Abstract
Transcription factors positively and/or negatively impact gene expression by recruiting coregulatory factors, which interact through protein-protein binding. Here we demonstrate that mouse pancreas size and islet β-cell function are controlled by the ATP-dependent Swi/Snf chromatin remodeling coregulatory complex that physically associates with Pdx1, a diabetes-linked transcription factor essential to pancreatic morphogenesis and adult islet cell function and maintenance. Early embryonic deletion of just the Swi/Snf Brg1 ATPase subunit reduced multipotent pancreatic progenitor cell proliferation and resulted in pancreas hypoplasia. In contrast, removal of both Swi/Snf ATPase subunits, Brg1 and Brm, was necessary to compromise adult islet β-cell activity, which included whole-animal glucose intolerance, hyperglycemia, and impaired insulin secretion. Notably, lineage-tracing analysis revealed Swi/Snf-deficient β-cells lost the ability to produce the mRNAs for Ins and other key metabolic genes without effecting the expression of many essential islet-enriched transcription factors. Swi/Snf was necessary for Pdx1 to bind to the Ins gene enhancer, demonstrating the importance of this association in mediating chromatin accessibility. These results illustrate how fundamental the Pdx1:Swi/Snf coregulator complex is in the pancreas, and we discuss how disrupting their association could influence type 1 and type 2 diabetes susceptibility.
Introduction
The mammalian pancreas consists of two functionally distinct compartments: the exocrine pancreas containing acinar and ductal cells essential for secreting digestive enzymes, and the endocrine pancreas containing hormone-secreting α- (glucagon), β- (insulin), δ- (somatostatin), ε- (ghrelin), and pancreatic polypeptide cells of the islets of Langerhans that are essential for regulating glucose homeostasis. All of these pancreatic cells derive from a common pool of progenitor cells at mouse embryonic day (e)8.5 that express the pancreas and duodenum homeobox 1 (Pdx1) transcription factor, a critical regulator of pancreas development, later β-cell formation, and adult islet β-cell function. In fact, pancreas agenesis occurs in mice and humans that lack PDX1 (1,2), whereas heterozygous mutations cause type 2 diabetes (T2D) because of islet β-cell dysfunction (3).
Embryonic Pdx1+ pancreatic progenitor cells rapidly divide and acquire the expression of other transcription factors essential to organ expansion and lineage diversification, including Ptf1a (4) and Sox9 (5). These Pdx1+Ptf1a+Sox9+ cells form the highly proliferative multipotent pancreatic progenitor cell (MPC) pool that differentiates into the distinct exocrine, ductal, and islet cell types (6). Notably, pancreas mass is restricted in mice by the MPC pool size (7), which is affected by early embryonic genetic removal of Pdx1, Ptf1a, or Sox9 (2,4,5). Because of considerable variations in pancreas mass (and therefore variable β-cell number) between humans (8), understanding how the transcriptional activities of these essential MPC regulators control growth rate and final organ size is of significant importance. It has been proposed that dissimilarities in pancreas mass influence diabetes susceptibility, a proposal supported by the reduced pancreas size of autoantibody-positive individuals with type 1 diabetes (9).
In the postnatal pancreas, Pdx1 is produced at much higher levels in islet β-cells than in other pancreas cell types (10). Conditional removal of Pdx1 from these cells in mice leads to a profound loss of β-cell function and identity, because these cells rapidly transdifferentiate to glucagon+ and insulin− α-like cells (11). This remarkable control derives not only from the positive actions of Pdx1 on target gene transcription but also from its repression of key α-cell functional genes in β-cells, such as MafB and Gcg. As a result of these novel and fundamental properties, Pdx1 is viewed as one of the most critical pancreas-enriched transcription factors (6).
Transcription factors like Pdx1 predominantly control gene expression through the recruitment of coregulators, often operating as large multiprotein complexes. These coregulators affect transcriptional activity positively and/or negatively, for example, by displacing nucleosomes, epigenetically modifying histones/DNA, and affecting recruitment of the RNA polymerase II transcriptional machinery. Because of limited knowledge of the coregulators influencing Pdx1 activity, an unbiased chemical crosslinking, immunoprecipitation, and mass spectrometry analysis was used to identify interacting proteins in rodent β-cell lines. Numerous proteins with an array of cellular functions were found to associate with Pdx1, including the ATP-dependent Swi/Snf chromatin remodeling complex (12). Significantly, Pdx1 was the principal islet β-cell–enriched transcription factor binding to Swi/Snf in mouse β-cell lines (12).
The multisubunit Swi/Snf complex uses the energy of ATP hydrolysis, through the actions of the two mutually exclusive Brg1 (i.e., also referred to as Smarca4) and Brm (i.e., Smarca2) ATPase subunits (13,14) (Fig. 1A), to disrupt DNA-nucleosome contacts and influence DNA accessibility. We previously discovered that in vitro knockdown of Brg1 in rodent β-cell lines negatively affected Pdx1 target gene expression (e.g., Ins, MafA, and Glut2). Moreover, Pdx1:Swi/Snf interactions were not only acutely enhanced in islet β-cells by increased glucose concentrations but were also reduced in human T2D β-cells (12). Here we show that the Pdx1:Brg1/Swi/Snf complex is critical for mouse MPC proliferation, with embryonic conditional deletion of only the Brg1 gene encoding ATPase resulting in a ∼50% smaller pancreas. In contrast, removing both Brg1 and Brm was necessary to impact postnatal β-cells, causing severe changes in expression of Ins and other β-cell regulatory genes, a hallmark feature of T2D β-cells. Collectively, our results suggest that Pdx1:Swi/Snf is required for controlling the growth rate of the embryonic pancreas, and thus its final postnatal size, and for maintaining β-cell identity in adult islets.
Figure 1.
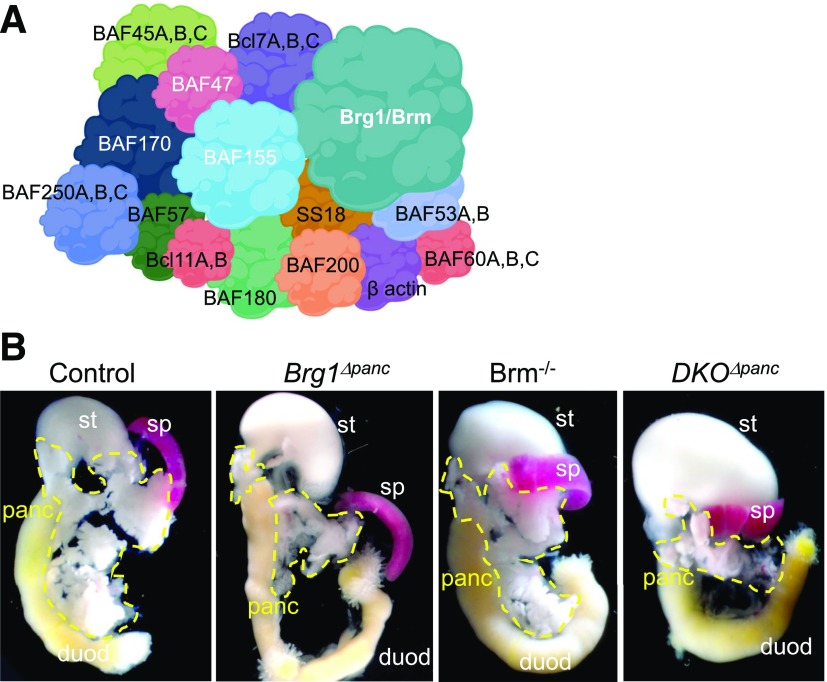
Pancreas size is reduced upon embryonic deletion of the mouse Brg1 Swi/Snf ATPase subunit. A: Schematic illustrating the composition of the mammalian Swi/Snf complex, with the core of the coregulator containing the Brg1 or Brm ATPase and the BAF47, BAF155, and BAF170 subunits (white font). Swi/Snf complexes also contain 8–10 other BAFs (Brm- or Brg1-associated factors) and accessory subunits, which vary depending on the tissue and developmental stage (14). B: Representative image of the pancreas (panc), stomach (st), spleen (sp), and duodenum (duod) in P1 control, Brg1Δpanc, Brm−/−, and DKOΔpanc mice.
Research Design and Methods
Animals
Ptf1a-Cre (15) and MIP-CreERT (16) mice were used to remove the Loxp sites surrounding exons 17 and 18 of the Brg1 locus (Brg1f/f [17]) and the Stop cassette in the Rosa26-Loxp-Stop-Loxp-tdTomato lineage reporter (R26LSL-tdTomato [18]). Brm−/− mice were generated using homologous recombination to insert the neomycin gene into Brm exon 4 (19). The following genotypes were used for the developmental studies: control, Ptf1a-Cre;Brg1f/+ or Ptf1a-Cre;Brg1f/+;Brm+/−; experimental, Brg1Δpanc (Ptf1a-Cre;Brg1f/f), Brm−/− (Ptf1a-Cre;Brm−/−), and DKOΔpanc (Ptf1a-Cre;Brg1f/f;Brm−/−). Noon of the day of the vaginal plug discovery was designated day e0.5. For BrdU injections, 100 mg of BrdU (B5002; Sigma-Aldrich) per kilogram of pregnant dam body weight was injected 30 min before embryo harvest.
