Abstract
We identified autoantibodies (AAb) reacting with a variant IA-2 molecule (IA-2var) that has three amino acid substitutions (Cys27, Gly608, and Pro671) within the full-length molecule. We examined IA-2var AAb in first-degree relatives of type 1 diabetes (T1D) probands from the TrialNet Pathway to Prevention Study. The presence of IA-2var–specific AAb in relatives was associated with accelerated progression to T1D in those positive for AAb to GAD65 and/or insulin but negative in the standard test for IA-2 AAb. Furthermore, relatives with single islet AAb (by traditional assays) and carrying both IA-2var AAb and the high-risk HLA-DRB1*04-DQB1*03:02 haplotype progress rapidly to onset of T1D. Molecular modeling of IA-2var predicts that the genomic variation that alters the three amino acids induces changes in the three-dimensional structure of the molecule, which may lead to epitope unmasking in the IA-2 extracellular domain. Our observations suggest that the presence of AAb to IA-2var would identify high-risk subjects who would benefit from participation in prevention trials who have one islet antibody by traditional testing and otherwise would be misclassified as “low risk” relatives.
Introduction
Type 1 diabetes (T1D) is an autoimmune disease that results from the targeted destruction of pancreatic β-cells by autoreactive T cells (1,2). The development of T1D is associated with the occurrence of autoantibodies (AAb) to pancreatic islet antigens that can be used as predictive biomarkers of disease progression (3).
AAb associated with T1D are mainly directed against proteins that are involved in the secretory pathway of insulin, including insulin, glutamic acid decarboxylase (GAD65), islet tyrosine phosphatase-like protein (IA-2), and zinc transporter 8 SLC30A8 (ZnT8).
The presence of AAb to IA-2 is associated with a high risk of T1D development (4–7). Screening for T1D-associated AAb allows for identification of asymptomatic, high-risk individuals (8) and for natural history studies of disease in cadaveric donors (9).
The neuroendocrine molecule IA-2 is a transmembrane glycoprotein of the tyrosine phosphatase–like protein family that is localized to the insulin-secretory granules of the pancreatic β-cell (10). IA-2 (PTPRN) encodes a 979–amino acid protein containing three domains: the N-terminal extracellular (or luminal) domain (amino acids 1–556), the transmembrane domain (amino acids 557–600), and the COOH-terminal intracellular (or cytoplasmic) domain (amino acids 601–979) containing a juxtamembrane (JM) domain (amino acids 601–686) and a protein tyrosine phosphatase domain (amino acids 687–979). T1D-associated AAb to IA-2 are mainly directed against epitopes within the cytoplasmic domain (11). Recently, we described antibody responses to the extracellular domain of IA-2 in subjects with T1D (12).
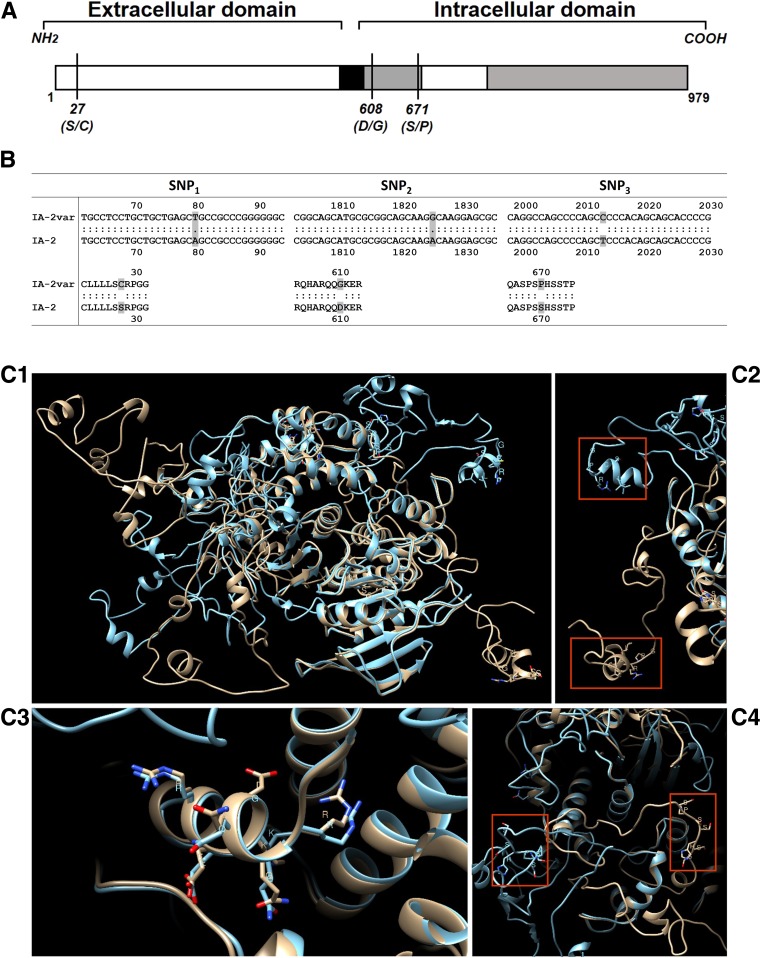
IA-2 isoforms can be formed by differential splicing of RNA transcripts leading to exon skipping, for example, isoforms lacking exon 13 (AA 557–629), which produces a secreted form lacking the transmembrane/JM domain, or lacking exon 14 (AA 653–693) (13,14). In the construct containing the full-length IA-2 molecule, originally provided by Dr. George Eisenbarth (University of Colorado, Denver, CO), we identified four single nucleotide polymorphisms (SNPs) within IA-2 (PTPRN), three of which were nonsynonymous SNPs at amino acid residues 27, 608, and 671, localized in the extracellular and in the JM domain of IA-2. This molecule was termed IA-2 variant (IA-2var).
The TrialNet Pathway to Prevention Study has screened >180,000 relatives for islet AAb (to insulin, GAD65, IA-2, and islet cell antibodies [ICAs]) (15) and has significantly improved our understanding of the disease progression through identifying individuals at early stages of disease for prevention trials. Individuals with multiple islet AAb with normoglycemia are considered to be at stage 1 T1D, those with multiple AAb and dysglycemia are at stage 2 T1D, and those who have developed the clinical symptoms of T1D are at stage 3 (16). In this study, first-degree relatives (FDRs) of T1D index case subjects from the TrialNet Pathway to Prevention Study were evaluated to determine the predictive value of IA-2var AAb and to assess possible immunological implications. We investigated antibody responses to the IA-2var and performed three-dimensional in silico structural comparisons of native IA-2 and IA-2var in order to determine the structural implications of the polymorphisms on antibody binding. These novel IA-2var AAb were evaluated for their ability to more precisely predict those individuals at high risk for developing T1D.
