Abstract
MicroRNAs (miRNAs) are a family of endogenous, small (approximately 22 nucleotides in length), noncoding, functional RNAs. With the development of molecular biology, the research of miRNA biological function has attracted significant interest, as abnormal miRNA expression is identified to contribute to serious human diseases such as cancers. Traditional methods for miRNA detection do not meet current demands. In particular, nanomaterial-based methods, nucleic acid amplification-based methods such as rolling circle amplification (RCA), loop-mediated isothermal amplification (LAMP), strand-displacement amplification (SDA) and some enzyme-free amplifications have been employed widely for the highly sensitive detection of miRNA. MiRNA functional research and clinical diagnostics have been accelerated by these new techniques. Herein, we summarize and discuss the recent progress in the development of miRNA detection methods and new applications. This review will provide guidelines for the development of follow-up miRNA detection methods with high sensitivity and specificity, and applicability to disease diagnosis and therapy.
Keywords: MicroRNAs, Detection method, Amplification strategies, Nanomaterial-based methods, Nucleic acid amplification-based methods
1. Introduction
MiRNAs are small non-coding RNAs of about 19–24 nucleotides, which inhibit the translation of proteins by binding to 3′-UTR of the target mRNA. Some studies have also found that miRNAs can interact directly with proteins to regulate gene expression [1] or influence the epigenetic mechanisms [2]. As a non-invasive blood biomarker, miRNAs are relatively stable and easy to detect.
Considering the critical role that miRNAs play in gene regulation and biological function, the specific and sensitive detection of miRNAs is becoming more and more important. However, the short sequence lengths, large variability in per-cell copy number, and high sequence similarity within families of expressed miRNAs conspire to make them challenging analytical targets [3]. Over the past decades, significant efforts have been invested to develop new detection methods [[4], [5], [6], [7]]. Previous review articles have provided valuable information about the evolution of various analytical methods for miRNA determination [8,9]. In this review article, we summarize the recent progress in miRNA analysis. Current miRNA detection techniques can be divided into two categories: traditional methods and new technology methods. Traditional methods for miRNA detection include northern blotting [[10], [11], [12]], microarray analysis [13,14] and quantitative polymerase chain reaction (qPCR) [15,16]. For new technology methods, in order to improve the sensitivity and selectivity of miRNA detection, these methods always rely on signal amplification strategies, such as nanoparticle-based amplification [[17], [18], [19], [20], [21]], isothermal exponential amplification [[22], [23], [24]], rolling circle amplification (RCA) [[25], [26], [27]], hybridization chain reaction (HCR) [[28], [29], [30]] and combination of these amplification strategies [[31], [32], [33]]. Herein, we summarize some of them and discuss their advantages and limitations for improving miRNA detection design.
2. Traditional methods
Methods for quantification and visualization of aberrant miRNA expression are urgently needed for early clinical diagnosis. Owing to their small sizes, high sequence homology among family members, and low abundance, it is a challenging work. In traditional methods, northern blotting is regarded as the gold standard, although it is poorly sensitive (nanomoles level), time-consuming and requires a large number of RNA samples. The microarray-based technique has the advantage of high throughput and multiplexing capacity, but poor sensitivity and a lengthy hybridization time restrict its wide applications. Moreover, qPCR, including stem-loop reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) and poly(A)-tailed RT-qPCR, are the most widely used miRNA analysis techniques, both of which have the advantage of high sensitivity and specificity. Unfortunately, these techniques are complex and require special laboratory skills. In addition, false-positives may be generated during the amplification process.
2.1. Northern blotting
Northern blotting is the standard and most widely used method for detecting miRNAs. It can be used not only for the detection of mature miRNAs, but also its precursors. It does not need specialized equipment. The basic principle is as follows: the RNA sample is digested by a restriction endonuclease, separated by agarose gel electrophoresis, denatured and transferred to a nitrocellulose film or nylon membrane according to its position in the gel, and fixed followed by reaction with the isotope or other marker labeled probes [34,35]. After washing the free probe, miRNAs can be detected by autoradiography or other suitable techniques.
The relative molecular size and relative abundance of miRNAs can be detected by northern blotting, but it also has many shortcomings, such as semi-quantitative, low throughput, cumbersome, time-consuming, and easy degradation of RNA. As a result, it requires a stringent experimental condition. Northern blot has low sensitivity, by which low molecular weight RNAs cannot be detected. Specific probes labeled with radioisotopes will help to increase the sensitivity but will increase the risk of the reaction at the same time.
In view of the above shortcomings, there are many improvements emerging in recent years. Locked nucleic acid (LNA) is a kind of oligonucleotide derivative, which is a rigid structure formed by linking the 2′ and 4′ carbons on the ribose through the methylene group. This kind of nucleic acid can increase the melting temperature (Tm value) of the primer or probe. As a probe, LAN greatly improves the thermal stability of hybridization sequences, hybridization efficiency and sensitivity of miRNA determination [[36], [37], [38]]. Varallyay et al. [12] used LNA instead of DNA oligonucleotide probes to improve the thermal stability of the complex between target miRNA and oligonucleotide probes, reducing dissociation, improving the experimental sensitivity and shortening the experimental time. Kim et al. [11] used non-radiolabeled probes to reduce the risk of the experiment. Pall and Hamilton used 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) to cross-link RNA to the appropriate membrane to increase the speed of the experiment [39]. Despite these improvements, northern blotting is inadequate as a quantitative detection of miRNAs for the lack of sensitivity and the inability to detect low levels of RNAs in some cells.
2.2. Real-time qPCR
Real-time qPCR has become a routine and reliable technique for detecting miRNA expression because of its large dynamic range, high sensitivity, and high sequence specificity [40]. Real-time qPCR can be considered as a single gold standard among miRNA detection techniques. In such methods, target miRNA was firstly transferred into cDNA by reverse transcription. Afterward, the PCR was performed to achieve real-time fluorescence detection. cDNA synthesis by reverse transcription can commonly use stem-loop primer [15,41,42], poly(T) adapter [16,43] or a gene-specific primer (GSP) that includes a tail sequence [44], while two fluorescent methods are used for monitoring miRNA qPCR [45]: TaqMan probe method [15,16,41] and SYBR Green fluorescent dye method [44]. SYBR Green is an intercalating fluorescent dye between dsDNAs which binds to dsDNAs and increases the fluorescence signal by 800–1000 times. However, SYBR Green can't recognize nonspecific products such as primer dimers, which reduces the accuracy of qPCR quantification of specific amplification products. So the dissociation curve analysis is used to monitor qPCR product homogeneity. The Tm value of dsDNA depends on its length and base composition, while the number of inflection points on the melting curve indicates the number of PCR products, including primer dimers. Therefore, the acceptable dissociation curve should have only one inflection point, while multiple inflection points indicate the presence of nonspecific products. TaqMan probes are oligonucleotide probes labeled with a reporter group at its 5′ end and a fluorescent quenching group at the 3′ end, whose binding sites are between the two primers. Primer dimers or other nonspecific amplicons in this method will not produce any fluorescent signal, and it has a higher specificity and reproducibility than SYBR Green method.
The real-time qPCR method is highly sensitive, but its false-positive anxiety and difficulty in primer design limit its use. The exact quantification of real-time qPCR depends on the interconnection of multiple steps, and each step needs to be optimized. To obtain meaningful and repeatable results, several parameters such as RNA extraction, RNA integrity control, cDNA synthesis, primer design, amplicon detection, and data normalization must be taken into account [46].
Mohammadi-Yeganeh et al. [15] developed a new and cost-effective two-step real-time qPCR for miRNA detection. The first step was to reverse transcriptase miRNAs by stem-loop real-time primers. The second step was to amplify the reverse transcripts using the TaqMan real-time qPCR assay. This stem-loop structure can be used not only to detect miRNAs that are not found by microarrays but also to detect other small non-coding RNA, such as piRNA and siRNAs. This method combines computer prediction with bioassay for the first time, enabling high throughput screening of a wide range of miRNA.
Gan et al. [40] compared the detection of miRNAs between northern blotting and real-time qPCR. In northern blotting, the amount of RNA can be directly quantified on a membrane by gel electrophoresis. And it has been shown to be superior in studying the degradation and transcription of RNA. However, northern blotting has requirements for the length of cDNA probe and the amount of RNA. So it is not suitable for weak gene detection or large-scale screening. The main advantages of real-time qPCR compared to northern blotting are greater sensitivity and specificity, as well as a broader quantification range. Its main problems are RNA template variability, inappropriate experimental design, inconsistent data analysis, and inappropriate data normalization.
2.3. Microarray technology
The microarray is the most widely used method for rapid and high throughput detection of miRNAs [13,47,48]. For this method, the sample RNA is produced by reverse transcription using a labeled probe, and these fluorophores or biotin-labeled cDNAs are detected using solid-phase oligonucleotides having the same sequence as the target miRNA. The labeled cDNA sample is loaded into each well followed by a series of washing steps to remove free DNAs. If the hybridized cDNA is biotinylated, the streptavidin-labeled fluorophore can be labeled; and if the cDNA has been labeled with the fluorophore, the fluorescence intensity of each well can be measured directly. The fluorescence intensity of each well can be used to determine the expression level of miRNAs.
