Abstract
Surfactant protein-A (SP-A) is an important mediator of pulmonary immunity. A specific genetic variation in SP-A2, corresponding to a glutamine (Q) to lysine (K) amino acid substitution at position 223 of the lectin domain, was shown to alter the ability of SP-A to inhibit eosinophil degranulation. Since a large subgroup of asthmatics have associated eosinophilia, often accompanied by inflammation associated with delayed clearance, our goal was to define how SP-A mediates eosinophil resolution in allergic airways and whether genetic variation affects this activity. Wild-type (WT), SP-A knock out (SP-A KO) and humanized (SP-A2 223Q/Q, SP-A2 223K/K) C57BL/6 mice were challenged in an allergic OVA model and parameters of inflammation examined. Peripheral blood eosinophils were isolated to assess the effect of SP-A genetic variation on apoptosis and chemotaxis. Five days post-challenge, SP-A KO and humanized SP-A2 223K/K mice have persistent eosinophilia in bronchoalveolar lavage fluid compared to WT and SP-A2 223Q/Q mice, suggesting an impairment in eosinophil resolution. In vitro, human SP-A containing either the 223Q or the 223K allele was chemoattractant for eosinophils while only 223Q resulted in decreased eosinophil viability. Our results suggest that SP-A aids in the resolution of allergic airway inflammation by promoting eosinophil clearance from lung tissue through chemotaxis, independent of SP-A2 Q223K, and by inducing apoptosis of eosinophils, which is altered by the polymorphism.
Keywords: asthma, surfactant protein-A, resolution, eosinophils, allergy, inflammation, lung
INTRODUCTION
The four surfactant proteins, A, B, C and D, are best characterized for their roles in pulmonary surfactant. Surfactant protein A (SP-A) is the most abundant protein of the four types and is a member of the collectin superfamily, which are known participants in innate immune defense (1). Previous studies have shown that SP-A has various roles in the host response against inhaled insults and is most well known as an opsonin to enhance phagocytic uptake of pathogens (1). Additionally, SP-A has been shown to bind to Mycoplasma pneumoniae (Mp), which is frequently associated with asthma exacerbations (2), resulting in inhibition of its growth in vitro (3, 4). Pastva et al found that mice lacking SP-A had enhanced Th2 associated indices of inflammation 24 hours after challenge as compared to WT mice in the ovalbumin (OVA) model (5). Along this line of evidence, previous studies have shown that SP-A isolated from asthmatics is dysfunctional in attenuating IL-8 and Muc5AC production in response to Mycoplasma pneumoniae infection as compared to SP-A isolated from non-asthmatic individuals (2).
Humans have two functional SP-A genes, SP-A1 and SP-A2, that together organize into a complex octadecamer (6, 7). Several allelic variations of the SP-A genes have been identified and similarly linked to varying responses to pulmonary infections and control of inflammation (8). In particular, others have shown that the presence of a K at position 223 was associated with higher rates of respiratory syncytial virus (RSV) infection among infants (9), while a Q at this position was associated with protection against respiratory distress syndrome (RDS) (10). Although we are not aware of any studies that specifically evaluate the possible alterations to the native full-length SP-A oligomeric structure due to SP-A2 genetic variation, it has been shown that stably transfected cell lines expressing single gene SP-A variants form oligomers that are of similar patterns and orders of magnitude (7). Likewise, patterns of oligomerization were not different between SP-A purified from normal individuals when compared to those purified from asthmatic individuals (2). This suggests that the association of the polymorphism at position 223 and altered function is not likely due to changes in SP-A structure but may be due to altered functionality in the context of asthma.
Eosinophilia is the increased presence of eosinophils in the airway and peripheral blood (11) and is a well-documented phenotype in the lungs of a large subgroup of asthmatics (12). Release of pre-formed granules from eosinophils leads to damage of the airway mucosa and remodeling (13). Therapies that aim to minimize eosinophilic inflammation aid in the reduction of symptoms associated with allergic airway disease (14). In addition, studies have shown that obese asthmatics have more severe tissue eosinophilia (15, 16) and that they also have decreased levels of SP-A compared to lean normal and lean asthmatic individuals (17). Moreover, in a mouse model of allergic airway inflammation, administration of exogenous SP-A promoted the resolution of tissue eosinophilia (17). We have also demonstrated that SP-A inhibits degranulation of eosinophils (18) and that this ability is altered by the genetic variation in SP-A2 position 223 (6), thus suggesting a differential role in the modulation of eosinophilic inflammation for SP-A2.
Here, we set out to determine if genetic variation in human SP-A2, Q223K, would alter the resolution of allergic airways disease by specifically mediating eosinophil activities. Using a combination of isolated human SP-A, eosinophils and mouse models that express human SP-A2, our studies suggest two novel functions for SP-A: 1) as a chemoattractant for eosinophils, which is not dependent on genetic variation at position 223, and 2) as an inducer of eosinophil apoptosis, which is dependent on position 223. Taken together, we show that SP-A is an important contributing factor leading to the resolution of eosinophilia in allergen-challenged mice and specific genetic variation (rs1965708) that is present in the human population alters this activity.
MATERIALS AND METHODS
Human SP-A Extraction
SP-A was isolated and purified from the bronchoalveolar lavage fluid of patients with alveolar proteinosis by butanol extraction methods as previously described (17, 19). The final concentration of SP-A was determined as 1.2 mg/ml and endotoxin levels were less than 0.01 pg/ml SP-A (Pierce™ LAL Chromogenic Endotoxin Quantitation Kit, ThermoFisher Scientific, Rockfield IL). Human SP-A extracted by our group is available and is currently being shipped to research laboratories both nationally and internationally.
Mouse Models
All experiments were done in accordance with University of Arizona on IACUC approved animal protocols. Humanized mice transgenic for SP-A2 were generated as previously described (20). SP-A KO mice were generated as previously described (21) backcrossed 14 generations onto the C57BL/6 background and bred in-house. Wild-type (WT) mice were purchased from Jackson Laboratories (Bar Harbor, ME) and bred in-house for experiments. Depending on the timing of induction of allergic inflammation in female mice using the OVA model, it has been reported that estrogen has dual effects (22). To eliminate this factor, we, therefore, used male age-matched (6 – 8 weeks, weighing 20 – 25 g) WT, SP-A KO, SP-A2 223Q/Q and SP-A2 223K/K mice on a C57BL/6J background. IL-5 transgenic mice were generously provided by the late Dr. James J. Lee (23) and were bred in house for isolation of eosinophils for our in vitro studies. All mice were housed under 12-hour light-dark cycle with free access to standard chow and water.
