zhang Y, Liu S, Mickanin C, Feng Y, Charlat O, Michaud GA, et al. , RNF146 is a poly(ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling, Nat Cell Biol 13 (2011) 623–629. doi: 10.1038/ncb2222. [DOI] [PubMed] [Google Scholar]
- [22].Callow MG, Tran H, Phu L, Lau T, Lee J, Sandoval WN, et al. , Ubiquitin ligase RNF146 regulates tankyrase and Axin to promote Wnt signaling, PLoS ONE 6 (2011) e22595. doi: 10.1371/journal.pone.0022595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zhou Z-D, Chan CH-S, Xiao Z-C, Tan E-K, Ring finger protein 146/Iduna is a poly(ADP- ribose) polymer binding and PARsylation dependent E3 ubiquitin ligase, Cell Adh Migr 5 (2011) 463–471. doi: 10.4161/cam.5.6.18356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Chi NW, Lodish HF, Tankyrase is a golgi-associated mitogen-activated protein kinase substrate that interacts with IRAP in GLUT4 vesicles, J Biol Chem 275 (2000) 38437–38444. doi: 10.1074/jbc.M007635200. [DOI] [PubMed] [Google Scholar]
- [25].Sbodio JI, Chi N-W, Identification of a tankyrase-binding motif shared by IRAP, TAB182, and human TRF1 but not mouse TRF1. NuMA contains this RXXPDG motif and is a novel tankyrase partner, J Biol Chem 277 (2002) 31887–31892. doi: 10.1074/jbc.M203916200. [DOI] [PubMed] [Google Scholar]
- [26].Guettler S, Larose J, Petsalaki E, Gish G, Scotter A, Pawson T, et al. , Structural basis and sequence rules for substrate recognition by tankyrase explain the basis for cherubism disease, 147 (2011) 1340–1354. doi: 10.1016/j.cell.2011.10.046. [DOI] [PubMed] [Google Scholar]
- [27].Levaot N, Voytyuk O, Dimitriou I, Sircoulomb F, Chandrakumar A, Deckert M, et al. , Loss of Tankyrase-Mediated Destruction of 3BP2 Is the Underlying Pathogenic Mechanism of Cherubism, 147 (2011) 1324–1339. doi: 10.1016/j.cell.2011.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Li X, Han H, Zhou M-T, Yang B, Ta AP, Li N, et al. , Proteomic Analysis of the Human Tankyrase Protein Interaction Network Reveals Its Role in Pexophagy, Cell Rep 20 (2017) 737–749. doi: 10.1016/j.celrep.2017.06.077. [DOI] [PubMed] [Google Scholar]
- [29].Bhardwaj A, Yang Y, Ueberheide B, Smith S, Whole proteome analysis of human tankyrase knockout cells reveals targets of tankyrase-mediated degradation, Nature Communications 8 (2017) 2214. doi: 10.1038/s41467-017-02363-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Aravind L, The WWE domain: a common interaction module in protein ubiquitination and ADP ribosylation, Trends Biochem Sci 26 (2001) 273–275. [DOI] [PubMed] [Google Scholar]
- [31].Eisemann T, McCauley M, Langelier M-F, Gupta K, Roy S, Van Duyne GD, et al. , Tankyrase-1 Ankyrin Repeats Form an Adaptable Binding Platform for Targets of ADP-Ribose Modification, Structure 24 (2016) 1679–1692. doi: 10.1016/j.str.2016.07.014. [DOI] [PubMed] [Google Scholar]
- [32].Morrone S, Cheng Z, Moon RT, Cong F, Xu W, Crystal structure of a Tankyrase-Axin complex and its implications for Axin turnover and Tankyrase substrate recruitment, Proc Natl Acad Sci USA 109 (2012) 1500–1505. doi: 10.1073/pnas.1116618109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Li B, Qiao R, Wang Z, Zhou W, Li X, Xu W, et al. , Crystal structure of a tankyrase 1-telomere repeat factor 1 complex, Acta Crystallogr F Struct Biol Commun 72 (2016) 320–327. doi: 10.1107/S2053230X16004131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Pollock K, Ranes M, Collins I, Guettler S, Identifying and Validating Tankyrase Binders and Substrates: A Candidate Approach, Methods Mol Biol 1608 (2017) 445–473. doi: 10.1007/978-1-4939-6993-7_28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Chen B, Dodge ME, Tang W, Lu J, Ma Z, Fan C-W, et al. , Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer, 5 (2009) 100–107. doi: 10.1038/nchembio.