Although the microarray can analyze thousands of samples a day, the cost is very high [14]. And the method also faces many challenges as follows: too short miRNAs and low copy number miRNA cannot be detected, and the specificity of analyzing the miRNA with similar sequences is not so good.
3. Newly developed methods
3.1. Nanomaterial-based miRNA detection
Recently, various nanomaterials have been applied within studies for miRNA detection, such as gold nanoparticles (AuNPs) [18,19,49] [50], magnetic nanoparticles [17,51], silver nanoclusters (AgNCs) [[52], [53], [54]], and quantum dots (QDs) [55]. Due to their high surface area, excellent electrical conductivity, and remarkable chemical stability, nanomaterials can be strong tools to improve performances of classical detection methods. Moreover, their powerful versatility of cellular transfection, excellent photostability and low immunogenicity should be taken into account when it refers to in vivo imaging. However, their inherent cytotoxicity and self-aggregation inside living cells are still problems for the stable application. Further efforts are made to overcome these deficiencies and explore the application of nanobiosensors. In this review, we summarize amplification methods based on nanomaterials (Table 1).
Table 1.
MiRNA amplification methods based on nanomaterials.
| Nanobio-sensors | Features | Application | Analyst | LOD |
|---|---|---|---|---|
| Nanostructured gold | Strong surface-plasmon resonance absorptions; High extinction coefficients; Target-triggered aggregation of AuNPs, resulting in a color change of the AuNP solution from red to purple. |
Immobilization of AuNPs on laccase-loaded poly-dopamine NPs | miRNA-21 | 70 pM [50] |
| AuNPs functionalization with thiolated DNA oligonucleotides | miRNA-10b | 100 aM [18] | ||
| Surface functionalization of Multi-walled carbon nanotubes with AuNPs | miRNA-155 | 33.4 fM [56] | ||
| Cysteine capped gold nanoclusters | miRNA-155 | 60 fM [57] | ||
| Nanostructured silver | High biocompatibility; Excellent photostability; Tunable luminescence, and subnanometer size; Enhanced fluorescence placed in close proximity with guanine-rich sequences. | DNA-Templated AgNCs | miRNA-21 | 38 pM [52] |
| Gold and silver nanorod/thionine/complementary DNA composite | miRNA-155 | 1 pM [58] | ||
| A specific architecture of nitrogen-doped functionalized graphene, AgNPs, and polyaniline | miRNA-21 | 0.2 fM [54] | ||
| Nanostructured copper | Rapid and easy synthesis; Excellent non-toxicity and biocompatibility |
Poly (thymine)-templated fluorescent CuNPs | miRNA-141 | 0.27 fM [20] |
| Oligonucleotide-templated copper nanoclusters | miRNA-155 | 2.2 pM [59] | ||
| MoS2 nanosheets decorated with a copper ferrite (CuFe2O4) | miRNA-205 | 0.48 pM [60] | ||
| Carbon nanomaterials | Low cost, high surface area, excellent electrical conductivity, remarkable chemical stability, and strong mechanical strength; Better performance in hardness and heat resistance. |
N-Carboxymethyl chitosan (NCS)/Mo2C nano-complex | miRNA-21 | 0.34 fM [61] |
| Multiwall carbon nanotubes/graphene oxide nanoribbons | miRNA-21 | 0.034 fM [62] |
Nanostructured gold, such as gold nanoparticles (AuNPs) and gold clusters, possessing strong surface-plasmon resonance absorptions and high extinction coefficients, has been found most widely applied in the development of analytical methods [56,57]. AuNPs-based colorimetric assays, without the aid of advanced instrumentation, offer a cost-effective, rapid, and convenient option for miRNA detection. AuNPs play important roles in the fabrication of sensing substrate to bind target capture probes through Au–S bond. Target-triggered aggregation of AuNPs, resulting in a color change of the AuNPs solution from red to purple, in response to the surface plasmon resonance absorption of dispersed and aggregated nanoparticles, serves as the basis for the optical detection. Persano et al. [18] described a new technology for specific and highly sensitive colorimetric miRNA detection in biological samples, which is based on the isothermal nicking enzyme amplification reaction and subsequent hybridization of the amplification product with gold nanoparticles and magnetic microparticles (barcode system). Similar to this, Li et al. [19] reported an ultrasensitive miRNA detection via isothermal exponential amplification reaction (EXPAR)-assisted AuNP amplification, which was inspired by the design of Park et al. [20]. Based on the method, they achieved the quantification of miRNAs in a linear range from 50 fM to 10 nM with a detection limit of 46 fM. What's more, this method has the ability to discriminate a single-nucleotide difference between homologous miRNAs.
Silver nanoclusters (AgNCs), as new signal transducers, have received massive interest because of their intrinsic merits of high biocompatibility, excellent photostability, tunable luminescence, and subnanometer size [52,58]. Different from AuNPs, the use of DNA-AgNCs avoids the covalent connection between AgNCs and indicators such as quantum dots [63]. The reason is that these AgNCs can be “lightened up” with 500-fold enhanced fluorescence once they are placed in close proximity with guanine-rich sequences [64]. Some enzyme-involved isothermal amplification strategies have been developed to achieve sensitive detection of miRNAs [27,65]. However, AgNCs are sensitive to surrounding protein enzymes and concomitant enzymatic reaction conditions, which may result in poor repeatability. To overcome the drawback, enzyme-free amplification procedures like CHA [66] and SDA [67] were introduced into the AgNCs related detection. In this way, the sensitivity and selectivity of the methods were improved with better stability.
Nanostructured copper such as copper nanoparticles (CuNPs), which have an advantage over AuNPs and AgNCs due to their rapid and easy synthesis, is also widely concerned [60,68]. Besides, CuNPs exhibit excellent non-toxicity and biocompatibility, which meet the need for biological sample analysis. Park et al. [20] devised a novel method for rapid and ultrasensitive detection of miRNAs by employing target-assisted isothermal exponential amplification (TAIEA) combined with poly (thymine)-templated fluorescent CuNPs as signaling probes. Borghei et al. [59] successfully detected miR-155 in spiked human serum samples via oligonucleotide-templated copper nanoclusters based on the shifts in the fluorescence emissions.
Recently, carbon nanomaterials have also been widely applied in the detection of miRNA, mainly including graphene and carbon nanotubes (CNTs) [62]. They have better properties than other materials in hardness, mechanical, heat resistance, and electrical conductivity [69]. A lot of carbon nanomaterials-based miRNA sensors have been exploited these years. However, the efficiency in the biological environment of these sensors is satisfying due to the poor solubility of elemental carbon. For these reasons, graphene oxide (GO) becomes a more widely used electrode material, whose surface possesses many carboxylic acid and hydroxyl groups [70] to improve its water-solubility. Many strategies have incorporated GO into the design of miRNA detection biosensors [58,71,72]. To meet the need of immobilization of biological molecules with high biological activity remained, CNTs were used because of their large surface area ratio and intertube attraction energy [73]. Wang et al. [62] immobilized ssDNA capture probes on the surface of multiwall carbon nanotubes/graphene oxide nanoribbons (MWCNTs/GONRs) modified electrode, together with the duplex-specific nuclease (DSN) assisted target recycling, and achieved monitoring of miRNA-21 with a limit of 0.034 fM.
Apart from what has been indicated above, there are an increasing number of modified nanoparticle complex (TiO2-α-Fe2O3 heterojunction, NCS/Mo2C nano-complex, etc) with different features, which offer new strategies for detection of miRNAs and other biomolecules in various systems [18,57,61,74].
3.2. Nucleic acid amplification techniques
To improve the sensitivity of miRNA detection methods, nucleic acid amplification techniques are usually utilized. In particular, numerous new technologies based on isothermal amplification have emerged due to their wider applicability to the point of care testing devices compared to conventional PCR-based assays. Nucleic acid amplification techniques are needed for the low content of miRNAs in tissues and cells. A multitude of nucleic acid amplification methods have been exploited, such as rolling circle amplification (RCA), duplex-specific nuclease (DSN)-based amplification, loop-mediated isothermal amplification (LAMP), exponential amplification reaction (EXPAR), strand-displacement amplification (SDA) and some enzyme-free amplifications. They can be easily applied to the real-time based assay by employing SYBR Green as fluorescent DNA-intercalating dyes. However, SYBR Green dyes can decrease amplification efficiency by the dose-dependent manner and are vulnerable to nonspecific amplification [75]. In order to detect amplified products specifically, some probes have been exploited. As we know, TaqMan probes [76] were under consideration for direct validation of amplification for a period of time, but they were dependent on Taq exonuclease activity, while many DNA polymerases did not possess this activity. In contrast to TaqMan, molecular beacons (MBs) [77] and one-step strand displacement (OSD) reporters [78] can get a better performance for the detection of sequence-specific nucleic acid amplicons with false-positive products excluded.
