Abstract
Knowledge of seasonal shifts in the bacterial community composition among different mulberry (Morus L.) cultivars will facilitate to develop the biocontrol phytopathogens strategy using endophytic bacteria. The present study investigated the endophytic bacterial communities of four mulberry cultivars that have different resistance to mulberry fruit sclerotiniosis using Illumina-based sequencing of the 16S rRNA gene fragment in spring and autumn. The results indicated that spring samples harbor higher bacterial operational taxonomic units (OTUs), α-diversity, and bacterial community complexity in comparison with autumn samples. The taxonomic composition analysis showed that the majority of endophytes were composed of Proteobacteria (genus level: Methylobaterium) and Actinobacteria in spring, while sequences classified as Proteobacteria (genus level: Pantoea and Pseudomonas) were abundant in autumn. Analysis of β-diversity also revealed endophytic bacteria were divided into two main groups by season. By comparison among different mulberry cultivars, we found that Pantoea, Methylobaterium, and Pseudomonas were the three major bacterial genera in all cultivars, while their relative abundances varied with cultivars and appeared no obvious relationship with resistance level of mulberry fruit sclerotiniosis. The complex correlation of the endophytic communities in susceptible mulberry cultivars was higher than that of the resistant cultivars. Overall, the findings suggested that season plays a key role in determining the mulberry endophytic bacterial communities, followed by host cultivar, and Proteobacteria was the predominant phylum in both seasons and different mulberry cultivars.
Keywords: Endophytic bacteria, Microbial diversity, Mulberry cultivar, Seasonal variation
Graphical Abstract
Highlights
-
•
Season played a key role in determining mulberry endophytic bacterial communities.
-
•
Mulberry endophytic bacterial variation was not obviously related with cultivars.
-
•
Mulberry recruited beneficial endophytes as potential biocontrol agents.
1. Introduction
Plant endophytes, a component of the microbiota of a host, are often described as non-pathogenic microorganisms residing in the living tissues of plants and that cause no apparent harm to their host [1,2]. Endophytic bacteria can colonize both monocotyledonous and dicotyledonous plants and reside there for long periods of time [3]. Plant hosts range from woody tree species, such as apple [4] and poplar tree [5] to herbaceous crop plants, such as maize [6] and rice [7]. It is becoming more apparent that plants and endophytes have both adapted to use their close association for mutual benefit. The plant microbiome appears to be a key determinant of plant health and productivity by providing a plethora of functional substances. Some endophytes may improve nutrient bioavailability as well as an increase in host tolerance to abiotic stresses. There is evidence that they also influence crop yield and quality [7]. Notably, endophytic bacteria have the advantage of colonizing, proliferating, and moving within plant tissues and thus are excellent candidates as biocontrol agents [8]. In turn, the host plant provides a protected living space, as well as a constant supply of nutrients for the endophytic bacteria [9,10].
Mulberry trees (Morus L.), the sole food plant of the silkworm (Bombyx mori), are widely cultivated throughout subtropical and temperate regions of the world [11]. Mulberry fruits (sorosis) also contain a variety of nutrient and nutraceutical substances [12] such as anthocyanin, rutin, and quercetin [13]. This makes mulberry fruits a valuable byproduct of the silkworm industry. Unfortunately, the productivity of mulberry fruits has been greatly threatened in different mulberry-planting zones by sclerotiniosis. This disease is mainly caused by soil-borne fungal pathogens in the family Sclerotiniaceae such as Ciboria shiraiana [14], Ciboria carunculoides [15], Sclerotinia sclerotiorum [16], and Scleromitrula shiraiana [17]. Yu et al. [18] reported that different mulberry cultivars had significantly different impacts on the soil microbial community, resulting in the differences in the level of resistance to sclerotiniosis. Little information is available, however, about the distribution of endophytic bacteria in different mulberry cultivars and their potential role in resistance to sclerotiniosis. Hence, the endophytic bacteria of four mulberry cultivars with different resistance levels to sclerotiniosis, two resistant cultivars ‘Chuan Sang No.7637’ (Morus alba L.) and ‘Changguo Sang’ (Morus laevigata Wall.), and two susceptible cultivars ‘Xin Lunjiao’ (Morus atropurpurea Roxb.) and ‘Hong Guo No.2’ (Morus atropurpurea Roxb.) [19], will be analyzed in present study.
It is well known that biotic and abiotic characteristics could influence the assembly of the endophytic community [20,21]. Yang et al. [22] demonstrated that the distribution of endophytic bacteria in leaves of peony varied with the different cultivars, and this phenomenon was also detected in the roots. Also, endophytic composition of two transgenic maize genotypes was varied from the near-isogenic non-transgenic maize genotypes [3]. Furthermore, array of studies have documented that climate, moisture, and temperature are important environmental properties that influence the population density and ecology of the microbial community [[23], [24], [25]]. For example, the effect of the temperature sensitivity on microbial respiration was reported to be season-dependent, being much greater in the dormant season than in the growing season [26]. Similarly, seasonal changes in the concentration of soluble sugars, proteins, amino acids, organic acids, and other plant nutrients also affect microbial communities. Thus far, the effect of climate on seasonal changes in microbial communities has been mainly assessed in forest or grassland soils [27,28], while few studies have focused on the relationship between plant endophytes and climate or season. A better understanding of seasonal variation in the composition of endophytic bacteria may help to elucidate their function in the growth and health of their hosts and their potential application in sustainable systems of agricultural production.
Endophytic bacteria have been typically isolated and analyzed using traditional culture-dependent approaches. This approach is highly dependent on the types of media used for isolation and the incubation conditions [29,30]. Moreover, culture-dependent biodiversity studies of the endophytic community are somewhat limited since it has been estimated that the bacteria were identified no >1% of the bacterial species present using conventional cultivation techniques [31]. In recent years, high-throughput sequencing techniques, also referred to as next-generation sequencing (NGS), have provided new insights into the composition of the bacterial microflora in different plants, such as rice [31], banana [32], and bean [33], etc. This approach has the ability to detect non-culturable bacterial taxa, as well taxa that are in low abundance because they grow slowly and are missed by traditional culture-based protocols [34]. Amplification of a region of bacterial 16S rRNA using universal primer sets, a process referred to as metabarcoding, followed by sequencing on an Illumina Miseq platform has revolutionized the study of bacterial communities and provided higher phylogenetic resolution, making it an excellent tool for conducting microbial diversity studies [7,30,35].
Multiple factors can influence the composition of the microbiome. In particular, host inter- and intraspecific host genetics and climate play key roles in determining variations in the diversity and species abundance within different bacterial communities [36]. In the present study, metabarcoding and Illumina-based sequencing technology was used to characterize the endophytic bacterial community in four mulberry cultivars in two different seasons (spring and autumn). The data collected can serve as a reference for selecting and evaluating potential biocontrol bacterial agents for use in treating mulberry sclerotiniosis.
2. Materials and Methods
2.1. Mulberry Sample Preparation
Four mulberry cultivars, ‘Changguo Sang’ (CGS) and ‘Chuan Sang No.7637’ (CSQ), both of which are resistant to mulberry fruit sclerotiniosis, and ‘Hong Guo No.2’ (HGE) and ‘Xin Lunjiao’ (XLJ), both of which are susceptible to mulberry fruit sclerotiniosis, were used in the study. ‘Changguo Sang’ was sampled at Southwest University experimental farm (29°49′1′′ N, 106°24′57′′ E), and the remaining three cultivars were sampled at the Chongqing Sericulture Science and Technology Research Institute experimental farm (29°50′39′′ N, 106°25′55′′ E). The two sampling sites are located near the Jialing River in Chongqing, China, and share the same climatic conditions. The branch samples were collected from two-year-old mulberry trees of four cultivars in April (represent spring) and September (represent autumn), 2016. Healthy branches approximately 50.0 cm in length and 1.5–2.0 cm in diameter, were collected, from six plant individuals in each cultivar. Branches were washed in running tap water to remove surface debris and then cut into several segments (about 7.0 cm). Ten surface-sterilized segments in each cultivar were randomly selected, pooled, and served as one replicate for the further endophyte enrichment. Three replicates in each of all samples (4 cultivars × 2 seasons) were performed in this study.
