ABSTRACT
Budding yeasts are distributed across a wide range of habitats, including as human commensals. However, under some conditions, these commensals can cause superficial, invasive, and even lethal infections. Despite their importance to human health, little is known about the ecology of these opportunistic pathogens, aside from their associations with mammals and clinical environments. During a survey of approximately 1000 non-clinical samples across the United States of America, we isolated 54 strains of budding yeast species considered opportunistic pathogens, including Candida albicans and Candida (Nakaseomyces) glabrata. We found that, as a group, pathogenic yeasts were positively associated with fruits and soil environments, whereas the species Pichia kudriavzevii (syn. Candida krusei syn. Issatchenkia orientalis) had a significant association with plants. Of the four species that cause 95% of candidiasis, we found a positive association with soil. These results suggest that pathogenic yeast ecology is more complex and diverse than is currently appreciated and raises the possibility that these additional environments could be a point of contact for human infections.
Keywords: pathogen, Candida albicans, Candida glabrata, ecology, Candida tropicalis, wild yeasts
Here we describe the isolation of more than 50 strains of budding yeast opportunistic pathogens from natural settings, suggesting that they have more complex ecology than is currently appreciated.
INTRODUCTION
Budding yeasts of the subphylum Saccharomycotina are distributed across a wide range of habitats (Kurtzman, Fell and Boekhout 2011; Buzzini, Lachance and Yurkov 2017; Opulente et al. 2018). These habitats can range from wild niches, including soil, flowers, and trees, to environments where they are considered commensals, such as in insects and animals, including the human microbiota (Kurtzman, Fell and Boekhout 2011; Gabaldón and Fairhead 2018). However, under rare circumstances, these commensal yeasts can cause candidiasis, which is an infection caused by yeasts often assigned to the genus Candida and considered opportunistic pathogens (Gabaldón, Naranjo-Ortíz and Marcet-Houben 2016). While there are rare reports of several other Saccharomycotina species being the agents of candidiasis, including Saccharomyces cerevisiae (Strope et al. 2015), the species Candida albicans, Candida (Nakaseomyces) glabrata, Candida parapsilosis, and Candida tropicalis are responsible for approximately 95% of infections (Pfaller and Diekema 2007; Diekema et al. 2012; Gabaldón, Naranjo-Ortíz and Marcet-Houben 2016). Other non-hybrid yeast species recognized as opportunistic pathogens include Pichia kudriazevii (syn. Candida krusei syn. Issatchenkia orientalis), Candida dublinensis, Candida orthopsilosis, Meyerozyma (Candida) guilliermondii, and Clavispora (Candida) lusitaniae (Kurtzman, Fell and Boekhout 2011). Pichia kudriavzevii is the fifth leading cause of yeast infections, is resistant to fluconazole and has reduced susceptibility to other antifungal treatments (Pelletier et al. 2005). Infections by the other species considered opportunistic pathogens are rare, but each has caused multiple infections in humans (Pfaller et al. 2010; Gabaldón, Naranjo-Ortíz and Marcet-Houben 2016). These nine species have been considered by leading taxonomists to be the most important budding yeast opportunistic pathogens (Kurtzman, Fell and Boekhout 2011).
While most budding yeast opportunistic pathogens are currently classified in the genus Candida, these pathogenic species belong to several phylogenetically distinct clades and are genetically diverse (Gabaldón, Naranjo-Ortíz and Marcet-Houben 2016). Despite this diversity, one commonality is that little is known about their ecology; it is unclear whether their primary ecological niche is endothermic animals and clinical settings, or whether they are adapted to additional environments. While disease-causing species have been recovered from a variety of habitats, such as food, air, and clothing, these rare isolates are generally interpreted as contamination, rather than being sourced from an environmental yeast habitat, such as soil, trees, flowers, or insects (Van Uden, Faia and Assis-Lopes 1956; Di Menna 1958; Kurtzman, Fell and Boekhout 2011; Bensasson et al. 2019). Therefore, it is unknown whether these species are able to survive for extended periods of time outside of clinical settings and endothermic animals. In other words, do these species have ecological niches beyond endothermic animals as commensals and clinical settings as pathogens? If so, do different species or groups have different habitats?
