Abstract
Interleukin 9 (IL-9) is an important mediator of allergic disease that is critical for mast cell driven diseases. IL-9 is produced by many cell types including T cells, basophils, and mast cells. Yet, how IL-9 is regulated in mast cells or basophils is not well characterized. In this report we tested the effects of deficiency of a mouse Il9 gene regulatory element (Il9 CNS-25) in these cells in vivo and in vitro. In mast cells stimulated with IL-3 and IL-33, the Il9 CNS-25 enhancer is a potent regulator of mast cell Il9 gene transcription and epigenetic modification at the Il9 locus. Our data show preferential binding of STAT5 and GATA1 to CNS-25 over the Il9 promoter in mast cells, and that T cells and mast cells have differing requirements for the induction of IL-9 production. Il9 CNS-25 is required for IL-9 production from T cells, basophils, and mast cells in a food allergy model, and deficiency in IL-9 expression results in decreased mast cell expansion. In a Nippostrongylus brasiliensis infection model we observed a similar decrease in mast cell accumulation. Although decreased mast cells correlated with higher parasite egg burden and delayed clearance in vivo, T cell-deficiency in IL-9 also likely contributes to the phenotype. Thus, our data demonstrate IL-9 production in mast cells and basophils in vivo requires Il9 CNS-25, and that Il9 CNS-25-dependent IL-9 production is required for mast cell expansion during allergic intestinal inflammation.
Introduction
Interleukin-9 (IL-9) is a pleiotropic cytokine that impacts allergic inflammation, and mast cell expansion and function (1, 2, 3). IL-9 is produced by several cell types including a specialized subset of T helper cells termed Th9 cells, innate lymphoid cells, and mast cells themselves (1, 2, 4). Much of our current understanding of IL-9 regulation comes from analysis of T cells (5, 6). IL-9 regulation in mast cells or basophils has not been studied in detail.
Mast cells are tissue resident cells of the innate immune system. They are one of the primary components of IgE-mediated inflammation in diseases such as food allergy, asthma, and helminth infections (7, 4, 8). Mast cells can be found in virtually all tissues, especially those in contact with the environment like skin and mucosa. Basal mast cell numbers in vivo are limited, but during disease development, mast cells accumulate in vivo (8). The process of mast cell expansion during disease is not well understood but there is evidence that IL-9 is responsible in mouse models of asthma and food allergy (1, 2). In the house dust mice (HDM) asthma model, mast cell numbers increase in response to increased IL-9 levels in the lung (1, 3). Recent work by Chen et al. (2), showed that in an OVA food allergy model, the induction of mucosal mast cells producing high concentrations of IL-9 (MMC9), plays an important role in susceptibility to IgE-mediated food allergy. Although some work has been done examining IL-9 production in mast cells, much of this was done in vitro (9, 10, 11).
Like mast cells, basophils are innate immune cells that circulate and are predominantly found in the blood (12). Basophils also contribute to allergic responses and have some functional overlap with mast cells, including IgE-mediated degranulation responses (12). Although IL-9 production in basophils has been observed, regulation in these cells has not been studied.
We recently described the importance of a DNA regulatory region (CNS-25) in the Il9 gene locus (5). We have demonstrated that this region regulated IL-9 production in T cells, and that animals lacking CNS-25 have reduced mast cell numbers and airway reactivity in the A. fumigatus-induced asthma model (5). We further observed mast cells and basophil IL-9 production was affected by Il9 CNS-25-deficiency using acute stimulation models, and in vitro derived mast cells and basophils. The effects of Il9 CNS-25-deficiency on IL-9 production from mast cells and basophils in vivo, and particularly in models where intestine is the target organ of inflammation, are lacking.
In this study we demonstrate that Il9 CNS-25 is required for appropriate IL-9 production in a food allergy model in basophils, and in the two major IL-9-secreting populations in the intestine, T cells and MMC9 cells. In a detailed analysis of the Il9 gene in cultured mast cells, we find that the locus is more activated in mast cells than in T cells, that activity is dependent on Il9 CNS-25, and that the locus is poised to be activated in response to the cytokine environment. We observed that the effects of Il9 CNS-25-deficiency on MC precursors is genetic background-dependent. Importantly, we observed that intestinal mastocytosis in both food allergy and N. brasiliensis infection model is dependent on Il9 CNS-25. Together, these data indicated that Il9 CNS-25-deficiency has as profound an effect on IL-9-producing mast cells and basophils as on T cells, and that Il9 CNS-25-dependent IL-9 production is required for mast cell expansion in allergic intestinal inflammation.
Materials and Methods
Mice
CNS-25-deleted mice (Il9ΔCNS-25) were generated as previously described (5) and used either on a C57BL/6 background or were backcrossed five generations to the BALB/c genetic background, and were maintained under specific pathogen-free conditions. C57BL/6 and BALB/cJ female and male mice were purchased from the Jackson Laboratory and used at the age of 6–12 weeks old. All experiments were performed with the approval of the Indiana University Institutional Animal Care and Use Committee.
In vitro cell differentiation
Naive CD4+CD62L+ T cells (purified using MACS (Miltenyi Biotec)) were activated with plate-bound anti-CD3 (2 μg ml−1 145–2C11; BioXCell) and soluble anti-CD28 (0.5 μg ml−1; BD Pharmingen) in complete culture media (RPMI 1640, ThermoFisher Scientific) containing 10% Fetal bovine serum (FBS, Atlanta Biologicals), 1% antibiotics (penicillin and streptomycin / stock; Pen 5000 μ ml−1, Strep 5000 μg ml−1 ), 1 mM sodium pyruvate, 1 mM L-Glutamine, 2.5 ml of Non-essential amino acids (Stock; 100 X), 5mM HEPES (all from LONZA) and 57.2 μM 2-Mercapoethanol (Sigma-Aldrich) with additional cytokines (all from PeproTech) and antibodies (all from BioXCell) to generate Th2 cells (20 ng ml−1 IL-4; and 10 μg ml−1 anti-IFNγ XMG) or Th9 cells (20 ng ml−1 IL-4, 2 ng ml−1 hTGF-β1, 10 μg ml−1 anti-IFNγ XMG). Cells were grown at 37°C under 5% CO2 and were expanded after 3 days with original concentration of cytokines in fresh medium. To generate mast cells, basophils, and eosinophils; bone marrow cells were isolated and RBCs were lysed using ACK Lysis Buffer (Thermo Fisher Scientific). Cells were cultured in RPMI 1640 medium (Invitrogen Life Technologies, Carlsbad, CA) containing 10% FBS, 2 mM l-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, 1 mM sodium pyruvate, and 1 mM HEPES. Media was supplemented with IL-3 (10ng/ml) and SCF (30ng/ml) for mast cells and 20 ng of IL-3 for basophils. After 7 days of culture, Basophils were isolated using anti-PE CD49b microbeads with a purity of more than 80% as determined by flow cytometry. Mast cells were allowed to mature for 21 days with purity of more than 90% determined by the co-expression of FcϵR1 and c-kit via flow cytometry. Unless specified, mature mast cells, basophils were set to 1×106 cells per ml in 10ng/ml of both IL-3 and SCF and stimulated with 50ng/ml of IL-33 for 4 hours (mRNA) or 16 hours (ELISA). Eosinophil culture was adopted from (13). Briefly, bone marrow cells were isolated and RBCs were lysed using ACK lysis buffer. Cells were cultured in RPMI 1640 medium (Invitrogen Life Technologies, Carlsbad, CA) containing 10% FBS, 2 mM l-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, 1 mM sodium pyruvate, 1 mM HEPES, and 50 μM 2-Mercapoethanol. Cells were cultured at 1×106/ml in 100ng/ml of SCF and FLT3-Ligand (Biolegend) for 5 days. On day 5, cells were washed and cultured in cRPMI plus 10ng/ml of IL-5 (Biolegend) thereafter. Eosinophils used on day 14 of culture were 95% Siglec F+.