The adult studies consisted of these genotypes: control, MIP-CreERT;Brg1f/+;Brm+/−;R26LSL-tdTomato/+; and experimental, Brg1Δβ;Brm+/− (MIP-CreERT;Brg1f/f;Brm+/−; R26LSLtdTomato/+), Brg1Δβ/+;Brm−/− (MIP-CreERT; Brg1f/+; Brm−/−; R26LSLtdTomato/+), and βDKO (MIP-CreERT; Brg1f/f; Brm−/−; R26LSLtdTomato/+). CreERT-mediated recombination of Brg1f/f and the R26LSLtdTomato was achieved by administration of 4 mg tamoxifen (T5648; Sigma-Aldrich) by oral gavage three times over a 5-day period.
Intraperitoneal Glucose Tolerance Test and Serum Insulin Measurements
Mice (n = 5–12) were given intraperitoneal injection of d-glucose (2 mg/g body wt) after a 6-h fast. Blood glucose was measured using a FreeStyle glucometer (Abbott Diabetes Care). Serum insulin was measured by radio immunoassay at the Vanderbilt Hormone Assay and Analytical Services Core.
Glucose-Stimulated Insulin Secretion
Secreted insulin from isolated control and βDKO mice (n = 8–10) islets was performed as described previously (20). The outcome was presented as the fold change between the percentage of secreted insulin (relative to insulin content) at 16.7 mmol/L glucose and the percentage of secreted insulin (relative to insulin content) at 2.8 mmol/L glucose. Islet insulin content was calculated as the concentration of insulin per islet in each reaction (ng/mL/islet).
Tissue Preparation and Immunostaining
Whole embryos and adult pancreata were fixed in 4% (v/v) paraformaldehyde, embedded, and sectioned to 6 μm. Immunofluorescence staining was performed as previously described (21) with the antibodies listed in Supplementary Table 1. Embryos were cut on the transverse (cross-sections) plane throughout the pancreatic epithelium, and manual cell counting was performed on antibody-stained sections prepared every 60 µm (e12.5) or 90 µm (e15.5) from the superior to inferior region.
Proximity Ligation Assay
The assay was performed on e12.5 sections following the manufacturer’s protocol (Sigma-Aldrich) with goat transcription factors Pdx1 (1:20,000) (AB47383; Abcam), Ptf1a (1:2,000) (from C.V.E.W.) or rabbit Sox9 (1:500) (AB5535; Millipore) in combination with rabbit ATPase Brg1 (1:400) (sc-10768; Santa Cruz Biotechnology), goat Brg1 (1:500) (AF5738; R & D Systems), mouse Brm (1:500) (sc-17828; Santa Cruz Biotechnology), or rabbit Brm (1:500) (ab1559; Abcam) antibodies. Immunofluorescence Z-Stack images were acquired on a Zeiss Axioimager M2 fluorescence scope and processed using ImageJ software.
Flow Cytometry, RNA Purification, and Quality Control of Sorted β-Cells
Isolated islets were dispersed into a single-cell suspension (Accumax; A7089; Sigma-Aldrich), stained with DAPI, and sorted by gating for Tomato+DAPI+ cells by FACS at the Vanderbilt Flow Cytometry Core. RNA was isolated from FACS-purified β-cells (control: 10,215 ± 1,589 cells [n = 3], βDKO: 16,267 ± 3,032 cells [n = 3]) using the Maxwell 16 LEV simplyRNA Tissue Kit (TM351; Promega), and then DNAse was treated and analyzed on an Agilent 2100 Bioanalyzer. Only samples with an RNA Integrity Number >8.0 were used for cDNA synthesis and library preparation.
RNA Sequencing and Analysis
cDNA libraries were constructed from RNA isolated from FACS-purified control and βDKO islet β-cells, and paired-end sequencing of three replicates was performed on an Illumina NovaSeq6000 (150 nucleotide reads). The generated FASTQ files were processed and interpreted using the Genialis visual informatics platform (https://www.genialis.com). Sequence quality checks were determined using raw and trimmed reads with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc), and Trimmomatic (22) was used to trim adapters and filter out poor-quality reads. Trimmed reads were then mapped to the University of California, Santa Cruz, mm10 reference genome using the HISAT2 aligner (23). Gene expression levels were quantified with HTSeq-count (24), and differential gene expression analyses were performed with DESeq2 (25). Poorly expressed genes, which have expression count summed over all samples of <10, were filtered out from the differential expression analysis input matrix. RNA expression analysis of selected candidates was performed with the quantitative (q)PCR primers provided in Supplementary Table 2.
Chromatin Immunoprecipitation Assays
Chromatin was prepared from 1,000 to 1,400 control or βDKO islets and chromatin immunoprecipitation (ChIP) was conducted with Pdx1 (AB47383), Nkx2.2 (HPA003468; Sigma-Aldrich), or IgG antibodies as described previously (21) (n = 3). The chromatin was sheared to ∼200–300 base pairs (bp). qPCR was performed with immunoprecipitated DNA over Ins2 (i.e., −95/−35 bp) and Nkx6.1 (−884/−720 bp). Binding enrichment is presented as the fold enrichment of the transcription factor signal on Ins2 or Nkx6.1 over the control β-actin signal relative to IgG. Primer sequences are available in Supplementary Table 2.
Statistical Analysis
Statistical significance was determined using the two-tailed Student t test. Data are presented as the mean ± SEM. A threshold of P < 0.05 was used to declare significance.
Study Approval
All animal studies were reviewed and approved by the Vanderbilt University Institutional Animal Care and Use Committee. Mice were housed and cared for according to the Vanderbilt Department of Animal Care and the Institutional Animal Care and Use Committee/Office of Animal Welfare Assurance standards and guidelines.
Results
Embryonic Pancreas–Specific Removal of the Brg1 ATPase Results in Pancreatic Hypoplasia
To evaluate the mechanistic basis by which Swi/Snf controls pancreas mass in vivo, we crossed mice producing a Cre recombinase driven by the Ptf1a locus (Ptf1a-Cre [15]) with mice containing Loxp sites surrounding exons 17 and 18 of the Brg1 gene (i.e., Brg1Δpanc [17]) or constitutive Brm-null (Brm−/− [19]) alleles or in combination to create a developmental double knockout (DKOΔpanc). Brm−/− animals are viable, fertile, and have no overt physiological or morphological phenotypes (19). Brg1 removal was observed in ∼50% of e12.5 pancreatic epithelial cells in Brg1Δpanc and DKOΔpanc mutants. Brg1 levels did not increase in Brm−/− embryos (Supplementary Fig. 1) and Brm did not in Brg1Δpanc mutants (Supplementary Fig. 2), signifying no compensatory upregulation between these alternative ATPase Swi/Snf subunits.
At postnatal day 1 (P1), Brg1Δpanc mice showed a severe reduction in pancreas size that was not observed in Brm−/− mice or exacerbated in DKOΔpanc (Fig. 1B). However, the size of the spleen, liver, and kidneys was unaffected in the Swi/Snf ATPase mutants (Fig. 1B) (data not shown). The incomplete inactivation of Brg1 within the MPC pool (Supplementary Fig. 1) led to the presence of nonrecombined escaper cells, with Brg1 absent from most P1 acinar cells (in which Ptf1a expression is enriched and thereby increasing the likelihood of inactivation of Brg1f/f alleles) but present in islet hormone+ Ptf1a− cells (4) (Supplementary Fig. 3). The preservation of Brg1 in DKOΔpanc islets presumably allows normal control of blood glucose levels postnatally, because β-cells totally deficient in all Swi/Snf activity have a significant impairment (described below). These observations suggest that the pancreatic hypoplasia reported in the 3-week-old Brg1Δpanc mutants (26) results from a reduction in embryonic MPC numbers.