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hong SJ, Li H, Becker KG, Dawson VL, Dawson TM, Identification and analysis of plasticity-induced late-response genes, Proc Natl Acad Sci USA 101 (2004) 2145–2150. doi: 10.1073/pnas.0305170101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wang Z, Michaud GA, Cheng Z, Zhang Y, Hinds TR, Fan E, et al. , Recognition of the iso- ADP-ribose moiety in poly(ADP-ribose) by WWE domains suggests a general mechanism for poly(ADP-ribosyl)ation-dependent ubiquitination, Genes Dev (2012). doi: 10.1101/gad.182618.111. [DOI] [PMC free article] [PubMed]
- [38].Kang HC, Lee Y-I, Shin J-H, Andrabi SA, Chi Z, Gagné J-P, et al. , Iduna is a poly(ADP- ribose) (PAR)-dependent E3 ubiquitin ligase that regulates DNA damage, Proc Natl Acad Sci USA 108 (2011) 14103–14108. doi: 10.1073/pnas.1108799108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Andrabi SA, Kang HC, Haince J-F, Lee Y-I, Zhang J, Chi Z, et al. , Iduna protects the brain from glutamate excitotoxicity and stroke by interfering with poly(ADP-ribose) polymer-induced cell death, Nat Med 17 (2011) 692–699. doi: 10.1038/nm.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].David KK, Andrabi SA, Dawson TM, Dawson VL, Parthanatos, a messenger of death, Front Biosci 14 (2009) 1116–1128. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Wang Y, Dawson VL, Dawson TM, Poly(ADP-ribose) signals to mitochondrial AIF: a key event in parthanatos, Exp. Neurol 218 (2009) 193–202. doi: 10.1016/j.expneurol.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Yu S-W, Wang H, Dawson TM, Dawson VL, Poly(ADP-ribose) polymerase-1 and apoptosis inducing factor in neurotoxicity, Neurobiol. Dis 14 (2003) 303–317. [DOI] [PubMed] [Google Scholar]
- [43].Mariotti L, Templeton CM, Ranes M, Paracuellos P, Cronin N, Beuron F, et al. , Tankyrase Requires SAM Domain-Dependent Polymerization to Support Wnt-β-Catenin Signaling, Molecular Cell 63 (2016) 498–513. doi: 10.1016/j.molcel.2016.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Riccio AA, McCauley M, Langelier M-F, Pascal JM, Tankyrase Sterile αMotif Domain Polymerization Is Required for Its Role in Wnt Signaling, Structure 24 (2016) 1573–1581. doi: 10.1016/j.str.2016.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].DaRosa PA, Klevit RE, Xu W, Structural basis for tankyrase-RNF146 interaction reveals noncanonical tankyrase-binding motifs, Protein Sci 27 (2018) 1057–1067. doi: 10.1002/pro.3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Daniels CM, Ong S-E, Leung AKL, The Promise of Proteomics for the Study of ADP- Ribosylation, Molecular Cell 58 (2015) 911–924. doi: 10.1016/j.molcel.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Li N, Zhang Y, Han X, Liang K, Wang J, Feng L, et al. , Poly-ADP ribosylation of PTEN by tankyrases promotes PTEN degradation and tumor growth, Genes Dev (2014). doi: 10.1101/gad.251785.114. [DOI] [PMC free article] [PubMed]
- [48].Ma L, Wang X, Jia T, Wei W, Chua M-S, So S, Tankyrase inhibitors attenuate WNT/β-catenin signaling and inhibit growth of hepatocellular carcinoma cells, Oncotarget 6 (2015) 25390–25401. doi: 10.18632/oncotarget.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Nusse R, Clevers H, Wnt/β-Catenin Signaling, Disease, and Emerging Therapeutic Modalities, Cell 169 (2017) 985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- [50].Klaus A, Birchmeier W, Wnt signalling and its impact on development and cancer, Nat Rev Cancer 8 (2008) 387–398. doi: 10.1038/nrc2389. [DOI] [PubMed] [Google Scholar]
