Abstract
Introduction:
For decades, the role of glycans and glycoproteins in the progression of breast cancer and other cancers have been evaluated. Through extensive studies focused on elucidating the biological functions of glycosylation, researchers have been able to implicate alterations in these functions to tumor formation and metastasis.
Areas Covered:
In this review we summarize how changes in glycosylation are associated with tumorigenesis, with emphasis on breast cancers. An overview of the changes in N-linked and O-linked glycans associated with breast cancer tumors and biofluids are described. Recent advances in glycomics are emphasized in the context of continuing to decipher the glycosylation changes associated with breast cancer progression.
Expert Opinion:
While changes in glycosylation have been studied in breast cancer for many years, the clinical relevance of these studies has been limited. This reflects the inherent biological and clinical heterogeneity of breast cancers. Glycomics analysis lags behind the advances in genomics and proteomics, but new approaches are emerging. A summary of known glycosylation changes associated with breast cancer is necessary to implement new findings in the context of clinical outcomes and therapeutic strategies. A better understanding of the dynamics of tumor and immune glycosylation is critical to improving emerging immunotherapeutic treatments.
Keywords: biomarkers, breast cancer, glycan, glycosylation, mass spectrometry
1. Glycosylation and Breast Cancer – Past to Present
The role of the extracellular carbohydrate coat that surrounds cells, termed the glycocalyx, has been evaluated in breast tumor formation and progression since 1952 [1]. Approximately half of all human proteins are glycosylated, and the majority of FDA-approved cancer biomarkers are comprised of glycoproteins or carbohydrate antigens [2, 3, 4, 5, 6, 7]. Glycoproteins can be ideal biomarkers because they enter circulation from tissues or blood cells through active secretion or leakage, making them assessable for analysis through serum [8]. Glycans serve as one of the initial points of contact during cell to cell interactions, so therefore changes in glycan biosynthesis due to disease can be more apparent than disease related changes to proteins [6]. In the 1960s, it was noted that plant lectins, proteins that bind carbohydrate structural motifs, displayed a heightened binding affinity for tumor cells compared to non-tumor cells, indicating the presence of specific mucopolysaccharides on tumor cells [9]. By the 1980s, biochemical studies of human breast cancers using lectin receptor assays demonstrated that these could be used to predict tumor differentiation and therapy response [10]. This era also brought the first evidence that the specific activities of enzymes involved in glycosylation (i.e., glycosyltransferases) were differentially expressed between normal and tumor cells [11]. In tumor cells, the activities of sialyl- and fucosyltransferases were elevated while the activities of galactosyl- and N-acetylglucosaminyltransferases were reduced when compared to normal cells [11]. Shortly after, it became widely accepted that analyzing glycosylation changes could be used for the discovery of relevant breast cancer biomarkers [2, 12, 13, 14, 15]. It was found that O-linked Thomsen-Friedenreich and Tn epitopes were autoimmunogenic pancarcinoma antigens that played an important role in the invasion of breast cancer cells [16, 17]. While these antigens were rarely expressed in benign tissues, they were found to be immunoreactive in approximately 90% of all carcinomas [16]. CA15-3, a highly glycosylated mucin-1 (MUC1) epitope, was also found to be highly upregulated in breast cancers, and quickly became one of the first serum biomarkers for breast cancer [2, 13, 18, 19]. Despite later discoveries finding that this test lacked clinical specificity and sensitivity [2, 13], these findings spurred continued mechanistic evaluation of glycosylation modifications with breast cancers, including the discovery that alpha 2,6 sialylation affects the adhesion capabilities of breast cancer cells [20]. More recent studies have now mapped the tissue distributions and histopathology localizations of N-linked glycans directly in clinical breast cancer tissues [21, 22].
While it is clear that changes in glycosylation are integrally linked with breast cancer development and progression, there remains much to be delineated in regards to how glycosylation changes affect tumor growth, responses to therapy, and tumor-stroma interactions with the immune system. This review will address the role of glycosylation in breast cancer carcinogenesis and progression, with an emphasis on the clinical, diagnostic potential and functional roles of O-linked and N-linked glycoconjugates. The biochemical basis of glycosylation and potential clinical relevance of breast cancer glycosylation will be summarized, and the role of glycans in possible therapeutic and biomarker uses will be presented. An overview of the current clinical and diagnostic properties of breast cancers and introduction to glycosylation are provided, followed by specific examples of the types of glycosylation changes that are increasingly being identified.
2. Breast Cancer Sub-types and Pathophysiology
Breast cancer remains the leading cause of cancer deaths in women worldwide. The chances of a woman developing this disease in her lifetime have increased significantly over the past few decades from 1 in 11 in 1975 to 1 in 8 in 2016 [23]. While the mortality rate from breast cancers confined to the breast or associated draining lymph nodes is low, the majority of breast cancer deaths are due to the spread of the disease. When breast cancer spreads to other vital organs such as the lung, liver, brain and bones, it can result in an impairment of the organ’s ability to function and ultimately death [24]. Despite the significant increase in early detection and screening technologies, such as the use of mammography, the mortality rate remains fairly unchanged [23]. In 2019 it is estimated that 268,600 new breast cancers will be diagnosed, and 6.9% of all cancer related deaths will result from breast cancer. Furthermore, once a breast cancer is metastatic, the patient’s five-year relative survival declines rapidly to 27.4% [25]. This indicates that while we have made substantial progress in the field of breast cancer research, there is still much more to be discovered in order to reduce the mortality rates of the disease.
Breast cancer is a heterogeneous disease that incorporates several distinct entities with remarkably different biological characteristics and clinical behavior [26]. Because of this, breast cancer is classified in several different manners, one of which being a pathology-based determination of hormone receptor status. Currently, four immunohistochemistry stains are used to classify breast cancer: estrogen receptor (ER), progesterone receptor (PR), human receptor tyrosine-protein kinase erbB-2 (HER2) and Ki-67. Of note, HER2 has multiple sites of N-glycosylation and their presence are linked with function [27]. Based on the presence of these receptors, breast cancer can be characterized as Luminal A, Luminal B, HER-2 positive and triple negative (TNBC) [28]. Among these subtypes, HER-2 positive breast cancer is common in younger women and has a natural history characterized by poor prognosis, high rate of recurrence and mortality without appropriate treatment. While targeted therapies exist for breast cancers that are positive for ER, PR and HER2 (such as endocrine therapies and HER2 targeted therapies), these therapies are ineffective in patients with triple negative breast cancer [29]. Despite triple negative breast cancers only representing approximately 10-15% of all breast cancers, these tumors are characterized by occurrence in a younger patient population, high proliferative activity and a relatively poor outcome even if treated with aggressive multi-agent chemotherapy [29, 30].
While no targeted therapies for triple negative breast cancer exist, there are targeted treatments available for patients with ER and HER2 positive breast cancers. Hormonal therapies, including aromatase inhibitors and selective ER modulators are actively used to treat patients with ER positive breast cancers [31]. Monoclonal antibodies, such as Trastuzumab [32], and receptor tyrosine kinase inhibitors, such as Lapatinib [33], are used to treat patients with HER2 positive breast cancers. However, all of these targeted therapies have associated issues with acquired resistance. Despite the fact it is known that breast cancer is a highly heterogeneous disease, the majority of patients diagnosed with a specific breast cancer subtype receive the same treatment, implying that all breast cancers within a given subtype are identical, even though it has been repeatedly proven that they are not [30, 34, 35, 36, 37]. This demonstrates the strong need for a push within the field for a greater understanding of breast cancer heterogeneity, more specific biomarkers, and greater alternatives for personalized medicine.
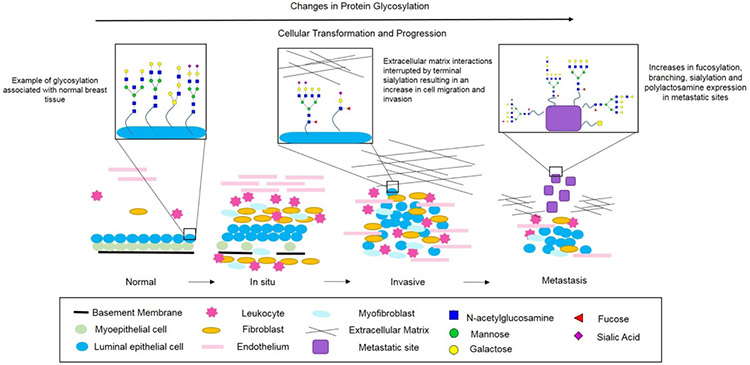
Breast cancer initiation can occur due to mutations or abnormal changes in genes that regulate cell growth and homeostasis [38]. Further disease progression is driven due to the accumulation of additional genetic changes combined with clonal expansion and selection [38, 39]. The most common form of breast cancer originates in the cells lining the milk ducts (ductal cancer), as well as in the milk production glands (lobular cancer) [40]. Depending on location and tumor grade, cancer detected in these regions are referred to as either lobular carcinoma in-situ (LCIS), ductal carcinoma in-situ (DCIS), invasive lobular carcinoma (ILC), or invasive ductal carcinoma (IDC). These diagnoses are determined by analysis of a biopsy and pathologist annotation. On a cellular level, normal breast ducts are composed of a basement membrane with a layer of luminal epithelial cells and a layer of myoepithelial cells. The stroma of these normal ducts contains various leukocytes, fibroblasts, myofibroblasts and endothelial cells. Normal cells can undergo benign proliferative changes or atypical hyperplasia, however, when a transforming event occurs that causes the cell to become potentially cancerous and these genetic/epigenetic changes accumulate, in situ carcinoma is formed. In situ carcinoma is characterized by the initiation of basement membrane degradation, a decrease in the number of myoepithelial cells, and an increase in the number of stromal cells. In situ carcinomas are non-invasive forms of breast cancer that are often referred to as precursor lesions. When a cell becomes invasive, there is a complete loss of the basement membrane and myoepithelial cells. This is also when the cell is able to invade surrounding tissue. Eventually, as the mutated cells accumulate and become metastatic, they are able to migrate and invade distant organs [38, 39, 41]. These molecular changes at the cell and tissue level are all linked with changes in glycosylation of individual glycoproteins in the cell membrane and extracellular matrix, and are summarized in this context in Figure 1, and further described in the next section.
Figure 1:
Schematic Overview of Normal, in situ, Invasive and Metastatic Breast Carcinoma Progression. Normal breast ducts contain a basement membrane with a layer of myoepithelial cells and a layer of luminal epithelial cells with various endothelial cells, fibroblasts and leukocytes in the stroma. Glycans on normal breast epithelium are basic bi- and tri-antennary structures typically lacking expression of core-fucosylated structures. As in situ carcinoma develops, the basement membrane begins to degrade and the number of myoepithelial cells decreases. Additionally, the number of stromal immune cells increases. A cancer is considered invasive when the basement membrane is no longer present, allowing the tumor cells to invade surrounding tissues, enter the blood stream and eventually form distant metastatic sites. As a cancer becomes invasive and metastatic, N-glycans expressed display increases in branching, sialylation and fucosylation. Additionally, O-glycans decrease in chain length and the expression of Tn antigen, Thomsen-Friedenreich antigen, sialyl-T antigen and sialyl-Tn antigen increases.
3. Glycosylation Overview and Breast Cancer
Complex carbohydrates are involved in almost all basic functions of multicellular organisms and are collectively localized in the glycocalyx that surrounds cells, i.e., the dense meshwork of glycoproteins, glycolipids and glycosaminoglycans attached to membrane proteins, lipids and extracellular matrix proteins. This glycocalyx on the outer layer of the cell surface functions to indicate “self” vs. “non-self” to the immune system and activate the innate immune system response mechanisms when necessary [33]. Some of the structural and modulatory functions of glycans include physical protection, expulsion of pathogens, diffusion barriers, and glycoprotein folding. Glycans are also involved in extrinsic recognition of bacterial and viral pathogens, as well as the intrinsic recognition of self-associated molecular patterns (SAMPs) and intercellular signaling [42]. The presence or absence of glycans has been shown to affect the membrane half-life of many membrane receptor proteins [43] including, glucose transporters [44], cytokine receptors [45], transforming growth factor beta (TGFβ) receptor [46], epidermal growth factor receptor (EGFR) [47], and GABAA receptors [48]. It is clear that significant changes in the molecular density of the glycans and glycoproteins in the glycocalyx are required to allow for cell mobility and migration associated with cancer progression (Figure 1).