Understanding the ecology of pathogenic yeasts is critical to human health for multiple reasons. First, mortality from infections by these yeasts remains high, and candidiasis is the fourth most common hospital-associated bloodstream infection (Morgan et al. 2005; Gabaldón and Fairhead 2018; Revie et al. 2018). Second, in addition to the nine already considered important opportunistic pathogens, Candida auris recently emerged as a multi-drug resistant opportunistic pathogen, with additional strains exhibiting resistance to many anti-fungal drugs (Dolande et al. 2017; Revie et al. 2018). Finally, it is unclear whether there might be environmental reservoirs that could act as contact points for their primary hosts, or whether the niches of pathogenic yeasts are indeed exclusively endothermic animals and clinical settings (Daszak, Cunningham and Hyatt 2000).
Based on previous literature, we would predict opportunistic pathogenic yeasts are not present in wild (i.e. non-endothermic, non-clinical) habitats. Therefore, we would expect that opportunistic pathogenic yeasts would not be isolated through enrichment protocols from wild habitats. In general, yeast enrichment protocols are predicted to select against yeasts that are in low abundance within an environment through repeated bottleneck events and through competition with yeasts that are in higher abundance in the environment and better adapted to culturing outside of their hosts (Fingerman, Dombrowski and Sniegowski 2002; Sampaio and Gonçalves 2008; Sylvester et al. 2015). Contrary to this expectation, we found opportunistic pathogenic yeasts isolated from multiple non-clinical environments across the United States of America. We hypothesize that these opportunistic pathogenic yeasts have niches outside of clinical and endothermic environments and are associated with specific environmental substrates. Furthermore, we hypothesize that pathogenic yeasts that are more commonly found in nature and only rarely cause candidiasis (i.e. P. kudriazevii, C. dublinensis, C. orthopsilosis, M. guilliermondii, and Cl. lusitaniae) differ from those yeasts that are known to cause 95% of candidiasis (i.e. C. albicans, C. glabrata, C. parapsilosis, and C. tropicalis). To test these hypotheses, we used data collected during a survey of approximately 1000 non-clinical, natural substrates, from which we isolated over 50 yeast strains from species considered to be opportunistic pathogens.
METHODS
Sample collection, yeast isolation and quality control
Yeasts were collected throughout the United States from multiple substrates by members of the Hittinger Lab, as well as citizen scientists (Table S1, Supporting Information). Statistical analyses did not detect bias in pathogenic isolates coming from any individual collector (χ2 = 0.072, P-value = 1, Table S2, Supporting Information) or among individuals isolating yeasts from samples (χ2 = 1.63, P-value = 1, Table S3, Supporting Information). All samples were collected using sterile bags and did not come into contact with the humans collecting them. Samples were processed by different members of the lab using published yeast enrichment and isolation protocols (Sylvester et al. 2015). Negative controls were included to ensure no contamination had occurred. After enrichment, yeast species were identified by internal transcribed spacer (ITS) sequencing, as previously described (Sylvester et al. 2015). Table S1 (Supporting Information) contains GenBank accessions for ITS sequences.
Statistical analyses
We removed strains from our analyses that were isolated from areas that are highly trafficked by humans, such as samples that were isolated from compost piles. In some cases, we isolated the same opportunistic pathogen out of a single sample; we collapsed multiple strains isolated from the same sample (including when isolated at different temperatures) to one representative strain since the strains may be closely related. In other words, for all analyses, a species was only counted once per sample, even if it was isolated multiple times from a sample. For analyses where we tested for associations between species and substrates, we removed any species that was isolated fewer than three times since there would be a lack of power to detect associations. In other words, any species that we isolated only once or twice was dropped from analyses that tested for associations between species and substrates.