Induction of Allergic Airways by use of Ovalbumin (OVA) in mice
For the time point experiments, sensitization was accomplished by intraperitoneal (i.p.) injections of OVA (50 μg per mouse, Sigma, St. Louis MO) in 150 μl of alum (40 mg/ml, ThermoFisher Scientific, Rockfield IL) on days 0 and 7. Challenge was given by three consecutive intranasal (i.n.) administrations of 100 μg OVA in PBS on days 14, 15 and 16. For the rescue experiments, the protocol was slightly modified to accommodate optimal delivery of replacement SP-A as previously described (17). Briefly, mice were given i.p. injections of 30 μg OVA in alum on days 0 and 14, and challenged with 1% OVA aerosol on days 21, 22 and 23 via a Nouvag Ultrasonic 2000 Nebulizer (Nouvag AG, Goldach, Switzerland). Subsequently, on day 24, SP-A was given oropharyngeally at 25 μg in 50 μL saline per mouse. Control mice for the rescue experiment were sensitized and challenged with OVA but received vehicle (saline) in place of SP-A.
Bronchoalveolar Lavage Fluid (BALF) and Lung Tissue
One, three and five days after the terminal intranasal challenge, mice were euthanized by anesthetic overdose (Urethane, 250 mg/ml, 1.5g/kg, Sigma, St. Louis MO). The trachea of each mouse was exposed and cannulated with a 19G catheter. Airways and lungs were washed with 1.5 ml of PBS (100 μM EDTA). Lungs were excised and removed. The left lung was fixed in 10% buffered formalin and transferred to 70% ethanol after 3 days for histologic analysis. The right lung was snap frozen in liquid nitrogen and kept at −80°C until assayed. Total cell counts were quantified using a Countess™ II FL Automated Cell Counter (Life Technologies, Carlsbad CA) and differential leukocyte counts were assessed on cytocentrifuged slides at a seeding density of 200,000 cells per slide stained with the Easy III™ rapid differential staining kit (Azer Scientific, Morgantown PA) using standard morphological identification of cell types.
Histological Analysis
Upon necropsy, the left lung lobe from each mouse was fixed in 10% formalin and embedded in paraffin. Mid-sagittal lung sections (4 μm thick) were stained with Sirius Red stain. Briefly, paraffin-embedded lungs were de-paraffinized using xylene and ethanol. Slides were then counterstained in hematoxylin for 3 minutes, rinsed in water and 100% ethanol, and incubated in Sirius Red for 1 hour following a previously described protocol (24). Histological slides at 400x magnification were photographed and scored by at least two blinded individuals. The average of these measurements were graphed for each mouse for statistical analysis.
Gene Expression by Real-time qRT-PCR
To evaluate mRNA expression of cyclophilin (internal control; sense: AGC ACT GGA GAG AAA GGA TTT GG, antisense: TCT TCT TGC TGG TCT TGC CAT T) and eosinophil-associated ribonuclease (EAR) (sense: CGA CTT TGT CTC CTG CTG, antisense: TGT CCC ATC CAA GTG AAC), real-time qRT-PCR was performed. Total RNA was extracted using TRIzol reagent (Invitrogen, San Diego CA) and reverse transcribed with the iScript cDNA synthesis kit (Bio-Rad Laboratories, Hercules CA). Targets were amplified using their respective primers and SYBR Green Supermix in the Bio-Rad CFX system (Bio-Rad Laboratories, Hercules CA). Relative CT was used to compare gene expression levels using 2-ΔΔCT method.
Determination of Proteins by ELISA
Human SP-A (BioVendor, Brno, Czech Republic), mouse eotaxin-1 (CCL11, eBioscience, Vienna, Austria) and eotaxin-2 (CCL24, R&D Systems, Minneapolis MN) assays were performed on BALF according to manufacturers’ protocols. Samples for CCL11 and CCL24 were diluted 1:2 and samples for human SP-A were assayed undiluted. Briefly, BALF samples were incubated on a 96-well plate coated with the capture antibody and detected with a Streptavidin-HRP:biotin-conjugated secondary antibody complex on a plate reader (BioTek Instruments, Winooski VT).
Eosinophil and leukocyte isolation
IL-5 transgenic mice were euthanized and blood collected by cardiac puncture through the left ventricle. Spleens were excised and subsequently minced. Red blood cells (RBCs) from blood or spleens were lysed and eosinophils isolated by negative selection as previously described (18). Purity of each preparation was verified by cytospin and an Easy III™ rapid differential staining kit (Azer Scientific, Morgantown PA) to be greater than 95%. Leukocytes were isolated from 20 mL of blood taken from asthmatic volunteers on an IRB approved protocol at the University of Arizona. Volunteers consisted of three female, non-Hispanic, white asthmatics, between the ages of 30 to 59, and were either not on medication or on combination Albuterol/Ipratoprium or Albuterol/Symbicort therapies. All blood samples were placed into tubes containing anticoagulant, kept at 4°C and processed within 6 hours of patients’ blood draw to avoid loss of viability. After density gradient centrifugation, RBCs were lysed and total leukocytes were used for flow cytometry experiments detailed below.
Determination of Proteins by Western Blotting
Purified eosinophils from blood or spleen were incubated with SP-A in RPMI 1640 at 37°C and 5% CO2 for 16 hours. For collection, 200 μl of Radioimmunoprecipitation Assay (RIPA) buffer (Teknova, Hollister CA) with protease inhibitors (Roche, Basel, Switzerland) was used for cell lysis and protein extraction. Proteins from cell lysates were quantified using a Pierce™ BCA protein assay kit (ThermoFisher Scientific, Rockfield IL) and equal amounts of lysate were loaded onto Mini-Protean® TGX™ precast gels (Bio-Rad Laboratories, Hercules CA). Antibodies for detecting cleaved caspase-3 and GAPDH were used according to manufacturer’s recommendations (Cell Signaling, Danvers MA). Western blots were imaged using a Chemidoc™ Imaging System and densitometry was quantified using Image Lab Software (Bio-rad Laboratories, Hercules CA).
Chemotaxis by Transmigration Plate Assay
The migration of purified eosinophils in response to SP-A in vitro was assessed using 5 μm polycarbonate membrane inserts in 24-well tissue culture plates (Costar, Corning NY) as previously described (25) with minor modifications. Briefly, inserts were incubated in RPMI 1640 (10% FBS) for one hour, after which, media was removed and eosinophils (1 × 106 cells per well with 30 ng/ml recombinant IL-5 in 200 μl) were added onto the inserts. Eosinophils were allowed to migrate for 90 minutes. Migrated cells (i.e., cells that reached the bottom chamber and on the underside of the inserts) were collected and counted using a Countess™ II FL Automated Cell Counter (Life Technologies, Carlsbad CA). Migration index was calculated as fold of total cells migrated in test well over vehicle control. Eotaxin-2 (Peprotech, Pittsburgh PA) was used as a positive chemoattractant control.
Assessment of Eosinophil Viability
Viability by Trypan Blue.
Uptake of Trypan blue by eosinophils was performed by mixing equal volumes of the dye and cell suspension. Live and dead cells were evaluated and counted using a Countess™ II FL Automated Cell Counter (Life Technologies, Carlsbad CA).
Cytotoxicity Assay by Real-Time Impedance Tracing.