In vitro cell stimulation
Cultured basophils, mast cells, and eosinophils were set to a concentration of 1×106/mL in complete RPMI. For stimulation, cells were treated with the following factors IL-3 (10ng/ml), IL-33 (50ng/ml), LPS (1μg/ml), TGF-beta (25ng/ml), TSLP (25ng/ml ), IL-25 (25ng/ml), and GMCSF (25ng/ml) for 4 hours for qRT-PCR, 16 hours for ELISA, and 24 hours for ChIP assay. For antigen stimulation cells were primed with DNP specific IgE (0.5ug/ml clone C38–2) for 16 hours, unbound IgE was washed out and cells were stimulated with DNP-HSA (50ng/ml) for 4 hours for qRT-PCR. Eosinophils were stimulated with IL-1-beta (20ng/ml), LPS (50ng/ml), and IL-33 (50ng/ml) for four hours for qRT-PCR.
Basophil and mast cell frequencies
Bone marrow cells were isolated as described above. The frequencies of basophils and mast cells and their progenitors were gated as follows. Mature mast cells in bone marrow and peritoneum were gated as FcϵR1 and c-kit double positive. Mature basophils in blood and peritoneum were gated as FcϵR1 and CD49b double positive. Bone marrow mast cell progenitors (MCp) were gated as lineage negative (CD5, B220, CD11b, CD27, anti-Gr-1(Ly6G/C), Ly6C, Sca-1, Ter119, CD19, NK1.1), FcϵR1lowc-Kit+ST2+β7+. Basophil mast cell progenitor (BMCp) in the spleen was gated as lineage negative (B220, CD3, Ly6C/G, NK1.1, GR1, Ter119, CD5) c-Kit+FcγRII/IIIhiβ7hiST2+.
OVA induced food allergy model
Mice, either Il9ΔCNS-25 or Il9WT on BALBc background (except for Supplement Figure 1E which were on C57BL/6 background), were interperitoneally (i.p.) injected with 100μg of chicken ovalbumin, “OVA,” (Sigma-Aldrich) and 1mg of imject-alum (Thermo Fisher Scientific) in a total volume of 200μl Q.S. with PBS. After immunization as shown in Figure 1A, mice were gavaged with 50mg of OVA in 200μl of water 6 times before sacrifice on day 29. Mice were starved of food and water for 1 hour before and 30 minutes after each gavage. Naive animals were gavaged with 50mg of OVA in 200μl water on day 28. Incidence of diarrhea was measured by observing mice for 1 hour after gavage on day 28. Lamina propria cells were isolated using Lamina Propria Disassociation Kit, mouse (Ref: 130–097-410) from Miltenyi Biotech. Techniques were performed according to manufacturer’s instructions. Blood was isolated via cardiac puncture on day 29 and stimulated with PMA (5ng/ml, Sigma Aldrich) and ionomycin (500ng/ml, EMD Millipore) for 2 hours followed by monensin (2μM, BioLegend) for a total of 6 hours.
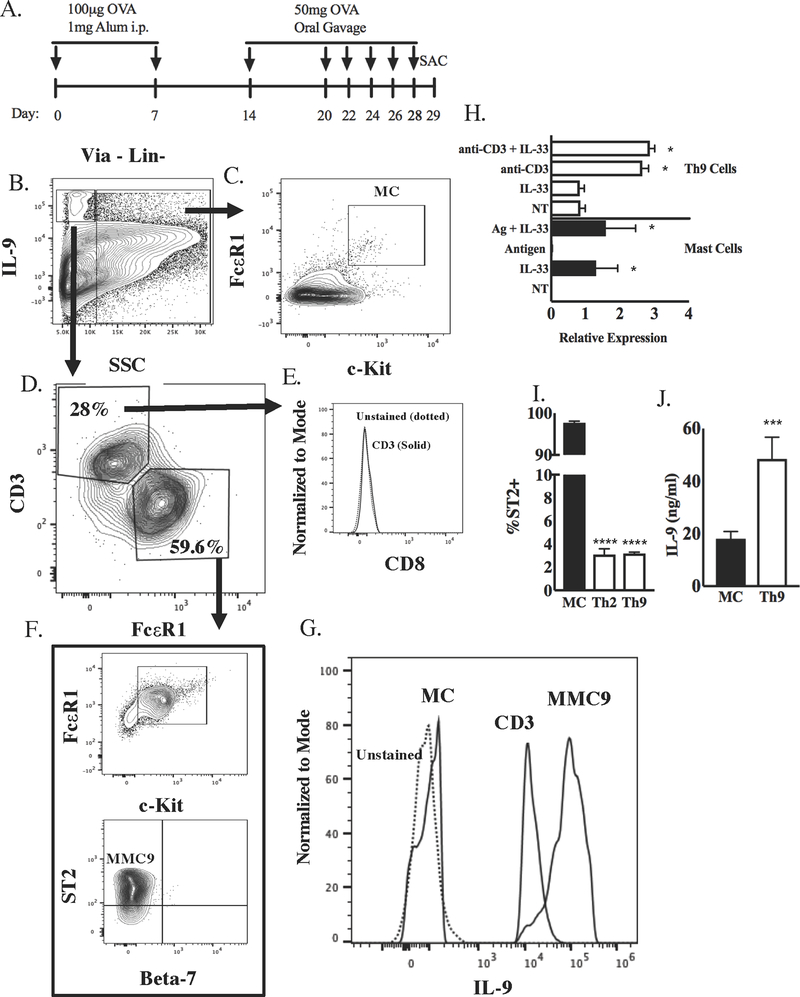
Figure 1. Mast cells and T cells are the main IL-9 producing cells in lamina propria during OVA food allergy model.
A, Schematic of OVA food allergy model using BALB/c mice. Lamina propria cells were isolated on day 29 and stimulated with PMA (5ng/ml) and ionomycin (500ng/ml) for 6 hours to analyze IL-9+ populations. B, Gating strategy for analysis of IL-9+ SSC-low and SSC-high populations of lamina propria cells. C, The IL-9-low SSC-high population was gated for expression of c-Kit and FcεR1 (mature mast cells, mMC). D, The IL-9-high SSC-low population was gated for CD3 and FcεR1 expression. E, CD8 expression on the CD3+ population in (D). F, The FcεR1+ population in (D) was further analyzed for expression of c-Kit, ST2, and integrin β7, characteristics of the MMC9 population. G, Comparative IL-9 staining in MMC9, T cells, and mMC gated from (B-D). H, Il9 induction from in vitro-generated Th9 cells (cultured with IL-4 and TGF-beta and stimulated with anti-CD3 and/or IL-33) and in bone marrow derived mast cells (BMMC; cultured with IL-3 and SCF and stimulated with Antigen and/or IL-33) was measured via qPCR. p value indicates comparison of treatment group with NT in the same cell culture (*, p value < 0.05). I, Percent ST2 expression on bone marrow-derived MCs compared to in vitro culture-generated Th2, and Th9 cells was measured via flow cytometry. J, ELISA was used to measure IL-9 release in Th9 cultures stimulated with anti-CD3 compared to mast cell cultures stimulated with IL-33. Data are presented as the mean ± SEM of three independent experiments (n=3/group).
