Abstract
Background
The incidence of esophageal adenocarcinoma (EAC) has been increasing over the last 40 years. While Barrett’s esophagus is a known risk factor for the development of EAC, the role of the microflora in the development of EAC is still largely unknown and is being investigated further by multiple centers. Our goal was to identify trends in microflora composition along various aspects of the upper gastrointestinal tract in patients with Barrett’s esophagus.
Methods
After obtaining institutional review board approval, 12 patients agreed to participate in the study. While endoscopy was performed for surveillance Barrett’s monitoring, additional biopsies of esophageal mucosa were taken from the (I) proximal esophagus, (II) mid-esophagus, (III) distal esophagus, and (IV) Barrett’s esophagus. Additional swabs were also taken from the uvula and the endoscope used during the procedure. The swabs from the uvula and endoscope were obtained prior to the endoscope entering the stomach, to prevent exposing the endoscope to the acidic environment of the stomach. The most common bacterial elements were identified by amplifying sample DNA using a panel of 5 “universal” fusion primer pairs. The 400–500 base pair fragments created an overlap which covered 95% of the bacterial 16s gene.
Results
Throughout the esophagus, 34 bacterial genera were found which had a relative abundance of >1.0. Streptococcal genera were prevalent in all aspects of the esophagus, ranging from 16% to 70% of the bacterial community. Haemophilus genera were uniquely abundant in the Barrett’s esophageal tissue but relatively absent elsewhere in the upper gastrointestinal tract. Overall, the percentage of Gram-positive organisms was much higher in the proximal than distal esophagus. The microflora pattern obtained from the uvula and endoscopic swabs did not correlate with any of the tissue biopsies along any aspect of the esophagus.
Conclusions
In patients with Barrett’s esophagus, Streptococcal genera are widespread throughout the esophagus. Gram-positive genera tend to decrease as a percentage of overall flora distally. Obtaining a simple swab of the oropharynx or endoscope itself appears to be a poor substitute for tissue biopsy of esophageal mucosa when evaluating microflora patterns.
Keywords: Microbiome, esophagoscopy, genomics
Introduction
Esophageal adenocarcinoma (EAC) is one of the few solid organ tumors which has increased in incidence over the last 40 years (1). An increased prevalence of obesity in adults, an aging population and dietary factors may explain the increased numbers of EAC currently (2-4). Despite the increased incidence, overall prognosis remains poor because most cases are discovered with locally advanced or distant metastatic disease (5,6).
Creating a comprehensive method to perform screening endoscopies for esophageal cancer has proven difficult. Currently the major risk factor which warrants screening is the presence of Barrett’s esophagus, which is defined as metaplastic change of the esophageal mucosa from squamous to simple columnar histology (7). Despite being a risk factor for the development of EAC, only 1 in 860 patients with Barrett’s esophagus will ultimately develop EAC (8). Given this very low rate of cancer development, only very few patients with Barrett’s esophagus ever receive screening endoscopy (9,10).
The role of the microflora in the development of Barrett’s esophagus has been increasingly studied (11-14). Other solid organ tumors have been shown to be related to changes in microflora composition (15,16), but the role of microflora changes in the esophagus is still poorly understood. Our goal was to identify the microflora composition and genomic patterns in patients with Barrett’s esophagus, and to determine whether oral swabs have the same patterns as esophageal tissue biopsies.
Methods
Endoscopy
After obtaining institutional review board approval, 12 patients agreed and were included in the study. Patients were scheduled to receive routine surveillance endoscopy for Barrett’s esophagus as clinically indicated. During the endoscopy, extra biopsies of the esophagus were taken from the (I) proximal esophagus, (II) mid-esophagus, (III) distal esophagus, and (IV) Barrett’s esophagus. These additional biopsies were collected for research analysis. Swabs were then taken of the uvula and of the endoscope itself.
Tissue swabs
A tissue swab was taken from its sterile packaging and applied to (I) the uvula and (II) the endoscope. The swab of the endoscope was taken after the endoscope was passed through the oropharynx to the distal esophagus but before tissue biopsies were performed and before the endoscope entered the stomach, to prevent the endoscope from being exposed to the acidic environment of the stomach. Prior to using the endoscope, it was sterilized and kept in a sterile bin to prevent contamination before it was inserted into the esophagus.