Pdx1 Binds to Brg1 and Brm1 in MPCs and Not Ptf1a or Sox9
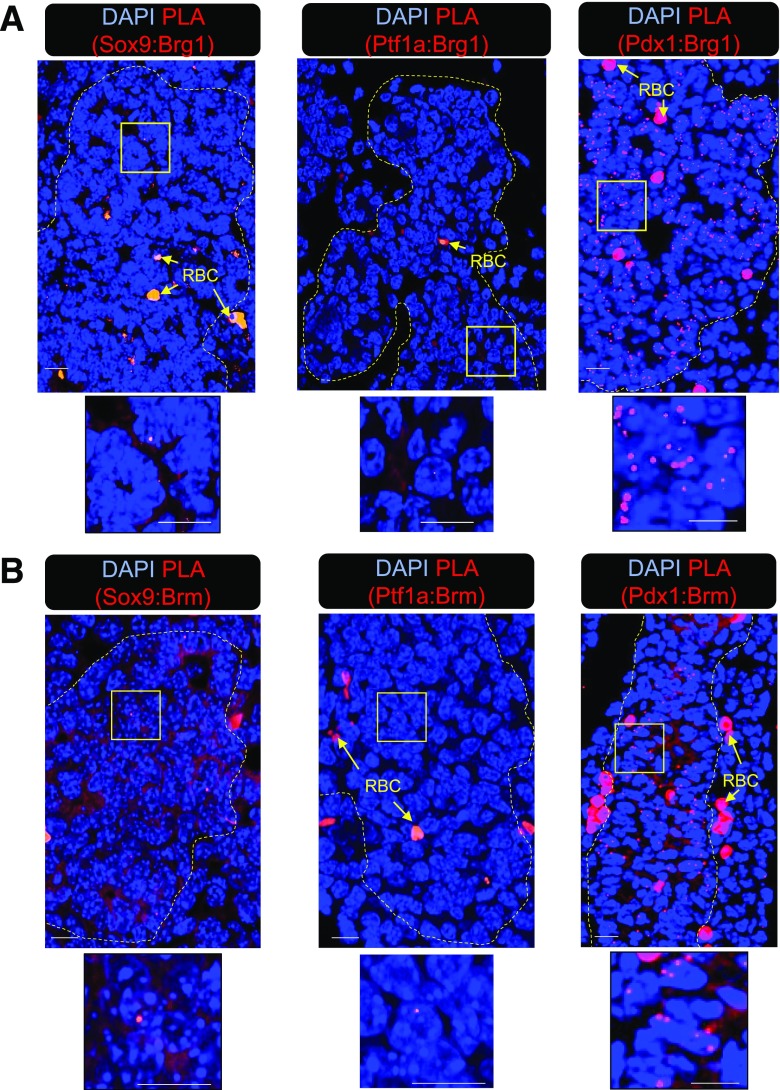
The proximity ligation assay (PLA) was used to evaluate the ability of Pdx1, Ptf1a, and Sox9 to interact with Brg1 and Brm in the developing pancreas, wherein a punctate fluorescent signal is generated if the physical distance between interacting proteins is within 30–40 nm. Pdx1:Brg1 and Pdx1:Brm signals were clearly detectable in e12.5 pancreatic epithelium, but scant binding was found between Ptf1a and Sox9 with either Swi/Snf ATPase subunits (Fig. 2). These results imply that Swi/Snf actions in MPCs are mediated principally through Pdx1.
Figure 2.
Pdx1, but not Sox9 or Ptf1a, interacts with Brg1 and Brm in wild-type e12.5 pancreatic epithelium. PLA was performed with antibodies specific for Brg1 (A) or Brm (B) and Sox9, Ptf1a, or Pdx1. Distinct fluorescence PLA signals were easily visible in the Pdx1:Brg1 and Pdx1:Brm experiments, but were nearly absent in the Brg1:Sox9, Brg1:Ptf1a, Brm:Sox9, and Brm:Ptf1a assays. The yellow square demarks the magnified area displayed below, and the dashed yellow marks outline the pancreatic epithelium. RBC, red blood cell. Scale bar = 10 μm.
All Pancreatic Cell Lineages Are Reduced in e15.5 Brg1Δpanc Mutants
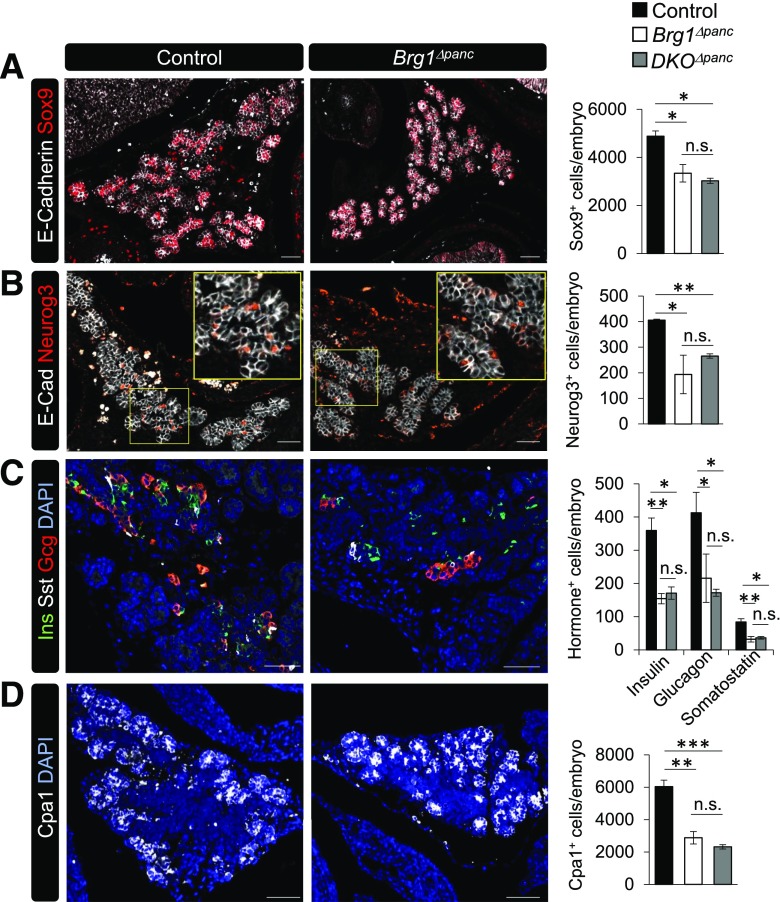
To determine whether the absence of Pdx1-recruited Brg1/Swi/Snf impacted the loss of specific pancreatic cell lineages, we immunostained and quantitated serially cut control and mutant e15.5 sections for cell type markers characteristic of this developmental stage. Expression of Sox9 at e15.5 denotes a population of bipotent progenitor cells (27), and neurogenin 3 (Neurog3) marks the subset of cells destined to become islet endocrine cells (28). We found that the number of cells expressing Sox9 and Neurog3 was reduced by ∼50% in Brg1Δpanc pancreata, which was not decreased further in the DKOΔpanc mutant (Fig. 3A and B). Moreover, the number of insulin+, glucagon+, and somatostatin+ cells, along with carboxypeptidase 1+ (Cpa1) acinar cells, was also reduced by ∼50% in the Brg1Δpanc and DKOΔpanc mutants (Fig. 3C and D). In addition, ductal branching was less expansive in Brg1Δpanc than in control embryos, which was expected due to reduced Brg1Δpanc pancreatic cell numbers (Supplementary Fig. 4). Together, these results demonstrate that all pancreatic lineages are negatively influenced by the loss of the embryonic Pdx1:Brg1/Swi/Snf complex.
Figure 3.
All pancreatic cell lineages are reduced in e15.5 Brg1Δpanc and DKOΔpanc epithelium. Control, Brg1Δpanc, and DKOΔpanc mutant embryos were stained with antibodies specific for E-cadherin and Sox9 (A), E-cadherin and Neurog3 (B), insulin (Ins), somatostatin (Sst), and glucagon (Gcg) (C), or Cpa1 (D). The yellow square in B marks the magnified area in the panel. DAPI nuclear staining is also provided in C and D. Cell type counting was performed on sections 90-µm apart that spanned the entire pancreatic region (n = 3). *P < 0.05. **P < 0.01. ***P < 0.001. n.s., not significant. Scale bar = 50 µm.