Real-time monitoring of eosinophil detachment and cell death were assessed by measuring electrical impedance using the xCELLigence Real-Time Cell Analyzer (ACEA Biosciences, San Diego CA) as previously described (26, 27). Briefly, media was placed in 96-well gold electrode coated plates (E-plates, ACEA Biosciences), allowed to equilibrate and a background reading was obtained. Eosinophils were then seeded at 1 × 106 cells/100 μl and allowed to settle for ~5 hours. SP-A was added at various concentrations and changes in electrical impedance were measured over time. Impedance measurements, presented as a normalized “Cell Index,” are calculated as detailed (26, 27). Under these conditions, a loss of Cell Index is associated with eosinophil detachment and cytotoxicity. Individual traces of cytotoxicity over time were averages of 2 – 3 technical replicates; standard deviations were eliminated for clarity. Quantification of cytoxicity was accomplished by measuring the area under the curve (AUC) after normalization of cell index. Graphical AUC measurements include baseline correction for untreated cells to best display changes.
Flow Cytometry Analysis.
For the direct treatment of cells with SP-A, leukocytes from human blood were isolated by density gradient centrifugation using Histopaque 1077 (Sigma, St. Louis MO). After 16 hours incubation with SP-A, human leukocytes were labeled with Siglec-8-PE (BioLegend, San Diego CA). For the in vivo rescue mouse experiments, mouse BALF cells were collected and incubated with the following fluorescent antibodies: CD11b-PE-Cy7 (BD Biosciences, San Diego CA) and CCR3-APC (BioLegend). Apoptotic cells were labelled using a FITC Annexin V Apoptosis Detection Kit (BD Biosciences, San Diego CA). Flow cytometry was performed on an Attune NXT Flow Cytometer (ThermoFisher Scientific, Rockfield IL). Apoptotic eosinophils were identified as Siglec-8+, Annexin V+ and PI− (human) or CD11b+, CCR3+, Annexin V+ and PI− (mouse). Data were processed and analyzed using FlowJo 10.5.3.
Statistical Analysis
All statistical analyses were done using Graphpad Prism software. Since there were four genetically distinct groups of mice, one-way ANOVA was used to assess global differences between groups, followed by multiple t-tests with Bonferroni’s correction for multiple comparisons.
RESULTS
Genetic variation in SP-A2 alters eosinophil resolution in allergic airways
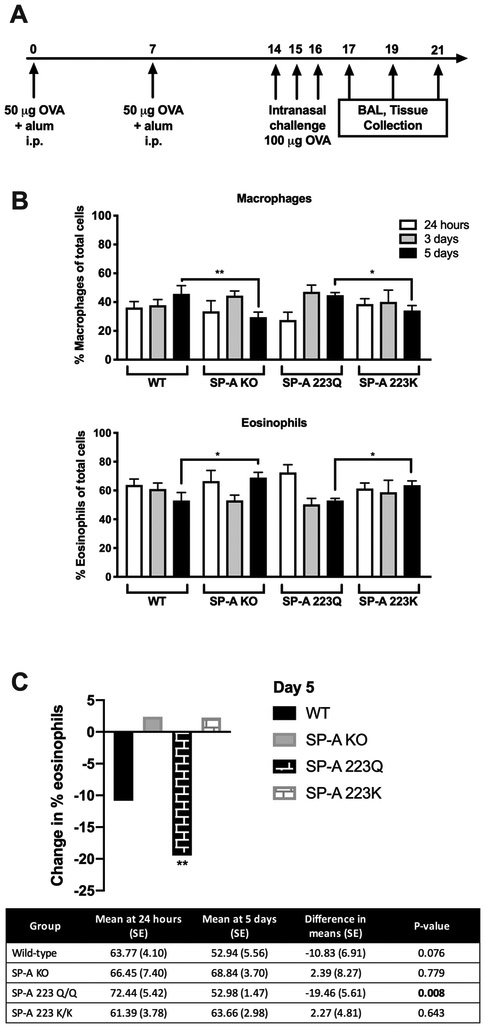
To determine whether the genetic variation at position 223 of SP-A2 plays a role in mediating the immune response to allergen challenge, we used the OVA-sensitization and challenge protocol (Fig. 1A). At 24 hours post terminal challenge, an increased presence of eosinophils was observed in the bronchoalveolar lavage fluid (BALF) in all groups; mice harboring the 223Q allele (223Q mice) exhibiting the most robust influx of eosinophils (Fig. 1B, Supplemental Fig. 1C). However, at 5 days post terminal challenge, mice deficient in SP-A (KO mice) and those with the 223K allele (223K mice) remained in a state of significantly enhanced eosinophilia (Fig. 1B, Supplemental Fig. 1C). When eosinophil recruitment and resolution is quantified over time, the 223Q mice had the highest frequency of eosinophils immediately after OVA challenge, but they also had the largest net decrease in eosinophils compared to all other groups by day 5 (Fig. 1C, Supplemental Fig. 1C). In contrast, eosinophil numbers in the BALF of 223K mice remained relatively unchanged from 24 hours to 5 days (Fig. 1C, Supplemental Fig. 1C).
Figure 1. Assessment of BALF eosinophilia over time.
A) OVA model of allergic airways. B) Cell distribution in BALF at 24 hours, 3 days and 5 days post-terminal challenge. C) Net change in eosinophil frequencies over time. Table shows difference in means at 24 hours and 5 days, unpaired Student’s t-test. One-way ANOVA with Bonferonni’s correction for multiple comparisons, *p<0.05, **p<0.01 Data (mean ± SEM) are from at least two independent experiments with n = 3-5 mice/group.
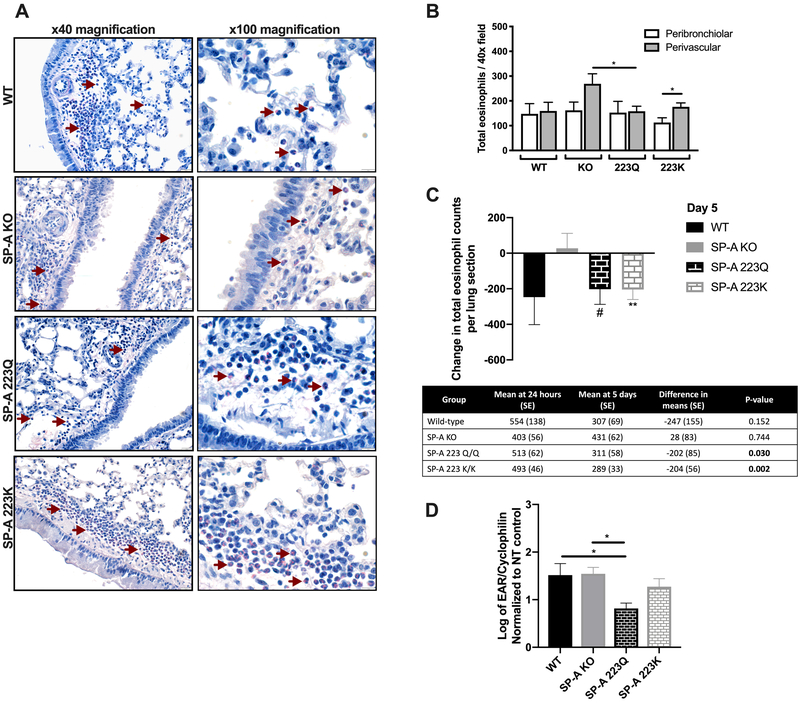
To assess eosinophil infiltration in mouse lung tissue, histochemical staining was performed with Sirius Red stain. Focal points of accumulated peribronchiolar and perivascular eosinophils were identified and quantified (Fig. 2A). There was a notable increase in the mean perivascular eosinophil counts compared to peribronchial eosinophil counts in OVA-challenged 223K mice and an increased trend in OVA-challenged KO mice (p = 0.06) (Fig. 2B), which could suggest an altered communication between these two compartments. Additionally, there was an increased trend in the perivascular eosinophil counts in the KO mice compared to the WT mice (p = 0.06) and a significant increase compared to the 223Q mice. Although overall eosinophil numbers in the lung tissue between groups were not different (Fig. 2A and 2B), mice deficient in SP-A had persistent tissue eosinophilia at 5 days post challenge, whereas both the 223Q and 223K mice had significantly decreased tissue eosinophil counts, in comparison to their respective eosinophil counts at 24 hours (Fig. 2C). An examination of eosinophil-associated ribonuclease (EAR) mRNA as a marker of lung eosinophil activation showed that the 223Q mice had the lowest expression of EAR among the four groups (Fig. 2D).