In order to determine whether there were associations between substrates and species, we permuted species and substrate presence/absence matrices (n = 1000 permutations) using a swap algorithm. Permutations were performed using the R package vegan (v. 1.6–2). To determine whether there was a positive association between species and substrates, we counted the number of times a given species was isolated from a given substrate. For example, C. tropicalis was isolated seven times from soil. These values were also calculated for the permuted data, and the mean of the permuted counts was used to determine the expected isolation count. The strength of each association was calculated by subtracting the observed values from the average expected values (from the permuted data). A positive association between a substrate and a species would have a positive value for the strength of association; the larger the value, the stronger the association with the substrate. We calculated the binomial confidence intervals to determine significant positive associations using the R package Hmisc (v. 4.1–1) and corrected for multiple tests across associations with Benjamini-Hochberg corrections. We also calculated whether, in general, pathogenic yeasts were isolated from specific substrates more often using the same method described above. In this analysis, we included all species, regardless of whether they were only isolated once or twice. Finally, to detect whether there were differences between pathogens that cause 95% of candidiasis infections and all other opportunistic yeast pathogens, we categorized them into two groups: Group 1 consisted of the yeasts that cause 95% of infections (i.e. C. albicans, C. glabrata, C. parapsilosis, and C. tropicalis), whereas the other opportunistic pathogenic yeasts (i.e. P. kudriazevii, C. dublinensis, C. orthopsilosis, M. guilliermondii, and Cl. lusitaniae) were classified into Group 2. After this classification, we tested for associations between these groups and substrates using the methods described above. Additionally, we tested for differences in isolation numbers on both plants and soil using pairwise Χ2 analyses and corrected for multiple tests with Benjamini-Hochberg corrections. We did not include the fungi substrate since there were not many isolates to compare between the two groups. All analyses were done in the statistical language R (v. 3.3.0). All graphs were made using the R package ggplot2 (v. 2.2.1).
RESULTS/DISCUSSION
Multiple non-clinical isolations of known pathogenic yeasts
We extensively sampled non-clinical substrates across the United States of America to enrich for and isolate yeasts. In total, we collected approximately 1000 samples and have isolated about 5000 strains of yeasts. Across our sampling regime, we isolated 54 strains of species that are considered opportunistic pathogens (Fig. 1, Table S1, Supporting Information). Recently and independently, three strains of C. albicans were isolated from oak bark in New Forest in the United Kingdom (Bensasson et al. 2019). Collectively, these results suggest that pathogenic yeast ecology is more complex and diverse than is currently appreciated and raises the possibility that these additional environments could be a point of contact for infections.
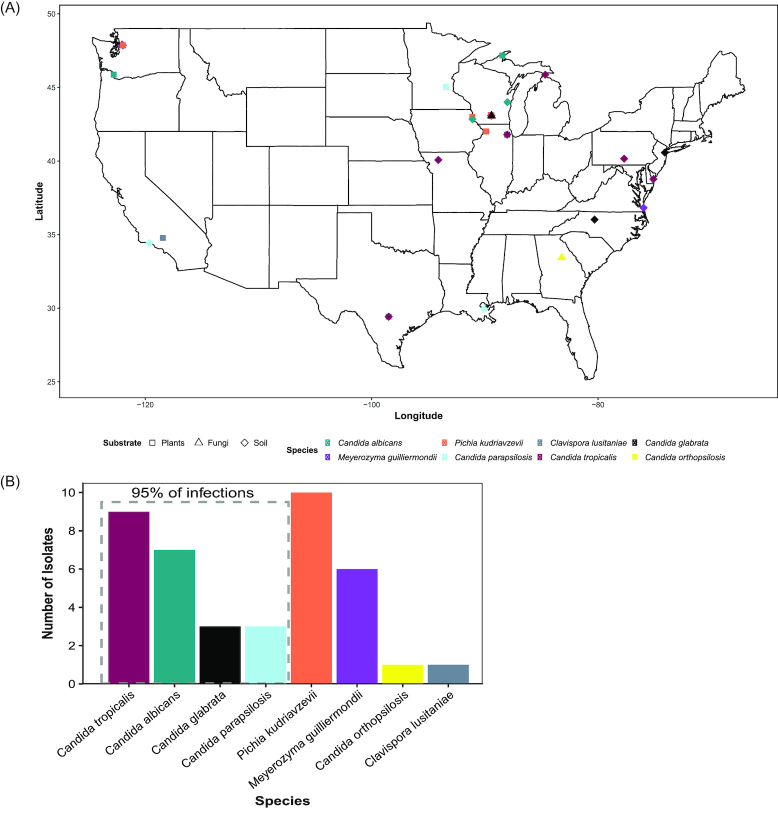
Figure 1.
