Abstract
Aim:
The present study was carried out to determine load of total bacteria, Escherichia coli and Salmonella spp. in dairy farm and its environmental components. In addition, the antibiogram profile of the isolated bacteria having public health impact was also determined along with identification of virulence and resistance genes by polymerase chain reaction (PCR) under a one-health approach.
Materials and Methods:
A total of 240 samples of six types (cow dung – 15, milk – 10, milkers’ hand wash – 10, soil – 10 water – 5, and vegetables – 10) were collected from four dairy farms. For enumeration, the samples were cultured onto plate count agar, eosin methylene blue, and xylose-lysine deoxycholate agar and the isolation and identification of the E. coli and Salmonella spp. were performed based on morphology, cultural, staining, and biochemical properties followed by PCR.The pathogenic strains of E. coli stx1, stx2, and rfbO157 were also identified through PCR. The isolates were subjected to antimicrobial susceptibility test against 12 commonly used antibiotics by disk diffusion method. Detection of antibiotic resistance genes ereA, tetA, tetB, and SHV were performed by PCR.
Results:
The mean total bacterial count, E. coli and Salmonella spp. count in the samples ranged from 4.54±0.05 to 8.65±0.06, 3.62±0.07 to 7.04±0.48, and 2.52±0.08 to 5.87±0.05 log colony-forming unit/g or ml, respectively. Out of 240 samples, 180 (75%) isolates of E. coli and 136 (56.67%) isolates of Salmonella spp. were recovered through cultural and molecular tests. Among the 180 E. coli isolates, 47 (26.11%) were found positive for the presence of all the three virulent genes, of which stx1 was the most prevalent (13.33%). Only three isolates were identified as enterohemorrhagic E. coli. Antibiotic sensitivity test revealed that both E. coli and Salmonella spp. were found highly resistant to azithromycin, tetracycline, erythromycin, oxytetracycline, and ertapenem and susceptible to gentamycin, ciprofloxacin, and imipenem. Among the four antibiotic resistance genes, the most observable was tetA (80.51-84.74%) in E. coli and Salmonella spp. and SHV genes were the lowest one (22.06-25%).
Conclusion:
Dairy farm and their environmental components carry antibiotic-resistant pathogenic E. coli and Salmonella spp. that are potential threat for human health which requires a one-health approach to combat the threat.
Keywords: one-health, antibiotic resistance genes, dairy farm, Escherichia coli, carbapenem resistance, Salmonella spp, virulence
Introduction
Antibiotic resistance is a global public health concern [1]. Antibiotic-resistant bacteria as the etiology of infection have been expanding at an alarming rate [2]. It is stated that almost 10 million people will die per year due to antimicrobial resistance () infections [3]. At present, drug-resistant microorganisms are broadly circulating in the environmental settings of the earth, and their negative effect has significantly risen in the past few years [4]. Haphazard use of antibiotics and absence of knowledge are the most imperative variables for the rise, selection, and spread of antibiotic-resistant organisms in the environment [5]. If such things happen continuously, it will bring a disaster to human being. At present, many of the antimicrobial agents are utilized in food animal production for controlling diseases and mostly used as growth promoter that is continuously disseminating in human food chain leads serious health problem in human and animals [6]. Cattle in dairy farm could be a potential source for the contamination of the farm environment and farm products by antibiotic-resistant Escherichia coli and Salmonella spp. present in cow dung. Moreover, these resistance elements can transfer to the people working on the farm directly from contaminated soil, water, and milk to cause serious human health problems [7].
E. coli is known as dangerous pathogens in the dairy farm sector worldwide as it causes significant economic losses [8]. There are several strains in E. coli and most of them are harmless, but a few of them cause serious foodborne infection in human [9]. Farm animals, especially cattle, asymptomatically carry Shiga toxin-producing E. coli (STEC) and enterohemorrhagic E. coli (EHEC). These pathogens are zoonotic in nature and can transmit to human from farm through contaminated milk, meat, water, and direct contact with animals or their environmental equipment [10,11]. Salmonella spp. is the most ubiquitous organisms in nature and major foodborne zoonotic pathogen, it is also one of the pathogens listed in the WHO priority pathogen list. Dairy cattle act as a reservoir of Salmonella spp. that cause salmonellosis in human [12]. Salmonella spp. can transmit through feces from infected cattle and their environment. In the past few years, Salmonella serotypes have become resistant to frequently used antibiotics that increased the treatment cost in food animal production [13]. Livestock manure contains microbial constituents, which make it a potential source of pathogenic microorganisms for animals and human. About 151.3 million tons of fresh farm animal manure are produced in Bangladesh annually that are mostly used as biofertilizer in agriculture land [14]. Several bacterial pathogens such as E. coli, Campylobacter, Salmonella, Listeria, Coxiella, and Mycobacterium have been recovered from manure that could be antibiotic-resistant and zoonotic in nature [15]. These pathogens can enter into the food chain when manure used as fertilizer in agriculture for crop, vegetables, and fruit production to interferer consumers health [16].
The emergence of antibiotic-resistant bacteria and their resistance genes has turned into a serious growing issue in current medication. There is lack of adequate surveillance data on the occurrence of antibiotic-resistant bacteria in livestock farming system in Bangladesh, especially in dairy cattle and the farm environment focusing one-health.
The present study was therefore designed using a one-health approach to determine the load of total bacteria, E. coli and Salmonella spp. in dairy cattle and farm environmental components as well as to determine their virulence genes, antibiogram phenotype, and genotype having public health significance.
Materials and Methods
Ethical approval and informed consents
No ethical approval was required; however, during the collection of samples; verbal permission was taken from the farm owners and farm workers.
Study area
The study was conducted on different dairy farms of Mymensingh district of Bangladesh namely, Research Animal farm Bangladesh Agricultural University (BAU), BAU dairy farm, dairy farm of Sutiakhali and Boira. These farms were selected on their use of cow dung as fertilizer for vegetable production. Cow dung, milk, milker’s hand wash, soil, and water were selected for sampling. In addition, vegetables grown in agriculture field within the farm where cow dung used as fertilizer were also collected.
Sample collection
A total of 240 samples of six items were collected from four dairy farms where each of the farms contributed 60 samples consist of 15 cow dung, 10 milk, 10 milkers’ hand wash, 10 soil, 5 water, and 10 vegetables. All the samples were taken aseptically by utilizing sterile zipper bag. Just after defecation, cow dung samples were collected. Sterile plastic spoon, container, and falcon tube were used for the collection of soil, milk, and water samples, respectively. Milker’s hand wash samples were also collected by washing the hand with phosphate-buffered saline and vegetable samples red spinach (Amaranthus gangeticus), Malabar spinach (Basella alba), green chili (Capsicum annum), and tomato (Solanum lycopersicum) from the vegetable production land. After collection, they were transported to the microbiology laboratory, Department of Microbiology and Hygiene, BAU as soon as possible in an ice box. Bacteriological examinations were done promptly before undesirable changes develop.