- [51].Li C, Zheng X, Han Y, Lv Y, Lan F, Zhao J, XAV939 inhibits the proliferation and migration of lung adenocarcinoma A549 cells through the WNT pathway, Oncol Lett 15 (2018) 8973–8982. doi: 10.3892/ol.2018.8491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Xu D, Liu J, Fu T, Shan B, Qian L, Pan L, et al. , USP25 regulates Wnt signaling by controlling the stability of tankyrases, Genes Dev 31 (2017) 1024–1035. doi: 10.1101/gad.300889.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Zhu X, Xing R, Tan R, Dai R, Tao Q, The RNF146 E3 ubiquitin ligase is required for the control of Wnt signaling and body pattern formation in Xenopus, Mech. Dev 147 (2017) 28–36. doi: 10.1016/j.mod.2017.08.001. [DOI] [PubMed] [Google Scholar]
- [54].Ueki Y, Tiziani V, Santanna C, Fukai N, Maulik C, Garfinkle J, et al. , Mutations in the gene encoding c-Abl-binding protein SH3BP2 cause cherubism, Nat Genet 28 (2001) 125–126. doi: 10.1038/88832. [DOI] [PubMed] [Google Scholar]
- [55].Lo B, Faiyaz-Ul-Haque M, Kennedy S, Aviv R, Tsui L-C, Teebi AS, Novel mutation in the gene encoding c-Abl-binding protein SH3BP2 causes cherubism, Am. J. Med. Genet. A 121A (2003) 37–40. doi: 10.1002/ajmg.a.20226. [DOI] [PubMed] [Google Scholar]
- [56].Imai Y, Kanno K, Moriya T, Kayano S, Seino H, Matsubara Y, et al. , A missense mutation in the SH3BP2 gene on chromosome 4p16.3 found in a case of nonfamilial cherubism, Cleft Palate Craniofac. J 40 (2003) 632–638. doi: 10.1597/1545-1569_2003_040_0632_ammits_2.0.co_2. [DOI] [PubMed] [Google Scholar]
- [57].Campbell CI, Samavarchi-Tehrani P, Barrios-Rodiles M, Datti A, Gingras A-C, Wrana JL, The RNF146 and tankyrase pathway maintains the junctional Crumbs complex through regulation of angiomotin, J Cell Sci 129 (2016) 3396–3411. doi: 10.1242/jcs.188417. [DOI] [PubMed] [Google Scholar]
- [58].Wang W, Li N, Li X, Tran MK, Han X, Chen J, Tankyrase Inhibitors Target YAP by Stabilizing Angiomotin Family Proteins, Cell Rep 13 (2015) 524–532. doi: 10.1016/j.celrep.2015.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Xu H, Luo P, Zhao Y, Zhao M, Yang Y, Chen T, et al. , Iduna protects HT22 cells from hydrogen peroxide-induced oxidative stress through interfering poly(ADP-ribose) polymerase- 1-induced cell death (parthanatos), Cell Signal 25 (2013) 1018–1026. doi: 10.1016/j.cellsig.2013.01.006. [DOI] [PubMed] [Google Scholar]
- [60].Langelier M-F, Planck JL, Roy S, Pascal JM, Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1, Science 336 (2012) 728–732. doi: 10.1126/science.1216338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Benjamin RC, Gill DM, Poly(ADP-ribose) synthesis in vitro programmed by damaged DNA. A comparison of DNA molecules containing different types of strand breaks, J Biol Chem 255 (1980) 10502–10508. [PubMed] [Google Scholar]
- [62].Satoh MS, Poirier GG, Lindahl T, Dual function for poly(ADP-ribose) synthesis in response to DNA strand breakage, Biochemistry 33 (1994) 7099–7106. [DOI] [PubMed] [Google Scholar]
- [63].Kitazawa M, Hatta T, Ogawa K, Fukuda E, Goshima N, Natsume T, Determination of Rate- Limiting Factor for Formation of Beta-Catenin Destruction Complexes Using Absolute Protein Quantification, J Proteome Res 16 (2017) 3576–3584. doi: 10.1021/acs.jproteome.7b00305. [DOI] [PubMed] [Google Scholar]
- [64].Beck M, Schmidt A, Malmstroem J, Claassen M, Ori A, Szymborska A, et al. , The quantitative proteome of a human cell line, Mol. Syst. Biol 7 (2011) 549. doi: 10.1038/msb.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Nagaraj N, Wiśniewski JR, Geiger T, Cox J, Kircher M, Kelso J, et al. , Deep proteome and transcriptome mapping of a human cancer cell line, Mol. Syst. Biol 7 (2011) 548. doi: 10.1038/msb.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Wiśniewski JR, Vildhede A, Norén A, Artursson P, In-depth quantitative analysis and comparison of the human hepatocyte and hepatoma cell line HepG2 proteomes, J Proteomics 136 (2016) 234–247. doi: 10.1016/j.jprot.2016.01.016. [DOI] [PubMed] [Google Scholar]
