Abstract
DNA binding with one finger (Dof) proteins constitute a ubiquitous plant-specific transcription factor (TF) family associated with diverse biological processes, including ripening. We conducted a genome-wide analysis of durian (Durio zibethinus Murr.) and identified 24 durian Dofs (DzDofs), 15 of which were expressed in fruit pulp. Gene expression analysis revealed differential expression of DzDofs during ripening in two commercial durian cultivars from Thailand, Monthong and Chanee. Comparing the expression levels of fruit pulp-expressed DzDofs between cultivars revealed ten potential cultivar-dependent Dofs, among which DzDof2.2 showed a significantly greater fold increase at every ripening stage in Chanee than in Monthong. The prediction of DzDof2.2’s function based on its orthologue in Arabidopsis revealed its possible role in regulating auxin biosynthesis. We observed significantly higher auxin levels during ripening of Chanee than Monthong which concurred with the greater expression of auxin biosynthetic genes. Transient expression of DzDof2.2 in Nicotiana benthamiana significantly upregulated the expression levels of auxin biosynthetic genes. Higher expression levels of DzDof2.2 in Chanee would enhance auxin levels through transcriptional regulation of auxin biosynthetic genes. Higher auxin levels in Chanee could activate auxin-mediated transcription, contributing to its faster ripening compared to Monthong through earlier initiation of the ethylene response (auxin-ethylene crosstalk).
Subject terms: Plant molecular biology, Natural variation in plants
Introduction
Transcription factors (TFs) are pivotal regulators of the gene expression networks that control plant growth, development, and responses to environmental stimuli. The functional characterization of TFs can provide new insights into how plants grow and respond to numerous biotic and abiotic stressors. The DNA binding with one finger (Dof) proteins constitute a ubiquitous, plant-specific TF family. All Dof TFs harbour a highly conserved domain of about 50–52 amino acids (known as the Dof domain), which is always found at their N-terminus. The Dof domain has a Cys2/Cys2 Zn2+ finger structure that specifically binds to the cis-element of AAAG in the promoter of its targeted genes1,2. The C-terminuses of Dofs are highly variable, and act as transcriptional regulators of the genes responsive to them3. Dof TFs are associated with a wide range of biological processes, such as seed germination, light responses, flowering, carbon metabolism, plant hormonal signalling, and responses to both biotic and abiotic stressors3–5. The first isolation of a Dof TF was carried out in maize6. Since then, many studies have documented the Dof TFs in other plants, such as Arabidopsis7, tomato5, rice8, cucumber9, pepper10, banana11, and kiwifruit12. In a study by Skirycz et al.13, Arabidopsis Dof1.1 (AtDof1.1, OBP2) was shown to regulate glucosinolate biosynthesis. Moreover, auxin levels in leaves and seedlings of OBP2 over-expressing lines were higher than those in wild-type plants. AtDof5.4 (OBP4) was reported to be involved in transcriptional regulation of the cell cycle14. In addition, higher expression levels of OBP4 under exposure to abiotic stressors (salt and heat) and exogenous auxin were also documented14. However, our knowledge of the actual roles of Dof TFs during the ripening of climacteric fruit is relatively limited. Notably, Feng et al.11 identified four ethylene-inducible Dofs that were expressed at increasingly high levels during banana ripening. This was the first report of a possible role for Dof TFs in climacteric ripening. In a study by Zhang et al.12, the role of a kiwifruit Dof (AdDof3) in starch degradation during ripening was investigated. AdDof3 was shown to physically interact with the AdBAM3L (β-amylase) promoter, a key gene involved in starch degradation.
Durian (Durio zibethinus Murr.) is an economically important tropical fruit crop native to Southeast Asia, which has recently begun to be distributed globally. Durian is distinctive for its strong and unique odour, formidable spiny husk, and overwhelming flavour. It plays a vital role as an important export commodity in the Southeast Asian region. Thailand is the top exporter of durian across this region. There are more than 200 durian cultivars, but only a few cultivars are commercially grown and are in high demand in the international market. In Thailand, two cultivars, Monthong (D. zibethinus Murr. cv. ‘Monthong’) and Chanee (D. zibethinus Murr. cv. ‘Chanee’) are cultivated commercially in several provinces, particularly in the eastern and southern parts of Thailand15. Notably, these two cultivars exhibit distinctive sensory characteristics from one another. Monthong is a less sweet cultivar with a mild odour and light yellowish pulp, whereas Chanee has an overpowering aroma, sweeter taste, and dark yellowish pulp. Generally, Thai farmers harvest durian at the commercially mature stage, which occurs around 90 and 105 days after anthesis for Chanee and Monthong, respectively. Harvesting is done at the mature stage to facilitate fruit handling and transportation, protect the fruits from pathogen infection on the tree, and enhance their sensory characteristics. Chanee ripens four days after harvesting at the mature stage, whereas Monthong has a longer post-harvest ripening period that requires five days after harvesting at the mature stage. Given that durian is a climacteric fruit, its shelf life is restricted once the ripening process starts. This phenomenon could negatively affect the handling and transportation of durian and cause major economic losses. Therefore, gaining a deeper understanding of the ripening process in durian is of the utmost importance to control its post-harvest ripening. Despite the rising importance of durian as an economic fruit crop, genomic research on this species is almost non-existent, especially with regard to its Dof TFs. Our study aimed to conduct a genome-wide analysis to identify the Dof TFs in the durian genome, which has recently been published16. Further, we performed some routine in silico analyses, including gene structure analysis, identification of conserved motifs, and phylogenetic analysis. Finally, we analysed the expression patterns of durian Dofs (DzDofs) during post-harvest ripening in both the Monthong and Chanee cultivars and identified ripening-associated and cultivar-dependent DzDofs. The findings of our study deepen our knowledge of the role of Dof TFs during post-harvest ripening of climacteric durian fruits, and how differential expression patterns of some Dofs could contribute to differences in the ripening behaviours of the Monthong and Chanee cultivars.
Results
Identification of 24 DzDof transcription factors
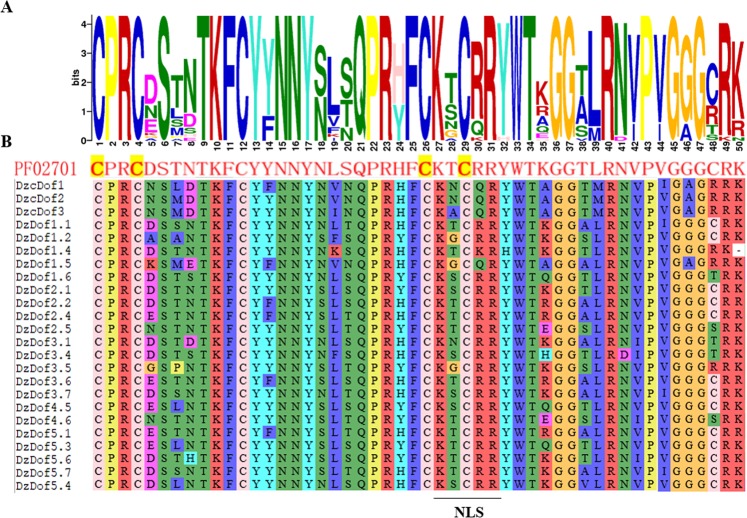
Our genome-wide attempts to identify DzDofs revealed a total of 24 genes encoding transcription factors of the DzDof family in Musang King genome, which was equal to the number of previously annotated DzDofs in the durian genome16 (Table 1). The sizes of the full-length coding sequences of DzDof genes ranged from 519 to 1560 bp. The deduced amino acid lengths of proteins made from DzDofs varied from 172 to 519 amino acids, with the isoelectric point (pI) of the resultant proteins predicted to range from 4.88 to 9.65 and their molecular masses from 19.19 to 56.59 kD (Table 1). Notably, DzDof2.4 and DzDof5.1 showed the highest levels of sequence similarity (89.85%), whereas DzDof1.2 and durian cyclic Dof1 (DzcDof1) were the least similar (22.82%) (Supplementary Table S1). Multiple sequence alignment of DzDofs revealed a highly conserved DNA binding domain of 50 amino acids at the N-terminus, structured as a Cys2/Cys2 Zn2+ finger (Fig. 1). This conserved motif, designated as the Dof domain, is the defining characteristic of Dof TFs. Moreover, a nuclear localization signal (NLS) was also detected in all DzDofs by in silico analyses. This sequence, which partly overlapped with the Dof domain, indicated the point of the nuclear localization of DzDofs.
Table 1.