Opportunistic pathogenic species of yeasts in the subphylum Saccharomycotina are found throughout the United States of America on a variety of non-clinical substrates. (A) Map displaying where opportunistic pathogens were isolated. The colors represent the species isolated, and the shapes of the points represent the substrate from which they were isolated. (B) Bar graph of unique strains of each species.
The strains we isolated came from 40 different samples; in 14 cases, multiple strains of a single species were isolated from a single sample, often from different isolation temperatures (Table S1, Supporting Information). Once those were collapsed into a single representative species for each sample, we had 40 unique strains, 55% of which were from the four species that cause 95% of candidiasis infections (Fig. 2). The species isolated most was P. kudriavzevii, making up 25% (n = 10) of our isolates of opportunistic pathogenic species. P.kudriavzevii has been previously isolated from non-clinical settings several times across a wide range of environments, including fermentations, soil, and fruits (Kurtzman, Fell and Boekhout 2011; Douglass et al. 2018), and its prevalence in our environmental isolations is consistent with previous studies.
Figure 2.
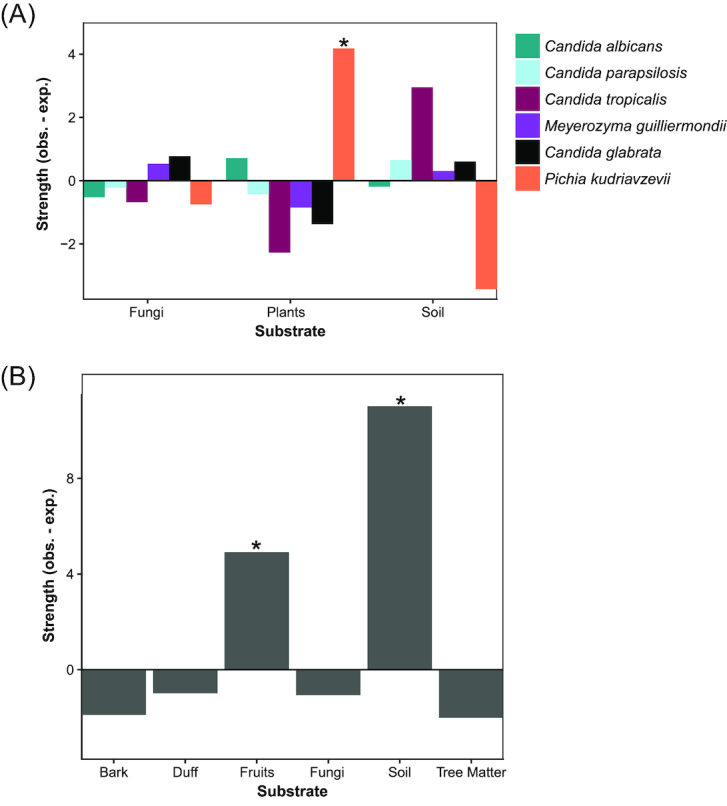
Pathogenic yeast species are associated with non-clinical/non-endothermic environments. (A) Pichia kudriavzevii was significantly associated with plants (Padj. = 0.018). Bar graphs representing the strength (difference between observed and expected counts) of associations between pathogenic species and isolation substrates. Positive values indicate cases where the observed number of isolations were greater than random expectations without associations (the average of the permuted data). The strength indicates how strong of an association there was between species and substrates. (B) As a group (i.e. using all species we isolated, but only a single isolate per environmental sample), opportunistic pathogens were associated with soil (Padj. < 0.001) and fruits (Padj. = 0.021). Bar graph representing the strength of association between pathogenic yeasts as a group and specific isolation environments.