Sample processing
Solid (cow dung, soil, and vegetables) and liquid (milk, milker’s hand wash, and water) samples were measured, respectively, in gram and ml. For cow dung and soil samples, 10 g sample and 90 ml 0.1% peptone water were taken in a beaker and mixed well to have the initial dilution. Vegetable samples collected were chopped into small pieces with a sterile knife and mixed homogeneously. A 25 g of these chopped vegetables was taken into a flask containing 225 ml 0.1% peptone water, and vigorously shook to homogenize [17]. Ten ml sample and 90 ml diluent were taken to prepare the initial dilution for a liquid sample. Finally, ten-fold serial dilution was made from all the initial dilutions for the bacterial count.
Bacteriological analysis
Initially, a ten-fold dilution of the sample was prepared in 0.1% peptone water in Eppendorf tube. Earlier, a plate count agar (PCA) was divided into four parts and marked separately. Four consecutive dilutions within the range of 10-1-10-6 were taken based on sample types for the four separate parts. Three drops of 10 µl from each dilution were inoculated into each part of the PCA plate separately and incubated at 37°C for 24 h for development of single colonies . After incubation, colonies were counted from three drops of a particular dilution where the average colony count of those three drops was 3-30/10 µl [18]. The results of the total bacterial count were expressed as colony-forming unit (CFU)/g or ml of sample. Similar methods were also applied for counting of E. coli and Salmonella spp. using eosin methylene blue (EMB) agar and xylose-lysine deoxycholate (XLD) agar, respectively.
Isolation and identification of bacteria
For obtaining pure culture, bacterial growth on EMB and XLD agar was further streaked on their respective media and incubated overnight at 37°C. Bacteria were identified on the basis of colony characteristics, morphological characteristics by Gram’s staining and biochemical characteristics, namely basic sugar fermentation test, methyl red test, Voges-Proskauer test, and indole test [19]. Final confirmation was done through molecular characterization by polymerase chain reaction (PCR). For PCR, DNA was extracted from pure culture using boiling methods following the procedures of Mahmud et al. [20]. PCR for genus-specific E. coli and Salmonella spp. was performed using previously studied primers (Table-1) following the standard protocol [21,22]. STEC were detected by PCR targeting stx1 and stx2 genes, while EHEC detection was based on detection of rfbO157 gene according to published methods [23,24].
Table 1.
List of primers used.
| Target genes | Primer sequence(5’- 3’) | Approximate band size(bp) | Annealing temperature(°C) | References |
|---|---|---|---|---|
| E.coli 16S rRNA | F: GACCTCGGTTTAGTTCACAGA R: CACACGCTGACGCTGACCA |
585 | 55 | [21] |
| invA | F: ATCAGTACCAGTCGTCTTATCTTGAT R: TCTGTTTACCGGGCATACCAT |
211 | 58 | [22] |
| st×1 | F: ACAATCAGGCGTCGCCAGCGCACTTGCT R: TGTTGCAGGGATCAGTGGTACGGGGATGC |
606 | 58 | [23] |
| st×2 | F: CCACATCGGTGTCTGTTATTAACCACACC R: GCAGAACTGCTCTGGATGCATCTCTGGTC |
372 | 58 | [23] |
| rfbO157 | F: AAGATTGCGCTGAAGCCTTTG R: CATTGGCATCGTGTGGACAG |
497 | 66 | [24] |
| ereA | F: GCCGGTGCTCATGAACTTGAG R: CGACTCTATTCGATCAGAGGC |
419 | 52 | [27] |
| tetA | F: GGTTCACTCGAACGACGTCA R: CTGTCCGACAAGTTGCATGA |
577 | 57 | [28] |
| tetB | F: CCTCAGCTTCTCAACGCGTG R: GCACCTTGCTGATGACTCTT |
634 | 56 | [28] |
| SHV | F: TCGCCTGTGTATTATCTCCC R: CGCAGATAAATCACCACAATG |
768 | 52 | [27] |
E. coli=Escherichia coli
Antimicrobial susceptibility test
All the isolated Salmonella spp. and E. coli were subjected to antimicrobial susceptibility test against 12 commonly used antibiotics, i.e., azithromycin (15 µg), chloramphenicol (30 µg), ciprofloxacin (5 µg), erythromycin (15 µg), gentamycin (10 µg), kanamycin (30 µg), neomycin (30 µg), oxytetracycline (30 µg), ertapenem (10 µg), meropenem (10 µg), imipenem (10 µg), and tetracycline (30 µg) following the disk diffusion methods described by Bauer et al. [25]. Finally, the zone of growth inhibition was compared with standards provided by the Clinical and Laboratory Standards Institute [26] to identify the resistant isolates.
Molecular detection of antibiotic resistance genes
Isolates of E. coli and Salmonella spp. that showed resistance to erythromycin, tetracycline, and beta-lactam (ertapenem, meropenem, and imipenem) phenotypically were further screened for the detection of ereA, tetA, and tetB and SHV resistance genes, respectively. The primers used for the detection of resistance genes are listed in Table-1 [27,28].
Statistical analysis
SPSS software version 20.0 (IBM, USA) was used to analyze the data. Frequency and mean were estimated using descriptive analysis.
Results
Microbial load
The microbial analysis revealed that the average total viable bacterial count (TVC) ranged from 4.54±0.05 to 8.65±0.06 log CFU/g or mlmean±standard deviation (SD) among all the samples analyzed. The highest TVC was observed in soil and lowest in water samples of Sutiakhali dairy farm. The highest E. coli count was found as 7.04±0.48 log CFU/g±SD in cow dung samples of Boira dairy farm and lowest as 3.62±0.07 log CFU/ml±SD in water samples of Sutiakhali dairy farm. The highest Salmonella spp. count was found 5.87±0.05 log CFU/g±SD and lowest 2.52±0.08 log CFU/ml±SD in soil and water samples of Sutiakhali dairy farm, respectively (Table-2).
Table 2.