The segments were surface-sterilized as described by Strobel et al. [37]. Briefly, the small segments were completely immersed in 75% ethanol for 30 s, and then adhering alcohol was set aflame using an alcohol lamp. The effectiveness of the surface sterilization was confirmed by making imprints of disinfected segments on potato agar medium plates and culturing the plates. Only those mulberry tissues with no microbial growth detected on the plates after incubation were used for the following endophyte enrichment.
2.2. Endophyte Enrichment
The process of endophyte enrichment was utilized the method described by Wang et al. [38]. Approximately 5 g of surface-sterilized bark tissues were chopped and homogenized in sterile distilled water using a sterilized plant tissue homogenizer. The suspension was filtered through two layers of gauze and centrifuged at 200 ×g for 5 min at 4 °C. The supernatant was then transferred to a new sterile tube, subsequently NaCl and 10% sodium dodecyl sulfate (SDS) were added to reach a concentration of 0.9% and 0.063% (W/V), respectively. The mixture was gently mixed and then incubated at 4 °C for 1 h. After settling, the upper phase was transferred to a new tube and centrifuged (5000 ×g, 10 min, 4 °C). The resulting precipitate was resuspended in 200 mL of sterile distilled water and the two reagents NaCl and SDS were subsequently added in the same way as described above. The procedure was repeated until at least 100 mg of precipitate could be obtained and resuspended in 1 mL of TE buffer (10 mM Tris-HCl, pH 8.0; 1 mM EDTA, pH 8.0).
2.3. DNA Extraction and 16S rRNA Gene Sequencing
DNA was extracted using a protocol described by Murray et al. [39] and Maropola et al. [40]. Lysozyme and RNaseA were added to the suspension containing the enriched microorganisms and mixed gently. The sample mixtures were incubated at 37 °C for 10 min, and then treated with 6 μL Proteinase K and 60 μL 10% SDS at 55 °C for 20 min until the liquid became clear. Subsequently, 200 μL 5 M NaCl was added and mixed. An equal volume of CTAB extraction buffer [2% (W/V) cetyltrimethylammonium bromide, CTAB; 100 mM Tris-HCl, 1.4 M NaCl, 20 mM EDTA, 1.5% polyvinyl-pyrrolidone, PVP; 0.5% 2-mercaptoethanol] was added and mixed by inverting the tube several times, followed by incubation in a 65 °C water bath for 20–45 min. An equal volume of phenol/chloroform/isoamyl alcohol solution (25:24:1, v/v/v) was added. The tubes were then thoroughly mixed by inversion and subsequently centrifuged at 16,000 ×g for 15 min. Afterwards, the supernatant was transferred to a new sterile tube and an equal volume of chloroform/isoamyl alcohol solution (24:1, v/v) was added. The upper phase was then collected and transferred to a new tube. Following centrifugation (16,000 ×g, 15 min), 3 M NaAc representing 10% of the total volume and 2.5 folds pre-cooled ethanol were added. The samples were then stored at −20 °C for 12 h and subsequently centrifuged (16,000 ×g, 15 min). The supernatant was discarded and the remaining DNA pellet was washed twice with 70% ethanol, and centrifugation (16,000 ×g, 5 min) between each washing. The DNA pellet was air dried and then resuspended in 30 μL of TE buffer and stored at −20 °C. The final DNA concentration and purification were determined by NanoDrop 2000 UV–vis spectrophotometer (Thermo Scientific, Wilmington, USA), and DNA quality was checked by 1% agarose gel electrophoresis.
The primer set 338F (5′-ACTCCTACGGGAGGCAGCA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) targeting the V3-V4 variable regions of the bacterial 16S rRNA gene were used to classify bacteria [41,42]. Each 20 μL PCR mixture contained 4 μL of 5 × FastPfu Buffer, 2 μL of 2.5 mM dNTPs, 0.8 μL of each Primer (5 μM), 0.4 μL of FastPfu Polymerase and 10 ng of template DNA. The PCR reactions were conducted using the following program: 3 min of denaturation at 95 °C, 27 cycles of 30 s at 95 °C, 30s for annealing at 55 °C, and 45 s for elongation at 72 °C, and a final extension at 72 °C for 10 min. Products were purified and recovered by agarosegel electrophoresis. The recovered products were quantified with Pico Green using a QuantiFluor™-ST (Promega, USA), and equimolar concentrations of PCR products for each sample were pooled. The PCR products were extracted from a 2% agarose gel and further purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) and quantified using QuantiFluor™-ST (Promega, USA) according to the manufacturer's protocol. Purified amplicons were pooled in equimolar and paired-end sequenced (2 × 300) on an Illumina MiSeq platform (Illumina, San Diego, USA) according to the standard protocols by Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China). Complete data sets were submitted to the NCBI Short Read Archive (SRA) database (Accession Number: SRP165615).
2.4. Bioinformatic and Statistical Analysis
To facilitate compare the differences of bacterial community structure in different mulberry samples, we created different groups. Considering the influence of seasons and cultivars on the bacterial community structure, a total of 8 groups were generated as follows: 1) SQ: ‘Chuan Sang No.7637’ in spring; 2) SC: ‘Changguo Sang’ in spring; 3) SH: ‘Hong Guo No.2’ in spring; 4) SX: ‘Xin Lunjiao’ in spring; 5) AQ: ‘Chuan Sang No.7637’ in autumn; 6) AC: ‘Changguo Sang’ in autumn; 7) AH: ‘Hong Guo No.2’ in autumn; and 8) AX: ‘Xin Lunjiao’ in autumn. Each of group consists of three biological replicates.
Additionally, 4 groups were built if only taking account into a factor of cultivars. We combined spring and fall community (each of community consists of three biological replicates) together of each cultivar as follows: 1) CSQ: ‘Chuan Sang No.7637’ in the spring and autumn; 2) CGS: ‘Changguo Sang’ in the spring and autumn; 3) XLJ: ‘Xin Lunjiao’ in the spring and autumn; and 4) HGE: ‘Hong Guo No.2’ in the spring and autumn.
Forward and reverse sequences were merged by overlapping paired-end reads using FLASH when the original DNA fragments were shorter than twice the length of reads. Sequencing reads were assigned to each sample according to the unique barcode of each sample. The raw tags were further strictly filtered using the method of Bokulich et al. [43] to obtain clean reads. The quality of the clean reads was analyzed using Quantitative Insights Into Microbial Ecology (QIIME 1.9.1) software package and amplicons matching non-target chloroplast and mitochondria were filtered using Usearch (version 7.0). The obtained sequences were clustered into operational taxonomic units (OTUs) using the UPARSE pipeline (http://www.drive5.com/uparse/) at 97% sequence similarity. Representative sequences for each OTU were selected and the Ribosomal Database Project (RDP) classifier was used to assign taxonomic data to each representative sequence [44,45]. Based on the taxonomic results, alpha diversity was calculated including Chao, Shannon and Simpson indices using Mothur software (version v.1.30.1). Rarefaction curves were also generated using Mothur at a 97% identity level based on these three metrics. Inter-relationships among bacterial communities from different samples were visualized using principal co-ordinates analysis (PCoA) and non-metric multidimensional scaling (NMDS) based on the distance matrix, with calculation of the Bray-Curtis algorithm. The Venn diagram was generated using R script (version R-3.3.1). The histogram was created using Microsoft Excel 2010. A network analysis was performed to assess the complexity of the interactions among the microbial taxa using Networkx software. Finally, community differences for the 5 most abundant bacterial genera distributions were evaluated using the Kruskal-Wallis H test and a one-way analysis of variance (ANOVA), with p values <0.05 considered statistically significant.