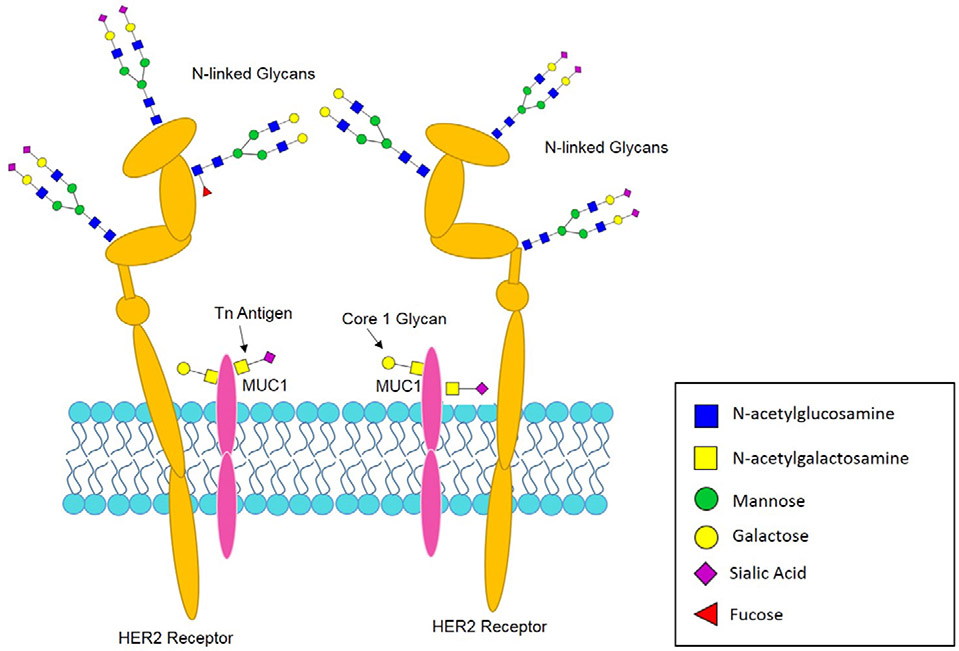
Glycosylation is a term used to describe the biosynthetic enzymatic process that involves the sequential removal and addition of individual carbohydrates to proteins and lipids [49]. For glycoproteins, the attached glycans are classified as either N-linked (Asn) or O-linked (Ser/Thr) based off the amino acid residue to which they are attached. N-linked glycosylation of newly synthesized proteins occurs co-translationally in the endoplasmic reticulum. N-linked glycans are further processed by a highly regulated sequential series of glycosidases and glycosyltransferases in the ER and Golgi apparatus. Individual glycosyltransferases may exhibit overlapping specificities in some instances [50, 51]. These enzymes typically transfer single sugar residues from nucleotide-sugar donors to protein and sugar acceptors, which results in glycan elongation forming a vast array of glycan structures [52]. N-linked glycans most commonly contain mannose, galactose, N-acetylglucosamine, fucose, and sialic acid sugars, and less commonly N-acetylgalactosamine and glucose. O-linked glycans are predominantly comprised of shorter structures with N-acetylgluosamine, galactose, N-acetylgalactosmine, sialic acid and fucose. The presence of sulfate and phosphate groups are also possible. Examples of glycoproteins with N-linked and O-linked modifications and carbohydrate antigens associated with cancer are shown in Figure 2 [49]. For a summary of the complex biosynthetic and processing reactions associated with N-linked and O-linked glycans, see the freely available on-line reference Essentials in Glycobiology [53].
Figure 2:
HER2 Receptor and MUC1 as Example Glycoproteins. A schematic diagram indicating the presence of N- and O- linked glycans on common cell surface glycoproteins found in breast cancer.
In cancer, oncogenic transformation associated with changes in glycosylation was first documented over seven decades ago [54, 55, 56]. Numerous studies have shown that tumor cells exhibit altered glycosylation patterns when compared to their non-malignant equivalents [15, 49, 57, 58, 59, 60, 61, 62, 63, 64]. In 1983 Hakomori and Kannagi published results indicating that there were two main mechanisms behind the tumor-associated changes in carbohydrate structures. The first mechanism was referred to as incomplete synthesis, and the second was referred to as neo-synthesis. The incomplete synthesis process was mainly found in early stage cancers, and was trademarked by the impairment of normal complex glycan synthesis in normal epithelial cells. In turn, this led to the biosynthesis of truncated glycan structures. The neo-synthesis process is mainly observed in advanced stage cancers, and is classified by the cancer-associated induction of particular genes involved in carbohydrate expression [65, 66]. Direct analysis of N-glycans in multiple tumor tissue types has detected these same types of molecular changes [21, 67]. Cumulative studies have defined changes in glycan expression to five main biological events or changes: 1) under/over expression of glycosyltransferases [68, 69, 70, 71], 2) changes in tertiary peptide backbone composition, 3) inconsistency of receptor substrates, 4) availability or abundance of sugar nucleotide donors and cofactors [72], and 5) expression and localization of pertinent glycosyltransferases in the Golgi [56, 61, 73]. The most common cancer-associated glycosylation changes are O-glycan truncation, sialylation, fucosylation, and N-glycan branching [56, 74, 75, 76, 77, 78]. These modifications have been directly linked to playing an important role in the progression of cancer, metastasis, and epithelial-mesenchymal transition (EMT) [75], and will each be further described in the context of breast cancer progression and biomarker development.
3.1. O-linked Glycosylation in Breast Cancer
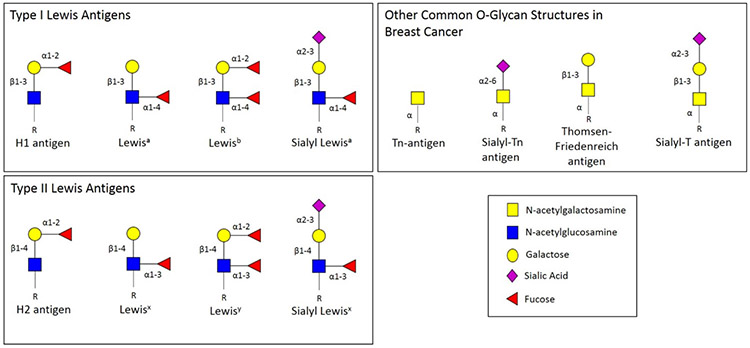
The most common types of O-glycan structures detected in breast cancer and many other cancer types is Tn-antigen, sialyl-Tn, and Thomsen-Friedenreich antigen (see Figure 3) [79, 80, 81, 82, 83, 84, 85, 86, 87, 88]. These structures are most commonly associated with mucin-glycoproteins, and the enzymes responsible for mucin-type O-glycosylation initiation also exhibit altered expression in cancer. These polypeptide GalNAc transferases control O-glycan occupancy sites, and the density with which these sites can be occupied [89, 90, 91]. Furthermore, multiple enzymes competing for the same substrates can also result in O-glycan truncation [92]. Elevation of Mucin-1 (MUC-1) has been noted in breast cancers for many years. MUC-1 is a large transmembrane protein that is carrier of aberrant O-glycosylation in tumor cells. Changes in aberrant O-glycosylation of MUC-1 result in the exposure of the protein core, allowing cells to adhere to distant tissues and resulting in metastasis [75, 90]. Furthermore, when MUC-1 is under-glycosylated, this is associated with higher tumor grades and poor prognosis [93].
Figure 3:
Lewis Antigens Terminal oligosaccharide structures used to classify particular antigens based on the presence of an alpha 1-4 linked fucose (Lewis A, B) or an alpha 1-3 linked fucose (Lewis X, Y) to the GlcNAc monosaccharide. Additionally, sialylated forms of Lewis A and Lewis X structures are also shown, in addition to other common O-glycan structures in breast cancer such as Tn-antigen, sialyl-Tn antigen, Thomsen-Friedenreich antigen and sialyl-T antigen.
Changes in mucin-type glycosylation have been observed in over 90% of breast cancers. Examples of these breast cancer associated changes include increased expression of Tn antigen and the loss of core 2 glycans [94, 95]. Additional alterations in the number, core structure and sialylation of O-glycans have also been linked to breast cancer [92, 96, 97, 98]. Truncated mucin-type O-glycans are often seen with terminating sialic acid residues due to the up-regulation of sialyltransferases in many breast cancers, especially those with positive estrogen receptor status [99]. ER+ cancers carry mainly core 1 based glycans on their O-linked glycoproteins, while ER- breast cancers primarily carry core 2-based glycans. The increased presence of core 2 glycans in ER- breast cancers is most likely a result of the overexpression of core 1 synthase glycoprotein-N-acetylgalactosamine 3-β-galactosyltransferase 1 (C1GALT1) and core 2 glucosaminyl (N-acetyl) transferase 1 (GCNT1) in these tumors [99]. In these cancers, the core 2 structures typically carry sialyl Lewisx glycans, which are not found on core 2 structures in normal breast tissue.
Alterations in breast cancer mucin-type O-linked glycosylation can result in tumor growth and progression through a variety of mechanisms. In the immune system, changes in mucin-type O-linked glycosylation can produce novel interactions between immune cells and lectins. This is demonstrated through the binding of sialylated glycans to sialic acid-binding immunoglobulin-type lectins (siglecs) on monocytes, macrophages and NK cells. Examples of this specific mechanism include the binding of sialylated MUC1 to siglec-9 on monocytes and macrophages, the binding of sialylated LacNAc (found on core 1 or core 2 branches) to siglecs-7 on NK cells, and the binding of Tn and sialylated Tn antigens to macrophage galactose-specific lectin on dendritic cells and macrophages. Furthermore, the expression of sialyl Lewisx can result binding to selectins on endothelial cells and various core glycans which can dictate how the cancer cells metastasize and respond to epidermal growth factor (EGF) binding [58, 88, 100, 101, 102]. Sialyl Lewisx antigens on leukocytes can contribute to inflammatory response as a result of their interaction with E-selectin on endothelial cells. Interactions between selectins and sialyl Lewisx glycans are crucial for immune cell trafficking, indicating that the cancer is exploiting a normal cellular process to aid in metastasis [58, 102]. This interaction exposes sialyl Lewisx antigens at the cell surface, a mechanism that malignant cells capitalize upon for extravasation from the blood circulation and metastasis [103].
3.2. Sialylation in Breast Cancer
The addition of sialic acid to N-linked and O-linked glycoproteins is common in breast cancer. Specific sialylated structures such as Thomsen-Friedenreich antigens, sialyl Lewis antigens, sialyl α2,6-lactosaminyl structures, and polysialic acids that have been shown to mediate cell to cell interactions are altered in cancer cells [104]. This arises from altered sialyltransferase expression that is a result of degradation during transferase biosynthesis [105]. Examples of these antigens include sialyl Lewisa and sialyl Lewisx. Both of these antigens have been documented with high expression in a variety of carcinomas, with sialyl Lewisx expression correlated with poor survival [106, 107]. This is most likely due to the fact that these antigens promote cancer cell adhesion and metastasis by serving as ligands for adhesion receptors expressed in activated endothelial cells [92, 108]. Alternatively, studies analyzing the role of neuraminidase-1 (Neu-1) have shown that the desialylation activity of the glycosidase regulates cancer growth [109]. Furthermore, when Neu-1 is selectively inhibited in in TNBC models, this results in an increase in E-cadherin expression and a decrease in N-cadherin expression, limiting EMT [110].
The presence of cell surface sialic acid patterns can be recognized as ‘self’ by the immune system, and therefore are described as self-associated molecular patterns (SAMPs). Siglecs are sialic acid-binding lectins that are expressed mainly on immune cells that recognize these SAMPs and produce signals to negatively regulate the immune system. Similar to other immune check point receptors such as programmed cell death 1 (PD1), most siglecs contain immunoreceptor tyrosine-based inhibition motifs (ITIMs), demonstrating how hypersialylation seen in cancer can induce the important role of siglecs in cancer immune suppression [58, 111, 112, 113, 114, 115, 116].
In breast cancer, sialyltransferase expression is altered, resulting in cancer associated sialylation increases. MALDI-TOF-MS analysis of the glycosylation profiles of breast cancer cell lines, mouse models and total human serum have all revealed an increase in sialylated glycans in breast cancer that are not found in corresponding normal controls [59, 117, 118]. Furthermore, enzymatic activity of ST3Gal I (β-galactosidase α2,3-sialyltransferase I) is elevated in breast cancer tissues when compared to normal breast tissue, while that of ST3Gal II is decreased [119, 120, 121]. Additionally, increase in ST3Gal I enzymatic activity correlates with the tumor grade and is implicated in the increased expression of sialylated Thomsen-Friedenreich antigen in breast cancer cell lines [105, 122]. This finding was validated by the increased expression of α2,3 sialic acid seen in grade 3 and 4 cancers when compared to that of grade 1 and 2 cancers [59]. This same study compared 50 primary breast tumor cases without lymph node metastasis and 50 pair-matched breast cancer primary tumors with associated lymph node metastasis, and found a higher level of α2,3 sialic acid residues in the pair-matched tumors compared to the primary tumors without metastasis. This demonstrates the high metastatic potential associated with increases in α2,3 sialic acid expression in breast cancer [59].
Various breast cancer cell lines have also been analyzed for sialylation changes. In MDA-MB-231 and MCF-7 cell lines, expression of ST6Gal II (β-galactosidase α2,6-sialyltransferase II) was analyzed using real-time PCR, Western blots and immunohistochemistry. High levels of ST6Gal II expression were associated with the invasive phenotype of the cell lines both in vivo and in vitro [123]. This further validated an analysis from 12 years prior that found α2,6 sialylation on the cell surface of MDA-MB-435 breast cancer tumor cells contributed to their cell-cell and cell-extracellular matrix adhesion capabilities [19]. Sialylated gangliosides, in particular the di-sialylated form GD2, have recently been associated with breast cancer stem cell function [124] and highly expressed in TNBC tissues [125], representing an emerging class of sialylated biomarker candidates. By combining in vitro and in vivo cell line data with patient samples, it can be concluded that sialic acids play a major role in breast cancer formation and metastasis.
3.3. Fucosylation in Breast Cancer
Fucosyltransferases catalyze the addition of fucose moieties to either terminal ends of the glycan structure in N-linked and O-linked structures, or to the core N-acetylglucosamine residues attached to asparagine in N-linked glycoproteins. Structurally, there are two main differences between fucose and other six-carbon sugars: a lack of a hydroxyl group on the C-6 carbon, and an L-configuration. Fucosyltransferases (FUT) are the enzymes responsible for adding fucose residues onto oligosaccharides. This reaction requires that substrate GDP-fucose, which can be synthesized from either that GDP-mannose-dependent de novo pathway or the free fucose-dependent salvage pathway in mammalian cells [126]. Fucose residues can be attached via α1,2-, α1,3-, or α1,4- linkages for outer arm modifications, or α1,6- for core fucosylation linkages. FUT1 or FUT2 are responsible for α1,2-linkages, and FUT3 or FUT4 are responsible for α1,4-linkages. α1,3-linkages can be formed from either FUT3, FUT4, FUT6, FUT7, FUT9, FUT10 or FUT11. Finally, core fucose modifications are facilitated by FUT8. There are no differences in masses for these fucose linkages, despite the fact that each are responsible for the synthesis of different antigens [80, 126].