Dof transcription factors identified in durian (Durio zibethinus Murr.).
| Gene name | Corresponding gene ID | ORF length (bp) | Protein length (aa) | MW (kD) | pI value |
|---|---|---|---|---|---|
| Durian cyclic Dof1 (DzcDof1) | XM_022890912.1 | 1443 | 480 | 52.01 | 6.37 |
| Durian cyclic Dof2 (DzcDof2) | XM_022884508.1 | 1560 | 519 | 56.59 | 5.95 |
| Durian cyclic Dof3 (DzcDof3) | XM_022882662.1 | 1458 | 485 | 53.17 | 6.77 |
| DzDof1.1 | XM_022885861.1 | 996 | 331 | 35.09 | 9.02 |
| DzDof1.2 | XM_022897927.1 | 927 | 308 | 34.52 | 4.88 |
| DzDof1.4 | XM_022864763.1 | 930 | 309 | 34.38 | 9.45 |
| DzDof1.5 | XM_022921573.1 | 519 | 172 | 19.19 | 9.15 |
| DzDof1.6 | XM_022916439.1 | 729 | 242 | 24.66 | 8.41 |
| DzDof2.1 | XM_022896728.1 | 870 | 289 | 31.97 | 6.88 |
| DzDof2.2 | XM_022874863.1 | 957 | 318 | 34.01 | 9.41 |
| DzDof2.4 | XM_022888412.1 | 1029 | 342 | 35.97 | 8.73 |
| DzDof2.5 | XM_022903860.1 | 840 | 279 | 30.53 | 9.27 |
| DzDof3.1 | XM_022865170.1 | 750 | 249 | 27.08 | 8.74 |
| DzDof3.4 | XM_022918231.1 | 714 | 237 | 24.97 | 9.27 |
| DzDof3.5 | XM_022889354.1 | 840 | 279 | 30.68 | 6.70 |
| DzDof3.6 | XM_022908803.1 | 1014 | 337 | 36.07 | 9.08 |
| DzDof3.7 | XM_022895569.1 | 987 | 328 | 34.98 | 8.85 |
| DzDof4.5 | XM_022921611.1 | 948 | 315 | 34.48 | 9.65 |
| DzDof4.6 | XM_022877869.1 | 873 | 275 | 30.19 | 8.46 |
| DzDof5.1 | XM_022891906.1 | 1029 | 342 | 35.91 | 9.45 |
| DzDof5.3 | XM_022868588.1 | 987 | 328 | 35.68 | 9.46 |
| DzDof5.4 | XM_022920120.1 | 945 | 314 | 34.26 | 6.82 |
| DzDof5.6 | XM_022881477.1 | 915 | 304 | 33.87 | 6.25 |
| DzDof5.7 | XM_022885261.1 | 981 | 326 | 35.36 | 9.02 |
Figure 1.
Multiple sequence alignment of the conserved Dof domains of 24 DzDofs. (A) Sequence representation LOGO obtained from multiple sequence alignment of the DzDof motifs. (B) Multiple sequence alignment of DzDof motifs. The nuclear localization signal (NLS) motif is also indicated in the alignment. PF02701 is the protein family ID of the zinc finger Dof domain retrieved from http://pfam.xfam.org/family/zf-Dof. DzcDof is durian cyclic Dof.
Phylogenetic analysis of DzDofs
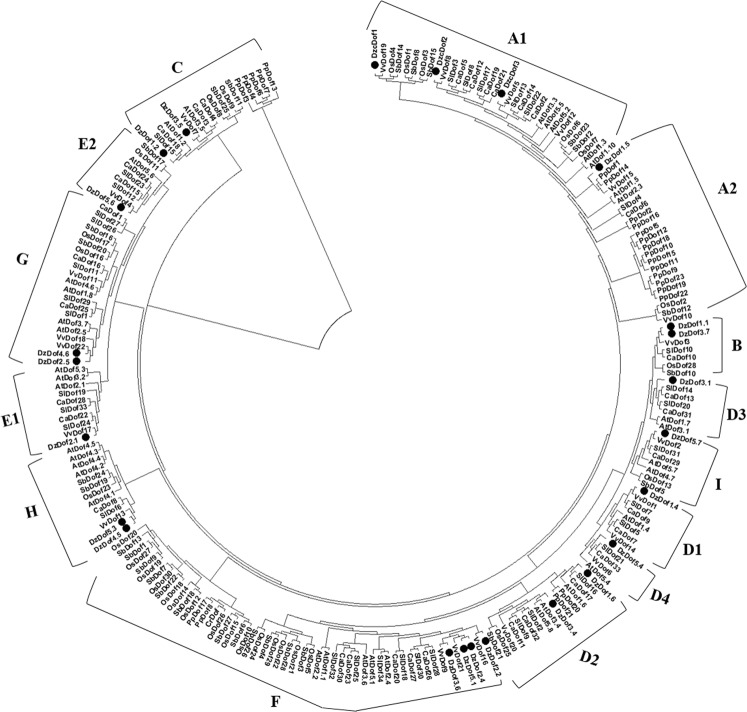
To investigate the evolutionary relationships among DzDof TFs and those from eight other representative species (dicots: Arabidopsis thaliana, Solanum lycopersicum, Capsicum annuum, and Vitis vinifera; monocots: Oryza sativa and Sorghum bicolor; non-vascular species: Chlamydomonas reinhardtii and Physcomitrella patens subsp. patens), a neighbour-joining (NJ) phylogenetic tree was constructed by aligning their full-length protein sequences, with 1000 bootstrap replicates. The phylogenetic tree divided the DzDof homologues into nine groups (A to I), which could be further classified into 14 subgroups: A1, A2, B, C, D1, D2, D3, D4, E1, E2, F, G, H, and I (Fig. 2). Group A was the largest clade, containing 56 members, while group B was the smallest clade and harboured only seven members. Interestingly, most of the Dofs from O. sativa and S. bicolor were placed in group F, highlighting the fact that these two species belonged to the grass family. Moreover, the only Dof TF from C. reinhardtii was classified into group F. Many Dofs from S. lycopersicum and C. annuum were clustered as pairs, which might point to some levels of domain consensus before the divergence of tomato and pepper. Fourteen pairs of DzDofs with V. vinifera Dofs (VvDofs) were shown while only one pair of DzDof with Arabidopsis Dof (AtDof) was found. All durian cyclic Dofs (DzcDofs) were grouped together in subclade A1.
Figure 2.
Phylogenetic tree of Dof gene families. Durian DzDofs were aligned with the Dof gene families of eight other representative species (dicots: Arabidopsis thaliana, Solanum lycopersicum, Capsicum annuum, and Vitis vinifera; monocots: Oryza sativa and Sorghum bicolor; non-vascular species: Chlamydomonas reinhardtii and Physcomitrella patens subsp. patens). The full-length protein sequences with 1000 bootstrap replicates were aligned with MEGA X software (with a JTT model and pairwise gap deletion) to construct the phylogenetic tree, using a bootstrap test of phylogeny with the minimum evolution test and default parameters43.
Conserved motifs and gene structure analysis
Gene structure analysis revealed no introns in all DzDofs (Supplementary Fig. S1). This structural feature (having few (0–2) introns) might be correlated with the important functions of Dof, as similar findings were previously reported for Arabidopsis and rice Dofs9. Moreover, to gain better insights into the conserved motifs of DzDofs, the MEME program was used to analyse putative motifs. As shown in Supplementary Fig. S2, ten distinct motifs were identified. Motif 1 (50 amino acids), known as the Dof domain, was detected at the N-terminus of all DzDofs. All members clustered into one group showed similar motif compositions, which suggested functional similarities among the members of the same group (Supplementary Fig. S3).
Tissue-specific expression of DzDofs
Analysing publicly available transcriptome data from different tissues of the durian cv. Musang King revealed a total of 15 DzDofs, including DzcDof1, DzcDof2, DzcDof3, DzDof1.4, DzDof1.5, DzDof2.1, DzDof2.2, DzDof2.4, DzDof2.5, DzDof3.1, DzDof4.6, DzDof5.1, DzDof5.3, DzDof5.4, and DzDof5.6, which were expressed in the fruit pulp at ripe stage (Supplementary Fig. S4). The remaining nine DzDofs (DzDof1.1, DzDof1.2, DzDof1.6, DzDof3.4, DzDof3.5, DzDof3.6, DzDof3.7, DzDof4.5, and DzDof5.7) were not expressed in the fruit pulp at ripe stage. However, their expressions could be detected in the other tissues (Supplementary Fig. S4).