Associations between pathogenic yeasts and non-clinical environments
While P. kudriazevii and M. guilliermondii have been often isolated from non-clinical/ectothermic environments (Brandão et al. 2017; Moubasher, Abdel-Sater and Zeinab 2018), the other species we isolated have only been rarely isolated from non-clinical settings. It is unknown whether these species are actively growing in these environments, but these non-clinical environments could potentially act as a secondary niche for these species. Furthermore, these environments could be an additional source of contact between these species and their primary hosts, potentially as the organism passes between hosts. To determine whether pathogenic yeasts were associated with particular non-clinical environments, we classified our samples in two ways. We used a more general description of whether each strain was isolated from plants, soil, or fungi; and then, when the specific information was available, a more specific description of the substrate (e.g. fruits, leaves, sand) was given. This method was used to determine whether specific parts of a substrate were more important for pathogenic yeasts than others.
Samples varied significantly in terms of the number of species isolated from an environment (Table S1, Supporting Information). Across all species with multiple isolation events, species were not limited to a single substrate. For example, C. albicans was isolated from fruits, soil, and plant matter. Pichia kudriazevii was significantly associated with isolation from plants (Padj = 0.0185) (Fig. 2A, Table S4, Supporting Information). We detected no other significant associations among substrates at the general descriptive level for the other species in our data.
Due to the smaller sample sizes at the specific descriptive levels for our substrates, we did not test for associations between them and species. However, at this level, our opportunistic pathogen isolates collectively exhibited significant associations with fruits (Padj. = 0.021) and soil (Padj. < 0.001) (Fig. 2B, Table S5, Supporting Information). The association between fruits and our isolates was most likely driven by our P. kudriavzevii isolates, which were, in our more general analysis, positively associated with plants. This association could be the result of animals acting as a vector to transport these opportunistic pathogenic species to fruits they regularly visit. The association with soil was mostly driven by C. tropicalis, but other species also contributed; 39% (n = 7) of our soil isolates were C. tropicalis. Our results, combined with the independent isolation of three strains of C. albicans from oaks in the United Kingdom (Bensasson et al. 2019), suggest that these alternative niches could potentially act as a secondary contact site between these pathogenic yeasts and their human hosts.
The most prevalent candidiasis agents are associated with soil
The ecological complexity of these opportunistic pathogens may go beyond species level differences, so we assessed whether yeasts might vary by pathogenicity as well. One notable ecological difference between prevalent causes of candidiasis (Group 1) and the important minor contributors (Group 2) is that some of the yeasts in Group 2 are frequently found in non-clinical settings (e.g. P. kudriavzevii and M. guillermondii). Therefore, we hypothesized that there would be differences in ecological associations between these two groups, so we tested for associations between each group and our isolation environments. We found that Group 1 was associated with soil environments (Padj = 0.024, Fig. 3, Table S6, Supporting Information). However, we did not detect associations between Group 2 and any specific substrate. We also quantified whether there were significant differences in isolation numbers for each substrate between Group 1 and Group 2. We found a significant difference in isolation numbers on soil (Padj. = 0.036, Χ = 5.56, Fig. 3, Table S7, Supporting Information). Collectively, these results suggest that there are ecological differences between these groups. These results further suggest that those pathogenic yeasts more commonly found in the environment are found across many different niches.
Figure 3.
