Abstract
Our recent study has demonstrated that increased connectivity in the cerebello-thalamo-cortical (CTC) circuitry is a state-independent neural trait that can potentially predict the onset of psychosis. One possible cause of such “trait” abnormality would be genetic predisposition. Here, we tested this hypothesis using multi-paradigm functional magnetic resonance imaging (fMRI) data from two independent twin cohorts. In a sample of 85 monozygotic (MZ) and 52 dizygotic (DZ) healthy twin pairs acquired from the Human Connectome Project, we showed that the connectivity pattern of the identified CTC circuitry was more similar in the MZ twins (r = 0.54) compared with that in the DZ twins (r = 0.22). The structural equation modeling analysis revealed a heritability estimate of 0.52 for the CTC connectivity, suggesting a moderately strong genetic effect. Moreover, using an independent schizophrenia cotwin sample (10 discordant MZ cotwins, 30 discordant DZ cotwins, and 32 control cotwins), we observed a significant linear relationship between genetic distance to schizophrenia and the connectivity strength in the CTC circuitry (i.e., schizophrenia MZ cotwins > schizophrenia DZ cotwins > control twins, P = 0.045). The present data provide converging evidence that increased connectivity in the CTC circuitry is likely to be a heritable trait that is associated with the genetic risk of schizophrenia.
Subject terms: Schizophrenia, Clinical genetics
Introduction
Psychotic disorders such as schizophrenia possess a strong genetic basis. Empirical data from research on twins and relatives have revealed an estimated heritability of ~80% and a 10-fold increase of risk in first-degree relatives of patients1,2, making the study of genetic mechanisms critically important to understand such complex disorders. At the neural level, the genetic risk of schizophrenia embodies the dysregulation of behavior-specific neural circuits, which in turn increases the risk of developing corresponding behavioral defects3. Such neurobiological changes (so-called “intermediate phenotypes”4 or “endophenotypes”5) have offered a valuable avenue to understand the pathogenesis of psychosis and to probe for effective prediction and intervention strategies.
Our recent study has demonstrated that increased connectivity in the cerebello-thalamo-cortical (CTC) circuitry is a state-independent “trait” alteration that may potentially predict the onset of psychosis and distinguish the patients from the healthy population6. Such alteration can be reliably detected across multiple functional magnetic resonance imaging (fMRI) paradigms in individuals both at clinical high risk (CHR) and with chronic illness, suggesting a “trait” brain functional abnormality that is robustly present across the entire course of psychotic disorders. These findings highly suggest that increased connectivity in the CTC circuitry may be caused by genetic predisposition to psychosis.
In this study, we tested this hypothesis using two independent twin data sets with multi-paradigm fMRI data. The first data set was part of the Human Connectome Project (HCP, ref. 7), including a total of 85 monozygotic (MZ) healthy twin pairs and 52 dizygotic (DZ) healthy twin pairs. Using this sample, we examined the heritability of the connectivity of the CTC circuitry using structural equation modeling (SEM). The second data set was drawn from the Swedish Schizophrenia Twin Study (STAR, ref. 8), which included 40 schizophrenia discordant cotwins (10 MZ, 30 DZ) and 32 healthy control twins. We specifically tested the relationship between genetic distance to schizophrenia and the CTC connectivity in this sample. We expected that the CTC connectivity is heritable in healthy twin pairs and is associated with degree of genetic risk of schizophrenia in schizophrenia cotwins (schizophrenia MZ cotwins > schizophrenia DZ cotwins > control twins).
Methods
Subjects
Two independent twin data sets were used in this study: the HCP twin sample7 and the STAR sample8. The HCP data included a total of 137 healthy twin pairs that have completed all eight fMRI paradigms used in the project (resting state, gambling, emotional processing, motor, working memory, relational processing, language processing, and social cognition) and have genotyping data available. Among these twin pairs, 85 were MZ (age 29.32 ± 3.39 years, 70 males) and 52 were DZ (age 28.50 ± 3.50 years, 52 males). The STAR sample comprised of 40 schizophrenia discordant cotwins (10 MZ, age 43.79 ± 11.94 years, 7 males; 30 DZ, age 51.11 ± 10.07 years, 16 males) and 32 healthy control twins (46.37 ± 6.92 years, 14 males), each assessed with a battery of two fMRI paradigms (working memory, emotional processing). The zygosity in both data sets was determined by genotyping data from subjects’ blood samples. The STAR participants were recruited through the Swedish Twin Registry and were evaluated by psychiatrists using the Structured Clinical Interview for DSM-IV9. The exclusion criteria for control twins included prior history of neurological or psychiatric disorders, head injury, substance dependence in the last 6 months, mental retardation, and inability to read or comprehend Swedish. All schizophrenia discordant cotwins were free of any psychotic disorders according to the DSM-IV criteria. The participants provided written informed consents and the research protocol was approved by the Regional Ethical Review Board in Stockholm. For sample details, see Tables 1 and 2.
Table 1.