Histology
Small intestines were flushed with PBS and 1 cm of jejunum was cut, fixed in formalin for at least 24 hours. Full length colons were flushed with PBS, cut open to rectangle and rolled onto toothpicks and placed in histology cassettes that were submerged in formalin for at least 24 hours. Tissues were stained using standard H&E, PAS, CAE protocols.
Helminth Infection
The life cycle of N. brasiliensis was maintained as previously described (14). Infective larvae (L3) were prepared from mouse fecal cultures. 650 N. brasiliensis L3 were injected sub-cutaneously into either Il9ΔCNS-25 or Il9WT mice on C57BL/6 background. Mice were monitored for eggs per gram of feces (EPG) by collecting feces by weight and resuspending in flotation solution (Saturated NaCl solution) based on 1 gram feces to 60mL of flotation solution, followed by counting two chambers in a McMaster Counting Slide, taking the average, and multiplying by 400 to obtain EPG (15),
Flow Cytometry
For flow cytometry analysis, cells were surface stained, fixed in 4% paraformaldehyde for 15 minutes. For intracellular staining, cells were subsequently fixed and stained using BD Bioscience Fixation/Permeabilization Solution Kits according to manufacturer’s protocol. Eosinophils in lamina propria were gated as viability dye negative, CD45+, Gr-1- CD11b+ Siglec-F+. Lineage markers cocktail for lamina propria cells contained antibodies for CD11b, CD11c, B220, Gr-1, CD335. Mature basophils were gated as FcεR1 and CD49b double-positive. Bone marrow MC progenitors (MCp) were gated as lineage-negative (CD5, B220, CD11b, CD27, Gr-1, Ly6C, Sca1, Ter119, CD19, and NK1.1) FcεR1 low c-Kit+ST2+β7+. Basophil MCp in the spleen was gated as lineage-negative (B220, CD3, Ly6C/G, NK1.1, Gr-1, Ter119, and CD5) c-Kit+FcγRII/IIIhiβ7hiST2+.
For cytokine staining, homogenized tissue was stimulated with PMA (5ng/ml) and ionomycin (500ng/ml) for 2 hours followed by monensin (2μM) for a total of 6 hours. Cells were surface stained for 30 minutes in FACS buffer. After fixation with 4% formaldehyde for 10 min at room temperature (RT), cells were washed two times with FACS buffer (PBS with 0.5% BSA). Fixed cells were permeabilized with permeabilization buffer (eBioscience), and stained for cytokines fluorochrome-conjugated antibodies (1:200 dilution) at 4 °C in dark for 30 minutes. Stained cells were washed two times with FACS buffer and resuspended with 500 μl of FACS buffer for flow analysis. Fluorescent antibodies for flow cytometric analysis are listed in Supplemental Table 1.
For in-vitro surface staining, cells were surface stained for 30 minutes in FACS buffer and fixed in 4% formaldehyde.
Enzyme-linked immunosorbent assay (ELISA)
IL-9 (Biolegend), IL-6 (Biolegend), Mast cell protease −2 (MCPT-1 eBiosciences) were all used according to manufacturer’s instructions. Total IgE from N. brasiliensis infected animals was measured as follows: plates were coated with rat-anti mouse IgE (B1E3) (16), in borate buffered saline, blocked (PBS with 0.02% Tween20 and 2% FBS), detected with biotin rat anti-mouse IgE (R1E4) and Streptavidin-Alkaline phosphatase (Southern Biotech). Plates were developed with phosphate tablets 2 (Sigma-Aldrich) dissolved in substrate buffer (0.1g MgCl2.6H2O, 0.2 NaN3, 50mL diethanolamine, pH to 9.8 per 500mL). Absorbance was measured at 405nm - 650nm (17). IgE standard was purified mouse IgE anti-DNP antibody(16).
Quantitative real-time PCR
Total RNA was extracted from cells or tissues using TRIzole reagents (Thermo Fisher Scientific) and reverse transcribed using qScript cDNA Synthesis Mix (QuantaBio) according to manufacturer’s instructions. For qPCR, Taqman real time PCR assay (ThermoFisher Scientific) or SYBR green master mix (Applied Biosystems) was used for gene expression analysis. Gene expression was normalized to housekeeping gene expression (β2-microglobulin). Relative gene expression was calculated by the change-in-threshold (2−ΔCT) method. All experiments were performed in duplicate in two independent experiments and results are presented as standard error of means of biological replicates. Taqman probes listed in Supplemental Table 1 and eRNA sequences were used as previously described (7).
Lentivirus production and infection
Human embryonic kidney (HEK) 293T cells were grown with 10 ml of DMEM with 10% FBS and 1% antibiotics in a 100 mm tissue culture dish. When confluency reached 95~99%, cells were transfected with lentiviral vectors expressing dCas9-VP64 (Addgene 61422) or gRNAs (5) to activate CNS-25, shGATA1 to knockdown GATA1 (Origene #TL500772), PAX2 and PMDG.2 using lipofectamine 3000. After 6 hours, media was changed to the lentivirus packaging media; Opti-MEM®I-GlutaMAX (ThermoFisher Scientific) with 5% FBS and 0.2 mM sodium pyruvate, and incubated another 16 hours. After collecting first and second batch of virus containing media, virus titer was concentrated by Lenti-X-concentrator (Takara Bio).
Sterile non-tissue-culture treated 24 well plates were coated with 50 μg/ml of Retronectin (Takara Bio) and incubate at 4°C overnight. Plate were washed with PBS and blocked with PBS with 2% bovine serum albumin (BSA) at RT for 30 minutes. Blocking buffer was removed, lentivirus stock (100 μl) and 500 μl of culture media was added to the plate. Lentivirus was loaded to the plate using centrifugation with 2000 x g at 32°C for 2 hours, BMMCs were added to the plate and incubated for 6 days. Cells were harvested on day 7 for sorting by fluorescence activated cell sorter (FACS; BD FACSAria) based on EGFP and tRFP and activated with IL-3 and IL-33 overnight for gene expression analysis.
Chromatin immunoprecipitation
In vitro-differentiated Th9 cells or bone marrow derived mast cells were crosslinked for 15 min with 1% formaldehyde at RT with rotation. The reaction was quenched by adding 0.125 M glycine and incubated at RT for 5 min. Cells were lysed with cell lysis buffer, followed by nuclear lysis buffer. Nuclei were degraded and chromosomal DNA were fragmented to a size range of 200–500 bp through ultrasonic processor (Vibra-cell). Post sonication, supernatant was diluted 10-fold with ChIP dilution buffer. After pre-clearing, the supernatant was incubated with the ChIP antibodies at 4 °C overnight with rotation. Immunocomplexes were precipitated with Protein Agarose A or G beads at 4 °C for 2–4 hours. Immunocomplexes were washed with low salt, high salt, LiCl and two times with TE buffer. After elution followed by reverse crosslinks, DNA was purified and analyzed by qPCR. Once normalized to Input DNA, the amount of output DNA of each target protein was calculated by subtracting that of the IgG control. ChIP antibodies are listed in Supplemental Table 1. Primers used are previously defined (5).