Differential expression patterns of DzDofs during post-harvest ripening of the Monthong cultivar
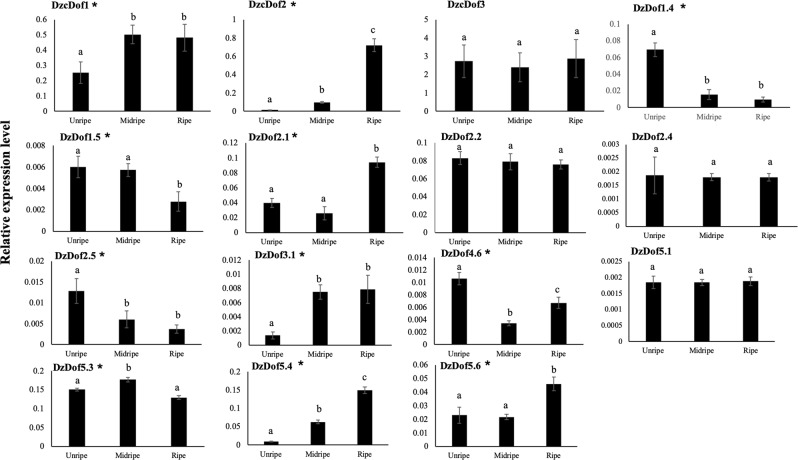
We examined the expression levels of 24 DzDofs at three different stages (unripe, midripe, and ripe) during post-harvest ripening. Using qRT-PCR, we observed that 15 of the 24 DzDofs were expressed in the fruit pulp, which were: DzcDof1, DzcDof2, DzcDof3, DzDof1.4, DzDof1.5, DzDof2.1, DzDof2.2, DzDof2.4, DzDof2.5, DzDof3.1, DzDof4.6, DzDof5.1, DzDof5.3, DzDof5.4, and DzDof5.6 (Fig. 3). This finding was similar to the fruit pulp-expressed DzDofs in Musang King cultivar. Notably, these 15 fruit pulp-expressed DzDofs showed different expression patterns during post-harvest ripening of durian fruit. Out of the 15 fruit pulp-expressed DzDofs, the expression patterns of 11 varied over the course of post-harvest ripening. Six DzDofs, including DzcDof1, DzcDof2, DzDof2.1, DzDof3.1, DzDof5.4, and DzDof5.6, were expressed at increasingly high levels as fruit ripening progressed in the Monthong cultivar. Moreover, four DzDofs, including DzDof1.4, DzDof1.5, DzDof2.5, and DzDof4.6, were expressed at decreasing levels over the course of fruit ripening. In addition, DzDof5.3 was expressed at significantly higher levels at the midripe compared to the unripe stage. However, its expression level declined from the midripe to the ripe stage. Taken together, these 11 DzDofs showed ripening-associated expression patterns, and their expression levels were further examined under ethylene treatment. In contrast, the expression of DzcDof3, DzDof2.2, DzDof2.4, and DzDof5.1 did not vary significantly during post-harvest ripening, and thus these were not considered to be ripening-associated TFs.
Figure 3.
Expression levels of DzDofs in durian pulp at three different stages (unripe, midripe, and ripe) during post-harvest ripening in the Monthong cultivar. Gene transcript level was quantified using qRT-PCR and was normalized with respect to the expression level of elongation factor 1 alpha (EF-1α), which was used as the reference gene. Error bars represent ± standard deviations (SD). For each DzDof, comparisons were made among its expression levels at three different stages. Three independent biological replicates were used. Three technical replicates were also performed for each biological replicate. Bars with different letters above them differed significantly according to Duncan’s multiple range test (P < 0.05). Eleven putative ripening-associated DzDofs are marked with asterisks (*).
Validation of ripening-associated DzDofs under three ripening treatments of the Monthong cultivar
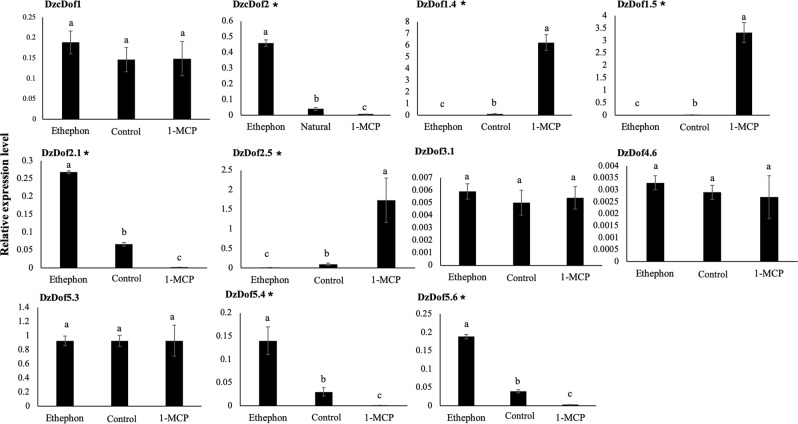
Ethylene plays a major role in climacteric fruit ripening. To further confirm the expression levels of the putative ripening-associated DzDofs under ethylene treatment, their transcript levels were measured under three different ripening conditions: natural, ethephon-induced, and 1-methylcyclopropene (1-MCP)-delayed ripening. As shown in Fig. 4, the expression levels of DzcDof2, DzDof2.1, DzDof5.4, and DzDof5.6 were significantly enhanced under ethylene treatment compared to those during natural ripening (control). However, under 1-MCP treatment, their transcript levels were dramatically decreased compared to those during natural ripening. Moreover, expression levels of DzDof1.4, DzDof1.5, and DzDof2.5 were dramatically suppressed under ethylene treatment, whereas 1-MCP treatment significantly enhanced their expression levels relative to those during natural ripening. These results provide convincing evidence regarding the ripening-associated roles of these seven DzDofs during fruit ripening. However, the expression of DzcDof1, DzDof3.1, DzDof4.6, and DzDof5.3 did not vary significantly under different ripening treatments, and therefore these were not considered to be ripening-associated TFs.
Figure 4.
Expression levels of 11 putative ripening-associated DzDofs under three different ripening treatments: natural (control), ethylene-induced, and 1-MCP-delayed ripening. Gene transcript level was quantified using qRT-PCR. Elongation factor 1 alpha (EF-1α) was used as the reference gene to normalize the expression levels of each other gene. Error bars indicate ± standard deviations (SD). For each DzDof, comparisons were made of its expression levels among three different ripening treatments. Three independent biological replicates were used. Three technical replicates were also performed for each biological replicate. Bars with different letters above them were significantly different according to Duncan’s multiple range test (P < 0.05). The DzDofs marked with asterisks (*) indicate the seven confirmed ripening-associated DzDofs.
Differential expression levels of fruit pulp-expressed DzDofs during post-harvest ripening of the Chanee cultivar
To examine the expression levels of DzDofs in another cultivar with different ripening behaviour and sensory characteristics, their expression levels were also measured during post-harvest ripening in the Chanee cultivar. The expression levels of DzDofs in Chanee followed a similar pattern to those in Monthong (Supplementary Fig. S5). DzcDof2, DzDof2.1, DzDof5.4, and DzDof5.6 were expressed at increasingly high levels during fruit ripening in the Chanee cultivar. Moreover, DzDof1.4, DzDof1.5, and DzDof2.5 were expressed at decreasing levels during ripening in this cultivar.
To further explore any potential association between the expression profiles of DzDofs and the different ripening characteristics of the Monthong and Chanee cultivars, the expression levels of Monthong DzDofs were compared to those in the Chanee cultivar. Among the 15 fruit pulp-expressed DzDofs, the expression profiles of 10 Dofs, including DzcDof2, DzDof1.4, DzDof2.1, DzDof2.2, DzDof2.5, DzDof3.1, DzDof4.6, DzDof5.3, DzDof5.4, and DzDof5.6, showed significant differences between the two cultivars in at least one stage of ripening (Supplementary Fig. S5). These ten DzDofs were thus considered potential cultivar-dependent TFs. However, the expression levels of the remaining DzDofs did not show any significant differences between the two cultivars during ripening. Among the cultivar-dependent DzDofs, DzDof5.4 had the greatest fold increase at the ripe stage in Chanee compared with that in Monthong. Moreover, DzDof2.2 was the only Dof whose expression level was higher in Chanee, and it showed a significant fold difference between the two cultivars at each ripening stage. This DzDof might be responsible for the different ripening characteristic of Chanee and Monthong, since its expression levels were significantly higher at each ripening stage in the Chanee cultivar than those in the Monthong cultivar. Hence, DzDof2.2 was further selected for functional prediction analysis via orthologue identification.
Orthologue identification and gene function prediction of the candidate cultivar-dependent DzDof2.2
Characterized orthologue identification is widely used as a powerful tool for functional prediction in genes17–19. It has been shown that orthologous proteins have similar biological functions in different species17. The protein sequence of DzDof2.2 was used as a query to find orthologous proteins in Arabidopsis. As a result, AtDof2.4, AtDof5.1, AtDof3.6 and AtDof1.1 (OBP2) were found to be the orthologues of DzDof2.2. This was consistent with the phylogenetic tree (Fig. 2), which clustered DzDof2.2 with the mentioned AtDofs together in the clade F. According to the functional characterization of AtDof1.113, significantly higher auxin levels were measured in the leaves and seedlings of AtDof1.1 over-expressing lines compared to those in wild-type plants. Accordingly, it might be possible that DzDof2.2 could also regulate auxin biosynthesis during ripening in durian. This finding prompted us to further measure endogenous auxin levels in the fruit pulps of both cultivars at different stages of ripening.