In cancer, terminal fucosylation is associated with the formation of Lewisx, Lewisy, sialyl Lewisx and sialyl Lewisa antigens. While the regular expression of these antigens are found in normal tissue, when their expression is altered they become associated with malignant phenotypes. In normal tissue, Lewis antigens play important roles in adhesion and communication with various cell types and the surrounding microenvironment. Overexpression of these antigens is usually due to genetic or epigenetic alterations that result in the upregulation of applicable fucosyltransferase (FUT) genes. This enables cancer cells to acquire the ability to proliferate, participate in EMT (resulting in the invasion of other cells and tissues), gain metastatic potential, and generate resistance to chemotherapy [57].
Changes in breast cancer fucosylation have been noted for many decades. In 1971, Rosato et. al. published a study to determine the concentration of protein-bound fucose in the serum of 300 patients with undiagnosed breast masses. This revealed a strong correlation between the presence of breast cancer and higher concentrations of protein bound fucose. Their study proposed the addition of fucose level evaluation to patient work up prior to the decision regarding whether or not a patient should undergo biopsy [127]. Several cell line studies have been reported to attempt to delineate the role of fucosylation in breast cancer tumorigenesis. When analyzing the effect of fucosylation on epidermal growth factor receptor (EGFR), studies found a strong association between increased EGFR core fucosylation and increased EGFR-mediated signaling. Increased EGFR core fucosylation resulted in increased EGFR dimerization and phosphorylation, which lead to increased signaling associated with tumor cell growth and malignancy [47, 92, 128, 129]. In 2017 Carrascal et. al. published a cell line study using an immortalized primary invasive ductal carcinoma (IDC) cell line treated with 2-fluorofucose, an inhibitor of fucosylation. By doing this, the group noticed a loss of sialyl Lewisx and sialyl Lewisa expression, and a reduced ability of the cells to adhere to E-selectin under hemodynamic flow conditions. Additionally, the cells exhibited reduced proliferation and migration rates, and reduced expression of growth factors such as basic fibroblast growth factor (FGF2), vascular endothelial growth factor, and transforming growth factor beta (TGFβ). Furthermore, signal regulating pathways were unable to activate, indicating that IDC fucosylation is linked to several malignant processes, including cell adhesion, migration, proliferation and growth factor expression [130]. In a separate study, Tu et. al. utilized gene expression profiling and western blots to determine the effects of FUT8-mediated core fucosylation of TGFβ on TGFβ-induced epithelial to mesenchymal transition (EMT). Studies showed an upregulation of FUT8 expression correlated with TGFβ-induced EMT that was linked to the invasive and migratory capabilities of the breast cancer cell lines used. This was validated through gain-of-function and loss-of-function studies where FUT8 was overexpressed or knocked down, providing an important link between FUT8-mediated core fucosylation of TGFβ and EMT [131].
In addition to cell line studies, breast cancer tumor tissue studies have been performed to attempt to establish the prognostic value of fucosylated epitopes. In 2013 a retrospective study was performed on 158 triple negative breast cancer patients to assess the expression of Lewisx antigen. Multivariate analysis indicated that positive Lewisx detection was an independent poor prognostic factor for recurrence-free survival and overall survival in patients younger than 50 years old [132]. A different study using immunohistochemistry to analyze the expression of Lewisx in 98 breast tumors found a strong expression pattern associated with the leading edge of the invading tumor, indicating the potential role of Lewisx in breast cancer invasion and metastasis [57, 133]. In an uncommon approach for glycosylation-related studies, an immunohistochemical staining analysis of FUT8 protein expression in breast cancer tissue microarrays was reported [134]. Low FUT8 protein expression was correlated with disease free survival, while high levels of FUT8 correlated with metastasis and tumor stage [134]. These cumulative studies establish a clear connection between fucosylation and breast cancer tumorigenesis and invasion, despite the lack of an exact mechanism behind this connection.
4. Glycan and Glycoprotein Biomarkers
The changes associated with glycan structures in breast cancer, as described in the previous sections and Figures 1 and 2, make them attractive targets as biomarkers for early detection or prognosis. A biomarker is a biologically important signature with unambiguous specificity for a distinct physiological condition [6, 135]. In breast cancer, tests such as Oncotype DX and Mammaprint use preset transcriptomic arrays to help dictate a patient’s adjuvant therapy plan after surgical resection [136, 137, 138, 139]. Predictive biomarkers are used to help assess whether or not a patient will benefit from a particular type of therapy. In breast cancer, HER2 and ER are used as predictive biomarkers. For example, if a patient has a breast cancer that is HER2+, they will likely be treated with a drug that inhibits the HER2 receptor, whereas a patient with a HER2- breast cancer would not [31, 140]. In recent research there has been a large focus on discovery of tissue and serum based biomarkers. Serum biomarkers are of particular interest because of their potential application towards “liquid biopsies” [141].
There have been three main serum glycoprotein immunoassay targets used for breast cancer detection: Cancer antigen 15-3 (CA 15-3) and CA 27-29 on MUC-1 and Carcinoembryonic antigen (CEA), an anchored glycoprotein involved in cell adhesion. However, these biomarkers clinically lacked sensitivity and fostered studies evaluating the glycans attached to the glycoprotein targets [5, 142]. In 2008, preliminary studies using high performance liquid chromatography (HPLC) and matrix-assisted laser desorption ionization time of flight mass spectrometry (MALDI-TOF MS) to analyze fluorescently tagged serum N-glycans from advanced breast cancer patients reported an increase in a mono-fucosylated tri-sialylated N-glycan in cancer patients [143]. Additional serum studies from breast cancer patients with lymph node metastasis, without lymph node metastasis and with benign breast disease showed increased levels of agalactosyl biantennary glycans and glycans containing sialyl Lewisx epitopes in patients with lymph node metastasis when compared to the other two patient groups [143]. An unexplored area for glycosylation biomarkers in the liquid biopsy space is that of cell and biofluid-derived exosome/extracellular vesicles [144]. While primarily associated with differential miRNA cargo, there are certainly glycoproteins present in these vesicles specifically derived from tumor cells. Little is known about differential glycosylation of exosomes, but they certainly represent an untapped source of that could be potential biomarker candidates.
Aberrant glycosylation changes have been linked to breast cancer metastasis because these glycosylation structures aid tumor cells in overcoming the blood brain barrier [75]. In 2011, de Leoz et. al. published a study analyzing glycosylation changes of serum glycoproteins in breast cancer mouse models and patient samples. Using matrix assisted laser desorption/ionization Fourier transform-ion cyclotron resonance mass spectrometry (MALDI-FTICR MS), they observed high mannose glycans were elevated in both breast cancer mouse models and patients when compared to their respective healthy controls [142]. Similar findings were also discovered while profiling the N-glycome of different breast cancer cell lines [62]. This demonstrates the potential clinical implications and utility of analyzing changes in serum glycosylation patterns for breast cancer biomarker discovery.
5. Methodology for Breast Cancer Glycosylation Analysis
The methodologies used for glycomic and glycoproteomic analysis have varied over the last 40 years, and have continued to improve. Much of the new capabilities for direct analysis of the glycans and glycoproteins have evolved with increasingly higher resolution and sensitive mass spectrometry approaches. A recent review highlights the many approaches that are being used [145]. In relation to breast cancer targets, these approaches have been limited. Two approaches recently published [21, 146] highlight the feasibility of glycan and glycoprotein analysis. In one approach [146], multiple frozen tissue specimens from triple negative and luminal (HER2-/ER+/PR+) tumor subtypes were homogenized and digested to the peptide level for subsequent capture of glycopeptides by an established hydrazine bead capture method. Hydrazine will react covalently with oxidized sugars on the glycopeptide. The attached peptides can then be released by treating with PNGase F, and then sequenced by LC-MS methods to identify the formerly glycosylated peptides [147]. Using this approach with label free LC-MS, over 2000 bead captured proteins were identified; 90 glycoproteins were differentially detected in higher abundance in triple negative tumors relative to luminal tumors, and 86 glycoproteins were in higher abundance in the luminal tumor tissues [146]. Further analysis indicated that a subset of 29 glycoproteins in higher abundance in the triple negative tumors could be used with public database transcriptomic data to distinguish triple negative from luminal tumors, as well as be predictive for patient survival [146].
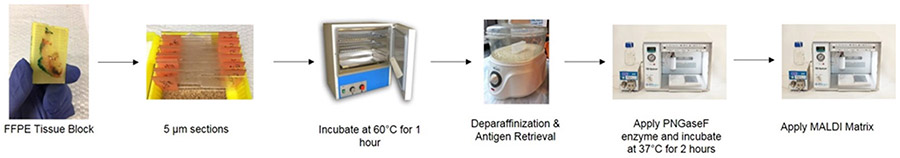
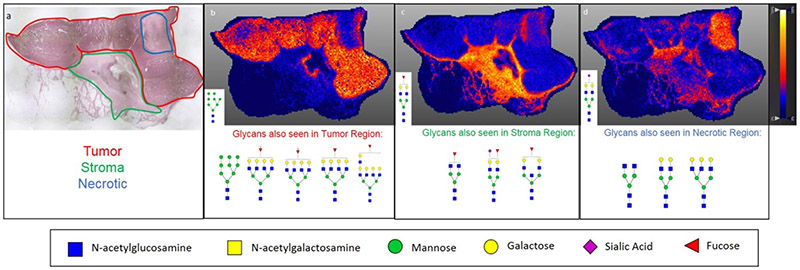
Recently, a new approach for utilizing imaging mass spectrometry for the analysis of glycosylation changes in formalin-fixed paraffin embedded (FFPE) tissue has been reported [148]. This approach is summarized in Figure 4, and the key is the spraying of a molecular coating of peptide N-glycosidase onto the tissue to specifically release N-glycans. This method not only allows for the discovery of N-linked glycosylation changes within a tissue, but it also allows these changes to be spatially localized and correlated with the tissue’s histopathology. Because it is effective with FFPE tissues, any pathology block prepared by standard clinical workflows can potentially be analyzed. Thus, N-linked glycans present in regions of tumor can be compared to adjacent regions of tissue and stroma. This approach has allowed the assessment of N-linked glycans of breast cancer xenograft tissue, FFPE pathology tissues and tumor tissue microarrays. It was found that high mannose and tetra-antennary glycans typically correlate with tumor regions, while some bi-antennary sialylated glycans correlate with regions of necrosis [22]. An example tissue and the types of glycans detected in specific regions are shown in Figure 5.
Figure 4:
Workflow for Preparation of FFPE Tissues for Glycan Analysis by MALDI IMS. FFPE tissue blocks are sliced into 5 ?m sections and placed onto glass slides. Once sectioned slides are prepared, they are placed at 60°C for 1 hour. Next, slides are placed through a series of deparaffinization washes and antigen retrieval. PNGaseF enzyme is then applied in a fine mist to the slides using a HTX TMSprayer to cleave glycans on the slides. Slides are then incubated at 37°C for 2 hours. After incubation, a MALDI compatible matrix is applied. At the conclusion of this process, slides can be stored under vacuum until ready for MALDI IMS analysis.
Figure 5:
Linking Histopathology with MALDI IMS Glycan Images. a) H&E stain of an invasive ductal carcinoma at 2X magnification with pathologist annotation of tumor (red), stroma (green), and necrotic (blue) regions. b) Distribution of Hex8HexNAc2 + 1Na (m/z 1743.5956) showing strong correlation with the tumor region of the tissue. c) Distribution of Hex5dHex1HexNAc2 + 1Na (m/z 1809.6838) throughout stroma region of the tissue. d) Distribution of Hex5HexNAc4NeuAc1 + 1Na throughout necrotic and stromal region of the tissue. Additionally, other glycan structures seen correlating with each region in b-d are shown below the respective image. Images were created using SCiLS Lab 2017a and previously described in Scott et. al. 2018.
6. Conclusions
In conclusion, studying glycosylation changes in breast cancer has the potential to lead to further mechanistic understandings of the disease, in addition to highlighting potential disease biomarkers and drug targets. Across decades of cumulative studies, it is clear that glycosylation changes in breast cancers are significant and detectable. Improvements in analytical instrumentation, especially mass spectrometers, and multi-omic approaches are beginning to lead to better glycan candidates for diagnostics and therapeutics. The ability to co-localize specific glycan structures with tissue specific regions and clinical subtypes through the use of IMS provides region-specific mechanistic insights not available with previous methodologies. Coupling this with advanced glycoproteomic strategies with well-annotated breast cancer samples with known pathology and clinical history will allow a new resurgence in biomarker candidates, in addition to furthering knowledge of the complex interactions of the glycocalyx within the tumor microenvironment.