3.2.1. RCA-based methods for miRNA detection
RCA has become increasingly popular in miRNA detection due to its simplicity, specificity, and high sensitivity. In most cases, miRNA works as a ligation template, and the padlock probe will respectively hybridize with the target miRNA which will be ligated by T4 RNA ligase or SplintR enzyme, forming a circular ssDNA, followed by extension around the circle with an external primer or miRNA itself as a primer [[79], [80], [81]], ultimately displacing the conjoined miRNA and continuing to produce long cascaded nucleic acid products. The nucleic acid products can be detected by a variety of signal readout techniques, such as fluorescence, colorimetry, electrochemistry, electrochemiluminescence, SERS, and LAMP. Interestingly, the concatamers are so massive that they can accumulate and be detected as discrete single molecules on surfaces.
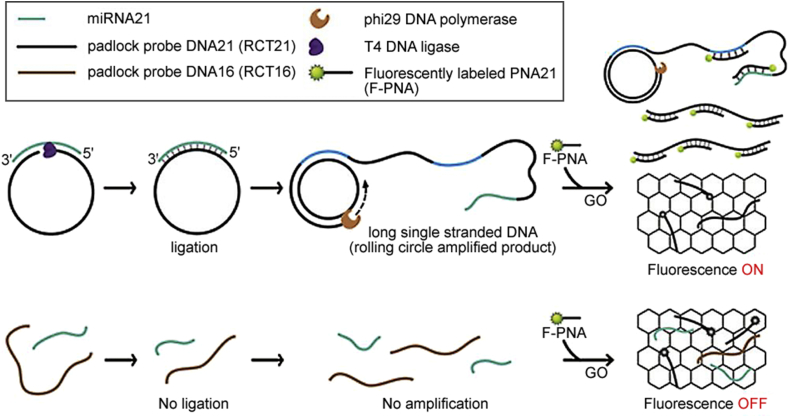
Due to its good specificity and sensitivity, many methods were designed for rapid and sensitive miRNA expression analysis based on RCA. For example, Yang et al. [25] described an electrochemical strategy for ultrasensitive and specific detection of microRNA based on multicomponent nucleic acid enzyme-mediated rolling circle amplification on a gold electrode. Hong et al. [26] presented a simple and convenient protocol for the quantitative detection of miRNA based on RCA, graphene oxide (GO), and fluorescently labeled peptide nucleic acid (F-PNA) (Fig. 1). How to design the probes would affect the amplification efficiency of the RCA experiment. Zhou et al. [82] designed a dumbbell probe, only in the presence of specific miRNA targets, RCA can be initiated. This strategy allowed quantification of miRNA with an extremely low concentration in RNA samples. Xu et al. [80] developed a sensitive and specific fluorescence method based on the combination of RCA and SDA for the detection of let-7a miRNA, which utilized multifunctional molecular beacon (MMB) to execute SDA without the help of any nucleic acid.
Fig. 1.
Schematic illustration of the miRNA detection process using the graphene oxide (GO)-based fluorometric assay combined with RCA-based miRNA amplification. (Reprint from Ref. [26] with permission from the American Chemical Society. Copyright 2016.)
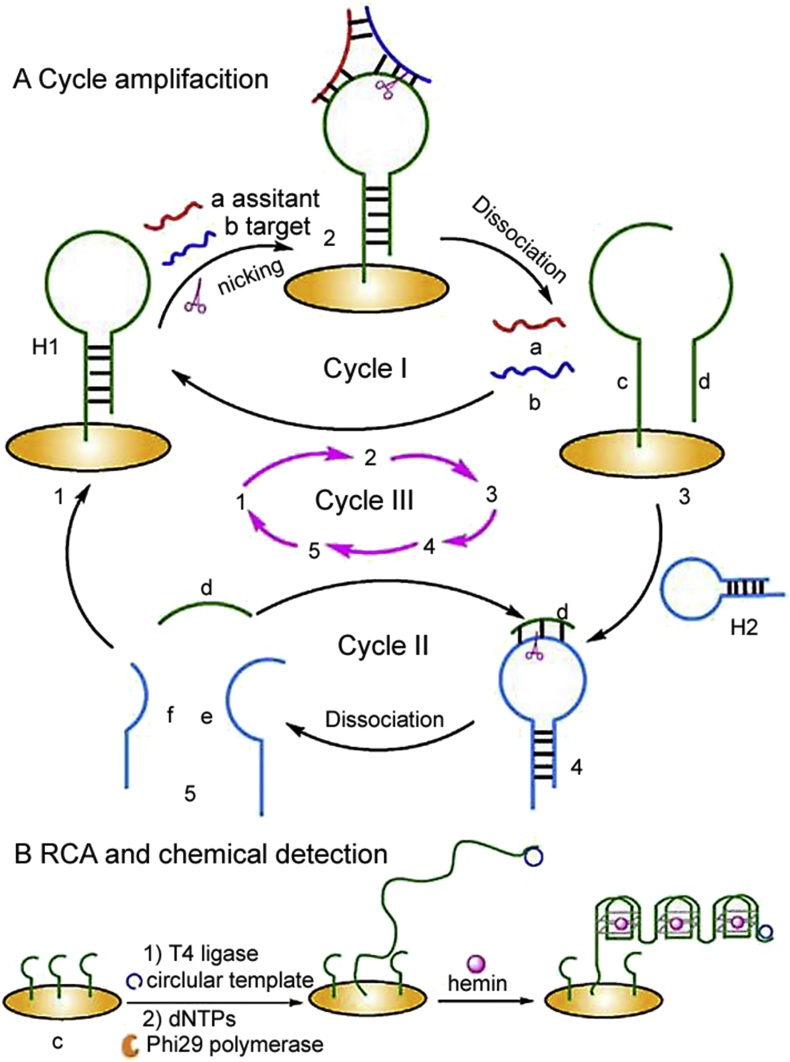
In order to improve the sensitivity and specificity of miRNA detection, nicking endonucleases (NEases) were introduced into the RCA process [83]. NEase recognized restriction enzyme sites and specifically cleaved one strand of double-stranded DNA or a DNA strand in a DNA/RNA hybrid strand [84]. Via introducing a palindromic fragment-integrated recognition site for nicking endonuclease Nt. AlWI into the padlock probe, the hybridization of target miRNA can continuously generate the nicked fragments which would be incorporated into strand-displacement amplification. Using this method, Xu et al. [81] detected target miRNA down to 5 pM successfully. Song et al. [85] also developed a biosensor for ultrasensitive let-7d miRNA detection based on NEase assisted template enhanced hybridization process cooperated with RCA. As shown in Fig. 2, in the presence of NEase and target miRNA, the“Y” structure of designed probe dissociated; consequently the RCA reaction can be started with the free part. Three cycles were included in the system, which would increase the sensitivity obviously. This paper also offers an idea about how to produce a reliable recognition site in the hybridization of miRNA and hairpin probe, which is introducing an assistant single-stranded DNA (ssDNA) to form a stable “Y” junction structure.
Fig. 2.
Cyclic scheme of assistant probe mediated nicking endonuclease signal amplification assay for miRNA detection through the formation of Y-shaped junction and G-quadruplex/hemin complex. (Reprint from Ref. [85] with permission from the Elsevier. Copyright 2016.)
3.2.2. DSN-based methods for miRNA detection
DSN can hydrolyze DNA in DNA/RNA or double-stranded DNA (dsDNA) in a specific length, regardless of the nucleotide sequence, and does not cleave ssDNA or RNA [86]. Based on this distinct characteristic of DSN, miRNAs can be recycled in the reaction, and thermal amplification was achieved. Le et al. [87] developed a miRNA-21 detection method with DSN-based amplification, which showed excellent sensitivity and selectivity (one base mismatch discrimination). DSN-based amplifications were compatible with various platform such as colorimetric, fluorescent and electrochemical assays.
Colorimetric assay usually analyzes the color change exhibited by the testing solution. The result can be monitored by the naked eye or a colorimeter. It provides a quick and easy choice for miRNA detection without the needs of any advanced instrumentation. Zhang et al. [88] designed an ssDNA chimeric probe which consisted of two main regions: sequence for target miRNA recognition at the 5′ end and CHA initiator at the 3'end. In the presence of target miRNA, a large number of ssDNA chimeric probes were cleaved to produce CHA initiator sequence fragments. The G-rich sequence produced in the CHA process bound hemin to bring a horseradish peroxidase activity, which catalyzed a colorimetric reaction. The application of DSN allowed the target miRNA to be recycled, as it can react with the remaining probes. The arranged method allowed quantitation of the sequence specificity of miRNA-21 with a detection limit of 9.2 fM in a dynamic range of 10 fM-1 nM, with an excellent ability to distinguish homological miRNAs.