Brg1-Deficient MPCs Have Reduced Proliferative Capacity
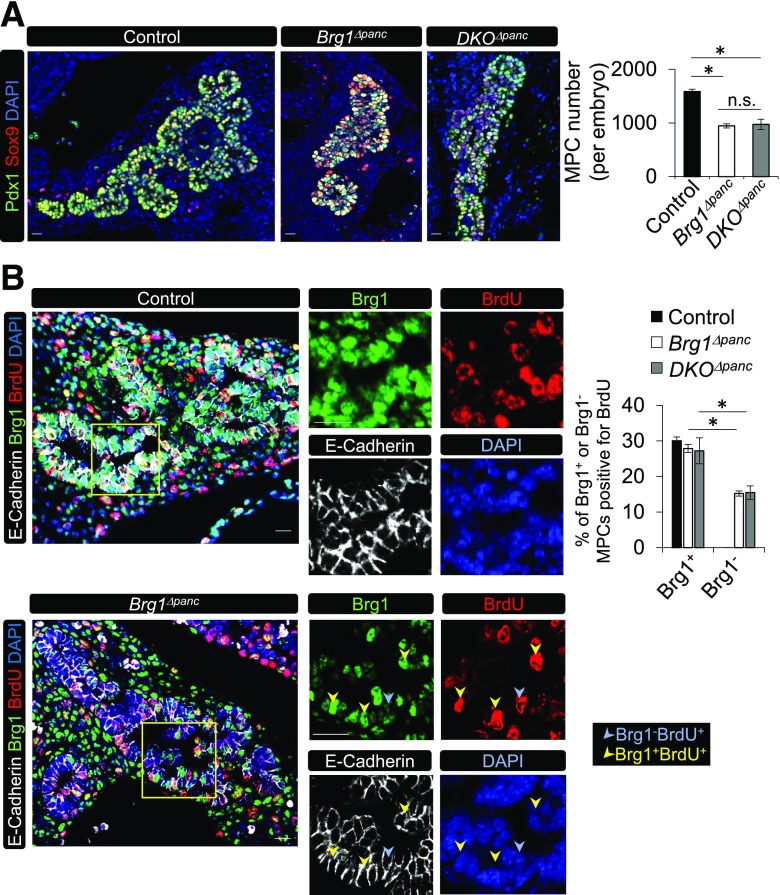
We next investigated whether the reduction in pancreatic cell type formation directly resulted from depletion of the embryonic Brg1Δpanc MPC pool. In agreement with this prediction, the number of coexpressing Pdx1 and Sox9 cells at e12.5 was reduced by ∼40% (Fig. 4A). However, there was no detectable difference in TUNEL+ cell numbers in the e12.5 control and Brg1Δpanc E-cadherin+ pancreatic epithelium (data not shown), suggesting that the decreased MPC population was not due to increased cell death. Pregnant dams were then injected with BrdU to assess the proliferation of Brg1-deficient MPCs. Control, Brg1Δpanc, and DKOΔpanc mutants incorporated BrdU into ∼30% of wild-type MPCs at e12.5, whereas incorporation in Brg1-deficient MPCs was only 14.4 ± 0.7% and 14.8 ± 1.8% in the Brg1Δpanc and DKOΔpanc mutants, respectively (Fig. 4B). These studies illustrate the novel function for Pdx1:Brg1/Swi/Snf in modulating MPC proliferation in the developing pancreas, which to our knowledge is the first transcriptional coregulator shown to influence MPC number and pancreas mass.
Figure 4.
MPC proliferation is dependent on the Brg1 ATPase subunit of Swi/Snf. A: A representative immunostaining image of Sox9+ and Pdx1+ cells in control, Brg1Δpanc, and DKOΔpanc pancreata at e12.5. Sox9+Pdx1+ MPC numbers were determined in sections obtained every 60 µm of the entire pancreatic region. B: E-cadherin, Brg1, and BrdU staining of pregnant dams injected with BrdU 30 min before the e12.5 embryos were harvested. Also provided is the percentage of Brg1+ or Brg1− cells that incorporated BrdU (n = 3). The yellow square illustrates the magnified area displayed in the panels on the right. *P < 0.05. Scale bar = 20 μm. DAPI nuclear staining is shown in A and B.
Impaired Adult Islet β-Cell Function Is Only Observed Upon Removal of Both the Brg1 and Brm ATPase Subunits of Swi/Snf
To test whether Pdx1:Swi/Snf also contributed to adult β-cell function, we crossed transgenic mice containing a tamoxifen-inducible, β-cell–specific Cre (mouse Ins1 enhancer/promoter [MIP]–driven CreERT [16]) and the Rosa26-Loxp-Stop-Loxp-tdTomato (R26LSL-tdTomato [18]) lineage reporter with Brg1f/f;Brm+/− (termed Brg1Δβ;Brm+/−), Brg1f/+;Brm−/− (i.e., Brg1Δβ/+;Brm−/−), or Brg1f/f;Brm−/− (i.e., βDKO) mice. All experimental and control animals contained the MIP-CreERT transgene and received tamoxifen, because this Cre line contains the human growth hormone minigene sequence that has been reported to independently augment islet β-cell mass, insulin content, and insulin secretion (29). Brg1 removal was induced in 4-week-old mice by three tamoxifen administrations every other day over a 5-day period. At 2 weeks after the last tamoxifen treatment, ∼70% of islet β-cells expressed the fluorescent Tomato lineage reporter and >90% of these βDKO cells lacked Brg1 (Supplementary Fig. 5).
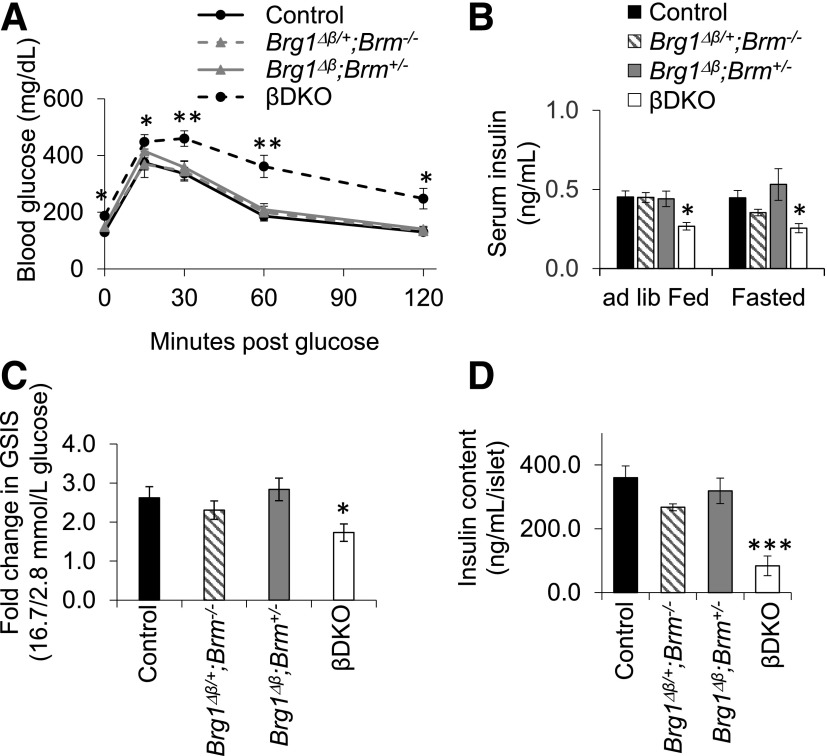
Brg1Δβ;Brm+/− and Brg1Δβ/+;Brm−/− mice were physiologically normal 2 weeks after the last tamoxifen administration, whereas βDKO animals suffered from fasting hyperglycemia, glucose intolerance, and reduced serum insulin levels (Fig. 5A and B). Glucose intolerance did not worsen in older mutant animals, and males and females both exhibited similar phenotypes (Supplementary Fig. 6). Glucose-stimulated insulin secretion (GSIS) was also compromised in size-matched islets isolated from βDKO animals, whereas Brg1Δβ;Brm+/− and Brg1Δβ/+;Brm−/− were unaffected (Fig. 5C). In addition, we observed a severe and specific reduction in βDKO islet insulin content (Fig. 5D), suggesting that their secretion deficiency results, at least in part, from limited hormone content.
Figure 5.
Severe defects in adult islet β-cell function are only produced in βDKO mice. Control, Brg1Δβ/+;Brm−/−, Brg1Δβ;Brm+/−, and βDKO mice were subjected to testing 2 weeks after the last tamoxifen treatment. All statistical comparisons are to controls. A: Fasting blood glucose and glucose tolerance were specifically compromised in βDKO mice during the intraperitoneal glucose tolerance test. B: Serum insulin levels were also only reduced in ad libitum (ad lib) fed or fasted βDKO animals. n = 5–9. *P < 0.05; **P < 0.01. C: GSIS was reduced in βDKO islets and not in Brg1Δβ/+;Brm−/− or Brg1Δβ;Brm+/−. n = 3–6. D: Islet insulin content was only decreased in the βDKO mutant (n = 4–8).