7. Expert Opinion
The breadth of glycomic analysis in breast cancer research is broad and currently resurging in the era of emerging immunotherapeutics. However, the cumulative results have been heterogeneous in scope, illustrated by the poor performance of MUC-1 as a biomarker. Challenges associated with breast cancer molecular and clinical heterogeneity have been an established issue in the field of breast cancer research for decades. Molecular heterogeneity of breast cancers has been described throughout the transcriptome, genome and proteome [149], thus heterogeneity within the glycome is not unexpected. Current breast cancer biomarkers are intended to maximize patient eligibility for targeted therapies, however they fail to factor in intratumoral heterogeneity. While some proteomic and genomic studies have attempted to identify markers for specific breast cancer subtypes [150, 151, 152, 153, 154, 155], clinical impact has been minimal. Another issue surrounding breast cancer in the clinic is the inability to predict metastasis based on a primary tumor. Currently there are patient-specific tests that utilize gene-expression panels to assess a patients’ risk of recurrence. Patients that come back with high risk of recurrence scores are recommended for chemotherapy to reduce their risk, while patients with low recurrence scores are often spared for chemotherapy regimens. While this type of test has added a whole new level of personalized medicine to breast cancer treatment, there are still many flaws. There is still much debate regarding what to do with patients who receive an “intermediate” risk score on these tests, and these tests are only applicable for ER+ cancers. There is still no way to predict metastasis in patients with HER2+ or triple negative breast cancers. This is one of the ways assessing tumor specific glycosylation patterns has the potential to add to current therapies. If an individual glycan is established as a specific disease state marker, this has the potential to be translated into a clinical assay through the development of a glycan-specific lectin stain. If a glycosylation pattern is established as a specific disease state marker, MALDI imaging mass spectrometry (IMS) can be performed on the already collected FFPE tissue from the patient. Whole tumor glycome analysis could provide information needed to develop a patient-specific treatment plan that could avoid common issues such as drug resistance, recurrence and metastasis.
Direct linkage of tumor associated glycans with their protein carriers is a missing component to developing glycome-based biomarkers. In parallel with continued improvements in the overall sensitivity and more peptide fragmentation/dissociation options on new high resolution mass spectrometers, there has been progress in direct glycopeptide analysis to determine both peptide and glycan sequences using multiple mass spectrometry approaches [156, 157, 158, 159, 160]. Historically, the majority of approaches have relied on removing glycans from their carrier proteins so that each can be analyzed separately. This is because direct analysis of intact glycopeptides by mass spectrometry is inherently challenging in that there is essentially a negative synergy in regards to the ionization and fragmentation parameters required. The optimal conditions for peptide fragmentation and sequencing are too harsh for glycans, and similarly, optimal glycan conditions do not have the requisite energy to fragment peptide bonds. This results in the requirement of more starting material and sub-optimal instrumentation settings for glycopeptides. Complex mixtures of glycoproteins are therefore even more challenging. Interpretation of mass spectra and tools for automated analysis of glycopeptides also lags behind resources available for standard proteomics [157, 161], but is rapidly improving. As highlighted [158], the continued improvements in mass spectrometry analysis, particularly at the glycopeptide level, will be key to coalesce the decades of glycomic analysis research in breast cancer around new disease glycoproteome-based biomarker and therapeutic targets.
Based on current technology and therapeutic trends, we predict that the roles of glycosylation in clinical breast cancer research will center around two main themes, tissue glycopeptide-based analysis and expanded glycan structural class characterizations, and immunotherapies. Recent advances in MALDI IMS have allowed for substantial breakthroughs in the field of clinical tissue glycosylation research in the past 5 years, a key being the spatial co-localization of glycans within FFPE tissues linked directly with histopathology and immunohistochemistry. As this field continues to evolve, the potentials for studying of N-linked glycosylation and other glycosylation structural classes increase exponentially over the next 5 years. With the development of mass spectrometers that allow for faster and higher resolution spatial analysis, the potential for 3-D analysis of glycan distribution within whole tumor blocks has become much more feasible [162]. Three dimensional analysis of whole tumor glycosylation will reveal further insights into the complex glycan network throughout the entire tumor, as opposed to just one representative section. These molecular 3D maps can also be integrated with clinical MRI images and emerging 3D histopathology approaches [163]. Another emerging analytical approach that could assist with deciphering the complexities of fucosylated and sialylated isomers is ion mobility mass spectrometry, an analytical technique that measures the mobility of gas-phase ions through an electric field in the presence of a buffer gas, and increasingly being applied to the separation of glycan isomers [164, 165]. In relation to glycosylated analytes, DESI (desorption electrospray ionization) and MALDI ionization coupled to ion mobility separations were recently reported for tissue imaging of multi-sialylated gangliosides and other glycosphingolipids [166]. It is expected that these tissue imaging MS workflows will continue to facilitate characterization of N-linked glycans, as well as O-glycans, glycosaminoglycans and glycolipids in the complex glycocalyx of breast cancers.
A major question to be addressed for breast cancers and all tumor types is what are the protein carriers of the diagnostic glycans; particularly in tumors with high polylactosamine expression? Currently, extensive glycoproteomic analysis of FFPE tissues clinical specimens is feasible [167], however, the majority of studies are done on biofluids where glycoprotein amounts are more abundant compared to focal regions of tissue. Much method development and optimization remains to be done for all facets of the mass spectrometry workflows, especially for glycopeptides. There are multiple new advancements in mass spectrometry approaches that address these long standing challenges [158], and their implementation will continue to expand rapidly in the next five years. A recent new tool just described for O-linked glycoproteins are two unique proteases that only cleave peptides modified with O-glycans [168, 169] This is a major advancement for O-glycan peptide analysis, as there is no homologous N-linked glycan specific enzyme like PNGase F for O-glycans. The O-glycan peptidase reagents allowed extensive proteomic MS identification and characterization of O-linked glycopeptides [168, 169]. In addition to application of these emerging glycan and glycopeptide analysis workflows to breast cancer tumor tissues, we also predict a complementary emphasis on the glycosylation analysis of tumor-associated immune cells and immunoglobulins. Lastly, an almost completely understudied area of glycosylation is that of the function of glycosaminoglycans (GAG) attached to extracellular matrix proteoglycans like syndecan, biglycan and glypican. The polymeric structures of heparan sulfate and chondroitin sulfate make them difficult to study, but they are known to be critical mediators of ECM and immune function [170]. Syndecan-1, a heparan sulfated proteoglycan, has been evaluated for decades as possible breast cancer biomarker in tissues, but only at the level of transcript and protein expression [171]. Analysis of the glycan composition of the GAG chains in the tumor microenvironment remains a largely open target of research with known clinical significance.
Immunotherapies targeting breast cancer, and the role of glycosylation in their success or failure, is the other area we predict where glycomics will have a major impact in the next five years. Deciphering the role of glycosylation in immune system function and tumor-immune interactions is becoming increasingly important. As new cancer immunotherapy strategies emerge that are potentially curative for some patients but ineffective in others, the primary question to address is why this occurs? Glycoproteins mediate much of the interactions between the immune system and tumors, but very little mechanistic information has been delineated. For breast cancers, the use of PD-L1/PD-1 pathway inhibitors in metastatic triple negative breast cancer (TNBC) has become increasingly popular despite limited efficacy [172]. The first trial, the Keynote-012 trial (), tested pembrolizumab in patients with heavily pre-treated advanced triple negative breast cancer. Of the 27 patients evaluated, 18.5% exhibited an overall response [173]. Additional trials () testing the efficacy of pembrolizumab for metastatic triple negative breast cancer treatments regimens have ranged from 13.6% to 23% response rates [174, 175]. While anti-PD-L1 therapies have shown increased efficacy in triple negative breast cancer patients with tumors expressing PD-L1, there is still need for significant improvement in response rates [172]. A link to the glycosylation status of PD-L1 has recently been published by Li et. al., who identified a mechanism of PD-L1 glycosylation in TNBC that promotes immune suppression due to an increase in interaction between PD-L1 and PD-1, indicating that glycosylated PD-L1 could potentially be a more effective clinical target [176]. Using multiple analytical strategies, additional studies have identified N-acetyllactosamine N-glycans on PD-L1 as the likely mediator of increased PD-1 binding [176, 177]. These findings demonstrate the importance of glycosylation in immunotherapy treatments, and highlights the potential for new studies analyzing glycosylation differences in patients who responded to immunotherapy and those who did not. We expect the two areas, improved glycopeptide analysis and immunotherapy improvement, to coincide. A better understanding of the role of N-acetyllactosasmine-glycoproteins like PD-L1 could provide mechanistic insights to why these therapies have varying efficacy, lead to the development of altered/new therapies, and help to drive patient treatment plans.
Table 1:
Table summarizing studies analyzing changes in glycosylation in breast cancer and significant findings over the years.
| Sample Type | Method | Change Observed | Reference |
|---|---|---|---|
| Tumor Tissues | IHC | Thomsen-Friedenreich and Tn epitopes play a role in breast cancer invasion | [17] |
| Serum | Immunoassays | MUC-1 is upregulated in breast cancer | [13,14,18] |
| MDA-MB-435 breast cancer cells | Cell transfection and RT-PCR | α2,6 sialylation affects adhesion capabilities of breast cancer cells | [19] |
| Tumor Tissues | MALDI-FTICR IMS | increase in N-glycan branching associated with advanced breast cancers | [21] |
| MDA-MB breast cancer cells | Cellular Assays | changes in MUC-1 glycosylation result in exposue of protein core which allows cells to adhere to distant tissues | [90] |
| Tumor Microarray | qRT-PCR and IHC | underglycosylated MUC-1 is associated with higher tumor grade and poor prognosis | [93] |
| Tumor Microarrays | CdSe Aqueous Quantum Dots | increased expression of Tn-antigen in breast cancer | [94] |
| Tumor Tissues | IHC | loss of core 2 O-glycans in breast cancer tumors | [95] |
| Tumor Microarrays | Gene Expression Assays | increase in truncated O-glycans with terminated sialic acids in ER+ breast cancers | [99] |
| Tumor Microarrays | IHC and RNA Analysis | ER+ cancers mainly carry core 1 O-glycans due to upregulation of C1GALT1, while ER- cancers mainly carry core 2 O-glycans due to upregulation of GCNT1 | [99] |
| Tumor Microarrays | Selectin Binding Assays and Flow Cytometry | sialyl Lewisx antigens on core 2 O-glycans only present on breast cancer tissues | [99] |
| Breast Cancer cell lines | Cellular Assays | expression of sialyl Lewisx can result in binding of selectins and various core glycans which can dictate how cells metastasize and respond to EGF binding | [58] |
| Breast Cancer cell lines | Cellular and Gene Expression Assays | Thomsen-Friedenreich antigens, sialyl Lewis antigens, sialyl α2,6-lactosaminyl structures and polysialic acids mediate cell-cell interactions and are altered in breast cancer | [104] |
| TNBC xenograft models | IHC | inhibiting Neu-1 limits EMT | [109] |
| Breast Cancer cell lines, mouse models and serum | MALDI-TOF MS | increase in sialylated glycans in breast cancer | [59, 117-118] |
| Xenograft models | IHC | ST3Gal I is increased in breast cancer while ST3Gal II is decreased | [121] |
| Breast Cancer cell lines | IHC | ST3Gal I enzymatic activity correlated with tumor grade and is implicated in increased expression of sialyl T antigen | [96] |
| Tumor Tissues | MAL Histochemistry | α2,3 sialic acids are expressed higher in grade III and VI breast cancers than grade I and II breast cancers | [59] |
| Breast Cancer cell lines | RT-PCR, Western Blots and IHC | ST6Gal II expression is associated with invasive phenotype in vivo and in vitro | [122] |
| MDA-MB-435 breast cancer cells | RT-PCR and Cellular Assays | α2,6 sialylation the surface of tumor cells contributes to cell-cell and cell-extracellular matrix adhesion capabilities | [19] |
| Breast Cancer Stem-Like Cells | Cellular Assays | Di-sialylated ganglioside GD2 is associated with breast cancer stem cell function | [124] |
| Tumor Tissues | IHC | Di-sialylated ganglioside GD2 is associated with triple negative breast cancer | [125] |
| Serum | Protein Quantification | protein bound fucose is present in higher concentrations in breast cancer patients | [127] |
| Breast Cancer cell lines | Cellular Assays, Western Blots and Lectin Blots | increased EGFR core fucosylation results in increased EGFR dimerization and phosphorylation, which leads to increased signaling associated with tumor growth and malignancy | [129] |
| Breast Cancer cell lines | Cellular Assays, RT-PCR, and Flow Cytometry | fucosylation in IDC is linked with malignant processes | [130] |
| Breast Cancer cell lines | Gene Expression Profiling and Western Blots | increased FUT8 expression correlated with TGFβ induced EMT | [131] |
| Tumor Tissues | Pathologic Analysis and IHC | Lewisx detection is an independent poor prognostic factor for recurrence free survival and overall survival in patients under 50 years old | [132] |
| Tumor Tissues | IHC | Lewisx is strongly associated with the leading edge of invading tumors, indicating its potential role in invasion in metastasis | [133] |
| Serum | HPLC and MALDI-TOF MS | increase in mono-fucosylated and tri-sialylated N-glycans in breast cancer | [143] |
| Serum | HPLC and MALDI-TOF MS | breast cancer patients with associated lymph node metastasis show increased expression of sialyl Lewisx containing glycans and agalactosyl biantennary glycans | [143] |
| Tumor Tissues | rt-PCR and Western Blot | Low expression of FUT8 is correlated with breast cancer disease free survival, while high expression of FUT8 is correlated with metastasis and high tumor stage | [134] |
| Xenograft models and Tumor Tissues | MALDI-FTICR MS | increase in high mannose glycans in breast cancers compared to healthy controls | [142] |
| Tumor Tissues | MALDI-FTICR IMS | high mannose and tetra-antennary glycans are associated with tumor region of breast cancer tissues, while bi-antennary and sialylated glycans are associated with necrotic regions | [22] |
| Tumor Tissues and Breast Cancer Cell Lines | IHC and Cellular Assays | N-acetyllactosamine N-glycans mediate binding of PD-L1 and PD-1, potentially effecting the efficacy of anti-PD-L1 therapies | [176] |
Article highlights.
Breast cancer is a heterogeneous disease that incorporates several distinct entities with remarkably different biological characteristics and clinical behavior.