Mean total bacteria, E.coli and Salmonella spp. counts in dairy cattle and farm environmental samples (log CFU/g or ml±SD).
| Sample(n) | Research animal farm | BAU dairy farm | Dairy farm in Sutiakhali | Dairy farm in Boira | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TVC | TEC | TSC | TVC | TEC | TSC | TVC | TEC | TSC | TVC | TEC | TSC | |
| Cow dung(15) | 7.93±0.044 | 6.98±0.041 | 5.74±0.066 | 7.96±0.047 | 7.00±0.042 | 5.78±0.069 | 7.99±0.045 | 7.03±0.042 | 5.83±0.062 | 7.99±0.054 | 7.04±0.048 | 5.82±0.084 |
| Milk(10) | 6.76±0.039 | 4.88±0.028 | 3.76±0.051 | 6.79±0.038 | 4.90±0.029 | 3.79±0.049 | 6.83±0.032 | 4.93±0.023 | 3.84±0.039 | 6.88±0.036 | 4.97±0.033 | 3.87±0.036 |
| Milker’s hand wash(10) | 6.57±0.067 | 5.75±0.047 | 4.58±0.060 | 6.63±0.062 | 5.80±0.042 | 4.64±0.083 | 6.68±0.052 | 5.83±0.044 | 4.69±0.061 | 6.71±0.044 | 5.85±0.040 | 4.71±0.057 |
| Soil(10) | 8.56±0.065 | 6.66±0.058 | 5.81±0.038 | 8.62±0.080 | 6.71±0.078 | 5.85±0.043 | 8.65±0.059 | 6.74±0.051 | 5.87±0.050 | 8.64±0.071 | 6.73±0.07 | 5.87±0.051 |
| Water(5) | 4.70±0.089 | 3.80±0.101 | 2.74±0.116 | 4.65±0.067 | 3.74±0.068 | 2.69±0.084 | 4.54±0.047 | 3.62±0.067 | 2.52±0.083 | 4.58±0.080 | 3.66±0.080 | 2.56±0.063 |
| Vegetables(10) | 6.84±0.045 | 5.57±0.087 | 4.57±0.091 | 6.87±0.047 | 5.59±0.101 | 4.63±0.091 | 6.87±0.044 | 5.53±0.048 | 4.63±0.064 | 6.91±0.039 | 5.60±0.095 | 4.68±0.076 |
TVC=Total viable bacterial count, TEC=Total E.coli count, TSC=Total Salmonella spp. count, CFU=Colony-forming unit, SD-Standard deviation, BAU=Bangladesh Agricultural University, E.coli=Escherichia coli
Isolation and identification of E. coli and Salmonella spp.
A total of 180 (75%) E. coli and 136 (56.67%) Salmonella spp. were isolated from the 240 samples through cultural and molecular tests (Figure-1). Among these, the highest E. coli (92.5%) and Salmonella spp. (72.5%) were detected in soil samples and lowest in water samples (Table-3).
Figure-1.
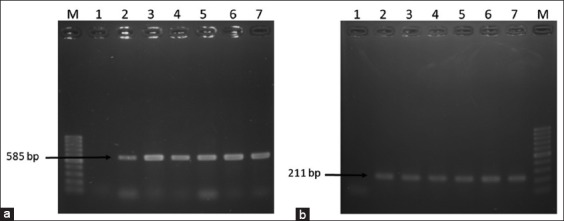
Polymerase chain reaction (PCR) amplification of 16S rRNA of Escherichia coli and invA gene of Salmonella spp. (a) PCR amplification of 16S rRNA of E. coli. Lane M: 100 bp DNA Marker, 1: Negative control, 2: Positive control, and 3-7: Representative E. coli isolates. (b) PCR amplification of invA gene of Salmonella spp. Lane M: 100 bp DNA Marker, 1: Negative control, 2-6: Representative Salmonella spp. isolates, and 7: Positive control.
Table 3.
Prevalence of E.coli and Salmonella spp. in dairy cattle and farm environment.
| Source of sample | Cow dung(%) | Milk(%) | Milker’s hand wash(%) | Soil(%) | Water(%) | Vegetables(%) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E.coli | Salmonella spp. | E.coli | Salmonella spp. | E.coli | Salmonella spp. | E.coli | Salmonella spp. | E.coli | Salmonella spp. | E.coli | Salmonella spp. | |
| RAF | 11/15 (73.33) | 9/15 (60) | 6/10 (60) | 4/10 (40) | 7/10 (70) | 5/10 (50) | 8/10 (80) | 6/10 (60) | 2/5 (40) | 1/5 (20) | 7/10 (70) | 5/10 (50) |
| BDF | 11/15 (73.33) | 10/15 (66.67) | 4/10 (40) | 3/10 (30) | 6/10 (60) | 4/10 (40) | 9/10 (90) | 6/10 (60) | 1/5 (20) | 1/5 (20) | 7/10 (70) | 4/10 (40) |
| DFS | 12/15 (80) | 10/15 (66.67) | 8/10 (80) | 5/10 (5) | 8/10 (80) | 6/10 (60) | 10/10 (100) | 8/10 (80) | 3/5 (60) | 2/5 (40) | 8/10 (80) | 5/10 (50) |
| DFB | 13/15 (86.67) | 13/15 (86.67) | 9/10 (90) | 6/10 (60) | 8/10 (80) | 7/10 (70) | 10/10 (100) | 9/10 (90) | 3/5 (60) | 2/5 (40) | 9/10 (90) | 5/10 (50) |
| Total | 47/60 (78.33) | 42/60 (70) | 27/40 (67.5) | 18/40 (45) | 29/40 (72.5) | 22/40 (55) | 37/40 (92.5) | 29/40 (72.5) | 9/20 (45) | 6/20 (30) | 31/40 (77.5) | 19/40 (47.5) |
RAF=Research animal farm, BDF=BAU dairy farm, DFS=Dairy farm in Sutiakhali, DFB=Dairy farm in Boira, E.coli=Escherichia coli, BAU=Bangladesh Agricultural University
Determination of virulent genes of E. coli
Among the 180 E. coli isolates, 47 (26.11%) were found positive for the presence of either one or all the three virulent genes (Figure-2), of which stx1 was the most prevalent (13.33%). Few isolates were also found positive for two of the virulence genes. About 4.44% isolates were found positive for both the stx1 and stx2 while two isolates (1.11%) were found to be positive for all the three virulent genes. Only three isolates (1.67%) were identified as EHEC based on the detection of rfbO157 gene. Among the six types of samples, cow dung was more contaminated with pathogenic E. coli strains than other collected samples. Distributions of virulent genes in the isolated E. coli in different samples are presented in Figure-3.
Figure-2.
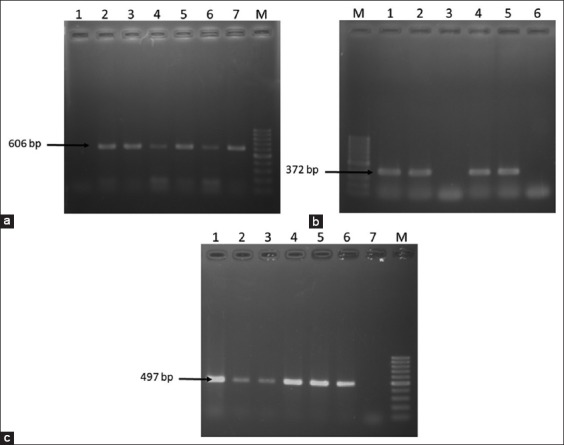
Polymerase chain reaction (PCR) amplification of virulence genes of Escherichia coli. (a) PCR amplification of stx1gene of E. coli. Lane M: 100 bp DNA Marker, 1: Negative control, 2-6: Representative E. coli isolates, and 7: Positive control. (b) PCR amplification of stx2 gene of E. coli. Lane M: 100 bp DNA Marker, 1: Positive control, 2-5: Representative E. coli isolates, and 6: Negative control. (c) PCR amplification of rfbO157 gene of E. coli. Lane M: 100 bp DNA Marker, 1-5: Representative E. coli isolates, 6: Positive control, and 7: Negative control.