Loss of Swi/Snf Activity in βDKO Islet β-Cells Severely Reduces Insulin Production Despite Retention of Many Important Enriched Transcription Factors
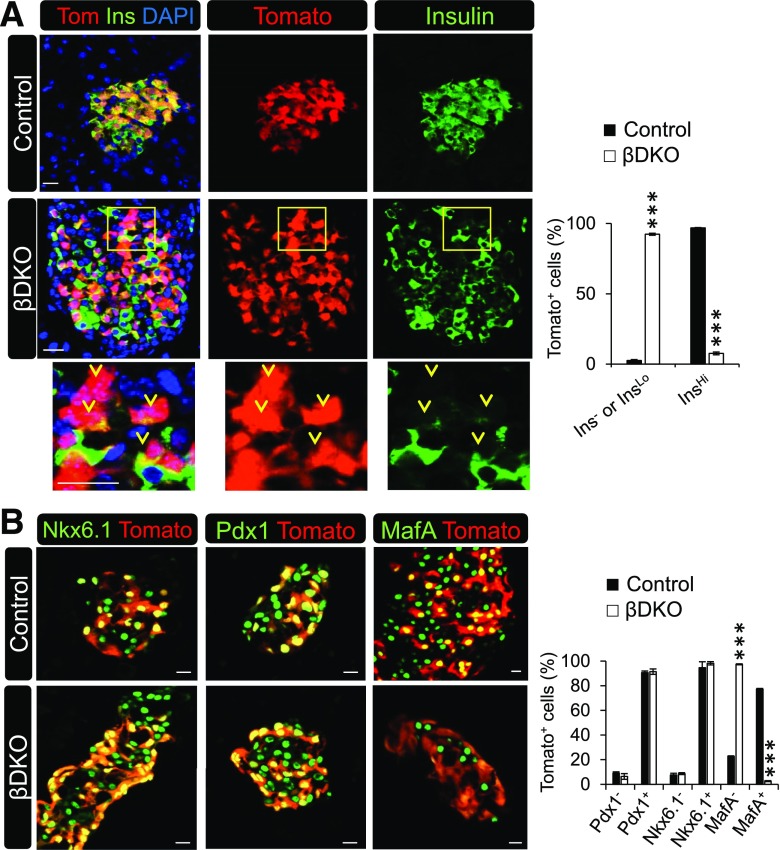
To gain insight into the cause of the significant decrease in insulin content in βDKO islets, we first took a candidate approach to monitor for the presence of various β-cell markers in control and βDKO pancreata. Most strikingly, nearly every Tomato-labeled cell in βDKO islets had little to no insulin immunoreactivity (Fig. 6A). There was also no overt change in islet cell mass or apoptosis in mutant islets, contrasting with the reduced pancreas mass of Brg1Δpanc mice caused by the developmental loss of this ATPase. As expected, Tomato-labeled Brg1Δβ;Brm+/− and Brg1Δβ/+;Brm−/− β-cells had no obvious defects in insulin production (Supplementary Fig. 7).
Figure 6.
βDKO β-cells have very reduced insulin protein levels yet retain Nkx6.1 and Pdx1. A: Representative confocal images of lineage-labeled control and βDKO Tomato+- and insulin+-stained islet cells. Most Tomato+ βDKO cells are insulinLo or insulin−, as illustrated in the lower panels by the yellow arrowheads of the magnified squared field. The graph depicts the percentage of Tomato+ insulin− or insulin+ cells to the total number of Tomato+ cells. B: Islet β-cell–enriched Pdx1 and Nkx6.1 are present in control and βDKO Tomato+ cells, whereas MafA is absent. Scale bar = 20 μm. The graph illustrates the percentage of Tomato+ transcription factor− or transcription factor+ cells to the total number of Tomato+ cells. ***P < 0.001.
Severe insulin deficiency is commonly observed upon β-cell ablation of key lineage-determining transcription factors such as Pdx1 (11), Nkx6.1 (30), Nkx2.2 (31), Pax6 (32), and Mnx1 (33), whereas removal of MafA impacts islet architecture and GSIS but has little effect on insulin content (34). Interestingly, Pdx1, Nkx6.1, Nkx2.2, Pax6, and Mnx1 levels were unaffected in Swi/Snf-deficient Tomato+ cells by immunofluorescence analysis, although MafA levels were significantly reduced (Fig. 6B and Supplementary Fig. 8). Moreover, βDKO Tomato+ cells did not produce the α-cell–specific glucagon hormone, an effect seen upon removal of Pdx1 from adult β-cells (11). In addition, somatostatin expression was not induced in this cell population (Supplementary Fig. 8), which occurs upon deletion of, for example, Mnx1 from β-cells (33). Overall, there was also no difference in the islet α- and δ-cell mass between control and βDKO islets (Supplementary Fig. 8).
Expression of Pdx1-Regulated Genes Involved in Cell Maturation, Insulin Production, and Insulin Secretion Is Affected in βDKO β-Cells
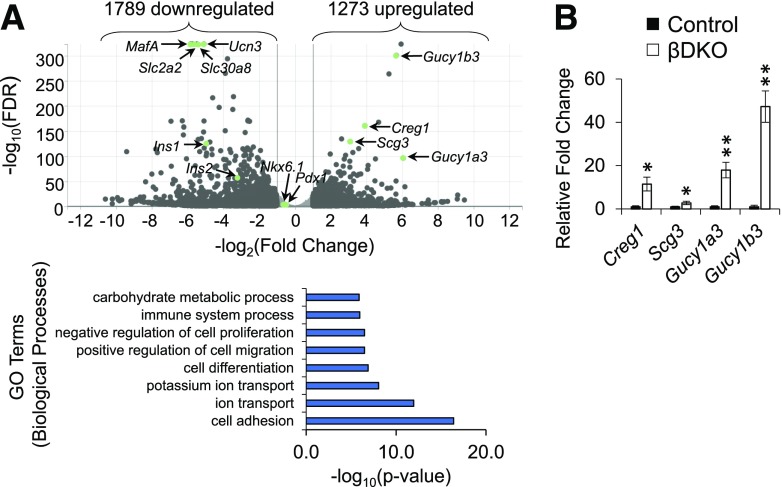
To more comprehensively define the molecular influence Swi/Snf has on β-cells, RNA sequencing was performed on FACS Tomato+ cells from control and βDKO islets. Using a ± twofold cutoff and false discovery rate of <0.05, there were 1,789 downregulated and 1,273 upregulated genes in βDKO β-cells (Fig. 7A). Gene ontology analysis (35,36) of these differentially expressed βDKO genes using Database for Annotation, Visualization and Integrated Discovery (DAVID) led to the identification of a very diverse array of biological pathways associated with Swi/Snf control, including cell adhesion, ion transport, cell differentiation, cell migration, cell proliferation, and carbohydrate metabolism (Supplementary Table 3). Expression of some of the most upregulated genes, including Creg1, Scg3, Gucy1a3, and Gucy1b3, was confirmed by qPCR analysis (Fig. 7B); however, the impact of these genes on β-cells is unclear.
Figure 7.
Swi/Snf affects a broad range of metabolic processes in islet β-cells. A: Top, volcano plot shows the most differentially expressed genes in βDKO β-cells. Bottom: The most significant biological pathways identified by gene ontology (GO) analysis. Swi/Snf contributed to very divergent regulatory processes, varying from cell adhesion (most significant) to ion transport, cell migration, and carbohydrate metabolism (least significant). Expression of Swi/Snf subunits BAF250A (Arid1a), BAF53B (Actl6b), and BAF60C (Smarcd3) was increased in βDKO β-cells by 2.11-, 2.08-, and 10.3-fold, respectively. FDR, false discovery rate. B: qPCR analysis from FACS-purified Tomato+ β-cells of various βDKO genes upregulated in the RNA sequencing. n = 3. *P < 0.05; **P < 0.01.
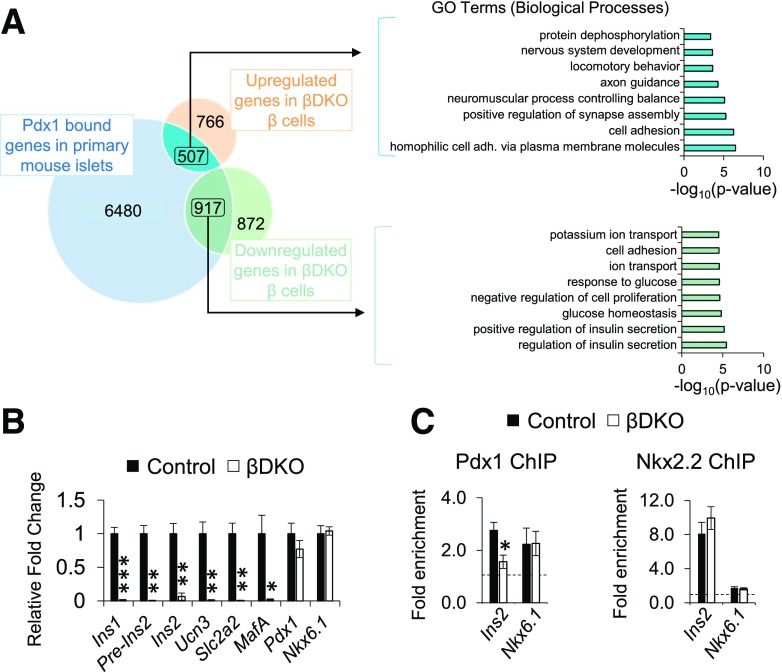
Greater insight into how Pdx1:Swi/Snf regulated gene expression was obtained upon comparing genes bound by Pdx1 in ChIP-sequencing analysis of mouse islets (37) to those upregulated or downregulated in βDKO β-cells. Genes upregulated and bound by Pdx1 (507/1,273) resided in biological pathways linked to cell adhesion and neuromuscular functions, whereas downregulated βDKO genes bound by Pdx1 (917/1,789) defined functional networks linked to insulin secretion and glucose homeostasis (Fig. 8A and Supplementary Table 4). Consequently, we concluded that Pdx1:Swi/Snf represents an essential positive regulator of β-cell function. Supportive evidence of these changes came from immunofluorescence and qPCR analysis of independently derived flow-sorted βDKO β-cells. These genes included those strongly linked to mature cell identity, such as Ins1, Ins2, Slc2a2 (Glut2), MafA, Slc30A8, and Ucn3 (Fig. 8B and Supplementary Fig. 9). However, their production was unaffected in Brg1Δβ;Brm+/− and Brg1Δβ/+;Brm−/− islets (Supplementary Figs. 7 and 10). Collectively, our analyses revealed that Swi/Snf-deficient β-cells have reduced expression of numerous Pdx1-regulated genes that are essential for sustaining β-cell identity.