The majority of all current FDA approved cancer biomarkers are glycoproteins, however there has been limited specificity and sensitivity of these biomarkers in breast cancer.
Glycosylation is the enzymatic process that involves the addition of single carbohydrates to proteins or lipids.
Alterations in glycan expression are due to changes in glycosyltransferase expression or localization, peptide composition, receptor substrates or sugar nucleotide availability.
Changes in MUC-1 glycosylation has been associated with metastatic potential in various breast cancer subtypes.
In ER- cancers, an increase in core 2 O-glycans expressing sialyl Lewisx has been documented and is thought to aid in metastasis.
Core fucosylation of EGFR results in increases in phosphorylation and dimerization, producing enhanced signaling that is associated with tumor growth and malignancy.
Recent studies have outlined the predictive potential of specific glycans in detecting poor clinical outcomes in breast cancer.
Acknowledgments
Funding
This paper was funded by the U.S. Department of Health and Human Services, National Institutes of Health, National Cancer Institute, grants: CA186799-01, CA207779; and the Smartstate SC Centers of Economic Excellence Endowed Centers of Excellence.
Footnotes
Declaration of interest
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Reviewer disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
References
- 1.Aub JC, Tieslau C, Lankester A. REACTIONS OF NORMAL AND TUMOR CELL SURFACES TO ENZYMES. I. WHEAT-GERM LIPASE AND ASSOCIATED MUCOPOLYSACCHARIDES. Proceedings of the National Academy of Sciences of the United States of America. 1963. October;50:613–9. PubMed PMID: 14077487; PubMed Central PMCID: PMCPMC221235. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barton JG, Bois JP, Sarr MG, et al. Predictive and prognostic value of CA 19-9 in resected pancreatic adenocarcinoma. Journal of gastrointestinal surgery : official journal of the Society for Surgery of the Alimentary Tract. 2009. November;13(11):2050–8. doi: 10.1007/s11605-009-0849-z. PubMed PMID: 19756875; eng. [DOI] [PubMed] [Google Scholar]
- 3.Drake RR. Glycosylation and Cancer: Moving Glycomics to the Forefront. Advances in cancer research. 2015;126:1–10. [DOI] [PubMed] [Google Scholar]
- 4.Kirwan A, Utratna M, O’Dwyer ME, et al. Glycosylation-Based Serum Biomarkers for Cancer Diagnostics and Prognostics. BioMed Research International. 2015. 10/05 04/02/received 05/28/revised 05/31/accepted;2015:490531. doi: 10.1155/2015/490531. PubMed PMID: PMC4609776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ludwig JA, Weinstein JN. Biomarkers in cancer staging, prognosis and treatment selection. Nat Rev Cancer. 2005. November;5(11):845–56. doi: 10.1038/nrc1739. PubMed PMID: 16239904; eng. [DOI] [PubMed] [Google Scholar]
- 6.Sawyers CL. The cancer biomarker problem. Nature. 2008. April/02/online;452:548. doi: 10.1038/nature06913. [DOI] [PubMed] [Google Scholar]
- 7.Song E, Mechref Y. Defining glycoprotein cancer biomarkers by MS in conjunction with glycoprotein enrichment. Biomarkers in medicine. 2015. September/02;9(9):835–844. doi: 10.2217/bmm.15.55. PubMed PMID: PMC4889013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grishman E Histochemical analysis of mucopolysaccharides occurring in mucus-producing tumors; mixed tumors of the parotid gland, colloid carcinomas of the breast, and myxomas. Cancer. 1952. July;5(4):700–7. PubMed PMID: 14935980; eng. [DOI] [PubMed] [Google Scholar]
- 9.Remmele W, Hildebrand U, Hienz HA, et al. Comparative histological, histochemical, immunohistochemical and biochemical studies on oestrogen receptors, lectin receptors, and Barr bodies in human breast cancer. Virchows Archiv A, Pathological anatomy and histopathology. 1986;409(2):127–47. PubMed PMID: 2424168; eng. [DOI] [PubMed] [Google Scholar]
- 10.Parodi AJ, Blank EW, Peterson JA, et al. Dolichol-bound oligosaccharides and the transfer of distal monosaccharides in the synthesis of glycoproteins by normal and tumor mammary epithelial cells. Breast cancer research and treatment. 1982;2(3):227–37. PubMed PMID: 6817834; eng. [DOI] [PubMed] [Google Scholar]
- 11.Feizi T Demonstration by monoclonal antibodies that carbohydrate structures of glycoproteins and glycolipids are onco-developmental antigens. Nature. 1985. March 7-13;314(6006):53–7. PubMed PMID: 2579340; eng. [DOI] [PubMed] [Google Scholar]
- 12.Desai PR. Immunoreactive T and Tn antigens in malignancy: role in carcinoma diagnosis, prognosis, and immunotherapy. Transfusion medicine reviews. 2000. October;14(4):312–25. doi: 10.1053/tmrv.2000.16229. PubMed PMID: 11055076; eng. [DOI] [PubMed] [Google Scholar]
- 13.Foster CS, Neville AM. Expression of breast epithelial differentiation antigens in human primary breast cancer. Journal of the National Cancer Institute. 1987. October;79(4):613–22. PubMed PMID: 2443736; eng. [PubMed] [Google Scholar]
- 14.Kirmiz C, Li B, An HJ, et al. A Serum Glycomics Approach to Breast Cancer Biomarkers. Molecular & Cellular Proteomics. 2007. January 1, 2007;6(1):43–55. doi: 10.1074/mcp.M600171-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Wolf MF, Ludwig A, Fritz P, et al. Increased expression of Thomsen-Friedenreich antigens during tumor progression in breast cancer patients. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 1988;9(4):190–4. PubMed PMID: 3420374; eng. [DOI] [PubMed] [Google Scholar]
- 16.Burchell J, Durbin H, Taylor-Papadimitriou J. Complexity of expression of antigenic determinants, recognized by monoclonal antibodies HMFG-1 and HMFG-2, in normal and malignant human mammary epithelial cells. Journal of immunology (Baltimore, Md : 1950). 1983. July;131(1):508–13. PubMed PMID: 6190927; eng. [PubMed] [Google Scholar]
- 17.Springer GF. Immunoreactive T and Tn epitopes in cancer diagnosis, prognosis, and immunotherapy. Journal of molecular medicine (Berlin, Germany). 1997. August;75(8):594–602. PubMed PMID: 9297627; eng. [DOI] [PubMed] [Google Scholar]
- 18.Duffy MJ, Evoy D, McDermott EW. CA 15-3: uses and limitation as a biomarker for breast cancer. Clinica chimica acta; international journal of clinical chemistry. 2010. December 14;411(23-24):1869–74. doi: 10.1016/j.cca.2010.08.039. PubMed PMID: 20816948; eng. [DOI] [PubMed] [Google Scholar]
- 19.Lin S, Kemmner W, Grigull S, et al. Cell surface alpha 2,6 sialylation affects adhesion of breast carcinoma cells. Exp Cell Res. 2002. May 15;276(1):101–10. doi: 10.1006/excr.2002.5521. PubMed PMID: 11978012; eng. [DOI] [PubMed] [Google Scholar]
- 20.Abd Hamid UM, Royle L, Saldova R, et al. A strategy to reveal potential glycan markers from serum glycoproteins associated with breast cancer progression. Glycobiology. 2008;18(12):1105–1118. doi: 10.1093/glycob/cwn095. [DOI] [PubMed] [Google Scholar]
- 21.Scott DA, Casadonte R, Cardinali B, et al. Increases in Tumor N-glycan Polylactosamines Associated with Advanced HER2 Positive and Triple Negative Breast Cancer Tissues. Proteomics Clinical applications. 2018. December 28:e1800014. doi: 10.1002/prca.201800014. PubMed PMID: 30592377; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scott DA, Norris-Caneda K, Spruill L, et al. Specific N-linked glycosylation patterns in areas of necrosis in tumor tissues. International Journal of Mass Spectrometry. 2018. 2018/01/September/;437:69–76. doi: 10.1016/j.ijms.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Howlader N NA, Krapcho M, Miller D, Bishop K, Kosary CL, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, Chen HS, Feuer EJ, Cronin KA (eds). SEER Cancer Statistics Review, 1975-2014. In: Institute NC, editor. SEER website2016. [Google Scholar]
- 24.Ibrahim E, Al-Gahmi AM, Zeenelin AA, et al. Basal vs. luminal A breast cancer subtypes: a matched case-control study using estrogen receptor, progesterone receptor, and HER-2 as surrogate markers. Medical oncology (Northwood, London, England). 2009;26(3):372–8. doi: 10.1007/s12032-008-9131-6. PubMed PMID: 19034706; eng. [DOI] [PubMed] [Google Scholar]
- 25.Cancer Stat Facts: Female Breast Cancer In: Surveillance EaERP, editor. Online: National Cancer Institute; 2019. [Google Scholar]
- 26.The Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumors. Nature. 2012. 09/23;490(7418):61–70. doi: 10.1038/nature11412. PubMed PMID: PMC3465532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peiris D, Spector AF, Lomax-Browne H, et al. Cellular glycosylation affects Herceptin binding and sensitivity of breast cancer cells to doxorubicin and growth factors [Article]. Scientific Reports. 2017. February/22/online;7:43006. doi: 10.1038/srep43006 https://www.nature.com/articles/srep43006#supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. The New England journal of medicine. 2010. November 11;363(20):1938–48. doi: 10.1056/NEJMra1001389. PubMed PMID: 21067385; eng. [DOI] [PubMed] [Google Scholar]
- 29.Yu T, Di G. Role of tumor microenvironment in triple-negative breast cancer and its prognostic significance. Chinese Journal of Cancer Research. 2017. January/20/received 04/12/accepted;29(3):237–252. doi: 10.21147/j.issn.1000-9604.2017.03.10. PubMed PMID: PMC5497211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liao HY, Zhang WW, Sun JY, et al. The Clinicopathological Features and Survival Outcomes of Different Histological Subtypes in Triple-negative Breast Cancer. Journal of Cancer. 2018;9(2):296–303. doi: 10.7150/jca.22280. PubMed PMID: 29344276; PubMed Central PMCID: PMCPMC5771337. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nahta R, Esteva FJ. Trastuzumab: triumphs and tribulations. Oncogene. 2007;26. doi: 10.1038/sj.onc.1210379. [DOI] [PubMed] [Google Scholar]
- 32.Burris HA, Hurwitz HI, Dees EC, et al. Phase I safety, pharmacokinetics, and clinical activity study of lapatinib (GW572016), a reversible dual inhibitor of epidermal growth factor receptor tyrosine kinases, in heavily pretreated patients with metastatic carcinomas. J Clin Oncol. 2005;23. doi: 10.1200/jco.2005.16.584. [DOI] [PubMed] [Google Scholar]
- 33.Lauc G, Pezer M, Rudan I, et al. Mechanisms of disease: The human N-glycome. Biochimica et biophysica acta. 2016. August;1860(8):1574–82. doi: 10.1016/j.bbagen.2015.10.016. PubMed PMID: 26500099; eng. [DOI] [PubMed] [Google Scholar]
- 34.Eccles SA, Aboagye EO, Ali S, et al. Critical research gaps and translational priorities for the successful prevention and treatment of breast cancer. Breast cancer research : BCR. 2013;15(5):R92–R92. doi: 10.1186/bcr3493. PubMed PMID: 24286369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fallahpour S, Navaneelan T, De P, et al. Breast cancer survival by molecular subtype: a population-based analysis of cancer registry data. CMAJ Open. 2017. Jul-Sep 09/25;5(3):E734–E739. doi: 10.9778/cmajo.20170030. PubMed PMID: PMC5621954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noone AM HN, Krapcho M, Miller D, Brest A, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, Chen HS, Feuer EJ, Cronin KA (eds). SEER Cancer Statistics Review (National Cancer Institute). based on November 2017. SEER data submission, posted to SEER website April 2018. 2017. [Google Scholar]
- 37.Network NCC. NCCN Clinical Practice Guidelines in Oncology. March 20, 2018: NCCN. [Google Scholar]
- 38.Polyak K Breast cancer: origins and evolution. The Journal of Clinical Investigation. 2007. 11/01/;117(11):3155–3163. doi: 10.1172/JCI33295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kleibl Z, Kristensen VN. Women at high risk of breast cancer: Molecular characteristics, clinical presentation and management. The Breast. 2016. 2016/08/January/;28:136–144. doi: 10.1016/j.breast.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 40.Apostolou P, Fostira F. Hereditary breast cancer: the era of new susceptibility genes. BioMed research international. 2013;2013:747318–747318. doi: 10.1155/2013/747318. PubMed PMID: 23586058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.SP T, RFJ S, Theodora G, et al. Molecular evolution of breast cancer. The Journal of Pathology. 2005;205(2):248–254. doi: doi: 10.1002/path.1691. [DOI] [PubMed] [Google Scholar]
- 42.Varki A Biological roles of glycans. Glycobiology. 2017. January;27(1):3–49. doi: 10.1093/glycob/cww086. PubMed PMID: 27558841; PubMed Central PMCID: PMCPMC5884436. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dennis JW, Lau KS, Demetriou M, et al. Adaptive Regulation at the Cell Surface by N-Glycosylation. Traffic. 2009;10(11):1569–1578. doi: doi: 10.1111/j.1600-0854.2009.00981.x. [DOI] [PubMed] [Google Scholar]