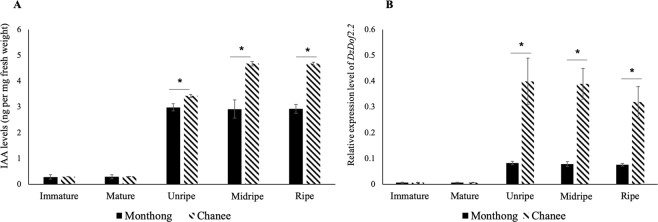
Higher indole-3-acetic acid (IAA) and auxin biosynthetic gene expression levels during the ripening of Chanee compared to Monthong
Since the cultivar-dependent DzDof2.2 could play a role in ripening by regulating auxin biosynthesis, endogenous levels of IAA were measured in the pulps of durian fruits at five different stages before (immature and mature) and during post-harvest ripening (unripe, midripe, and ripe) of both the Monthong and Chanee cultivars. As shown in Fig. 5A, during the immature and mature stages, IAA was detected at low levels, which were not significantly different between the two cultivars. However, after being harvested (from the mature to unripe stages), IAA levels significantly increased in both cultivars, with there being a significantly greater increase in Chanee than in Monthong. Moreover, IAA levels were significantly higher during the post-harvest ripening of Chanee compared to those in Monthong. The higher IAA levels in Chanee could contribute to its faster post-harvest ripening compared to that of Monthong through enhancing climacteric ethylene biosynthesis (auxin-ethylene crosstalk). Assessing transcript levels by qRT-PCR confirmed the higher expression levels of both ethylene biosynthetic genes, e.g. ACC synthase (ACS) and ACC oxidase (ACO), during ripening of Chanee compared to Monthong (Supplementary Fig. S6), suggesting a higher endogenous ethylene content in Chanee. Moreover, the expression levels of both genes peaked at the ripe stage, indicating the highest levels of climacteric ethylene biosynthesis at this stage during post-harvest ripening of durian fruit (Supplementary Fig. S6).
Figure 5.
Indole-3-acetic acid (IAA) and DzDof2.2 expression levels. (A) IAA levels were measured at five different stages (immature, mature, unripe, midripe, and ripe) in both the Monthong and Chanee cultivars. Comparisons were made between the two cultivars at each stage. An asterisk (∗) above the bars indicates a significant difference between cultivars (Student’s t-test, P < 0.05). (B) Expression levels of DzDof2.2 were measured at five different stages (immature, mature, unripe, midripe, and ripe) in both the Monthong and Chanee cultivars. Comparisons were made between the two cultivars at each stage. Three independent biological replicates were used. Three technical replicates were also performed for each biological replicate. An asterisk (∗) above the bars indicates a significant difference between cultivars (Student’s t-test, P < 0.05).
To assess the correlation between DzDof2.2 expression levels and IAA content, we analysed the expression levels of DzDof2.2 before ripening at the immature and mature stages, in addition to the previous results assessed during post-harvest ripening (Supplementary Fig. S5), in both cultivars. At these two stages, it was found that DzDof2.2 was expressed at low levels in both the Monthong and Chanee cultivars, without any significant difference between the two cultivars (Fig. 5B). However, there was a significant increase in the expression levels of DzDof2.2 from the mature to the unripe stage in both cultivars, with a significantly greater fold change in Chanee compared to that in Monthong (Fig. 5B). The consistency of the expression levels of DzDof2.2 with auxin levels before and during ripening could support the possible role of DzDof2.2 in controlling auxin biosynthesis.
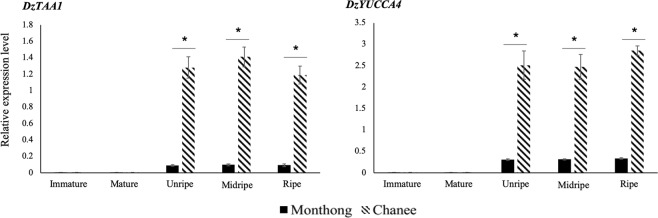
From the analysis of our in-house transcriptome data of Chanee fruit pulp, we identified two candidate auxin biosynthetic genes, including L-tryptophan aminotransferase 1 (TAA1) and the indole-3-pyruvate monooxygenase (YUCCA4) of the indole-3-pyruvic acid (IPyA) pathway. Interestingly, TAA1 and YUCCA4 had low expression levels during the immature and mature stages (before post-harvest ripening), with no significant differences between the two cultivars (Fig. 6). However, there was a dramatic increase in the expression levels of both genes after harvesting (from the mature to the unripe stage) in both the Monthong and Chanee cultivars. This phenomenon was consistent with the increased auxin levels observed after harvesting, which could occur as a stress response to mechanical injury during fruit harvesting. Notably, TAA1 and YUCCA4 were expressed at significantly higher levels during the post-harvest ripening of Chanee compared to that of the Monthong cultivar (Fig. 6). This observation was consistent with the higher IAA levels observed during ripening in the Chanee than in the Monthong cultivar. Interestingly, we scanned the promoter regions of TAA1 and YUCCA4 and identified 40 and 34 cis-regulatory motifs (AAAG or its reverse-oriented sequence CTTT, known as the binding site for Dof TFs)1,2, respectively (Supplementary Fig. S7). This observation could pinpoint the possible role of Dof TFs in the transcriptional regulation of auxin biosynthetic genes.
Figure 6.
Expression levels of auxin biosynthetic genes. Expression levels of two candidate auxin biosynthetic genes, L-tryptophan aminotransferase 1 (TAA1) (left panel) and the indole-3-pyruvate monooxygenase (YUCCA4) (right panel) of the indole-3-pyruvic acid (IPyA) pathway, were measured at five different stages (immature, mature, unripe, midripe, and ripe) in both the Monthong and Chanee cultivars. Comparisons were made between the two cultivars at each stage. Three independent biological replicates were used. An asterisk (∗) above the bars indicates a significant difference between cultivars (Student’s t-test, P < 0.05).
Functional characterization of DzDof2.2 through transient expression in Nicotiana benthamiana
To assess the function of DzDof2.2 in planta, it was transiently expressed in the leaves of N. benthamiana and the expression levels of major auxin biosynthetic genes from N. benthamiana, NbTAA1 and NbYUCCA3, were analysed. We observed a significantly higher expression levels of both NbTAA1 and NbYUCCA3 in the leaves transiently expressing DzDof2.2 when compared to the control (Fig. 7). This observation confirmed the transcriptional regulation of auxin biosynthetic genes by DzDof2.2 TF. Consistently, our in silico analysis identified cis-regulatory binding sites (AAAG/CTTT) for Dof TFs in the promoter regions of both NbTAA1 and NbYUCCA3 (Supplementary Fig. S8).
Figure 7.
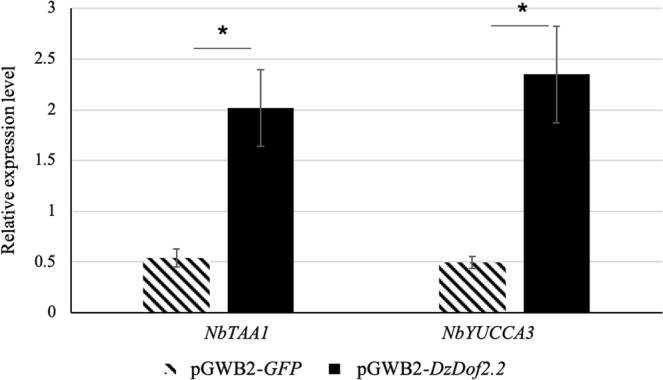
Expression levels of auxin biosynthetic genes in Nicotiana benthamiana transiently expressing DzDof2.2. Expression levels of two candidate auxin biosynthetic genes from N. benthamiana, L-tryptophan aminotransferase 1 (NbTAA1) and the indole-3-pyruvate monooxygenase (NbYUCCA3) of the indole-3-pyruvic acid (IPyA) pathway, were measured in N. benthamiana leaves infiltrated with pGWB2-DzDof2.2 (treatment) and pGWB2-GFP (control). Comparisons were made between the control and treatment. Three independent biological replicates were used. An asterisk (∗) above the bars indicates a significant difference (Student’s t-test, P < 0.05).