Sequencing
Sample sequencing was carried out using a fusion-polymerase chain reaction (PCR) method. Briefly, fusion-primers were designed in accordance with the manufacturer’s guidelines (Ion Amplification Library Preparation–Fusion Method, Life Technologies, Carlsbad, CA, USA) using Ion Xpress Barcodes linked to 16s gene primer pairs targeting hyper-variable regions 1–8 (17). Each 25 µL PCR was carried out using: 12.5 µL iQ supermixTM (Bio-Rad, Hercules, CA, USA), 1 µL of both forward and reverse (5 µM) primers, 9.5 µL nuclease-free water and 1 µL of DNA template. DNA from 12 bio-pooled samples (oral swab, n=7, endoscope swab, n=12, proximal esophagus, n=6, mid-esophagus, n=6, distal esophagus, n=5, Barrett’s esophagus, n=7) were used as a template for creation of subsequent fusion 16s libraries. PCR was completed in a c1000 thermocycler (Bio-Rad) using the following parameters: (Cycle 1), 95 C, 3 minutes; (Cycle 2), Step 1—95 C, 45 seconds; Step 2—primer-specific annealing temps., 45 seconds; Step 3—72 C 2 minutes, repeat 39×; Step 4—72 C for 7 minutes. PCR products were purified using Qiagen Qiaquick spin-columns and quantified using a spectrophotometer (Bio-Rad). PCR products were then diluted, mixed in equal proportion and sequenced on an Ion Torrent GeneStudio S5 System using Ion 520 sequencing kits together with 520 size chips following the manufacturer’s instructions (Life Technologies).
Bioinformatics for ion torrent
After generation, sequencing reads were filtered for quality and binned according to Ion Xpress barcode using Ion Torrent Suite software version 5.10.0. Sequencing reads in FASTQ format were further processed using web-based Galaxy software (18). First, raw FASTQ files were normalized using the FASTQ groomer tool function. Next, each barcoded read was trimmed to remove the primer sequence and subsequently filtered to the expected size of the 16s gene target. After this level of processing, the sequence reads were concurrently compared to the SILVA 16s database using bowtie 2 software (19,20). This yielded a call to genera level as well as the number of times each sequence matched the database (hit-rate). Where multiple calls to the same genera were made the number of hits were added accordingly. These numbers were then converted to percentage of total to give an overall ratio of the sequenced Barrett’s esophageal sample.
Results
Streptococcus
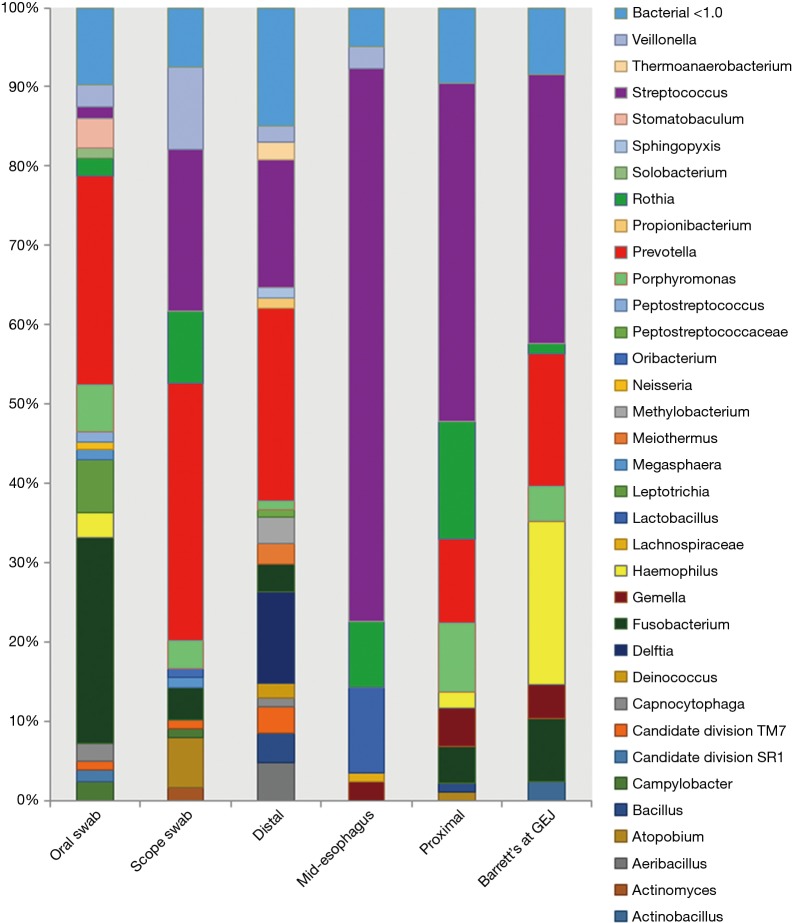
Throughout the esophagus, 34 bacterial genera were found which had a relative abundance of >1.0 (Figure 1). Streptococcal genera were prevalent at all points of the esophagus, ranging from 16% to 70% of the microflora composition. Streptococcus increased in prevalence distally along the esophagus.