Demographic data for the HCP sample
| Healthy MZ (N = 170, or 85 pairs) | Healthy DZ (N = 104, or 52 pairs) | P value | |
|---|---|---|---|
| Age (year) | 29.32 ± 3.39 | 28.50 ± 3.50 | 0.06 |
| Sex (M/F) | 70/100 | 52/52 | 0.15 |
| Head motion (FD, mm) | 0.17 ± 0.05 | 0.16 ± 0.05 | 0.10 |
Table 2.
Demographic and clinical data for the STAR sample
| Schizophrenia MZ cotwins (N = 10) | Schizophrenia DZ cotwins (N = 30) | Control twins (N = 32) | P value | |
|---|---|---|---|---|
| Age (year) | 43.79 ± 11.94 | 51.11 ± 10.07 | 46.37 ± 6.92 | 0.04 |
| Sex (M/F) | 7/3 | 16/14 | 14/18 | 0.34 |
| Head motion (FD, mm) | 0.13 ± 0.06 | 0.13 ± 0.07 | 0.10 ± 0.05 | 0.06 |
| DSM Diagnosis | 4 major depressive disorder, 1 acute stress disorder, 2 healthy, 3 unavailable | 6 major depressive disorder, 2 drug or alcohol dependence, 11 healthy, 11 unavailable | – | – |
Note that all schizophrenia cotwins are free of any psychotic disorders
Imaging paradigms
A total of eight fMRI paradigms were employed in the HCP project. For details on these paradigms see previously published work10,11. Notably, the resting-state paradigm was scanned with four sessions spanned across two consecutive days, and the other paradigms were scanned with two sessions on a single day. To balance the amount of data across paradigms and to avoid overrepresentation of resting state in the following cross-paradigm computations, we only used the resting-state data from the two sessions acquired on the first day. The STAR data set used two fMRI paradigms that have been described previously12,13. In brief, the spatial working memory task used an event-related design where subjects were asked to encode the location of three target circles and to indicate whether the ensuing probe circle appeared at the same location as one of the target circles. During the block-designed emotional face processing task, subjects were shown a series of emotional faces and geometric shapes and were instructed to match either the target faces or the target shapes.
Data acquisition
The HCP data were collected from a customized 3T Siemens Connectome scanner equipped with a 32-channel head coil. BOLD images were acquired using a multiband GRE-EPI sequence with the following parameters: TR = 720 ms, TE = 33.1 ms, FA = 52°, FOV = 208 × 180 mm2, 2 mm slice thickness, 72 slices, multiband factor = 8. Of note, unlike the commonly applied phase-encoding directions along the “anterior-posterior” axis, the HCP fMRI data were acquired using left-right and right-left phase-encoding directions to accelerate scan time and to minimize image distortion11. Each paradigm was scanned with two sessions, with each direction per session. In addition, T1-weighted and T2-weighted images were acquired using 3D-MPRAGE and 3D-T2-SPACE sequences with 0.7 mm voxel size and 224 × 224 mm2 FOV. The T1 images used a TR of 2400 ms and TE of 2.14 ms, and the T2 images used a TR of 3200 ms and TE of 565 ms.
The STAR data were acquired from a 1.5 T GE scanner equipped with a standard 8-channel head coil. GRE-EPI sequence was used for functional imaging: TR = 2500 ms, TE = 40 ms, FA = 90°, 3.5 mm slice thickness, voxel size 3.44 × 3.44 × 4.5 mm3. T2-weighted images were scanned using a spin-echo sequence with 4000 ms TR, 82 ms TE, and 4 mm slice thickness.
Data preprocessing
The HCP data were downloaded after the completion of imaging preprocessing (https://db.humanconnectome.org). The preprocessing procedure followed the standard HCP preprocessing pipeline including five major steps: PreFreeSurfer, FreeSurfer, PostFreeSurfer, fMRIVolume, and fMRISurface14. In brief, images were corrected for gradient nonlinearity induced distortion, head motion, and phase-encoding related distortion, registered to individuals’ T1-weighted images, and normalized to the MNI space. The STAR data were preprocessed using the standard pipeline implemented in the Statistical Parametric Mapping software (SPM12, https://www.fil.ion.ucl.ac.uk/spm/software/spm12/), including slice timing, head motion realignment, functional-structural image coregistration, MNI space normalization, and spatial smoothing. All preprocessed images were further scrutinized for head motion using the measure of frame-wise displacement (FD15) to ensure data quality. All subjects in both data sets had a mean FD < 0.5 mm.