Discussion
TFs play significant roles in regulating gene expression networks. A large body of evidence suggests that the identification and functional characterization of TFs should provide a better understanding of how plants develop, grow, and respond to environmental stimuli. Herein, we conducted a genome-wide analysis and identified 24 putative DzDofs in the durian genome (Musang King cv.). The number of identified DzDofs was lower than that found in banana (74 MaDof)11, rice (30 OsDof)20, Arabidopsis (36 AtDof)2, tomato (34 SiDof)5, Chinese cabbage (76 BraDof)21, cucumber (36 CsDof)9, and potato (35 StDof)4. Consistent with previous studies, the alignment of DzDof protein sequences revealed a highly conserved Dof domain, which was observed at the N-terminal regions of all 24 DzDofs (Fig. 1), suggesting that Dof TFs have been evolutionarily conserved among plants. Apart from the Dof domain, nine other distinct motifs were identified that were differentially distributed among DzDofs. In addition, as predicted by in silico analyses, all of the 24 deduced DzDofs harboured NLSs to help localize them to the nucleus. Digital expression profiles of DzDofs from Musang King cultivar revealed a total of 15 ripe fruit pulp-expressed Dofs (Supplementary Fig. S4). These 15 DzDofs were identical to those expressed during post-harvest ripening of both Monthong and Chanee cultivars, suggesting their functions in the pulp tissue. Dof TFs have been shown to play crucial roles in various physiological phenomena, such as the regulatory networks of plant responses to diverse biotic and abiotic stressors, seed germination, morphology, light responses, and fruit ripening3,11,22.
In our study, among the 15 fruit pulp-expressed Dofs in Monthong cultivar, seven ripening-associated DzDofs, including DzcDof2, DzDof1.4, DzDof1.5, DzDof2.1, DzDof2.5, DzDof5.4, and DzDof5.6, were identified based on their differential expression during post-harvest ripening in the Monthong cultivar (Fig. 3). The expression levels of DzcDof2, DzDof2.1, DzDof5.4, and DzDof5.6 were confirmed to be induced under ethephon and suppressed under 1-MCP treatment (Fig. 4). These ethylene-inducible DzDofs were expressed at increasing levels during ripening, and thus could act as either putative transcriptional activators or repressors of ripening. Zhang et al.12 identified an ethylene-induced Dof TF in kiwifruit (AdDof3) that acted as a ripening-activator TF and enhanced starch degradation during fruit ripening. On the other hand, an ethylene-induced banana Dof (MaDof23), which was expressed at increasing levels during ripening, was a transcriptional repressor. This Dof acted antagonistically to a ripening-activator ethylene response factor (MaERF9) to regulate banana fruit ripening11. Notably, the expression levels of the remaining ripening-associated DzDofs (DzDof1.4, DzDof1.5, and DzDof2.5) were dramatically suppressed under ethephon and induced under 1-MCP treatment. It is interesting that these ripening-associated DzDofs were expressed at decreasing levels during ripening. To the best of our knowledge, this is the first report describing this particular effect of 1-MCP treatment on Dof TFs. The 1-MCP treatment delays climacteric ripening by inhibiting ethylene-dependent signalling by competitively binding to ethylene receptors23,24. Besides this inhibitory effect, 1-MCP also downregulates the expression of ethylene biosynthetic genes25–27 (e.g., 1-aminocyclopropane-carboxylase (ACC) synthase (ACS) and ACC oxidase (ACO)), ethylene perception genes27–30 (e.g., ethylene receptors ETR and ERS), and ethylene signalling genes31,32 (e.g., ethylene response factor (ERF)). It is possible that 1-MCP upregulates the expression of these three DzDofs by inhibiting ethylene biosynthesis and signalling. This observation suggests that these DzDofs could be negatively regulated by ethylene, and act as putative transcriptional repressors of ripening. Consistent with our observations, the expression of a kiwifruit Dof (AdDof4) was suppressed by ethylene treatment and was thus considered to be a ripening-repressor TF12.
In addition, we compared the expression patterns of DzDofs between the two cultivars and found that DzDofs in the Chanee cultivar showed similar expression patterns, including identical ripening-associated DzDofs, to those in the Monthong cultivar. The similar expression patterns of DzDofs between Monthong and Chanee cultivars suggest that these DzDofs might have similar regulatory roles during fruit ripening in these two cultivars. However, although the expression patterns were similar between the two cultivars, comparisons of the expression levels of fruit pulp-expressed DzDofs between cultivars revealed a total of 10 potential cultivar-dependent Dofs. Among these, the expression levels of DzDof2.2 underwent a significantly greater fold change at each ripening stage in the Chanee than in the Monthong cultivar. Although the expression level of DzDof2.2 did not vary significantly during the unripe, midripe, and ripe stages, its expression level harboured a significant increase from mature (before ripening) to unripe stage (post-harvest ripening) in both cultivars with a greater fold increase in Chanee than in Monthong (Fig. 5B). Therefore, we hypothesized that DzDof2.2 would play a role during ripening of durian fruit and the difference in its expression levels between the two cultivars might contribute to the faster ripening and different sensory characteristics of the Chanee cultivar from those of the Monthong one.
To examine the possible function of DzDof2.2, we found its orthologues in Arabidopsis. Among the four identified orthologues, AtDof1.1 (OBP2) is involved in regulating glucosinolate biosynthesis. In addition, transgenic Arabidopsis lines over-expressing OBP2 had higher auxin concentrations in their leaves and seedlings compared to those in the wild type13. This phenomenon could suggest a role of DzDof2.2 in regulating auxin biosynthesis. Higher expression levels of DzDof2.2 could enhance auxin biosynthesis, leading to the higher auxin levels during ripening in the Chanee than in the Monthong cultivar (Fig. 5A). Notably, the increased expression levels of DzDof2.2 were consistent with the increased auxin levels from the mature to the unripe stage (Fig. 5). We further confirmed that the higher level of auxin in Chanee co-occurred with the increased expression of auxin biosynthetic genes. Four tryptophan-dependent auxin biosynthetic pathways have been well-characterized in Arabidopsis, including the indole-3-pyruvic acid (IPyA), tryptamine (TAM), indole-3-acetaldoxime (IAOx), and indole-3-acetamide (IAM) pathways33. Among these pathways, IPyA is considered the major auxin biosynthetic pathway in many plant species34. The main biosynthetic enzymes of the IPyA pathway in durian, DzTAA1 and DzYUCCA4, were expressed at significantly higher levels during post-harvest ripening of the Chanee cultivar compared to those in the Monthong cultivar. Moreover, there was a significant increase in the expression levels of both genes from the mature to the unripe stage, with a higher fold increase in Chanee compared to that in Monthong (Fig. 6). This expression profile was consistent with the changes in expression levels of DzDof2.2 and IAA levels. It has been well-documented that auxin plays a major role during the ripening of climacteric fruits, such as peach, tomato, and papaya35,36. Concomitant with the increase in climacteric ethylene biosynthesis during ripening of tomato and peach, elevated levels of auxin have also been measured35. Auxin can stimulate climacteric ethylene biosynthesis through upregulating the ACC synthase gene family (auxin-ethylene crosstalk)37–39. Consistently, we observed significantly higher expression levels of both ethylene biosynthetic genes (ACS and ACO) during post-harvest ripening of Chanee compared to Monthong, suggesting higher ethylene content in Chanee than in Monthong. It has been well documented that the expression levels of ethylene biosynthetic genes are positively correlated with the ethylene content during climacteric ripening40,41. Besides this well-known role of auxin in upregulating ethylene biosynthesis, auxin can also play a role of its own during climacteric ripening by transcriptionally modulating other ripening-related auxin-responsive genes35. Taken together, our results demonstrate that auxin is actively involved in durian fruit ripening.
Interestingly, the promoter regions of DzTAA1 and DzYUCCA4 were enriched with cis-regulatory binding sites (AAAG/CTTT) for Dof TFs. This observation could suggest the possible transcriptional regulation of these two genes by Dof TFs. Our results obtained from transient expression of DzDof2.2 in N. benthamiana leaves provided convincing evidence of the possible role of DzDof2.2 in transcriptional regulation of auxin biosynthetic genes during ripening in durian (Fig. 7). Consistent with the promoters of auxin biosynthetic genes from durian, we identified the occurrences of Dof TF binding sites in the promoter regions of NbTAA1 and NbYUCCA3 (Supplementary Fig. S8).
It is notable that, among the 10 identified cultivar-dependent DzDofs, DzDof5.4 had the greatest fold increase at the ripe stage in Chanee relative to that in Monthong (Supplementary Fig. S5). OBP4 (AtDof5.4) was the closest orthologue of DzDof5.4. Interestingly, exogenous IAA induced the expression of AtDof5.4. This might explain why the expression levels of DzDof5.4 peaked at the ripe stage in both cultivars, with a dramatically greater fold increase in Chanee compared to that in Monthong. The higher IAA levels during ripening, the greater expression levels of DzDof5.4 were thus observed in our study.