3. Results
3.1. Richness and Diversity
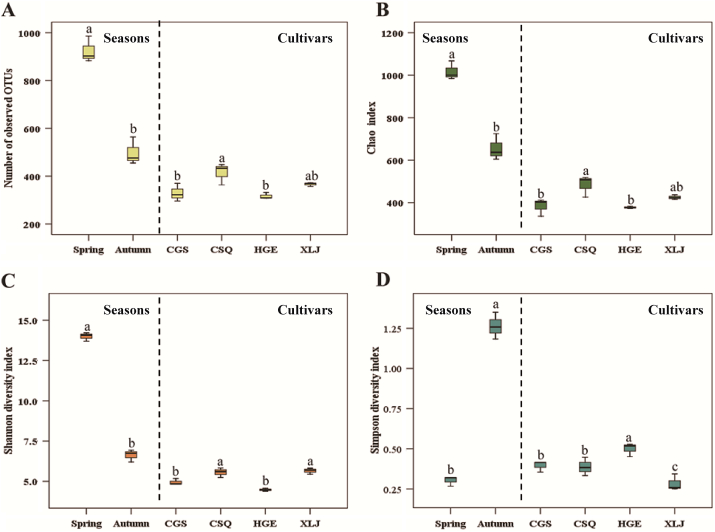
After filtering chimeric sequences and mismatches, a total of 824,427 sequences and 568 different operational taxonomic units (OTUs) were generated from all samples sequenced on an Illumina MiSeq at a 97% similarity level. The rarefaction curves approached the saturation plateau in all samples (Fig. S1), suggesting that our data were large enough to reflect the bacterial diversity present in the samples. The number of OTUs exhibited differences between the spring and autumn samples, where the number of spring OTUs (n = 922) was higher than in the autumn samples (n = 496). Additionally, the number of OTUs in XLJ (n = 364) and CSQ (n = 413) were higher than HGE (n = 314) and CGS (n = 327) (Fig. 1A). These differences were not linked to differences between the cultivars in their level of resistance to mulberry fruit sclerotiniosis. The analysis of richness and diversity indices revealed differences in all of the communities regardless of season or mulberry cultivar. In regards to season, Chao was significantly higher in spring (1018) than in autumn (654) (p < 0.05) (Fig. 1B). Moreover, spring samples exhibited higher diversity (Shannon, 14.00; Simpson, 0.30) than autumn samples (Shannon, 6.62; Simpson, 1.26) (Fig. 1C & D). In regards to cultivar, XLJ (Chao, 425; Shannon, 5.65) and CSQ (Chao, 484; Shannon, 5.56) exhibited higher richness and diversity than CGS (Chao, 384; Shannon, 4.95) and HGE (Chao, 378; Shannon, 4.47) (Fig. 1B & C). These results, however, were also not related to the resistance to mulberry fruit sclerotiniosis exhibited by the different cultivars.
Fig. 1.
Richness and diversity analysis of the endophytic bacterial communities in different seasons and in different mulberry cultivars. Richness based on the number of observed OTUs (A) and Chao index (B), and diversity based on Shannon index (C) and Simpson index (D). CSQ, CGS, XLJ and HGE represent bacterial communities from ‘Chuan Sang No.7637’, ‘Changguo Sang’, ‘Xin Lunjiao’ and ‘Hong Guo No.2’, respectively. Bars with the different letters indicate a significant difference between means by one-way analysis of variance (ANOVA) and least significant difference (LSD) tests (p < 0.05). Values represent the mean. Error bars indicate ± standard deviation.
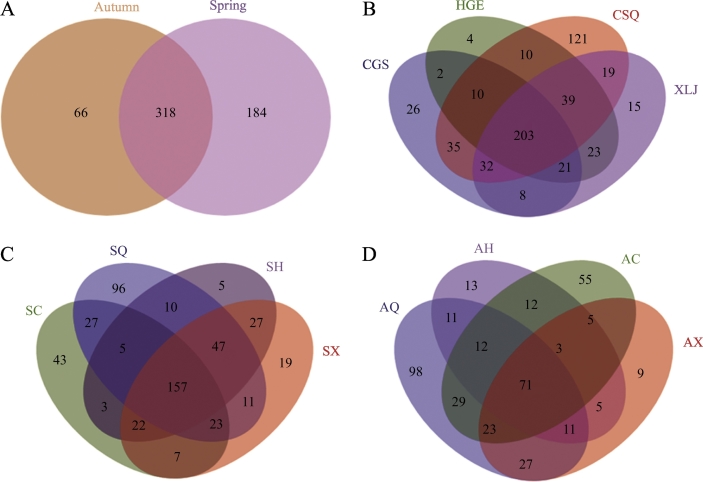
The number of common and unique bacterial OTUs in the different communities is presented in a Venn diagram (Fig. 2). The number of shared OTUs (n = 318) between spring and fall communities collectively (Fig. 2A) was higher than the number (n = 203) shared between different mulberry cultivars (Fig. 2B). Additionally, the number of OTUs shared between different cultivars in the spring (n = 157) (Fig. 2C) was higher than that it was in autumn (n = 71) (Fig. 2D). The number of unique OTUs in the susceptible mulberry cultivars (XLJ, 15; HGE, 4) was lower than the number in the resistant mulberry cultivars (CSQ, 121; CGS, 26). Additionally, the number of OTUs shared between the susceptible cultivars (n = 286) was slightly higher than that of the number shared between the resistant cultivars (n = 280) (Fig. 2B). These data suggest that season and mulberry cultivar are reasons for the observed variation in the composition of the mulberry endophytic bacterial OTUs.
Fig. 2.
Venn diagram of the number of OTUs obtained in different cultivars and in different seasons (spring and fall). Values represent the number of OTUs. (A) grouping by season. Spring and autumn represent spring bacterial communities and autumn bacterial communities, respectively. (B) grouping by cultivar. CSQ, CGS, XLJ and HGE represent bacterial communities from ‘Chuan Sang No.7637’, ‘Changguo Sang’, ‘Xin Lunjiao’, and ‘Hong Guo No.2’, respectively. (C) cultivars in spring. SC, SQ, SX and SH represent bacterial communities from ‘Changguo Sang’, ‘Chuan Sang No.7637’, ‘Xin Lunjiao’, and ‘Hong Guo No.2’ in spring, respectively. (D) cultivars in autumn. AC, AQ, AX and AH represent bacterial communities from ‘Changguo Sang’, ‘Chuan Sang No.7637’, ‘Xin Lunjiao’, and ‘Hong Guo No.2’ in autumn, respectively.
3.2. Taxonomic Composition Analysis
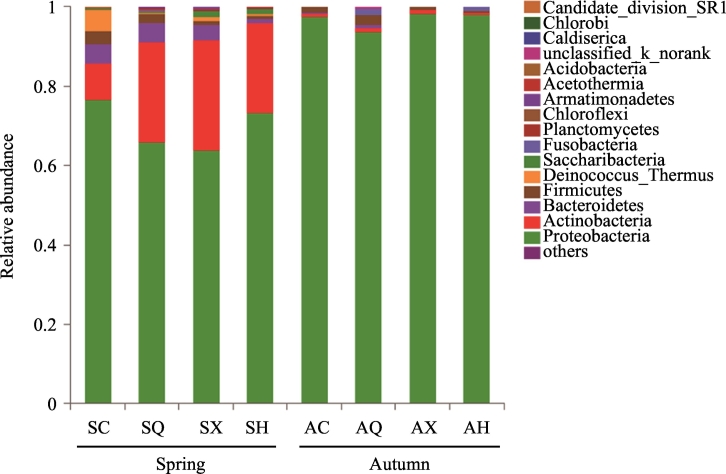
The obtained sequences were classified into 27 phyla, 47 classes, 96 orders, 170 families and 316 genera. Overall, the bacterial composition and distribution were not homogeneous in the spring and autumn samples. The most predominant bacterial phyla were Proteobacteria (69.35%) and Actinobacteria (22.65%) in the spring, collectively accounting for at least 90.00% of the total bacterial population. Only the Proteobacteria were predominant in autumn samples, however, having a percentage >97.00% (Fig. 3). At the order level, members of the Rhizobiales (48.12%) accounted for the highest number of reads in spring samples, with a few families including Methylobacteriaceae, Aurantimonadaceae, and Rhizobiaceae, etc. (Fig. S2). In contrast, Enterobacteriales (55.57%) and Pseudomonadales (37.40%) were the predominant orders in autumn, with primary families represented by Enterobacteriaceae and Pseudomonadaceae (Fig. S2).