Figure 1.
Relative abundance of microflora in esophageal biopsies and swabs in patients with Barrett’s esophagus.
Other genera
Prevotella was found in increased amounts in the proximal and distal esophagus. Delftia genera were found only in the distal esophagus. Fusobacterium was seen at high levels in the oropharynx but not elsewhere in the esophagus. Haemophilus was uniquely abundant in the Barrett’s mucosa but absent elsewhere (Figure 2).
Figure 2.
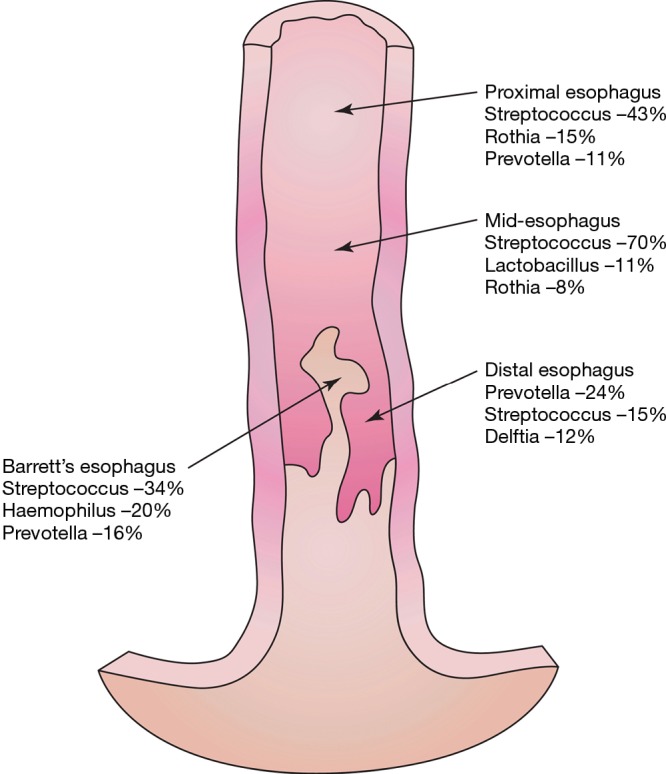
Most common bacteria at each point along esophagus.
Uvula and endoscope swabs
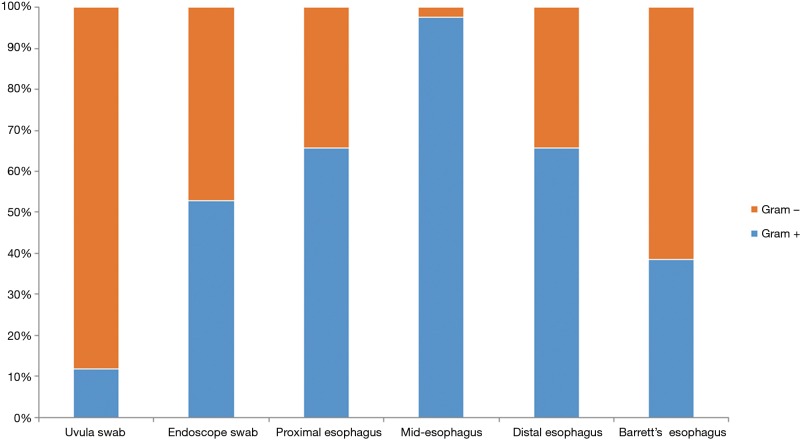
The microflora composition of the uvula and endoscope swabs differed significantly from the esophageal mucosal biopsies (Figure 3). In particular, the amount of gram negative organisms was grossly underestimated by the uvular swabs compared to all points along the esophagus (Figure 4). Though the endoscope swabs were a closer surrogate to the intraluminal environment of the esophagus, the Streptococcus level was much higher at most points along the esophagus than the endoscope swab.