Figure-3.
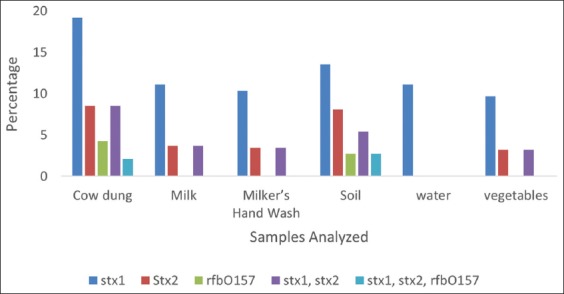
Distribution of stx1, stx2, and rfbO157 in isolated Escherichia coli.
Antimicrobial susceptibility test
Isolated E. coli and Salmonella spp. were subjected to antimicrobial susceptibility test against 12 commonly used antibiotics including three beta-lactam antibiotics, namely ertapenem, meropenem, and imipenem. From Table-4, it is evident that all the isolates of E. coli and Salmonella spp. were 100% resistance to azithromycin. E. coli was also found highly resistant to tetracycline (89.44%), erythromycin (88.89%), oxytetracycline (78.89%), and ertapenem (66.67%). Similarly, Salmonella spp. was found resistant to erythromycin (87.5%), followed by tetracycline (86.76%), oxytetracycline (75.73%), and ertapenem (50%) antibiotics. Both the isolates were highly susceptible to gentamycin, ciprofloxacin, and imipenem.
Table 4.
Antibiogram of the isolated E.coli and Salmonella spp.
| Antibiotic | E.coli | Salmonella spp. | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cow dung(47) | Milk(27) | Milker’s Hand Wash(29) | Soil(37) | Water (9) | Vegetables (31) | Total (180) | Cow dung(42) | Milk (18) | Milker’s Hand Wash (22) | Soil(29) | Water(6) | Vegetables (19) | Total (136) | |
| AZM | 47 (100) | 27 (100) | 29 (100) | 37 (100) | 9 (100) | 31 (100) | 180 (100) | 42 (100) | 18 (100) | 22 (100) | 29 (100) | 6 (100) | 19 (100) | 136 (100) |
| C | 19 (40.42) | 8 (29.63) | 9 (31.03) | 13 (35.13) | 3 (33.3) | 9 (29.03) | 61 (33.89) | 15 (35.71) | 5 (27.78) | 7 (31.82) | 9 (31.03) | 2 (33.33) | 5 (26.31) | 43 (31.62) |
| CIP | 10 (21.28) | 4 (14.81) | 4 (13.79) | 6 (16.22) | 1 (11.11) | 4 (12.90) | 29 (16.11) | 8 (19.05) | 2 (11.11) | 3 (13.64) | 4 (13.79) | 0 (0) | 2 (10.53) | 19 (13.97) |
| E | 44 (93.62) | 24 (88.89) | 25 (86.21) | 33 (89.19) | 8 (88.89) | 26 (83.87) | 160 (88.89) | 39 (92.86) | 15 (83.33) | 19 (86.36) | 26 (89.65) | 5 (83.33) | 15 (78.95) | 119 (87.5 |
| GEN | 6 (12.76) | 2 (7.41) | 2 (6.90) | 4 (10.81) | 0 (0) | 2 (6.45) | 16 (8.89) | 4 (9.52) | 1 (5.55) | 1 (4.54) | 2 (6.90) | 0 (0) | 1 (5.26) | 9 (6.62) |
| K | 20 (42.55) | 9 (33.33) | 9 (31.03) | 13 (35.13) | 2 (22.22) | 6 (19.35) | 59 (32.78) | 16 (38.09) | 5 (27.78) | 6 (27.27) | 9 (31.03) | 1 (16.67) | 2 (10.53) | 39 (28.68) |
| N | 21 (44.68) | 10 (37.04) | 9 (31.03) | 13 (35.13) | 2 (22.22) | 6 (19.35) | 61 (33.89) | 18 (42.86) | 6 (33.33) | 7 (31.82) | 11 (37.93) | 2 (33.33) | 3 (15.79) | 47 (34.56) |
| O | 40 (85.11) | 21 (77.78) | 22 (75.86) | 30 (81.08) | 7 (77.78) | 22 (70.97) | 142 (78.89) | 35 (83.33) | 13 (72.22) | 15 (68.18) | 22 (75.86) | 4 (66.67) | 14 (73.68) | 103 (75.73) |
| ETP | 35 (74.47) | 18 (66.67) | 17 (58.62) | 26 (70.27) | 6 (66.67) | 18 (58.06) | 120 (66.67) | 25 (59.52) | 9 (50) | 10 (45.45) | 16 (55.17) | 2 (33.3) | 6 (31.58) | 68 (50) |
| MEM | 16 (34.04) | 7 (25.92) | 7 (24.14) | 10 (27.03) | 2 (22.22) | 7 (22.58) | 49 (27.22) | 11 (26.19) | 4 (22.22) | 5 (22.73) | 7 (24.14) | 1 (16.67) | 3 (15.79) | 31 (22.79) |
| IPM | 11 (23.40) | 5 (18.52) | 5 (17.24) | 8 (21.62) | 1 (11.11) | 4 (12.90) | 34 (18.89) | 7 (16.67) | 2 (11.11) | 3 (13.64) | 4 (13.79) | 1 (16.67) | 1 (5.26) | 18 (13.23) |
| TE | 45 (95.74) | 24 (88.89) | 24 (82.76) | 34 (91.89) | 8 (88.89) | 26 (83.87) | 161 (89.44) | 39 (92.86) | 15 (83.33) | 18 (81.82) | 25 (86.21) | 5 (83.33) | 16 (84.21) | 118 (86.76) |
AZM=Azithromycin, C=Chloramphenicol, CIP=Ciprofloxacin, E=Erythromycin, GEN=Gentamycin, K=Kanamycin, N=Neomycin, O=Oxytetracycline, ETP=Ertapenem, MEM=Meropenem, IPM=Imipenem, TE=Tetracycline, E.coli=Escherichia coli
Molecular detection of antibiotic resistance genes
E. coli and Salmonella spp. that showed phenotypically resistance to erythromycin, tetracycline, and beta-lactam antibiotics were further screened for the detection of ereA, tetA, and tetB and SHV resistance genes (Figure-4). From Table-5, it is evident that tetA was the most prevalent resistance genes (80.51-84.47%%) among the four resistance genes both in E. coli and Salmonella spp. and SHV genes were the lowest one (22.06-25%). On a sample basis, bacteria isolated from cow dung harbored the highest resistance genes (50-53.25%) compared to other samples originated from the dairy farms.
