Abstract
Hundreds of cellular proteins require iron (Fe) cofactors for activity, and cells express systems for their assembly and distribution. Molecular details of the cytosolic iron pool used for iron cofactors are lacking, but iron chaperones of the poly rC-binding protein (PCBP) family play a key role in ferrous ion distribution. Here we show that, in cells and in vitro, PCBP1 coordinates iron via conserved cysteine and glutamate residues and a molecule of non-covalently bound glutathione (GSH). Proteomics analysis of PCBP1-interacting proteins identified BolA2, which functions, in complex with Glrx3, as a cytosolic [2Fe–2S] cluster chaperone. The Fe–GSH-bound form of PCBP1 complexes with cytosolic BolA2 via a bridging Fe ligand. Biochemical analysis of PCBP1 and BolA2, in cells and in vitro, indicates that PCBP1–Fe–GSH–BolA2 serves as an intermediate complex required for the assembly of [2Fe–2S] clusters on BolA2–Glrx3, thereby linking the ferrous iron and Fe–S distribution systems in cells.
Introduction
Iron cofactors are required for the activity of hundreds of proteins that perform critical functions within cells. The simplest cofactors are mononuclear or dinuclear iron centers directly coordinated within the active sites of enzymes or regulatory proteins found throughout the cell. The more complex cofactors, heme and iron–sulfur (Fe–S) clusters, are synthesized by dedicated cellular machines. The final step of heme synthesis occurs exclusively within mitochondria, while heme proteins are located in every subcellular compartment1. Fe–S clusters are assembled and repaired in both the mitochondria and cytosol, while Fe–S cluster proteins are additionally located in the nucleus2,3. Each of these cofactors requires compartment-specific distribution systems that protect the cofactors from chemical attack, facilitate their insertion into target proteins, and prevent their adventitious binding in non-target sites. Cofactor assembly and distribution systems have been studied, but substantial gaps in our knowledge of these systems persist.
In the cytosol of mammalian cells, delivery of Fe(II) ions to mono- and di-iron enzymes is facilitated by the poly r(C)-binding protein (PCBP) family4,5. PCBPs are adaptor proteins that bind multiple types of ligands, affecting their fates6–10. PCBP1 and PCBP2 bind to C-rich motifs in many types of RNAs and single stranded DNAs and can alter their processing. Structurally, PCBPs contain three conserved RNA-binding domains, termed heterogeneous nuclear ribonucleoprotein K-homology (KH) domains, which have specific roles in interactions with RNA and proteins. PCBP1 and PCBP2 also function as iron chaperones in the cytosolic/nuclear compartment11. In vitro studies demonstrate that PCBP1 can bind 3 ferrous ions with affinities in the low micromolar range, similar to the levels of kinetically exchangeable iron in the cytosol12. The molecular details of how PCBPs coordinate labile iron have not been determined. PCBP1 and, to a lesser extent, PCBP2 deliver ferrous iron to ferritin, an iron storage protein, and to specific, cytosolic iron enzymes13. These targets include the prolyl hydroxylases, which regulate hypoxia-inducible factor 1α, and deoxyhypusine hydroxylase, which hypusinates EIF5A14,15. Mice lacking PCBP1 in bone marrow exhibit anemia and defects in red blood cell heme synthesis due to impaired flux of iron through ferritin16. PCBP2 can specifically interact with membrane proteins, facilitating iron import through DMT117 and export through ferroportin18. PCBP2 may also capture iron released from heme oxygenase 1 and 219. HEK293 cells depleted of PCBP1 or PCBP2 exhibit low activity of cytosolic aconitase14, a labile [4Fe–4S] cluster enzyme. This suggests that PCBP1 or PCBP2 may also be directly or indirectly involved in synthesis or repair of Fe–S clusters.
Iron–sulfur cluster assembly occurs in the cytosol via a system distinct from that of the mitochondria20,21; numerous proteins required for the maturation and distribution of [4Fe–4S] clusters to cytosolic and nuclear enzymes have been identified2,3. However, the early steps that result in the assembly of a [4Fe–4S] cluster have remained murky. In yeast, the initial steps of [2Fe–2S] cluster assembly occur on core machinery located in the mitochondria; some product of this machinery is exported to the cytosol for cytosolic Fe–S assembly20. Mammalian cells exhibit dual localization of core components to both mitochondria and cytosol, which may facilitate de novo Fe–S assembly in the cytosol22. The earliest steps in cytosolic Fe–S cluster assembly require the incorporation of an inorganic sulfur species generated by cysteine desulfurase. This species is exported from mitochondria via the transporter ABCB7, although the chemical nature of this sulfur-containing species has not been defined and may be produced in the cytosol, as well23. Similarly, the source of iron for cytosolic Fe–S assembly has not been identified. The scaffolds upon which the simplest clusters, [2Fe–2S], are assembled likely involve cytosolic isoforms of the core Fe–S machinery. Another candidate scaffold for assembly of cytosolic Fe–S clusters involves the cytosolic monothiol glutaredoxin, Glrx3. Yeast cells lacking Glrx3 homologues exhibit severe defects in cytosolic [4Fe–4S] assembly, while depletion of Glrx3 in fish and human cells leads to more subtle defects in Fe–S assembly and iron homeostasis24–26. Recent evidence indicates that Glrx3 functions as a [2Fe–2S] cluster chaperone in human cells and requires the cytosolic BolA-like protein, BolA2, to coordinate [2Fe–2S] clusters and transfer them to an early-acting component of the cytosolic Fe–S assembly machinery27.
Here we have used affinity-capture and mass spectrometry to identify proteins that associate with PCBP1 in human cells. We identify BolA2 in complex with PCBP1 and demonstrate that PCBP1–BolA2 forms a distinct iron chaperone complex for cytosolic [2Fe–2S] cluster assembly.
Results
BolA2 forms a complex with PCBP1 without Glrx3
To determine the proteins interacting with PCBP1 in mammalian cells, we performed proteomic analysis on complexes isolated from HEK293 cells expressing an inducible, N-terminal Flag-tagged PCBP1. Affinity purification followed by mass spectrometry analysis of Flag-PCBP1 complexes revealed more than 200 interacting proteins, including RNA-binding proteins, ferritin, and some novel targets28. PCBP1 was associated with the cytosolic BolA-like protein, BolA2 (Supplementary Table 1). Iron supplementation of cells enhanced the PCBP1–BolA2 interaction, while high salt washes dissociated the PCBP1–BolA2 complex. Although Glrx3 is found in complex with BolA2 in HEK293 cells, Glrx3 was not found in immunoprecipitates of PCBP1. This association between PCBP1 and BolA2 was confirmed by reciprocal co-immunoprecipitation (co-IP) analysis using ectopically expressed proteins in HEK293 cells. IP using anti-Flag resin followed by Western blotting with anti-BolA2 antiserum revealed that BolA2 was readily detectable in immune complexes from cells expressing PCBP1-Flag, but not from cells containing an empty vector, confirming the specificity of the interaction (Fig. 1a). Similarly, PCBP1 was identified in complexes from cells expressing BolA2-Flag but was undetectable in cells containing empty vector (Fig. 1b). We constructed inducible cell lines that expressed BolA2-Flag at levels similar to endogenous BolA2. PCBP1 was readily detected in immune complexes from this cell line, whether PCBP1 was expressed at the endogenous level or overexpressed (Fig. 1c). Thus, these results confirm that PCBP1–BolA2 complexes form in mammalian cells. In contrast, similar experiments designed to detect PCBP2 in complex with BolA2 did not reproducibly demonstrate PCBP2 in precipitates of BolA2-Flag (Supplementary Figure 1), indicating that the BolA2 complex was specific to PCBP1.
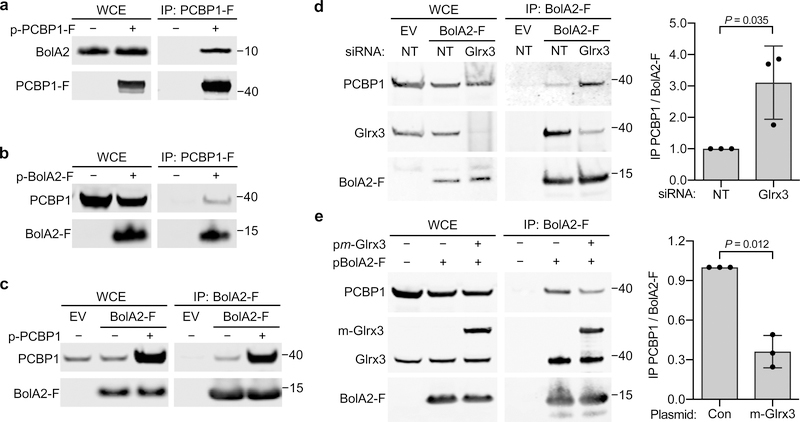
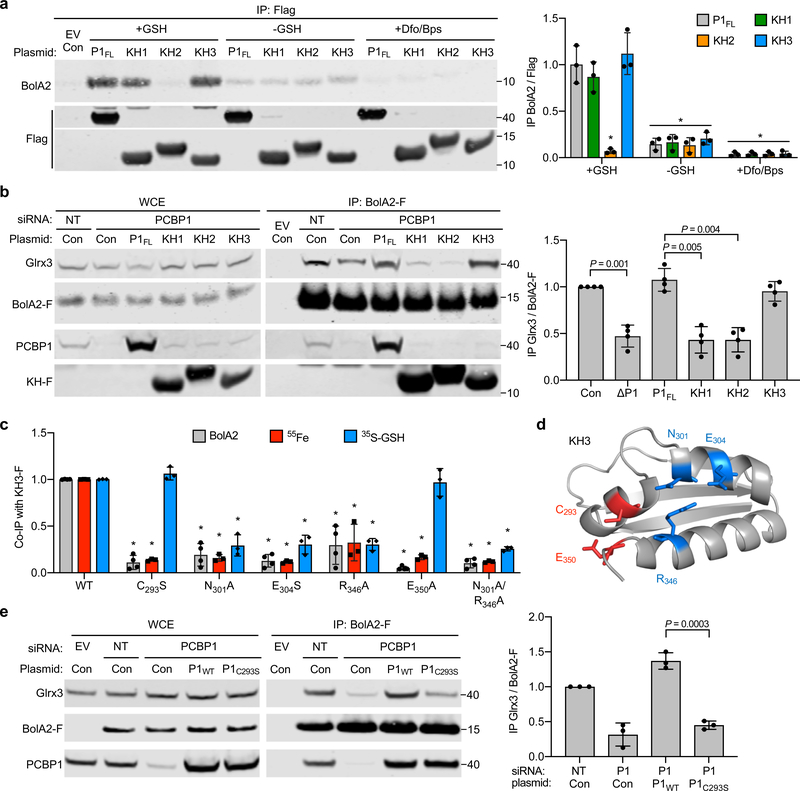
Fig. 1: BolA2 forms a complex with PCBP1 and binds in absence of Glrx3.