Figure 8.
Pdx1:Swi/Snf controls genes essential to islet β-cell function. A: Left, Venn diagram of upregulated and downregulated βDKO genes overlaid with Pdx1-bound genes obtained from ChIP experimentation of primary mouse islets (37). Right: Most of the pathways identified by gene ontology (GO) analysis of the 507 genes that were Pdx1 bound and upregulated in βDKO β-cells were associated with cell adhesion (adh.) and neuromuscular functions, whereas the 917 genes that were Pdx1 bound and downregulated in βDKO β-cells were associated with β-cell function, such as glucose homeostasis and insulin secretion. B: Expression of a key subset of islet β-cell regulators identified in βDKO RNA sequencing and Pdx1 ChIP studies was confirmed upon qPCR of FACS-purified Tomato+ β-cells. *P < 0.05; **P < 0.01; ***P < 0.001. C: Pdx1 ChIP binding to the Ins2 enhancer was decreased in βDKO compared with control islets, whereas there was no impact on binding to the Nkx6.1 enhancer. Nkx2.2 binding to the Ins2 and Nkx6.1 enhancer was unchanged between βDKO and control islets. The dashed line represents IgG enrichment on the target gene after normalization to β-actin. n = 3. *P < 0.05.
Pdx1 Binding to the Ins Enhancer Is Compromised in βDKO β-Cells
Rodent Ins1 and Ins2 gene expression is largely mediated by transcription factors that bind within cis-acting enhancer sequences found roughly between −340 and −90 bp upstream of the transcription start site (38), a region well conserved in the human INS gene (39). In addition to Pdx1 binding at the A3/A4 (−201 to −195 bp in Ins2) and proximal A1 (−63 to −59 bp) elements (40), enhancer activity is mediated by several islet-enriched transcription factors with vital roles in developing and adult β-cells, including Pax6 (−317 to −311 bp) (41), Nkx2.2 (−128 to −122 bp) (42), and MafA (−126 to −101 bp) (43).
We compared Pdx1 binding to the Ins2 and Nkx6.1 enhancers in ChIP experiments performed on control and βDKO islets, noting that Nkx6.1 was viewed as an internal control because expression was unaffected in βDKO islets (Fig. 8B). Nkx2.2 binding to both enhancers was also evaluated, with the Nkx2.2 and Pdx1 elements found roughly −807 to −796 bp upstream of the Nkx6.1 transcription start site (44)). Pdx1 binding was selectively reduced on the Ins2 gene in βDKO islets, whereas Nkx2.2 binding was unchanged, as was Pdx1 and Nkx2.2 to the Nkx6.1 enhancer (Fig. 8C). These data reveal that the presence of the Brg1/Brm-associated Swi/Snf complex was necessary for Pdx1 binding to the Ins gene enhancer.
Discussion
The binding of transcription factors to cis-acting DNA control elements is by itself not sufficient to regulate target gene expression (45). Accordingly, their associated coregulators serve as an essential layer of control by, for example, influencing chromatin structure, enabling interactions among transcription factors, and recruitment of other effectors of RNA polymerase II. Among the hundreds of positive- and negative-acting transcriptional coregulators, very few have been directly linked to β-cell–enriched transcription factor activity, despite strong evidence that Pdx1 (1,2,11), Nkx2.2 (31), Mnx1 (33), and Nkx6.1 (30) are essential to core programs of pancreas formation and β-cell activity. Here we evaluated the significance of Pdx1 recruitment of the Swi/Snf chromatin remodeling complex to pancreas formation developmentally and in islet β-cells. Conditional and constitutive mutants of the core ATPase subunits in mice were used to modulate Swi/Snf activity in vivo. Our results revealed that the Brg1 ATPase subunit regulates pancreas size by stimulating Pdx1:Swi/Snf-mediated MPC proliferation, while both Brg1 and Brm regulate expression of Pdx1-driven genes required for islet β-cell identity, including Ins.
Adult pancreas mass is limited by the size of the embryonic MPC pool (7). Here our data demonstrated that the inability of Pdx1 to recruit Brg1/Swi/Snf in Brg1Δpanc mice reduced acinar, ductal, and islet cell numbers and pancreas mass by ∼50%, without affecting formation of other organs. Moreover, we showed that this resulted from reduced proliferation of the MPC pool. Notably, only Pdx1 was found in the PLA to bind to Brg1 and Brm in MPCs, whereas no interactions were observed with the MPC-enriched and functionally important Ptf1a (4) or Sox9 (5) transcription factors. This evidence indicated that Swi/Snf regulation of MPC expansion was principally through Pdx1.
Interestingly, pancreas mass was only affected in Brg1Δpanc mice and not further in the double-ATPase DKOΔpanc mutant. This may simply mean that the Brm ATPase, which has a much less impactful global regulatory phenotype in relation to Brg1 (19,46), has no influence on pancreatogenesis. This possibility is supported by a recent report that found BRM transcript levels increased (∼17-fold) during directed differentiation of human embryonic stem cells from stage 5 MPC-like cells to stage 6 β-like cells in vitro (Supplementary Table 5, data mined from reference [47]). Further, this increase in BRM provides a plausible explanation about why eliminating both Brg1 and Brm in adult islet βDKO β-cells had such a penetrant effect. Alternatively, Pdx1 may be recruiting a distinct Swi/Snf-related complex, termed PBAF (polybromo-associated BAF), that is only regulated by Brg1 (48). Although many subunits are shared between PBAF and the Brg1- and Brm-regulated BAF (Brg1-/Brm-associated factor) complex, each were shown to possess unique regulatory properties in controlling vitamin D receptor–driven anti-inflammatory and prosurvival responses in islet β-cells (49).
More broadly, our results suggest that coregulators of Pdx1, Ptf1a, and Sox9 influence pancreas size, a global physical determinant linked to type 1 diabetes and T2D susceptibility (9,50). Future efforts should involve not only identifying coregulators affecting transcription factor activity but also the processes and factors that influence their recruitment to target loci. These would be expected to include posttranslational modification mechanisms that positively or negatively affect transcription factor:coregulator interactions. For example, phosphorylation of the p53 transcription factor increases CBP/p300 histone acetyltransferase association, amplifying transcriptional activity (51).
T2D is ultimately caused by the inability of islet β-cells to produce sufficient amounts of insulin to cause transport of blood glucose into insulin-resistant tissues to maintain normoglycemia. A hallmark of this disease is an increase in the number of “empty” β-cells, defined by their lack of insulin immunoreactivity (52). This was also a novel characteristic of βDKO β-cells (Fig. 6) and associated with a lack of Pdx1 binding to the endogenous Ins2 gene enhancer. These results are consistent with a recent report showing that DNA binding by the REST transcription factor relies on the remodeling activity of SWI/SNF in embryonic stem cells (53). In contrast to Pdx1, there was no apparent change in Nkx2.2 transcription factor binding to the Ins2 gene or in Pdx1, Nkx2.2, and Nkx6.1 nuclear protein levels. In fact, MafA was the only other core Ins regulator apparently absent in βDKO β-cells (Fig. 7). Because loss of MafA alone does not abolish Ins production in MafAΔpanc (34) or MafAΔβ (54) mice, insulin deficiencies in βDKO “empty” β-cells likely reflect the combined loss of the MafA protein and inability of Pdx1 to bind to the Ins enhancer. This proposal is supported by the ability of Pdx1 + MafA to reprogram human islet α-cells to β-like cells (55), or in transgenic mice, when combined with the embryonic islet cell determination factor, Neurog3, to produce β-like cells in the intestine of mice (56).