Figure 3.
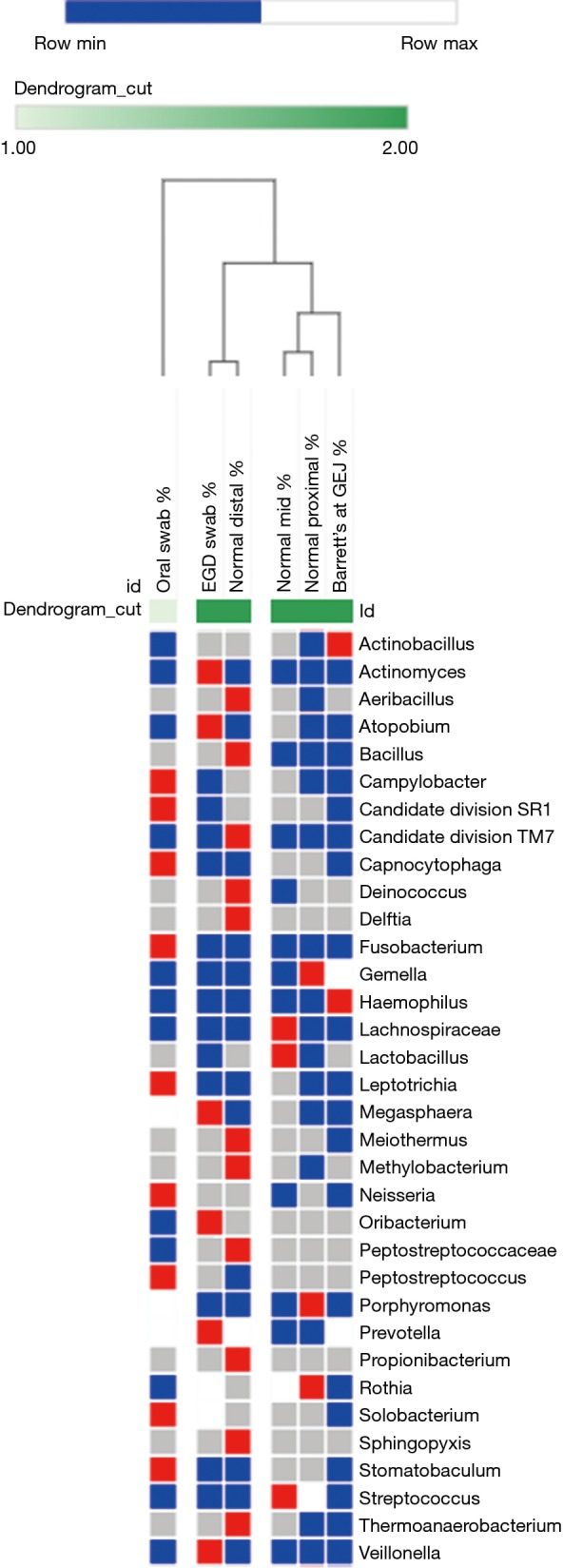
Cluster analysis of microflora at different points along esophagus in patients with Barrett’s esophagus.
Figure 4.
Percentage of Gram positive/negative organisms at different points along esophagus.
Discussion
Increasingly the role of the microflora in the development of esophageal disease is becoming more understood but is still poorly described. Because Barrett’s esophagus is one of the few known risk factors for esophageal cancer, we chose a cohort entirely of these patients. Our goal was to characterize the microflora along various points within the esophagus, and then to determine the best methods to detect these bacterial distributions.