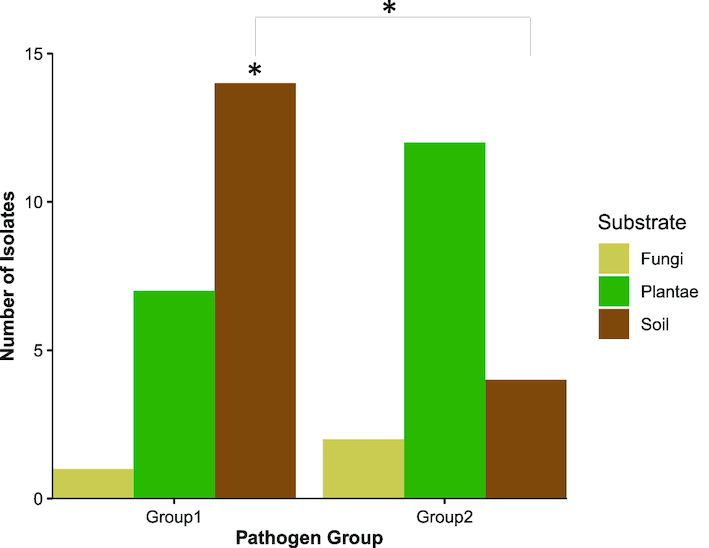
Pathogenic yeasts that cause 95% of candidiasis (Group 1) differed with other pathogenic yeasts (Group 2). Group 1 yeasts were positively associated with soil environments (Padj = 0.024, n = 14), and Group 1 yeasts were isolated more often from soil than Group 2 yeasts (P = 0.036, Χ = 5.56). Bar graphs representing the counts of Group 1 and Group 2 species isolated from different natural substrates. Group 1 included C. albicans, C. tropicalis, C. glabrata, and C. parapsilosis. Group 2 included P. kudriavzevii, M. guilliermondii, Cl. Lusitaniae, and C. orthopsilosis. Both groups contained a single representative isolate per environmental sample.
CONCLUSIONS
We isolated several opportunistic pathogenic species of yeasts from the subphylum Saccharomycotina from multiple non-clinical, non-endothermic environments across the United States of America. Among those strains isolated, over 50% represent the opportunistic pathogens that cause 95% of candidiasis. These species, C. albicans, C. tropicalis, C. parapsilosis, and C. glabrata, have been rarely isolated from non-clinical settings, and when they have been isolated from the environment, they have usually been interpreted as contaminants. Our extensive isolations from natural settings challenge this assumption, suggesting that opportunistic pathogens can persist in alternative niches and that their ecologies may be more complicated than is currently assumed. Non-clinical environments could be a short-term habitat as these yeasts are passed between their predominant hosts, endothermic animals.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the following citizen scientists or lab members for collecting samples or generating preliminary results: Aaron G. Barton, Amanda Beth Hulfachor, Russell L. Wrobel, Leslie Shown, The Zasadil Family, EmilyClare P. Baker, Drew T. Doering, Angela Sheddan, Sarah Wright, Bill Saucier, Bill Vagt, Annette Opulente and S. Smead and family. This material is based upon work supported by the National Science Foundation under Grant Nos. DEB-1253634 (to CTH), DEB-1442148 (to CTH) and DGE-1256259 (Graduate Research Fellowship to QKL); funded in part by Lakeshore Nature Preserve Student Engagement Grants (to MJ and RMS); and funded in part by the DOE Great Lakes Bioenergy Research Center (DOE Office of Science BER DE-SC0018409 to Timothy J. Donohue). QKL was also supported by the Predoctoral Training Program in Genetics, funded by the National Institutes of Health (5T32GM007133). CTH is a Pew Scholar in the Biomedical Sciences and Vilas Faculty Early Career Investigator, supported by the Pew Charitable Trusts and Vilas Trust Estate, respectively.
Conflicts of interest. None declared.