a,b, Binding between PCBP1 and BolA2 in cells. HEK293 cells were transfected with plasmid (p) negative control (−), PCBP1-Flag (PCBP1-F; a), or BolA2-Flag (BolA2-F; b) and anti-Flag immunoprecipitation (IP) performed. Whole cell extracts (WCE) and immune complexes were analyzed by immunoblot using antibodies for PCBP1, BolA2, and Flag-tag. c, Binding of endogenous PCBP1 with BolA2 in cells. Tetracycline-inducible stable cell lines expressing empty vector (EV) or BolA2-Flag (BolA2-F) were treated overnight with doxycycline, transfected with control (−) or PCBP1 plasmid, and analyzed as in b. d, Glrx3-independent formation of PCBP1–BolA2 complex in cells. BolA2-F or EV cell lines were transfected with non-targeting (NT) or Glrx3 siRNA and analyzed as in b. e, Glrx3 competes with PCBP1 for interaction with BolA2 in cells. HEK293 cells were transfected with control (Con), myc-Glrx3 (m-Glrx3), or BolA2-F plasmids and analyzed as in b. In d and e, WCE and immune complexes were analyzed by immunoblot using antibodies for PCBP1, BolA2 or Glrx3 and the relative ratio of co-precipitated PCBP1 to BolA2-F in IP was quantified (right). All experiments were repeated independently three times with similar results. Bar graphs represent means ± SD. Significant P values (P < 0.05), as determined by two-tailed unpaired t-test relative to control, are shown. Uncropped western blots are presented in Supplementary Figure 13.
In human cells, BolA2 complexes with the cytosolic monothiol glutaredoxin, Glrx327. To investigate whether Glrx3 was involved in PCBP1–BolA2 formation, we performed co-IP in cells depleted of Glrx3 using small interfering RNA (siRNA). BolA2-Flag complexes from cells depleted of Glrx3 contained 3-fold higher levels of PCBP1–BolA2 compared to cells treated with control siRNA (Fig. 1d). In contrast, cells overexpressing myc-tagged Glrx3 contained 60% lower levels of PCBP1–BolA2 compared to controls (Fig. 1e), suggesting that Glrx3 competes with PCBP1 for BolA2 binding. These results demonstrate that PCBP1 and BolA2 form a complex independent of Glrx3 and interaction between PCBP1 and BolA2 is enhanced in the absence of Glrx3.
BolA2 requires PCBP1 to bind iron without Glrx3
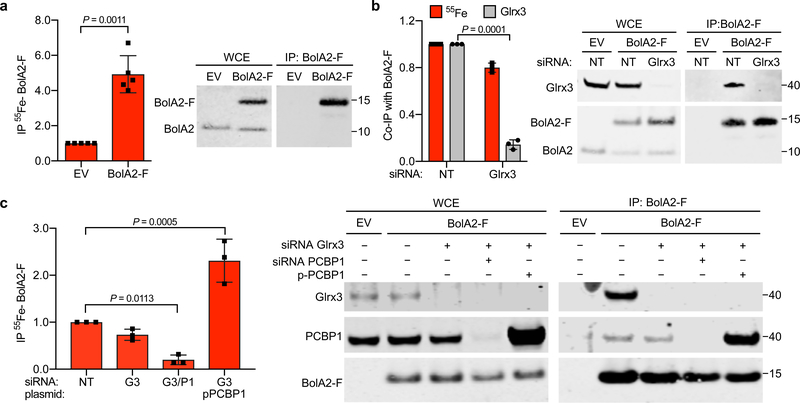
In human cells, the BolA2–[2Fe–2S]–Glrx3 chaperone complex serves as a rapidly expandable pool of Fe–S clusters and responds to changes in cellular iron status27. However, BolA2 has not been reported to bind iron independently. To directly measure the amount of iron associated with BolA2, we induced BolA2-Flag expression, labelled cells with 55FeCl3, and measured the amount of 55Fe co-precipitating with BolA2-Flag (Fig. 2a). We detected 55Fe in BolA2-Flag complexes at levels 5-fold greater than uninduced control cells, indicating that we could measure the 55Fe associated with BolA2. To test whether Glrx3 was required for iron binding to BolA2, we depleted Glrx3 and found the amount of 55Fe bound to BolA2-Flag was only modestly diminished (< 20% reduction) compared to Glrx3-expressing cells (Fig. 2b). To test whether PCBP1 was required for iron binding to BolA2, we measured the amount of 55Fe co-precipitated with BolA2-Flag from PCBP1-depleted or PCBP1-overexpressing cells under Glrx3-depleted conditions. The amount of 55Fe associated with BolA2-Flag was markedly reduced (>80% reduction) in cells co-depleted of PCBP1 and Glrx3 (Fig. 2c). In contrast, when PCBP1 was overexpressed in the absence of Glrx3, we measured a 300% increase in 55Fe bound to BolA2-Flag. These results suggested that iron binding to BolA2 largely depends on PCBP1 and does not require Glrx3.
Fig. 2: BolA2 requires PCBP1, but not Glrx3, to form an iron-binding complex.
a, Isolation of 55Fe-containing BolA2 complexes from cells. BolA2-F and EV inducible cell lines were labeled with 55FeCl3 for 16 h. WCE and anti-Flag immune complexes were analyzed by scintillation counting (left) and immunoblot (right). Non-specific (EV) and specific (BolA2-F) 55Fe-binding was normalized to whole cell 55Fe content and expressed relative to EV control. Data are means ± SD of n = 5 independent experiments. b, 55Fe binding to BolA2 complexes from cells lacking Glrx3. Inducible BolA2-F cells were treated with NT or Glrx3 siRNA and labeled with 55FeCl3. WCE and anti-Flag IPs were analyzed by scintillation counting and immunoblotting for Glrx3 and BolA2-F. Quantification (left) of specifically bound 55Fe (red bars) and ratio of co-precipitated Glrx3 to IP BolA2-F (grey bars) are normalized to NT control. Data are means ± SD of n = 3 independent experiments. c, PCBP1-dependent 55Fe binding to BolA2 complexes. Inducible BolA2-F cells were treated with non-targeting (−), Glrx3, or PCBP1 siRNAs and transfected with control (−) or PCBP1 plasmids. Cells were labeled with 55FeCl3, WCE and immune complexes were analyzed by scintillation counting for 55Fe (left) and immunoblotting for Glrx3, PCBP1 and BolA2-F (right). Specifically bound 55Fe was normalized to IP BolA2-F and expressed relative to NT control. Data are means ± SD of n = 3 independent experiments. For all experiments significant P values (P < 0.05), as determined by two-tailed unpaired t-test relative to control, are shown. Uncropped western blots are presented in Supplementary Figure 14.
PCBP1–BolA2 requires iron and glutathione for stability
In human cells, BolA2–Glrx3 complex formation depends on the incorporation of [2Fe–2S] clusters27. BolA2 forms a [2Fe–2S]-bridged heterotrimeric complex with Glrx3, with iron ions coordinated by a conserved cysteine and histidine from BolA2, a cysteine from Glrx3, and the cysteine moiety of noncovalently bound, reduced glutathione (GSH) on Glrx325,29. GSH is an abundant tripeptide, attaining concentrations of 2–10 mM in mammalian cells, and serves as the primary intracellular thiol redox buffer and cofactor for glutaredoxins and other antioxidant enzymes30. To understand complex formation between PCBP1 and BolA2, we overexpressed PCBP1 and BolA2-Flag and performed co-IP in the presence or absence of GSH and/or iron. PCBP1–BolA2 was stable to immunoprecipitation when isolated in buffer containing 5 mM GSH (Fig. 3a). In contrast, addition of iron chelators and/or omission of GSH destabilized the complex, resulting in a 70% decrease in PCBP1–BolA2. Moreover, addition of an alternative reducing agent, dithiothreitol, did not result in stabilization of PCBP1–BolA2 complexes (Supplementary Fig. 2). These data suggest that both iron and GSH are required for stable PCBP1–BolA2 complex formation.