Figure 2. Assessment of tissue eosinophilia over time.
A) Representative bright field images of eosinophils (red arrows indicate representative eosinophils with bright pink-stained cytoplasm) in lung tissue by Sirius red staining (left panel: 40x magnification, right panel: 100x magnification) and B) quantification of eosinophil counts at day 5. C) Net change in eosinophil frequencies over time. Table shows difference in means at 24 hours and 5 days, unpaired Student’s t-test, #p<0.05, **p<0.01 D) EAR mRNA in lung tissue at 5 days post-terminal challenge. One-way ANOVA with Bonferonni’s correction for multiple comparisons, *p<0.05. Data (mean ± SEM) are from at least two independent experiments with n = 3-5 mice/group.
Genetic variation in SP-A2 does not alter eotaxin expression
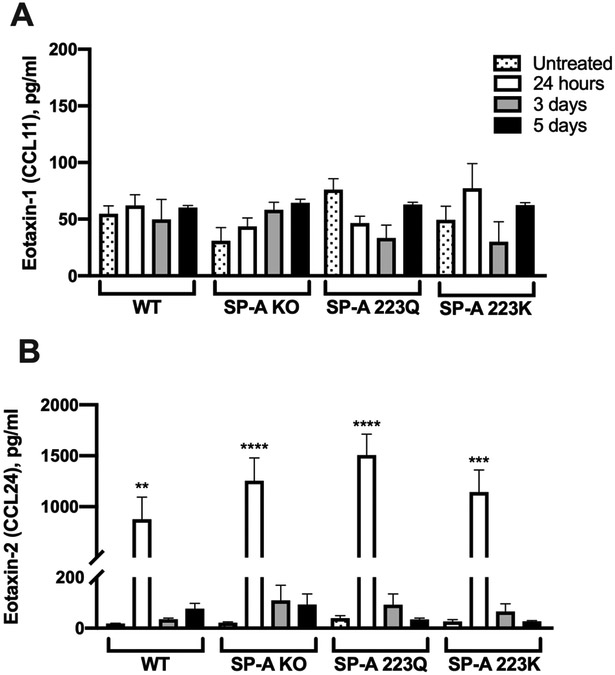
Since the presence of SP-A was associated with a decline in eosinophil counts from 24 hours to 5 days in the lung tissue (Fig. 2), we next sought to determine whether SP-A played a role in regulating the production of eotaxins. Eotaxins are potent inducers of eosinophil movement. Both eotaxin-1 and eotaxin-2 have been previously shown to be elevated in the OVA model: eotaxin-2 was significantly higher in BALF and tissue, while eotaxin-1 was significantly increased only in tissue, over their respective controls (28). We therefore investigated the levels of these two eotaxins in the BALF 24 hours, 3 and 5 days after OVA challenge. Similar to what others have found, eotaxin-1 was not significantly elevated in OVA-treated mice over their respective untreated controls (Fig. 3A). Additionally, there were no differences in eotaxin-1 observed between OVA-challenged groups during the time course of the study (Fig. 3A). Although eotaxin-2 levels in BALF were significantly elevated at 24 hours over untreated controls, these levels were quickly diminished by days 3 and 5 in all groups (Fig. 3B).
Figure 3. Examination of eotaxins in BALF after OVA challenge.
Analysis of protein concentrations by ELISA of A) eotaxin-1 or CCL11, and B) eotaxin-2 or CCL24. One-way ANOVA with Bonferroni’s correction for multiple comparisons. Data (mean ± SEM) are from at least two independent experiments with n = 3-5 mice/group.
SP-A is a chemoattractant for eosinophils
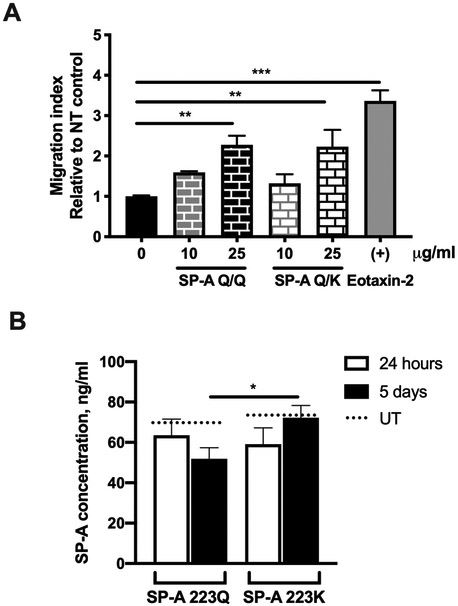
As there were no differences based on SP-A genotype in the levels of eotaxins in the BALF of OVA-challenged mice, we next sought to examine whether SP-A contributes to eosinophil movement by acting as a chemoattractant. Indeed, in an in vitro transmigration assay, SP-A, used at physiologic concentrations typically found in the lung, acted as a chemoattractant for eosinophils (Fig. 4A). Regardless of the polymorphism at position 223, this chemoattractant ability was comparable to the positive control, eotaxin-2.
Figure 4. Evaluation of the ability of SP-A to induce eosinophil migration in vitro.
A) Migration of mouse eosinophils was measured by a plate-based assay. Migration index calculated as number of live eosinophils in the bottom chamber over control. B) SP-A concentrations in BALF of OVA challenged mice, UT = untreated. One-way ANOVA with Bonferonni’s correction for multiple comparisons. *p<0.05, **p<0.01, ***p<0.001. Data (mean ± SEM) are from at least two independent experiments with n = 2-3 replicates/treatment or 3-5 mice/group.