Cross-paradigm connectivity and extraction of CTC network
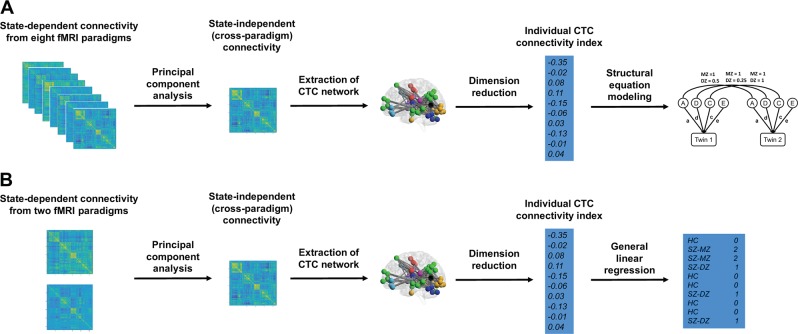
The overall processing pipeline of this study is presented in Fig. 1. The cross-paradigm connectivity (CPC) computation and extraction of CTC network followed our prior work6. Specifically, the mean time series for each of the 270 nodes in the expanded Power brain atlas6,13,16 were extracted from the preprocessed images. The extracted time series were further corrected for task-related coactivations (for task data), white matter and cerebrospinal fluid signals, the 24 head motion parameters (i.e., the six rigid-body parameters generated from the realignment step, their first derivatives, and the squares of these 12 parameters17,18), and the FD. The corrected time series were then temporally filtered (task data: 0.008 Hz high pass, rest data: 0.008–0.1 Hz band pass) to account for scanner and physiological noise. Subsequently, a 270 × 270 pairwise connectivity matrix was generated using Pearson correlation for each subject during each paradigm and scan session.
Fig. 1.
Data processing pipelines for two data sets (a: HCP sample; b: STAR sample)
Following prior suggestions11, we averaged the derived connectivity matrices across the two sessions for each paradigm to boost signals, generating eight paradigm-specific connectivity matrices for each subject in the HCP data and two paradigm-specific connectivity matrices for each subject in the STAR data. The matrices for each subject were vectorized, concatenated across paradigms, and then decomposed into a set of principal components (PCs) using singular value decomposition (SVD). The first PCs generated from the analysis represented the shared CPC and were thus extracted. From these CPC matrices we further extracted the identified CTC network based on our prior work6.
Heritability estimation of CTC connectivity
The HCP data were employed to quantify heritability of the CTC connectivity. We first calculated the similarities of the MZ and DZ twin pairs using intra-class correlations. A larger correlation coefficient between MZ pairs compared with that between DZ pairs indicates a potential genetic effect. Since the MZ correlation was larger than twice of the DZ correlation in our sample (see Results section), we estimated an ADE model using structural equation modeling (SEM). This is due to the fact that dominant genetic effects (D) would decrease the DZ correlation relative to the MZ correlation19,20. Specifically, using the SEM toolbox OpenMx implemented in R (https://openmx.ssri.psu.edu/), we decomposed the total variance of phenotype into additive genetic effects (A), dominant genetic effects (D), and unique environmental effects (E). In general, the MZ twins share 100% of additive and dominant genetic effects, while the DZ twins share 50% and 25% of additive and dominant genetic effects, respectively19,20. Based on this information, we estimated the squares of each path coefficients (a2, d2, e2) that reflect the proportion of phenotypic variation attributable to the A, D, E effects, respectively. This was done by iteratively maximizing the log-likelihood between observed and predicted covariance matrices. The goodness-of-fit of the estimated model was quantified by −2 times the log-likelihood (−2LL, ref. 19) as well as the Akaike Information Criterion (AIC, ref. 21), and was compared to the saturated model in which all covariances were treated as free parameters using Chi-squared test. A non-significant P value ( > 0.05) indicates a good model fit. Moreover, the full ADE model was further compared with nested AE, DE, and E models, and the most parsimonious model was determined by the one with the smallest −2LL and AIC values. Heritability (h2) was subsequently quantified as the proportion of phenotypic variance attributable to genetic effects in the most parsimonious model.
Association of CTC connectivity with schizophrenia genetic risk
We examined the association between CTC connectivity and genetic risk of schizophrenia in the STAR data. Specifically, a linear regression model was employed where CTC connectivity was included as dependent variable and genetic proximity to schizophrenia (i.e., schizophrenia MZ cotwins > schizophrenia DZ cotwins > control twins), age, sex, and FD were included as regressors. A significant regression coefficient for genetic proximity indicates that CTC connectivity is associated with schizophrenia genetic risk. Since the size of STAR sample was relatively small, we further included all twins in the HCP sample as controls in the regression model. To control for discrepancy between the two data sets, cohort was included as an additional regressor in the model for the combined sample.
Results
Heritability of CTC connectivity
There were no significant differences in the mean and variance of CTC connectivity between MZ and DZ twins in the HCP sample (P = 0.12 and 0.46, respectively). The intra-class correlation coefficients for CTC connectivity among MZ and DZ twin pairs in the HCP sample were 0.54 and 0.22, respectively, suggesting a genetic effect (Fig. 2). Since the correlation between the MZ twins was larger than twice of that between the DZ twins, we evaluated an ADE model using SEM. The results revealed a heritability of 0.52 (95% CI: 0.36–0.64). Specifically, 52% of the variance in CTC connectivity was attributable to additive genetic effects (a2 = 0.52), and 48% was attributable to unique environmental effects (e2 = 0.48). In contrast, dominant genetic effects were estimated to be negligible (d2 < 0.001). There was no significant difference in comparison between the ADE model and the saturated model (−2LLADE = 1388.18, AICADE = 848.18, −2LLSaturated = 1379.74, AICSaturated = 851.74, P = 0.2), indicating a good model fit for the present data. Moreover, the further comparison of the full ADE model with the nested models showed the smallest −2LL and AIC for the AE model (−2LLAE = 1388.18, AICAE = 846.18, P = 1), suggesting that the AE model is the most parsimonious and should be accepted as the model that best fits the data.