Fig. 3.
Relative abundance of endophytic bacteria from different communities at the phylum level. Taxa with an abundance <0.01 are included in “others”. The x-axis represents different communities (cultivars × season) and the y-axis represents the relative abundance of all communities. SC, SQ, SX and SH represent bacterial communities from ‘Changguo Sang’, ‘Chuan Sang No.7637’, ‘Xin Lunjiao’, and ‘Hong Guo No.2’ in spring, respectively. AC, AQ, AX and AH represent bacterial communities from ‘Changguo Sang’, ‘Chuan Sang No.7637’, ‘Xin Lunjiao’, and ‘Hong Guo No.2’ in autumn, respectively. Each column represents the mean of three biological replicates per cultivar.
No obvious difference in bacterial community variations was observed between mulberry cultivars at the phylum level (Fig. 3). The distribution within bacterial communities exhibited some differences, however, at the order level. Notably, Enterobacteriales (CGS, 13.60%; CSQ, 45.25%; XLJ, 17.81%; HGE, 47.41%) and Rhizobiales (CGS, 13.80%; CSQ, 21.67%; XLJ, 25.46%; HGE, 30.65%) were the major groups in all of the mulberry cultivars, but Pseudomonadales (56.69%) was most abundant in the CGS group (Fig. S2). At the family level, the most common families were Enterobacteriaceae (CSQ, 45.25%; HGE, 47.41%) and Methylobacteriaceae (CSQ, 11.91%; HGE, 26.12%) in CSQ and HGE, while CGS mainly harbored Pseudomonadaceae (53.41%), followed by Enterobacteriaceae (13.60%) and Methylobacteriaceae (10.75%) (Fig. S2). Additionally, the abundance of Enterobacteriaceae (17.80%), Methylobacteriaceae (21.38%), and Pseudomonadaceae (28.66%) was homogeneous in XLJ (Fig. S2). These analyses indicated that the composition of the mulberry endophytic bacteria was not associated with resistance to sclerotiniosis.
3.3. Core Genera Distribution
Among the different 568 OTUs across all the samples, a core endophytic microbiome was observed at the genus level. Pantoea and Pseudomonas were the most predominant genera in autumn, with a relative abundance ranging from 11.23% to 89.10% and 1.21% to 79.65%, respectively (Table 1). Methylobaterium was the most abundant genus in the spring, with a relative abundance ranging from 17.59% to 52.37% in different cultivars (Table 1). Notably, genera such as Novosphingobium, Friedmanniella, Modestobacter, and Lysinimonas were detected in spring, but not in the autumn. In contrast, taxa of the unclassified Enterobacteriaceae were only present among the different samples in autumn, but were not observed in spring. In general, the majority of bacteria were readily detected in spring, and the richness of bacterial genera was also higher in the spring except for Pantoea, Pseudomonas, and unclassified Enterobacteriaceae (Table 1).
Table 1.
The relative abundance of the top 20 core genera in each sample.
| Phylum | Genus | Relative abundance (%) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| SH | SX | SC | SQ | AH | AX | AC | AQ | ||
| Proteobacteria | Pantoea | – | – | 16.02 ± 7.56c | 1.31 ± 0.12d | 89.10 ± 4.90a | 22.61 ± 3.36bc | 11.23 ± 13.2c | 84.86 ± 7.21a |
| Pseudomonas | – | – | 1.15 ± 0.19d | 3.56 ± 2.81 cd | 1.28 ± 0.06d | 52.33 ± 0.48b | 79.65 ± 14.13a | 1.21 ± 0.66d | |
| Methylobaterium | 52.37 ± 2.43a | 41.65 ± 3.74a | 17.59 ± 15.23c | 23.95 ± 2.75b | 0.44 ± 0.012d | – | 2.72 ± 0.74d | – | |
| Enterobacter | 0.01 ± 0.01b | – | 0.62 ± 0.34b | 0.20 ± 0.32b | 5.22 ± 5.39a | 2.27 ± 2.71ab | 0.31 ± 0.53b | 2.35 ± 3.33ab | |
| Sphingomonas | 1.88 ± 0.23d | 2.96 ± 0.25bc | 2.58 ± 0.59c | 7.14 ± 0.29a | 0.03 ± 0.02e | – | – | 0.03 ± 0.06e | |
| Aureimonas | 1.14 ± 0.03bc | 0.79 ± 0.09d | 0.07 ± 0.12e | 5.24 ± 0.29a | 0.34 ± 0.07e | 0.19 ± 0.05e | 0.40 ± 0.07e | 0.85 ± 0.12 cd | |
| Stenotrophomonas | 0.63 ± 0.22c | 0.90 ± 0.16bc | 4.13 ± 2.92a | 0.38 ± 0.04c | – | – | – | 0.13 ± 0.23c | |
| Rhizobium | 2.10 ± 0.19c | 3.17 ± 0.41bc | 0.14 ± 0.03d | 5.61 ± 1.56a | – | – | 0.20 ± 0.05d | – | |
| Novosphingobium | 0.72 ± 0.10bc | 0.65 ± 0.13c | 0.16 ± 0.15c | 2.29 ± 0.75a | – | – | – | – | |
| Hymenobacter | 0.22 ± 0.08de | 1.14 ± 0.23c | 3.55 ± 0.91a | 1.81 ± 0.45bc | 0.17 ± 0.29e | – | 0.17 ± 0.17e | – | |
| Peseudomonadales;o_; g_ | 1.73 ± 1.10a | 2.88 ± 1.64a | 5.94 ± 8.06a | 4.75 ± 7.02a | 0.62 ± 0.17a | 1.08 ± 0.63a | 0.49 ± 0.17a | 0.86 ± 0.57a | |
| Aurantimonadaceae;f_; g_ | 3.55 ± 0.03bc | 2.51 ± 0.30 cd | 3.05 ± 2.38c | 5.75 ± 0.87a | – | – | 0.81 ± 0.13d | – | |
| Enterobacteriaceae;f_; g_ | – | – | – | – | – | 8.38 ± 2.90a | 0.21 ± 0.09b | – | |
| Variovorax | 2.42 ± 0.17a | 1.07 ± 0.27c | – | 1.73 ± 0.20b | – | – | – | 0.05 ± 0.09d | |
| Firmicutes | Microbacterium | 1.01 ± 0.43bc | 0.75 ± 0.06c | 0.20 ± 0.04c | 4.05 ± 1.17a | – | 0.27 ± 2.90c | – | – |
| Actinobacteria | Lysinimonas | 0.01 ± 0.00b | 0.01 ± 0.00b | 0.01 ± 0.00b | 0.02 ± 0.00a | – | – | – | – |
| Friedmanniella | 9.89 ± 1.23a | 8.20 ± 1.90a | 1.91 ± 0.45b | 2.52 ± 0.25b | – | – | – | – | |
| Modestobacter | 1.14 ± 0.19b | 2.96 ± 1.11a | 0.15 ± 0.14b | 3.36 ± 0.03a | – | – | – | – | |
| Kineococcus | 0.44 ± 0.16bc | 1.11 ± 0.46ab | 1.20 ± 0.52a | 1.68 ± 0.53a | 0.17 ± 0.07c | – | – | – | |
| Deinococcus-Thermus | Deinococcus | 0.56 ± 0.14c | 1.00 ± 0.17bc | 5.26 ± 3.51a | 0.26 ± 0.07c | – | – | – | 0.02 ± 0.04c |
“f_; g_” and “o_; g_” represent taxa not grouped into any known genera within these families and orders, respectively. SC, SQ, SX and SH represent bacterial communities from ‘Changguo Sang’, ‘Chuan Sang No.7637’, ‘Xin Lunjiao’, and ‘Hong Guo No.2’ in spring, respectively. AC, AQ, AX and AH represent bacterial communities from ‘Changguo Sang’, ‘Chuan Sang No.7637’, ‘Xin Lunjiao’, and ‘Hong Guo No.2’ in autumn, respectively. Data shown are means ± standard deviation of three replicates. Means with different letters are significantly different (p < 0.05).