It has been previously shown that surfactant protein levels in allergen-challenged WT mice were unchanged in the BALF compared to untreated controls (18). It has also been shown that both strains of the naïve humanized SP-A2 transgenic mice have similar SP-A levels in the BALF (20). To determine whether SP-A concentration after OVA challenge was altered and thus could be a contributing factor in eosinophil movement into the lung lumen, we examined the BALF of OVA-challenged 223Q and 223K mice for overall SP-A concentration. SP-A levels were similar in the BALF of 223Q and 223K mice at 24 hours post-terminal OVA challenge, both of which were not significantly different from untreated (UT) levels (20) (Fig. 4B). However, at day 5, 223K mice had significantly higher levels of SP-A compared to 223Q mice (Fig. 4B).
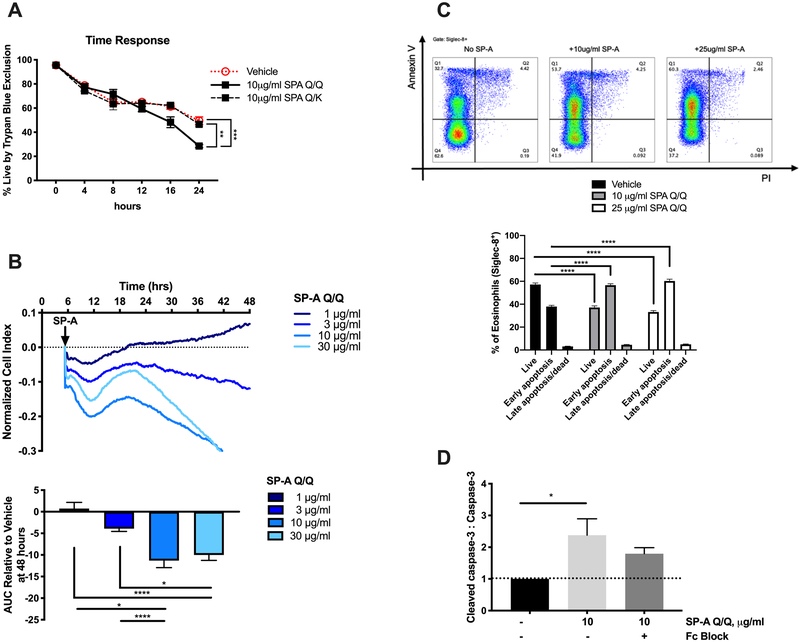
Genetic variation in SP-A2 alters the ability of SP-A to reduce eosinophil viability
It is known that SP-A can bind to eosinophils in a dose-dependent manner and that this binding is partially mediated by CD16/32 (FcγRIII/FcγRII) (18). Thus, we hypothesized that SP-A may be binding to eosinophils to initiate apoptosis and clearance. Here we examined whether the difference in eosinophil resolution was due to modulation of eosinophil cell death by SP-A. In fact, direct stimulation of purified eosinophils from IL-5 transgenic mice by isolated human SP-A homozygous for the Q allele resulted in significantly greater eosinophil death over a 24-hour period compared to heterozygous SP-A (Q/K) (Fig. 5A). To more directly assess the ability of SP-A to induce cytotoxicity in eosinophils, we evaluated SP-A addition using xCELLigence Real-Time Cell Analyzer (RTCA) (Fig. 5B). Addition of SP-A greater than 3 μg/ml resulted in concentration-dependent cytotoxicity that developed in minutes and persisted for up to 48 hours of measurement. To evaluate whether human eosinophils would respond similarly, leukocytes from the peripheral blood of asthmatic patients were stained for apoptosis after incubation with human SP-A. Flow cytometric analysis of these cells allowed us to identify the eosinophil (Siglec-8+) population and, further, detect a shift from live (Annexin V−, PI−) to early apoptotic (Annexin V+, PI−) within the gated eosinophil population (Fig. 5C). This phenomenon appears to be unique to eosinophils as SP-A did not have the same effect on neutrophils, another important inflammatory cell in airway inflammation and asthma (Supplemental Fig. 2). In addition, caspase-3 cleavage was also increased in SP-A-treated eosinophils compared to vehicle; however, this increase was not significantly abrogated by blocking the CD16/32 (FcγRIII/FcγRII) (Fig. 5D).
Figure 5. Evaluation of the ability of SP-A to induce eosinophil apoptosis in mouse and human eosinophils in vitro.
A) Time course of viability assessed by Trypan Blue and B) RTCA tracing and dose response of in vitro stimulation of mouse eosinophils by SP-A, AUC = area under the curve. C) Representative flow diagrams of human eosinophil apoptosis and cell death by Annexin V and PI and quantification after 16 hours incubation with SP-A; live = Annexin V−, PI−, early apoptosis = Annexin V+, PI−, late apoptosis/dead = Annexin V+, PI+ D) Densitometry of caspase-3 by Western blot of mouse eosinophils standardized to non-treated control. ANOVA with correction for multiple comparisons, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Data (mean ± SEM) are from at least two independent experiments with n = 2-3 replicates/treatment.
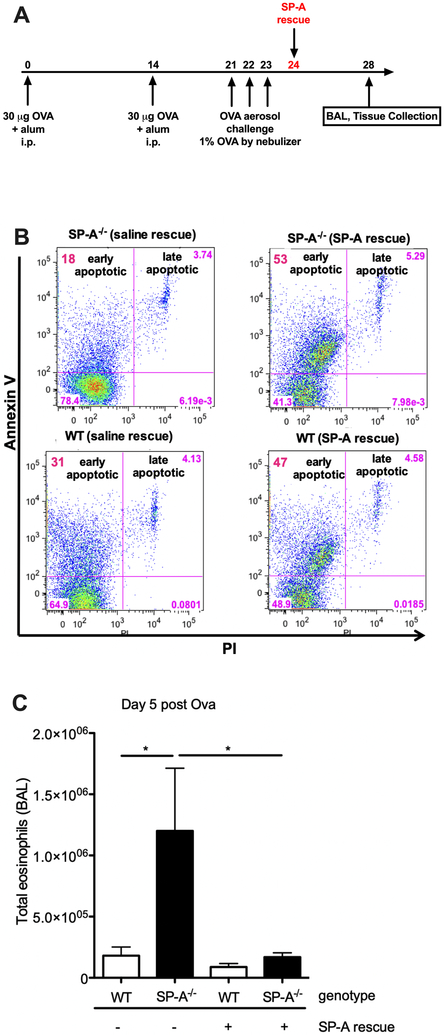
Treatment with exogenous SP-A enhances eosinophil apoptosis in allergic airways
In Fig. 5, we showed that either the lack of SP-A or presence of an altered isoform of SP-A led to prolonged eosinophil survival, which could explain the persistent eosinophilia in the OVA-challenged KO and 223K mice. We sought to determine whether replacement of SP-A in SP-A KO mice would rescue this effect. SP-A KO mice were given OVA and subsequently treated with exogenous human SP-A homozygous for the Q allele at position 223 one day after post-terminal aerosol challenge (Fig. 6A). BALF and lung tissue were collected 5 days after aerosol challenge (Fig. 6A). Indeed, SP-A KO mice given exogenous SP-A had an increased number of apoptotic eosinophils compared to those given saline (Fig. 6B). Conversely, total live eosinophils in the BALF were decreased in the SP-A KO mice given the rescue treatment (Fig. 6C). Although not statistically significant, WT mice given exogenous SP-A also had a trend towards increased apoptotic eosinophils and decreased live eosinophils in the BALF compared to their vehicle controls (Fig. 6B and 6C).