Fig. 2.
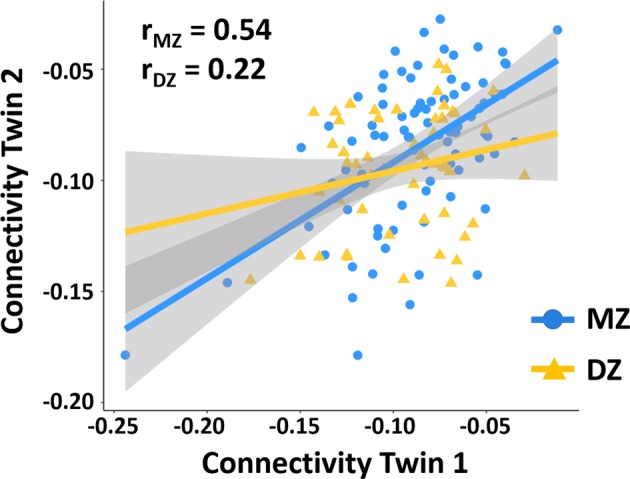
Intra-class correlations of CTC connectivity among the MZ and DZ twins in the HCP data. The correlation coefficient was much higher for the MZ twins compared to that for the DZ twins, suggesting a potential genetic effect
Association of CTC connectivity with schizophrenia genetic risk
The Levene test showed an equal variance of CTC connectivity between the three examined groups (P = 0.84). The regression analysis revealed a significant linear relationship between genetic proximity to schizophrenia and CTC connectivity in the STAR sample (P = 0.045, Fig. 3), suggesting that CTC connectivity is related to schizophrenia genetic risk. In particular, schizophrenia MZ cotwins showed the highest connectivity, followed by schizophrenia DZ cotwins, while control twins had the lowest connectivity, which is consistent with our prior findings that patients with schizophrenia are associated with higher connectivity in the CTC circuitry6. Moreover, the inclusion of HCP data as control subjects demonstrated even stronger effect (P = 0.01), which further supports the idea that CTC hyperconnectivity is likely to be a heritable trait that is associated with genetic predisposition to schizophrenia.
Fig. 3.
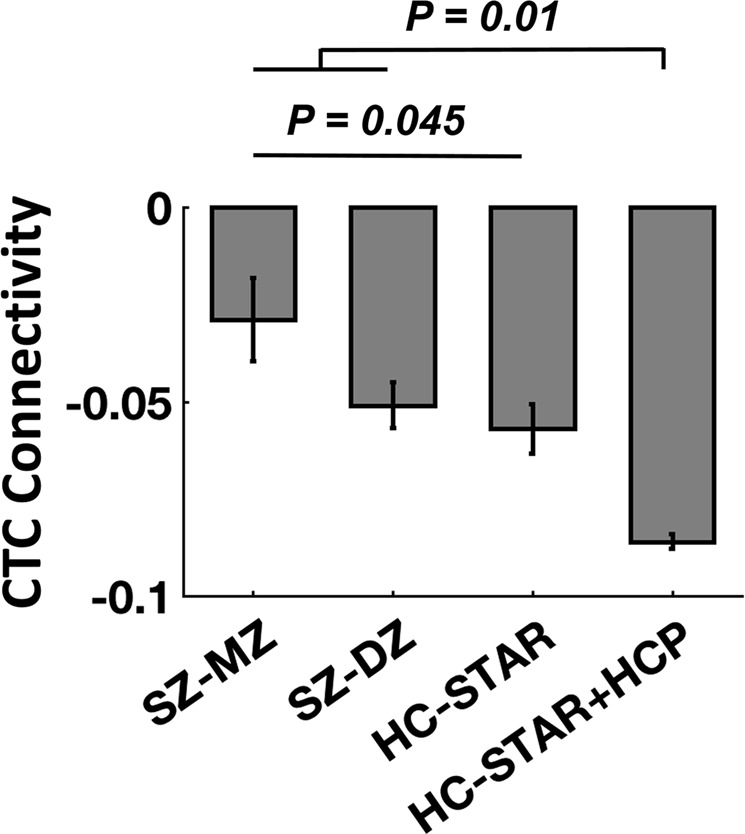
Linear regression model showed significant linear relationship between CTC connectivity and genetic proximity to schizophrenia (i.e., schizophrenia MZ cotwins > schizophrenia DZ cotwins > healthy controls), suggesting that CTC hyperconnectivity is associated with genetic risk of schizophrenia. Error bars stand for standard errors
Supplementary analysis on head motion
Previous study has reported that head motion is moderately heritable during resting-state fMRI scans22. As a result, it is possible that our results may be influenced by this confounding factor. To test this hypothesis, we further examined the intra-class correlation coefficients for head motion FD measures across the MZ and DZ twins, respectively, in the HCP sample. We found that the correlation coefficients for MZ and DZ twins in the current sample were very close (rMZ = 0.38, rDZ = 0.34), suggesting a very trivial genetic effect. This supplementary analysis indicates that the heritability in CTC connectivity is unlikely to be driven by the heritability in head motion.