Research Design and Methods
Subjects
The study population included normoglycemic FDRs of T1D index case subjects recruited in the TrialNet Pathway to Prevention Study (ClinicalTrials.gov identifier: NCT00097292) (17). All parents and children older than 18 years gave informed consent for the study, and the protocol was approved by the TrialNet Ancillary Studies Committee and by the institutional review board at each TrialNet site.
The TrialNet Pathway to Prevention Study has screened >180,000 FDRs of T1D patients for islet AAb. Individuals are tested for mIAA, IA-2, and GAD65 AAb as described below. Those positive for one or more of these autoantibodies are then tested for ZnT8 antibodies and ICAs. The subjects evaluated in the current study consist of 1,686 FDRs without diabetes (773 male and 913 female, mean ± SD age 17.1 ± 13.5 years). These relatives were selected based on the presence of single (n = 809), multiple (n = 576), or the absence of (n = 301) islet-related AAb. We tested one serum sample from the earliest available blood draw on each relative. We received coded samples from the TrialNet Pathway to Prevention Study. Diabetes was diagnosed according to the American Diabetes Association criteria (18) (Supplementary Tables 1 and 2).
A total of 566 FDRs progressed to T1D during the follow-up period (Table 1). The mean follow-up time was similar between those relatives who progressed to T1D (progressors) and those who did not (nonprogressors). The relatives who developed T1D during the follow-up time were significantly younger than those who did not (P < 0.0001) (Table 1).
Table 1.
Characteristics of FDR of T1D probands who progressed and who did not progress to diabetes during the follow-up
| Progressors (n = 566) | Nonprogressors (n = 1,120) | P | |
|---|---|---|---|
| Age at screening (years), median (range) | 9.3 (1–45.7) | 12.9 (1–51.2) | 0.0001 |
| Follow-up (years), median (range) | 8.1 (3.4–11.9) | 8.4 (4.5–11.8) | 0.09 |
| Sex, % male | 50.5 | 43.5 | 0.007 |
| Race, % Caucasian | 89 | 86.4 | 0.14 |
Islet AAb
GAD65, IAA, and IA-2/ICA512 (ICA512bdc construct) AAb were detected by radiobinding in the TrialNet Core laboratory at the Barbara Davis Center for Childhood Diabetes. ICAs were assayed by indirect immunofluorescence at the University of Florida, Gainesville, FL.
IA-2var AAb
The IA-2var full-length molecule was in vitro transcribed and translated in the presence of [35S]-methionine (PerkinElmer) using the TNT coupled rabbit reticulocyte system (Promega, Madison, WI). AAb against IA-2var were detected by a modified quantitative radioimmunoprecipitation assay (11) using 50% protein A-Sepharose to separate free [35S]-methionine from antibody-bound labeled products. Assays were run in triplicate, and the results were calculated as an index as previously described (19,20). The threshold for the IA-2var AAb assay was established as the 99th percentile of the AAb indexes using 178 healthy control sera (106 females and 72 males, mean age 34.6 ± 10.2 years, 76% Caucasian, cutoff index: 0.218). All AAb test results using the same samples were confirmed. The interassay variation for IA-2var AAb was 8.3% (n = 10), while the intra-assay variation was 2.4% (n = 15). The IA-2var AAb assay achieved ratings of 62% sensitivity and 99% specificity during testing for the 2016 Islet Autoantibody Standardization Program (IASP).
Nucleotide Sequence and SNP Verification
All IA-2–containing construct plasmids were sequence verified at the University of Michigan and Baylor College of Medicine sequencing cores using the following contiguous primers: Seq1 [CGCCCGGAGCTCGGAAAGATGCGGCGCCCG], Seq2 [GAGAACTGGGGAGGTGAC-TTGCAAAAGGGG], Seq3 [AGTGGGCAAAGGTGGAGCTGGG-GCCAGCTC], Seq4 [GTTCTGCTGAAGAG-GGCAGGGGCTTCAGCC], Seq5 [CCTGGCCACTCCTACGG-GGACCTTCCAGGG], Seq6 [TTCTCTAGCAGGACAGGTGTCA-CAGGGGGT], Seq7 [ACCCCCTGTGACACCTGTCCTGCTAGAGAA], Seq8 [CACGAAGTCCTTGTTCAAC-CGGGCAGAGGG], Seq9 [CCACCAGCGGGGTCAGCATGACG-ATGACGG], and Seq10 [GGCCAGATGAGGGTGCCTCCCTCTACCACG]. SNPs were analyzed and validated using the National Center for Biotechnology Information/Basic Local Alignment Search tool (NCBI/BLAST) and flanking primers. The full-length IA-2 construct containing the SNPs was kindly donated by Dr. George Eisenbarth.
Nucleotide sequencing data of the IA-2 full-length molecule identified four SNPs, three of which are nonsynonymous. Two nonsynonymous SNPs are localized within the JM domain (Asp608Gly and Ser671Pro), and the third SNP is localized within the extracellular domain (Ser27Cys) of IA-2 (Fig. 1A and B). The GeneBank accession number for IA-2var is MN071399.
Figure 1.
A: Schematic representation of IA-2var (GeneBank accession number: MN071399). The vertical lines represent the amino acid substitutions: SNP1 (S→C), SNP2 (D→G), and SNP3 (S→P). B: Sequence data derived from the IA-2var construct. A total of four SNPs were identified, three of which (shown) are nonsynonymous SNPs. C: Predicted three-dimensional structural models of native IA-2 (gold) and IA-2var (blue). C1: Predicted structures are overlay and matched with UCSF Chimera (version 1.13.1). C2: Zoom for region showing residues LLSSRPG for native IA-2 (gold) and SNP1 residues LLSCRPG for IA-2var (blue). C3: Zoom for region showing residues RQQDKER for native IA-2 (gold) and SNP2 residues RQQGKER for IA-2var (blue). C4: Zoom for region showing residues SPSSHSS for native IA-2 (gold) and SNP3 residues SPSPHSS for IA-2var (blue). Bolded amino acid one-letter abbreviations correspond to amino acid substitutions.