Streptococcal genera appear to be very prevalent in patients with Barrett’s esophagus. Although Streptococcus is seen in the esophagus in patients without esophageal disease, the levels in our study were much higher throughout the esophagus (21). And interestingly, the levels appear to decrease more sharply in patients along the distal esophagus than in patients without Barrett’s esophagus (22). Though previous studies have investigated the levels of Streptococcus in the distal esophagus, the gradient decrease along the esophagus may be more impactful in the development of Barrett’s esophagus.
Haemophilus genera were present in abundance in Barrett’s tissue but nowhere else along the esophagus. Most other studies have not shown this association, and have not compared the levels of Haemophilus along various aspects of the esophagus. Further studies are needed, but it appears that this genus may serve as a useful marker for Barrett’s esophagus or for patients with reflux disease who may be at increased risk of developing Barrett’s esophagus.
Another important goal for us was to develop a reliable method to measure the microbial composition of the esophagus. The current standard of care to screen patients with Barrett’s esophagus is by endoscopy and mucosal biopsy. But discovering a less invasive and more cost effective method to screen patients with Barrett’s esophagus would increase the screening rate, which is currently less than 5% nationally (23). From our studies it appears that both a uvular swab and a swab of the endoscope itself do not serve as surrogates for analysis of tissue biopsy. We expected that the microflora found on the uvular and endoscope swabs would have a slight difference from the proximal esophagus and then change gradually toward the gastroesophageal junction. But our study showed that the microflora found on both swabs were markedly different than the microflora of the esophageal biopsies. Further studies are needed to determine whether there is an inherent difference in microbial analysis seen in swabs versus tissue biopsies, and whether there is a less invasive method of analyzing the esophageal microbiome without tissue biopsy.
Our study had some limitations. Overall it was a small cohort of patients, but we decided to use a bio-pooling strategy to reduce potential variability. An average patient sample for each condition was synthesized by combining individual DNAs in equal ratios in a bio-pooling strategy. This reduced variability between individuals and gave an average microbiome for the esophageal region sampled. Microbiota of interest identified using this method were then evaluated using single-target qPCR to obtain absolute abundance and increase statistical power. Furthermore, our cohort was limited only to patients with Barrett’s esophagus. Given the number of biopsies we were taking during the procedure, we did not want to expose patients without esophageal disease undergoing endoscopy for another reason to this number of biopsies. But we feel that we have gained important information with our study results.
The role of the intraluminal microflora in patients with esophageal disease is still being investigated, but there do appear to be significant differences in patients with Barrett’s esophagus. Streptococcus drops more sharply toward the gastroesophageal junction, and Haemophilus species are abundant within the Barrett’s tissue itself. Further studies are needed to determine whether the microflora changes are a result of chronic acid inflammation or allow the esophageal mucosa to undergo the metaplastic transformation to Barrett’s esophagus.
Acknowledgments
Funding: Dr. Okereke’s work was supported by grants from the National Center for Advancing Translational Sciences (NCATS) of the National Institutes of Health, award numbers UL1TR001439 and KL2TR001441.