The relative abundance of the top five core genera was also compared among the four mulberry cultivars with different resistance to mulberry fruit sclerotiniosis and a few differences were observed (Fig. 4). The relative frequency of Pantoea was higher in HGE and CSQ than that in XLJ and CGS, while Pseudomonas was significantly lower in HGE and CSQ (p < 0.05). Additionally, the percentages of Methylobaterium and Friedmanniella in the susceptible mulberry cultivars (XLJ, HGE) were slightly higher than in the resistant mulberry cultivars (CGS, CSQ). Therefore, these primary bacterial genera in mulberry could simultaneously occur in the different resistance level, suggesting there was no obvious relationship between the distribution of major bacterial genera and resistance level of fruit sclerotiniosis.
Fig. 4.
Comparison in the abundance of the top 5 dominant bacterial genera in different seasons and mulberry cultivars. *** indicates a significant difference at p < 0.001, ** p < 0.01, * p < 0.05. The x-axis represents the genus mean proportions and the y-axis represents the top 5 dominant bacterial genera. CGS, CSQ, XLJ, and HGE represent bacterial communities from ‘Changguo Sang’, ‘Chuan Sang No.7637’, ‘Xin Lunjiao’, and ‘Hong Guo No.2’, respectively.
3.4. β-Diversity Analysis
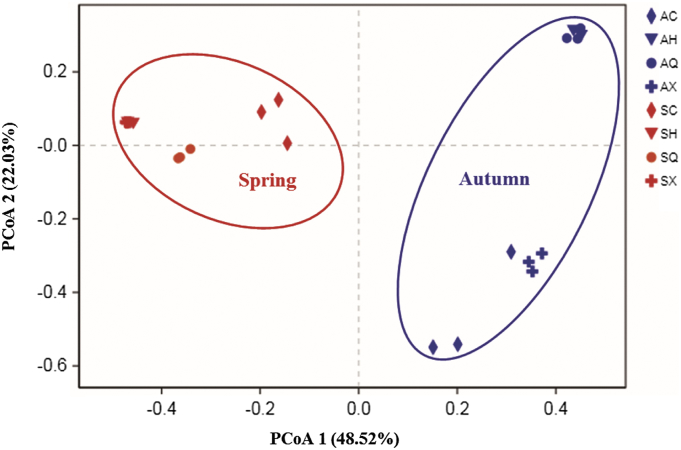
To further compare the relationship of endophytic bacteria populations among the four mulberry cultivars in spring and fall, PCoA analysis was conducted using the OTUs. The PCoA results graphically demonstrated that season was a strong factor in accounting for the observed variations in the composition of the endophytic bacterial community, in which spring samples were placed at a higher PCoA 2 value (22.03%), while autumn samples appeared a higher PCoA 1 value (48.52%) (Fig. 5). The result obtained by the PCoA analysis was also supported by NMDS (Fig. S3). Hierarchical clustering also demonstrated that there was a clustering of endophytic bacteria according to seasons, and that each replicate of a mulberry cultivar clustered together, except the replicates for AH and AQ (Fig. S4). These analyses revealed distinct differences in spring and autumn endophytic bacterial communities, while no clustering was evident due to ratings of susceptible or resistant cultivars.
Fig. 5.
PCoA plot of the relationship between samples on the basis of similarity in the community composition of bacterial OTUs. Two first components (PCoA 1 and PCoA 2) are plotted and represent 70.55% of the variation. SC, SQ, SX and SH represent bacterial communities from ‘Changguo Sang’, ‘Chuan Sang No.7637’, ‘Xin Lunjiao’, and ‘Hong Guo No.2’ in spring, respectively. AC, AQ, AX and AH represent bacterial communities from ‘Changguo Sang’, ‘Chuan Sang No.7637’, ‘Xin Lunjiao’, and ‘Hong Guo No.2’ in autumn, respectively.
3.5. Network Structure of Mulberry Endophytes
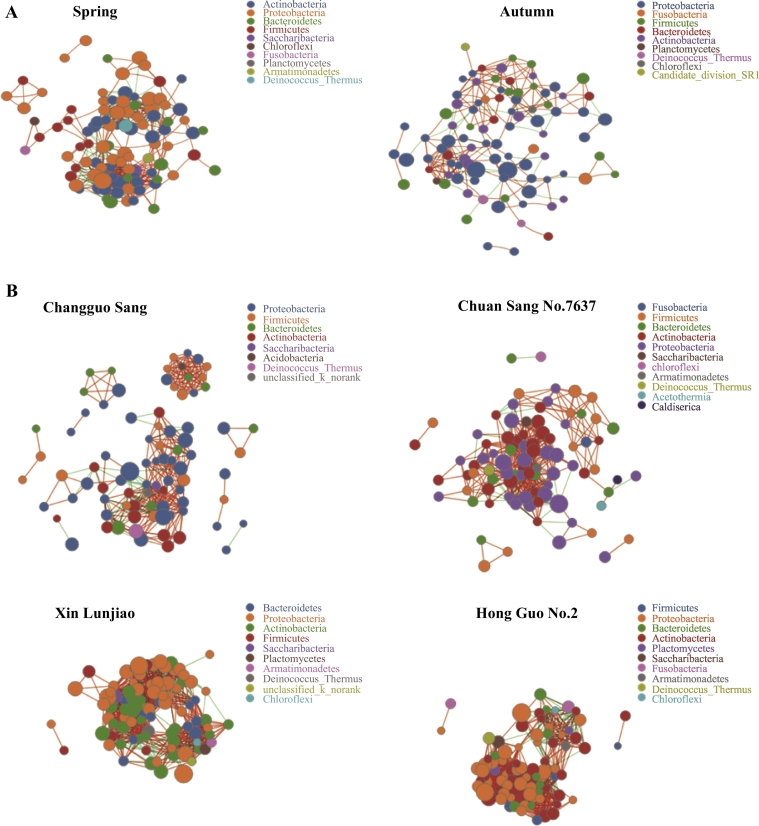
In order to explore the complexity of the interactions within the endophytic communities among the different mulberry cultivars in the two seasons, a correlation network analysis was conducted and its topological properties were calculated. The results of the correlation network analysis revealed a strong difference between the communities based on season. Spring samples exhibited a higher level of complexity and modular structure (Fig. 6A), with a greater number of connections per node (node average degree = 10.70), relative to the autumn samples (node average degree = 4.60) (Table 2). Furthermore, spring samples also presented the higher number of positive correlation (positive edges = 410) in comparison with the autumn samples (positive edges = 176). Additionally, the correlation network analysis of microbial communities for the four mulberry cultivars also revealed differences. The susceptible mulberry cultivars appeared to have a more complex correlation than the resistant mulberry cultivars (Fig. 6B). More specifically, the XLJ (node = 90, edge = 955) and HGE (node = 86, edge = 968) had more node and edge numbers than CSQ (node = 88, edge = 448) and CGS (node = 84, edge = 380) (Table 2). Simultaneously, positive correlation numbers in the susceptible mulberry cultivars (XLJ = 889, HGE = 836) were also higher than in the resistant mulberry cultivars (CSQ = 405, CGS = 326) (Table 2). In summary, the correlation network analyses indicated that the interaction degree of microbial community has been strongly influenced by seasons and cultivars resistance, and the spring samples and the susceptible mulberry cultivars possessed a greater microbial complexity and species abundance than autumn samples and the resistant mulberry cultivars.
Fig. 6.
Correlation network analysis of microbial communities in different seasons (A) and in different mulberry cultivars (B). The size of the node is proportional to the richness of bacteria. Node color corresponds to phylum classification. Edge color represents positive (red) and negative (blue) correlations, and the edge thickness is equivalent to the correlation values.
Table 2.