Fluorescence-based detection is the most common method utilized in biosensing because of its high sensitivity, simplicity, and diversity. It has great improvements in sensitivity and automation compared with the former colorimetric assays. A fluorescence biosensor model for miRNA quantification combines DSN with MB probes whose opposite arms are labeled with a fluorophore and a quench. In the presence of the target miRNA, the MB probe binds to the target and the stem-loop structure of MB is replaced by a more stable duplex structure, producing a fluorescence signal as a result of the broader distance between the fluorophore and quench. However, there are still several problems to be solved. DSN digests not only DNA in DNA/RNA but also dsDNA with poor selectivity. Lin et al. [89] found that DNA-MB was digested by DSN even in the absence of target miRNA, resulting in a 2.5-fold increase in fluorescence intensity. Backbone-modified (2-OMe-RNA modified on its stem) molecular beacons can remain undigested by DSN. This method obtained a linear response between 0.5 pM and 500 pM, with a lower detection limit of 0.4 pM. As a matter of fact, it will produce additional cost and complicated synthesizing procedure. Aiming to overcome the limitation, Ma et al. [90] presented a short-probe-based duplex-specific nuclease signal amplification strategy to detect miRNAs. By simply shortening the length of the DNA probe, the strategy remarkably improved the specificity without the loss of amplification efficiency at 37 °C; thereby, imaging in a living cell can be conducted.
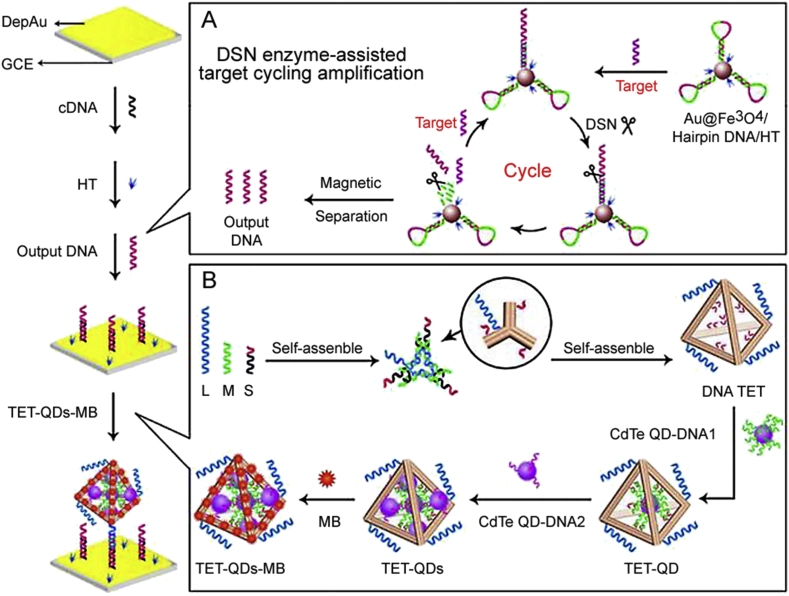
Electrochemical biosensor based miRNA detection is welcomed owing to its high sensitivity, low cost, wide applicability, and the operational ease of the electrochemical biosensors [86]. Monitoring of electrochemical reactions depends on changes in voltage or current caused by changes in the conductivity of the conductor or medium in the system [91]. Li et al. [92] constructed a photoelectrochemical biosensor with near-zero background noise for ultrasensitive miRNA-141 detection based on DNA tetrahedron (TET) as a nanocarrier for efficient immobilization of CdTe quantum dots (QDs)-Methylene Blue (MB) (TET-QDs-MB complex) as a signal probe. As illustrated in Fig. 3, massive output DNAs released from DSN enzyme-assisted target cycling amplification hybridized with cDNA on the modified electrode surface and then captured the TET-QDs-MB complex to produce a desirable photocurrent signal. miRNA-141 could be quantified within a wide linear range from 50 aM to 50 pM with a low detection limit of 17 aM.
Fig. 3.
Schematic diagrams of this proposed photoelectrochemical biosensor for miRNA-141 determination. (Reprint from Ref. [92] with permission from the American Chemical Society. Copyright 2018.)
There are many strategies based on another characteristic of DSN that will reveal the 3′-OH terminal of DNA. In this way, template-free extension reaction can be triggered with the help of terminal deoxynucleotidyl transferase (TdT) [93,94].
3.2.3. LAMP-based methods for miRNA detection
LAMP is an isothermal reaction commonly used for the amplification of DNAs and RNAs, which exhibits great sensitivity as a result of its exponential amplification feature. It uses 4–6 different primers to identify 6–8 different target sequences simultaneously, by which the selectivity is improved obviously.
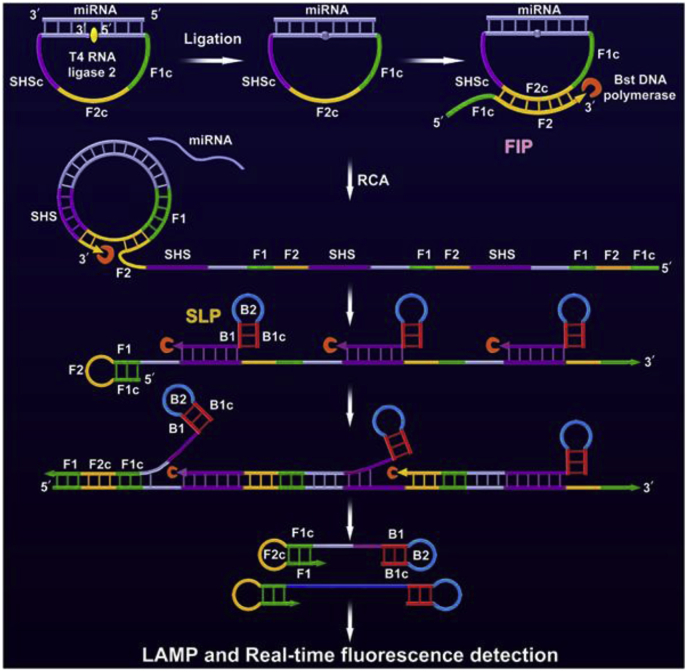
In most of the LAMP-based miRNAs quantification strategies, miRNAs work as triggers to start the reaction [95]. Only in the presence of the target miRNA, primes can conduct extension with the help of DNA polymerase and strand displacement DNA synthesis. In the LAMP-based miRNA detection, the design of probe can be quite complicated as the LAMP template contains 4–6 pre-defined sequences for stem-loop formation, which also decrease the sensitivity of the strategy due to the effect of the synergistic hybridization and extension of the multiple primers along the long template. Herein, Sun et al. [96] put forward a new idea that simplifies the probe design. In this work, a stem-loop template DNA and a stem-loop primer were designed instead of the original overlong template. A double stem-loop DNA, the starting material of subsequent LAMP, can be obtained in the presence of target miRNA. Tian et al. [79] also established a rapid and ultrasensitive RCA-LAMP method for the detection of miRNAs with a limit of 10 aM by rationally combining RCA with efficient LAMP (Fig. 4). In the presence of target miRNA, the RCA product, a repeated long DNA, can act as the template to generate a mass of double stem-loop DNAs with functional sequences for LAMP. Zhang et al. [7] indicated that SplintR ligase showed higher efficiency than T4 ligase in their recent work as an improvement to the RCA-LAMP.
Fig. 4.
Schematic illustration of the RCA-LAMP method for miRNA detection. (Reprint from Ref. [79] with permission from the Elsevier. Copyright 2018.)
Similar to PCR, a major disadvantage of LAMP is the use of indirect evaluation methods such as SYBR Green I dye, precipitation, and hydroxy naphthol blue dye, which are unable to distinguish desired products from nonspecific products, leading to false positive easily. In order to overcome the shortcoming, Jiang et al. [78] replaced the intercalating dye with a toehold-mediated strand exchange reaction termed one-step strand displacement (OSD) reporter, and successfully distinguished side products from true amplicons produced from templates. Later on, molecular beacons were applied in LAMP [97]. Furthermore, there is still brilliant space for retrofitting primers and detection probes to adapt to the detecting requirement.
3.2.4. SDA-based miRNA detection
SDA is a kind of isothermal reaction whose mechanisms refer to nicking, polymerase extension, and strand displacement. Since miRNAs (general role as a template) are recycled in the reaction based on polymerase extension-fueled strand displacement, it usually yields linear amplification. Shi et al. [98] reported an exponential SDA strategy within two cycles of nicking, polymerization, and displacement reactions triggered by target miRNA. Notably, as low as 16 zmol of the target miRNA was detected by this one-pot assay within 90 min.
Contributing to the high amplification efficiency and compatibility, various methods have been applied in vitro DNA or RNA analysis. Ou et al. [99] reported a simple and rapid fluorescence strategy for highly sensitive and specific detection of miRNA based on SDA-mediated entropy-driven circuit reaction, which exhibited a much wider linear range from 1 fM to 10 nM with a low detection limit of 0.18 fM. It highlighted the dual amplification effect and time-saving feature. Jia et al. [23] used a molecular beacon probe to capture the products generated from SDA; thus the second amplification was initiated, with a similar principle to the cycling probe technology, which was based on repeated digestion of the DNA–RNA hybrid by the RNase H. The limit of the quantification for this method reached 0.1 pM after one sample enrichment and two steps of signal amplification. With its isothermal and enzyme-free nature, qualitative or quantitative detections of nucleic acid especially miRNAs in vivo were attractive. Based on the photothermal effect of gold nanorods (AuNRs), Dai et al. [21] designed two programmable oligonucleotide hairpin probe functionalized AuNRs to develop a near-infrared (NIR) laser triggered SDA approach for sensitive miRNA imaging quantitative analysis in single living cells and multicellular tumor spheroids, which got rid of the participation of enzyme and provided a new way for sensitive real-time monitoring of intracellular miRNA.