To summarize, here, we reported a comprehensive analysis of durian Dof TFs. Out of 24 identified DzDofs, 15 were fruit pulp-expressed, among which seven DzDofs were identified as being ripening-associated for both the Chanee and Monthong cultivars. Notably, comparing the expression patterns of fruit pulp-expressed DzDofs between the Monthong and Chanee cultivars revealed 10 potential cultivar-dependent TFs, among which DzDof2.2 showed a significantly greater fold increase at every ripening stage in Chanee than in Monthong. We suggest that DzDof2.2 could transcriptionally control the expression of auxin biosynthetic genes. Higher expression levels of DzDof2.2 in Chanee could enhance its auxin levels during ripening through transcriptional regulation of auxin biosynthetic genes. Higher auxin levels in Chanee would upregulate ethylene biosynthesis by transcriptionally activating the ACC synthase gene family, and lead to an earlier ethylene response (auxin-ethylene crosstalk), and thus faster post-harvest ripening compared to that in the Monthong cultivar. Our results provide compelling evidence of the role of Dof TFs during the post-harvest ripening of durian. The results obtained herein should provide an important foundation for future studies involving gene cloning and the functional characterization of ripening-associated and cultivar-dependent DzDofs.
Materials and Methods
Plant materials and treatments
Durian (Durio zibethinus Murr.) cv. Monthong and Chanee fruits were collected from a commercial orchard located in Trat province in the eastern part of Thailand in early April 2017. Fruit samples of similar size and weight (~3–4 kg each) were harvested at two different stages: immature (at 70 days (for Chanee) and 85 days (for Monthong) after anthesis) and mature (at 90 days (for Chanee) and 105 days (for Monthong) after anthesis). Immature samples were peeled immediately after harvesting, whereas some mature samples were peeled immediately, and some were kept at room temperature (30 °C) until peeling. Five types of samples were used in this study: immature, mature, unripe (harvested at the mature stage and kept at room temperature for one day (both cultivars) and then peeled), midripe (harvested at the mature stage and kept at room temperature for two days (Chanee) or three days (Monthong) and then peeled), and ripe (harvested at the mature stage and kept at room temperature for four days (Chanee) or five days (Monthong) and then peeled). After peeling, two of the central pulps were collected from each fruit sample, following the method described by Pinsorn et al.15. Briefly, the first pulp was collected along with a seed, and was used to measure fruit firmness with a texture analyser (TA-XT2i; Stable Micro Systems, Godalming, UK) to assure that samples of the two cultivars were compared at the same ripening stages. A puncture test with a 6-mm probe, at a test speed of 2 mm/s and testing distance of 5 mm, was carried out on five random points in each pulp. Midripe and ripe pulps had a mean ± SD firmness of 3.4 ± 0.81 and 1.55 ± 0.45 N, respectively, in both cultivars. After this test, the second pulp was collected without a seed, immediately frozen in liquid nitrogen, and stored at −80 °C for further use.
To further investigate the role of ethylene during post-harvest ripening, three different ripening treatments were applied as follows: natural, ethephon-induced, and 1-methylcyclopropene (1-MCP)-delayed ripening. Mature durian samples were collected and treated with either ethephon (48% 2-chloroethylphosphonic acid; Alpha Agro Tech Co., Ltd., Thailand) or 1-MCP (0.19% 1-MCP tablet; BioLene Co., Ltd., China) for ethephon-induced and 1-MCP-delayed ripening, respectively. For ethephon treatment, the ethephon solution was applied to the upper area of each fruit stalk. For 1-MCP treatment, each fruit was placed inside a closed 20-L chamber. Then, one tablet of 1-MCP was placed into a beaker inside the chamber. Water (5 mL) was added to the beaker to generate gaseous 1-MCP and the chamber was immediately closed for 12 h at room temperature (30 °C) while the control samples were kept under similar conditions without 1-MCP. After treatment, control and treated samples were kept at room temperature (30 °C) until the ethephon-treated samples ripened. All samples from the three ripening conditions were then peeled. After that, two central pulps were collected from each sample and processed as mentioned previously. In this study, for each type of sample (immature, mature, unripe, midripe, and ripe, as well as natural ripening, ethephon-induced ripening, and 1-MCP-delayed ripening), three biological replicates were used. Each biological replicate was defined as one durian fruit harvested from a separate tree.
For the agroinfiltration experiment, N. benthamiana seeds were sown in pots containing peat moss and were grown under controlled conditions (temperature 25 °C and 16/8 h light/dark photoperiod; artificial light of 4,500 Lux). Two-week-old plants were transplanted individually into pots and were grown under similar conditions.
In silico identification and analysis of durian Dofs
The whole-genome sequence of durian cv. ‘Musang King’ was used to identify putative durian Dofs. First, the conserved Dof domain (PF02701) determined based on a Hidden Markov Model (HMM) was obtained from the Pfam protein database (http://pfam.xfam.org). The HMM profile of the Dof domain was then used as a query to search against in the Musang King genome database16. The amino acid sequences of DzDofs were further obtained and checked for the presence of the highly conserved motif (Dof domain) at the N-terminus. The isoelectric point (pI) and molecular weight of each of the identified DzDof proteins were then calculated using the web-based tool ExPASy (http://web.expasy.org/compute_pi/)42. Nuclear localization signal (NLS) analysis was also done using a web-based tool, found at: http://cello.life.nctu.edu.tw 9.
Phylogenetic analysis of DzDofs
To investigate phylogenetic relationships among DzDofs and Dofs from eight other representative species (dicots: Arabidopsis thaliana, Solanum lycopersicum, Capsicum annuum, and Vitis vinifera; monocots: Oryza sativa and Sorghum bicolor; non-vascular species: Chlamydomonas reinhardtii and Physcomitrella patens subsp. patens), a neighbour-joining (NJ) phylogenetic tree was constructed by aligning their full-length protein sequences with 1000 bootstrap replicates using MEGA X software (with a JTT model and pairwise gap deletion)43.
Gene structure analysis and identification of conserved motifs
To obtain a deeper understanding of DzDof structures, Gene Structure Display Server 2.0 (with default parameters) (http://gsds.cbi.pku.edu.cn) was used to characterize the exon-intron structures of DzDof-family genes. Motifs were identified using the MEME program (http://meme-suite.org)44, with the following parameter settings: motif length = 6–100; motif sites = 2–120; maximum number of motifs = 10; and the distribution of one single motif was “any number of repetitions”.
Tissue-specific expression analysis
In order to analyse the expression levels of DzDofs in different tissues, Illumina reads were obtained from a public repository database (SRA, Sequence Read Archive) as follows: SRX3188225 (root tissue), SRX3188222 (stem tissue), SRX3188226 (leaf tissue), and SRX3188223 (aril/pulp tissue). FASTQC software was used to analyse the total reads quality parameters (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). To filter low quality reads, the trimming process was performed using Trimmomatic V0.32 (http://www.usadellab.org/cms/?page=trimmomatic) (https://github.com/seqan/flexbar). Thereafter, transcriptome assembly was performed using Trinityrnaseq-Trinity-v2.4.0 (https://github.com/trinityrnaseq/trinityrnaseq/wiki/Running-Trinity)45. For expression profiling analysis, abundance estimation tool in Trinity package was used to align the input reads to the de novo assembled transcriptome to obtain raw counts of each contig. The raw read counts were then merged in a single read count matrix. This matrix was used as input for normalization to generate a Trimmed Mean of M-values (TMM) normalized matrix. Thereafter, the normalized total read counts (RC) were used to generate heatmap using MetaboAnalyst 4.0, an open source R-based program46. For the heatmap construction, the values were sum normalized, log2 transformed and autoscaled.
RNA isolation and quantitative real time polymerase chain reaction (qRT-PCR)
Total RNA was isolated from durian fruit pulp and purified using PureLink Plant RNA Reagent (Thermo Fisher Scientific™) according to the manufacturer’s instructions. Samples were treated with DNase I (Thermo Fisher Scientific™) to remove genomic DNA. The quality and quantity of RNA samples were assessed using agarose gel electrophoresis and an Eppendorf BioPhotometer D30 with A260/280 and A260/230 ratios between 1.8 to 2.0 and 2.0 to 2.2, respectively. One microgram of total RNA was reverse-transcribed to cDNA using a RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific™), in accordance with the manufacturer’s recommended protocol. All primers used in this study were designed using Primer 3 online (http://primer3.ut.ee/). The primers used are presented in Supplementary Table S2. qRT-PCR was performed to investigate patterns in the expression of DzDofs at the immature and mature stages (before ripening) and unripe, midripe and ripe stages (post-harvest ripening) of fruits of both the Monthong and Chanee cultivars. The PCR reaction was performed in a total volume of 20 μL containing 1 μL of diluted cDNA (1 ng of cDNA), 10 μL of 2x QPCR Green Master Mix LRox (biotechrabbit, Berlin, Germany), and 200 nM of each gene-specific primer. Amplification was carried out with a CFX95 Real-time System (Bio-Rad Laboratories Inc., California, USA) under the following conditions: initial activation at 95 °C for 3 min, followed by 40 cycles of denaturation at 95 °C for 15 s, then annealing at 56–60 °C for 30 s, and finally extension at 72 °C for 20 s. A melting curve analysis for testing specific products was carried out by heating the products from 55 °C to 95 °C at increments of 0.5 °C. A single product was obtained for each DzDof. All qRT-PCR experiments were performed with three independent biological replicates. The elongation factor 1 alpha (EF-1α) gene was used as a reference gene (See Supplementary Table S3 for primers). The relative expression level of each gene compared with that of the reference gene was calculated using the 2-ΔCt method47.