Figure-4.
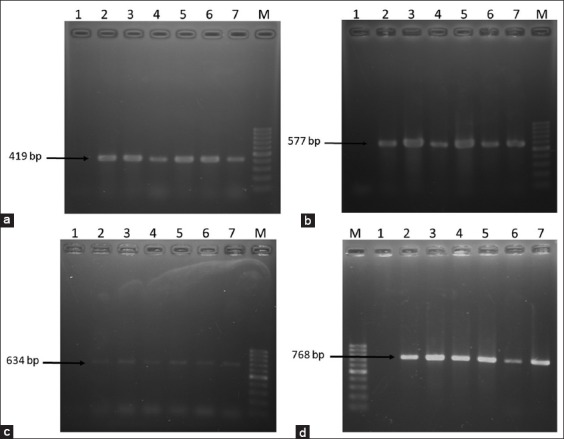
Polymerase chain reaction (PCR) amplification of antibiotic resistance genes of Escherichia coli and Salmonella spp. (a) PCR amplification of ereA gene of erythromycin resistant E. coli and Salmonella spp. Lane M: 100 bp DNA Marker, 1: Negative control, 2-3: Representative E. coli isolates, 4-5: Representative Salmonella spp. isolates, 6: Positive control for E. coli, and 7: Positive control for Salmonella spp. (b) PCR amplification of tetA gene of tetracycline resistant E. coli and Salmonella spp. Lane M: 100 bp DNA Marker, 1: Negative control, 2-3: Representative E. coli isolates, 4-5: Representative Salmonella spp. isolates, 6: Positive control for E. coli, and 7: Positive control for Salmonella spp. (c) PCR amplification of tetB gene of tetracycline resistant E. coli and Salmonella spp. Lane M: 100 bp DNA Marker, 1: Negative control, 2-3: Representative E. coli isolates, 4-5: Representative Salmonella spp. isolates, 6: Positive control for E. coli. and 7: Positive control for Salmonella spp. (d) PCR amplification of SHV gene of ertapenem, imipenem and meropenem resistant E. coli and Salmonella spp. Lane M: 100 bp DNA Marker, 1: Negative control,2: Positive control for E. coli, 3: Positive control for Salmonella spp., 4-5: Representative E. coli isolates, and 6-7: Representative Salmonella spp. isolates.
Table 5.
Distribution of antibiotic resistance genes of the isolated E.coli and Salmonella spp.
| Resistance gene | E.coli | ||||||
|---|---|---|---|---|---|---|---|
| Cow dung | Milk | Milker’s hand wash | Soil | Water | Vegetables | Total | |
| ereA | 19/44 (43.18) | 8/24 (33.33) | 8/25 (32.00) | 12/33 (36.36) | 3/8 (37.5) | 8/26 (30.77) | 58/160 (36.25) |
| tetA | 40/45 (88.89) | 20/24 (83.33) | 21/24 (87.5) | 28/34 (82.35) | 6/8 (75) | 21/26 (80.77) | 136/161 (84.47) |
| tetB | 21/45 (46.67) | 9/24 (37.5) | 8/24 (33.33) | 13/34 (38.23) | 3/8 (37.5) | 10/26 (38.46) | 64/161 (39.75) |
| SHV | 10/35 (28.57) | 4/18 (22.22) | 4/17 (23.53) | 7/26 (26.92) | 0/6 (0) | 5/18 (27.78) | 30/120 (25.00) |
| Total | 90/169 (53.25) | 41.90 (45.55) | 41/90 (45.55) | 60/127 (47.24) | 12/30 (40) | 44/96 (45.83) | 288/603 (47.76) |
| Resistance gene | Salmonella spp. | ||||||
| Cow dung | Milk | Milker’s hand wash | Soil | Water | Vegetables | Total | |
| ereA | 16/39 (41.02) | 5/15 (33.33) | 6/19 (31.58) | 9/26 (34.61) | 0/5 (0) | 4/15 (26.67) | 40/119 (33.61) |
| tetA | 33/39 (84.61) | 12/15 (80.00) | 14/18 (77.78) | 20/25 (80.00) | 4/5 (80.00) | 12/16 (75.00) | 95/118 (80.51) |
| tetB | 16/39 (41.02) | 5/15 (33.33) | 7/18 (38.89) | 9/25 (36.00) | 1/5 (20.00) | 5/16 (31.25) | 43/118 (36.44) |
| SHV | 6/25 (24.00) | 2/9 (22.22) | 2/10 (20.00) | 4/16 (25.00) | 0/2 (0) | 1/6 (16.67) | 15/68 (22.06) |
| Total | 71/142 (50) | 24/54 (44.44) | 29/65 (44.61) | 42/92 (45.65) | 5/17 (29.41) | 22/53 (41.51) | 193/423 (45.63) |
E.coli=Escherichia coli
Discussion
Antimicrobial agents are indiscriminately used in animal production system for disease prevention and control resulting in the development of resistance against these agents, particularly in zoonotic bacteria that can easily transfer to human through food chains [29]. Zoonotic antibiotic-resistant microorganisms such as E. coli and Salmonella spp. now have become the global issue as these organisms may bargain the capacity of different treatment regimens to address sickness and disease in human therapeutic settings [30]. In Bangladesh, not enough baseline data are available on the occurrence of antibiotic-resistant bacteria in dairy farm and farm environment to support the National Action Plan of the Government on AMR. To the best of our knowledge this is the first one-health based comprehensive research on the investigation of antibiotic-resistant E. coli and Salmonella spp. from dairy cattle, dairy farm environment and farm workers in Bangladesh having public health significance.
Cattle in the farm are continuously shedding cow dung and urine into the soil. Thus, soli of the dairy farm is getting heavily contaminated with bacteria of cow dung and urine. Similarly, the present study showed that among all the samples, the highest TVC was found in soil samples of Sutiakhali dairy Farm (Table-2). Since E. coli and Salmonella spp. are the part of natural intestinal flora, they were often found in higher number in cow dung. Cow dung contains huge microbial population including pathogenic bacteria that have a potential effect on human and animal health [31]. Contaminated drinking water is the common source of coliform bacteria in dairy farm [32]. In comparison to other collected samples, water samples were found less contaminated by E. coli. Hassan et al. [33] found the geometric mean of heterotrophic plate count of tap water from Mymensingh, Gazipur, and Sherpur district were 8.4×105, 2.5×106, and 6.8×105 CFU/100 ml that were higher than our study. Milk is the most important output of dairy farm for human. Here, we found the milk samples contaminated with E. coli and Salmonella spp. This contamination may be due to improper hygiene practice in dairy farm especially hygiene of milker’s hand as higher TVC, E. coli, and Salmonella spp. count were found in milker’s hand wash than milk sample. Khan et al. [34] also investigated milk sample of Boira area and found the TVC and TCC as 5.93 and 2.52 log CFU/ml, respectively, a slightly lower than our findings.