Overall, the conventional utilization of enzyme hinders SDA from wider application since enzyme-mediated reactions are restricted by many factors such as temperature, and ionic composition. Therefore, the developments of more transformational SDA methods and other enzyme-free amplifications are of crucial necessity.
3.2.5. Enzyme-free amplification
Enzyme-free amplification is a kind of toehold-mediated DNA strand displacement reaction which has attracted much attention in nucleic acid quantification due to its isothermal and enzyme-free merits. In the enzyme-free amplification reaction, miRNA usually acts as a trigger to initiate the strand displacement process by opening up one or more DNA hairpins driven by the negative free energy change of base pair formation. In recent years, many enzyme-free amplification methods have been developed for miRNA detection and imaging based on hybridization chain reaction (HCR) [100,101], catalytic hairpin assembly (CHA) [66,71], and entropy-driven catalysis [102,103].
As an enzyme-free, room temperature and rapid linear amplification technique, HCR has been proposed for short sequence oligonucleotides. Two different kinetically trapped hairpin DNA molecules (H1 and H2) that have partially complementary segments are triggered by a DNA initiator strand to start autonomously a cascade of hybridization events in which both hairpins sequentially nucleate and open to assemble into a long nicked double-stranded amplification polymer. Typically, HCR uses two hairpin DNA molecules in order to get a significant reduction of the background signal. HCR products are highly ordered DNA double helices, so that the signal molecules can be attached on the helices with precisely controlled density, which is shown to be beneficial for amplification efficiency. Moreover, the amplification efficiency using HCR does not rely on the circulation of target analytes, but on the alternate self-assembly of two hairpin probes, which makes this process particularly attractive for degradable targets such as miRNAs.
Zhou et al. [100] reported a simple label-free and enzyme-free electrochemical strategy based on non-lineal HCR for sensitive and highly specific detection of miRNA. The special design of Y-DNA helped the biosensor obtain the ability to distinguish between single base mutations. This biosensor also performed well in clinical serum samples. Miao et al. [28] put forward a novel colorimetric strategy for miRNA analysis based on HCR-mediated localized surface Plasmon resonance variation of silver nanoparticles. The stability variation of the colloid system can then be monitored by recording the corresponding UV–vis spectrum and initial miRNA level was thus determined. This sensing system was very simple which involved only four DNA strands. Based on the mechanism of HCR, Cai et al. [30] introduced a novel technique called allosteric hairpin DNA switch-HCR detection to improve the sensitivity of the detection for miRNA let-7a.
CHA has a good application prospect because of its powerful signal amplification capability, negligible background interference, and versatility that allows for combination with different signal strategies. Kim et al. [66] evaluated the concentration of miR-141 based on CHA and fluorescence enhancement of DNA-AgNCs nearby G-rich DNA successfully. However, the sensitivity of the strategy was restricted by the poor efficiency of hairpin assembly and the low binding ratio between target and dye-modified probe. Therefore, Zhen et al. [104] fixed the dye-modified probe on the surface of graphene oxide (GO) with a capture probe. Only in the presence of the H1–H2 probe produced from CHA triggered by target miRNA-21, the dye-modified probe can leave away from the GO surface. By monitoring the decrease of fluorescence anisotropy, miRNA-21 could be detected down to 47 pM with a dynamic range of 0–16 nM.
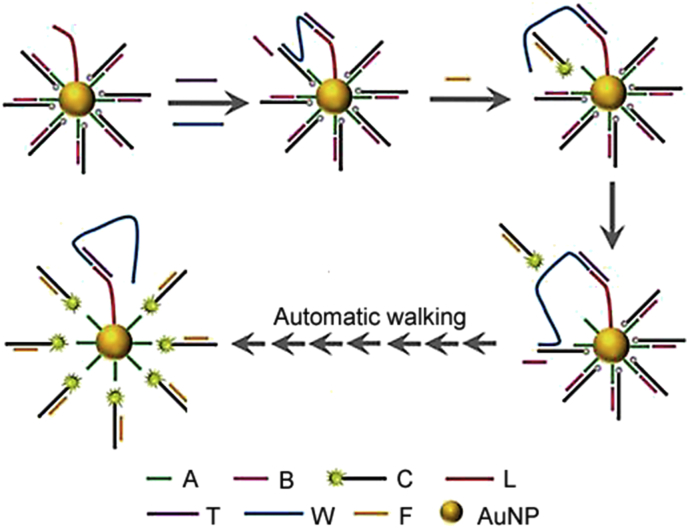
In contrast to CHA and HCR, entropy-driven catalytic reaction is driven forward thermodynamically by the entropic gain of the liberated molecules without the change of the total number of base pairs, which is faster, better understood, more stable than hairpin-based designs and can avoid unnecessary background interference due to complicated secondary structure of pseudoknots or kissing loops in DNA molecules [105]. Liang et al. [102] tailed the entropy-driven catalytic strategy into a nanomachine for automatic miRNAs imaging. As shown in Fig. 5, the nanomachine contains a DNA-coated AuNP assembly, a walking leg and a fuel. After walking leg was tethered to the AuNPs to form a walkable leg with effective intramolecular hybridization capacity, the entropy-driven catalytic reaction took place. This nanomachine was applied for intracellular imaging of miR-21 in four types of cell lines including cells with low expression profile effectively.
Fig. 5.
Schematic illustrating the operation of entropy-driven DNA nanomachine for miRNAs analysis. (Reprint from Ref. [102] with permission from the John Wiley & Sons, Inc. Copyright 2017.)
3.3. Combination of amplification strategies
To achieve a good performance of detection, scientists always combine several amplification methods. There are mainly three advantages of the combination strategies. First, it fulfills the enhancement of trace target molecules before analysis to improve the sensitivity of the method. For example, a simple and sensitive electrochemical miRNA biosensor was developed by combining the advantages of HCR with enzyme enhancement for signal amplification and sandwich type hybridization performed onto magnetic microcarriers [31]. Second, it avoids the use of traditional labels or enzymes, which simplifies the procedure and saves the cost. By combining SDA with RCA, a label-free method to detect miRNA was developed [33]. Third, the participation of electrochemical sensors and nanomaterial-based biosensors helps to reduce the background noise effectively. Some detection strategies for miRNAs using HCR were proposed by coupling to positively charged gold nanoparticles [32] or silver nanoparticles [28].
4. Summary and prospect
In the above methods, each of them has its own unique advantages as well as unavoidable shortcomings. At present, conventional detection methods such as qRT-PCR, northern blotting, and microarray are widely used to detect miRNA, but they have some limitations discouraging their use such as long processing time, laborious techniques, the sample size requirements, false-positive results, different sensitivity of the kit, and the instrument. However, considerable efforts have been made to improve miRNA detection and a variety of improved or new approaches have been developed, such as RCA and LAMP. In this review, we have provided insights into these methods.
Although numerous designs for miRNA detection have been reported, developing more efficient and practicable methods still remains necessary. In practical applications, several methods are often used together to increase the accuracy of the test results. miRNA detection is of considerable significance in disease diagnosis and study of miRNA function. With the rapid development of new techniques and new methods, scientists have more choices in the quantitative detection of miRNA. We need to combine specific experimental purposes and to balance the advantages and disadvantages of various detection methods to obtain the most economical and optimized experimental methods.
Acknowledgments
We acknowledge financial support from the National Natural Science Foundation of China (Grant 81573389) and the National Key R&D Program of China (2017YFC0908600).
Footnotes
Peer review under responsibility of Xi'an Jiaotong University.
Contributor Information
Sheng Cai, Email: caisheng@zju.edu.cn.
Su Zeng, Email: zengsu@zju.edu.cn.
Conflicts of interest
The authors declare that there are no conflicts of interest.