DNA from 108 randomly selected subjects was amplified with primers flanking exons 13 and 14 in the PTPRN gene (forward, 5′-CTCTGAAACCTCCCTATGCCAC; reverse, 5′-TCTCACCATCCCATTCCTTCAC). Amplicons were subjected to direct Sanger sequencing using the forward primer and analyzed using an Applied Biosystems 3130xl Genetic Analyzer.
Statistical Analysis
Data were analyzed using GraphPad Prism 7 (GraphPad Software, Inc., La Jolla, CA) and SPSS 23.0 (SPSS, Chicago, IL). Life table analysis was applied to estimate the cumulative risk of developing T1D with data censored according to length of follow-up. Survival curves were compared using the log-rank test. Exact log-rank test for unequal follow-up was also performed (21). Results confirmed highly satisfactory performance of the exact procedure conditioning on realized follow-up, particularly in the case of unequal follow-up (22).
Cox proportional hazards regression analysis was applied to examine the variables predictive of progression to T1D and to investigate the effect of these variables simultaneously. The results are given as relative risks (hazard ratios). The models included those variables that were considered to be potentially significant predictors of T1D onset, including age as well as combination of AAb. Biochemically detected islet AAb (IAA, GAD65, ICA512bdc, and IA-2var AAb) were examined as continuous and dichotomous variables. All variables were retained in the model.
χ2 or Fisher exact tests were used in order to compare proportions and evaluate statistically significant associations between two categorical variables. A P value <0.05 was considered statistically significant.
Structural Modeling and Binding Site Prediction
Three-dimensional structural models for native IA-2 (NM_002846.3) and IA-2var were obtained using the I-TASSER server (23). No restrictions or model templates were used for model prediction. The predicted molecular structure was selected based on the highest confidence value for both C-score (a confidence score for estimating the quality of predicted models by I-TASSER) and TM-score (the template modeling score and measures of similarity between two protein structures with different tertiary structures). An estimation of the intrinsically unstructured sequence of both proteins was performed with the protein disorder predictor PONDR-FIT and Disorder Atlas (24). Binding site comparisons were predicted via I-TASSER. The predicted binding sites in the modeled structure are evaluated based on the binding site score. The model structure with the highest C-score for both the native IA-2 and IA-2var was uploaded in UCSF Chimera (version 1.7rc) for visualization, amino acid sequence alignments, and structural superimposition.