REFERENCES
- Bensasson D, Dicks J, Ludwig JMet al.. Diverse lineages of Candida albicans live on old oaks. Genetics. 2019;211:277–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandão LR, Vaz ABM, Santo LCEet al.. Diversity and biogeographical patterns of yeast communities in antarctic, patagonian and tropical lakes. Fungal Ecol. 2017;28:33–43. [Google Scholar]
- Buzzini P, Lachance M-A, Yurkov A, Yeasts In Natural Ecosystems: Diversity. Cham, Switzerland: Springer, 2017. [Google Scholar]
- Daszak P, Cunningham AA, Hyatt AD. Emerging infectious diseases of wildlife–threats to biodiversity and human health. Science. 2000;287:443–449. [DOI] [PubMed] [Google Scholar]
- Diekema D, Arbefeville S, Boyken Let al.. The changing epidemiology of healthcare-associated candidemia over three decades. Diagn Microbiol Infect Dis. 2012;73:45–48. [DOI] [PubMed] [Google Scholar]
- Di Menna ME. Candida albicans from grass leaves. Nature. 1958;181:1287. [DOI] [PubMed] [Google Scholar]
- Dolande M, García N, Capote AMet al.. Candida auris: antifungal multi-resistant emerging yeast. Curr Fungal Infect Rep. 2017;11:197–202. [Google Scholar]
- Douglass AP, Offei B, Braun-Galleani Set al.. Population genomics shows no distinction between pathogenic Candida krusei and environmental Pichia kudriavzevii: one species, four names. PLOS Pathog. 2018;14:e1007138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingerman E, Dombrowski PG, Sniegowski PD. Saccharomyces cerevisiae and Saccharomyces paradoxus coexist in a natural woodland site in North America and display different levels of reproductive isolation from European conspecifics. FEMS Yeast Res. 2002;1:299–306. [DOI] [PubMed] [Google Scholar]
- Gabaldón T, Fairhead C. Genomes shed light on the secret life of Candida glabrata: not so asexual, not so commensal. Curr Genet. 2018;65:93–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabaldón T, Naranjo-Ortíz MA, Marcet-Houben M. Evolutionary genomics of yeast pathogens in the Saccharomycotina. FEMS Yeast Res. 2016;16:fow064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzman C, Fell JW, Boekhout T, The Yeasts: A Taxonomic Study. London, United Kingdom: Elsevier, 2011. [Google Scholar]
- Morgan J, Meltzer MI, Plikaytis BDet al.. Excess mortality, hospital stay, and cost due to candidemia: a case-control study using data from population-based candidemia surveillance. Infect Control Hosp Epidemiol. 2005;26:540–47. [DOI] [PubMed] [Google Scholar]
- Moubasher AH, Abdel-Sater MA, Zeinab SM. Diversity of floricolous yeasts and filamentous fungi of some ornamental and edible fruit plants in Assiut area, Egypt. Curr Res Environ Appl Mycol. 2018;8:135–61. [Google Scholar]
- Opulente DA, Rollinson EJ, Bernick-Roehr Cet al.. Factors driving metabolic diversity in the budding yeast subphylum. BMC Biol. 2018;16:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier R, Alarie I, Lagacé Ret al.. Emergence of disseminated candidiasis caused by Candida krusei during treatment with caspofungin: case report and review of literature. Med Mycol. 2005;43:559–64. [DOI] [PubMed] [Google Scholar]
- Pfaller MA, Diekema DJ, Gibbs DLet al.. Results from the ARTEMIS DISK global antifungal surveillance study, 1997 to 2007: a 10.5-year analysis of susceptibilities of Candida species to fluconazole and voriconazole as determined by CLSI standardized disk diffusion. J Clin Microbiol. 2010;48:1366–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev. 2007;20:133–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revie NM, Iyer KR, Robbins Net al.. Antifungal drug resistance: evolution, mechanisms and impact. Curr Opin Microbiol. 2018;45:70–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampaio JP, Gonçalves P. Natural populations of Saccharomyces kudriavzevii in Portugal are associated with oak bark and are sympatric with S. cerevisiae and S. paradoxus. Appl Environ Microbiol. 2008;74:2144–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strope PK, Skelly DA, Kozmin SGet al.. The 100-genomes strains, an S. cerevisiae resource that illuminates its natural phenotypic and genotypic variation and emergence as an opportunistic pathogen. Genome Res. 2015;25:762–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvester K, Wang Q-M, James Bet al.. Temperature and host preferences drive the diversification of Saccharomyces and other yeasts: a survey and the discovery of eight new yeast species. FEMS Yeast Res. 2015;15:fov002. [DOI] [PubMed] [Google Scholar]
- Van Uden N, Faia MDM., Assis-Lopes L. Isolation of Candida albicans from vegetable sources. Microbiology. 1956;15:151–53. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.