Figure 6. Evaluation of the effect of exogenous SP-A administration on eosinophils in SP-A deficient mice after OVA challenge.
A) Schematic of OVA challenge and SP-A rescue. B) Representative flow diagrams of eosinophil apoptosis and cell death by Annexin V and PI. C) Total live eosinophil counts in BALF 5 days post-terminal challenge. *p<0.05. Data (mean ± SEM) are representative of two independent experiments with n = 5 mice/group.
DISCUSSION
Here, we present novel findings from our investigations regarding the role of SP-A in mediating the resolution of eosinophilia using a well-established model of allergic airway disease. Not only did we discover that SP-A is chemoattractant for eosinophils at physiologic concentrations, we also found that SP-A can induce eosinophil apoptosis. In humans, SP-A is comprised of products from two SP-A genes, both of which have known polymorphisms within the human population. Of those polymorphisms associated with disease, variation at position 223, which changes a glutamine (Q) to a lysine (K) was of special interest to us given the findings that recombinant SP-A containing the 223K was less active against eosinophil degranulation as 223Q (6).
Using SP-A humanized mice that represent the genetic variation of interest, amino acid substitution of a Q for a K at position 223 in the CRD of SP-A2, we were able to discern differences with respect to their resolution of eosinophilia in an allergic model. We further show that SP-A isolated from humans containing either Q/Q or Q/K at position 223 was chemoattractant for eosinophils at relatively the same rate. In contrast, only SP-A containing Q/Q was able to effectively induce eosinophil apoptosis in vitro and mice expressing the Q/Q had significantly enhanced resolution of eosinophilia as compared to the K/K mice in vivo. Therefore, in some individuals, SP-A has the capacity to play dual roles during the resolution of allergic airway inflammation through mediation of eosinophil clearance from the lung tissue by directly promoting chemotaxis, which is independent of position SP-A2 Q223K, and aiding in apoptosis of eosinophils in the lumen, which is affected by the polymorphism at position SP-A2 Q223K.
Compared to 223Q mice, 223K mice had significantly more eosinophils in the BALF at 5 days post terminal OVA challenge, whereas the overall eosinophil numbers in the lung tissue were not different between groups. Although IL-5 plays a primary role in the expansion of the eosinophil cellular pool in the bone marrow and peripheral blood (29), eosinophils traffic from the lung tissue into the lung lumen predominantly by chemotaxis mechanisms during allergen-induced eosinophilia (30). Upon examination of chemotactic factors that could influence eosinophil movement from lung tissue into the bronchoalveolar compartment, eotaxin levels were not different between the different genotypes of mice. This eliminates the likelihood that the eosinophil differences detected between the groups were attributable to eotaxin production.
We next tested the ability of SP-A to be a chemoattractant for eosinophils. Human SP-D, which, like SP-A, has sequence homology to its murine counterpart, is known to inhibit eotaxin-induced migration of eosinophils (31). However, little is known about the role of SP-A in eosinophil chemotaxis. We discovered that SP-A was chemoattractant for eosinophils regardless of the polymorphism at position 223 of SP-A2. This further highlights the significant contribution of SP-A to the net decrease in eosinophil counts in the lung tissue of 223Q and 223K mice by day 5 post-OVA challenge. We assessed SP-A levels in the humanized mice to rule out the possibility that the humanized 223Q mice had more SP-A available and thus had a better resolution of eosinophilia. On the contrary, we discovered that SP-A was significantly increased in the BALF of 223K mice compared to 223Q mice at day 5 during resolution. Therefore, we cannot rule out that the increased levels of SP-A detected in the BALF of 223K mice contributes to the increased presence of eosinophils in those mice at day 5 during resolution. However, KO mice, which are completely deficient in SP-A, had similar persistent eosinophilia as the 223K mice at 5 days after allergen challenge, which supports the possibility of a separate non-chemotactic contributing mechanism to this phenomenon.
In this vein, we next examined the ability of SP-A to induce eosinophil apoptosis. It is well-recognized that eosinophilia has critical contributions to the pathophysiology of asthma and airway inflammation (32-34). It has been shown that collagen deposition and increased thickness of airway smooth muscle were absent in eosinophil-deficient (Δdbl GATA), allergen-challenged mice (32), which underlie the importance of the timely resolution and clearance of eosinophils during inflammation. In fact, numerous therapies targeting eosinophils, eosinophil-derived products or its trafficking mechanisms are currently available or are being investigated in clinical trials (34-36). We found that SP-A harboring the SP-A2 223Q allele has the ability to induce eosinophil apoptosis, while the presence of at least one 223K allele diminished this activity to the levels of the vehicle control. Although the influence of SP-A as a chemoattractant may play a small role by marginally increasing the migration of eosinophils into the lung lumen, our data indicate that SP-A as an inducer of apoptosis is the foremost contributing mechanism to the resolution of eosinophilia in the allergic airways of OVA-challenged mice. More importantly, SP-A was able to induce apoptosis in human eosinophils, attesting to the translational nature and the clinical relevance of these findings.
This work was further strengthened by our studies in which allergic mice were given “rescue” SP-A. By flow cytometry, SP-A KO mice had a greater proportion of live (Annexin V−, PI) eosinophils as compared to WT mice on day 5 during the resolution phase. When treated with exogenous SP-A, both WT and SP-A KO BALF samples had a shift from live (Annexin V−, PI+) to early apoptotic (Annexin V+, PI−). This resulted in a significant clearance of eosinophilia in KO mice as compared to their vehicle controls.
We have shown that SP-A has the ability to mediate release of eosinophil peroxidase (EPO) upon stimulation with a pulmonary pathogen, Mycoplasma pneumoniae (Mp) (18). And while in vitro stimulation of eosinophils with Mp in the presence of recombinant SP-A2 with Q present at position 223 significantly inhibited EPO release, SP-A2 with K present at position 223 was less effective (6). The significantly decreased EAR expression in the lungs of the OVA-treated 223Q mice, but not in the 223K mice, would suggest a potentially similar mediation by SP-A in limiting eosinophil activation. Importantly, the induction of apoptosis by SP-A, with 223Q being more active than SP-A with 223K, parallels our previous findings that SP-A 223Q is more protective against eosinophil degranulation (6). Overall, we believe the mechanism of action for SP-A 223Q is to bind to eosinophils and limit their degranulation while promoting their apoptosis and clearance from the lung.
One important limitation in our study is that the prevalence of SP-A homozygous for the K allele is less than 8% in the general population (37). As a result, we have had difficulty recruiting for this particular genotype from which to isolate human SP-A 223K/K. Therefore, there is a possibility that the chemoattractant capability will be altered when both alleles contain a K at position 223. Another important limitation is our inability to test our results observed in the OVA allergic model on another equally relevant allergic model, house dust mite (HDM). Two common house dust mite species, Dermatophagoides pteronyssinus (Der p) and Dermatophagoides farinae (Der f), both possess cysteine protease activity and have been shown to cleave native human SP-A purified from BALF (38). This occurred in a time- and dose-dependent manner, affecting several biological functions of SP-A, thus making it difficult to ascertain the activity differences due to degradation within the animal model.