Correlation network analysis of microbial communities in four mulberry cultivars in two seasons.
| Sampling time |
Mulberry cultivars |
|||||
|---|---|---|---|---|---|---|
| Spring | Autumn | CGS | CSQ | XLJ | HGE | |
| Node numbers | 97 | 93 | 84 | 88 | 90 | 86 |
| Edge numbers | 519 | 214 | 380 | 448 | 955 | 968 |
| Node average degreea | 10.70 | 4.60 | 9.05 | 10.18 | 21.22 | 22.51 |
| Positive edges | 410 | 176 | 326 | 405 | 889 | 836 |
| Negative edges | 109 | 38 | 54 | 43 | 66 | 132 |
The correlation network indices are calculated based on the top 100 genera. CGS, CSQ, XLJ, and HGE represent bacterial communities from ‘Changguo Sang’, ‘Chuan Sang No.7637’, ‘Xin Lunjiao’, and ‘Hong Guo No.2’, respectively.
The average number of connections per node in the network, that is, the node connectivity.
4. Discussion
Plants harbor a high diversity of endophytes, many of which provide numerous benefits that support growth and survival [7,8]. Plant cultivar and environmental conditions also seem to significantly affect the endophytic bacterial community present in a host [29,34]. Exploring the diversity of endophytic bacteria in a given plant species provides a base for understanding their function in the plant.
In the present study, the endophytic bacterial communities of four mulberry cultivars were characterized in the spring and autumn based on Illumina Miseq sequencing of the V3-V4 variable region of bacterial 16S rRNA gene. The ability of the Illumina platform to generate enormous data sets for each sampled community is a distinct advantage [46]. The endophyte enrichment protocol used in this study could introduce some biases in the obtained data, while the plateauing rarefaction curves (Fig. S1) indicated that the libraries were enough large to represent the entire endophytic bacterial diversity present in mulberry.
The mulberry endophytic bacterial community was shown to be highly dynamic. β-diversity analyses indicated that the season was the main determinant of the endophyte community structure in mulberry, followed by host cultivar. Studies of endophytes in elm [36], urban trees [47], and grape [43] have emphasized the key role that season plays in shaping the bacterial community. These studies found that endophytic colonization increased during rainy and warm seasons, which agrees with the findings of the present study. Seasonal variations in the endophyte community could be attributed to the optimal temperature for microbial growth or the changing physiology of the deciduous trees [48]. Wang et al. [26] reported that the succession of microbial communities that occurs in plants during the growing season can be explained by two potential mechanisms, the first of which is related to temporal changes in abiotic conditions such as moisture and temperature, and the second is the changes that occur in plant tissue exudates and photosynthetic products over the course of the growing season.
The physiological characteristics of host plants are dynamic, where many factors affect the species composition of microbial communities. Hardoim et al. [49] revealed that distinct genetic and morphological of host plant cultivar determined, to a large extent, the structure and composition of the different bacterial communities. Lucas et al. [33] investigated the rhizobacterial composition of different common bean (Phaseolus vulgaris) cultivars with variable levels of resistance to the fungal root pathogen Fusarium oxysporum, in which Pseudomonadaceae, Bacillaceae, Solibacteraceae, and Cytophagaceae were more abundant in the resistant cultivar. Whereas, results of this study revealed that Pseudomonadales (Pseudomonadaceae) was inclined to distribute in CGS (resistant cultivar) and XLJ (susceptible cultivar), while Enterobacteriales (Enterobacteriaceae) was mostly occurred in CSQ (resistant cultivar) and HGE (susceptible cultivar), suggesting the structure of mulberry endophytic bacteria seemed no obvious relationship with resistance level (Fig. S2). To further confirm this hypothesis, comparative analysis of core bacteria between different resistant levels was also explored at the genus level, and the result showed that community structures of endophyte appeared to be unrelated to the resistance of the cultivars to mulberry fruit sclerotiniosis (Fig. 4). Interestingly, we found a lower complexity in the network of the resistant cultivars (CGS, CSQ) (Fig. 6B). This phenomenon, however, was contrary with the observation of Lucas et al. [33], in which pointed out the rhizospheric microbiome of disease-resistant bean cultivar showed the highest level of complexity and modular structure than the disease-susceptible cultivar. Different from the rhizosphere environments, endophytic bacteria reside within the interior of plant tissues and whether there is a relationship between the resistance of plant cultivars and the complexity of endophytic community needs to be further explored in the future.
When analyzing the diversity of all of the samples combined at the phylum level, Proteobacteria was the most dominant phylum of endophytic bacteria (Fig. 3), regardless of season or cultivar. Proteobacteria was the dominant bacterial phylum in both spring and autumn suggesting that there is substantial overlap in key community members across different seasons, some of which may be involved in nitrogen fixation [50,51]. Additionally, Actinobacteria also was the second prevalent endophytic bacterial community in spring samples in our study (Fig. 3). This finding is in agreement with earlier reports on poplar [5] and tree peony [22]. Actinobacteria are common in soil can colonize host plant tissue through root hairs in rhizosphere, and then multiply extensively [52]. The presence of a large number of Actinobacteria could potentially enhance plant health by producing antifungal compounds, which may help to increase resistance to mulberry fruit sclerotiniosis in spring.
To use of natural antagonistic microorganisms as an alternative to chemical or physical methods has emerged as a promising strategy for plant disease control because they represent a low risk to human health and have low environmental impact. In recent studies, the cell suspension of Bacillus thuringiensis C25 and the wettable powder of Trichoderma sp. can be successfully used to control the mulberry fruit disease in fields, with the highest disease prevention efficacy reaching up to 84.02% [15,53]. Also, our group isolated an endophytic Bacillus subtilis 7PJ-16 with strong antagonistic activity from mulberry stem in previous work. This strain not only could effectively control mulberry fruit sclerotiniose in the field, but also promote mulberry seed germination as well as mulberry seedling growth under greenhouse conditions [9]. Furthermore, members of the genus Pantoea [54], Methylobacterium [55], and Pseudomonas [56], dominant genera noted in the present study (Table 1; Fig. 4), have been shown to be ideal candidates for biological control agents. Egamberdieva [56] revealed that Pseudomonas and Pantoea provided important benefits to plants by synthesizing phytohormones and improving host stress tolerance. We previously reported that Pantoea agglomerans SWg2 isolated from healthy mulberry roots possessed significant antagonistic activity against Pseudomonas syringae pv. Mori, which causes mulberry bacterial blight [57]. Methylobacterium species also possess the ability to nodulate and fix nitrogen in crops such as sugarcane (Saccharum L.) [51], crotalaria (Crotalaria L.) [55], and scotch pine (Pinus sylvestris L.) [58]. Additionally, Methylobacterium plays an important ecological role in the carbon cycle owing to their ability to metabolize various products of plant decomposition [59]. We hypothesize that mulberry may selectively recruit Pantoea and Pseudomonas species to help them adapt to the adverse environment in autumn by inducing plant systemic resistance and increased stress tolerance. In contrast, Methylobacterium was primarily active in the spring samples when the metabolic activity of mulberry was very high participated in the active metabolism of mulberry in the spring.
5. Conclusions
The current study investigated that endophytic bacterial community of four mulberry cultivars having different resistance to mulberry fruit sclerotiniosis in two seasons (spring and autumn) by high throughput sequencing analysis. The results demonstrated that diversity and community composition of endophytic bacteria in different mulberry cultivars were significantly influenced by season, followed by host cultivar. Proteobacteria was the predominant phylum in both seasons and different mulberry cultivars. Mulberry could recruit majority of endophytes composed of potential functional taxa such as Pantoea, Methylobaterium, and Pseudomonas, which have been shown to be potential candidates for biocontrol agents. The present study significantly enhances our understanding of the factors influencing the community structures of endophytic bacteria and also lays the foundation for conducting research on the use of mulberry endophytes as biological control agents against mulberry diseases. The specific resistance properties or functional traits of mulberry endophytes, however, need to be further explored. Furthermore, culture-dependent methods need to be used to screen the endophytic bacteria as potential biocontrol agents for the control of mulberry fruit scleroiniosis.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgements
The authors acknowledge the financial support of the National Natural Science Foundation of China (31870518 and 31601678) to Jie Xie, and the Fundamental Research Funds for the Central Universities (XDJK2019B047 and XDJK2018D020) to Jie Xie and Ting Ou. Help from Chongqing Sericulture Science Technology Research Institute in providing mulberry samples was greatly appreciated.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2019.07.018.
Contributor Information
Jia Liu, Email: jialiu1983@163.com.