Identification of orthologues
Orthologues of candidate DzDofs in Arabidopsis were identified using the web-based tool EnsemblPlants, release 40 (https://plants.ensembl.org/index.html), using the default parameters48.
Indole-3-acetic acid (IAA) measurement
Endogenous IAA levels in fruit pulps from both cultivars were measured. IAA was extracted from fruit pulp samples following the method described by Sinha and Basu49. Briefly, each pulp sample was freeze-ground into a fine powder. After this, 500 mg of the powder was mixed with 10 mL of chilled 80% ethanol, vortexed vigorously for 3 min, and kept at 4 °C for 2 h with occasional vortexing (at least five times). Afterwards, the mixture was centrifuged at 10,000 rpm (4 °C) for 15 min, and the supernatant was then used for IAA measurements. The IAA concentration (A535) was quantitatively measured via the colorimetric assay of Gordon and Weber50 using a standard curve prepared from serial dilutions of commercial IAA stock solutions (Duchefa Biochemie, The Netherlands), ranging from 7.5 to 500 μM and with an R2 of 0.99.
In silico identification and promoter analysis of auxin biosynthetic genes
Based on the auxin biosynthetic pathways identified in Arabidopsis, the indole-3-pyruvic pathway was selected as the candidate pathway for auxin biosynthesis34 in durian. The conserved domain of each biosynthetic enzyme (based on a HMM) was obtained from the Pfam protein database (http://pfam.xfam.org/) as follows: using the PF00155 for aminotransferase (TAA1) and the PF00743 for flavin-containing monooxygenases of the YUCCA family (YUCCA4). The HMM profile of each enzyme was then used as a query to search against our in-house transcriptome database and the Musang King genome (i.e., TAA1 (XM_022878297.1) and YUCCA4 (XM_022900772.1)). For qRT-PCR, primers were designed as mentioned previously (Supplementary Table S3). In order to analyse cis-regulatory elements in the promoter regions of auxin biosynthetic genes, the 2000-bp upstream promoter regions of these genes were scanned for the Dof domain (AAAG/CTTT) using the online tool PLACE (http://www.dna.affrc.go.jp/)36.
Expression analysis of ACC synthase (ACS) and ACC oxidase (ACO) in Monthong and Chanee cultivars
In order to compare the expression levels of ethylene biosynthetic genes during post-harvest ripening of Monthong and Chanee, the HMM profile of ACS and ACO (PF00155 from the Pfam protein database (http://pfam.xfam.org/)) was used as a query to search against our in-house transcriptome database and the Musang King genome (i.e., ACS (XM_022901720.1) and ACO (XM_022903266.1). To perform qRT-PCR, primers were designed as mentioned previously (see primers in Supplementary Table S3).
Transient expression of DzDof2.2 in N. benthamiana leaf
The full-length cDNA of DzDof2.2 was amplified from the cDNA using Phusion Hot Start II High-Fidelity DNA Polymerase (Thermo Fisher Scientific, USA) (primers are listed in Supplementary Table S4). Then, the PCR amplicon was introduced into pDONR207 vector using Gateway® BP Clonase® II (Thermo Fisher Scientific, USA). The putative DzDof2.2 from pDONR207-DzDof2.2 was inserted in to the pGWB2 expression vector using Gateway® LR Clonase® II (Thermo Fisher Scientific, USA) to produce pGWB2-DzDof2.2. The resulting pGWB2-DzDof2.2 as well as pGWB2-GFP (control) were individually inserted into the Agrobacterium tumefaciens strain GV3101 by electroporation. The positive A. tumefaciens colony containing each construct was grown in 25 mL of LB broth containing 25 mg L−1 gentamycin and 25 mg L−1 rifampicin at 30 °C with shaking at 250 rpm overnight. Cells were harvested by centrifugation at 4,000 × g for 10 min and resuspended in MM buffer (10 mM MES and 10 mM MgCl2, pH 5.6) to an optical density of 0.6 (OD600). For agroinfiltration, the agrobacterium solution containing either pGWB2-DzDof2.2 or pGWB2-GFP (control) was mixed with the agrobacterium solution harbouring the gene encoding the silencing inhibitor protein p19 at a ratio of 1:1. Thereafter, acetosyringone was added (100 mg L−1) and the mixed culture solution was incubated at room temperature for 3 h under dark conditions. Then, the solution was used to infiltrate the abaxial surface of three individual leaves per plant using a needleless 1-mL syringe. For each construct, three 4-week-old plants were used. After three days, the infiltrated leaves were collected, immediately frozen in liquid nitrogen and ground into a fine powder which was then used for RNA isolation and cDNA synthesis. The cDNA was further used as the template for qRT-PCR analysis. In order to design primers for N. benthamiana auxin biosynthetic genes, the Nicotiana tabacum genome (i.e., TAA1 (XM_016657015.1) and YUCCA3 (XM_016640518.1)) were used to blast against the genome sequence of N. benthamiana (SGN Nicotiana benthamiana ftp site; https://solgenomics.net/organism/Nicotiana_benthamiana/genome)51. Primers are listed in Supplementary Table S4. The 2000-bp upstream promoter regions of these genes were scanned for the Dof domain (AAAG/CTTT) using the online tool PLACE (http://www.dna.affrc.go.jp/)36.
Statistical analyses
All experiments were conducted using three biological replicates. Statistical comparisons of the mean values were conducted using one-way ANOVAs, followed by Duncan’s multiple range test or Student’s t-test at the 0.05 significance level using the statistical package for social sciences (SPSS) software, version 20. All data were represented as means ± standard deviations (SD).
Supplementary information
Acknowledgements
This research was supported by the Thailand Research Fund (RSA6080021) and Chulalongkorn University (GRU 6203023003-1) (to S.S.) and Ratchadapisek Somphot Fund for Postdoctoral Fellowship, Chulalongkorn University to (G.K.). We thank Tsuyoshi Nakagawa (Shimane University, Japan) for providing the Gateway® expression vector pGWB2, Sophien Kamoun (Sainsbury Laboratory, UK) for providing A. tumefaciens GV3101 carrying pJL3:p19 and Kittiya Tantisuwanichkul for assisting in ethephon and 1-MCP treatment.
Author Contributions
S.S. conceived the research. G.K. performed the experiments with the help of W.S. and P.P. L.S. analysed the RNA-Seq data. G.K. and S.S. designed the experiments, analysed the data, and wrote the manuscript. All authors have read and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-48601-7.
References
- 1.Yanagisawa S. Dof domain proteins: plant-specifc transcription factors associated with diverse phenomena unique to plants. Plant Cell Physiol. 2004;45:386–391. doi: 10.1093/pcp/pch055. [DOI] [PubMed] [Google Scholar]
- 2.Yanagisawa S. The Dof family of plant transcription factors. Trends Plant Sci. 2002;7:555–560. doi: 10.1016/S1360-1385(02)02362-2. [DOI] [PubMed] [Google Scholar]
- 3.Noguero M, Atif RM, Ochatt S, Tompson RD. The role of the DNA-binding One Zinc Finger (DOF) transcription factor family in plants. Plant Sci. 2013;209:32–45. doi: 10.1016/j.plantsci.2013.03.016. [DOI] [PubMed] [Google Scholar]
- 4.Venkatesh J, Park SW. Genome-wide analysis and expression profiling of DNA-binding with one zinc finger (Dof) transcription factor family in potato. Plant Physiol. Biochem. 2015;94:73–85. doi: 10.1016/j.plaphy.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 5.Cai X, et al. Genome-wide analysis of plant-specific Dof transcription factor family in tomato. J Integr. Plant Biol. 2013;55:552–566. doi: 10.1111/jipb.12043. [DOI] [PubMed] [Google Scholar]
- 6.Yanagisawa S, Izui K. Molecular cloning of two DNA-binding proteins of maize that are structurally different but interact with the same sequence motif. J. Biol. Chem. 1993;268:16028–16036. [PubMed] [Google Scholar]
- 7.Papi M, et al. Identification and disruption of an Arabidopsis zinc finger gene controlling seed germination. Genes Dev. 2000;14:28–33. [PMC free article] [PubMed] [Google Scholar]
- 8.Washio K. Functional dissections between GAMYB and Dof transcription factors suggest a role for protein-protein associations in the gibberellin-mediated expression of the RAmy1A gene in the rice aleurone. Plant Physiol. 2003;133:850–863. doi: 10.1104/pp.103.027334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wen C-l, et al. Identification and characterization of Dof transcription factors in the cucumber genome. Sci. Rep. 2016;6:23072. doi: 10.1038/srep23072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu Z, et al. Genome-wide identification and expression profile of dof transcription factor gene family in pepper (Capsicum annuum L.) Front. Plant Sci. 2016;7:574. doi: 10.3389/fpls.2016.00574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng B, et al. The banana fruit Dof transcription factor MaDof23 acts as a repressor and interacts with MaERF9 in regulating ripening-related genes. J. Exp. Bot. 2016;67:2263–2275. doi: 10.1093/jxb/erw032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang A, et al. Transcriptome analysis identifies a zinc finger protein regulating starch degradation in kiwifruit. Plant Physiol. 2018;178:850–863. doi: 10.1104/pp.18.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skirycz A, et al. DOF transcription factor AtDof1.1 (OBP2) is part of a regulatory network controlling glucosinolate biosynthesis in Arabidopsis. Plant J. 2006;47:10–24. doi: 10.1111/j.1365-313X.2006.02767.x. [DOI] [PubMed] [Google Scholar]
- 14.Ramirez-Parra E, et al. The transcription factor OBP4 controls root growth and promotes callus formation. New Phytol. 2017;213:1787–1801. doi: 10.1111/nph.14315. [DOI] [PubMed] [Google Scholar]
- 15.Pinsorn P, et al. Metabolic variation in the pulps of two durian cultivars: Unraveling the metabolites that contribute to the flavor. Food Chem. 2018;268:118–125. doi: 10.1016/j.foodchem.2018.06.066. [DOI] [PubMed] [Google Scholar]
- 16.Teh BT, et al. The draft genome of tropical fruit durian (Durio zibethinus) Nat. Genet. 2017;49:1633–1644. doi: 10.1038/ng.3972. [DOI] [PubMed] [Google Scholar]
- 17.Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–617. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 18.Hofmann, K. Protein classification & functional assignment in Trends guide to bioinformatics (eds Patterson, M. & Handel, M.) 18–21 (Elsevier Science 1998).