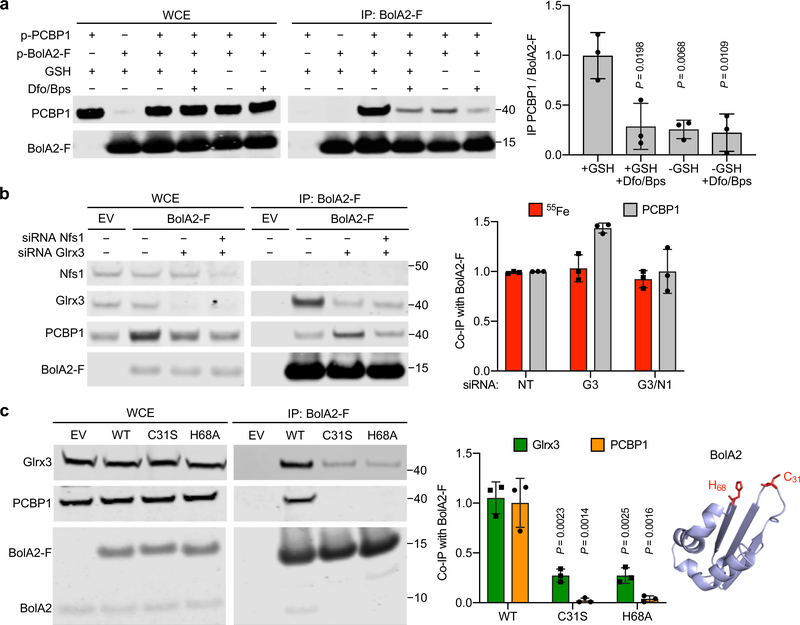
Fig. 3: Stabilization of PCBP1–BolA2 complex by iron and glutathione without inorganic sulfur.
a, Requirement of iron and glutathione for PCBP1–BolA2 complex stabilization. HEK293 cells were transfected with the indicated plasmids, then lysed in IP buffer +/− 5 mM glutathione (GSH) or 100 μM chelator mix (Dfo/Bps). WCE and anti-Flag IPs were analyzed by immunoblot using antibodies for PCBP1 and BolA2 (left) with quantitation (right). Data are means ± SD of n = 3 independent experiments. Significant P values (P < 0.05), as determined by two-tailed unpaired t-test relative to +GSH control, are shown. b, 55Fe bound to BolA2 complexes from cells lacking Nfs1. Inducible EV or BolA2-F cells were treated with NT, Glrx3, or Nfs1 siRNAs and labeled with 55FeCl3. WCE and anti-Flag IPs were analyzed by immunoblotting for Nfs1, Glrx3, PCBP1 and BolA2-F and by scintillation counting (left) with quantitation (right). Data are means ± SD of n = 3 independent experiments. c, Requirement of BolA2 iron-binding residues for interactions with Glrx3 and PCBP1. Stable cell lines expressing EV, wild type (WT), C31S or H68A mutants of BolA2-F were examined by IP (left) with quantitation (middle). Data are means ± SD of n = 3 independent experiments. Significant P values (P < 0.01), as determined by two-tailed unpaired t-test relative to WT control, are shown. Homology model of human BolA2 on murine structure (PDB: 1V9J, sequence identity 87%), conserved iron binding residues shown in red (right). Uncropped western blots are presented in Supplementary Figure 15.
We next determined whether inorganic sulfur was required for the PCBP1–BolA2 interaction. Nfs1 encodes the sole cysteine desulfurase in mammalian cells; cells lacking this enzyme are unable to produce sulfide for both mitochondrial and cytosolic Fe–S cluster synthesis31. Using siRNA, we depleted Nfs1 and/or Glrx3 in the inducible BolA2-Flag cell line, labelled cells with 55FeCl3 and measured the amount of 55Fe and PCBP1 co-precipitating with BolA2-Flag (Fig. 3b). The amounts of 55Fe co-precipitated with BolA2-Flag were similar in control-, Glrx3-, and Nfs1/Glrx3-depleted cells. Furthermore, the cells depleted of Nfs1 and Glrx3 contained similar levels of PCBP1–BolA2 as control cells, suggesting that inorganic sulfide or an Fe–S cluster was not required for iron binding to BolA2 or for PCBP1–BolA2 complex formation.
Because Glrx3 appears to compete with PCBP1 for binding and iron coordination with BolA2, we questioned whether the same site on BolA2 was involved in the formation of both complexes. Conserved Cys31 and His68 in human BolA2 are required for Fe–S cluster coordination with Glrx329. Both PCBP1 and Glrx3 were readily detectable in anti-Flag IPs from cells expressing wild type BolA2-Flag compared to an empty vector negative control (Fig. 3c). In BolA2-Flag mutants C31S and H68A, levels of co-precipitating Glrx3 and PCBP1 were reduced to 25% and 5% of wild type levels, respectively. These results suggest that the same iron-binding residues on BolA2 are necessary for the formation of the BolA2–Fe–PCBP1 as well as the BolA2–[2Fe–2S]–Glrx3 complex.
PCBP1 delivers iron to the BolA2–[2Fe–2S]–Glrx3 complex
Whether PCBP1 requires GSH or a protein partner such as BolA2 for iron coordination in cells remains to be determined. We transfected cells with plasmids encoding PCBP1-Flag and BolA2 or a control plasmid, labeled cells with 55FeCl3, and measured the amount of iron and BolA2 co-precipitating with PCBP1 (Fig. 4a). The amount of BolA2 co-precipitating with PCBP1 mirrored the overall level of BolA2 expression: cells overexpressing BolA2 exhibited abundant BolA2–PCBP1, while cells expressing low levels of BolA2 exhibited very low amounts of the complex. In contrast, similar amounts of 55Fe were associated with PCBP1-Flag from cells overexpressing BolA2 or not, suggesting that PCBP1 binds iron independently of BolA2. To determine the effect of GSH on iron binding to PCBP1, we measured the amount of 55Fe associated with PCBP1 when BolA2 was low and GSH was omitted from the lysis buffer. In the absence of GSH, the 55Fe bound to PCBP1 fell to 10% of the level present when buffer contained GSH (Fig. 4a). Collectively our data indicated that PCBP1 requires GSH, but not BolA2, to bind iron.
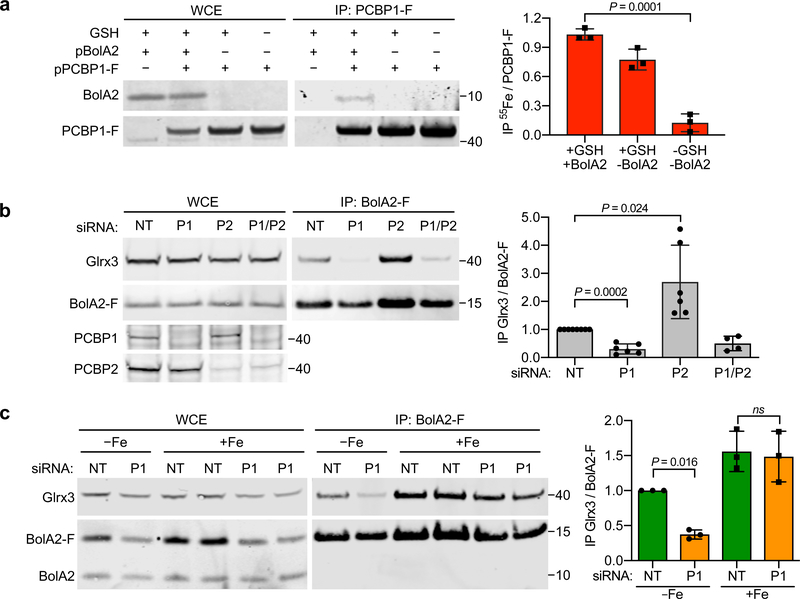
Fig. 4: Requirement of PCBP1 in delivery of iron to Grx3–[2Fe–2S]–BolA2 complex.
a, Requirement of GSH for iron binding to PCBP1. HEK293 cells were transfected with control, BolA2, or PCBP1-F plasmids and labelled with 55FeCl3. Anti-Flag IPs were performed +/− 5 mM GSH. WCE and anti-Flag IPs were analyzed by immunoblotting for BolA2 and PCBP1-F (left) and by scintillation counting for 55Fe quantitation (right). Data are means ± SD of n = 3 independent experiments. b, Loss of Glrx3–BolA2 complexes in cells lacking PCBP1. Inducible BolA2-F cells were treated with NT, PCBP1, or PCBP2 siRNA and analyzed by IP and immunoblotting for Glrx3 and BolA2 (left) with quantitation (right). Data are means ± SD of n = 6 independent experiments. c, Restoration of Glrx3–BolA2 complexes by iron supplementation in cells lacking PCBP1. Inducible BolA2-F cells were transfected with NT or PCBP1 siRNA and treated overnight with 50 μM FeCl3. WCE and anti-Flag IPs were analyzed by immunoblotting against Glrx3 and BolA2 (left) with quantitation (right). Data are means ± SD of n = 3 independent experiments. For all experiments significant P values (P < 0.05), as determined by two-tailed unpaired t-test relative to control, are shown. Uncropped western blots are presented in Supplementary Figure 16.
Unlike in vitro studies32 or in yeast cells33, in human cells, formation of a BolA2–Glrx3 complex requires the coordination of [2Fe–2S] clusters; in the absence of Fe–S clusters, BolA2 and Glrx3 do not form a stable complex27. Because PCBP1 is involved in the distribution of ferrous iron in the cytosol of mammalian cells11,14,15, we tested whether the iron chaperone activity of PCBP1 was required to form BolA2–[2Fe–2S]–Glrx3 complexes. Cells expressing or depleted of PCBP1 or the homologous PCBP2 were examined for BolA2–[2Fe–2S]–Glrx3 formation (Fig. 4b). Isolated BolA2-Flag immune complexes from cells lacking PCBP1 contained less than 30% of the Glrx3 found in control cells expressing PCBP1, indicating that PCBP1 is required for the formation of the [2Fe–2S] clusters needed to form the BolA2–[2Fe–2S]–Glrx3 complex and that endogenous PCBP2 cannot substitute for PCBP1. Surprisingly, cells lacking PCBP2 exhibited enhanced BolA2–[2Fe–2S]–Glrx3 complex formation. Co-depletion of both PCBP1 and PCBP2 resulted in impaired complex formation, indicating that PCBP1 is required for the enhanced complex formation seen in PCBP2-depleted cells. Overall, these data suggest that PCBP1 has a specific role in providing iron for the formation of the BolA2–[2Fe–2S]–Glrx3 complex. PCBP2 may impact this process by competing with PCBP1 for cytosolic iron.