- [67].Hein MY, Hubner NC, Poser I, Cox J, Nagaraj N, Toyoda Y, et al. , A human interactome in three quantitative dimensions organized by stoichiometries and abundances, Cell 163 (2015) 712–723. doi: 10.1016/j.cell.2015.09.053. [DOI] [PubMed] [Google Scholar]
- [68].Vivelo CA, Wat R, Agrawal C, Tee HY, Leung AKL, ADPriboDB: The database of ADP- ribosylated proteins, (2016). doi: 10.1093/nar/gkw706. [DOI] [PMC free article] [PubMed]
- [69].Rolli V, O’Farrell M, Ménissier-de J. Murcia, de Murcia G, Random mutagenesis of the poly(ADP-ribose) polymerase catalytic domain reveals amino acids involved in polymer branching, Biochemistry 36 (1997) 12147–12154. doi: 10.1021/bi971055p. [DOI] [PubMed] [Google Scholar]
- [70].Chen Q, Kassab MA, Dantzer F, Yu X, PARP2 mediates branched poly ADP-ribosylation in response to DNA damage, Nature Communications 9 (2018) 3233. doi: 10.1038/s41467-018-05588-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Huttlin EL, Bruckner RJ, Paulo JA, Cannon JR, Ting L, Baltier K, et al. , Architecture of the human interactome defines protein communities and disease networks, Nature 545 (2017) 505–509. doi: 10.1038/nature22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, et al. , The BioPlex Network: A Systematic Exploration of the Human Interactome, Cell 162 (2015) 425–440. doi: 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Yu D, Liu R, Yang G, Zhou Q, The PARP1-Siah1 Axis Controls HIV-1 Transcription and Expression of Siah1 Substrates, Cell Rep 23 (2018) 3741–3749. doi: 10.1016/j.celrep.2018.05.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Vyas S, Chesarone-Cataldo M, Todorova T, Huang Y-H, Chang P, A systematic analysis of the PARP protein family identifies new functions critical for cell physiology, Nature Communications 4 (2013) 2240. doi: 10.1038/ncomms3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Mao Z, Hine C, Tian X, Van Meter M, Au M, Vaidya A, et al. , SIRT6 promotes DNA repair under stress by activating PARP1, Science 332 (2011) 1443–1446. doi: 10.1126/science.1202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Li L, Zhao H, Liu P, Li C, Quanquin N, Ji X, et al. , PARP12 suppresses Zika virus infection through PARP-dependent degradation of NS1 and NS3 viral proteins, Sci Signal 11 (2018) eaas9332. doi: 10.1126/scisignal.aas9332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Metzger MB, Hristova VA, Weissman AM, HECT and RING finger families of E3 ubiquitin ligases at a glance, J Cell Sci 125 (2012) 531–537. doi: 10.1242/jcs.091777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].He F, Tsuda K, Takahashi M, Kuwasako K, Terada T, Shirouzu M, et al. , Structural insight into the interaction of ADP-ribose with the PARP WWE domains, FEBS Lett (2012). doi: 10.1016/j.febslet.2012.09.009. [DOI] [PubMed]
- [79].Krietsch J, Rouleau M, Pic E, Ethier C, Dawson TM, Dawson VL, et al. , Reprogramming cellular events by poly(ADP-ribose)-binding proteins, Mol. Aspects Med (2012). doi: 10.1016/j.mam.2012.12.005. [DOI] [PMC free article] [PubMed]