Results
Structural Modeling and Binding Site Prediction of IA-2var and IA-2 Native Proteins
We constructed three-dimensional molecular models of the native and IA-2var–containing proteins (Fig. 1C). The proteins share a common topology of α-helixes and β-sheets in the core as shown in the three-dimensional structural superimposition of both proteins, but large discrepancies do occur in their three-dimensional overlays (Fig. 1C1 [IA-2 native vs. IA-2var]). Furthermore, the extracellular domain (amino acid residues 1–577) of native IA-2 and IA-2var exhibits nonhomologous three-dimensional structural topology (Fig. 1C2). AAb binding site predictions illustrate nearly identical results for residues shared between native IA-2 and IA-2var (Supplementary Table 3). There are two predicted binding site regions for the native IA-2 sequence. The first predicted binding site involves amino acids residues 740–742, 909–915, 951, and 954. The second predicted binding site involves residues 819, 909–911, 913, 915, and 954. Only one binding site was predicted for the IA-2var sequence, involving amino acid residues 739–743, 909–915, 951, and 954 (Supplementary Table 3).
Variant-Specific AAb to IA-2 (IA-2var) Are Associated With High Risk of T1D Progression
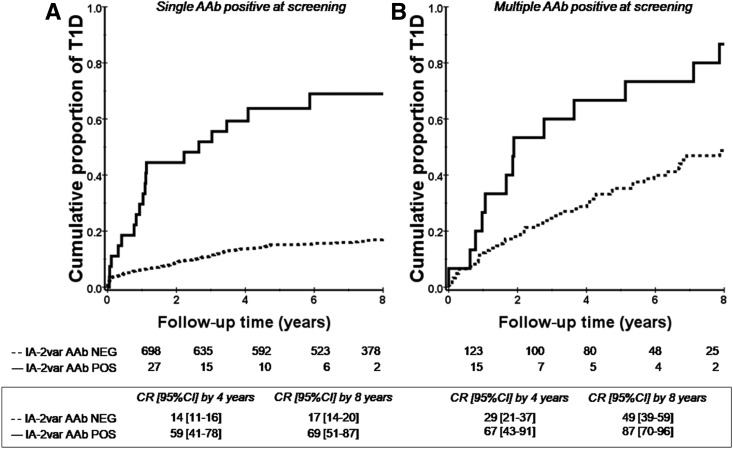
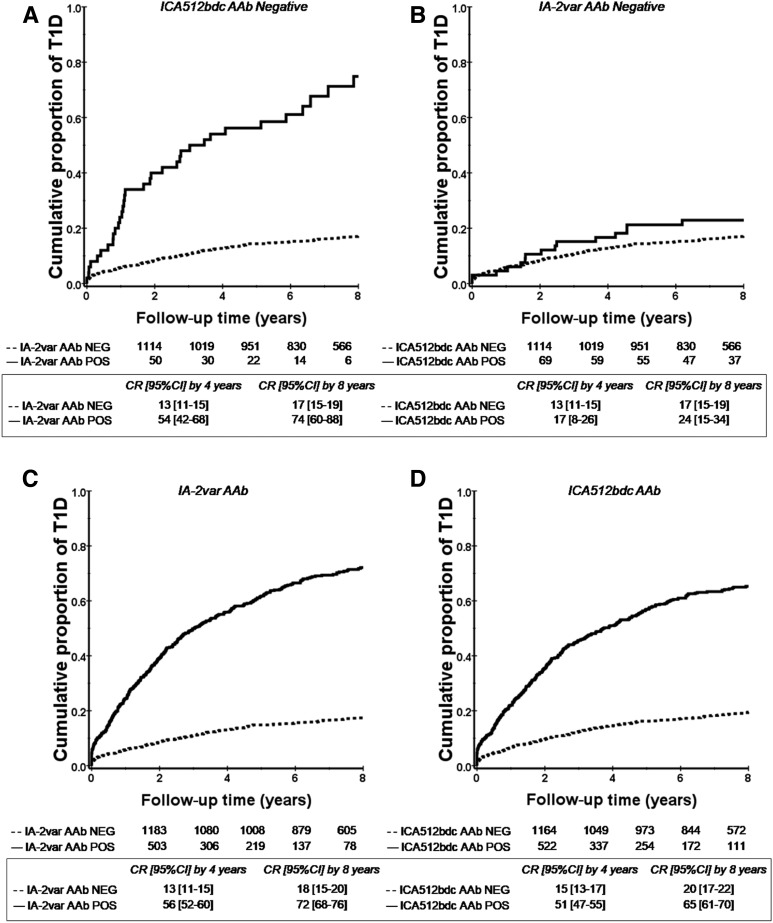
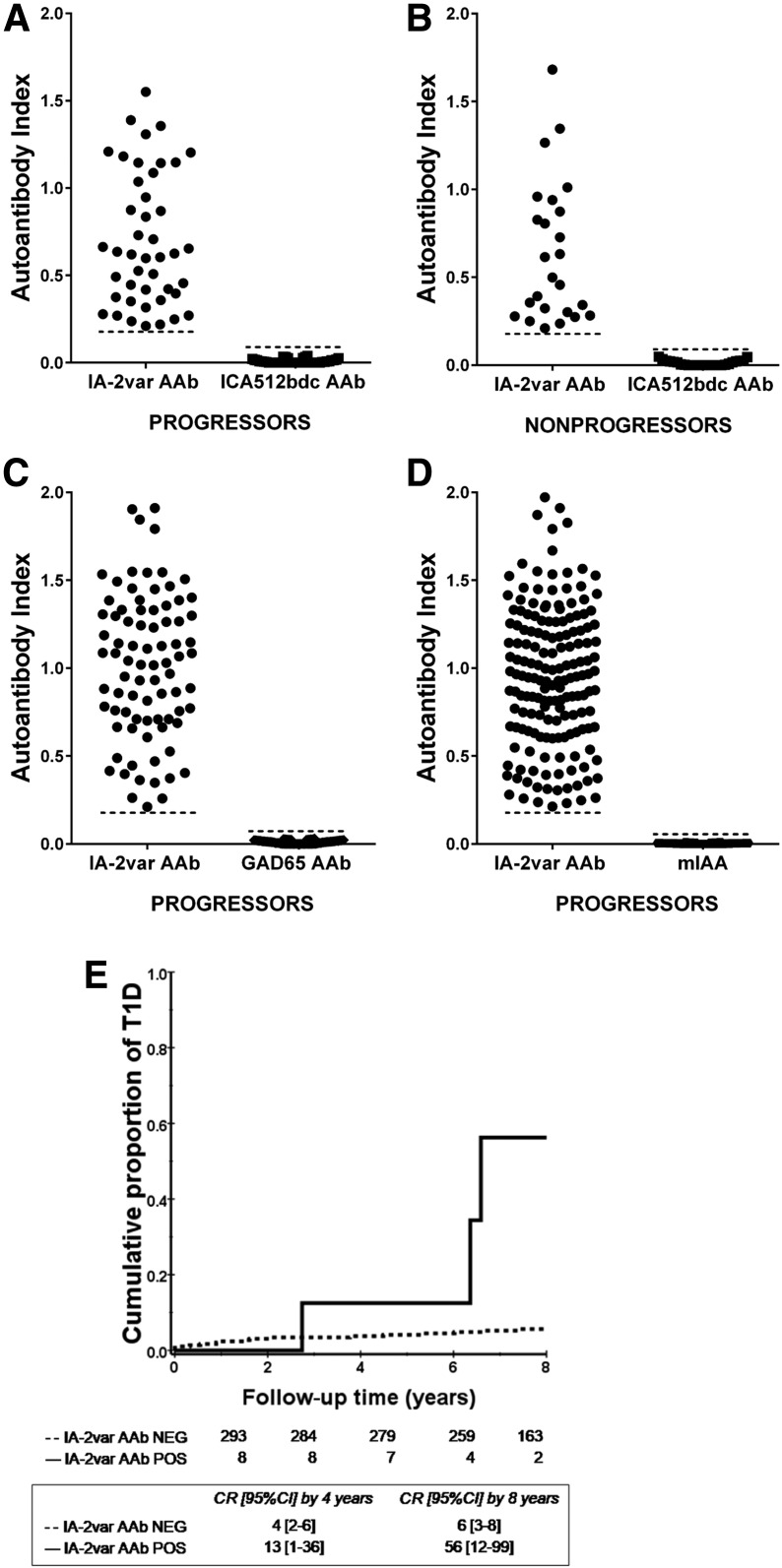
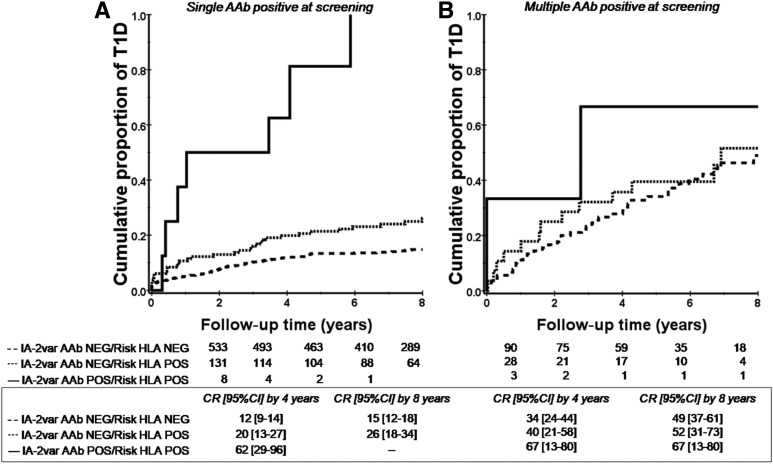
The presence of IA-2var–specific AAb improves risk stratification in subjects with single and multiple AAb at screening as defined by standard islet antibody testing (P < 0.0001 and P = 0.001) (Fig. 2A and B and Supplementary Fig. 1: this analysis includes ICA testing). In addition, the presence of IA-2var AAb in relatives who were negative for ICA512bdc AAb was associated with a higher T1D risk compared with those who were ICA512bdc AAb positive and IA-2var AAb negative (P < 0.0001 and 0.3, respectively) (Fig. 3A and B). IA-2var AAb were detected in 7.8% of the relatives who developed T1D (progressors) and tested negative for ICA512bdc AAb, 14.2% of those GAD65 AAb negative, and 29.7% of those mIAA negative (Fig. 4A–D). IA-2var AAb–positive subjects were younger than those testing negative (12.7 vs. 18.9 years of age, respectively). Of interest, we found that in a small subgroup of seronegative relatives at screening (negative for ICA512bdc, GAD65 AAb, mIAA), the presence of IA-2var AAb still conferred a higher risk of progression to overt disease compared with being negative for IA-2var AAb (P = 0.001) (Fig. 4E) (see also Supplementary Table 2 and Supplementary Fig. 8).