Supplementary analysis on ACE model
In this study, an ADE model was chosen to estimate heritability since the correlation of CTC connectivity between the MZ twins was larger than twice of that between the DZ twins. In a supplementary analysis we also estimated heritability using an ACE model. The ACE model showed a heritability of 0.38 (95% CI: 0.17–0.56). In particular, similar to the results from an ADE model, 38% of total variance was attributable to additive genetic effects (a2 = 0.38), 62% was attributable to unique environmental effects (e2 = 0.62), and common environmental effects were negligible (c2 < 0.001). This supplementary analysis again suggests that the variations in CTC connectivity are mainly caused by additive genetic effects and unique environmental effects.
Supplementary analysis on statistical power
Because the STAR sample in this study was relatively small, we performed a power analysis to determine whether the study was adequately powered to detect the observed effect sizes. The power to reject the null hypothesis of no group difference was 0.7 for the comparison between SZ-MZ cotwins and SZ-DZ cotwins (α = 0.05, one-tailed test). The power increased to 0.9 when comparing SZ-MZ cotwins and HC twins (α = 0.05, one-tailed test), suggesting that the present sample, although small, is likely to have been adequately powered.
Discussion
This study tested the hypothesis that the CTC hyperconnectivity is related to genetic predisposition to schizophrenia using two independent twin data sets. Overall, we observed a relatively high heritability of CTC connectivity in the HCP twin sample and a significant association between CTC hyperconnectivity and genetic proximity to schizophrenia. These findings provide converging evidence that CTC hyperconnectivity is likely to be a heritable trait that is related to the genetic risk of schizophrenia.
Given that increased connectivity in the CTC circuitry is a “trait” abnormality that can be reliably detected across different behavioral domains in psychosis, the strong relationship between this change and genetic risk of schizophrenia is within expectation. Notably, both the structure and the function of two key brain regions in this circuitry, the thalamus and cerebellum, have previously been found to be heritable23–29 and potentially related to schizophrenia genetic risk30,31. Several cortical functional systems involved in this circuitry, including the default-mode network, fronto-parietal network, and attentional network, were also found to be heritable in the healthy population32–34. Here, our findings parallel with these previous findings and further demonstrate that a widely distributed network covering cerebellum, thalamus and cerebral cortex is heritable and regulated by genetic risk of schizophrenia.
Since CTC hyperconnectivity may reflect a downstream phenomenon of N-methyl-D-aspartate receptor (NMDAR) deficits35–37, this alteration is presumably related to genes that are associated with glutamate and/or gamma-Aminobutyric acid (GABA) signaling. Large-scale genome-wide association studies (GWAS) have identified a set of common genetic variants in glutamatergic genes in patients with schizophrenia, including, for instance, GRM3, GRIN2A, GRIA1, SRR, CLCN3, among others38,39. Gene expression and brain imaging analyses suggested a converging effect of these genes on brain structure and function, especially in the prefrontal cortex40–43, hippocampus43, and thalamus43. Moreover, pathway analysis also revealed a significant association of genetic variants in NMDAR complex and glutamate metabolism pathways with the pathogenesis of schizophrenia44,45. In addition to common variants, rare genetic mutations such as copy number variation have also been found to be highly enriched for genes involved in the glutamatergic and GABAergic neurotransmission46–48. These lines of evidence suggest that neurotransmission-linked schizophrenia genetic factors may affect excitation–inhibition balance in the human brain that further sets an over-excitatory tone for the overall brain connectivity. Such altered brain connectivity, especially in a CTC circuitry, is therefore presented as a “trait” abnormality that can be reliably detected across various states and phases of schizophrenia.
Other genes that may relate to the connectivity in the CTC circuitry are those associated with dopamine signaling, given the essential role of this circuitry in error processing49,50. Notably, dopamine system plays a key role in error processing in the brain. It is assumed that the prediction errors, which reflect the mismatch between the priors and outcomes, are encoded by the phasic dopamine neuron firing in the ventral-tegmental area (VTA) of the midbrain51,52. The activity of the VTA dopamine neurons alters the dopamine input to the mesolimbic and mesocortical pathways, which in turn, modulates a variety of human behaviors such as cognition, reward processing, reinforcement learning, and social activity53–55. Previous studies have shown that dopamine-related genes such as DRD2, DRD4, COMT, and DAT influence error-related negativity56–58 and increase the risk for schizophrenia38,59. As a result, genetic variants involved in dopamine signaling may regulate error processing associated behaviors via the regulation of the CTC circuitry.