Circularized Chromosome Conformation Capture (i.e., 4C) and PDX1-binding enhancer elements were used as anchor bait sites in the human EndoC-βH1 pancreatic β-cell line (57,58), and the human INS locus was found to physically contact many distinct genes affecting β-cell secretory processes. Knockdown of INS mRNA levels in EndoC-βH1 cells demonstrated that expression of 259 genes was affected, with 45 residing in 4C contact regions and ∼40% associated with metabolic pathways. Moreover, these investigators proposed that the chromosomal interactions with the INS locus were conjoined with the transcriptional machinery at the various loci. If PDX1:Swi/Snf contributed to such control, we predicted some overlap between the βDKO-regulated genes and those identified in the human 4C INS knockdown analysis. However, not one of their 45 genes was differentially expressed in βDKO β-cells. Possible explanations for this difference could involve the unique variable number of tandem repeats regions upstream of the human INS enhancer (39) and/or simply the experimental context (i.e., human EndoC-βH1 cells vs. the mouse βDKO model). Alternatively, our ability to detect Nkx2.2 binding to the Ins2 enhancer in βDKO islets raises the possibility that 4C-detected interactions persist because they are regulated by transcription factors binding independently of Pdx1.
Genome-wide association studies have found numerous genomic loci linked to T2D (59), but the Swi/Snf subunits have not been associated directly to diabetes pathogenesis. Notably, our results suggest that only genetic variants leading to a complete loss of Swi/Snf activity would yield a βDKO diabetic phenotype. However, a recent study found that genomic deletions and rearrangements in Swi/Snf subunits exist in approximately one-third of pancreatic cancers containing alterations in known tumor-associated genes (e.g., MYC, KRAS, CDKN2A, TGFBR2, MAP2K4, and SMAD4) (60). Specifically in regards to the ATPase subunits, heterozygous and homozygous mutations in the BRG1 or BRM genes occur in 9.6% and 2.6%, respectively, of human pancreatic cancer samples. Moreover, combined mutations in BRG1 and BRM were found in several pancreatic cancer cell lines, but double mutations were absent from primary tumor samples, although the latter involved a limited sample number (60). Notably, ∼80% of individuals with pancreatic cancer often present with new-onset T2D or impaired glucose tolerance at diagnosis (61). Although these studies do not directly link the prevalence of Swi/Snf subunit mutations to pancreatic cancer–associated diabetes, loss-of-function mutations in BRG1 and BRM provide a potential intersection for pancreatic cancer and T2D, especially given the influence Swi/Snf has on mature β-cell function.
Functional heterogeneity within the β-cell population was first described more than two decades ago (62,63), with recent observations identifying distinct normal and T2D β-cell populations that differ in their molecular composition and glucose-stimulated insulin secretion properties (64). Interestingly, Pdx1, MafA, and Nkx6.1 levels appear similar between these β-cell subtypes, raising an intriguing possibility that variations in transcription factor:coregulator interactions could be affecting activity. This possibility is supported by our previous findings that Pdx1:Swi/Snf interactions were not observed in all healthy human islet β-cells in the PLA (12), which adds another level of heterogeneity to a subpopulation of human β-cells. Such observations emphasize the importance of investigating how coregulator recruitment by endocrine cell–enriched transcription factors contribute to human β-cell functional heterogeneity under normal physiological conditions.
Collectively, our study provides fundamental insight into the role of Pdx1:Swi/Snf complexes in pancreas organogenesis and in maintaining principal features of the mature β-cell state. The translational significance of our findings is underscored by the knowledge that in humans, a subset of T2D β-cells lose their ability to produce insulin and that overall pancreas size, with otherwise normal cell-type proportional allocations, is a risk factor in the development of diabetes. Furthermore, our findings that Swi/Snf activity is crucial for driving Ins expression and that Pdx1:Swi/Snf interactions are negatively affected in human T2D (12) indicate that therapies that enhance such interactions are an attractive target in T2D. Upon broader consideration, we propose that transcriptional coregulator recruitment is essential to the formation and function of the other islet cell types (α, β, δ, ε, and pancreatic polypeptide). In this context, our studies are currently focused on additional Pdx1-interacting proteins, such as the multisubunit nucleosome remodeler and deacetylase (NuRD) complex, the Myst2 histone acetyltransferase, and the Tif1β corepressor.
Supplementary Material
Article Information
Acknowledgments. The authors thank Dr. Xin Tong and Dr. Emily Walker (both from Vanderbilt University) for critically reading the manuscript.
Funding. This work was supported by grants from the American Diabetes Association (1-16-PDF-109 to J.M.S.), National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases (K01-DK-115633 to J.M.S. and R01-DK-050203 to R.S.), and National Cancer Institute (R01-CA-172045 to M.H.). The authors thank the Vanderbilt Flow Cytometry Shared Resource (National Cancer Institute P30-CA-68485), Vanderbilt Technologies for Advanced Genomics core (VANTAGE), supported in part by Clinical and Translational Science Awards (5UL1-RR-024975-03), the Vanderbilt Ingram Cancer Center (National Cancer Institute P30-CA-68485), the Vanderbilt Vision Center (National Eye Institute P30-EY-08126), and National Institutes of Health/National Center for Research Resources (G20-RR-030956), and the Vanderbilt Hormone Assay and Analytical Services Core (National Institute of Diabetes and Digestive and Kidney Diseases DK-020593) for core laboratory and technical assistance.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. J.M.S., J.-H.L., D.P., M.G., A.B.O., F.M., and N.R. designed, executed, and analyzed experiments. J.M.S. and R.S. wrote the manuscript. A.B., M.A.M., M.H., C.V.E.W., and R.S. designed and analyzed experiments. All authors reviewed the manuscript. R.S. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Data and Resource Availability. Raw and analyzed RNA sequencing data sets have been deposited in GEO (accession number GSE128945). All noncommercially available resources generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Prior Presentation. Parts of this study were presented in abstract form at the 76th Scientific Sessions of the American Diabetes Association, New Orleans, LA, 10–14 June 2016, and at the 77th Scientific Sessions of the American Diabetes Association, San Diego, CA, 9–13 June 2017.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db19-0349/-/DC1.
J.M.S. is currently affiliated with the Department of Pediatrics, Indiana University School of Medicine, Indianapolis, IN.
References
- 1.Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature 1994;371:606–609 [DOI] [PubMed] [Google Scholar]
- 2.Offield MF, Jetton TL, Labosky PA, et al. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development 1996;122:983–995 [DOI] [PubMed] [Google Scholar]
- 3.Stoffers DA, Ferrer J, Clarke WL, Habener JF. Early-onset type-II diabetes mellitus (MODY4) linked to IPF1. Nat Genet 1997;17:138–139 [DOI] [PubMed] [Google Scholar]
- 4.Krapp A, Knöfler M, Ledermann B, et al. The bHLH protein PTF1-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes Dev 1998;12:3752–3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seymour PA, Freude KK, Tran MN, et al. SOX9 is required for maintenance of the pancreatic progenitor cell pool. Proc Natl Acad Sci U S A 2007;104:1865–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan FC, Wright C. Pancreas organogenesis: from bud to plexus to gland. Dev Dyn 2011;240:530–565 [DOI] [PubMed] [Google Scholar]
- 7.Stanger BZ, Tanaka AJ, Melton DA. Organ size is limited by the number of embryonic progenitor cells in the pancreas but not the liver. Nature 2007;445:886–891 [DOI] [PubMed] [Google Scholar]
- 8.Saisho Y, Butler AE, Meier JJ, et al. Pancreas volumes in humans from birth to age one hundred taking into account sex, obesity, and presence of type-2 diabetes. Clin Anat 2007;20:933–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell-Thompson M, Wasserfall C, Montgomery EL, Atkinson MA, Kaddis JS. Pancreas organ weight in individuals with disease-associated autoantibodies at risk for type 1 diabetes. JAMA 2012;308:2337–2339 [DOI] [PubMed] [Google Scholar]
- 10.Guz Y, Montminy MR, Stein R, et al. Expression of murine STF-1, a putative insulin gene transcription factor, in beta cells of pancreas, duodenal epithelium and pancreatic exocrine and endocrine progenitors during ontogeny. Development 1995;121:11–18 [DOI] [PubMed] [Google Scholar]
- 11.Gao T, McKenna B, Li C, et al. Pdx1 maintains β cell identity and function by repressing an α cell program. Cell Metab 2014;19:259–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKenna B, Guo M, Reynolds A, Hara M, Stein R. Dynamic recruitment of functionally distinct Swi/Snf chromatin remodeling complexes modulates Pdx1 activity in islet β cells. Cell Rep 2015;10:2032–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang W, Côté J, Xue Y, et al. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J 1996;15:5370–5382 [PMC free article] [PubMed] [Google Scholar]