Detection of E. coli and Salmonella spp. in the samples analyzed originating from various farms was not unexpected, since E. coli and Salmonella spp. are ubiquitous in nature. In this study, we did not identify the isolated Salmonella at species level, but their presence is alarming. However, detail study on identification of these Salmonella at species level in underway in another study. They are the part of intestinal microflora of animals and birds. Barlow et al. [30], Rodriguez-Rivera et al. [35], Navajas-Benito et al. [36], Jajarmi et al. [37], and Batabyal et al. [38] also reported the presence of E. coli and Salmonella spp. in dairy farm samples. The occurrence of higher prevalence of E. coli and Salmonella spp. in the dairy farm may be due to improper management of cow dung resulting transmission of E. coli and Salmonella spp. into dairy farm environment especially milk and water [39]. Vegetable samples analyzed were also found positive for E. coli and Salmonella spp. This may be linked with the use of untreated cow dung as the organic fertilizer for vegetable production in nearby agriculture land. Mukherjee et al. [40] reported that fresh cow dung or improper treatment of cow dung may be the cause of transmission of E. coli and Salmonella spp. to vegetables on farm.
STEC and EHEC are major human pathogen associated with foodborne illness [37]. In this study, STEC and EHEC were detected from various samples (Figure-3). The prevalence of these pathogens was highest in cow dung. Pathogenic E. coli are commonly found in animal feces but their prevalence in current study was quite higher than the previous study. In Bangladesh a study reported, the occurrence of 10% STEC [41] in the cattle fecal samples and in India it was reported as 19% [42]. The observed variations in the occurrence of STEC and EHEC among these studies might be due to different geographic locations and variations in farm management.
Antibiotic resistance is a serious health issue globally. This study focused on distribution and occurrence of antibiotic-resistant E. coli and Salmonella spp. in dairy farm environment having public health importance. Twelve commonly used antibiotics including carbapenem group were tested in the antibiogram study. All the E. coli and Salmonella spp. were found 100% resistant to azithromycin. Earlier Islam et al. [43] observed lower resistance against azithromycin in E. coli isolated from milk in Bangladesh. In the present study, gentamycin, ciprofloxacin, imipenem, meropenem, kanamycin, chloramphenicol, and neomycin were found most effective whereas erythromycin, tetracycline, and ertapenem were found less effective against E. coli and Salmonella spp. These observed variations in the sensitivity pattern may be linked with the variations of concentration and frequency of the use of these antibiotics in Bangladesh that need further investigation.
Carbapenem group of antibiotics (ertapenem, imipenem, and meropenem) is major choice of antibiotic for treating disease caused by multidrug-resistant E. coli infections in human. In veterinary practice, these drugs are not commonly used in Bangladesh yet. Islam et al. [44] found imipenem sensitive E. coli isolates in clinical samples of human origin in Bangladesh. On the other hand, Mamun et al. [45] found E. coli isolated from rectal swab of healthy cattle as highly sensitive to carbapenem group of antibiotics study. For the 1st time in Bangladesh, the present study showed that E. coli and Salmonella spp. isolated from dairy farm are resistant to carbapenem group of antibiotics, e.g. ertapenem (50-66.67%), meropenem (up to 27.79%) and imipenem (13.23-18.89%). Carbapenem-resistant Enterobacteriaceae are listed as the critical group of priority pathogen as defined by the WHO [46].
This may be due to the transmission of carbapenem resistance bacteria from human particularly farm workers and visitors to dairy cattle and farm environment.
Among the resistance genes, tetA and tetB responsible for resistance against tetracycline were found as the most prevalent. However, this was not unexpected, since tetracycline is one of the most widely used antibiotics in Bangladesh. Previously, Navajas-Benito et al. [37] also reported the prevalence of a higher number of tetracycline resistance genes (11/15) in E. coli associated with dairy farm. On the other hand, the prevalence of SHV and ereA genes was found comparatively low in this study as reported elsewhere [47,48].
Conclusion
The present study identified the widespread occurrence of antibiotic-resistant E. coli and Salmonella spp. in dairy cattle and farm environment. Many of these isolates were also found pathogenic in nature. From farm, these resistant pathogens can transmit to human through the food chain (contaminated milk and vegetables) or through direct and indirect contact. Practice of good farm management including hygiene and manure treatment need to be established to reduce the chance of transmission of antibiotic-resistant bacteria to human along with judicial use of antibiotics in the dairy cattle. In addition, one-health approaches are need to be adopted to control AMR in humans, animals and environment.
Authors’ Contributions
MTR, MAS, and AMMTR designed the study. MAS, AAMS, and RS did laboratory work assisted by MTR. AMMTR, and MAS wrote the manuscript and analyzed. MTR, MBR and SMLK critically checked and improved the manuscript. All authors read and approved the final manuscript.
Acknowledgments
The authors are very grateful to the dairy farm owners for access to the samples. The authors also acknowledge the part of financial support from Bangladesh Agricultural University Research System (BAURES; Project No: 2017/236/BAU) and the Ministry of Education (Project No. 2018/692/MOE), Government of People’s Republic of Bangladesh to conduct the research. Md. Abdus Sobur was supported by NST Research Fellowship from Ministry of Science and Technology, Government of People’s Republic of Bangladesh.
Competing Interests
The authors declare that they have no competing interests.
Publisher’s Note
Veterinary World remains neutral with regard to jurisdictional claims in published institutional affiliation.
References
- 1.Tule A, Hassani U. Colonization with antibiotic-resistant E. coli in commensal fecal flora of newborns. Int. J. Curr. Microbiol. Appl. Sci. 2017;6(5):1623–1629. [Google Scholar]
- 2.Fair R.J, Tor Y. Antibiotics and bacterial resistance in the 21stcentury. Perspect. Med. Chem. 2014;6:14459. doi: 10.4137/PMC.S14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mckenna M. The Coming Cost of Superbugs:10 Million Deaths Per Year. 2014. [Last accesed on 10.01.2019]. Available from: http://www.wired.com/2014/12/oneill-rptamr .