Figure 2.
IA-2var AAb are associated with high risk of T1D progression in relatives with single or multiple islet AAb at screening. A: Progression to T1D in relatives who were negative for ICA512bdc AAb and were selected for having a single AAb (GAD65 AAb or IAA) in the absence (dashed line) or presence (solid line) of IA-2var AAb (P < 0.0001). B: Progression to T1D in relatives who were negative for ICA512bdc AAb and were selected for having multiple AAb (GAD65 AAb and IAA) in the absence (dashed line) or presence (solid line) of IA-2var AAb (P = 0.001). CR, cumulative risk; NEG, negative; POS, positive.
Figure 3.
IA-2var AAb improve prediction of T1D in relatives testing negative for the conventional IA-2 AAb assay (ICA512bdc AAb). A: Relatives selected for being negative for ICA512bdc AAb in the absence (dashed line) or presence (solid line) of IA-2var AAb (P < 0.0001). B: Relatives who were negative for IA-2var AAb in the absence (dashed line) or presence (solid line) of AAb against ICA512bdc (P = 0.3). C and D: Progression to T1D in relatives in relation to the absence (dashed line) or presence (solid line) of IA-2var AAb (C) and progression to T1D in relatives in the absence (dashed line) or presence (solid line) of ICA512bdc AAb (D). CR, cumulative risk; NEG, negative; POS, positive.
Figure 4.
IA-2var AAb can be detected in relatives who are negative for traditional islet AAb at screening (ICA512bdc, GAD65, and mIAA). A: 7.8% (44 of 566) of the relatives who developed T1D (progressors) were negative for ICA512bdc but negative for IA-2var AAb. B: 2.2% (25 out of 1,120) of ICA512bdc AAb–negative nonprogressors were IA-2var AAb positive. C and D: 14.2% (81 of 566) of progressors were IA2var AAb positive and GAD65 AAb negative (C), and 29.7% (168 of 566) of the progressors were IA-2var AAb positive but mIAA negative (D). E: In seronegative relatives at screening (negative for ICA512bdc, GAD65 AAb, IAA), the presence of IA-2var AAb (solid line) still conferred a higher risk of progression to T1D compared with those negative for IA-2var AAb (dashed line) (P = 0.001). CR, cumulative risk; NEG, negative; POS, positive.
FDRs of T1D probands testing positive for single islet AAb at screening demonstrated that the presence of additional IA-2var AAb occurred in those who progressed rapidly to clinical T1D, particularly in those who carry the high-risk HLA-DRB1*04-DQB1*0302 haplotype (P < 0.0001) (Fig. 5A [see also Supplementary Fig. 2A and B: analysis with ICA testing]). This finding suggests that including IA-2var AAb in future first-line screening will more accurately identify those high-risk individuals with a greater risk of progression to T1D.
Figure 5.
IA-2var AAb and HLA DQ and DR genotypes confer a significant T1D risk in relatives with single and multiple islet AAb tested by traditional antibody assays. All of these subjects lacked ICA512bdc AAb. A: Relatives with single islet AAb (GAD65 AAb or IAA) carrying both IA-2var AAb and the HLA-DRB1*04-DQB1*0302 haplotype (P < 0.0001) (solid line). Relatives with single islet AAb (GAD65 AAb or IAA) carrying the HLA-DRB1*04-DQB1*0302 haplotype who were negative for IA-2var AAb (dotted line). Relatives with single islet AAb (GAD65 AAb or IAA) who were negative for IA-2var AAb and lacked the HLA-DRB1*04-DQB1*0302 haplotype (dashed line). B: Relatives with multiple islet AAb (GAD65 AAb and IAA) carrying both IA-2var AAb and the HLA-DRB1*04-DQB1*0302 haplotype (P = 0.02) (solid line). Relatives with multiple islet AAb (GAD65 AAb and IAA) carrying the HLA-DRB1*04-DQB1*0302 haplotype who were negative for IA-2var AAb (dotted line). Relatives with multiple islet AAb (GAD65 AAb and IAA) who were negative for IA-2var and lacked the HLA-DRB1*04-DQB1*0302 haplotype (dashed line). CR, cumulative risk; NEG, negative; POS, positive.
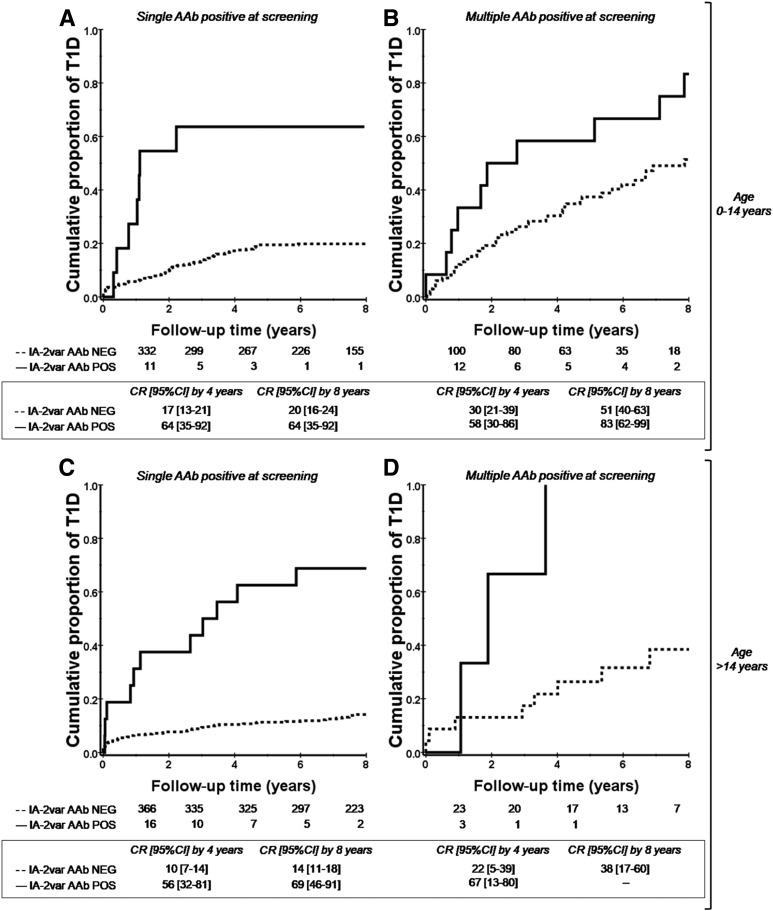
Further analyses were performed using Cox proportional hazards regression models to assess whether presence of IA-2var AAb, after adjusting for multiple AAb and age, remains as an independent risk factor for T1D. Models included IA-2var AAb and the presence of multiple AAb and their interaction as main effects for the subgroup of ICA512bdc AAb–negative FDRs of T1D index case subjects. The main effect of presence of IA-2var AAb was statistically significant, suggesting that there is an increased risk of T1D progression in the presence of AAb to IA-2var independent of age (Fig. 6) or other AAb (see also Supplementary Fig. 3A and C: analysis with ICA testing). Thus, the risk conferred by IA-2var AAb in the presence of multiple AAb appears to be in addition to other factors (whether additive or multiplicative in effect cannot be determined by these analyses). After models were constructed without an interaction term and adjusted for the risk of multiple AAb, the hazard ratio for IA-2var AAb for age at collection <14 years was 1.56 (P = 1.57 × 10−4 [95% CI 1.238, 1.964]). As stated above, survival analysis revealed that the presence of IA-2var AAb conferred a high risk of T1D progression in FDRs positive for single or multiple islet AAb by traditional antibody testing (Fig. 2A and B). There was no evidence of an interaction between the presence of multiple (other than IA-2var) AAb and IA-2var risk factors (P > 0.85). Supplementary Figs. 4 and 5 show the prevalence of IA-2var AAb in patients newly diagnosed with T1D.