One major limitation of our study is the relatively small sample size, especially for schizophrenia cotwins. Although we showed that this small sample is still likely to have reasonable statistical power, further fMRI studies with larger twin samples are encouraged to replicate these findings. Second, different fMRI paradigms were used in two studied samples. While including different paradigms would strengthen the argument that CTC hyperconnectivity is a heritable “trait” rather than a “state-dependent” change, such differences may to certain degree influence the outcome individual “trait” networks between samples. Third, while we only included schizophrenia discordant cotwins without psychotic disorders in this study, some of the cotwins were diagnosed with other types of mental disorders (in particular major depressive disorder, see Table 2). As a result, our findings may to certain degree be affected by this confounding factor. Last but not least, while we provided direct evidence that CTC hyperconnectivity is related to the genetic risk of schizophrenia and discussed potential associations between such phenotype and neurotransmitter signaling, how common variants in glutamate and dopamine-related genes would affect the connectivity in the CTC circuitry is still an open question. Future imaging genetic studies are encouraged to investigate such relationships.
In sum, using multi-paradigm fMRI data from two independent twin cohorts, our study shows the first evidence that increased connectivity in the CTC circuitry is likely to be a heritable trait related to the genetic risk of schizophrenia. These findings echo our previous work on CTC hyperconnectivity as a biological trait for psychosis prediction and overall highlight the critical role of CTC circuitry in the pathogenesis of schizophrenia.
Acknowledgements
This work was supported by the Brain and Behavior Research Foundation NARSAD Young Investigator Grant (No. 27068) to Dr. Cao, by the National Institute of Health (NIH) grant U01 MH081902 to Dr. Cannon, by gifts from the Staglin Music Festival for Mental Health and International Mental Health Research Organization to Dr. Cannon, and by the ALF (regional agreement on medical training and clinical research between Stockholm County Council and the Karolinska Institutet) grants 20100305 and 20090183 to Dr. Hultman. We thank the study participants, research staff members, and the Swedish Board of Health and Welfare for study support.
Conflict of interest
Dr. Cannon has served as a consultant for Boehringer-Ingelheim Pharmaceuticals and Lundbeck A/S. The other authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch. Gen. Psychiatry. 2003;60:1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- 2.Gejman PV, Sanders AR, Kendler KS. Genetics of schizophrenia: new findings and challenges. Annu Rev. Genom. Hum. Genet. 2011;12:121–144. doi: 10.1146/annurev-genom-082410-101459. [DOI] [PubMed] [Google Scholar]
- 3.Cao H, Dixson L, Meyer-Lindenberg A, Tost H. Functional connectivity measures as schizophrenia intermediate phenotypes: advances, limitations, and future directions. Curr. Opin. Neurobiol. 2016;36:7–14. doi: 10.1016/j.conb.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 4.Meyer-Lindenberg A, Weinberger DR. Intermediate phenotypes and genetic mechanisms of psychiatric disorders. Nat. Rev. Neurosci. 2006;7:818–827. doi: 10.1038/nrn1993. [DOI] [PubMed] [Google Scholar]
- 5.Cannon TD, Keller MC. Endophenotypes in the genetic analyses of mental disorders. Annu Rev. Clin. Psychol. 2006;2:267–290. doi: 10.1146/annurev.clinpsy.2.022305.095232. [DOI] [PubMed] [Google Scholar]
- 6.Cao H, et al. Cerebello-thalamo-cortical hyperconnectivity as a state-independent functional neural signature for psychosis prediction and characterization. Nat. Commun. 2018;9:3836. doi: 10.1038/s41467-018-06350-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Essen DC, et al. The WU-Minn Human Connectome Project: an overview. Neuroimage. 2013;80:62–79. doi: 10.1016/j.neuroimage.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johansson Viktoria, Hultman Christina M., Kizling Isabelle, Martinsson Lennart, Borg Jacqueline, Hedman Anna, Cannon Tyrone D. The schizophrenia and bipolar twin study in Sweden (STAR) Schizophrenia Research. 2019;204:183–192. doi: 10.1016/j.schres.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.First, M. B., Spitzer R. L., Gibbon, M., & Williams, J. B. W. Structured Clinical Interview for DSM-IV-TR Axis I Disorders, Research Version, Patient edition (SCID-I/P) (Biometrics Research, New York State Psychiatric Institute, New York, 2002).