Previous work shows that the loss of iron chaperone activity associated with PCBP1 depletion can be rescued by supplementing cells with excess iron at levels sufficient to raise the cytosolic pool of iron 14,15. We tested the effects of iron supplementation on BolA2–Glrx3 formation and found that again, BolA2–[2Fe–2S]–Glrx3 complex formation was enhanced in control cells; furthermore, complex formation in cells lacking PCBP1 could be restored by iron supplementation, indicating that the iron chaperone activity of PCBP1 promoted [2Fe–2S] cluster formation on Glrx3 and BolA2 (Fig. 4c).
KH3 domain promotes [2Fe–2S] formation on BolA2–Glrx3
Structurally, PCBPs contain three KH domains: KH1 and KH2 in the amino terminus and KH3 in the carboxyl terminus, which are separated by an unstructured variable domain. We mapped the domain(s) of PCBP1 involved in interactions with BolA2. PCBP1 variants containing individual Flag-tagged domains were expressed in cells and examined for their capacity to co-precipitate BolA2 (Fig. 5a and Supplementary Fig. 3). Interestingly, both the KH1 and KH3 domains co-precipitated BolA2 at levels similar to the full-length PCBP1, while the KH2 domain exhibited no interaction. Co-precipitation of BolA2 with the KH1 or KH3 domain was lost in the absence of GSH or iron, as was observed earlier with full-length PCBP1 (Fig. 3a). Thus, iron- and GSH-dependent BolA2 binding occurs with both the KH1 and KH3 domains of PCBP1.
Fig. 5: KH3 domain of PCBP1 binds BolA2 and restores BolA2–Glrx3 complex formation.
a, Iron- and glutathione-dependent interaction of BolA2 with KH1 and KH3 of PCBP1. HEK293 cells were transfected with plasmids for Flag-tagged, full length PCBP1 (P1FL) or individual KH domains (KH1, KH2, and KH3). Anti-Flag IP was performed in IP buffer +/− GSH and Dfo/Bps. IPs were analyzed by immunoblotting for BolA2 and Flag (left). Ratios of co-precipitated BolA2 to each KH-F is normalized to full length PCBP1 (right). Data are means ± SD of n = 3 independent experiments. Statistical significance level was calculated by one-way ANOVA analysis (F = 39.05, d.f. = 29, P < 0.0001) with a Dunnett’s multiple comparison test, *P ≤ 0.0001 compared to P1FL+GSH. b, Requirement of KH3 domain to form Glrx3–BolA2 complex. Inducible BolA2-F cells were treated with NT or PCBP1 siRNA, then transfected with siRNA-resistant plasmids as indicated. WCE and anti-Flag IPs were analyzed by immunoblotting against Glrx3, PCBP1 and Flag (left) with quantitation (right). Data are means ± SD of n = 4 independent experiments. Significant P values (P < 0.05), as determined by two-tailed unpaired t-test, are shown. c, Distinct iron and GSH binding sites on KH3 domain required for BolA2 interaction. Cells expressing BolA2 and KH3-Flag variants, as indicated, were labeled with 55FeCl3. Anti-Flag IPs were directly analyzed for BolA2 (see Supplementary Fig. 4) or 55Fe binding. Alternatively, washed complexes were incubated with 100 μM 35S-GSH and analyzed for GSH binding. Ratio of co-precipitated BolA2 or 55Fe or 35S-GSH to each KH3-F variant is normalized to WT. Data are means ± SD of n = 3 independent experiments. Statistical significance level was calculated by one-way ANOVA analysis (F = 43.91, d.f. = 20, P < 0.0001) with a Dunnett’s multiple comparison test, *P ≤ 0.0001 compared to WT. d, Ribbon structure of KH3 domain (PDB: 1WVN) illustrating ligands for GSH binding (blue sticks) and iron binding (red sticks). e, Requirement of Cys293 residue on PCBP1 to form Glrx3–BolA2 complex. Inducible BolA2-F cells were treated with NT or PCBP1 siRNA, then transfected with siRNA-resistant plasmids as indicated. WCE and anti-Flag IPs were analyzed by immonoblot against Glrx3, PCBP1 and Flag (left) with quantitation (right). Data are means ± SD of n = 3 independent experiments. Significant P values (P < 0.05), as determined by two-tailed unpaired t-test, are shown. Uncropped western blots are presented in Supplementary Figure 17.
We next determined whether a single KH domain could substitute for the full-length PCBP1 in facilitating BolA2–[2Fe–2S]–Glrx3 complex formation. We depleted endogenous PCBP1, expressed siRNA-resistant PCBP1 or KH-Flag domains, then measured the amount of Glrx3 co-precipitating with BolA2-Flag. As observed in Figure 4b, the BolA2–Glrx3 complex was destabilized upon depletion of endogenous PCBP1 (Fig. 5b) and overexpression of full-length PCBP1 restored the BolA2–Glrx3 complex to non-targeting control levels. In contrast, neither KH1 nor KH2 domains restored the BolA2–Glrx3 complex in the absence of PCBP1, despite the capacity of KH1 to bind to BolA2. Instead, KH3 overexpression was sufficient to restore BolA2–Glrx3 complex formation to the level of full-length PCBP1. These data indicated that the carboxyl-terminal KH3 domain of PCBP1 was functionally sufficient to promote formation of the BolA2–[2Fe–2S]–Glrx3 complex in cells lacking endogenous PCBP1.
We then conducted a structure-function analysis of the BolA2–KH3 complex using site-directed mutagenesis of the KH3 domain. After examining the crystal structure of KH3, we engineered substitution mutations in (i) a conserved cysteine (Cys293) reported to undergo iron-dependent homocysteinylation34, (ii) residues identified by modeling GSH binding on KH3, (iii) residues in the vicinity of Cys293 that could serve as N-, O-, and S-based Fe(II) ligands35, and (iv) residues coordinating RNA in the structure of a KH3–RNA complex36. Wild-type and mutant versions of KH3 were expressed in cells and examined for their capacity to co-precipitate BolA2 (Fig. 5c,d and Supplementary Fig. 4). KH3 mutants carrying substitutions in residues coordinating RNA (Gly296, Arg297, Gly299, Arg306, Arg325) demonstrated BolA2 binding at levels similar to wild-type KH3, suggesting that the binding site for BolA2 was distinct from the RNA-binding site. In contrast, KH3 mutants C293S, N301A, E304S, R346A, E350A, N301A/R346A bound BolA2 at levels 5–30% of wild-type KH3. We further analyzed the 55Fe binding activity of these KH3 mutants and found that all of the amino acid substitutions that impaired BolA2 binding also reduced iron coordination to levels 10–30% of wild-type KH3 (Fig. 5c). To directly assess residues potentially involved in GSH binding, KH3 IPs were labeled in vitro with 35S-GSH. Wild-type KH3 bound GSH at 5-fold greater levels than control IPs, indicating that isolated KH3 can directly bind GSH. All of the KH3 mutants predicted to affect GSH coordination (Asn301, Glu304, Arg346) exhibited reduced levels (< 30% of wild-type) of 35S-GSH-binding. In contrast, predicted iron-binding mutants C293S and E350A exhibited no defect in GSH binding. Mutation of non-conserved Cys355 did not affect BolA2 coordination (Supplementary Fig. 4), suggesting the iron-binding function of Cys293 was specific and not a general feature of sulhydryl residues on KH3. Together these data suggested that KH3 can directly coordinate both GSH and iron at adjacent sites on the domain and that both ligands are required for coordination of BolA2.
Finally, we determined whether the C293S mutation in full-length PCBP1 could block the formation of the BolA2–[2Fe–2S]–Glrx3 complex. We expressed wild-type or mutant PCBP1C293S in PCBP1-depleted cells and measured the formation of the BolA2–Glrx3 complex (Fig. 5e). Mutant PCBP1C293S did not support complex formation, as levels of co-precipitated Glrx3 were similar to those of PCBP1-depleted control cells. Thus, the Fe–GSH binding site on the KH3 domain of PCBP1 was necessary and sufficient to facilitate formation of the BolA2–[2Fe–2S]–Glrx3 complex.
Allosteric binding of iron and GSH to KH3
Our data obtained from cells suggested that both iron and GSH were involved in the formation of the BolA2–KH3 complex. To determine the effects of each ligand on the binding of the other, we purified recombinant KH3 and BolA2 (Supplementary Fig. 5) and performed in vitro analyses of ligand binding. To measure the allosteric effects of Fe(II) on the binding of GSH by the PCBP1 KH3 domain, we used microscale thermophoresis of fluorescently labeled KH3 (Fig. 6a). Titration of GSH in buffers without iron indicated an apparent KD = 390 ± 50 μM, while titration of GSH in buffer containing 10 μM Fe(II) demonstrated a ∼5-fold higher affinity of GSH for KH3, with a KD = 80 ± 10 μM. We confirmed the effect of iron on in vitro GSH binding by KH3 using the 35S-GSH assay (Supplementary Fig. 6). Wild-type KH3 directly bound GSH in the absence of added iron and increased GSH binding 2-fold in the presence of Fe(II), while mutant KH3C293S exhibited wild-type levels of GSH binding that did not increase with addition of iron, thus supporting the allosteric activity of iron.