Figure 6.
The presence of IA-2var AAb conferred a high risk of T1D progression in both relatives younger and older than 14 years of age who were positive at screening for single or multiple islet AAb by traditional antibody testing. All of these subjects lacked ICA512bdc AAb. A: Progression to T1D in relatives younger than 14 years of age who were positive for single islet AAb at screening (GAD65 AAb or IAA) in the absence (dashed line) or presence (solid line) of IA-2var AAb (P < 0.0001). B: Progression to T1D in relatives younger than 14 years of age who were positive for multiple islet AAb at screening in the absence (dashed line) or presence (solid line) of IA-2var AAb (P = 0.04). C: Progression to T1D in relatives older than 14 years of age who were positive for single islet AAb at screening (GAD65 AAb or IAA) in the absence (dashed line) or presence (solid line) of IA-2var AAb (P < 0.0001). D: Progression to T1D in relatives older than 14 years of age who were positive for multiple islet AAb at screening in the absence (dashed line) or presence (solid line) of IA-2var AAb (P = 0.03). CR, cumulative risk; NEG, negative; POS, positive.
We sequenced genomic DNA from 108 randomly chosen individuals to determine whether differences existed in the IA-2 nucleotide sequence at the predicted antibody binding sites. We identified five SNPs in exons 13 and 14 of PTPRN (Supplementary Table 4). Three are synonymous SNPs (rs144452294, rs17847405, and rs17847406), and two are novel nonsynonymous SNPs in the codons encoding amino acid positions 568 and 621. Consistent with previous studies, rs17847406 had the highest minor allele frequency (MAF) in our population, with 18 of 108 (16.7%) subjects having at least one copy of the C allele: 16 heterozygotes and 2 homozygous CC. The other four SNPs were infrequent (MAF < 0.05), with only 1 or 2 (of 108) subjects being heterozygous. Neither of the nonsynonymous SNPs in the codons encoding amino acid positions 608 and 671, that are both present in the AAb probe, was detected in these subjects.
The molecular modeling, antibody binding site predictions, and IA-2 gene (PTPRN) SNP analysis suggest that antibodies for IA-2 and those for IA-2var likely recognize different conformational epitopes. These epitopes may be unmasked by the amino acid differences in IA-2var.
Discussion
We identified a new variant of the neuroendocrine autoantigen IA-2 reacting with sera from subjects at risk for progressing to clinical T1D. IA-2var–specific AAb are strikingly associated with accelerated progression to T1D both in younger and older FDRs of T1D index case subjects in the TrialNet Pathway to Prevention Study who have single, multiple, or no islet AAb as defined by the standard AAb assays (25,26). This result regarding the ability of IA-2var to provide increased resolution of risk of T1D reinforces our observations that were conducted using a smaller sample from the Children’s Hospital of Pittsburgh population (27).
There are at least three explanations for the increased predictive value and sensitivity for the new IA-2var antibody test: 1) the IA-2var construct contains residues 1–256 and 556–600 that are absent from the probes used for the existing IA-2 antibody assays and could contain additional epitopes; 2) one or more of the IA-2 amino acid residues (Cys27, Gly608, and Pro671) may directly enhance AAb binding, contrasting with other previously reported IA-2 mutants (28,29); and 3) molecular modeling of IA-2var predicts that the amino acid substitutions induce changes in the three-dimensional structure of the molecule, which may lead to epitope unmasking.
Evidence suggesting the presence of masked epitopes within the cytoplasmic domain of IA-2 has been reported in a study that mapped the immunodominant epitopes of the ICA512bdc construct (30). This study identified two immunodominant regions, IA-2 residues 761–964 and 929–979. Although none of the amino acids in IA-2var lie within these regions, it should be noted that substitutions in the JM domain can interfere with IA-2 homo- and heterodimerization (31). This could conceivably explain how the IA-2var substitutions may lead to exposure of neo-epitopes (32,33). Whether the amino acid residues Cys27, Gly608, and Pro671have an impact on the function of the molecule remains to be established. However, evolutionary changes to the ancestral PTP domain at two other amino acids have made IA-2 and IA-2β enzymatically inactive (34,35). These changes influence enzymatic activity (36).
Although IA-2 (PTPRN) SNP genotyping was performed in a relatively small sample (n = 108), two of the three SNPs are infrequent (MAF < 0.05) in the population. This suggests that the majority of subjects with IA-2var AAb do not in fact express IA-2var but that the IA-2var probe is mimicking a conformation of IA-2 that is adopted under the pathological conditions that lead to AAb generation.