Statistical analysis
Two-tailed Student’s t test or one-way analysis of variance was used to generate p-value data for all data. Post hoc Tukey test was used for multiple comparisons.
Results
IL-9 production in mast cells
We recently identified Il9 CNS-25 as an enhancer of the Il9 gene locus important for the regulation of IL-9 production in T cells and mast cells (5). However, our previous work examined mast cell IL-9 production in vitro or following acute in vivo stimulation, and not during an active allergen sensitization in vivo. Given the importance of IL-9-producing mucosal mast cells (MMC9s) in a food allergy (2), we first examined IL-9-producing cells in this model. We subjected wild type BALB/c mice to an OVA induced food allergy model. Mice were immunized twice with OVA and Alum and then orally challenged with OVA six times (Fig 1A). Using flow cytometry, we gated IL-9-positive cells (both SSC-low and SSC-high) in the lamina propria following stimulation with PMA (5ng/ml) and ionomycin (500ng/ml) for 6 hours. We further gated on c-Kit and FcεR1 in the SSC-high population to identify mature mast cells (mMC) (Fig. 1B–C). The SSC-low population predominantly expressed CD3 and were negative for CD8 (Fig. 1B, D, E) or were FcεR1-positive and co-expressed c-Kit and ST2 but not integrin β7, the phenotype of MMC9 cells (Fig. 1B, D, F). We then compared IL-9 production in these gated populations and observed MMC9 had the highest IL-9 content, with IL-9 production in T cells slight lower, and minimal IL-9 detected in mMC (Fig. 1G). These results demonstrate that the two major IL-9+ populations in this model are MMC9s and T cells.
To further analyze these differences, we compared the conditions that generate IL-9-producing mast cells and T cells in vitro, using in vitro derived cells because it was not feasible to purify sufficient in vivo populations for the subsequent detailed analyses. Compared to Th9 cells (naïve CD4 T cells differentiated with TGFβ and IL-4; representative cultures in Supplemental Fig. 1A) that expressed Il9 following antigen receptor stimulation, bone marrow-derived mast cells (BMMC; representative cultures in Supplemental Fig. 1B) did not express Il9 following antigen crosslinking of receptor bound IgE (Fig 1H). IL-33 was a potent stimulator of Il9 expression in mast cells, but not Th9 cells, and this was consistent with a lack of IL-33 receptor expression in Th9 cells (Fig 1H–I). However, in contrast to the MMC9 cells that produced more IL-9 than T cells, IL-33-stimulated BMMC secreted less IL-9 than Th9 cells (Fig 1J). Overall these results indicate that there are distinct pathways leading to IL-9 production in T cells and mast cells.
Mast cell transcription factor binding at Il9 CNS-25
To begin to define mechanisms of IL-9 production by mast cells we cultured BMMCs in IL-3 and SCF and stimulated them with various factors. As shown in Figure 2A, LPS and IL-33 induced Il9 expression when cell cultures had IL-3 present. We found that in the absence of IL-3, IL-33 was unable to induce Il9 but could induce other cytokines such as Il6 (Fig 2B). We observed parallel cytokine dependence for the induction of Il9 and Il6 in C57BL/6 and BALB/c BMMC (Fig. 2B and Supplemental Fig. 1C). This suggested that STAT5 activation by IL-3 (18) was needed to induce Il9 expression in mast cells. Indeed, BMMC cultured with IL-3 alone, but not IL-33 alone, induced STAT5 phosphorylation (Fig 2C), and STAT5 was significantly bound to the Il9 locus at the promoter and the CNS-25 element (Fig 2D). Notably, addition of IL-33, that was required for IL-9 production, did not alter STAT5 binding (Fig 2D). Although GM-CSF and IgE cross linking can activate STAT5 in mast cells (18)(19), GM-CSF, but not antigen induced IgE cross linking, was able to substitute for IL-3 in this assay (Fig 2A).
Figure 2. Stat5 and Gata1 bind to Il9 gene locus in BMMCs.
Real-time PCR analysis of Il9 expression in BMMCs stimulated with various factors (A) or with and without IL-3 and/or IL-33 (B) for 4 hours. C, Stat5 phosphorylation in BMMCs stimulate with and without IL-3 and/or IL-33 for 15 minutes (flow cytometry). ChIP analysis of Stat5 (D) and Gata1 (E) binding to Il9 gene locus in BMMCs stimulated with IL-3 ± IL-33. F, shRNA was used to knockdown Gata1 in BMMCs. Cells were then stimulated with IL-3 + IL-33 and Gata1, Il9, and Il6 levels were measured via qPCR. Each bar is normalized to beta microglobulin 2 (β2m) what is set to 1. * p value < 0.05 (A, C-F), **** p value <0.0001 (B). Data are presented as the mean ± SEM of three (A-B) and two (C-F) independent experiments (n=4/group).
In T cells, GATA3 binds to the Il9 locus at enhancer elements including CNS-25 (5). In mast cells, GATA1, but not GATA3 is expressed at significant levels and was suggested to regulate IL-9 (20, 9). To determine if GATA1 bound Il9 in mast cells in a similar pattern, we performed ChIP assays. We observed GATA1 predominantly bound to the Il9 CNS-25 region, but not to other regulatory elements (Fig 2E). Additionally, knockdown of GATA1 lowered IL-33+IL-3 induced Il9 but not Il6 levels, suggesting that GATA1 is critical for IL-9 production in mast cells (Fig 2F).
Il9 CNS-25-dependent regulation in mast cells and basophils
IL-9 production in cultured mast cells is dependent on Il9 CNS-25 (5). We also found that BM-derived basophils can produce IL-9 in response to IL-33+IL-3 and Il9ΔCNS-25 basophils have diminished IL-9 production with no change in antigen induced degranulation (Supplemental Fig. 2A–B). Eosinophils are also able to express Il9 in some conditions (22), but we were unable to detect intracellular IL-9 in the eosinophil population in the OVA allergy model (data not shown). Minimal Il9 mRNA can be detected in eosinophil cultures (Supplemental Fig. 2C) and this level was very low compared to Th9 cells, mast cells, and basophil cultures (Supplemental Fig. 2D).
To further examine the Il9 locus in mast cells we analyzed RNA polymerase II and enhancer RNA (eRNA, short non-coding RNA transcribed near enhancers) at the CNS-25 element (Fig 3A–C). RNA pol II was enriched at the Il9 CNS-25 region (Fig 3B) and this correlated with eRNA, particularly distal to CNS-25 that was higher in mast cells than T cells (Fig 3C). RNA pol II binding at the Il9 promoter was enhanced by IL-33 treatment (Fig 3B).
Figure 3. CNS-25 regulates histone modification at distal sites in the Il9 gene locus in BMMCs.