Jie Xie, Email: healthjie@163.com.
Appendix A. Supplementary data
Supplementary material
References
- 1.Hallmann J., Quadt-Hallmann A., Mahaffee W.F., Kloepper J.W. Bacterial endophytes in agricultural crops. Can J Microbiol. 1997;43:895–914. [Google Scholar]
- 2.Strobel G., Daisy B., Castillo U., Harper J. Natural products from endophytic microorganisms. J Nat Prod. 2004;67:257–268. doi: 10.1021/np030397v. [DOI] [PubMed] [Google Scholar]
- 3.de Silva D.A.F., Cotta S.R.C., Vollú R.E., Jurelevicius D.A., Marques J.M. Endophytic microbial community in two transgenic maize genotypes and in their near-isogenic non-transgenic maize genotype. BMC Microbiol. 2014;14:332–340. doi: 10.1186/s12866-014-0332-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu J., Abdelfattah A., Norelli J., Burchard E., Schena L. Apple endophytic microbiota of different rootstock/scion combinations suggests a genotype-specific influence. Microbiome. 2018;6:18–28. doi: 10.1186/s40168-018-0403-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beckers B., De Beeck Op, Weyens N., Boerjan W., Vangronsveld J. Structural variability and niche differentiation in the rhizosphere and endosphere bacterial microbiome of field-grown poplar trees. Microbiome. 2017;5:25–41. doi: 10.1186/s40168-017-0241-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Silva K.J., de Armas R.D., Cláudio R.F.S.S., Ogliari J.B. Communities of endophytic microorganisms in different developmental stages from a local variety as well as transgenic and conventional isogenic hybrids of maize. World J Microbiol Biotechnol. 2016;32:180–189. doi: 10.1007/s11274-016-2149-6. [DOI] [PubMed] [Google Scholar]
- 7.Edwards J., Johnson C., Santos-Medellína C., Lurie E., Podishetty N.K. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc Natl Acad Sci U S A. 2016:E911–E920. doi: 10.1073/pnas.1414592112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lodewyckx C., Vangronsveld J., Porteous F., Moorea E.R.B., Taghavi S. Endophytic bacteria and their potential application. Crit Rev Plant Sci. 2002;21:583–606. [Google Scholar]
- 9.Xu W.F., Ren H.S., Ou T., Lei T., Wei J.H. Genomic and functional characterization of the endophytic Bacillus subtilis 7PJ-16 strain, a potential biocontrol agent of mulberry fruit sclerotiniose. Microb Ecol. 2019;77:651–663. doi: 10.1007/s00248-018-1247-4. [DOI] [PubMed] [Google Scholar]
- 10.Mendes R., Kruijt M., de Bruijn I., Dekkers E., van der Voort M. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science. 2011;332:1097–1100. doi: 10.1126/science.1203980. [DOI] [PubMed] [Google Scholar]
- 11.Kumar V., Gupta V.P. Scanning electron microscopy on the perithecial development of Phyllactinia corylea on mulberry-II, Sexual stage. J Phytopathol. 2004;152:169–173. [Google Scholar]
- 12.Ji X.L., Lu G.B., Gai Y.P., Zheng C.C., Mu Z.M. Biological control against bacterial wilt and colonization of mulberry by an endophytic Bacillus subtilis strain. FEMS Microbiol Ecol. 2008;65:565–573. doi: 10.1111/j.1574-6941.2008.00543.x. [DOI] [PubMed] [Google Scholar]
- 13.Kim T.W., Kwon Y.B., Lee J.H., Youm J.K., Lee H.S. A study on the antidiabetic effect of mulberry fruits. Korean J Seric Sci. 1996;38:100–107. [Google Scholar]
- 14.Hu X.J., Roberts D.P., Jiang M.L., Zhang Y.B. Decreased incidence of disease caused by Sclerotinia sclerotiorum and improved plant vigor of oilseed rape with Bacillus subtilis Tu-100. Appl Microbiol Biotechnol. 2005;68:802–807. doi: 10.1007/s00253-005-1938-x. [DOI] [PubMed] [Google Scholar]
- 15.Razia S., Kangmin K. Bacillus thuringiensis C25 suppresses popcorn disease caused by Ciboria shiraiana in mulberry (Morus australis L.) Biocontrol Sci Technol. 2016;26:145–162. [Google Scholar]
- 16.Boland G.J., Hall R. Index of plant hosts of Sclerotinia sclerotiorum. Can J Plant Pathol. 1994;16:93–108. [Google Scholar]
- 17.Hong S.K., Kim W.G., Sung G.B., Nam S.H. Identification and distribution of two fungal species causing sclerotial disease on mulberry fruits in Korea. Mycobiology. 2007;35:87–90. doi: 10.4489/MYCO.2007.35.2.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu C., Hu X.M., Deng W., Li Y., Han G.M. Soil fungal community comparison of different mulberry genotypes and the relationship with mulberry fruit sclerotionsis. Sci Rep. 2016;6:28365–28374. doi: 10.1038/srep28365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang C.S., Tang X.P., Lei T. Study on the difference of resistance to mulberry sclerotinia disease in different fruit mulberry varieties. Newslett Sericult Sci. 2012;32:10–12. [Google Scholar]
- 20.Blodgett J.T., Swart W.J., Lnow SVdM, Weeks WJ. Soil amendents and water influence the incidence of endophytic fungi in Amaranthus hybrids in South Africa. Appl Soil Ecol. 2007;35:311–318. [Google Scholar]
- 21.Abid U., Adnan A., Luo Q.Q., Aamir H.K., Hakim M. Microbiome diversity in cotton rhizosphere under normal and drought conditions. Microb Ecol. 2018;77:429–439. doi: 10.1007/s00248-018-1260-7. [DOI] [PubMed] [Google Scholar]
- 22.Yang R.X., Liu P., Ye W.Y. Illumina-based analysis of endophytic bacterial diversity of tree peony (Paeonia Sect. Moutan) roots and leaves. Braz J Microbiol. 2017;48:695–705. doi: 10.1016/j.bjm.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bradford M.A., Davies C.A., Frey S.D., Maddox T.R., Melillo J.M. Thermal adaptation of soil microbial respiration to elevated temperature. Ecol Lett. 2008;11:1316–1327. doi: 10.1111/j.1461-0248.2008.01251.x. [DOI] [PubMed] [Google Scholar]
- 24.Nico S., Davide A., Camilla C., Adriano B., Massimo P. Diversity and cyclical seasonal transitions in the bacterial community in a large and deep perialpine lake. Microb Ecol. 2018;76:125–143. doi: 10.1007/s00248-017-1120-x. [DOI] [PubMed] [Google Scholar]
- 25.Emmerling C., Udelhoven T., Schröder D. Response of soil microbial biomass and activity to agricultural de-intensification over a 10 year period. Soil Biol Biochem. 2001;33:2105–2114. [Google Scholar]
- 26.Wang S., Tang J.Y., Ma J., Li X.D., Li Y.H. Moss habitats distinctly affect their associated bacterial community structures as revealed by the high-throughput sequencing method. World J Microbiol Biotechnol. 2018;34:45–58. doi: 10.1007/s11274-018-2436-5. [DOI] [PubMed] [Google Scholar]
- 27.Wang G.H., Xu Y.X., Jin J., Liu J.D., Zhang Q.Y. Effect of soil type and soybean genotype on fungal community in soybean rhizosphere during reproductive growth stages. Plant and Soil. 2009;317:135–144. [Google Scholar]
- 28.Kuffner M., Hai B., Rattei T., Melodelima C., Schloter M. Effects of season and experimental warming on the bacterial community in a temperate mountain forest soil assessed by 16S rRNA gene pyrosequencing. FEMS Microbiol Ecol. 2012;82:551–562. doi: 10.1111/j.1574-6941.2012.01420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pettersson M., Baath E. Temperature-dependent changes in the soil bacterial community in limed and unlimed soil. FEMS Microbiol Ecol. 2003;45:13–21. doi: 10.1016/S0168-6496(03)00106-5. [DOI] [PubMed] [Google Scholar]
- 30.Shi Y.W., Kai L., Chun L., Wang L., Zhao Z.Y. Illumina-based analysis of bacterial diversity related to halophytes Salicornia europaea and Sueada aralocaspica. J Microbiol. 2015;53:678–685. doi: 10.1007/s12275-015-5080-x. [DOI] [PubMed] [Google Scholar]