References
- 1.Castell-Auvi A., Cedo L., Movassat J. Procyanidins modulate microRNA expression in pancreatic islets. J. Agric. Food Chem. 2013;61:355–363. doi: 10.1021/jf303972f. [DOI] [PubMed] [Google Scholar]
- 2.Ranjbar R., Karimian A., Aghaie Fard A. The importance of miRNAs and epigenetics in acute lymphoblastic leukemia prognosis. J. Cell. Physiol. 2019;234:3216–3230. doi: 10.1002/jcp.26510. [DOI] [PubMed] [Google Scholar]
- 3.Huang Y., Zou Q., Wang S.P. The discovery approaches and detection methods of microRNAs. Mol. Biol. Rep. 2011;38:4125–4135. doi: 10.1007/s11033-010-0532-1. [DOI] [PubMed] [Google Scholar]
- 4.de Planell-Saguer M., Rodicio M.C. Detection methods for microRNAs in clinic practice. Clin. Biochem. 2013;46:869–878. doi: 10.1016/j.clinbiochem.2013.02.017. [DOI] [PubMed] [Google Scholar]
- 5.Tian T., Wang J., Zhou X. A review: microRNA detection methods. Org. Biomol. Chem. 2015;13:2226–2238. doi: 10.1039/c4ob02104e. [DOI] [PubMed] [Google Scholar]
- 6.Shen Y., Tian F., Chen Z. Amplification-based method for microRNA detection. Biosens. Bioelectron. 2015;71:322–331. doi: 10.1016/j.bios.2015.04.057. [DOI] [PubMed] [Google Scholar]
- 7.Zhang X., Liu Y., Yang Y. Ligation-promoted hyperbranched rolling circle amplification enables ultrasensitive detection of microRNA in clinical specimens. Sens. Actuators B Chem. 2018;277:634–639. [Google Scholar]
- 8.Kalogianni D.P., Kalligosfyri P.M., Kyriakou I.K. Advances in microRNA analysis. Anal. Bioanal. Chem. 2018;410:695–713. doi: 10.1007/s00216-017-0632-z. [DOI] [PubMed] [Google Scholar]
- 9.Cheng Y., Dong L., Zhang J. Recent advances in microRNA detection. Analyst. 2018;143:1758–1774. doi: 10.1039/C7AN02001E. [DOI] [PubMed] [Google Scholar]
- 10.Akmal M., Baig M.S., Khan J.A. Suppression of cotton leaf curl disease symptoms in Gossypium hirsutum through over expression of host-encoded miRNAs. J. Biotechnol. 2017;263:21–29. doi: 10.1016/j.jbiotec.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 11.Kim S.W., Li Z., Moore P.S. A sensitive non-radioactive northern blot method to detect small RNAs. Nucleic Acids Res. 2010;38:e98. doi: 10.1093/nar/gkp1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Varallyay E., Burgyan J., Havelda Z. MicroRNA detection by northern blotting using locked nucleic acid probes. Nat. Protoc. 2008;3:190–196. doi: 10.1038/nprot.2007.528. [DOI] [PubMed] [Google Scholar]
- 13.Li W., Ruan K. MicroRNA detection by microarray. Anal. Bioanal. Chem. 2009;394:1117–1124. doi: 10.1007/s00216-008-2570-2. [DOI] [PubMed] [Google Scholar]
- 14.Cissell K.A., Deo S.K. Trends in microRNA detection. Anal. Bioanal. Chem. 2009;394:1109–1116. doi: 10.1007/s00216-009-2744-6. [DOI] [PubMed] [Google Scholar]
- 15.Mohammadi-Yeganeh S., Paryan M., Samiee S.M. Development of a robust, low cost stem-loop real-time quantification PCR technique for miRNA expression analysis. Mol. Biol. Rep. 2013;40:3665–3674. doi: 10.1007/s11033-012-2442-x. [DOI] [PubMed] [Google Scholar]
- 16.Niu Y., Zhang L., Qiu H. An improved method for detecting circulating microRNAs with S-Poly(T) Plus real-time PCR. Sci. Rep. 2015;5:15100. doi: 10.1038/srep15100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oishi M., Sugiyama S. An efficient particle-based DNA circuit system: catalytic disassembly of DNA/PEG-modified gold nanoparticle-magnetic bead composites for colorimetric detection of miRNA. Small. 2016;12:5153–5158. doi: 10.1002/smll.201601741. [DOI] [PubMed] [Google Scholar]
- 18.Persano S., Guevara M.L., Wolfram J. Label-free isothermal amplification assay for specific and highly sensitive colorimetric miRNA detection. ACS Omega. 2016;1:448–455. doi: 10.1021/acsomega.6b00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li R.-D., Yin B.-C., Ye B.-C. Ultrasensitive, colorimetric detection of microRNAs based on isothermal exponential amplification reaction-assisted gold nanoparticle amplification. Biosens. Bioelectron. 2016;86:1011–1016. doi: 10.1016/j.bios.2016.07.042. [DOI] [PubMed] [Google Scholar]
- 20.Park K.W., Batule B.S., Kang K.S. Rapid and ultrasensitive detection of microRNA by target-assisted isothermal exponential amplification coupled with poly (thymine)-templated fluorescent copper nanoparticles. Nanotechnology. 2016;27:425502. doi: 10.1088/0957-4484/27/42/425502. [DOI] [PubMed] [Google Scholar]
- 21.Dai W., Dong H., Guo K. Near-infrared triggered strand displacement amplification for MicroRNA quantitative detection in single living cells. Chem. Sci. 2018;9:1753–1759. doi: 10.1039/c7sc04243d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Y., Li D., Cheng W. Chemiluminescence imaging for microRNA detection based on cascade exponential isothermal amplification machinery. Anal. Chim. Acta. 2016;936:229–235. doi: 10.1016/j.aca.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 23.Jia H., Bu Y., Zou B. Signal amplification of microRNAs with modified strand displacement-based cycling probe technology. Analyst. 2016;141:6297–6302. doi: 10.1039/c6an01024e. [DOI] [PubMed] [Google Scholar]
- 24.Huang J.-F., Zhao N., Xu H.-Q. Sensitive and specific detection of miRNA using an isothermal exponential amplification method using fluorescence-labeled LNA/DNA chimera primers. Anal. Bioanal. Chem. 2016;408:7437–7446. doi: 10.1007/s00216-016-9829-9. [DOI] [PubMed] [Google Scholar]
- 25.Yang J., Tang M., Diao W. Electrochemical strategy for ultrasensitive detection of microRNA based on MNAzyme-mediated rolling circle amplification on a gold electrode. Microchim. Acta. 2016;183:3061–3067. [Google Scholar]
- 26.Hong C., Baek A., Hah S.S. Fluorometric detection of microRNA using isothermal gene amplification and graphene oxide. Anal. Chem. 2016;88:2999–3003. doi: 10.1021/acs.analchem.6b00046. [DOI] [PubMed] [Google Scholar]
- 27.Deng R., Tang L., Tian Q. Toehold-initiated rolling circle amplification for visualizing individual microRNAs in situ in single cells. Angew. Chem. Int. Ed. 2014;53:2389–2393. doi: 10.1002/anie.201309388. [DOI] [PubMed] [Google Scholar]
- 28.Miao J., Wang J., Guo J. A plasmonic colorimetric strategy for visual miRNA detection based on hybridization chain reaction. Sci. Rep. 2016;6:32219. doi: 10.1038/srep32219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwarzkopf M., Pierce N.A. Multiplexed miRNA northern blots via hybridization chain reaction. Nucleic Acids Res. 2016;44:e129. doi: 10.1093/nar/gkw503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cai S., Cao Z., Lau C. Label-free technology for the amplified detection of microRNA based on the allosteric hairpin DNA switch and hybridization chain reaction. Analyst. 2014;139:6022–6027. doi: 10.1039/c4an01178c. [DOI] [PubMed] [Google Scholar]
- 31.Torrente-Rodriguez R.M., Campuzano S., Montiel V.R.-V. Sensitive electrochemical determination of miRNAs based on a sandwich assay onto magnetic microcarriers and hybridization chain reaction amplification. Biosens. Bioelectron. 2016;86:516–521. doi: 10.1016/j.bios.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 32.Miao X., Ning X., Li Z. Sensitive detection of miRNA by using hybridization chain reaction coupled with positively charged gold nanoparticles. Sci. Rep. 2016;6:32358. doi: 10.1038/srep32358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng X., Niu L., Wei D. Label-free detection of microRNA based on coupling multiple isothermal amplification techniques. Sci. Rep. 2016;6:35982. doi: 10.1038/srep35982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pall G.S., Codony-Servat C., Byrne J. Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by northern blot. Nucleic Acids Res. 2007;35:e60. doi: 10.1093/nar/gkm112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Torres A.G., Fabani M.M., Vigorito E. MicroRNA fate upon targeting with anti-miRNA oligonucleotides as revealed by an improved Northern-blot-based method for miRNA detection. RNA. 2011;17:933–943. doi: 10.1261/rna.2533811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shu P., Wu C., Liu W. The spatiotemporal expression pattern of MicroRNA in the developing mouse nervous system. J. Biol. Chem. 2018;294:3444–3453. doi: 10.1074/jbc.RA118.004390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fabani M.M., Abreu-Goodger C., Williams D. Efficient inhibition of miR-155 function in vivo by peptide nucleic acids. Nucleic Acids Res. 2010;38:4466–4475. doi: 10.1093/nar/gkq160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lanford R.E., Hildebrandt-Eriksen E.S., Petri A. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C Virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pall G.S., Hamilton A.J. Improved northern blot method for enhanced detection of small RNA. Nat. Protoc. 2008;3:1077–1084. doi: 10.1038/nprot.2008.67. [DOI] [PubMed] [Google Scholar]