A, Schematic of the Il9 gene locus. Zoomed boxed region shows regions against which eRNA primers were designed. ChIP analysis of RNA Polymerase II binding to Il9 gene locus in BMMC stimulated with IL-3 + IL-33 (B) (* p value < 0.05). Real-time PCR analysis of eRNA levels (C) in stimulated BMMCs and Th9 cells (* p value < 0.05). D, Histone modification markers in BMMCs and Th9 cells (ChIP). E, Histone modification markers in BMMCs from Il9WT and Il9ΔCNS-25 at CNS0 and CNS1. F, inflammatory gene expression in Il9WT and Il9ΔCNS-25 BMMCs stimulated with IL-33. G, Il9 and Il6 mRNA levels in BMMC transduced with dCas9-VP64 and gRNA for hMYOD or CNS-25. Cytokine mRNA levels normalized to Beta-2M (qPCR). D – G: *p < 0.05, ** p < 0.01, *** p < 0.001 **** p <0.0001. Data are presented as the mean ± SEM of two (B-E) and three (F-G) independent experiments (n=4/group).
We then compared histone modifications at the Il9 locus in mast cells and Th9 cells. The enrichment of active histone modifications (mono- and tri-methyl H3K4, acetylated H3K27) detected at the Il9 gene locus in BMMCs was generally higher than that detected in Th9 cells. This was particularly true for the CNS-25 region (Fig 3D). This was surprising considering Il9 transcription and production were higher in Th9 than in BMMC (Fig 1). In the absence of CNS-25, active histone modification markers are reduced at the Il9 gene promoter (CNS1) and enhancer (CNS0) region (Fig 3E). The effects of Il9 CNS-25-deficiency are specific because BMMCs from Il9ΔCNS-25 animals show reduced Il9 transcription, but transcription levels of Tnf, Il13, Il1b, Il6 and Mcpt2 that were comparable to BMMCs from Il9WT animals (Fig 3F). Moreover, antigen-induced degranulation in BMMCs from the Il9ΔCNS-25 animals was comparable to that of BMMCs from Il9WT animals (Supplemental Fig. 2E). These results suggest that Il9 CNS-25-deficiency does not impact mast cell development or the capacity to produce cytokines, either directly or indirectly.
We then used an approach to directly test the ability of activators bound to Il9 CNS-25 in mast cells to induce Il9 expression (23, 24). Retrovirus expressing a dCas9-VP64 fusion protein (24) was co-transduced with gRNAs that directed dCas9 binding to Il9 CNS-25 or a control gene (Fig 3G). Il9 transcription was significantly induced with the addition of guide RNAs targeting CNS-25, while Il6 transcription was not affected (Fig 3G). Thus, targeting of a transcriptional activator to Il9 CNS-25 in mast cells can increase transcription from the locus. Together, these results suggest that the Il9 locus in mast cells is in a more active configuration than in T cells and is poised to be induced by activators including cytokines and transcriptional activators recruited to the locus.
Il9 CNS-25 regulates IL-9 production in mast cells and basophils in a food allergy model
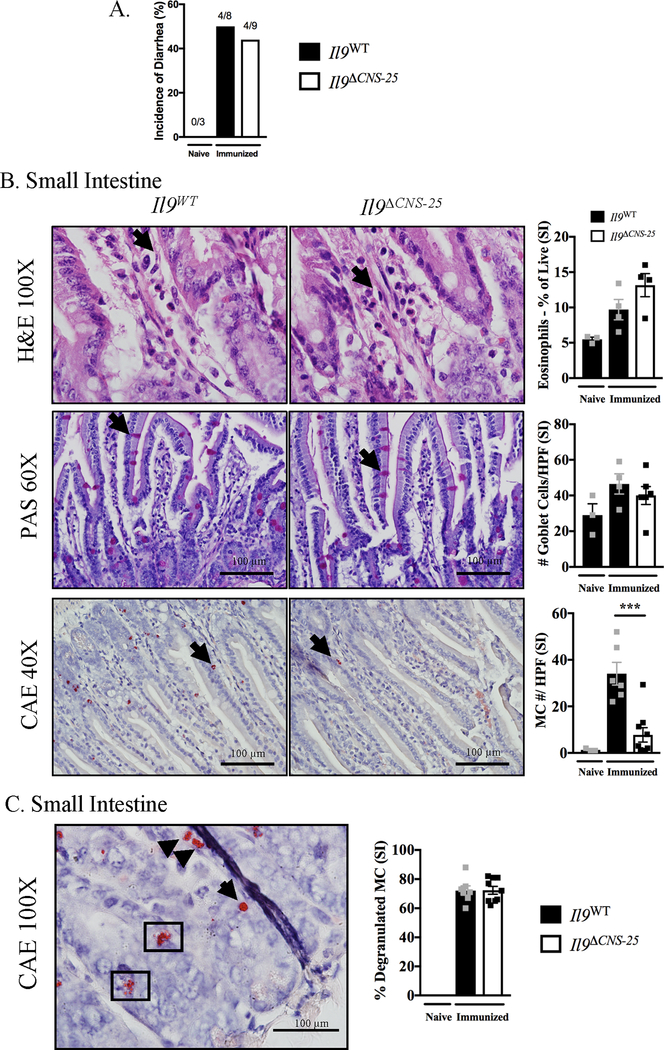
We next defined the requirement for Il9 CNS-25 in the development of IL-9-producing mast cells in vivo. We subjected Il9ΔCNS-25 and Il9WT BALB/c littermate control mice to the OVA induced food allergy model as shown in Figure 1A. We found that Il9ΔCNS-25 animals develop diarrhea and OVA specific IgE at levels comparable to Il9WT animals (Fig 4A and Supplemental Fig. 3A). As shown in representative hematoxylin and eosin (H&E) and Periodic Acid-Schiff (PAS) staining of the jejunum, there is no difference in cell infiltrate, percentage of eosinophils, and goblet cell hyperplasia between Il9ΔCNS-25 and Il9WT animals (Fig 4B). Chloroacetate esterase (CAE) staining to detect mast cells showed a significant decrease in the number of cells in the jejunum of Il9ΔCNS-25 animals compared to Il9WT animals although there was no difference in percent of the mast cells present in the tissue that had degranulated (Fig 4C), suggesting that although there are fewer mast cells in the Il9ΔCNS-25 mice, they are capable to activation in response to stimuli. Of note, mast cell expansion in the immunized control animals was seen in the small intestine but not the colon (Fig 4B and Supplemental Fig. 3B).
Figure 4. Il9ΔCNS-25 animals have reduced mast cell number in OVA food allergy model.
Il9WT and Il9ΔCNS-25 animals were subjected to OVA food allergy model as shown in Figure 1A. Incidence of diarrhea (A) was measured and small intestine tissue (B) was stained as follows: H&E (eosinophils), PAS (goblet cells) and CAE (mast cells) (*** p < 0.001). Representative images of Il9WT and Il9ΔCNS-25 animals. Quantification of eosinophils derived from flow cytometry staining. C, Representative image of CAE stained granulated (arrow) and degranulated (boxed) mast cells and percent degranulated mast cells in small intestine. Data are presented as the mean ± SEM of three independent experiments (n=4/group).