- 4.Prestinaci F, Pezzotti P, Pantosti A. Antimicrobial resistance:A global multifaceted phenomenon. Pathog. Glob. Health. 2015;109(7):309–318. doi: 10.1179/2047773215Y.0000000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li B, Webster T.J. Bacteria antibiotic resistance:New challenges and opportunities for implant-associated orthopedic infections. J. Orthop. Res. 2018;36(1):22–32. doi: 10.1002/jor.23656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hao H, Cheng G, Iqbal Z, Ai X, Hussain H.I, Huang L, Dai M, Wang Y, Liu Z, Yuan Z. Benefits and risks of antimicrobial use in food-producing animals. Front. Microbiol. 2014;5 doi: 10.3389/fmicb.2014.00288. Article no. 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang Q, Wang W, Regev-Yochay G, Lipsitch M, Hanage W.P. Antibiotics in agriculture and the risk to human health:How worried should we be?Evol. Appl. 2015;8(3):240–247. doi: 10.1111/eva.12185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allocati N, Masulli M, Alexeyev M, Di Ilio C. Escherichia coli in Europe:An overview. Int. J. Environ. Res. Public Health. 2013;10(12):6235–6254. doi: 10.3390/ijerph10126235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heredia N, García S. Animals as sources of food-borne pathogens: A review. Anim. Nutr. 2018;4(3):250–255. doi: 10.1016/j.aninu.2018.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amézquita-López B.A, Soto-Beltrán M, Lee B.G, Yambao J.C, Quiñones B. Isolation, genotyping and antimicrobial resistance of Shiga toxin-producing Escherichia coli. J. Microbiol. Immunol. Infect. 2018;51(4):425–434. doi: 10.1016/j.jmii.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 11.Bonanno L, Loukiadis E, Mariani-Kurkdjian P, Oswald E, Garnier L, Michel V, Auvray F. Diversity of Shiga toxin-producing Escherichia coli (STEC) O26:H11 strains examined via Stx subtypes and insertion sites of Stx and EspK bacteriophages. Appl. Environ. Microbiol. 2015;81(11):3712–3721. doi: 10.1128/AEM.00077-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halimi H.A, Seifi H.A, Rad M. Bovine salmonellosis in Northeast of Iran:Frequency, genetic fingerprinting and antimicrobial resistance patterns of Salmonella spp. Asian Pac. J. Trop. Biomed. 2014;4(1):1–7. doi: 10.1016/S2221-1691(14)60199-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nair D.V.T, Venkitanarayanan K, Johny A.K. Antibiotic-resistant Salmonella in the food supply and the potential role of antibiotic alternatives for control. Foods. 2018;7(10):167. doi: 10.3390/foods7100167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Integrated Livestock Manure Management Policy. Government of the People's Republic of Bangladesh, Ministry of Fisheries and Livestock. 2005. [Last accessed on 02.01.2019]. pp. 1–2. Available from: https://www.mofl.portal.gov.bd/sites/default/files/files/mofl.portal.gov.bd/page/221b5a19_4052_4486_ane71_18f1ff6863c1/ILMM%20Policy.pdf .
- 15.Argudin M, Deplano A, Meghraoui A, Dodemont M, Heinrichs A, Denis O, Nonhoff C, Roisin S. Bacteria from animals as a pool of antimicrobial resistance genes. Antibiotics. 2017;6(2):12. doi: 10.3390/antibiotics6020012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manyi-Loh C, Mamphweli S, Meyer E, Okoh A. Antibiotic use in agriculture and its consequential resistance in environmental sources:Potential public health implications. Molecules. 2018;23(4):795. doi: 10.3390/molecules23040795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weldezgina D, Muleta D. Bacteriological contaminants of some fresh vegetables irrigated with Awetu river in Jimma town, Southwestern Ethiopia. Adv. Biol. 2016;2016:1526764. [Google Scholar]
- 18.Thomas P, Sekhar A.C, Upreti R, Mujawar M.M, Pasha S.S. Optimization of single plate-serial dilution spotting (SP-SDS) with sample anchoring as an assured method for bacterial and yeast cfu enumeration and single colony isolation from diverse samples. Biotechnol. Rep. 2015;8:45–55. doi: 10.1016/j.btre.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bergey D.H, Buchanan R.E, Gibbons N.E American Society for Microbiology. Bergey's Manual of Determinative Bacteriology. Baltimore: Williams and Wilkins Co; 1974. [Google Scholar]
- 20.Mahmud S, Nazir K.N.H, Rahman M.T. Prevalence and molecular detection of fluoroquinolone-resistant genes (qnrA and qnrS) in Escherichia coli isolated from healthy broiler chickens. Vet. World. 2018;11(12):1720–1724. doi: 10.14202/vetworld.2018.1720-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schippa S, Iebba V, Barbato M, Di Nardo G, Totino V, Checchi M.P, Longhi C, Maiella G, Cucchiara S, Conte M.P. A distinctive'microbialsignature'in celiac pediatric patients. BMC Microbiol. 2010;10(1):175. doi: 10.1186/1471-2180-10-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shanmugasundaram M, Radhika M, Murali H.S, Batra H.V. Detection of Salmonella enterica serovar Typhimurium by selective amplification of fliC, fljB, iroB, invA, rfbJ, STM2755, STM4497 genes by polymerase chain reaction in a monoplex and multiplex format. World J. Microbiol. Biotechnol. 2009;25(8):1385–1394. [Google Scholar]
- 23.Talukdar P.K, Rahman M, Rahman M, Nabi A, Islam Z, Hoque M.M, Endtz H.P, Islam M.A. Antimicrobial resistance, virulence factors and genetic diversity of Escherichia coli isolates from household water supply in Dhaka, Bangladesh. PLoS One. 2013;8(4):e61090. doi: 10.1371/journal.pone.0061090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Desmarchelier P.M, Bilge S.S, Fegan N, Mills L, Vary J.C, Tarr P.I. A PCR specific for Escherichia coli O157 based on the rfb locus encoding O157 lipopolysaccharide. J. Clin. Microbiol. 1998;36(6):1801–1804. doi: 10.1128/jcm.36.6.1801-1804.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bauer A, Kirby W, Sherris J.C, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 1966;45(4):493–496. [PubMed] [Google Scholar]
- 26.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. 26th ed. Wayne, Pennsylvania: CLSI Supplement M100s. Clinical and Laboratory Standards Institute; 2016. [Google Scholar]
- 27.Van T.T.H, Chin J, Chapman T, Tran L.T, Coloe P.J. Safety of raw meat and shellfish in Vietnam:An analysis of Escherichia coli isolations for antibiotic resistance and virulence genes. Int. J. Food Microbiol. 2008;124(3):217–223. doi: 10.1016/j.ijfoodmicro.2008.03.029. [DOI] [PubMed] [Google Scholar]