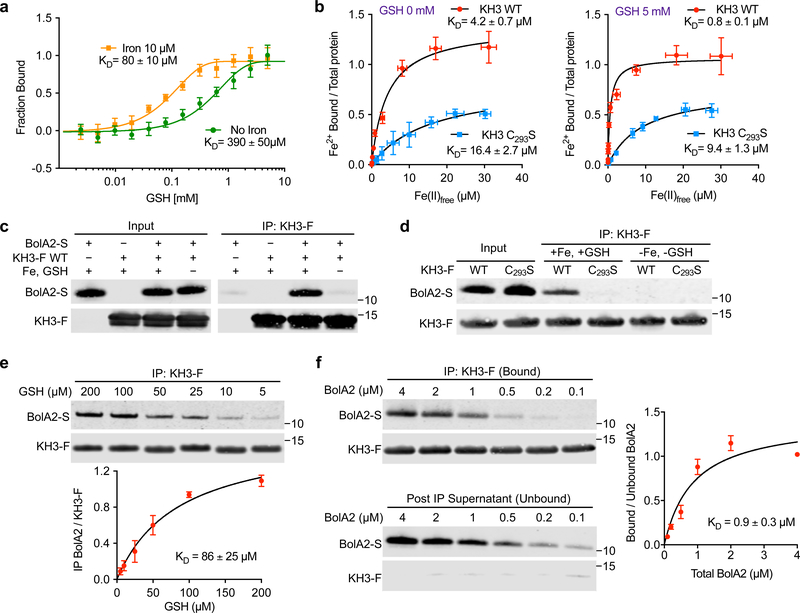
Fig. 6: Reconstitution of KH3–BolA2 complex with iron and GSH in vitro.
a, GSH binding affinity for KH3 domain increased by iron. Purified, fluorescently labeled KH3 domain (200 nM) was mixed with increasing concentrations of GSH, without added iron (green) or with 10 μM FeSO4 (orange) and apparent dissociation constants (KD) were determined for GSH using microscale thermophoresis. Data are means ± SEM of n = 4 independent experiments. b, Increased Fe(II) binding affinities of WT and C293S KH3 in presence of GSH. Dissociation constants (KD) of WT (red) and C293S (blue) KH3 for Fe(II) were determined using Mag-fura-2 competition, +/− 5 mM GSH. Data are means ± SD of n = 4 independent experiments. c,d, Requirement of iron and glutathione for reconstitution of KH3–BolA2 complex in vitro. Recombinant, purified BolA2-Strep (BolA2-S, 1 μM) and WT (c) or C293S KH3-Flag (KH3-F, 1 μM; d) were mixed in IP buffer +/− 5 mM GSH or 100 μM FeSO4 and anti-Flag IP was performed. IP input and immune complexes were analyzed by immunoblot against BolA2 and Flag-tag. Blots are representative of n = 3 independent experiments with similar results. e, GSH-dependent binding of KH3 to BolA2 in vitro. Purified BolA2-S and KH3-F were mixed in IP buffer containing 100 μM FeSO4 with the indicated concentrations of GSH and analyzed by anti-Flag IP and immunoblotting. Ratio of BolA2 to IP KH3-F was plotted as a function of GSH concentration and the data was fitted by Michaelis-Menten analysis to obtain the KD. Data are means ± SD of n = 3 independent experiments. f, Apparent binding affinity of BolA2 to KH3 domain. Purified KH3-F (4 μM) was mixed with indicated concentrations of BolA2-S (4–0.1 μM) in IP buffer containing 5 mM GSH and 100 μM FeSO4. Anti-Flag IP and Western blotting was performed. Ratio of bound to unbound BolA2 was plotted as a function of total BolA2 concentration and the data was fitted by Michaelis-Menten analysis to obtain the KD. Data are means ± SD of n = 3 independent experiments. Uncropped western blots are presented in Supplementary Figure 18.
To precisely characterize the iron coordination by the purified KH3 domain of PCBP1, we determined its Fe(II)-binding affinities and the allosteric effects of GSH using the competitive iron-binding chromophore Mag-Fura 2 (MF2). Iron binding by MF2 was monitored spectrophotometrically37,38 and glutathione at physiological concentrations did not interfere with Fe binding (Supplementary Fig. 7a). We then titrated Fe(II) into a mixture of MF2 and purified KH3 (Supplementary Fig. 7b), calculated the free Fe(II) and Fe–KH3/total KH3 levels, and determined the KD and apparent stoichiometry for the Fe–KH3 complex (Fig. 6b). Wild-type KH3 bound Fe(II) in the absence of glutathione at KD = 4.2 ± 0.7 μM. GSH increased the affinity of KH3 for Fe(II) by ~5-fold, with KD = 0.8 ± 0.1 μM in 5 mM GSH and a stoichiometry of 1:1. We confirmed these measurements by performing similar analyses using the chromophore Fura-4f, which exhibits a higher affinity for Fe(II) (Supplementary Fig. 8). The C293S KH3 variant showed significantly lower Fe(II) binding affinities, with KD = 16.4 ± 2.7 μM at 0 mM GSH and KD = 9.4 ± 1.3 μM at 5 mM GSH. Fe binding to the C293S variant exhibited a metal:protein stoichiometry nearer to 0.5:1, suggesting that this mutant KH3 may coordinate Fe(II) as a dimer. Thus, GSH binding can allosterically stabilize the Fe–KH3 complex. Furthermore, the affinity of Fe(II) for KH3 in presence of GSH is sufficient to promote Fe(II)–PCBP1 complex formation in the cytosol, as the concentration of chelatable iron in the cytosol has been measured in the low micromolar range (0.5–2 μM)12,39,40.
We determined whether the identified factors were sufficient to reconstitute the KH3–BolA2 complex in vitro. We mixed purified KH3-Flag and BolA2 in buffer with or without Fe(II) and GSH and determined the amount of BolA2 bound to KH3-Flag after capture on the anti-Flag resin (Fig. 6c). The in vitro reconstitution closely mimicked complex formation in cells: BolA2 was co-precipitated with KH3-Flag only in the presence of Fe and GSH. Reconstitution of the KH3–BolA2 complex without any added source of sulfur supported our observation that the KH3–BolA2 complex coordinates iron, but not an iron–sulfur cluster. Similar experiments using BolA2 and KH3-Flag variant C293S failed to reconstitute the KH3–BolA2 complex, even with addition of Fe and GSH (Fig. 6d), indicating that iron coordination by Cys293 of the KH3 domain was required for complex formation with BolA2. We determined the stoichiometry of the KH3–BolA2 complex using size exclusion chromatography (Supplementary Fig. 9). In the absence of GSH and iron, a mixture of KH3 and BolA2 migrated as monomers, at the expected molecular sizes of 15 and 12 KDa, respectively. In mixtures of BolA2 and KH3 that included GSH and iron, a complex of BolA2–KH3 was detected at the molecular size of 27 KDa, indicating formation of a 1:1 BolA2–KH3 heterodimer.
To quantitatively examine the effect of GSH on BolA2–KH3 complex formation, we performed in vitro reconstitution and quantitative pull-down assays across a broad range of GSH concentrations (Fig. 6e and Supplementary Fig. 10). The affinity of GSH for the BolA2–KH3 complex was KD = 86 ± 25 μM, which was similar to the affinity of GSH for KH3 alone in the presence of iron (Fig. 6a). Because BolA2 binding did not affect the apparent affinity of GSH for the BolA2–KH3 complex, these data suggest that the GSH bound to KH3 does not directly interact with the BolA2 protein. Furthermore, these data support a model in which a molecule of GSH, noncovalently bound to KH3, contributes directly to the coordination of iron in KH3 by serving as an iron ligand, and making the KH3-bound iron sterically available for coordination with BolA2.
Finally, we determined the affinity of BolA2 for the intact, preformed KH3–Fe–GSH complex. We mixed KH3-Flag with Fe(II) and GSH, then added BolA2 in a range of concentrations (Fig. 6f). The apparent dissociation constant for KH3–BolA2 complex was KD = 0.9 ± 0.3 μM. Because the affinities of Fe and GSH for KH3 and KH3 for BolA2 determined in these studies are concentrations within the physiological range present in cells, the iron chaperone complexes defined in these in vitro studies likely represent complexes that form in cells.
Discussion
The studies presented here reveal functions of PCBP1 in complex with BolA2. Together, they form an iron chaperone complex, transferring iron in the early steps of cytosolic Fe–S cluster assembly, thus linking the cytosolic labile iron pool to the cytosolic Fe–S cluster assembly process. These studies also demonstrate that PCBP1 can bind iron in the cytosol as an Fe(II)–GSH complex. Previous in silico analyses of potential small-molecule ligands for iron suggested that over 95% of the kinetically exchangeable cytosolic iron pool is likely to be coordinated by the free thiol in reduced GSH41. Here we show that the KH3 domain (and likely the KH1 domain) of PCBP1 binds Fe and GSH with 1:1 stoichiometries and affinities that, at equilibrium, would support complex formation in the cytosol. Allosteric effects of each ligand on the binding of the other support the coordination of an Fe–GSH complex on KH3 in which the free sulfhydryl of GSH functions as an iron ligand, along with Cys293 and Glu350, to stabilize bound iron. GSH coordination appears to be necessary for the Fe(II) to coordinate the Cys31 and His68 ligands of BolA2, as the iron-bound form of KH3 without GSH did not bind BolA2 in vitro. In silico docking analysis between KH3 and BolA2 suggested a possible electrostatic protein–protein interaction wherein the putative iron-binding residues on BolA2 and KH3 are positioned at the modeled interface of the two protomers (Supplementary Fig. 10)42. The coordination of Fe–GSH on KH3 may confer selectivity for Fe(II), as cytosolic manganese, although present in concentrations similar to Fe(II), binds GSH with > 100-fold lower affinity than Fe(II)43. Thus, the coordination of Fe–GSH complexes on PCBP1 potentially accounts for its metal specificity as an iron chaperone and suggests that much of the cytosolic iron pool is in the form of PCBP1–Fe–GSH complexes.