Our results show that SP-A aids in the resolution of allergic airways by inducing eosinophil apoptosis. We have reason to believe this induction of apoptosis may be specific for eosinophils. We did not observe an induction of apoptosis by SP-A for isolated neutrophils (Supplemental Fig. 2). In fact, SP-A has been shown to protect against lung epithelial cell apoptosis in bleomycin-induced acute lung injury (39), further supporting the notion that induction of apoptosis may be limited to eosinophils. Additionally, although binding of SP-A to eosinophils was previously shown to be partially mediated by CD16/32 (FcγRIII/FcγRII) (18), the induction of apoptosis on eosinophils does not seem to be facilitated through this receptor. Thus, further studies, which are beyond the scope of this manuscript, will involve the identification of the receptor(s) and downstream signaling processes responsible for this event.
Our findings raise the potential merit in the use of derivatives of specific SP-A variants (ie. SP-A2 223Q/Q) as replacement therapy. It also warrants consideration of the utility of SP-A genotyping in future precision medicine initiatives for better treatment of lung diseases. Further studies with patient cohorts are needed to investigate which specific asthma endotypes would benefit from this type of therapeutic intervention. While the presence of at least one K allele is relatively low (20 – 25%) in the general population, it is enriched (35%) within the African American subgroup (37). Of note, the prevalence of SP-A homozygous for the K allele is over 12% in the African American subgroup, compared to less than 8% in the general population (37). According to the Office of Minority Health of the United States Department of Health and Human Services, nearly 2.6 million non-Hispanic blacks are reported to have asthma and African Americans were three times more likely to die from causes related to asthma than Whites. Although other factors, such as socioeconomic status, play a role in these disparities, this also suggests that there is value in the consideration of specific subgroups who are at increased risk in future investigations for SP-A differences in association with eosinophilic asthma.
Supplementary Material
KEY POINTS.
Surfactant protein-A (SP-A) aids in the resolution of allergic airway inflammation
SP-A promotes eosinophil clearance through chemotaxis and apoptosis
Genetic variation alters the ability of SP-A to induce eosinophil apoptosis
Acknowledgements
We thank the late Dr. James J. Lee for the generous gift of the IL-5 transgenic mice. We also thank Sarah David and Chelsea Large (Asthma and Airway Disease Research Center, Tucson AZ) for the recruitment of volunteers, Akarsh Manne for technical assistance, as well as, the laboratory of Dr. Francesca Polverino for the use of their microscope to obtain high resolution images of our histological slides.
Grant Support: This work was funded by NIH HL-125602 (Ledford), U19 AI-125357 (Kraft), ADHS16–162519 (Kraft) and AI-135935 (Ledford).
REFERENCES
- 1.Wright JR 2005. Immunoregulatory functions of surfactant proteins. Nat Rev Immunol 5: 58–68. [DOI] [PubMed] [Google Scholar]
- 2.Wang Y, Voelker DR, Lugogo NL, Wang G, Floros J, Ingram JL, Chu HW, Church TD, Kandasamy P, Fertel D, Wright JR, and Kraft M. 2011. Surfactant protein A is defective in abrogating inflammation in asthma. Am J Physiol Lung Cell Mol Physiol 301: L598–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kannan TR, Provenzano D, Wright JR, and Baseman JB. 2005. Identification and characterization of human surfactant protein A binding protein of Mycoplasma pneumoniae. Infect Immun 73: 2828–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Piboonpocanun S, Chiba H, Mitsuzawa H, Martin W, Murphy RC, Harbeck RJ, and Voelker DR. 2005. Surfactant protein A binds Mycoplasma pneumoniae with high affinity and attenuates its growth by recognition of disaturated phosphatidylglycerols. J Biol Chem 280: 9–17. [DOI] [PubMed] [Google Scholar]
- 5.Pastva AM, Mukherjee S, Giamberardino C, Hsia B, Lo B, Sempowski GD, and Wright JR. 2011. Lung effector memory and activated CD4+ T cells display enhanced proliferation in surfactant protein A-deficient mice during allergen-mediated inflammation. J Immunol 186: 2842–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dy ABC, Tanyaratsrisakul S, Voelker DR, and Ledford JG. 2018. The Emerging Roles of Surfactant Protein-A in Asthma. Journal of Clinical and Cellular Immunology 9: 553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang G, Bates-Kenney SR, Tao JQ, Phelps DS, and Floros J. 2004. Differences in biochemical properties and in biological function between human SP-A1 and SP-A2 variants, and the impact of ozone-induced oxidation. Biochemistry 43: 4227–4239. [DOI] [PubMed] [Google Scholar]
- 8.Pastva AM, Wright JR, and Williams KL. 2007. Immunomodulatory roles of surfactant proteins A and D: implications in lung disease. Proc Am Thorac Soc 4: 252–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Löfgren J, Rämet M, Renko M, Marttila R, and Hallman M. 2002. Association between surfactant protein A gene locus and severe respiratory syncytial virus infection in infants. J Infect Dis 185: 283–289. [DOI] [PubMed] [Google Scholar]
- 10.Marttila R, Haataja R, Rämet M, Pokela ML, Tammela O, and Hallman M. 2003. Surfactant protein A gene locus and respiratory distress syndrome in Finnish premature twin pairs. Ann Med 35: 344–352. [DOI] [PubMed] [Google Scholar]
- 11.Fahy JV 2009. Eosinophilic and neutrophilic inflammation in asthma: insights from clinical studies. Proc Am Thorac Soc 6: 256–259. [DOI] [PubMed] [Google Scholar]
- 12.Jacobsen EA, Lee NA, and Lee JJ. 2014. Re-defining the unique roles for eosinophils in allergic respiratory inflammation. Clin Exp Allergy 44: 1119–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park YM, and Bochner BS. 2010. Eosinophil Survival and Apoptosis in Health and Disease. Allergy, Asthma & Immunology Research 2: 87–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green RH, Brightling CE, McKenna S, Hargadon B, Parker D, Bradding P, Wardlaw AJ, and Pavord ID. 2002. Asthma exacerbations and sputum eosinophil counts: a randomised controlled trial. Lancet 360: 1715–1721. [DOI] [PubMed] [Google Scholar]