- 44.Ohtsubo K, Takamatsu S, Minowa MT, et al. Dietary and Genetic Control of Glucose Transporter 2 Glycosylation Promotes Insulin Secretion in Suppressing Diabetes. Cell. 2005. 2005/12/29/;123(7):1307–1321. doi: 10.1016/j.cell.2005.09.041. [DOI] [PubMed] [Google Scholar]
- 45.Partridge EA, Le Roy C, Di Guglielmo GM, et al. Regulation of Cytokine Receptors by Golgi N-Glycan Processing and Endocytosis. Science. 2004;306(5693):120–124. doi: 10.1126/science.1102109. [DOI] [PubMed] [Google Scholar]
- 46.Schachter H The search for glycan function: Fucosylation of the TGF-β1 receptor is required for receptor activation. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(44):15721–15722. doi: 10.1073/pnas.0507659102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu YC, Yen HY, Chen CY, et al. Sialylation and fucosylation of epidermal growth factor receptor suppress its dimerization and activation in lung cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2011. July 12;108(28):11332–7. doi: 10.1073/pnas.1107385108. PubMed PMID: 21709263; PubMed Central PMCID: PMCPMC3136320. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lo WY, Lagrange AH, Hernandez CC, et al. Glycosylation of {beta}2 subunits regulates GABAA receptor biogenesis and channel gating. J Biol Chem. 2010. October 8;285(41):31348–61. doi: 10.1074/jbc.M110.151449. PubMed PMID: 20639197; PubMed Central PMCID: PMCPMC2951209. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Munkley J, Elliott DJ. Hallmarks of glycosylation in cancer. Oncotarget. 2016. 03/17 01/28/received 03/02/accepted;7(23):35478–35489. doi: 10.18632/oncotarget.8155. PubMed PMID: PMC5085245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuzmanov U, Kosanam H, Diamandis EP. The sweet and sour of serological glycoprotein tumor biomarker quantification. BMC medicine. 2013. February 07;11:31. doi: 10.1186/1741-7015-11-31. PubMed PMID: 23390961; PubMed Central PMCID: PMCPMC3751898. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spiro RG. Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology. 2002. April;12(4):43r–56r. PubMed PMID: 12042244; eng. [DOI] [PubMed] [Google Scholar]
- 52.Potapenko IO, Haakensen VD, Luders T, et al. Glycan gene expression signatures in normal and malignant breast tissue; possible role in diagnosis and progression. Molecular oncology. 2010. April;4(2):98–118. doi: 10.1016/j.molonc.2009.12.001. PubMed PMID: 20060370; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Varki AC, Esko RD, JD. et al. , editors. Essentials of Glycobiology 3rd Edition. 3 ed. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 2017. [Google Scholar]
- 54.Abbott KL, Aoki K, Lim J-M, et al. Targeted Glycoproteomic Identification of Biomarkers for Human Breast Carcinoma. Journal of proteome research. 2008. 2008/April/01;7(4):1470–1480. doi: 10.1021/pr700792g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ladenson RP, Schwartz SO, Ivy AC. Incidence of the blood groups and the secretor factor in patients with pernicious anemia and stomach carcinoma. The American journal of the medical sciences. 1949. February;217(2):194–7. PubMed PMID: 18109280; eng. [DOI] [PubMed] [Google Scholar]
- 56.Taniguchi N, Kizuka Y. Glycans and cancer: role of N-glycans in cancer biomarker, progression and metastasis, and therapeutics. Advances in cancer research. 2015;126:11–51. doi: 10.1016/bs.acr.2014.11.001. PubMed PMID: 25727145; eng. [DOI] [PubMed] [Google Scholar]
- 57.Blanas A, Sahasrabudhe NM, Rodríguez E, et al. Fucosylated Antigens in Cancer: An Alliance toward Tumor Progression, Metastasis, and Resistance to Chemotherapy. Frontiers in Oncology. 2018. 02/23 11/29/received 02/05/accepted;8:39. doi: 10.3389/fonc.2018.00039. PubMed PMID: PMC5829055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Burchell JM, Beatson R, Graham R, et al. O-linked mucin-type glycosylation in breast cancer [ 10.1042/BST20170483]. Biochemical Society Transactions. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cui H, Lin Y, Yue L, et al. Differential expression of the alpha2,3-sialic acid residues in breast cancer is associated with metastatic potential. Oncology reports. 2011. May;25(5):1365–71. doi: 10.3892/or.2011.1192. PubMed PMID: 21344161; eng. [DOI] [PubMed] [Google Scholar]
- 60.Josic D, Martinovic T, Pavelic K. GLYCOSYLATION AND METASTASES. Electrophoresis. 2018. September 24. doi: 10.1002/elps.201800238. PubMed PMID: 30246896; eng. [DOI] [PubMed] [Google Scholar]
- 61.Kim YJ, Varki A. Perspectives on the significance of altered glycosylation of glycoproteins in cancer. Glycoconjugate journal. 1997. August;14(5):569–76. PubMed PMID: 9298689; eng. [DOI] [PubMed] [Google Scholar]
- 62.Liu X, Nie H, Zhang Y, et al. Cell surface-specific N-glycan profiling in breast cancer. PloS one. 2013;8(8):e72704. doi: 10.1371/journal.pone.0072704. PubMed PMID: 24009699; PubMed Central PMCID: PMCPMC3751845. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ray MRJ, David M. Hallmarks of Metastasis. Lung Cancer Metastasis. 2009:29–46. [Google Scholar]
- 64.Rodrigues JG, Balmaña M, Macedo JA, et al. Glycosylation in cancer: Selected roles in tumour progression, immune modulation and metastasis. Cellular Immunology. 2018. 2018/03/20/. doi: 10.1016/j.cellimm.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 65.Hakomori S, Kannagi R. Glycosphingolipids as tumor-associated and differentiation markers. Journal of the National Cancer Institute. 1983. August;71(2):231–51. PubMed PMID: 6576183; eng. [PubMed] [Google Scholar]
- 66.Kannagi R, Yin J, Miyazaki K, et al. Current relevance of incomplete synthesis and neo-synthesis for cancer-associated alteration of carbohydrate determinants--Hakomori’s concepts revisited. Biochimica et biophysica acta. 2008. March;1780(3):525–31. doi: 10.1016/j.bbagen.2007.10.007. PubMed PMID: 17980710; eng. [DOI] [PubMed] [Google Scholar]
- 67.Drake RR, West CA, Mehta AS, et al. MALDI mass spectrometry imaging of N-linked glycans in tissues. Advances in Experimental Medicine and Biology 2018. p. 59–76. [DOI] [PubMed] [Google Scholar]
- 68.Buckhaults P, Chen L, Fregien N, et al. Transcriptional regulation of N-acetylglucosaminyltransferase V by the src oncogene. J Biol Chem. 1997. August 1;272(31):19575–81. PubMed PMID: 9235963; eng. [DOI] [PubMed] [Google Scholar]
- 69.Hatano K, Miyamoto Y, Nonomura N, et al. Expression of gangliosides, GD1a, and sialyl paragloboside is regulated by NF-kappaB-dependent transcriptional control of alpha2,3-sialyltransferase I, II, and VI in human castration-resistant prostate cancer cells. International journal of cancer. 2011. October 15;129(8):1838–47. doi: 10.1002/ijc.25860. PubMed PMID: 21165949; eng. [DOI] [PubMed] [Google Scholar]
- 70.Kumamoto K, Goto Y, Sekikawa K, et al. Increased expression of UDP-galactose transporter messenger RNA in human colon cancer tissues and its implication in synthesis of Thomsen-Friedenreich antigen and sialyl Lewis A/X determinants. Cancer research. 2001. June 1;61(11):4620–7. PubMed PMID: 11389099; eng. [PubMed] [Google Scholar]
- 71.Pinho SS, Oliveira P, Cabral J, et al. Loss and recovery of Mgat3 and GnT-III Mediated E-cadherin N-glycosylation is a mechanism involved in epithelial-mesenchymal-epithelial transitions. PloS one. 2012;7(3):e33191. doi: 10.1371/journal.pone.0033191. PubMed PMID: 22427986; PubMed Central PMCID: PMCPMC3302839. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Arnold JN, Saldova R, Hamid UM, et al. Evaluation of the serum N-linked glycome for the diagnosis of cancer and chronic inflammation. Proteomics. 2008. August;8(16):3284–93. doi: 10.1002/pmic.200800163. PubMed PMID: 18646009; eng. [DOI] [PubMed] [Google Scholar]
- 73.Hakomori S Glycosylation defining cancer malignancy: new wine in an old bottle. Proceedings of the National Academy of Sciences of the United States of America. 2002. August 6;99(16):10231–3. doi: 10.1073/pnas.172380699. PubMed PMID: 12149519; PubMed Central PMCID: PMCPMC124893. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Drake RR, Jones EE, Powers TW, et al. Altered glycosylation in prostate cancer. Advances in cancer research. 2015;126:345–82. doi: 10.1016/bs.acr.2014.12.001. PubMed PMID: 25727153; eng. [DOI] [PubMed] [Google Scholar]
- 75.Kölbl AC, Andergassen U, Jeschke U. The Role of Glycosylation in Breast Cancer Metastasis and Cancer Control. Frontiers in Oncology. 2015. October/13 08/26/received 09/24/accepted;5:219. doi: 10.3389/fonc.2015.00219. PubMed PMID: PMC4602128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Taparra K, Tran PT, Zachara NE. Hijacking the Hexosamine Biosynthetic Pathway to Promote EMT-Mediated Neoplastic Phenotypes. Frontiers in Oncology. 2016. April/18 02/22/received 03/27/accepted;6:85. doi: 10.3389/fonc.2016.00085. PubMed PMID: PMC4834358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lau KS, Partridge EA, Grigorian A, et al. Complex N-glycan number and degree of branching cooperate to regulate cell proliferation and differentiation. Cell. 2007. April 6;129(1):123–34. doi: 10.1016/j.cell.2007.01.049. PubMed PMID: 17418791; eng. [DOI] [PubMed] [Google Scholar]
- 78.Kudelka MR, Ju T, Heimburg-Molinaro J, et al. Simple sugars to complex disease--mucin-type O-glycans in cancer. Advances in cancer research. 2015;126:53–135. doi: 10.1016/bs.acr.2014.11.002. PubMed PMID: 25727146; PubMed Central PMCID: PMCPMC5812724. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.David L, Nesland JM, Clausen H, et al. Simple mucin-type carbohydrate antigens (Tn, sialosyl-Tn and T) in gastric mucosa, carcinomas and metastases. APMIS Supplementum. 1992;27:162–72. PubMed PMID: 1520525; eng. [PubMed] [Google Scholar]
- 80.Drake RR, Powers TW, Jones EE, et al. MALDI Mass Spectrometry Imaging of N-Linked Glycans in Cancer Tissues. Advances in cancer research. 2017;134:85–116. doi: 10.1016/bs.acr.2016.11.009. PubMed PMID: 28110657; PubMed Central PMCID: PMCPMC6020705. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gill DJ, Chia J, Senewiratne J, et al. Regulation of O-glycosylation through Golgi-to-ER relocation of initiation enzymes. J Cell Biol. 2010. May 31;189(5):843–58. doi: 10.1083/jcb.201003055. PubMed PMID: 20498016; PubMed Central PMCID: PMCPMC2878949. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Berois N, Gattolliat CH, Barrios E, et al. GALNT9 gene expression is a prognostic marker in neuroblastoma patients. Clin Chem. 2013. January;59(1):225–33. doi: 10.1373/clinchem.2012.192328. PubMed PMID: 23136245; eng. [DOI] [PubMed] [Google Scholar]
- 83.Itzkowitz S, Kjeldsen T, Friera A, et al. Expression of Tn, sialosyl Tn, and T antigens in human pancreas. Gastroenterology. 1991. June;100(6):1691–700. PubMed PMID: 1850375; eng. [DOI] [PubMed] [Google Scholar]
- 84.Powers TW, Holst S, Wuhrer M, et al. Two-dimensional N-glycan distribution mapping of hepatocellular carcinoma tissues by MALDI-imaging mass spectrometry [Article]. Biomolecules. 2015;5(4):2554–2572. doi: 10.3390/biom5042554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Radhakrishnan P, Dabelsteen S, Madsen FB, et al. Immature truncated O-glycophenotype of cancer directly induces oncogenic features. Proceedings of the National Academy of Sciences of the United States of America. 2014. September 30;111(39):E4066–75. doi: 10.1073/pnas.1406619111. PubMed PMID: 25118277; PubMed Central PMCID: PMCPMC4191756. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Reis CA, David L, Seixas M, et al. Expression of fully and under-glycosylated forms of MUC1 mucin in gastric carcinoma. International journal of cancer. 1998. August 21;79(4):402–10. PubMed PMID: 9699534; eng. [DOI] [PubMed] [Google Scholar]
- 87.Ricardo S, Marcos-Silva L, Pereira D, et al. Detection of glyco-mucin profiles improves specificity of MUC16 and MUC1 biomarkers in ovarian serous tumours. Molecular oncology. 2015. February;9(2):503–12. doi: 10.1016/j.molonc.2014.10.005. PubMed PMID: 25454345; PubMed Central PMCID: PMCPMC5528651. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sewell R, Backstrom M, Dalziel M, et al. The ST6GalNAc-I sialyltransferase localizes throughout the Golgi and is responsible for the synthesis of the tumor-associated sialyl-Tn O-glycan in human breast cancer. J Biol Chem. 2006. February 10;281(6):3586–94. doi: 10.1074/jbc.M511826200. PubMed PMID: 16319059; eng. [DOI] [PubMed] [Google Scholar]
- 89.Bhide GP, Colley KJ. Sialylation of N-glycans: mechanism, cellular compartmentalization and function [journal article]. Histochemistry and Cell Biology. 2017. February 01;147(2):149–174. doi: 10.1007/s00418-016-1520-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ciborowski P, Finn OJ. Non-glycosylated tandem repeats of MUC1 facilitate attachment of breast tumor cells to normal human lung tissue and immobilized extracellular matrix proteins (ECM) in vitro: potential role in metastasis. Clinical & experimental metastasis. 2002;19(4):339–45. PubMed PMID: 12090474; eng. [DOI] [PubMed] [Google Scholar]