- 40.Gan Y.-B., Zhou Z.-J., An L.-J. A comparison between northern blotting and quantitative real-time PCR as a means of detecting the nutritional regulation of genes expressed in roots of arabidopsis thaliana. Agric. Sci. China. 2011;10:335–342. [Google Scholar]
- 41.Czimmerer Z., Hulvely J., Simandi Z. A versatile method to design stem-loop primer-based quantitative PCR assays for detecting small regulatory RNA molecules. PLoS One. 2013;8:e55168. doi: 10.1371/journal.pone.0055168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Londin E., Loher P., Telonis A.G. Analysis of 13 cell types reveals evidence for the expression of numerous novel primate- and tissue-specific microRNAs. Proc. Natl. Acad. Sci. U.S.A. 2015;112:E1106–E1115. doi: 10.1073/pnas.1420955112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi R., Chiang V.L. Facile means for quantifying microRNA expression by real-time PCR. Biotechniques. 2005;39:519–525. doi: 10.2144/000112010. [DOI] [PubMed] [Google Scholar]
- 44.Raymond C.K., Roberts B.S., Garrett-Engele P. Simple, quantitative primer-extension PCR assay for direct monitoring of microRNAs and short-interfering RNAs. RNA. 2005;11:1737–1744. doi: 10.1261/rna.2148705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Varkonyi-Gasic E., Wu R., Wood M. Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods. 2007;3:12. doi: 10.1186/1746-4811-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Benes V., Castoldi M. Expression profiling of microRNA using real-time quantitative PCR, how to use it and what is available. Methods. 2010;50:244–249. doi: 10.1016/j.ymeth.2010.01.026. [DOI] [PubMed] [Google Scholar]
- 47.Calin G.A., Croce C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 48.Wang Y., Zou L., Wu T. Identification of mRNA-miRNA crosstalk in human endothelial cells after exposure of PM2.5 through integrative transcriptome analysis. Ecotoxicol. Environ. Saf. 2019;169:863–873. doi: 10.1016/j.ecoenv.2018.11.114. [DOI] [PubMed] [Google Scholar]
- 49.Alhasan A.H., Kim D.Y., Daniel W.L. Scanometric microRNA array profiling of prostate cancer markers using spherical nucleic acid-gold nanoparticle conjugates. Anal. Chem. 2012;84:4153–4160. doi: 10.1021/ac3004055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen T., Xu Y., Wei S. A signal amplification system constructed by bi-enzymes and bi-nanospheres for sensitive detection of norepinephrine and miRNA. Biosens. Bioelectron. 2019;124-125:224–232. doi: 10.1016/j.bios.2018.10.030. [DOI] [PubMed] [Google Scholar]
- 51.Hosseinzadeh E., Ravan H., Mohammadi A. Target-triggered three-way junction in conjugation with catalytic concatemers-functionalized nanocomposites provides a highly sensitive colorimetric method for miR-21 detection. Biosens. Bioelectron. 2018;117:567–574. doi: 10.1016/j.bios.2018.06.051. [DOI] [PubMed] [Google Scholar]
- 52.Pan M., Liang M., Sun J. Lighting up fluorescent silver clusters via target-catalyzed hairpin assembly for amplified biosensing. Langmuir. 2018;34:14851–14857. doi: 10.1021/acs.langmuir.8b01576. [DOI] [PubMed] [Google Scholar]
- 53.Liu R., Wang Q., Li Q. Surface plasmon resonance biosensor for sensitive detection of microRNA and cancer cell using multiple signal amplification strategy. Biosens. Bioelectron. 2017;87:433–438. doi: 10.1016/j.bios.2016.08.090. [DOI] [PubMed] [Google Scholar]
- 54.Salahandish R., Ghaffarinejad A., Omidinia E. Label-free ultrasensitive detection of breast cancer miRNA-21 biomarker employing electrochemical nano-genosensor based on sandwiched AgNPs in PANI and N-doped graphene. Biosens. Bioelectron. 2018;120:129–136. doi: 10.1016/j.bios.2018.08.025. [DOI] [PubMed] [Google Scholar]
- 55.Foda M.F., Huang L., Shao F. Biocompatible and highly luminescent near-infrared CuInS2/ZnS quantum dots embedded silica beads for cancer cell imaging. ACS Appl. Mater. Interfaces. 2014;6:2011–2017. doi: 10.1021/am4050772. [DOI] [PubMed] [Google Scholar]
- 56.Ma H., Xue N., Li Z. Ultrasensitive detection of miRNA-155 using multi-walled carbon nanotube-gold nanocomposites as a novel fluorescence quenching platform. Sens. Actuators B Chem. 2018;266:221–227. [Google Scholar]
- 57.Borghei Y.S., Hosseini M., Ganjali M.R. Oxidase-like catalytic activity of Cys-AuNCs upon visible light irradiation and its application for visual miRNA detection. Sens. Actuators B Chem. 2018;273:1618–1626. [Google Scholar]
- 58.Tian R., Ning W., Chen M. High performance electrochemical biosensor based on 3D nitrogen-doped reduced graphene oxide electrode and tetrahedral DNA nanostructure. Talanta. 2019;194:273–281. doi: 10.1016/j.talanta.2018.09.110. [DOI] [PubMed] [Google Scholar]
- 59.Borghei Y.-S., Hosseini M., Ganjali M.R. Label-free fluorescent detection of microRNA-155 based on synthesis of hairpin DNA-templated copper nanoclusters by etching (top-down approach) Sens. Actuators B Chem. 2017;248:133–139. [Google Scholar]
- 60.Chand R., Ramalingam S., Neethirajan S. A 2D transition-metal dichalcogenide MoS2 based novel nanocomposite and nanocarrier for multiplex miRNA detection. Nanoscale. 2018;10:8217–8225. doi: 10.1039/c8nr00697k. [DOI] [PubMed] [Google Scholar]
- 61.Tian L., Qi J., Ma X. A facile DNA strand displacement reaction sensing strategy of electrochemical biosensor based on N-carboxymethyl chitosan/molybdenum carbide nanocomposite for microRNA-21 detection. Biosens. Bioelectron. 2018;122:43–50. doi: 10.1016/j.bios.2018.09.037. [DOI] [PubMed] [Google Scholar]
- 62.Wang J., Lu J., Dong S. An ultrasensitive electrochemical biosensor for detection of microRNA-21 based on redox reaction of ascorbic acid/iodine and duplex-specific nuclease assisted target recycling. Biosens. Bioelectron. 2019;130:81–87. doi: 10.1016/j.bios.2019.01.031. [DOI] [PubMed] [Google Scholar]
- 63.Yue R., Li Z., Wang G. Logic sensing of microRNA in living cells using DNA-programmed nanoparticle network with high signal gain. ACS Sens. 2019;4:250–256. doi: 10.1021/acssensors.8b01422. [DOI] [PubMed] [Google Scholar]
- 64.Yeh H.-C., Sharma J., Shih I.-M. A fluorescence light-up Ag nanocluster probe that discriminates single-nucleotide variants by emission color. J. Am. Chem. Soc. 2012;134:11550–11558. doi: 10.1021/ja3024737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Deng R., Zhang K., Sun Y. Highly specific imaging of mRNA in single cells by target RNA-initiated rolling circle amplification. Chem. Sci. 2017;8:3668–3675. doi: 10.1039/c7sc00292k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim H., Kang S., Park K.S. Enzyme-free and label-free miRNA detection based on target-triggered catalytic hairpin assembly and fluorescence enhancement of DNA-silver nanoclusters. Sens. Actuators B Chem. 2018;260:140–145. [Google Scholar]
- 67.Zhang J., Li C., Zhi X. Hairpin DNA-templated silver nanoclusters as novel beacons in strand displacement amplification for microRNA detection. Anal. Chem. 2016;88:1294–1302. doi: 10.1021/acs.analchem.5b03729. [DOI] [PubMed] [Google Scholar]
- 68.Qing Z., He X., He D. Poly(thymine)-Templated selective formation of fluorescent copper nanoparticles. Angew. Chem. Int. Ed. 2013;52:9719–9722. doi: 10.1002/anie.201304631. [DOI] [PubMed] [Google Scholar]
- 69.Chen Y.X., Huang K.J., Niu K.X. Recent advances in signal amplification strategy based on oligonucleotide and nanomaterials for microRNA detection-a review. Biosens. Bioelectron. 2018;99:612–624. doi: 10.1016/j.bios.2017.08.036. [DOI] [PubMed] [Google Scholar]
- 70.Gao Y.-P., Wu X., Huang K.-J. Two-dimensional transition metal diseleniums for energy storage application: a review of recent developments. CrystEngComm. 2017;19:404–418. [Google Scholar]
- 71.Bao J., Hou C., Zhao Y. An enzyme-free sensitive electrochemical microRNA-16 biosensor by applying a multiple signal amplification strategy based on Au/PPy-rGO nanocomposite as a substrate. Talanta. 2019;196:329–336. doi: 10.1016/j.talanta.2018.12.082. [DOI] [PubMed] [Google Scholar]