It has been reported that in relatives with IAA, seroconversion occurs more often in early childhood (25). In the current study, we found that IA-2var AAb are associated with a high risk of progression of overt T1D in both relatives younger and older than 14 years of age with single and multiple AAb at screening. One strength of this study is the sample size of the cohort; a limitation is that FDRs were not screened from birth. Hence, the age at which seroconversion has occurred is unknown. This may potentially lead to underestimation of the duration of AAb positivity. However, the focus of this study is to describe a new variant of the neuroendocrine autoantigen IA-2 reacting with sera from subjects at risk for progressing to T1D and to determine the predictive value of a new biomarker, IA-2var AAb, that improves prediction of T1D in FDRs of T1D probands carrying single and multiple islet AAb. In addition, we found that relatives with single islet AAb (by traditional assays) and carrying both IA-2var AAb and the high-risk HLA-DRB1*04-DQB1*03:02 haplotype progress rapidly to overt T1D. This has significant relevance to prevention trial design. Of interest, although based on small numbers, the presence of IA-2var AAb in seronegative subjects conferred a higher risk of T1D development. This observation suggests that other immunological abnormalities (i.e., T-cell responses) may be present in seronegative relatives (by conventional AAb testing) carrying IA-2var AAb (19).
Another limitation of this study is the lack of data on AAb directed toward the intracellular domain of IA-2 (IA-2ic), which is considered the immunodominant region of the molecule.
In summary, AAb responses directed to the IA-2var are associated with accelerated progression to T1D in relatives with no serologic response to other antigenic determinants of IA-2. The use of IA-2var AAb enhances the predictive value of T1D progression in relatives with both single and multiple islet AAb at screening, albeit there is still a possibility of missing some relatives at risk. This observation suggests that IA-2var AAb may allow for better characterization of the risk of progression to clinical T1D, improve staging accuracy of presymptomatic T1D (16), and in turn identify individuals who might benefit in future prevention trials. Our findings may also provide new clues in the search for pathogenic IA-2 epitopes associated with disease progression. Further studies are required to assess the mechanisms by which variant-specific AAb of variable affinity develop and their possible role in the chain of events leading to clinical T1D development.
Supplementary Material
Article Information
Acknowledgments. The authors thank the TrialNet Coordinating Center for providing the results available through the TrialNet Pathway to Prevention Study. The authors also acknowledge the support of the Type 1 Diabetes TrialNet Study Group, which identified study participants and provided samples and follow-up data for this study.
Funding. This work was supported by the McNair Medical Institute at The Robert and Janice McNair Educational Foundation; the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health (NIH) (grant R01 DK53456); and JDRF (grant 17-2012-688).
The Type 1 Diabetes TrialNet Study Group is a clinical trials network funded by the NIH through the National Institute of Diabetes and Digestive and Kidney Diseases, the National Institute of Allergy and Infectious Diseases, and the Eunice Kennedy Shriver National Institute of Child Health and Human Development, through cooperative agreements U01 DK061010, U01 DK061034, U01 DK061042, U01 DK061058, U01 DK085465, U01 DK085453, U01 DK085461, U01 DK085466, U01 DK085499, U01 DK085504, U01 DK085509, U01 DK103180, U01 DK103153, U01 DK085476, U01 DK103266, U01 DK103282, U01 DK106984, U01 DK106994, U01 DK107013, U01 DK107014, UC4 DK097835, UC4 DK106993, and JDRF.
The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the NIH or JDRF.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. M.J.A.-C. and S.L.P. performed the AAb assays. M.J.A.-C. and M.P. wrote the manuscript. M.P.M., S.S., A.D.V., C.F.V., R.G., D.J.B., S.H., C.J.G., L.Y., H.W.D., A.W.M., S.S.R., and M.P. edited the manuscript. M.J.A.-C., S.H., and M.P. performed the statistical analyses. H.W.D. performed the IA-2var genotyping. S.S. performed the molecular modeling of the IA-2var. M.P. oversaw all aspects of experimental design, data analysis, and literature research and contributed the bulk of funding to this project. M.P. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Prior Presentation. Preliminary data relative to this study were presented in abstract form at the 76th Scientific Sessions of the American Diabetes Association, New Orleans, LA, 10–14 June 2016.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db18-1351/-/DC1.
References
- 1.Roep BO, Peakman M. Diabetogenic T lymphocytes in human type 1 diabetes. Curr Opin Immunol 2011;23:746–753 [DOI] [PubMed] [Google Scholar]
- 2.Coppieters KT , Dotta F, Amirian N, et al. Demonstration of islet-autoreactive CD8 T cells in insulitic lesions from recent onset and long-term type 1 diabetes patients. J Exp Med 2012;209:51–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jacobsen LM, Haller MJ, Schatz DA. Understanding pre-type 1 diabetes: the key to prevention. Front Endocrinol (Lausanne) 2018;9:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lan MS, Lu J, Goto Y, Notkins AL. Molecular cloning and identification of a receptor-type protein tyrosine phosphatase, IA-2, from human insulinoma. DNA Cell Biol 1994;13:505–514 [DOI] [PubMed] [Google Scholar]
- 5.Arvan P, Pietropaolo M, Ostrov D, Rhodes CJ. Islet autoantigens: structure, function, localization, and regulation. Cold Spring Harb Perspect Med 2012;2:a007658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Achenbach P, Warncke K, Reiter J, et al. . Stratification of type 1 diabetes risk on the basis of islet autoantibody characteristics. Diabetes 2004;53:384–392 [DOI] [PubMed] [Google Scholar]
- 7.Steck AK, Vehik K, Bonifacio E, et al.; TEDDY Study Group . Predictors of progression from the appearance of islet autoantibodies to early childhood diabetes: The Environmental Determinants of Diabetes in the Young (TEDDY). Diabetes Care 2015;38:808–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu P, Krischer JP; Type 1 Diabetes TrialNet Study Group . Prognostic classification factors associated with development of multiple autoantibodies, dysglycemia, and type 1 diabetes—a recursive partitioning analysis. Diabetes Care 2016;39:1036–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wasserfall C, Montgomery E, Yu L, et al. . Validation of a rapid type 1 diabetes autoantibody screening assay for community-based screening of organ donors to identify subjects at increased risk for the disease. Clin Exp Immunol 2016;185:33–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hermel JM, Dirkx R Jr, Solimena M. Post-translational modifications of ICA512, a receptor tyrosine phosphatase-like protein of secretory granules. Eur J Neurosci 1999;11:2609–2620 [DOI] [PubMed] [Google Scholar]