- 10.Barch DM, et al. Function in the human connectome: task-fMRI and individual differences in behavior. Neuroimage. 2013;80:169–189. doi: 10.1016/j.neuroimage.2013.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith SM, et al. Resting-state fMRI in the Human Connectome Project. Neuroimage. 2013;80:144–168. doi: 10.1016/j.neuroimage.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cannon TD, et al. Dorsolateral prefrontal cortex activity during maintenance and manipulation of information in working memory in patients with schizophrenia. Arch. Gen. Psychiatry. 2005;62:1071–1080. doi: 10.1001/archpsyc.62.10.1071. [DOI] [PubMed] [Google Scholar]
- 13.Cao Hengyi, McEwen Sarah C, Forsyth Jennifer K, Gee Dylan G, Bearden Carrie E, Addington Jean, Goodyear Bradley, Cadenhead Kristin S, Mirzakhanian Heline, Cornblatt Barbara A, Carrión Ricardo E, Mathalon Daniel H, McGlashan Thomas H, Perkins Diana O, Belger Aysenil, Seidman Larry J, Thermenos Heidi, Tsuang Ming T, van Erp Theo G M, Walker Elaine F, Hamann Stephan, Anticevic Alan, Woods Scott W, Cannon Tyrone D. Toward Leveraging Human Connectomic Data in Large Consortia: Generalizability of fMRI-Based Brain Graphs Across Sites, Sessions, and Paradigms. Cerebral Cortex. 2018;29(3):1263–1279. doi: 10.1093/cercor/bhy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glasser MF, et al. The minimal preprocessing pipelines for the Human Connectome Project. Neuroimage. 2013;80:105–124. doi: 10.1016/j.neuroimage.2013.04.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage. 2012;59:2142–2154. doi: 10.1016/j.neuroimage.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Power JD, et al. Functional network organization of the human brain. Neuron. 2011;72:665–678. doi: 10.1016/j.neuron.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Power JD, et al. Methods to detect, characterize, and remove motion artifact in resting state fMRI. Neuroimage. 2014;84:320–341. doi: 10.1016/j.neuroimage.2013.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Satterthwaite TD, et al. An improved framework for confound regression and filtering for control of motion artifact in the preprocessing of resting-state functional connectivity data. Neuroimage. 2013;64:240–256. doi: 10.1016/j.neuroimage.2012.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grasby KL, Verweij KJH, Mosing MA, Zietsch BP, Medland SE. Estimating heritability from twin studies. Methods Mol. Biol. 2017;1666:171–194. doi: 10.1007/978-1-4939-7274-6_9. [DOI] [PubMed] [Google Scholar]
- 20.Rijsdijk FV, Sham PC. Analytic approaches to twin data using structural equation models. Brief. Bioinform. 2002;3:119–133. doi: 10.1093/bib/3.2.119. [DOI] [PubMed] [Google Scholar]
- 21.Akaike H. Akaike’s information criterion. International encyclopedia of statistical science. Springer 2011, p 25.
- 22.Couvy-Duchesne B, et al. Heritability of head motion during resting state functional MRI in 462 healthy twins. Neuroimage. 2014;102:424–434. doi: 10.1016/j.neuroimage.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Renteria ME, et al. Genetic architecture of subcortical brain regions: common and region-specific genetic contributions. Genes Brain Behav. 2014;13:821–830. doi: 10.1111/gbb.12177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roshchupkin GV, et al. Heritability of the shape of subcortical brain structures in the general population. Nat. Commun. 2016;7:13738. doi: 10.1038/ncomms13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macare C, Meindl T, Nenadic I, Rujescu D, Ettinger U. Preliminary findings on the heritability of the neural correlates of response inhibition. Biol. Psychol. 2014;103:19–23. doi: 10.1016/j.biopsycho.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 26.Guadalupe T, Mathias SR. vanErp TGM, Whelan CD, Zwiers MP, Abe Y et al. Human subcortical brain asymmetries in 15,847 people worldwide reveal effects of age and sex. Brain Imaging Behav. 2017;11:1497–1514. doi: 10.1007/s11682-016-9629-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brouwer RM, et al. Genetic influences on individual differences in longitudinal changes in global and subcortical brain volumes: results of the ENIGMA plasticity working group. Hum. Brain Mapp. 2017;38:4444–4458. doi: 10.1002/hbm.23672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blokland GA, et al. Genetic effects on the cerebellar role in working memory: same brain, different genes? Neuroimage. 2014;86:392–403. doi: 10.1016/j.neuroimage.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Z, et al. Neural mechanism and heritability of complex motor sequence and audiovisual integration: a healthy twin study. Hum. Brain Mapp. 2018;39:1438–1448. doi: 10.1002/hbm.23935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Collin G, et al. Impaired cerebellar functional connectivity in schizophrenia patients and their healthy siblings. Front Psychiatry. 2011;2:73. doi: 10.3389/fpsyt.2011.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cooper D, Barker V, Radua J, Fusar-Poli P, Lawrie SM. Multimodal voxel-based meta-analysis of structural and functional magnetic resonance imaging studies in those at elevated genetic risk of developing schizophrenia. Psychiatry Res. 2014;221:69–77. doi: 10.1016/j.pscychresns.