- 91.Goldstein LJ, Gray R, Badve S, et al. Prognostic utility of the 21-gene assay in hormone receptor-positive operable breast cancer compared with classical clinicopathologic features. J Clin Oncol. 2008. September 1;26(25):4063–71. doi: 10.1200/jco.2007.14.4501. PubMed PMID: 18678838; PubMed Central PMCID: PMCPMC2654377. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications [Review]. Nat Rev Cancer. 2015. 09//print;15(9):540–555. doi: 10.1038/nrc3982. [DOI] [PubMed] [Google Scholar]
- 93.Ghosh S, May MJ, Kopp EB. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annual review of immunology. 1998;16:225–60. doi: 10.1146/annurev.immunol.16.1.225. PubMed PMID: 9597130; eng. [DOI] [PubMed] [Google Scholar]
- 94.Au GH, Mejias L, Swami VK, et al. Quantitative assessment of Tn antigen in breast tissue micro-arrays using CdSe aqueous quantum dots. Biomaterials. 2014. March;35(9):2971–80. doi: 10.1016/j.biomaterials.2013.12.034. PubMed PMID: 24411673; eng. [DOI] [PubMed] [Google Scholar]
- 95.Girling A, Bartkova J, Burchell J, et al. A core protein epitope of the polymorphic epithelial mucin detected by the monoclonal antibody SM-3 is selectively exposed in a range of primary carcinomas. International journal of cancer. 1989. June 15;43(6):1072–6. PubMed PMID: 2471698; eng. [DOI] [PubMed] [Google Scholar]
- 96.Burchell J, Poulsom R, Hanby A, et al. An alpha2,3 sialyltransferase (ST3Gal I) is elevated in primary breast carcinomas. Glycobiology. 1999. December;9(12):1307–11. PubMed PMID: 10561455; eng. [DOI] [PubMed] [Google Scholar]
- 97.Lloyd KO, Burchell J, Kudryashov V, et al. Comparison of O-linked carbohydrate chains in MUC-1 mucin from normal breast epithelial cell lines and breast carcinoma cell lines. Demonstration of simpler and fewer glycan chains in tumor cells. J Biol Chem. 1996. December 27;271(52):33325–34. PubMed PMID: 8969192; eng. [DOI] [PubMed] [Google Scholar]
- 98.Muller S, Alving K, Peter-Katalinic J, et al. High density O-glycosylation on tandem repeat peptide from secretory MUC1 of T47D breast cancer cells. J Biol Chem. 1999. June 25;274(26):18165–72. PubMed PMID: 10373415; eng. [DOI] [PubMed] [Google Scholar]
- 99.Julien S, Ivetic A, Grigoriadis A, et al. Selectin ligand sialyl-Lewis x antigen drives metastasis of hormone-dependent breast cancers. Cancer research. 2011. December 15;71(24):7683–93. doi: 10.1158/0008-5472.Can-11-1139. PubMed PMID: 22025563; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Beatson R, Maurstad G, Picco G, et al. The Breast Cancer-Associated Glycoforms of MUC1, MUC1-Tn and sialyl-Tn, Are Expressed in COSMC Wild-Type Cells and Bind the C-Type Lectin MGL. PloS one. 2015;10(5):e0125994. doi: 10.1371/journal.pone.0125994. PubMed PMID: 25951175; PubMed Central PMCID: PMCPMC4423978. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gill DJ, Tham KM, Chia J, et al. Initiation of GalNAc-type O-glycosylation in the endoplasmic reticulum promotes cancer cell invasiveness. Proceedings of the National Academy of Sciences of the United States of America. 2013. August 20;110(34):E3152–61. doi: 10.1073/pnas.1305269110. PubMed PMID: 23912186; PubMed Central PMCID: PMCPMC3752262. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Marth JD, Grewal PK. Mammalian glycosylation in immunity. Nature reviews Immunology. 2008. November;8(11):874–87. doi: 10.1038/nri2417. PubMed PMID: 18846099; PubMed Central PMCID: PMCPMC2768770. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cazet A, Julien S, Bobowski M, et al. Tumour-associated carbohydrate antigens in breast cancer. Breast cancer research : BCR. 2010;12(3):204–204. doi: 10.1186/bcr2577. PubMed PMID: 20550729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dall’Olio F, Chiricolo M. Sialyltransferases in cancer. Glycoconjugate journal. 2001. Nov-Dec;18(11-12):841–50. PubMed PMID: 12820717; eng. [DOI] [PubMed] [Google Scholar]
- 105.Harduin-Lepers A, Krzewinski-Recchi MA, Colomb F, et al. Sialyltransferases functions in cancers. Frontiers in bioscience (Elite edition). 2012. January 1;4:499–515. PubMed PMID: 22201891; eng. [DOI] [PubMed] [Google Scholar]
- 106.Baldus SE, Zirbes TK, Monig SP, et al. Histopathological subtypes and prognosis of gastric cancer are correlated with the expression of mucin-associated sialylated antigens: Sialosyl-Lewis(a), Sialosyl-Lewis(x) and sialosyl-Tn. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 1998;19(6):445–53. PubMed PMID: 9817972; eng. [DOI] [PubMed] [Google Scholar]
- 107.Amado M, Carneiro F, Seixas M, et al. Dimeric sialyl-Le(x) expression in gastric carcinoma correlates with venous invasion and poor outcome. Gastroenterology. 1998. March;114(3):462–70. PubMed PMID: 9496936; eng. [DOI] [PubMed] [Google Scholar]
- 108.Rosen SD, Bertozzi CR. The selectins and their ligands. Current opinion in cell biology. 1994. October;6(5):663–73. PubMed PMID: 7530461; eng. [DOI] [PubMed] [Google Scholar]
- 109.Haxho F, Neufeld RJ, Szewczuk MR. Neuraminidase-1: a novel therapeutic target in multistage tumorigenesis. Oncotarget. 2016;7(26):40860–40881. doi: 10.18632/oncotarget.8396. PubMed PMID: 27029067; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Haxho F, Allison S, Alghamdi F, et al. Oseltamivir phosphate monotherapy ablates tumor neovascularization, growth, and metastasis in mouse model of human triple-negative breast adenocarcinoma Breast cancer (Dove Medical Press; ). 2014;6:191–203. doi: 10.2147/bctt.S74663. PubMed PMID: 25525387; PubMed Central PMCID: PMCPMC4266271. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Beatson R, Tajadura-Ortega V, Achkova D, et al. The mucin MUC1 modulates the tumor immunological microenvironment through engagement of the lectin Siglec-9. Nature immunology. 2016. November;17(11):1273–1281. doi: 10.1038/ni.3552. PubMed PMID: 27595232; PubMed Central PMCID: PMCPMC5257269. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hudak JE, Canham SM, Bertozzi CR. Glycocalyx engineering reveals a Siglec-based mechanism for NK cell immunoevasion. Nature chemical biology. 2014. January;10(1):69–75. doi: 10.1038/nchembio.1388. PubMed PMID: 24292068; PubMed Central PMCID: PMCPMC3893890. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jandus C, Boligan KF, Chijioke O, et al. Interactions between Siglec-7/9 receptors and ligands influence NK cell-dependent tumor immunosurveillance. J Clin Invest. 2014. April;124(4):1810–20. doi: 10.1172/jci65899. PubMed PMID: 24569453; PubMed Central PMCID: PMCPMC3973073. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Laubli H, Pearce OM, Schwarz F, et al. Engagement of myelomonocytic Siglecs by tumor-associated ligands modulates the innate immune response to cancer. Proceedings of the National Academy of Sciences of the United States of America. 2014. September 30;111(39):14211–6. doi: 10.1073/pnas.1409580111. PubMed PMID: 25225409; PubMed Central PMCID: PMCPMC4191788. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Macauley MS, Crocker PR, Paulson JC. Siglec-mediated regulation of immune cell function in disease. Nature reviews Immunology. 2014. October;14(10):653–66. doi: 10.1038/nri3737. PubMed PMID: 25234143; PubMed Central PMCID: PMCPMC4191907. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Xiao H, Woods EC, Vukojicic P, et al. Precision glycocalyx editing as a strategy for cancer immunotherapy. Proceedings of the National Academy of Sciences of the United States of America. 2016. September 13;113(37):10304–9. doi: 10.1073/pnas.1608069113. PubMed PMID: 27551071; PubMed Central PMCID: PMCPMC5027407. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ma X, Dong W, Su Z, et al. Functional roles of sialylation in breast cancer progression through miR-26a/26b targeting ST8SIA4 [Original Article]. Cell Death &Amp; Disease. 2016. December/29/online;7:e2561. doi: 10.1038/cddis.2016.427 https://www.nature.com/articles/cddis2016427#supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tian Y, Esteva FJ, Song J, et al. Altered expression of sialylated glycoproteins in breast cancer using hydrazide chemistry and mass spectrometry. Molecular & Cellular Proteomics. 2012. doi: 10.1074/mcp.M111.011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Brockhausen I Mucin‐type O‐glycans in human colon and breast cancer: glycodynamics and functions. EMBO reports. 2006;7(6):599–604. doi: 10.1038/sj.embor.7400705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Brockhausen I, Yang J-M, Burchell J, et al. Mechanisms Underlying Aberrant Glycosylation of MUC1 Mucin in Breast Cancer Cells. European Journal of Biochemistry. 1995;233(2):607–617. doi: doi: 10.1111/j.1432-1033.1995.607_2.x. [DOI] [PubMed] [Google Scholar]
- 121.Picco G, Julien S, Brockhausen I, et al. Over-expression of ST3Gal-I promotes mammary tumorigenesis. Glycobiology. 2010;20(10):1241–1250. doi: 10.1093/glycob/cwq085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Videira PA, Correia M, Malagolini N, et al. ST3Gal.I sialyltransferase relevance in bladder cancer tissues and cell lines. BMC Cancer. 2009. 2009/October/07;9(1):357. doi: 10.1186/1471-2407-9-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ren D, Jia L, Li Y, et al. ST6GalNAcII mediates the invasive properties of breast carcinoma through PI3K/Akt/NF-kappaB signaling pathway. IUBMB Life. 2014. April;66(4):300–8. doi: 10.1002/iub.1268. PubMed PMID: 24756995; eng. [DOI] [PubMed] [Google Scholar]
- 124.Nguyen K, Yan Y, Yuan B, et al. ST8SIA1 Regulates Tumor Growth and Metastasis in TNBC by Activating the FAK-AKT-mTOR Signaling Pathway. Molecular cancer therapeutics. 2018. December;17(12):2689–2701. doi: 10.1158/1535-7163.Mct-18-0399. PubMed PMID: 30237308; PubMed Central PMCID: PMCPMC6279518. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Orsi G, Barbolini M, Ficarra G, et al. GD2 expression in breast cancer. Oncotarget. 2017. May 9;8(19):31592–31600. doi: 10.18632/oncotarget.16363. PubMed PMID: 28415563; PubMed Central PMCID: PMCPMC5458232. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Becker DJ, Lowe JB. Fucose: biosynthesis and biological function in mammals. Glycobiology. 2003;13(7):41R–53R. doi: 10.1093/glycob/cwg054. [DOI] [PubMed] [Google Scholar]
- 127.Rosato FE, Seltzer M, Mullen J, et al. Serum fucose in the diagnosis of breast cancer. Cancer. 1971;28(6):1575–1579. doi: doi:. [DOI] [PubMed] [Google Scholar]
- 128.Takahashi M, Kuroki Y, Ohtsubo K, et al. Core fucose and bisecting GlcNAc, the direct modifiers of the N-glycan core: their functions and target proteins. Carbohydrate Research. 2009. 2009/August/17/;344(12):1387–1390. doi: 10.1016/j.carres.2009.04.031. [DOI] [PubMed] [Google Scholar]
- 129.Wang X, Gu J, Ihara H, et al. Core fucosylation regulates epidermal growth factor receptor-mediated intracellular signaling. J Biol Chem. 2006. February 3;281(5):2572–7. doi: 10.1074/jbc.M510893200. PubMed PMID: 16316986; eng. [DOI] [PubMed] [Google Scholar]
- 130.Carrascal MA, Silva M, Ramalho JS, et al. Inhibition of fucosylation in human invasive ductal carcinoma reduces E-selectin ligand expression, cell proliferation, and ERK1/2 and p38 MAPK activation. Molecular oncology. 2018. May;12(5):579–593. doi: 10.1002/1878-0261.12163. PubMed PMID: 29215790; PubMed Central PMCID: PMCPMC5928367. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tu C-F, Wu M-Y, Lin Y-C, et al. FUT8 promotes breast cancer cell invasiveness by remodeling TGF-β receptor core fucosylation. Breast cancer research : BCR. 2017;19(1):111–111. doi: 10.1186/s13058-017-0904-8. PubMed PMID: 28982386; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Koh YW, Lee HJ, Ahn JH, et al. Expression of Lewis X is associated with poor prognosis in triple-negative breast cancer. American journal of clinical pathology. 2013. June;139(6):746–53. doi: 10.1309/ajcp2e6qndidpttc. PubMed PMID: 23690116; eng. [DOI] [PubMed] [Google Scholar]
- 133.Brooks SA, Leathem AJ. Expression of the CD15 antigen (Lewis x) in breast cancer. The Histochemical journal. 1995. September;27(9):689–93. PubMed PMID: 8557532; eng. [PubMed] [Google Scholar]
- 134.Yue L, Han C, Li Z, et al. Fucosyltransferase 8 expression in breast cancer patients: A high throughput tissue microarray analysis. Histology and histopathology. 2016. May;31(5):547–55. doi: 10.14670/hh-11-693. PubMed PMID: 26596733; eng. [DOI] [PubMed] [Google Scholar]