- 72.Lee J., Kim Y.-K., Lee S. Graphene oxide-based NET strategy for enhanced colorimetric sensing of miRNA. Sens. Actuators B Chem. 2019;282:861–867. [Google Scholar]
- 73.Jacobs C.B., Peairs M.J., Venton B.J. Review: carbon nanotube based electrochemical sensors for biomolecules. Anal. Chim. Acta. 2010;662:105–127. doi: 10.1016/j.aca.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 74.Wang Y., Shi H., Cui K. Hierarchical hematite/TiO2 nanorod arrays coupled with responsive mesoporous silica nanomaterial for highly sensitive photoelectrochemical sensing. Biosens. Bioelectron. 2018;117:515–521. doi: 10.1016/j.bios.2018.06.030. [DOI] [PubMed] [Google Scholar]
- 75.Gudnason H., Dufva M., Bang D.D. Comparison of multiple DNA dyes for real-time PCR: effects of dye concentration and sequence composition on DNA amplification and melting temperature. Nucleic Acids Res. 2007;35:e127. doi: 10.1093/nar/gkm671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen C., Ridzon D.A., Broomer A.J. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tyagi S., Kramer F. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 78.Jiang Y.S., Bhadra S., Li B.L. Robust strand exchange reactions for the sequence-specific, real-time detection of nucleic acid amplicons. Anal. Chem. 2015;87:3314–3320. doi: 10.1021/ac504387c. [DOI] [PubMed] [Google Scholar]
- 79.Tian W., Li P., He W. Rolling circle extension-actuated loop-mediated isothermal amplification (RCA-LAMP) for ultrasensitive detection of microRNAs. Biosens. Bioelectron. 2018;128:17–22. doi: 10.1016/j.bios.2018.12.041. [DOI] [PubMed] [Google Scholar]
- 80.Xu H., Wu D., Zhang Y. RCA-enhanced multifunctional molecule beacon-based strand-displacement amplification for sensitive microRNA detection. Sens. Actuators B Chem. 2018;258:470–477. [Google Scholar]
- 81.Xu H., Zhang Y., Zhang S. Ultrasensitive assay based on a combined cascade amplification by nicking-mediated rolling circle amplification and symmetric strand-displacement amplification. Anal. Chim. Acta. 2019;1047:172–178. doi: 10.1016/j.aca.2018.10.004. [DOI] [PubMed] [Google Scholar]
- 82.Zhou Y., Huang Q., Gao J. A dumbbell probe-mediated rolling circle amplification strategy for highly sensitive microRNA detection. Nucleic Acids Res. 2010;38:e156. doi: 10.1093/nar/gkq556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang Z.-Y., Li F., Zhang Y. Sensitive detection of cancer gene based on a nicking-mediated RCA of circular DNA nanomachine. Sens. Actuators B Chem. 2017;251:692–698. [Google Scholar]
- 84.Mittal S., Thakur S., Mantha A.K. Bio-analytical applications of nicking endonucleases assisted signal-amplification strategies for detection of cancer biomarkers -DNA methyl transferase and microRNA. Biosens. Bioelectron. 2019;124:233–243. doi: 10.1016/j.bios.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 85.Song T., Guo X., Li X. Label-free electrochemical detection of RNA based on “Y” junction structure and restriction endonuclease-aided target recycling strategy. J. Electroanal. Chem. 2016;781:251–256. [Google Scholar]
- 86.Qiu X., Zhang H., Yu H. Duplex-specific nuclease-mediated bioanalysis. Trends Biotechnol. 2015;33:180–188. doi: 10.1016/j.tibtech.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 87.Le B.H., Nguyen T.T., Joo H.N. Large-Stokes-shift-based folded DNA probing systems targeting DNA and miRNA 21 with signal amplification. Bioorg. Med. Chem. 2018;26:4881–4885. doi: 10.1016/j.bmc.2018.08.027. [DOI] [PubMed] [Google Scholar]
- 88.Zhang H., Wang K., Bu S. Colorimetric detection of microRNA based on DNAzyme and nuclease-assisted catalytic hairpin assembly signal amplification. Mol. Cell. Probes. 2018;38:13–18. doi: 10.1016/j.mcp.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 89.Lin X., Zhang C., Huang Y. Backbone-modified molecular beacons for highly sensitive and selective detection of microRNAs based on duplex specific nuclease signal amplification. Chem. Commun. 2013;49:7243–7245. doi: 10.1039/c3cc43224f. [DOI] [PubMed] [Google Scholar]
- 90.Ma Y., Chen J., Chen D. Short-probe-based duplex-specific nuclease signal amplification strategy enables imaging of endogenous microRNAs in living cells with ultrahigh specificity. Talanta. 2018;186:256–264. doi: 10.1016/j.talanta.2018.04.071. [DOI] [PubMed] [Google Scholar]
- 91.Chang B.-Y., Park S.-M. Electrochemical impedance spectroscopy. Annu. Rev. Anal. Chem. 2010;3:207–229. doi: 10.1146/annurev.anchem.012809.102211. [DOI] [PubMed] [Google Scholar]
- 92.Li M., Xiong C., Zheng Y. Ultrasensitive photoelectrochemical biosensor based on DNA tetrahedron as nanocarrier for efficient immobilization of CdTe QDs-methylene blue as signal probe with near-zero background noise. Anal. Chem. 2018;90:8211–8216. doi: 10.1021/acs.analchem.8b01641. [DOI] [PubMed] [Google Scholar]
- 93.Guo J., Yuan C., Yan Q. An electrochemical biosensor for microRNA-196a detection based on cyclic enzymatic signal amplification and template-free DNA extension reaction with the adsorption of methylene blue. Biosens. Bioelectron. 2018;105:103–108. doi: 10.1016/j.bios.2018.01.036. [DOI] [PubMed] [Google Scholar]
- 94.Xu F., Luo L., Shi H. Label-free and sensitive microRNA detection based on a target recycling amplification-integrated superlong poly(thymine)-hosted copper nanoparticle strategy. Anal. Chim. Acta. 2018;1010:54–61. doi: 10.1016/j.aca.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 95.Li C., Li Z., Jia H. One-step ultrasensitive detection of microRNAs with loop-mediated isothermal amplification (LAMP) Chem. Commun. 2011;47:2595–2597. doi: 10.1039/c0cc03957h. [DOI] [PubMed] [Google Scholar]
- 96.Sun Y., Tian H., Liu C. One-step detection of microRNA with high sensitivity and specificity via target-triggered loop-mediated isothermal amplification (TT-LAMP) Chem. Commun. 2017;53:11040–11043. doi: 10.1039/c7cc06140d. [DOI] [PubMed] [Google Scholar]
- 97.Liu W., Huang S., Liu N. Establishment of an accurate and fast detection method using molecular beacons in loop-mediated isothermal amplification assay. Sci. Rep. 2017;7:40125. doi: 10.1038/srep40125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shi C., Liu Q., Ma C. Exponential strand-displacement amplification for detection of microRNAs. Anal. Chem. 2014;86:336–339. doi: 10.1021/ac4038043. [DOI] [PubMed] [Google Scholar]
- 99.Ou S., Xu T., Liu X. Rapid and ultrasensitive detection of microRNA based on strand displacement amplification-mediated entropy-driven circuit reaction. Sens. Actuators B Chem. 2018;255:3057–3063. [Google Scholar]
- 100.Zhou L., Wang Y., Yang C. A label-free electrochemical biosensor for microRNAs detection based on DNA nanomaterial by coupling with Y-shaped DNA structure and non-linear hybridization chain reaction. Biosens. Bioelectron. 2019;126:657–663. doi: 10.1016/j.bios.2018.11.028. [DOI] [PubMed] [Google Scholar]
- 101.Nie Y., Yuan X., Zhang P. Versatile and ultrasensitive electrochemiluminescence biosensor for biomarker detection based on non-enzymatic amplification and aptamer-triggered emitter release. Anal. Chem. 2019;91:3452–3458. doi: 10.1021/acs.analchem.8b05001. [DOI] [PubMed] [Google Scholar]
- 102.Liang C.P., Ma P.Q., Liu H. Rational engineering of a dynamic, entropy-driven DNA nanomachine for intracellular microRNA imaging. Angew. Chem. Int. Ed. Enql. 2017;56:9077–9081. doi: 10.1002/anie.201704147. [DOI] [PubMed] [Google Scholar]
- 103.Zhang N., Shi X.-M., Guo H.-Q. Gold nanoparticle couples with entropy-driven toehold-mediated DNA strand displacement reaction on magnetic beads: toward ultrasensitive energy-transfer-based photoelectrochemical detection of miRNA-141 in real blood sample. Anal. Chem. 2018;90:11892–11898. doi: 10.1021/acs.analchem.8b01966. [DOI] [PubMed] [Google Scholar]
- 104.Zhen S.J., Xiao X., Li C.H. An enzyme free DNA circuit-assisted graphene oxide enhanced fluorescence anisotropy assay for microRNA detection with improved sensitivity and selectivity. Anal. Chem. 2017;89:8766–8771. doi: 10.1021/acs.analchem.7b00955. [DOI] [PubMed] [Google Scholar]
- 105.Zhang D.Y., Turberfield A.J., Yurke B. Engineering entropy-driven reactions and networks catalyzed by DNA. Science. 2007;318:1121–1125. doi: 10.1126/science.1148532. [DOI] [PubMed] [Google Scholar]