- 14.Mani U, S AS, Goutham RNA, Mohan SS. SWI/SNF Infobase-An exclusive information portal for SWI/SNF remodeling complex subunits. PLoS One 2017;12:e0184445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawaguchi Y, Cooper B, Gannon M, Ray M, MacDonald RJ, Wright CVE. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat Genet 2002;32:128–134 [DOI] [PubMed] [Google Scholar]
- 16.Tamarina NA, Roe MW, Philipson L. Characterization of mice expressing Ins1 gene promoter driven CreERT recombinase for conditional gene deletion in pancreatic β-cells. Islets 2014;6:e27685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sumi-Ichinose C, Ichinose H, Metzger D, Chambon P. SNF2beta-BRG1 is essential for the viability of F9 murine embryonal carcinoma cells. Mol Cell Biol 1997;17:5976–5986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Madisen L, Zwingman TA, Sunkin SM, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 2010;13:133–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reyes JC, Barra J, Muchardt C, Camus A, Babinet C, Yaniv M. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2α). EMBO J 1998;17:6979–6991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spaeth JM, Hunter CS, Bonatakis L, et al. The FOXP1, FOXP2 and FOXP4 transcription factors are required for islet alpha cell proliferation and function in mice. Diabetologia 2015;58:1836–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spaeth JM, Gupte M, Perelis M, et al. Defining a novel role for the Pdx1 transcription factor in islet β-cell maturation and proliferation during weaning. Diabetes 2017;66:2830–2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30:2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 2015;12:357–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2015;31:166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.von Figura G, Fukuda A, Roy N, et al. The chromatin regulator Brg1 suppresses formation of intraductal papillary mucinous neoplasm and pancreatic ductal adenocarcinoma. Nat Cell Biol 2014;16:255–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seymour PA. Sox9: a master regulator of the pancreatic program. Rev Diabet Stud 2014;11:51–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gradwohl G, Dierich A, LeMeur M, Guillemot F. neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc Natl Acad Sci U S A 2000;97:1607–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brouwers B, de Faudeur G, Osipovich AB, et al. Impaired islet function in commonly used transgenic mouse lines due to human growth hormone minigene expression. Cell Metab 2014;20:979–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor BL, Liu FF, Sander M. Nkx6.1 is essential for maintaining the functional state of pancreatic beta cells. Cell Rep 2013;4:1262–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gutiérrez GD, Bender AS, Cirulli V, et al. Pancreatic β cell identity requires continual repression of non-β cell programs. J Clin Invest 2017;127:244–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitchell RK, Nguyen-Tu M-S, Chabosseau P, et al. The transcription factor Pax6 is required for pancreatic β cell identity, glucose-regulated ATP synthesis, and Ca2+ dynamics in adult mice. J Biol Chem 2017;292:8892–8906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan FC, Brissova M, Powers AC, Pfaff S, Wright CVE. Inactivating the permanent neonatal diabetes gene Mnx1 switches insulin-producing β-cells to a δ-like fate and reveals a facultative proliferative capacity in aged β-cells. Development 2015;142:3637–3648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hang Y, Yamamoto T, Benninger RKP, et al. The MafA transcription factor becomes essential to islet β-cells soon after birth. Diabetes 2014;63:1994–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57 [DOI] [PubMed] [Google Scholar]
- 36.Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khoo C, Yang J, Weinrott SA, et al. Research resource: the pdx1 cistrome of pancreatic islets. Mol Endocrinol 2012;26:521–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fromont-Racine M, Bucchini D, Madsen O, et al. Effect of 5′-flanking sequence deletions on expression of the human insulin gene in transgenic mice. Mol Endocrinol 1990;4:669–677 [DOI] [PubMed] [Google Scholar]
- 39.Hay CW, Docherty K. Comparative analysis of insulin gene promoters: implications for diabetes research. Diabetes 2006;55:3201–3213 [DOI] [PubMed] [Google Scholar]
- 40.Ohlsson H, Karlsson K, Edlund T. IPF1, a homeodomain-containing transactivator of the insulin gene. EMBO J 1993;12:4251–4259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sander M, Neubüser A, Kalamaras J, Ee HC, Martin GR, German MS. Genetic analysis reveals that PAX6 is required for normal transcription of pancreatic hormone genes and islet development. Genes Dev 1997;11:1662–1673 [DOI] [PubMed] [Google Scholar]
- 42.Cissell MA, Zhao L, Sussel L, Henderson E, Stein R. Transcription factor occupancy of the insulin gene in vivo. Evidence for direct regulation by Nkx2.2. J Biol Chem 2003;278:751–756 [DOI] [PubMed] [Google Scholar]
- 43.Zhao L, Guo M, Matsuoka TA, et al. The islet beta cell-enriched MafA activator is a key regulator of insulin gene transcription. J Biol Chem 2005;280:11887–11894 [DOI] [PubMed] [Google Scholar]
- 44.Watada H, Mirmira RG, Leung J, German MS. Transcriptional and translational regulation of β-cell differentiation factor Nkx6.1. J Biol Chem 2000;275:34224–34230 [DOI] [PubMed] [Google Scholar]
- 45.Mouchiroud L, Eichner LJ, Shaw RJ, Auwerx J. Transcriptional coregulators: fine-tuning metabolism. Cell Metab 2014;20:26–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bultman S, Gebuhr T, Yee D, et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell 2000;6:1287–1295 [DOI] [PubMed] [Google Scholar]
- 47.Veres A, Faust AL, Bushnell HL, et al. Charting cellular identity during human in vitro β-cell differentiation. Nature 2019;569:368–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hodges C, Kirkland JG, Crabtree GR. The many roles of BAF (mSWI/SNF) and PBAF complexes in cancer. Cold Spring Harb Perspect Med 2016;6:a026930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wei Z, Yoshihara E, He N, et al. Vitamin D switches BAF complexes to protect β cells. Cell 2018;173:1135–1149.e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Philippe M-F, Benabadji S, Barbot-Trystram L, Vadrot D, Boitard C, Larger E. Pancreatic volume and endocrine and exocrine functions in patients with diabetes. Pancreas 2011;40:359–363 [DOI] [PubMed] [Google Scholar]
- 51.Lambert PF, Kashanchi F, Radonovich MF, Shiekhattar R, Brady JN. Phosphorylation of p53 serine 15 increases interaction with CBP. J Biol Chem 1998;273:33048–33053 [DOI] [PubMed] [Google Scholar]
- 52.Cinti F, Bouchi R, Kim-Muller JY, et al. Evidence of β-cell dedifferentiation in human type 2 diabetes. J Clin Endocrinol Metab 2016;101:1044–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barisic D, Stadler MB, Iurlaro M, Schübeler D. Mammalian ISWI and SWI/SNF selectively mediate binding of distinct transcription factors. Nature 2019;569:136–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scoville DW, Cyphert HA, Liao L, et al. MLL3 and MLL4 methyltransferases bind to the MAFA and MAFB transcription factors to regulate islet β-cell function. Diabetes 2015;64:3772–3783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matsuoka TA, Kawashima S, Miyatsuka T, et al. Mafa enables Pdx1 to effectively convert pancreatic islet progenitors and committed islet α-cells into β-cells in vivo. Diabetes 2017;66:1293–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen Y-J, Finkbeiner SR, Weinblatt D, et al. De novo formation of insulin-producing “neo-β cell islets” from intestinal crypts. Cell Rep 2014;6:1046–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jian X, Felsenfeld G. Insulin promoter in human pancreatic β cells contacts diabetes susceptibility loci and regulates genes affecting insulin metabolism. Proc Natl Acad Sci U S A 2018;115:E4633–E4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu Z, Wei G, Chepelev I, Zhao K, Felsenfeld G. Mapping of INS promoter interactions reveals its role in long-range regulation of SYT8 transcription. Nat Struct Mol Biol 2011;18:372–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mahajan A, Taliun D, Thurner M, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet 2018;50:1505–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shain AH, Giacomini CP, Matsukuma K, et al. Convergent structural alterations define SWItch/Sucrose NonFermentable (SWI/SNF) chromatin remodeler as a central tumor suppressive complex in pancreatic cancer. Proc Natl Acad Sci U S A 2012;109:E252–E259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang F, Herrington M, Larsson J, Permert J. The relationship between diabetes and pancreatic cancer. Mol Cancer 2003;2:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Giordano E, Bosco D, Cirulli V, Meda P. Repeated glucose stimulation reveals distinct and lasting secretion patterns of individual rat pancreatic B cells. J Clin Invest 1991;87:2178–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kiekens R, In ’t Veld P, Mahler T, Schuit F, Van De Winkel M, Pipeleers D. Differences in glucose recognition by individual rat pancreatic B cells are associated with intercellular differences in glucose-induced biosynthetic activity. J Clin Invest 1992;89:117–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dorrell C, Schug J, Canaday PS, et al. Human islets contain four distinct subtypes of β cells. Nat Commun 2016;7:11756. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.