The Cys293 residue that is critical for iron binding on KH3 was previously identified as a site that modulates the RNA binding activity of PCBP1. In folate-deficient placental cells, PCBP1 (also called hnRNP E1) is reversibly homocysteinylated on Cys293, which enhances its affinity for folate receptor mRNA and increases the expression of folate receptor34,44,45. In these studies, iron and glutathione reduces the affinity of PCBP1 for mRNA, which is likely due to the coordination of Fe–GSH. Thus the RNA-binding and iron chaperone activities of PCBP1 may be coordinately regulated in cells.
Our genetic loss-of-function studies indicate that PCBP1 is required for the formation of the [2Fe–2S] clusters that bridge the Glrx3–[2Fe–2S]–BolA2 complex. In human cells, Glrx3 and BolA2 form a complex only when they coordinate the bridging [2Fe–2S] clusters. Although both the KH1 and the KH3 domains of PCBP1 bound to BolA2 in an Fe- and GSH-dependent manner, only KH3 could support the formation of [2Fe–2S] clusters on the BolA2–Glrx3 complex in cells. Sequence comparison of KH1 and KH3 reveals that Cys293 in KH3 is not conserved in KH1, although residues that could contribute O− ligands are present. Thus, the sulfhydryl from Cys293 may be specifically required to facilitate transfer of Fe(II) or Fe–GSH from PCBP1–BolA2 to BolA2–Glrx3 (Supplementary Fig. 11). The origin and chemical form of the sulfur species that contributes sulfide to the nascent BolA2–[2Fe–2S]–Glrx3 complex is not known, but could be initially coordinated by either BolA2, Glrx3, or both. An alternative model posits that the PCBP1–Fe–GSH–BolA2 complex could serve to repair or restore a damaged or partially assembled cluster on Glrx3; these models are not mutually exclusive and both could be occurring in cells.
The Glrx3–[2Fe–2S]2–BolA22 complex can function as an Fe–S cluster chaperone, binding and transferring its [2Fe–2S] clusters to the apo- form of Ciapin1 (also called anamorsin). Ciapin1 coordinates a pair of Fe–S clusters and functions in the early steps of cytosolic [Fe–S] assembly29,46. Cluster transfer from Glrx3–[2Fe–2S]2–BolA22 to Ciapin1 has been observed in vitro and in cells. We show that PCBP1 provides an entry point for iron in the assembly or repair of cytosolic Fe–S clusters and that the PCBP1–Fe–GSH–BolA2 complex serves as an intermediate for the transfer of iron to Glrx3–BolA2. Whether the PCBP1–Fe–GSH–BolA2 complex can transfer iron to other targets or is required for the efficient capture of iron from other sources is an important question that awaits further investigation.
Online Methods
Plasmids
Plasmids pcDNA5-PCBP1 and pcDNA5-Flag-PCBP1 were created by amplifying PCBP1 from pYES2-Flag-PCBP113 and subcloning the resulting fragment into pcDNA5/FRT/TO vectors (Thermo Fisher Scientific) containing 2xFlag at N-terminus using Gibson Assembly (New England Biolabs). Plasmids pcDNA5-BolA2, pcDNA5-BolA2-Flag, and pcDNA3–6x-myc-Glrx3 were obtained as mentioned previously27. Plasmids for expression of individual K-Homology domains of PCBP1 were created by deleting the different regions from pcDNA5-Flag-PCBP1: KH1 (Δ301–1089), KH2 (Δ85–309, Δ537–1089), KH3 (Δ85–879), KH1–2 (Δ559–1089), KH2–3 (Δ85–309), and ΔVr (Δ568–861) using Q5 Site-Directed Mutagenesis kit (New England Biolabs). Single substitution variants of PCBP1, KH3 and BolA2 were introduced into pcDNA5-Flag-PCBP1: C293S or into pcDNA5-Flag-KH3: C293S, G296A, R297A, G299A, N301A, E304S, R306A, R325A, S332A, R346A, E350A, C355S, N301A/R346A or into pcDNA5-BolA2-Flag: C31S, H68A using Q5 Site-Directed Mutagenesis kit (New England Biolabs). All plasmids were confirmed by DNA sequencing.
Cell Culture and Stable Cell Lines
HEK293 cells were grown in DMEM supplemented with 10% FBS, 50 U/ml penicillin G, and 50 μg/ml streptomycin. Flp-In T-Rex 293 cell lines (Thermo Fisher Scientific) containing stably integrated pcDNA5/FRT/TO vectors were constructed according to the manufacturer’s instructions using pcDNA5-Flag-PCBP1 or pcDNA5-BolA2-Flag and maintained in DMEM supplemented with 10% tetracycline-free FBS (HyClone serum), 50 μg/ml hygromycin, and 15 μg/ml blasticidin. Where indicated, doxycycline was used at 1 μg/ml or iron was used as ferric ammonium citrate.
Protein Depletion by siRNA
Cells were treated with 5–20 nM of the targeting Stealth siRNA (Supplementary Table 2) according to manufacturer’s protocol. Non-targeting, scrambled sequence pools of siRNA were used as a control. Unless otherwise stated, cells were treated with 2 sequential siRNA treatments spaced by 24 hours using Lipofectamine RNAiMax transfection reagent (Invitrogen) and harvested 4 days after the initial transfection.
Co-immunoprecipitations and Immunoblotting
HEK293 cell were transfected with 5–10 μg indicated plasmids using Lipofectamine 2000 transfection reagent (Invitrogen) and harvested 24 h post transfection. Stable cell line expressing indicated Flag epitope-tagged proteins were treated with 1 μg/ml doxycycline to induce expression of the respective gene for 24 h. Cells were lysed in IP buffer (100 mM Tris-HCl pH7.5, 40 mM KCl, 0.1% Nonidet P40, 5% glycerol, 5 mM GSH, 1x protease inhibitor cocktail, Roche), and clarified whole cell extracts were subjected to immunoprecipitation using anti-Flag agarose for 30 minutes at 4°C. Following 4 washes with IP buffer, immune complexes were eluted in 2X LDS sample buffer (Invitrogen) containing 10% β-mercaptoethanol at 95 °C for 10 min and analyzed by immunoblotting. Membranes were incubated with the indicated primary antibodies, followed by detection using IR dye-conjugated secondary antibodies (1:10,000, Li-Cor). Antibodies used for Western blotting were rat anti-Flag (1:2,000, BioLegend), chicken anti-PCBP1 (1:10,000)11, chicken anti-BolA2 (1:2,000)27, rabbit anti-Glrx3 (1:10,000), and rabbit anti-Nfs1 (1:1,000)47. Detection and quantitative analysis were performed using Odyssey CLx (Li-Cor) software. For quantification ratio of target protein to anti-Flag protein were calculated and normalized to control. Where indicated, glutathione was removed from IP buffer or iron was used at 10–100 μM as ferrous ammonium sulfate or iron chelators were used at 100 μM as a mixture of desferrioxamine B (Dfo)/ bathophenanthrolinedisulfonic acid (BPS).
Mass spectrometry analysis
HEK293 cell lines expressing Flag-PCBP1 or the empty vector were grown in medium supplemented with 1 μg/ml doxycycline and 100 μM ferric ammonium citrate or 100 μM desferrioxamine B. Six 15-cm plates were harvested, and Flag immunoprecipitates were prepared as described previously48. High-salt washes were performed by increasing sodium chloride in the wash buffer to 500 μM. Immune complexes were eluted with Flag peptide, and the eluates were precipitated in 10% cold trichloroacetic acid. The precipitated eluates underwent tryptic digestion and mass spectrometry analysis as described previously48,49. Proteomics dataset is available at MassIVE Repository (https://massive.ucsd.edu/) using the MSV000083887 accession.
55Fe or 35S-GSH labeling
HEK293 cells were transfected with indicated plasmids or treated with 1 μg/mL doxycycline and 2–5 μM 55FeCl3 (PerkinElmer) for 16 h. Cells were washed three times with PBS, including first wash with PBS + 10 mM EDTA, 2 mM BPS, and 50 μM Sodium ascorbate. Whole cell extracts were prepared using IP buffer. Immunoprecipitation was then performed using anti-Flag agarose for 30 min at 4°C with gentle tumbling. Immune complexes were washed 4 times with IP buffer and retained 55Fe was quantitated by liquid scintillation counting (LSC500, Beckman Coulter). IPs from cells transfected with control plasmid or not treated with doxycycline were used to determine nonspecific 55Fe background. Co-precipitating 55Fe was normalized to total cellular 55Fe accumulation and Flag-tag protein level in whole cell extracts or IP fractions.
For 35S-glutathione (35S-GSH) labelling, whole cell extracts were prepared from cells expressing FLAG-tagged proteins and captured on anti-FLAG resin. Immobilized FLAG-tagged proteins were labelled with 100 μM 35S-GSH (~1 μCi, PerkinElmer) and incubated for 20 min at 4°C with gentle tumbling. Beads were washed 1 time with IP buffer containing 100 μM GSH and retained 35S-GSH was quantitated by liquid scintillation counting (LSC500, Beckman Coulter). Bound 35S-GSH was normalized to unbound 35S-GSH and Flag-tag protein level in whole cell extracts or IP fractions.