- 19.Chen R, Jeong S. Functional prediction: Identification of protein orthologs and paralogs. Protein Sci. 2000;9:2344–2353. doi: 10.1110/ps.9.12.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lijavetzky D, Carbonero P, Vicente-Carbajosa J. Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evol. Biol. 2003;3:17. doi: 10.1186/1471-2148-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma J, Li MY, Wang F, Tang J, Xiong AS. Genome-wide analysis of Dof family transcription factors and their responses to abiotic stresses in Chinese cabbage. BMC Genomics. 2015;16:33. doi: 10.1186/s12864-015-1242-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gupta S, et al. Insights into structural and functional diversity of Dof (DNA binding with one fnger) transcription factor. Planta. 2015;241:549–562. doi: 10.1007/s00425-014-2239-3. [DOI] [PubMed] [Google Scholar]
- 23.Sisler EC, Blankenship SM, Guest M. Competition of cyclooctenes and cyclooctadienes for ethylene binding and activity in plants. Plant Growth Regul. 1990;9:157–164. doi: 10.1007/BF00027443. [DOI] [Google Scholar]
- 24.Hall AE, Findell JL, Schaller GE, Sisler EC, Bleecker AB. Ethylene perception by the ERS1 protein in Arabidopsis. Plant Physiol. 2000;123:1449–1457. doi: 10.1104/pp.123.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the Raf family of protein kinases. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-B. [DOI] [PubMed] [Google Scholar]
- 26.Tatsuki M, Endo A. Analyses of expression patterns of ethylene receptor genes in apple (Malus domestica Borkh.) fruits treated with or without 1-methylcyclopropene (1-MCP) J. Japan. Soc. Hort. Sci. 2006;75:481–487. doi: 10.2503/jjshs.75.481. [DOI] [Google Scholar]
- 27.Costa F, et al. Use of homologous and heterologous gene expression profiling tools to characterize transcription dynamics during apple fruit maturation and ripening. BMC Plant Biol. 2010;10:229. doi: 10.1186/1471-2229-10-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.El-Sharkawy I, et al. Isolation and characterization of four ethylene signal transduction elements in plums (Prunus salicina L.) J. Exp. Bot. 2007;58:3631–3643. doi: 10.1093/jxb/erm213. [DOI] [PubMed] [Google Scholar]
- 29.Tatsuki M, Endo A, Ohkawa H. Influence of time from harvest to 1-MCP treatment on apple fruit quality and expression of genes for ethylene biosynthesis enzymes and ethylene receptors. Postharvest Biol. Technol. 2007;43:28–35. doi: 10.1016/j.postharvbio.2006.08.010. [DOI] [Google Scholar]
- 30.Tatsuki M, Hayama H, Nakamura Y. Apple ethylene receptor protein concentrations are affected by ethylene, and differ in cultivars that have different storage life. Planta. 2009;230:407–417. doi: 10.1007/s00425-009-0953-z. [DOI] [PubMed] [Google Scholar]
- 31.Yang X, Song J, Campbell-Palmer L, Fillmore S, Zhang Z. Effect of ethylene and 1-MCP on expression of genes involved in ethylene biosynthesis and perception during ripening of apple fruit. Postharvest Biol. Technol. 2013;78:55–66. doi: 10.1016/j.postharvbio.2012.11.012. [DOI] [Google Scholar]
- 32.Wang A, Tan D, Takahashi A, Li TZ, Harada T. MdERFs, two ethylene-response factors involved in apple fruit ripening. J. Exp. Bot. 2007;58:3743–3748. doi: 10.1093/jxb/erm224. [DOI] [PubMed] [Google Scholar]
- 33.Stepanova AN, et al. The Arabidopsis YUCCA1 Flavin Monooxygenase Functions in the Indole-3-Pyruvic Acid Branch of Auxin Biosynthesis. Plant Cell. 2011;23:3961–3973. doi: 10.1105/tpc.111.088047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cook SD, et al. Auxin biosynthesis: Are the indole-3-acetic acid and phenylacetic acid biosynthesis pathways mirror images? Plant Physiol. 2016;171:1230–1241. doi: 10.1104/pp.16.00454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trainotti L, Tadiello A, Casadoro G. The involvement of auxin in the ripening of climacteric fruits comes of age: the hormone plays a role of its own and has an intense interplay with ethylene in ripening peaches. J. Exp. Bot. 2007;58:3299–3308. doi: 10.1093/jxb/erm178. [DOI] [PubMed] [Google Scholar]
- 36.Liu K, et al. Genome-wide analysis and characterization of Aux/IAA family genes related to fruit ripening in papaya (Carica papaya L.) BMC Genomics. 2017;18:351. doi: 10.1186/s12864-017-3722-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abel S, Theologis A. Early genes and auxin action. Plant Physiol. 1996;111:9–17. doi: 10.1104/pp.111.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen YF, Etheridge N, Schaller GE. Ethylene signal transduction. Ann. Bot. 2005;95:901–915. doi: 10.1093/aob/mci100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bleecker AB, Kende H. Ethylene: a gaseous signal molecule in plants. Annu. Rev. Cell Dev. Biol. 2000;16:1–18. doi: 10.1146/annurev.cellbio.16.1.1. [DOI] [PubMed] [Google Scholar]
- 40.Alexander L, Grierson D. Ethylene biosynthesis and action in tomato: a model for climacteric fruit ripening. J. Exp. Bot. 2002;53:2039–2055. doi: 10.1093/jxb/erf072. [DOI] [PubMed] [Google Scholar]
- 41.Ságio SA, et al. Identification and expression analysis of ethylene biosynthesis and signaling genes provides insights into the early and late coffee cultivars ripening pathway. Planta. 2014;239:951–63. doi: 10.1007/s00425-014-2026-1. [DOI] [PubMed] [Google Scholar]
- 42.Gasteiger, E. Protein identifcation and Analysis Tools on the ExPASy Server in Proteomics Protocols Handbook (ed. Walker, John M.) pp. 571–607 (Humana, 2005).
- 43.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Timothy L, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:202–208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haas BJ, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chong J, et al. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018;46:486–494. doi: 10.1093/nar/gky310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;4:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 48.Tulpan D, Leger S. The plant orthology browser: An orthology and gene order visualizer for plant comparative genomics. Plant Genome. 2017;10:1. doi: 10.3835/plantgenome2016.08.0078. [DOI] [PubMed] [Google Scholar]
- 49.Sinha BK, Basu PS. Indole-3-acetic acid metabolism in root nodules of Pongamia pinnata (L.) Biochem Physiol Pflanz. 1981;176:218–227. doi: 10.1016/S0015-3796(81)80074-1. [DOI] [Google Scholar]
- 50.Gordon SA, Weber RP. Colorimetric estimation of indoleacetic acid. Plant Physiol. 1951;26:192–195. doi: 10.1104/pp.26.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bombarely A, et al. A Draft genome sequence of Nicotiana benthamiana to enhance molecular plant-microbe biology research. Mol Plant Microbe Interact. 2012;25:1523–1530. doi: 10.1094/MPMI-06-12-0148-TA. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.