- 135.Kailemia MJ, Park D, Lebrilla CB. Glycans and glycoproteins as specific biomarkers for cancer [journal article]. Analytical and Bioanalytical Chemistry. 2017. January 01;409(2):395–410. doi: 10.1007/s00216-016-9880-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chang MC, Souter LH, Kamel-Reid S, et al. Clinical utility of multigene profiling assays in early-stage breast cancer. Current oncology (Toronto, Ont). 2017. October;24(5):e403–e422. doi: 10.3747/co.24.3595. PubMed PMID: 29089811; PubMed Central PMCID: PMCPMC5659165. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Habel LA, Shak S, Jacobs MK, et al. A population-based study of tumor gene expression and risk of breast cancer death among lymph node-negative patients. Breast cancer research : BCR. 2006;8(3):R25. doi: 10.1186/bcr1412. PubMed PMID: 16737553; PubMed Central PMCID: PMCPMC1557737. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Penault-Llorca FM, Filleron T, Asselain B, et al. Prediction of recurrence with the Oncotype DX recurrence score in node-positive, HR-positive, breast cancer patients treated with adjuvant chemotherapy: Results from PACS01 trial. Journal of Clinical Oncology. 2014. 2014/May/20;32(15_suppl):11052–11052. doi: 10.1200/jco.2014.32.15_suppl.11052. [DOI] [Google Scholar]
- 139.Xin L, Liu Y-H, Martin TA, et al. The Era of Multigene Panels Comes? The Clinical Utility of Oncotype DX and MammaPrint. World Journal of Oncology. 2017. 05/04 03/29/accepted;8(2):34–40. doi: 10.14740/wjon1019w. PubMed PMID: PMC5649994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Apuri S. Neoadjuvant and Adjuvant Therapies for Breast Cancer. Southern medical journal. 2017. October;110(10):638–642. doi: 10.14423/smj.0000000000000703. PubMed PMID: 28973704; eng. [DOI] [PubMed] [Google Scholar]
- 141.Sokolenko AP, Imyanitov EN. Molecular Diagnostics in Clinical Oncology. Frontiers in molecular biosciences. 2018;5:76. doi: 10.3389/fmolb.2018.00076. PubMed PMID: 30211169; PubMed Central PMCID: PMCPMC6119963. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.de Leoz MLA, Young LJT, An HJ, et al. High-Mannose Glycans are Elevated during Breast Cancer Progression. Molecular & Cellular Proteomics : MCP. 2011. November/19 07/06/received 11/15/revised;10(1):M110.002717. doi: 10.1074/mcp.M110.002717. PubMed PMID: PMC3013453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Pierce A, Saldova R, Abd Hamid UM, et al. Levels of specific glycans significantly distinguish lymph node-positive from lymph node-negative breast cancer patients. Glycobiology. 2010;20(10):1283–1288. doi: 10.1093/glycob/cwq090. [DOI] [PubMed] [Google Scholar]
- 144.Jia Y, Chen Y, Wang Q, et al. Exosome: emerging biomarker in breast cancer. Oncotarget. 2017. June 20;8(25):41717–41733. doi: 10.18632/oncotarget.16684. PubMed PMID: 28402944; PubMed Central PMCID: PMCPMC5522217. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ruhaak LR, Xu G, Li Q, et al. Mass Spectrometry Approaches to Glycomic and Glycoproteomic Analyses. Chem Rev. 2018. September 12;118(17):7886–7930. doi: 10.1021/acs.chemrev.7b00732. PubMed PMID: 29553244; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hill JJ, Tremblay TL, Fauteux F, et al. Glycoproteomic comparison of clinical triple-negative and luminal breast tumors. Journal of proteome research. 2015. March 6;14(3):1376–88. doi: 10.1021/pr500987r. PubMed PMID: 25658377; eng. [DOI] [PubMed] [Google Scholar]
- 147.Zhang H, Li XJ, Martin DB, et al. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nature biotechnology. 2003. June;21(6):660–6. doi: 10.1038/nbt827. PubMed PMID: 12754519; eng. [DOI] [PubMed] [Google Scholar]
- 148.Powers TW, Neely BA, Shao Y, et al. MALDI imaging mass spectrometry profiling of N-glycans in formalin-fixed paraffin embedded clinical tissue blocks and tissue microarrays. PloS one. 2014;9(9):e106255. doi: 10.1371/journal.pone.0106255. PubMed PMID: 25184632; PubMed Central PMCID: PMCPMC4153616. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Turashvili G, Brogi E. Tumor Heterogeneity in Breast Cancer. Frontiers in medicine. 2017;4:227–227. doi: 10.3389/fmed.2017.00227. PubMed PMID: 29276709; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wang J, Ma Z, Carr SA, et al. Proteome Profiling Outperforms Transcriptome Profiling for Coexpression Based Gene Function Prediction. Mol Cell Proteomics. 2017. January;16(1):121–134. doi: 10.1074/mcp.M116.060301. PubMed PMID: 27836980; PubMed Central PMCID: PMCPMC5217778. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Johansson HJ, Socciarelli F, Vacanti NM, et al. Breast cancer quantitative proteome and proteogenomic landscape. Nature Communications. 2019. 2019/April/08;10(1):1600. doi: 10.1038/s41467-019-09018-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Matsunuma R, Chan DW, Kim B-J, et al. DPYSL3 modulates mitosis, migration, and epithelial-to-mesenchymal transition in claudin-low breast cancer. Proceedings of the National Academy of Sciences. 2018;115(51):E11978–E11987. doi: 10.1073/pnas.1810598115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Griffith OL, Spies NC, Anurag M, et al. The prognostic effects of somatic mutations in ER-positive breast cancer. Nature Communications. 2018. 2018/September/04;9(1):3476. doi: 10.1038/s41467-018-05914-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mertins P, Mani DR, Ruggles KV, et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature. 2016. June 2;534(7605):55–62. doi: 10.1038/nature18003. PubMed PMID: 27251275; PubMed Central PMCID: PMCPMC5102256. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Mardamshina M, Geiger T. Next-Generation Proteomics and Its Application to Clinical Breast Cancer Research. The American journal of pathology. 2017;187(10):2175–2184. doi: 10.1016/j.ajpath.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 156.Banazadeh A, Veillon L, Wooding KM, et al. Recent advances in mass spectrometric analysis of glycoproteins. Electrophoresis. 2017. January;38(1):162–189. doi: 10.1002/elps.201600357. PubMed PMID: 27757981; PubMed Central PMCID: PMCPMC5221507. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hu H, Khatri K, Klein J, et al. A review of methods for interpretation of glycopeptide tandem mass spectral data. Glycoconjugate journal. 2016. June;33(3):285–96. doi: 10.1007/s10719-015-9633-3. PubMed PMID: 26612686; PubMed Central PMCID: PMCPMC4882288. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ruhaak LR, Xu G, Li Q, et al. Mass Spectrometry Approaches to Glycomic and Glycoproteomic Analyses. Chemical Reviews. 2018. 2018/March/19. doi: 10.1021/acs.chemrev.7b00732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Sanda M, Zhang L, Edwards NJ, et al. Site-specific analysis of changes in the glycosylation of proteins in liver cirrhosis using data-independent workflow with soft fragmentation. Anal Bioanal Chem. 2017. January;409(2):619–627. doi: 10.1007/s00216-016-0041-8. PubMed PMID: 27822650; PubMed Central PMCID: PMCPMC5370557. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Thaysen-Andersen M, Packer NH. Advances in LC–MS/MS-based glycoproteomics: Getting closer to system-wide site-specific mapping of the N- and O-glycoproteome. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 2014. 2014/09/01/;1844(9):1437–1452. doi: 10.1016/j.bbapap.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 161.Klein J, Carvalho L, Zaia J. Application of network smoothing to glycan LC-MS profiling. Bioinformatics (Oxford, England). 2018. October 15;34(20):3511–3518. doi: 10.1093/bioinformatics/bty397. PubMed PMID: 29790907; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Vos DRN, Jansen I, Lucas M, et al. Strategies for managing multi-patient 3D mass spectrometry imaging data. J Proteomics. 2019. February 20;193:184–191. doi: 10.1016/j.jprot.2018.10.008. PubMed PMID: 30343012; eng. [DOI] [PubMed] [Google Scholar]
- 163.Liang Y, Wang F, Zhang P, et al. Development of a Framework for Large Scale Three-Dimensional Pathology and Biomarker Imaging and Spatial Analytics. AMIA Joint Summits on Translational Science proceedings AMIA Joint Summits on Translational Science. 2017;2017:75–84. PubMed PMID: 28815110; eng. [PMC free article] [PubMed] [Google Scholar]
- 164.Gray CJ, Thomas B, Upton R, et al. Applications of ion mobility mass spectrometry for high throughput, high resolution glycan analysis. Biochimica et biophysica acta. 2016. August;1860(8):1688–709. doi: 10.1016/j.bbagen.2016.02.003. PubMed PMID: 26854953; eng. [DOI] [PubMed] [Google Scholar]
- 165.Fenn LS, McLean JA. Structural separations by ion mobility-MS for glycomics and glycoproteomics. Methods in molecular biology (Clifton, NJ). 2013;951:171–94. doi: 10.1007/978-1-62703-146-2_12. PubMed PMID: 23296531; PubMed Central PMCID: PMCPMC5102027. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Skraskova K, Claude E, Jones EA, et al. Enhanced capabilities for imaging gangliosides in murine brain with matrix-assisted laser desorption/ionization and desorption electrospray ionization mass spectrometry coupled to ion mobility separation. Methods (San Diego, Calif). 2016. July 15;104:69–78. doi: 10.1016/j.ymeth.2016.02.014. PubMed PMID: 26922843; eng. [DOI] [PubMed] [Google Scholar]
- 167.Hinneburg H, Korac P, Schirmeister F, et al. Unlocking Cancer Glycomes from Histopathological Formalin-fixed and Paraffin-embedded (FFPE) Tissue Microdissections. Mol Cell Proteomics. 2017. April;16(4):524–536. doi: 10.1074/mcp.M116.062414. PubMed PMID: 28122943; PubMed Central PMCID: PMCPMC5383776. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Malaker SA, Pedram K, Ferracane MJ, et al. The mucin-selective protease StcE enables molecular and functional analysis of human cancer-associated mucins. Proceedings of the National Academy of Sciences of the United States of America. 2019. April 9;116(15):7278–7287. doi: 10.1073/pnas.1813020116. PubMed PMID: 30910957; PubMed Central PMCID: PMCPMC6462054. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Yang W, Ao M, Hu Y, et al. Mapping the O-glycoproteome using site-specific extraction of O-linked glycopeptides (EXoO). Molecular systems biology. 2018. November 20;14(11):e8486. doi: 10.15252/msb.20188486. PubMed PMID: 30459171; PubMed Central PMCID: PMCPMC6243375. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Tzanakakis G, Neagu M, Tsatsakis A, et al. Proteoglycans and Immunobiology of Cancer-Therapeutic Implications. Frontiers in immunology. 2019;10:875. doi: 10.3389/fimmu.2019.00875. PubMed PMID: 31068944; PubMed Central PMCID: PMCPMC6491844. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Soliman NA, Yussif SM, Shebl AM. Syndecan-1 could be added to hormonal receptors and HER2/neu in routine assessment of invasive breast carcinoma, relation of its expression to prognosis and clinicopathological parameters. Pathology, research and practice. 2019. May;215(5):977–982. doi: 10.1016/j.prp.2019.02.003. PubMed PMID: 30738694; eng. [DOI] [PubMed] [Google Scholar]
- 172.Vikas P, Borcherding N, Zhang W. The clinical promise of immunotherapy in triple-negative breast cancer. Cancer management and research. 2018;10:6823–6833. doi: 10.2147/CMAR.S185176. PubMed PMID: 30573992; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Nanda R, Chow LQ, Dees EC, et al. Pembrolizumab in Patients With Advanced Triple-Negative Breast Cancer: Phase Ib KEYNOTE-012 Study. J Clin Oncol. 2016. July 20;34(21):2460–7. doi: 10.1200/jco.2015.64.8931. PubMed PMID: 27138582; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Adams S, Loi S, Toppmeyer D, et al. Phase 2 study of pembrolizumab as first-line therapy for PD-L1–positive metastatic triple-negative breast cancer (mTNBC): Preliminary data from KEYNOTE-086 cohort B. Journal of Clinical Oncology. 2017. 2017/May/20;35(15_suppl):1088–1088. doi: 10.1200/JCO.2017.35.15_suppl.1088. [DOI] [Google Scholar]
- 175.Adams S, Schmid P, Rugo HS, et al. Phase 2 study of pembrolizumab (pembro) monotherapy for previously treated metastatic triple-negative breast cancer (mTNBC): KEYNOTE-086 cohort A. Journal of Clinical Oncology. 2017. 2017/May/20;35(15_suppl):1008–1008. doi: 10.1200/JCO.2017.35.15_suppl.1008. [DOI] [Google Scholar]
- 176.Li C-W, Lim S-O, Chung EM, et al. Eradication of Triple-Negative Breast Cancer Cells by Targeting Glycosylated PD-L1. Cancer Cell. 2018;33(2):187–201.e10. doi: 10.1016/j.ccell.2018.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Salatino M, Girotti MR, Rabinovich GA. Glycans Pave the Way for Immunotherapy in Triple-Negative Breast Cancer. Cancer Cell. 2018;33(2):155–157. doi: 10.1016/j.ccell.2018.01.015. [DOI] [PubMed] [Google Scholar]