Protein Expression and Purification
K-homology domain (KH3-Flag) of human PCBP1 was amplified from pcDNA5-KH3-Flag, cloned into a pCDF-6xHis-SUMO fusion construct vector and transformed into BL21star(DE3)pLysS Escherichia coli (Invitrogen) competent cells containing ampicillin resistance. Competent cells were grown in 1L LB media at 37°C to an OD600 of 0.6, induced with 1 mM IPTG and grown for 4 h at 37°C. Cells were collected, resuspended in 20 ml of buffer B (100 mM Tris pH 7.5, 150 mM NaCl, 5 mM GSH, 40 mM imidazole, 10% glycerol, and 1 mM tris-2-carboxyethyl-phosphine (TCEP)), containing 25 U/ml Benzonase nucleases (Sigma) and EDTA-free protease inhibitor cocktail (Roche). Cells were lysed with a multi shot constant system (Pressure Bioscience). The extract was spin down at 25,000 rpm for 30 min and loaded onto an AKTA HisTrap HP nickel column (GE Healthcare). Separation was carried out using a linear gradient starting with buffer B and ending with buffer E (100 mM Tris pH 7.5, 150 mM NaCl, 5 mM GSH, 400 mM imidazole, 10% glycerol, 1 mM TCEP). Protein eluted between 100 – 250 mM imidazole. The 6xHis-SUMO tag was cleaved by adding SUMO protease (Invitrogen) according to the manufacturer’s instructions. Protein was buffer-exchanged with buffer B using Ultra-3 Centricon (Millipore) filtration devices and pass through the nickel column again to separate the tag from the untagged protein. Protein purity was analyzed by 4–12% SDS-PAGE followed by Coomassie Brilliant Blue staining and Western blot using an anti-Flag antibody.
Human BolA2-Strep tagged construct was generated by inserting Strep-tag II sequence at C-terminal in pETDuet-1-BolA2Δ1−66 (gift from Caryn Outten). pETDuet-1-BolA2Δ1–66-Strep plasmid was transformed into BL21star(DE3)pLysS E. coli (Invitrogen) competent cells. Competent cells were grown in 1 L of LB medium at 30 °C until the OD600 reached 0.6 and induce with 1 mM IPTG for 18 h. Strep-Tactin purification was performed according to manufacturer’s protocol. Briefly, cells were resuspended in 20 mL buffer W (100 mM Tris pH 8.0, 150 mM NaCl, 1 mM EDTA, 10% glycerol, and 1 mM TCEP) and lysed with a multi shot constant system. The extract was spin down at 25,000 rpm for 30 min and loaded onto gravity flow Strep-Tactin Superflow high capacity column (IBA). Protein were eluted with buffer D (100 mM Tris pH 8.0, 150 mM NaCl, 1 mM EDTA, 2.5 mM Desthiobiotin, 10% glycerol, and 1 mM TCEP). Buffer exchange was completed using Ultra-3 Centricon (Millipore) filtration devices. Protein purity was analyzed by 4–12% SDS-PAGE followed by Coomassie Brilliant Blue staining and Western blot using an anti-BolA2 antibody.
In vitro Reconstitution
Purified KH3-Flag and BolA2-Strep (0.1 – 4 μM) were mixed in 200 μL IP buffer (100 mM Tris-HCl pH7.5, 40 mM KCl, 0.1% Nonidet P40, 5% glycerol, 1 mM TCEP), containing 100 μM ferrous ammonium sulfate, Fe-Chelator (100 μM Desferoxamine, 100 μM BPS), or glutathione (0 – 5 mM) as indicated. The in vitro mix were incubated with anti-Flag agarose for 20 minutes at 4°C with gentle tumbling. Beads were washed 3 times with IP buffer to remove uncaptured proteins before eluting with 2X LDS sample buffer (Invitrogen) containing 10% β-mercaptoethanol at 95 °C for 10 min. The input and elute samples were subjected to immunoblot analysis and quantitation.
For 35S-GSH labelling of KH3-Flag, purified 5 μM KH3-Flag was labelled with 100 μM 35S-GSH (~1 μCi, PerkinElmer) in IP buffer and incubated with anti-FLAG resin for 20 min at 4°C. Beads were washed 1 time with IP buffer containing 100 μM GSH and retained 35S-GSH was quantitated by liquid scintillation counting (LSC500, Beckman Coulter). Bound 35S-GSH was normalized to unbound 35S-GSH and KH3-Flag protein levels, and in vitro mix without KH3-Flag was used as a control.
Size Exclusion Chromatography (SEC)
SEC was performed on HiPrep 16/60 Sephacryl S-100 high resolution column using AKTA purifier system with simultaneous UV/vis detection (GE Healthcare). The column was calibrated using the bovine serum albumin (66 kDa), carbonic anhydrase (29 kDa), cytochrome c oxidase (12 kDa), and aprotinin (6 kDa) with the SEC buffer (100 mM Tris, pH 7.5, 40 mM KCl, and 1 mM TCEP). Purified KH3-Flag and BolA2-Strep (50 μM) were mixed in presence of 100 μM ferrous ammonium sulfate and 1 mM glutathione or 500 μM ammonium sulfate, and analyzed by SEC on column pre-equilibrated with the SEC buffer. The elution profiles were followed by absorption at 280 nm and protein identities in elution fractions were checked with SDS-PAGE and Western blotting.
Microscale Thermophoresis (MST)
Purified His-SUMO-KH3 domain was washed with buffer containing 100 mM Tris pH 7.5, 300 mM NaCl, 5 mM ammonium sulfate, 10% glycerol, 1 mM TCEP to avoid the intramolecular interactions and remove nucleic acid contamination. His-SUMO-KH3 domain were fluorescently labeled using the Monolith His-Tag Labeling Kit RED-tris-NTA 2nd Generation as recommended by the supplier (NanoTemper Technologies, Germany). The fluorescently labelled protein (200 nM) was titrated with serial dilutions of glutathione (from 5 mM to 0.1 μM), in presence or absence of 10 μM FeSO4, in buffer containing 50 mM Tris, pH 7.5, 40 mM KCl, 0.2 mM TCEP, and 0.05% Tween20. Thermophoresis assays were performed using Monolith NT.115 at 25°C (LED power 60%, IR laser power 40%) in premium coated capillaries. Three independent experiments were recorded at 680 nm. The thermophoresis data were processed by MO Affinity Analysis (NanoTemper) and Prism7 (GraphPad) software to estimate the KD values.
Iron binding affinity
Iron binding affinities were determined using ratiometric metal indicator mag-fura-2 (MF2) and fura-4F (Invitrogen) as described previously37,38. Both MF2 and fura-4F forms a 1:1 complex with most divalent cations. The apo indicator has an absorbance maximum at 366 nm (MF2) or 380 nm (fura-4F) which shifts toward 325 nm concurrent with metal binding. For determination of metal binding affinity to protein, we tracked the absorbance decrement of the MF2 or fura-4F, in presence of protein, upon addition of metal ion. KH3-Flag protein (10 μM) and indicator (10 μM) were titrated with 1 mM FeSO4 in Chelex treated buffer 50 mM HEPES (pH 7.3), 150 mM NaCl, 1 mM TCEP, with or without 5 mM GSH. Concentrations of free indicator was determined using extinction coefficients for MF2 (ε366 29,900 M−1 cm−1) and fura-4F (ε380 21,000 M−1 cm−1). The variable Q and P are solved for using equations 1 and 2 respectively.
| (equation 1) |
| (equation 2) |
Q is equal to the ratio of free to metal-bound indicator, ε, εI, and εL are the observed, free indicator, and metal-bound indicator extinction coefficients respectively. P is the concentration of protein-metal complex, Mt is the total metal concentration, Ka is the association constant for the metal-indicator complex, and Ft is the total indicator concentration. Ratio of metal-bound KH3 to total KH3 was plotted as a function of free iron concentration and the data was fitted by Michaelis-Menten analysis to obtain the dissociation constant (KD) and stoichiometry (N). Reported errors for KD and stoichiometry are asymptotic standard errors provided by the Prism7 (GraphPad).
Statistics
Standard deviation (SD) and standard error of the mean (SEM) were calculated, and statistical significance was determined by two-tailed unpaired t-test or one-way ANOVA analysis using Prism7 (GraphPad) software.
Data Availability
The proteomics dataset (Supplementary Table 1) is available at MassIVE Repository (https://massive.ucsd.edu/) with the accession number MSV000083887.
Supplementary Material
Acknowledgements
This work was supported by the Intramural Research Program of the National Institutes of Diabetes and Degestive and Kidney Diseases and the Office of Dietary Supplements, Office of the Director, National Institutes of Health. J.A.W. is supported by NIH grants GM089778 and GM112763. We thank C. Outten (University of South Carolina) for plasmids, and T. Stemmler (Wayne State University) for plasmids and helpful discussions.
Footnotes
Competing interest
The authors declare no competing interests.