- 11.Kawasaki E, Yu L, Gianani R, et al. . Evaluation of islet cell antigen (ICA) 512/IA-2 autoantibody radioassays using overlapping ICA512/IA-2 constructs. J Clin Endocrinol Metab 1997;82:375–380 [DOI] [PubMed] [Google Scholar]
- 12.Acevedo-Calado M, James EA, Morran MP, et al. . Identification of unique antigenic determinants in the amino terminus of IA-2 (ICA512) in childhood and adult autoimmune diabetes: new biomarker development. Diabetes Care 2017;40:561–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park YS, Kawasaki E, Kelemen K, et al. . Humoral autoreactivity to an alternatively spliced variant of ICA512/IA-2 in type I diabetes. Diabetologia 2000;43:1293–1301 [DOI] [PubMed] [Google Scholar]
- 14.Diez J, Park Y, Zeller M, et al. . Differential splicing of the IA-2 mRNA in pancreas and lymphoid organs as a permissive genetic mechanism for autoimmunity against the IA-2 type 1 diabetes autoantigen. Diabetes 2001;50:895–900 [DOI] [PubMed] [Google Scholar]
- 15.Battaglia M, Anderson MS, Buckner JH, et al. . Understanding and preventing type 1 diabetes through the unique working model of TrialNet. Diabetologia 2017;60:2139–2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Insel RA, Dunne JL, Atkinson MA, et al. . Staging presymptomatic type 1 diabetes: a scientific statement of JDRF, the Endocrine Society, and the American Diabetes Association. Diabetes Care 2015;38:1964–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vehik K, Beam CA, Mahon JL, et al.; TrialNet Natural History Study Group. Development of autoantibodies in the TrialNet Natural History Study. Diabetes Care 2011;34:1897–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.American Diabetes Association 2. Classification and diagnosis of diabetes: Standards of Medical Care in Diabetes—2017. Diabetes Care 2017;40(Suppl. 1):S11–S24 [DOI] [PubMed] [Google Scholar]
- 19.Pietropaolo M, Becker DJ, LaPorte RE, et al. . Progression to insulin-requiring diabetes in seronegative prediabetic subjects: the role of two HLA-DQ high-risk haplotypes. Diabetologia 2002;45:66–76 [DOI] [PubMed] [Google Scholar]
- 20.Pietropaolo M, Yu S, Libman IM, et al. . Cytoplasmic islet cell antibodies remain valuable in defining risk of progression to type 1 diabetes in subjects with other islet autoantibodies. Pediatr Diabetes 2005;6:184–192 [DOI] [PubMed] [Google Scholar]
- 21.Heinze G, Gnant M, Schemper M. Exact log-rank tests for unequal follow-up. Biometrics 2003;59:1151–1157 [DOI] [PubMed] [Google Scholar]
- 22.Mehta C, Patel N. StatXact 5: Statistical Software for Exact Nonparametric Inference. Cambridge, MA, Cytel Software Corporation, 2001 [Google Scholar]
- 23.Vincent M, Whidden M, Schnell S. Quantitative proteome-based guidelines for intrinsic disorder characterization. Biophys Chem 2016;213:6–16 [DOI] [PubMed] [Google Scholar]
- 24.Vincent M, Schnell S. Disorder Atlas: web-based software for the proteome-based interpretation of intrinsic disorder predictions [article online], 2017. Available from https://www.biorxiv.org/content/10.1101/060699v2. Accessed 26 March 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bosi E, Boulware DC, Becker DJ, et al.; Type 1 Diabetes TrialNet Study Group . Impact of age and antibody type on progression from single to multiple autoantibodies in type 1 diabetes relatives. J Clin Endocrinol Metab 2017;102:2881–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bingley PJ, Boulware DC, Krischer JP; Type 1 Diabetes TrialNet Study Group . The implications of autoantibodies to a single islet antigen in relatives with normal glucose tolerance: development of other autoantibodies and progression to type 1 diabetes. Diabetologia 2016;59:542–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morran MP, Casu A, Arena VC, et al. . Humoral autoimmunity against the extracellular domain of the neuroendocrine autoantigen IA-2 heightens the risk of type 1 diabetes. Endocrinology 2010;151:2528–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dromey JA, Weenink SM, Peters GH, et al. . Mapping of epitopes for autoantibodies to the type 1 diabetes autoantigen IA-2 by peptide phage display and molecular modeling: overlap of antibody and T cell determinants. J Immunol 2004;172:4084–4090 [DOI] [PubMed] [Google Scholar]
- 29.Weenink SM, Lo J, Stephenson CR, et al. . Autoantibodies and associated T-cell responses to determinants within the 831-860 region of the autoantigen IA-2 in type 1 diabetes. J Autoimmun 2009;33:147–154 [DOI] [PubMed] [Google Scholar]
- 30.Farilla L, Tiberti C, Luzzago A, et al. . Application of phage display peptide library to autoimmune diabetes: identification of IA-2/ICA512bdc dominant autoantigenic epitopes. Eur J Immunol 2002;32:1420–1427 [DOI] [PubMed] [Google Scholar]
- 31.Størling J, Overgaard AJ, Brorsson CA, et al. . Do post-translational beta cell protein modifications trigger type 1 diabetes? Diabetologia 2013;56:2347–2354 [DOI] [PubMed] [Google Scholar]
- 32.James EA, Pietropaolo M, Mamula MJ. Immune recognition of β-cells: neoepitopes as key players in the loss of tolerance. Diabetes 2018;67:1035–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strollo R, Rizzo P, Spoletini M, et al. . HLA-dependent autoantibodies against post-translationally modified collagen type II in type 1 diabetes mellitus. Diabetologia 2013;56:563–572 [DOI] [PubMed] [Google Scholar]
- 34.Gross S, Blanchetot C, Schepens J, et al. . Multimerization of the protein-tyrosine phosphatase (PTP)-like insulin-dependent diabetes mellitus autoantigens IA-2 and IA-2beta with receptor PTPs (RPTPs). Inhibition of RPTPalpha enzymatic activity. J Biol Chem 2002;277:48139–48145 [DOI] [PubMed] [Google Scholar]
- 35.Wishart MJ, Dixon JE. Gathering STYX: phosphatase-like form predicts functions for unique protein-interaction domains. Trends Biochem Sci 1998;23:301–306 [DOI] [PubMed] [Google Scholar]
- 36.Magistrelli G, Toma S, Isacchi A. Substitution of two variant residues in the protein tyrosine phosphatase-like PTP35/IA-2 sequence reconstitutes catalytic activity. Biochem Biophys Res Commun 1996;227:581–588 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.