- 28.Momtaz H, Farzan R, Rahimi E, Safarpoor Dehkordi F, Souod N. Molecular characterization of Shiga toxin-producing Escherichia coli isolated from ruminant and donkey raw milk samples and traditional dairy products in Iran. Sci. World J 2012. 2012:13. doi: 10.1100/2012/231342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szmolka A, Nagy B. Multidrug-resistant commensal Escherichia coli in animals and its impact on public health. Front. Microbiol. 2013;4:258. doi: 10.3389/fmicb.2013.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barlow R.S, McMillan K.E, Duffy L.L, Fegan N, Jordan D, Mellor G.E. Prevalence and antimicrobial resistance of Salmonella and Escherichia coli from Australian cattle populations at slaughter. J. Food Prot. 2015;78(5):912–920. doi: 10.4315/0362-028X.JFP-14-476. [DOI] [PubMed] [Google Scholar]
- 31.Pandey P, Chiu C, Miao M, Wang Y, Settles M, Del Rio N.S, Castillo A, Souza A, Pereira R, Jeannotte R. 16S rRNA analysis of diversity of manure microbial community in dairy farm environment. PLoS One. 2018;13(1):e0190126. doi: 10.1371/journal.pone.0190126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khan M, Abro S.H, Taj M.K, Abro R, Baloch H, Rind R, Rind M.R, Tunio S.A. Bacterial contamination of drinking water used at dairy farms in Quetta, Balochistan. Pure Appl. Biol. 2016;5(4):714–718. [Google Scholar]
- 33.Hassan M.S, Kabir S.M.L, Sarker Y.A, Rahman M.T. Bacteriological assessment of tap water collected from different markets of Mymensingh, Gazipur and Sherpur districts of Bangladesh with special focus on the molecular detection and antimicrobial resistance of the isolated Escherichia coli. Asian Australas. J. Food Saf. Secur. 2018;2(1):21–28. [Google Scholar]
- 34.Khan M.T.G, Zinnah M.A, Siddique M.P, Rashid M.H.A, Islam M.A, Choudhury K.A. Physical and microbial qualities of raw milk collected from Bangladesh agricultural university dairy farm and the surrounding villages. Bangladesh J. Vet. Med. 2008;6(2):217–221. [Google Scholar]
- 35.Rodriguez-Rivera L.D, Cummings K.J, Loneragan G.H, Rankin S.C, Hanson D.L, Leone W.M, Edrington T.S. Salmonella prevalence and antimicrobial susceptibility among dairy farm environmental samples collected in Texas. Foodborne Pathog. Dis. 2016;13(4):205–211. doi: 10.1089/fpd.2015.2037. [DOI] [PubMed] [Google Scholar]
- 36.Navajas-Benito E.V, Alonso C.A, Sanz S, Olarte C, Martínez-Olarte R, Hidalgo-Sanz S, Somalo S, Torres C. Molecular characterization of antibiotic resistance in Escherichia coli strains from a dairy cattle farm and its surroundings. J. Sci. Food Agric. 2016;97(1):362–365. doi: 10.1002/jsfa.7709. [DOI] [PubMed] [Google Scholar]
- 37.Jajarmi M, Fooladi A.A.I, Badouei M.A, Ahmadi A. Virulence genes, Shiga toxin subtypes, major O-serogroups, and phylogenetic background of Shiga toxin-producing Escherichia coli strains isolated from cattle in Iran. Microb. Pathog. 2017;109:274–279. doi: 10.1016/j.micpath.2017.05.041. [DOI] [PubMed] [Google Scholar]
- 38.Batabyal K, Banerjee A, Pal S, Dey S, Joardar S.N, Samanta I, Isore D.P, Singh A.D. Detection, characterization, and antibiogram of extended-spectrum beta-lactamase Escherichia coli isolated from bovine milk samples in West Bengal, India. Vet. World. 2018;11(10):1423–1427. doi: 10.14202/vetworld.2018.1423-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pangloli P, Dje Y, Oliver S.P, Mathew A, Golden D.A, Taylor W.J, Draughon F.A. Evaluation of methods for recovery of Salmonella from dairy cattle, poultry, and swine farms. J. Food Prot. 2003;66(11):1987–1995. doi: 10.4315/0362-028x-66.11.1987. [DOI] [PubMed] [Google Scholar]
- 40.Mukherjee A, Speh D, Dyck E, Diez-Gonzalez F. Preharvest evaluation of coliforms, Escherichia coli, Salmonella, and Escherichia coli O157:H7 in organic and conventional produce grown by Minnesota farmers. J. Food Prot. 2004;67(5):894–900. doi: 10.4315/0362-028x-67.5.894. [DOI] [PubMed] [Google Scholar]
- 41.Islam M.A, Mondol A.S, De Boer E, Beumer R.R, Zwietering M.H, Talukder K.A, Heuvelink A.E. Prevalence and genetic characterization of Shiga toxin-producing Escherichia coli isolates from slaughtered animals in Bangladesh. Appl. Environ. Microbiol. 2008;74(17):5414–5421. doi: 10.1128/AEM.00854-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Das S.C, Khan A, Panja P, Datta S, Sikdar A, Yamasaki S, Takeda Y, Bhattachyarya S.K, Ramamurthy T, Nair G.B. Dairy farm investigation on Shiga toxin-producing Escherichia coli (STEC) in Kolkata, India with emphasis on molecular characterization. Epidemiol. Infect. 2005;133(4):617–626. doi: 10.1017/s0950268805004000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Islam M.A, Kabir S.M.L, Seel S.K. Molecular detection and characterization of Escherichia coli isolated from raw milk sold in different markets of Bangladesh. Bangladesh J. Vet. Med. 2016;14(2):271–275. [Google Scholar]
- 44.Islam M.A, Huq M, Nabi A, Talukdar P.K, Ahmed D, Talukder K.A, Craviota A, Endtz H.P. Occurrence and characterization of multidrug-resistant New Delhi metallo-β-lactamase-1-producing bacteria isolated between 2003 and 2010 in Bangladesh. J. Med. Microbiol. 2013;62(1):62–68. doi: 10.1099/jmm.0.048066-0. [DOI] [PubMed] [Google Scholar]
- 45.Mamun M, Hassan J, Nazir K.H.M, Islam A, Zesmin K, Rahman B, Rahman M.T. Prevalence and molecular detection of quinolone-resistant E. coli in rectal swab of apparently healthy cattle in Bangladesh. Int. J. Trop. Dis. Health. 2017;24(2):1–7. [Google Scholar]
- 46.Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet D.L, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, Ouellette M. Discovery, research, and development of new antibiotics:the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018;18(3):318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 47.Ball T.A, Monte D.F, Aidara-Kane A, Matheu-Alvarez J, Ru H, Thakur S, Horovitz J, Ejobi F, Lacher D.W, Fedorka-Cray P.J. Phenotypic and genotypic characterization of Escherichia coli and Salmonella enterica from dairy cattle farms in the Wakiso district, Uganda:A cross-sectional study. Foodborne Pathog. Dis. 2019;16(1):54–59. doi: 10.1089/fpd.2018.2528. [DOI] [PubMed] [Google Scholar]
- 48.Messele Y.E, Abdi R.D, Tegegne D.T, Bora S.K, Babura M.D, Emeru B.A, Werid G.M. Analysis of milk-derived isolates of E. coli indicating drug resistance in central Ethiopia. Trop. Anim. Health Prod. 2018;51(3):661–667. doi: 10.1007/s11250-018-1737-x. [DOI] [PubMed] [Google Scholar]