Reference
- 1.Hamza I & Dailey HA One ring to rule them all: trafficking of heme and heme synthesis intermediates in the metazoans. Biochim Biophys Acta 1823, 1617–1632, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ciofi-Baffoni S, Nasta V & Banci L Protein networks in the maturation of human iron-sulfur proteins. Metallomics 10, 49–72, (2018). [DOI] [PubMed] [Google Scholar]
- 3.Lill R et al. The role of mitochondria and the CIA machinery in the maturation of cytosolic and nuclear iron-sulfur proteins. Eur J Cell Biol 94, 280–291, (2015). [DOI] [PubMed] [Google Scholar]
- 4.Philpott CC & Jadhav S The ins and outs of iron: Escorting iron through the mammalian cytosol. Free Radic Biol Med 133, 112–117, (2019). [DOI] [PubMed] [Google Scholar]
- 5.Yanatori I & Kishi F DMT1 and iron transport. Free Radic Biol Med 133, 55–63, (2019). [DOI] [PubMed] [Google Scholar]
- 6.Chaudhury A, Chander P & Howe PH Heterogeneous nuclear ribonucleoproteins (hnRNPs) in cellular processes: Focus on hnRNP E1’s multifunctional regulatory roles. RNA 16, 1449–1462, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makeyev AV & Liebhaber SA The poly(C)-binding proteins: a multiplicity of functions and a search for mechanisms. RNA 8, 265–278, (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ostareck-Lederer A & Ostareck DH Control of mRNA translation and stability in haematopoietic cells: the function of hnRNPs K and E1/E2. Biol Cell 96, 407–411, (2004). [DOI] [PubMed] [Google Scholar]
- 9.Ostareck-Lederer A & Ostareck DH Precision mechanics with multifunctional tools: how hnRNP K and hnRNPs E1/E2 contribute to post-transcriptional control of gene expression in hematopoiesis. Curr Protein Pept Sci 13, 391–400, (2012). [DOI] [PubMed] [Google Scholar]
- 10.Philpott CC, Ryu MS, Frey A & Patel S Cytosolic iron chaperones: Proteins delivering iron cofactors in the cytosol of mammalian cells. J Biol Chem 292, 12764–12771, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi H, Bencze KZ, Stemmler TL & Philpott CC A cytosolic iron chaperone that delivers iron to ferritin. Science 320, 1207–1210, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Epsztejn S, Kakhlon O, Glickstein H, Breuer W & Cabantchik I Fluorescence analysis of the labile iron pool of mammalian cells. Anal Biochem 248, 31–40, (1997). [DOI] [PubMed] [Google Scholar]
- 13.Leidgens S et al. Each member of the poly-r(C)-binding protein 1 (PCBP) family exhibits iron chaperone activity toward ferritin. J Biol Chem 288, 17791–17802, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frey AG et al. Iron chaperones PCBP1 and PCBP2 mediate the metallation of the dinuclear iron enzyme deoxyhypusine hydroxylase. Proc Natl Acad Sci U S A 111, 8031–8036, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nandal A et al. Activation of the HIF prolyl hydroxylase by the iron chaperones PCBP1 and PCBP2. Cell Metab 14, 647–657, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ryu MS, Zhang D, Protchenko O, Shakoury-Elizeh M & Philpott CC PCBP1 and NCOA4 regulate erythroid iron storage and heme biosynthesis. J Clin Invest 127, 1786–1797, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yanatori I, Yasui Y, Tabuchi M & Kishi F Chaperone protein involved in transmembrane transport of iron. Biochem J 462, 25–37, (2014). [DOI] [PubMed] [Google Scholar]
- 18.Yanatori I, Richardson DR, Imada K & Kishi F Iron Export through the Transporter Ferroportin 1 Is Modulated by the Iron Chaperone PCBP2. J Biol Chem 291, 17303–17318, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yanatori I, Richardson DR, Toyokuni S & Kishi F The iron chaperone poly(rC)-binding protein 2 forms a metabolon with the heme oxygenase 1/cytochrome P450 reductase complex for heme catabolism and iron transfer. J Biol Chem 292, 13205–13229, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Braymer JJ & Lill R Iron-sulfur cluster biogenesis and trafficking in mitochondria. J Biol Chem 292, 12754–12763, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maio N & Rouault TA Iron-sulfur cluster biogenesis in mammalian cells: New insights into the molecular mechanisms of cluster delivery. Biochim Biophys Acta 1853, 1493–1512, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rouault TA & Maio N Biogenesis and functions of mammalian iron-sulfur proteins in the regulation of iron homeostasis and pivotal metabolic pathways. J Biol Chem 292, 12744–12753, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Srinivasan V, Pierik AJ & Lill R Crystal structures of nucleotide-free and glutathione-bound mitochondrial ABC transporter Atm1. Science 343, 1137–1140, (2014). [DOI] [PubMed] [Google Scholar]
- 24.Haunhorst P et al. Crucial function of vertebrate glutaredoxin 3 (PICOT) in iron homeostasis and hemoglobin maturation. Mol Biol Cell 24, 1895–1903, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, Mapolelo DT, Randeniya S, Johnson MK & Outten CE Human glutaredoxin 3 forms [2Fe–2S]-bridged complexes with human BolA2. Biochemistry 51, 1687–1696, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muhlenhoff U et al. Cytosolic monothiol glutaredoxins function in intracellular iron sensing and trafficking via their bound iron-sulfur cluster. Cell Metab 12, 373–385, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frey AG, Palenchar DJ, Wildemann JD & Philpott CC A Glutaredoxin.BolA Complex Serves as an Iron-Sulfur Cluster Chaperone for the Cytosolic Cluster Assembly Machinery. J Biol Chem 291, 22344–22356, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li F, Bullough KZ, Vashisht AA, Wohlschlegel JA & Philpott CC Poly(rC)-Binding Protein 2 Regulates Hippo Signaling To Control Growth in Breast Epithelial Cells. Mol Cell Biol 36, 2121–2131, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banci L, Camponeschi F, Ciofi-Baffoni S & Muzzioli R Elucidating the Molecular Function of Human BOLA2 in GRX3-Dependent Anamorsin Maturation Pathway. J Am Chem Soc 137, 16133–16143, (2015). [DOI] [PubMed] [Google Scholar]
- 30.Franco R, Schoneveld OJ, Pappa A & Panayiotidis MI The central role of glutathione in the pathophysiology of human diseases. Arch Physiol Biochem 113, 234–258, (2007). [DOI] [PubMed] [Google Scholar]
- 31.Biederbick A et al. Role of human mitochondrial Nfs1 in cytosolic iron-sulfur protein biogenesis and iron regulation. Mol Cell Biol 26, 5675–5687, (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H et al. The yeast iron regulatory proteins Grx3/4 and Fra2 form heterodimeric complexes containing a [2Fe–2S] cluster with cysteinyl and histidyl ligation. Biochemistry 48, 9569–9581, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumanovics A et al. Identification of FRA1 and FRA2 as genes involved in regulating the yeast iron regulon in response to decreased mitochondrial iron-sulfur cluster synthesis. J Biol Chem 283, 10276–10286, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang YS et al. Evidence Favoring a Positive Feedback Loop for Physiologic Auto Upregulation of hnRNP-E1 during Prolonged Folate Deficiency in Human Placental Cells. J Nutr 147, 482–498, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trott O & Olson AJ AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31, 455–461, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sidiqi M et al. Structure and RNA binding of the third KH domain of poly(C)-binding protein 1. Nucleic Acids Res 33, 1213–1221, (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patel SJ et al. Fine-tuning of Substrate Affinity Leads to Alternative Roles of Mycobacterium tuberculosis Fe2+-ATPases. J Biol Chem 291, 11529–11539, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walkup GK & Imperiali B Fluorescent chemosensors for divalent zinc based on zinc finger domains. Enhanced oxidative stability, metal binding affinity, and structural and functional characterization. Journal of the American Chemical Society 119, 3443–3450, (1997). [Google Scholar]
- 39.Esposito BP, Epsztejn S, Breuer W & Cabantchik ZI A review of fluorescence methods for assessing labile iron in cells and biological fluids. Anal Biochem 304, 1–18, (2002). [DOI] [PubMed] [Google Scholar]
- 40.Petrat F, de Groot H, Sustmann R & Rauen U The chelatable iron pool in living cells: a methodically defined quantity. Biol Chem 383, 489–502, (2002). [DOI] [PubMed] [Google Scholar]
- 41.Hider RC & Kong XL Glutathione: a key component of the cytoplasmic labile iron pool. Biometals 24, 1179–1187, (2011). [DOI] [PubMed] [Google Scholar]
- 42.Kozakov D et al. The ClusPro web server for protein-protein docking. Nat Protoc 12, 255–278, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ba LA, Doering M, Burkholz T & Jacob C Metal trafficking: from maintaining the metal homeostasis to future drug design. Metallomics 1, 292–311, (2009). [DOI] [PubMed] [Google Scholar]
- 44.Antony A et al. Translational upregulation of folate receptors is mediated by homocysteine via RNA-heterogeneous nuclear ribonucleoprotein E1 interactions. J Clin Invest 113, 285–301, (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang YS et al. Incrimination of heterogeneous nuclear ribonucleoprotein E1 (hnRNP-E1) as a candidate sensor of physiological folate deficiency. J Biol Chem 286, 39100–39115, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Banci L et al. N-terminal domains mediate [2Fe–2S] cluster transfer from glutaredoxin-3 to anamorsin. Nat Chem Biol 11, 772–778, (2015). [DOI] [PubMed] [Google Scholar]
- 47.Land T & Rouault TA Targeting of a human iron-sulfur cluster assembly enzyme, nifs, to different subcellular compartments is regulated through alternative AUG utilization. Mol Cell 2, 807–815, (1998). [DOI] [PubMed] [Google Scholar]
- 48.Vashisht AA et al. Control of iron homeostasis by an iron-regulated ubiquitin ligase. Science 326, 718–721, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wohlschlegel JA Identification of SUMO-conjugated proteins and their SUMO attachment sites using proteomic mass spectrometry. Methods Mol Biol 497, 33–49, (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The proteomics dataset (Supplementary Table 1) is available at MassIVE Repository (https://massive.ucsd.edu/) with the accession number MSV000083887.