- [80].Teloni F, Altmeyer M, Readers of poly(ADP-ribose): designed to be fit for purpose, 44 (2016) 993–1006. doi: 10.1093/nar/gkv1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Ahel I, Ahel D, Matsusaka T, Clark AJ, Pines J, Boulton SJ, et al. , Poly(ADP-ribose)- binding zinc finger motifs in DNA repair/checkpoint proteins, Nature 451 (2008) 81–85. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- [82].Oberoi J, Richards MW, Crumpler S, Brown N, Blagg J, Bayliss R, Structural basis of poly(ADP-ribose) recognition by the multizinc binding domain of checkpoint with forkhead-associated and RING Domains (CHFR), J Biol Chem 285 (2010) 39348–39358. doi: 10.1074/jbc.M110.159855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Solberg NT, Waaler J, Lund K, Mygland L, Olsen PA, Krauss S, TANKYRASE Inhibition Enhances the Antiproliferative Effect of PI3K and EGFR Inhibition, Mutually Affecting β-CATENIN and AKT Signaling in Colorectal Cancer, Mol Cancer Res 16 (2018) 543–553. doi: 10.1158/1541-7786.MCR-17-0362. [DOI] [PubMed] [Google Scholar]
- [84].Mariotti L, Pollock K, Guettler S, Regulation of Wnt/β-catenin signalling by tankyrase-dependent poly(ADP-ribosyl)ation and scaffolding, Br. J. Pharmacol 174 (2017) 4611–4636. doi: 10.1111/bph.14038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Zhong Y, Katavolos P, Nguyen T, Lau T, Boggs J, Sambrone A, et al. , Tankyrase Inhibition Causes Reversible Intestinal Toxicity in Mice with a Therapeutic Index < 1, Toxicol Pathol 44 (2016) 267–278. doi: 10.1177/0192623315621192. [DOI] [PubMed] [Google Scholar]
- [86].Norum JH, Skarpen E, Brech A, Kuiper R, Waaler J, Krauss S, et al. , The tankyrase inhibitor G007-LK inhibits small intestine LGR5+ stem cell proliferation without altering tissue morphology, Biol. Res 51 (2018) 3. doi: 10.1186/s40659-017-0151-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Uitdehaag JCM, Verkaar F, Alwan H, de Man J, Buijsman RC, Zaman GJR, A guide to picking the most selective kinase inhibitor tool compounds for pharmacological validation of drug targets, Br. J. Pharmacol 166 (2012) 858–876. doi: 10.1111/j.1476-5381.2012.01859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Schichl YM, Resch U, Lemberger CE, Stichlberger D, de Martin R, Novel phosphorylation-dependent ubiquitination of tristetraprolin by mitogen-activated protein kinase/extracellular signal-regulated kinase kinase kinase 1 (MEKK1) and tumor necrosis factor receptor-associated factor 2 (TRAF2), J Biol Chem 286 (2011) 38466–38477. doi: 10.1074/jbc.M111.254888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Uzunova K, Göttsche K, Miteva M, Weisshaar SR, Glanemann C, Schnellhardt M, et al. , Ubiquitin-dependent proteolytic control of SUMO conjugates, J Biol Chem 282 (2007) 34167–34175. doi: 10.1074/jbc.M706505200. [DOI] [PubMed] [Google Scholar]
- [90].Bhogaraju S, Kalayil S, Liu Y, Bonn F, Colby T, Matic I, et al. , Phosphoribosylation of Ubiquitin Promotes Serine Ubiquitination and Impairs Conventional Ubiquitination, Cell 167 (2016) 1636–1649. 10.1016/j.cell.2016.11.019. [DOI] [PubMed] [Google Scholar]
- [91].Yang C-S, Jividen K, Spencer A, Dworak N, Ni L, Oostdyk LT, et al. , Ubiquitin Modification by the E3 Ligase/ADP-Ribosyltransferase Dtx3L/Parp9, Molecular Cell 66 (2017) 503–516. 10.1016/j.molcel.2017.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Cho-Park PF, Steller H, Proteasome Regulation by ADP-Ribosylation, 153 (2013) 614–627. doi: 10.1016/j.cell.2013.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Krastev DB, Pettitt SJ, Campbell J, Song F, Tanos BE, Stoynov SS, et al. , Coupling bimolecular PARylation biosensors with genetic screens to identify PARylation targets, Nature Communications 9 (2018) 2016. doi: 10.1038/s41467-018-04466-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Ando Y, Elkayam E, McPherson RL, Dasovich M, Cheng S-J, Voorneveld J, et al. , ELTA: Enzymatic Labeling of Terminal ADP-Ribose, Molecular Cell (2019). doi: 10.1016/j.molcel.2018.12.022. [DOI] [PMC free article] [PubMed]