We then tested how IL-9 production was affected by the absence of Il9 CNS-25 in this model. Il9ΔCNS-25 MMC9 isolated from the lamina propria of immunized animals (gated as in Fig. 1) showed reduced IL-9 expression compared to Il9WT (Fig 5A). This is similar to the effect of Il9ΔCNS-25 deficiency on IL-9 production from BMMC (Supplemental Fig. 1A) (5). Additionally, the lamina propria of the Il9ΔCNS-25 animals had reduced CD3+CD4+IL-9+ Th9 cells (Fig 5A). Il9 but not Il4 gene expression was reduced in intestinal tissue of Il9ΔCNS-25 animals (Fig 5B). Interestingly, although the percentages of IL-9-positive T cells are decreased in Il9ΔCNS-25 mutant mice, the percentages of MMC9 cells are not altered by Il9ΔCNS-25 deficiency (Fig. 5A). Rather, the amount of IL-9 per cell (IL-9 MFI) is reduced in the MMC9 population. This further supports distinct effects of Il9ΔCNS-25 deficiency in mast cells and T cells.
Figure 5. Il9ΔCNS-25 animals have reduced IL-9 expression in T cells, mast cell, and basophils.
A, lamina propria cells from food allergic Il9WT and Il9ΔCNS-25 animals were isolated and stimulated for 6 hours. IL-9 expression in mast cells (expressed as MFI) and CD4+ T cells (expressed as % Th9 cells) are shown (*p < 0.05, ** p < 0.01). B, Il9 and Il4 mRNA expression in small intestinal tissue (*p < 0.05). Data are presented as the mean ± SEM of three independent experiments (n=4/group).
In addition to mast cells, we found that basophils also produce IL-9 during the food allergy response (Supplemental Fig. 3C). Accordingly, blood basophils of immunized Il9ΔCNS-25 animals showed reduced IL-9 levels (indicated as MFI) compared to Il9WT (Supplemental Fig. 3C). Additionally, immunized Il9ΔCNS-25 animals had lower IL-9+ T helper cells in the blood and in the mesenteric lymph nodes (Supplemental Fig. 3C–D).
Our previous report using mice on the C57BL/6 genetic background demonstrated that Il9ΔCNS-25 mice have reduced mast cell and basophils progenitors (5). Although C57BL/6 mice do not develop the MMC9 population in this model (2), when Il9ΔCNS-25 animals from C57BL/6 background were subjected to the OVA food allergy model we found that, compared to Il9WT counterparts, they have reduced IL-9+ blood basophils (with similar levels of total basophils), reduced intestinal mast cells, and diminished Il9 expression in intestinal tissues (Supplemental Fig. 3E), all consistent with observations in the BALB/c background mice. The intestinal mast cell numbers in animals on the C57BL/6 background were much lower than detected in BALB/c background animals. BALB/c Il9ΔCNS-25 animals had comparable mast cell progenitors (MCp) in the bone marrow and similar blood and mast cell basophil shared progenitor (MC/Bp) in the spleen (Supplemental Fig. 3F), indicating that the effect of Il9 CNS-25-deficiency on progenitor populations varied with the genetic background of the mice, possibly because the mast cell niche is not saturated in C57BL/6 mice and can more easily expand during a pathological response.
Il9 CNS-25-dependent N. brasiliensis immunity
To demonstrate the effects of Il9 CNS-25 deficiency in a mast cell-dependent model of immunity (4, 7), we infected C57BL/6 Il9ΔCNS-25 animals with L3 larvae of Nippostrongylus brasiliensis (Fig 6A). Il9ΔCNS-25 animals had higher worm burden, indicated by eggs recovered per gram of feces (EPG), on day 6 and 7 (Fig 6B). Eight days after infection, mesenteric lymph node cells from Il9ΔCNS-25 animals produced significantly lower amounts of Il4 and Il9, compared to cells from wild type mice (Fig 6C). Cytokine mRNA levels in stimulated spleen cells were comparable (Fig 6C). Mcpt1 levels in small intestine were significantly reduced in Il9ΔCNS-25 animals with no significant change in Mcpt2 and Rnase2 (Eosinophil-derived neurotoxin) levels (Fig 6D). Protease levels in the lung on day 8 post infection were unchanged and showed an overall lower expression compared to small intestine (Fig 6D). Intracellular IL-9, but not IL-4, was significantly reduced in CD3+CD4+T cells from mesenteric lymph nodes on day 8 post infection (Fig 6E). Total Serum IgE levels were enhanced in Il9ΔCNS-25 animals compared to Il9WT (Fig 6F) and this might be linked to delayed clearance of the worms and antigen persistence, rather than a repressive effect of IL-9 on IgE production. ELISA performed on mucus from Il9ΔCNS-25 animals showed reduced MCPT1 levels in the jejunum (Fig 6G). PAS staining of small intestine showed no change in number of goblet cells (Fig 6H). CAE staining of small intestine showed significantly diminished mast cell numbers with no change in mast cell degranulation in Il9ΔCNS-25 (Fig 6I). Thus, although the phenotypes in vivo are likely the result of diminished IL-9 production from multiple cells types including T cells and mast cells, these data demonstrate that CNS-25 deletion in mice reduces mastocytosis in the intestine during helminth infection, leading to impaired worm clearance.
Figure 6. Il9ΔCNS-25 animals have reduced parasite clearance, mast cell numbers, and Il9 expression in Nippostrongylus brasiliensis infected animals.
Il9WT and Il9ΔCNS-25 animals were subjected to mouse helminth infection model (A) and eggs per gram (EPG) was counted on days 6, 7, and 8 (B). C -D, Spleen, mesenchymal lymph nodes, small intestine, and lungs from Il9WT and Il9ΔCNS-25 animals isolated on day 8 and stimulated for 4 hours and gene expression was measured via qRT-PCR. Intracellular IL-9 and IL-4 levels in CD3+CD4+ cells from stimulated MLN (E) were measured via flow cytometery and plasma IgE levels (F) were measured by ELISA. G, MCPT-1 levels in mucus from jejunum of Il9WT and Il9ΔCNS-25 animals measured by ELISA. H, small intestine was stained with PAS for goblet cell count. I, CAE staining of jejunum was used to measure mast cells and percent degranulated mast cells in Il9WT and Il9ΔCNS-25 animals. (*p < 0.05, ** p < 0.01). Data are presented as the mean ± SEM of two independent experiments (n=5/group).
Discussion
Increasing evidence shows that IL-9 plays a critical role in the development and maintenance of allergic diseases. In particular, studies of allergic asthma and food allergies show that IL-9 regulates mast cell hyperplasia during disease development (1, 25, 26). In the HDM model of allergic disease, blockade of IL-9 causes a reduction in mast cell numbers in the lung (1). Recent studies have found that IL-9 production regulates an important step in the development of food allergies (2). The precise source of IL-9 in this context is still not clear. Based on our recent description of an Il9 enhancer we have characterized the requirement for Il9 CNS-25 for in vivo and in vitro IL-9 production from mast cells and basophils and defined the requirement for Il9 CNS-25 in allergic intestinal inflammation.