Ethical Statement: Institutional review board approval was obtained prior to the study being conducted. Informed consent was obtained from each patient in accordance to institutional review board regulations.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.National Cancer Institute: Surveillance, Epidemiology and End Results Program. Available online: https://seer.cancer.gov/statfacts/html/esoph.html
- 2.Wheeler JB, Reed CE. Epidemiology of esophageal cancer. Surg Clin North Am 2012;92:1077-87. 10.1016/j.suc.2012.07.008 [DOI] [PubMed] [Google Scholar]
- 3.Krishna SG, Hussan H, Cruz-Monserrate Z, et al. A review of the impact of obesity on common gastrointestinal malignancies. Integr Cancer Sci Ther 2017;4. doi: . 10.15761/ICST.1000223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turcotte S, Duranceau A. Gastroesophageal reflux and cancer. Thorac Surg Clin 2005;15:341-52. 10.1016/j.thorsurg.2005.03.003 [DOI] [PubMed] [Google Scholar]
- 5.Bharat A, Crabtree T. Management of advanced-stage operable esophageal cancer. Surg Clin North Am 2012;92:1179-97. 10.1016/j.suc.2012.07.012 [DOI] [PubMed] [Google Scholar]
- 6.Alsop BR, Sharma P. Esophageal Cancer. Gastroenterol Clin North Am 2016;45:399-412. 10.1016/j.gtc.2016.04.001 [DOI] [PubMed] [Google Scholar]
- 7.Gibson MK, Dhaliwal AS, Clemons NJ, et al. Barrett's esophagus: cancer and molecular biology. Ann N Y Acad Sci 2013;1300:296-314. 10.1111/nyas.12252 [DOI] [PubMed] [Google Scholar]
- 8.Hvid-Jensen F, Pedersen L, Drewes AM, et al. Incidence of adenocarcinoma among patients with Barrett's esophagus. N Engl J Med 2011;365:1375-83. 10.1056/NEJMoa1103042 [DOI] [PubMed] [Google Scholar]
- 9.Gupta M, Iyer PG. Screening for Barrett's Esophagus. Gastroenterol Clin North Am 2015;44:265-83. 10.1016/j.gtc.2015.02.003 [DOI] [PubMed] [Google Scholar]
- 10.Choi SE, Hur C. Screening and surveillance for Barrett's esophagus: current issues and future directions. Curr Opin Gastroenterol 2012;28:377-81. 10.1097/MOG.0b013e328353d58e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Snider EJ, Freedberg DE, Abrams JA. Potential Role of the Microbiome in Barrett's Esophagus and Esophageal Adenocarcinoma. Dig Dis Sci 2016;61:2217-25. 10.1007/s10620-016-4155-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang L, Chaudhary N, Baghdadi J, et al. Microbiome in reflux disorders and esophageal adenocarcinoma. Cancer J 2014;20:207-10. 10.1097/PPO.0000000000000044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blackett KL, Siddhi SS, Cleary S, et al. Oesophageal bacterial biofilm changes in gastro-oesophageal reflux disease, Barrett's and oesophageal carcinoma: association or causality? Aliment Pharmacol Ther 2013;37:1084-92. 10.1111/apt.12317 [DOI] [PubMed] [Google Scholar]
- 14.Pei Z, Bini EJ, Yang L, et al. Bacterial biota in the human distal esophagus. Proc Natl Acad Sci U S A 2004;101:4250-5. 10.1073/pnas.0306398101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zackular JP, Baxter NT, Iverson KD, et al. The gut microbiome modulates colon tumorigenesis. MBio 2013;4:e00692-13. 10.1128/mBio.00692-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang LL, Yu XJ, Zhan SH, et al. Participation of microbiota in the development of gastric cancer. World J Gastroenterol 2014;20:4948-52. 10.3748/wjg.v20.i17.4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klindworth A, Pruesse E, Schweer T, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 2013;41:e1. 10.1093/nar/gks808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blankenberg D, Gordon A, Von Kuster G, et al. Manipulation of FASTQ data with Galaxy. Bioinformatics 2010;26:1783-5. 10.1093/bioinformatics/btq281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yilmaz P, Parfrey LW, Yarza P, et al. The SILVA and "All-species Living Tree Project (LTP)" taxonomic frameworks. Nucleic Acids Res 2014;42:D643-8. 10.1093/nar/gkt1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012;9:357-9. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y, Lin Z, Lin Y, et al. Streptococcus and Prevotella are associated with the prognosis of oesophageal squamous cell carcinoma. J Med Microbiol 2018;67:1058-68. 10.1099/jmm.0.000754 [DOI] [PubMed] [Google Scholar]
- 22.Yang L, Lu X, Nossa CW, et al. Inflammation and intestinal metaplasia of the distal esophagus are associated with alterations in the microbiome. Gastroenterology 2009;137:588-97. 10.1053/j.gastro.2009.04.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.di Pietro M, Chan D, Fitzgerald RC, et al. Screening for Barrett's Esophagus. Gastroenterology 2015;148:912-23. 10.1053/j.gastro.2015.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]