- 31.Tholozan J.L., Cappelier J.M., Tissier J.P., Delattre G., Federighi M. Physiological characterization of viable-but-nonculturable Campylobacter jejuni cell. Appl Environ Microbiol. 1999;65:1110–1116. doi: 10.1128/aem.65.3.1110-1116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nurul S.M.S., Goh S.Y., Noni A., Rofina Y.O., Chan K.G. Diversity of microbiota associated with symptomatic and nonsymptomatic bacterial wilt-diseased banana plants determined using 16S rRNA metagenome sequencing. World J Microbiol Biotechnol. 2017;33:168–178. doi: 10.1007/s11274-017-2336-0. [DOI] [PubMed] [Google Scholar]
- 33.Lucas W.M., Jos M.R., de Mattias H., Rodrigo M., Siu M.T. Influence of resistance breeding in common bean on rhizosphere microbiome composition and function. ISME J. 2018;12:212–224. doi: 10.1038/ismej.2017.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi Y.W., Yang H.M., Zhang T., Sun J., Lou K. Illumina-based analysis of endophytic bacterial diversity and space-time dynamics in sugar beet on the north slope of Tianshan mountain. Appl Microbiol Biotechnol. 2014;98:6375–6385. doi: 10.1007/s00253-014-5720-9. [DOI] [PubMed] [Google Scholar]
- 35.Smith D.R., Quinlan A.R., Peckham H.E., Makowsky K., Tao W. Rapid whole-genome mutational profiling using next-generation sequencing technologies. Genome Res. 2008;18:1638–1642. doi: 10.1101/gr.077776.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mocali S., Bertelli E., Cello F.D., Mengoni A., Sfalanga A. Fluctuation of bacteria isolated from elm tissues during different seasons and from different plant organs. Res Microbiol. 2003;154:105–114. doi: 10.1016/S0923-2508(03)00031-7. [DOI] [PubMed] [Google Scholar]
- 37.Strobel G., Li J.Y., Ford E., Worapong J., Baird G.I. Pestalotiopsis jesteri, sp. nov. an endophyte from Fragraea bodenii, a common plant in the southern highlands of Papua New Guinea. Mycotaxon. 2000;76:257–266. [Google Scholar]
- 38.Wang H.X., Geng Z.L., Zeng Y., Shen Y.M. Enriching plant microbiota for a metagenomic library construction. Environ Microbiol. 2008;10:2684–2691. doi: 10.1111/j.1462-2920.2008.01689.x. [DOI] [PubMed] [Google Scholar]
- 39.Murray M.G., Thompson W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maropola M.K.A., Ramond J.B., Trindade M. Impact of metagenomic DNA extraction procedures on the identifiable endophytic bacterial diversity in Sorghum bicolor (L. Moench) J Microbiol Methods. 2015;112:104–117. doi: 10.1016/j.mimet.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 41.Mori H., Maruyama F., Kato H., Toyoda A., Dozono A. Design and experimental application of a novel non-degenerate universal primer set that amplifies prokaryotic 16S rRNA genes with a low possibility to amplify eukaryotic rRNA genes. DNA Res. 2014;21:217–227. doi: 10.1093/dnares/dst052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu N., Tan G.C., Wang H.Y., Gai X.P. Effect of biochar additions to soil on nitrogen leaching, microbial biomass and bacterial community structure. Eur J Soil Biol. 2016;74:1–8. [Google Scholar]
- 43.Bokulich N.A., Thorngated J.H., Richardsone P.M., Mills D.A. Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc Natl Acad Sci U S A. 2014;111:E139–E148. doi: 10.1073/pnas.1317377110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun J., Zhang Q., Zhou J., Wei Q. Illumina amplicon sequencing of 16S rRNA tag reveals bacterial community development in the rhizosphere of apple nurseries at a replant disease site and a new planting site. PLoS One. 2014;9 doi: 10.1371/journal.pone.0111744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bodenhausen N., Horton M.W., Bergelson J. Bacterial communities associated with the leaves and the roots of Arabidopsis thaliana. PLoS One. 2013;8 doi: 10.1371/journal.pone.0056329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Friedman J., Alm E.J. Inferring correlation networks from genomic survey data. PLoS Comput Biol. 2012;8 doi: 10.1371/journal.pcbi.1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shu Y.S., Roberta F. Seasonal variation of bacterial endophytes in urban trees. Front Microbiol. 2015;6:427–440. doi: 10.3389/fmicb.2015.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adams P.D., Kloepper J.W. Effect of host genotype on indigenous bacterial endophytes of cotton (Gossypium hirsutum L.) Plant and Soil. 2002;240:181–189. [Google Scholar]
- 49.Hardoim P.R., Andreote F.D., Reinhold-Hurek B., Sessitsch A., van Overbeek L.S. Rice root-associated bacteria: insights into community structures across 10 cultivars. FEMS Microbiol Ecol. 2011;77:154–164. doi: 10.1111/j.1574-6941.2011.01092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun L., Qiu F.B., Zhang X.X., Dai X., Dong X.Z. Endophytic bacterial diversity in rice (Oryza sativa L.) roots estimated by 16S rDNA sequence analysis. Microb Ecol. 2008;55:415–424. doi: 10.1007/s00248-007-9287-1. [DOI] [PubMed] [Google Scholar]
- 51.Quecine M.C., Araújo W.L., Rossetto P.B., Ferreira A., Tsui S. Sugarcane growth promotion by the endophytic bacterium Pantoea agglomerans 33.1. Appl Environ Microbiol. 2012;78:7511–7518. doi: 10.1128/AEM.00836-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kay E., Bertolla F., Vogel T.M., Simonet P. Opportunistic colonization Ralstonia solanacearum infected plants by Acinetobacter sp. and its natural competence development. Microb Ecol. 2002;43:291–297. doi: 10.1007/s00248-002-2007-y. [DOI] [PubMed] [Google Scholar]
- 53.Ye M.Q., Yue H.L., Luo G.Q., Yang Q., Kuang Z.S. Effect of a fungal pathogen, Trichoderma hamatum, on growth and germination of Ciboria carunculoides under laboratory conditions. Pak J Zool. 2014;46:1377–1384. [Google Scholar]
- 54.Sammer U.F., Reiher K., Spiteller D., Wensing A., VÖlksch B. Assessment of the relevance of the antibiotic 2-amino-3-(oxirane-2,3-dicarboxamido)-propanoyl- valine from Pantoea agglomerans biological control strains against bacterial plant pathogens. Microbiol Open. 2012;1:438–449. doi: 10.1002/mbo3.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abdoulaye S.Y., Giraud E., Jourand P., Garcia N., Willems A. Methylotrophic Methylobacterium bacteria nodulate and fix nitrogen in symbiosis with legumes. J Bacteriol. 2001;183:214–220. doi: 10.1128/JB.183.1.214-220.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Egamberdieva D. Alleviation of salt stress by plant growth regulators and IAA producing bacteria in wheat. Acta Physiol Plant. 2009;31:861–864. [Google Scholar]
- 57.Xie J., Shu P., Strobel G., Chen J., Wei J.H. Pantoea agglomerans SWg2 colonizes mulberry tissues, promotes disease protection and seedling growth. Biol Control. 2017;113:9–17. [Google Scholar]
- 58.Pirttilä A.M., Laukkanen H., Pospiech H., Raili Myllylä R., Hohtola A. Detection of intracellular bacteria in buds of scotch pine (Pinus sylvestris L.) by in situ hybridization. Appl Environ Microbiol. 2000;66:3037–3077. doi: 10.1128/aem.66.7.3073-3077.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pini F., Frascella A., Santopolo L., Bazzicalupo M., Biondi E.G. Exploring the plant-associated bacterial communities in Medicago sativa L. BMC Microbiol. 2012;12:78–88. doi: 10.1186/1471-2180-12-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material