The signals that activate IL-9 production appear to be distinct in T cells and mast cells. Whereas antigen receptor stimulation is the primary activator in T cells, cytokines, specifically IL-3 and IL-33, and TLR ligands, activate IL-9 in mast cells and basophils. This is also true of MMC9 cells studied ex vivo (2). The signaling pathways leading to IL-9 induction in mast cells are still unclear but likely involve STAT5 downstream of IL-3 and NF-κB downstream of IL-33 or TLR. STAT5 possibly establishes competence at the locus and allows the IL-33 signal to increase RNA pol II occupancy and transcription. This is interesting because other cytokine genes, like IL-6, did not require the STAT5-activating signal for induction by IL-33. The inability of IgE receptor crosslinking to mediate either of these signals suggests that mast cells respond with IL-9 production to a restricted set of stimuli.
There are a number of significant differences in how the Il9 locus is regulated in T cells and mast cells. Although Il9 CNS-25 is clearly required for IL-9 production by T cells and mast cells, both in vivo and in vitro, activating chromatin modifications (H3K27ac) and enhancer modifications (H3K4me1) were quantitatively higher in bone marrow derived mast cells than Th9 cells, despite transcription being higher in Th9 cells. We also observed high H3K4me3 modifications, usually associated with promoters, at CNS-25 in BMMC, contrasting basal amounts in Th9 cells. This correlated with increased eRNA adjacent to CNS-25 in mast cells compared to Th9 cells. It was also interesting that while GATA3 is bound to CNS-25 and CNS-6 in Th9 cells (5), GATA1 is predominantly bound only to CNS-25 in mast cells. The Il9 locus in mast cells was induced by cytokine treatment and by Cas9-mediated gene activation. These distinctions suggest that the Il9 locus is at a different activation state in mast cells and may be more poised for activation than the locus is in Th9 cells.
Another interesting feature highlighted in this work is how CNS-25-deficiency affects the IL-9-producing population. In Th9 cells, deletion of CNS-25 results in a smaller of percentage of cells becoming IL-9 secretors (5) (Fig. 5). This suggests that in Th9 cells, the CNS-25 element increases the number of cells acquiring an IL-9-secreting phenotype and is thus a competence factor for expression. In mast cells and basophils however, deletion of CNS-25 results in less IL-9 per cell (reflected in IL-9 staining MFI) but not a decrease in the percentage of IL-9-secreting cells. This suggests that in mast cells and basophils, CNS-25 is not a competence factor but rather an efficiency factor, determining how much IL-9 is made per cell. This might be a result of a more active locus in mast cells, particularly reflecting the distinct chromatin modifications at the locus between mast cells and T cells.
Although we clearly defined CNS-25-dependent mast cell IL-9 production in the food allergy model, there was no change in the incidence of diarrhea between wild type and mutant mice and we did not detect any differences in core body temperature (Fig 4B). Previous reports of food allergy responses have utilized IL-9/IL-9R deficient animals where there is a complete loss of IL-9 signaling, and diminished diarrhea response (2, 25, 26). The Il9ΔCNS-25 animals have a partial reduction of IL-9 levels and therefore it likely has enough IL-9 signaling to reach the threshold required for a diarrhea response comparable to Il9WT mice (5). In agreement with this is the comparable cell infiltrate and goblet cell hyperplasia seen in the jejunum of Il9ΔCNS-25 compared to Il9WT animals. Although eosinophils were difficult to identify on H&E stained slides, we show by flow cytometry that Il9ΔCNS-25 and Il9WT have the same proportions of eosinophils in the lamina propria (Fig 4C). The Il9ΔCNS-25 animals did have a significantly diminished number of mast cell numbers in the jejunum. Previous reports suggest that IL-9 regulates the MMC9 population in an autocrine manner (2). We speculate that the reduction of jejunum mast cells in Il9ΔCNS-25 animals is due to reduced IL-9 levels in mast cells and not T cells, though discriminating the function of these two populations will require additional study. We observed that rate of degranulation as detected by histological staining was similar in our studies, although we did not measure degranulation products directly. Clearly, aspects of inflammation in this model require IL-9, and diminished amounts might still be sufficient to generate some responses.
In helminth infections, mast cells play a critical effector role by causing expulsion of adult worms (4, 7). Reitz et al. recently demonstrated that BALB/c and C57BL/6 background IL-9R-deficient mice have higher worm burden during helminth infections. Additionally, Il9−/− mice also show higher worm burden and reduced mast cell numbers (4), suggesting that IL-9 is required for the mast cell-dependent worm expulsion. We found that Il9ΔCNS-25 have delayed N. brasiliensis clearance compared to Il9WT. The mucosal phenotype in mast cells is defined by the high expression of MCPT-1 and MCPT-2. Consequently, MCPT-1 and MCPT-2 are the primary mast cell proteases present in small intestine tissue during inflammation (27). Il9ΔCNS-25 animals have reduced Mcpt1 expression in small intestine and reduced MCPT-1 concentrations in mucus recovered from small intestines (Fig. 6D and G). Similar to the phenotype seen in OVA food allergy response, in the N. brasiliensis helminth model, ll9ΔCNS-25 animals had reduced mast cell accumulation. These results indicate that CNS-25 deletion has a functional effect on worm clearance during helminth infection where mast cells are critical effector cells, although it is still likely that T cells contribute to the IL-9-dependent pathology in this model.
In this report we demonstrate the requirement for the Il9 CNS-25 regulatory element in mast cells and basophil IL-9 production. We show distinct regulatory features of Il9 in mast cells and T cells and show that the Il9 CNS-25 element is required for MMC9, IL-33-stimulated mast cell, and basophil IL-9 production in vivo and in cultured cells. Additionally, we report that CNS-25-dependent IL-9 regulates mast cell numbers in food allergy and helminth infection where mast cells are important mediators of disease. Thus, Il9 CNS-25-dependent IL-9 production contributes to pathology in type 2 intestinal inflammation.
Supplementary Material
Key Points.
The Il9 CNS-25 is required for IL-9 production in mast cells and basophils
There is differential control of the Il9 gene between mast cells and T cells
Intestinal mast cell expansion requires Il9 CNS-25-dependent IL-9
Acknowledgements
The authors thank Drs. Alexander Dent, Baohua Zhou, and other members of the Kaplan lab for helpful comments and review of this manuscript.
This work was supported by PHS grants from the National Institutes of Health R01 AI057459, R01 AI129241, and R03 AI135356 to M.H.K. A.A.Q. was supported by T32 DK007519. Core facility usage was also supported by IU Simon Cancer Center Support Grant P30 CA082709 and U54 DK106846. Support provided by the Herman B Wells Center was in part from the Riley Children’s Foundation.
Abbreviations
- mMC
mature mast cell
- MMC9
mucosal mast cell producing IL-9
- MCp
mast cell precursor
Footnotes
Conflicts of Interest: The authors declare no conflicts of interest.