- 15.van der Wiel E, Ten Hacken NH, van den Berge M, Timens W, Reddel HK, and Postma DS. 2014. Eosinophilic inflammation in subjects with mild-to-moderate asthma with and without obesity: disparity between sputum and biopsies. Am J Respir Crit Care Med 189: 1281–1284. [DOI] [PubMed] [Google Scholar]
- 16.Desai D, Newby C, Symon FA, Haldar P, Shah S, Gupta S, Bafadhel M, Singapuri A, Siddiqui S, Woods J, Herath A, Anderson IK, Bradding P, Green R, Kulkarni N, Pavord I, Marshall RP, Sousa AR, May RD, Wardlaw AJ, and Brightling CE. 2013. Elevated sputum interleukin-5 and submucosal eosinophilia in obese individuals with severe asthma. Am J Respir Crit Care Med 188: 657–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lugogo N, Francisco D, Addison KJ, Manne A, Pederson W, Ingram JL, Green CL, Suratt BT, Lee JJ, Sunday ME, Kraft M, and Ledford JG. 2017. Obese Asthmatics Have Decreased Surfactant Protein-A Levels: Mechanisms and Implications. J Allergy Clin Immunol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ledford JG, Mukherjee S, Kislan MM, Nugent JL, Hollingsworth JW, and Wright JR. 2012. Surfactant protein-A suppresses eosinophil-mediated killing of Mycoplasma pneumoniae in allergic lungs. PLoS One 7: e32436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McIntosh JC, Mervin-Blake S, Conner E, and Wright JR. 1996. Surfactant protein A protects growing cells and reduces TNF-alpha activity from LPS-stimulated macrophages. Am J Physiol 271: L310–319. [DOI] [PubMed] [Google Scholar]
- 20.Ledford JG, Voelker DR, Addison KJ, Wang Y, Nikam VS, Degan S, Kandasamy P, Tanyaratsrisakul S, Fischer BM, Kraft M, and Hollingsworth JW. 2015. Genetic Variation in SP-A2 Leads to Differential Binding to Mycoplasma pneumoniae Membranes and Regulation of Host Responses. Journal of Immunology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korfhagen TR, Bruno MD, Ross GF, Huelsman KM, Ikegami M, Jobe AH, Wert SE, Stripp BR, Morris RE, Glasser SW, Bachurski CJ, Iwamoto HS, and Whitsett JA. 1996. Altered surfactant function and structure in SP-A gene targeted mice. Proc Natl Acad Sci U S A 93: 9594–9599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riffo-Vasquez Y, Ligeiro de Oliveira AP, Page CP, Spina D, and Tavares-de-Lima W. 2007. Role of sex hormones in allergic inflammation in mice. Clin Exp Allergy 37: 459–470. [DOI] [PubMed] [Google Scholar]
- 23.Lee NA, McGarry MP, Larson KA, Horton MA, Kristensen AB, and Lee JJ. 1997. Expression of IL-5 in thymocytes/T cells leads to the development of a massive eosinophilia, extramedullary eosinophilopoiesis, and unique histopathologies. J Immunol 158: 1332–1344. [PubMed] [Google Scholar]
- 24.Meyerholz DK, Griffin MA, Castilow EM, and Varga SM. 2009. Comparison of histochemical methods for murine eosinophil detection in an RSV vaccine-enhanced inflammation model. Toxicol Pathol 37: 249–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borchers MT, Ansay T, DeSalle R, Daugherty BL, Shen H, Metzger M, Lee NA, and Lee JJ. 2002. In vitro assessment of chemokine receptor-ligand interactions mediating mouse eosinophil migration. J Leukoc Biol 71: 1033–1041. [PubMed] [Google Scholar]
- 26.Flynn AN, Hoffman J, Tillu DV, Sherwood CL, Zhang Z, Patek R, Asiedu MN, Vagner J, Price TJ, and Boitano S. 2013. Development of highly potent protease-activated receptor 2 agonists via synthetic lipid tethering. FASEB J 27: 1498–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeng C, Nguyen C, Boitano S, Field JA, Shadman F, and Sierra-Alvarez R. 2018. Cerium dioxide (CeO2) nanoparticles decrease arsenite (As(III)) cytotoxicity to 16HBE14o- human bronchial epithelial cells. Environ Res 164: 452–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ben-Yehuda C, Bader R, Puxeddu I, Levi-Schaffer F, Breuer R, and Berkman N. 2008. Airway eosinophil accumulation and eotaxin-2/CCL24 expression following allergen challenge in BALB/c mice. Exp Lung Res 34: 467–479. [DOI] [PubMed] [Google Scholar]
- 29.Foster PS, Mould AW, Yang M, Mackenzie J, Mattes J, Hogan SP, Mahalingam S, Mckenzie AN, Rothenberg ME, Young IG, Matthaei KI, and Webb DC. 2001. Elemental signals regulating eosinophil accumulation in the lung. Immunol Rev 179: 173–181. [DOI] [PubMed] [Google Scholar]
- 30.Pope SM, Zimmermann N, Stringer KF, Karow ML, and Rothenberg ME. 2005. The eotaxin chemokines and CCR3 are fundamental regulators of allergen-induced pulmonary eosinophilia. J Immunol 175: 5341–5350. [DOI] [PubMed] [Google Scholar]
- 31.von Bredow C, Hartl D, Schmid K, Schabaz F, Brack E, Reinhardt D, and Griese M. 2006. Surfactant protein D regulates chemotaxis and degranulation of human eosinophils. Clin Exp Allergy 36: 1566–1574. [DOI] [PubMed] [Google Scholar]
- 32.Humbles AA, Lloyd CM, McMillan SJ, Friend DS, Xanthou G, McKenna EE, Ghiran S, Gerard NP, Yu C, Orkin SH, and Gerard C. 2004. A critical role for eosinophils in allergic airways remodeling. Science 305: 1776–1779. [DOI] [PubMed] [Google Scholar]
- 33.Adamko D, Lacy P, and Moqbel R. 2004. Eosinophil function in allergic inflammation: from bone marrow to tissue response. Curr Allergy Asthma Rep 4: 149–158. [DOI] [PubMed] [Google Scholar]
- 34.Rothenberg ME, and Hogan SP. 2006. The eosinophil. Annu Rev Immunol 24: 147–174. [DOI] [PubMed] [Google Scholar]
- 35.Flood-Page P, Menzies-Gow A, Phipps S, Ying S, Wangoo A, Ludwig MS, Barnes N, Robinson D, and Kay AB. 2003. Anti-IL-5 treatment reduces deposition of ECM proteins in the bronchial subepithelial basement membrane of mild atopic asthmatics. J Clin Invest 112: 1029–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bel EH, and Ten Brinke A. 2017. New Anti-Eosinophil Drugs for Asthma and COPD: Targeting the Trait! Chest 152: 1276–1282. [DOI] [PubMed] [Google Scholar]
- 37.Database of Single Nucleotide Polymorphisms (dbSNP). National Center for Biotechnology Information, National Library of Medicine, Bethesda, MD. [Google Scholar]
- 38.Deb R, Shakib F, Reid K, and Clark H. 2007. Major house dust mite allergens Dermatophagoides pteronyssinus 1 and Dermatophagoides farinae 1 degrade and inactivate lung surfactant proteins A and D. J Biol Chem 282: 36808–36819. [DOI] [PubMed] [Google Scholar]
- 39.Goto H, Ledford JG, Mukherjee S, Noble PW, Williams KL, and Wright JR. 2010. The role of surfactant protein A in bleomycin-induced acute lung injury. Am J Respir Crit Care Med 181: 1336–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.