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 32.Glahn DC, et al. Genetic control over the resting brain. Proc. Natl Acad. Sci. USA. 2010;107:1223–1228. doi: 10.1073/pnas.0909969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miranda-Dominguez O, et al. Heritability of the human connectome: a connectotyping study. Netw. Neurosci. 2018;2:175–199. doi: 10.1162/netn_a_00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan J, Wu Y, Fossella JA, Posner MI. Assessing the heritability of attentional networks. BMC Neurosci. 2001;2:14. doi: 10.1186/1471-2202-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coyle JT. NMDA receptor and schizophrenia: a brief history. Schizophr. Bull. 2012;38:920–926. doi: 10.1093/schbul/sbs076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cohen SM, Tsien RW, Goff DC, Halassa MM. The impact of NMDA receptor hypofunction on GABAergic neurons in the pathophysiology of schizophrenia. Schizophr. Res. 2015;167:98–107. doi: 10.1016/j.schres.2014.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gonzalez-Burgos G, Lewis DA. NMDA receptor hypofunction, parvalbumin-positive neurons, and cortical gamma oscillations in schizophrenia. Schizophr. Bull. 2012;38:950–957. doi: 10.1093/schbul/sbs010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Z, et al. Genome-wide association analysis identifies 30 new susceptibility loci for schizophrenia. Nat. Genet. 2017;49:1576–1583. doi: 10.1038/ng.3973. [DOI] [PubMed] [Google Scholar]
- 40.Egan MF, et al. Variation in GRM3 affects cognition, prefrontal glutamate, and risk for schizophrenia. Proc. Natl Acad. Sci. USA. 2004;101:12604–12609. doi: 10.1073/pnas.0405077101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’Connor JA, Hemby SE. Elevated GRIA1 mRNA expression in Layer II/III and V pyramidal cells of the DLPFC in schizophrenia. Schizophr. Res. 2007;97:277–288. doi: 10.1016/j.schres.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fromer M, et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat. Neurosci. 2016;19:1442–1453. doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salmi M, et al. Transient microstructural brain anomalies and epileptiform discharges in mice defective for epilepsy and language-related NMDA receptor subunit gene Grin2a. Epilepsia. 2018;59:1919–1930. doi: 10.1111/epi.14543. [DOI] [PubMed] [Google Scholar]
- 44.Jia P, Wang L, Meltzer HY, Zhao Z. Common variants conferring risk of schizophrenia: a pathway analysis of GWAS data. Schizophr. Res. 2010;122:38–42. doi: 10.1016/j.schres.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang X, et al. Common and rare genetic risk factors converge in protein interaction networks underlying schizophrenia. Front Genet. 2018;9:434. doi: 10.3389/fgene.2018.00434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fromer M, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pocklington AJ, et al. Novel findings from CNVs implicate inhibitory and excitatory signaling complexes in schizophrenia. Neuron. 2015;86:1203–1214. doi: 10.1016/j.neuron.2015.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.XiangWei W, Jiang Y, Yuan H. De novo mutations and rare variants occurring in NMDA receptors. Curr. Opin. Physiol. 2018;2:27–35. doi: 10.1016/j.cophys.2017.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Buckner RL. The cerebellum and cognitive function: 25 years of insight from anatomy and neuroimaging. Neuron. 2013;80:807–815. doi: 10.1016/j.neuron.2013.10.044. [DOI] [PubMed] [Google Scholar]
- 50.Andreasen NC, Pierson R. The role of the cerebellum in schizophrenia. Biol. Psychiatry. 2008;64:81–88. doi: 10.1016/j.biopsych.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schultz W, Dayan P, Montague PR. A neural substrate of prediction and reward. Sci. (New Y., NY) 1997;275:1593–1599. doi: 10.1126/science.275.5306.1593. [DOI] [PubMed] [Google Scholar]
- 52.Schultz W, Dickinson A. Neuronal coding of prediction errors. Annu Rev. Neurosci. 2000;23:473–500. doi: 10.1146/annurev.neuro.23.1.473. [DOI] [PubMed] [Google Scholar]
- 53.Holroyd CB, Coles MGH. The neural basis of human error processing: reinforcement learning, dopamine, and the error-related negativity. Psychol. Rev. 2002;109:679–709. doi: 10.1037/0033-295X.109.4.679. [DOI] [PubMed] [Google Scholar]
- 54.Goldman-Rakic PS. The cortical dopamine system: role in memory and cognition. Adv. Pharm. 1998;42:707–711. doi: 10.1016/S1054-3589(08)60846-7. [DOI] [PubMed] [Google Scholar]
- 55.Schultz W. Predictive reward signal of dopamine neurons. J. Neurophysiol. 1998;80:1–27. doi: 10.1152/jn.1998.80.1.1. [DOI] [PubMed] [Google Scholar]
- 56.Meyer A, et al. Additive effects of the dopamine D2 receptor and dopamine transporter genes on the error-related negativity in young children. Genes Brain Behav. 2012;11:695–703. doi: 10.1111/j.1601-183X.2012.00812.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Biehl SC, et al. Dopamine transporter (DAT1) and dopamine receptor D4 (DRD4) genotypes differentially impact on electrophysiological correlates of error processing. PLoS ONE. 2011;6:e28396. doi: 10.1371/journal.pone.0028396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Manoach, D. S. & Agam, Y. Neural markers of errors as endophenotypes in neuropsychiatric disorders. Fronti Hum. Neurosci.7, 350 (2013). [DOI] [PMC free article] [PubMed]
- 59.Egan MF, et al. Effect of COMT Val108/158 Met genotype on frontal lobe function and risk for schizophrenia. Proc. Natl Acad. Sci. USA. 2001;98:6917–6922. doi: 10.1073/pnas.111134598. [DOI] [PMC free article] [PubMed] [Google Scholar]