References
- 1.Sehra S, Yao W, Nguyen ET, Glosson-Byers NL, Akhtar N, Zhou B, and Kaplan MH 2015. TH9 cells are required for tissue mast cell accumulation during allergic inflammation. J. Allergy Clin. Immunol 136: 433–40.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen C-Y, Lee J-B, Liu B, Ohta S, Wang P-Y, Kartashov AV, Mugge L, Abonia JP, Barski A, Izuhara K, Rothenberg ME, Finkelman FD, Hogan SP, and Wang Y-H 2015. Induction of Interleukin-9-Producing Mucosal Mast Cells Promotes Susceptibility to IgE-Mediated Experimental Food Allergy. Immunity 43: 788–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kearley J, Erjefalt JS, Andersson C, Benjamin E, Jones CP, Robichaud A, Pegorier S, Brewah Y, Burwell TJ, Bjermer L, Kiener PA, Kolbeck R, Lloyd CM, Coyle AJ, and Humbles AA 2011. IL-9 governs allergen-induced mast cell numbers in the lung and chronic remodeling of the airways. Am. J. Respir. Crit. Care Med 183: 865–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Licona-Limón P, Henao-Mejia J, Temann AU, Gagliani N, Licona-Limón I, Ishigame H, Hao L, Herbert DR, and Flavell RA 2013. Th9 Cells Drive Host Immunity against Gastrointestinal Worm Infection. Immunity 39: 744–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koh B, Abdul Qayum A, Srivastava R, Fu Y, Ulrich BJ, Janga SC, and Kaplan MH 2018. A conserved enhancer regulates Il9 expression in multiple lineages. Nat Commun 9: 4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiao X, Fan Y, Li J, Zhang X, Lou X, Dou Y, Shi X, Lan P, Xiao Y, Minze L, and Li XC 2018. Guidance of super-enhancers in regulation of IL-9 induction and airway inflammation. J. Exp. Med 215: 559–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reitz M, Hartmann W, Rüdiger N, Orinska Z, Brunn M-L, and Breloer M 2018. Interleukin-9 promotes early mast cell-mediated expulsion of Strongyloides ratti but is dispensable for generation of protective memory. Sci Rep 8: 8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang T, Finn DF, Barlow JW, and Walsh JJ 2016. Mast cell stabilisers. Eur. J. Pharmacol 778: 158–168. [DOI] [PubMed] [Google Scholar]
- 9.Stassen M, Klein M, Becker M, Bopp T, Neudörfl C, Richter C, Heib V, Klein-Hessling S, Serfling E, Schild H, and Schmitt E 2007. p38 MAP kinase drives the expression of mast cell-derived IL-9 via activation of the transcription factor GATA-1. Mol. Immunol 44: 926–933. [DOI] [PubMed] [Google Scholar]
- 10.Stassen M, Müller C, Arnold M, Hültner L, Klein-Hessling S, Neudörfl C, Reineke T, Serfling E, and Schmitt E 2001. IL-9 and IL-13 production by activated mast cells is strongly enhanced in the presence of lipopolysaccharide: NF-kappa B is decisively involved in the expression of IL-9. J. Immunol 166: 4391–4398. [DOI] [PubMed] [Google Scholar]
- 11.Wiener Z, Falus A, and Toth S 2004. IL-9 increases the expression of several cytokines in activated mast cells, while the IL-9-induced IL-9 production is inhibited in mast cells of histamine-free transgenic mice. Cytokine 26: 122–130. [DOI] [PubMed] [Google Scholar]
- 12.Miyake K, & Karasuyama H 2017. Emerging roles of basophils in allergic inflammation. Allergology International, 66(3), 382–391. [DOI] [PubMed] [Google Scholar]
- 13.Dyer KD, Moser JM, Czapiga M, Siegel SJ, Percopo CM, and Rosenberg HF 2008. Functionally competent eosinophils differentiated ex vivo in high purity from normal mouse bone marrow. J. Immunol 181: 4004–4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Camberis M, Le Gros G, and Urban J 2003. Animal model of Nippostrongylus brasiliensis and Heligmosomoides polygyrus. Curr Protoc Immunol Chapter 19: Unit 19.12. [DOI] [PubMed] [Google Scholar]
- 15.Martin RK, Damle SR, Valentine YA, Zellner MP, James BN, Lownik JC, Luker AJ, Davis EH, DeMeules MM, Khandjian LM, Finkelman FD, Urban JF, and Conrad DH 2018. B1 Cell IgE Impedes Mast Cell-Mediated Enhancement of Parasite Expulsion through B2 IgE Blockade. Cell Rep 22: 1824–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keegan AD, Fratazzi C, Shopes B, Baird B, and Conrad DH 1991. Characterization of new rat anti-mouse IgE monoclonals and their use along with chimeric IgE to further define the site that interacts with FcϵRII and FcϵRI. Mol. Immunol 28: 1149–1154. [DOI] [PubMed] [Google Scholar]
- 17.Damle SR, Martin RK, Cross JV, and Conrad DH 2017. Macrophage migration inhibitory factor deficiency enhances immune response to Nippostrongylus brasiliensis. Mucosal Immunol 10: 205–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shelburne CP, McCoy ME, Piekorz R, Sexl V, Roh K-H, Jacobs-Helber SM, Gillespie SR, Bailey DP, Mirmonsef P, Mann MN, Kashyap M, Wright HV, Chong HJ, Bouton LA, Barnstein B, Ramirez CD, Bunting KD, Sawyer S, Lantz CS, and Ryan JJ 2003. Stat5 expression is critical for mast cell development and survival. Blood 102: 1290–1297. [DOI] [PubMed] [Google Scholar]
- 19.Barnstein BO, Li G, Wang Z, Kennedy S, Chalfant C, Nakajima H, Bunting KD, and Ryan JJ 2006. Stat5 expression is required for IgE-mediated mast cell function. J. Immunol 177: 3421–3426. [DOI] [PubMed] [Google Scholar]
- 20.Ando T, Xiao W, Gao P, Namiranian S, Matsumoto K, Tomimori Y, Hong H, Yamashita H, Kimura M, Kashiwakura J-I, Hata TR, Izuhara K, Gurish MF, Roers A, Rafaels NM, Barnes KC, Jamora C, Kawakami Y, and Kawakami T 2014. Critical role for mast cell Stat5 activity in skin inflammation. Cell Rep 6: 366–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho I-C, Tai T-S, and Pai S-Y 2009. GATA3 and the T-cell lineage: essential functions before and after T-helper-2-cell differentiation. Nat. Rev. Immunol 9: 125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gounni AS, Nutku E, Koussih L, Aris F, Louahed J, Levitt RC, Nicolaides NC, and Hamid Q 2000. IL-9 expression by human eosinophils: regulation by IL-1beta and TNF-alpha. J. Allergy Clin. Immunol 106: 460–466. [DOI] [PubMed] [Google Scholar]
- 23.Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, Nureki O, and Zhang F 2015. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517: 583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, Thakore PI, Glass KA, Ousterout DG, Leong KW, Guilak F, Crawford GE, Reddy TE, and Gersbach CA 2013. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat. Methods 10: 973–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forbes EE, Groschwitz K, Abonia JP, Brandt EB, Cohen E, Blanchard C, Ahrens R, Seidu L, McKenzie A, Strait R, Finkelman FD, Foster PS, Matthaei KI, Rothenberg ME, and Hogan SP 2008. IL-9- and mast cell-mediated intestinal permeability predisposes to oral antigen hypersensitivity. J. Exp. Med 205: 897–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osterfeld H, Ahrens R, Strait R, Finkelman FD, Renauld J-C, and Hogan SP 2010. Differential roles for the IL-9/IL-9 receptor alpha-chain pathway in systemic and oral antigen-induced anaphylaxis. J. Allergy Clin. Immunol 125: 469–476.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benedé S, and Berin MC 2018. Mast cell heterogeneity underlies different manifestations of food allergy in mice. PLoS ONE 13: e0190453. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.