Abstract
Nuclear receptor-binding SET domain protein 2 (NSD2) is a histone H3 lysine 36 (H3K36)-specific methyltransferase enzyme that is overexpressed in a number of cancers, including multiple myeloma. NSD2 binds to S-adenosyl-L-methionine (SAM) and nucleosome substrates to catalyze the transfer of a methyl group from SAM to the ε-amino group of histone H3K36. Equilibrium binding isotope effects and density functional theory calculations indicate that the SAM methyl group is sterically constrained in complex with NSD2, and that this steric constraint is released upon nucleosome binding. Together, these results show that nucleosome binding to NSD2 induces a significant change in the chemical environment of enzyme-bound SAM.
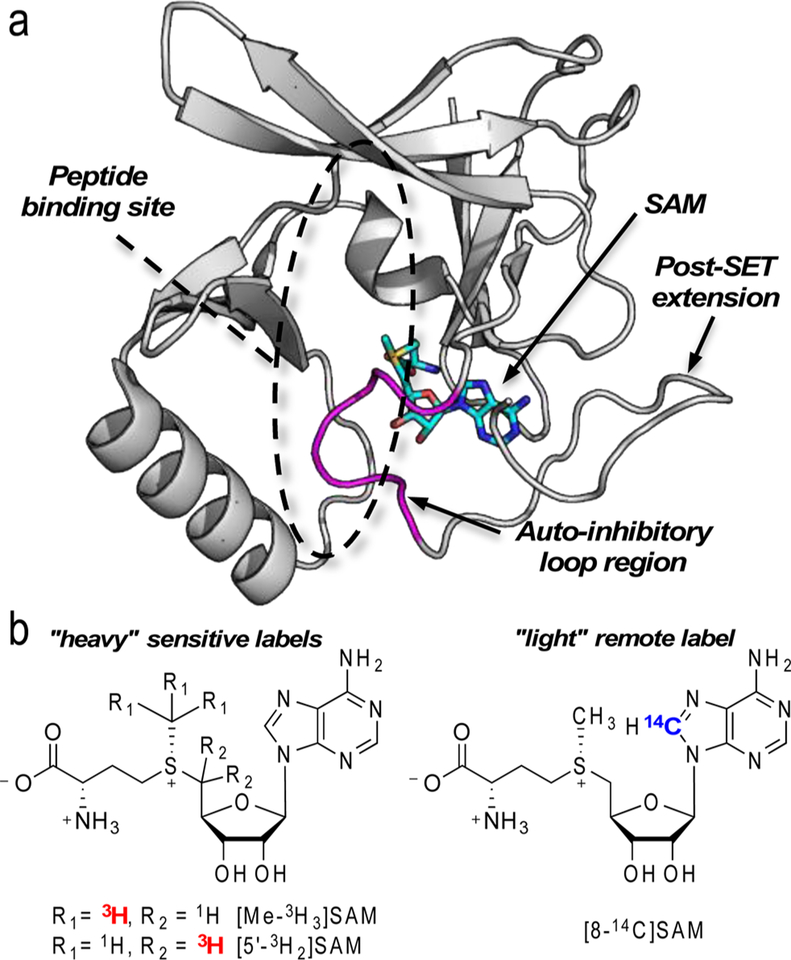
Methylation of lysine residues by protein lysine methyltransferase (PKMT) enzymes is an essential protein post-translational modification involved in transcriptional regulation, DNA damage response, and chromatin remodeling.1,2 In humans, nearly all of these enzymes contain a catalytic SET domain that is responsible for the site-specific transfer of the methyl (Me) group of S-adenosyl-L-methionine (SAM) onto the ε-amino group of protein lysine residues.3 Despite the structural similarity of their catalytic domains, these enzyme differ in both substrate specificity and the number of Me groups transferred.4,5 In particular, the human PKMT nuclear receptor-binding SET domain 2 (NSD2) catalyzes dimethylation of histone H3 lysine 36 (H3K36) of nucleosome substrates.6,7 Recently, it was shown that basic C-terminal post-SET extension of NSD2 is required for the methylation of H3K36 in nucleosome substrates.8 Similar to the homologous histone H3K36 PKMT enzyme NSD1, the NSD2 post-SET extension is attached to the catalytic SET domain via an autoinhibitory loop region, which occupies the peptide substrate-binding site in the available structure of NSD1 (PDB code: 3OOI; Figure 1a).9 NSD2 is overexpressed in a number of cancers, including multiple myeloma,10,11 making it a potential target for new cancer therapeutics. Despite interest in NSD2, there is a lack of structural information, specifically regarding how binding of nucleosome might influence the catalytic activity of NSD2 or the binding environment of the SAM substrate.
Figure 1.
Structure of NSD1 and labeled SAM used for BIE measurement. (a) Structure of NSD1 (PDB code: 3OOI) bound to SAM (cyan) showing the SET domain, autoinhibitory loop (magenta), and post-SET region. (b) BIEs were measured using [Me-3H3]SAM or [5′−3H2]SAM and [8-14 C]SAM.
We recently determined the transition-state structure for the reaction catalyzed by NSD2 via the measurement of multiple kinetic isotope effects (KIEs).12 Such V/K KIEs measured under competitive conditions can include contributions from all isotope-sensitive steps, up to and including the first irreversible step, which includes changes in bonding environment resulting from substrate-binding interactions. To better understand the substrate-binding interactions required for SAM binding to NSD2, we use equilibrium binding isotope effect (BIE) measurements and computational modeling to investigate changes in the chemical environment of NSD2-bound SAM resulting from the binding of nucleosome substrate.
Equilibrium BIEs directly report on differences in the bonding environment experienced by enzyme substrates in the free and enzyme-bound states.13 Specifically, BIEs result from the preferential binding of either a light or heavy isotope containing substrate to the protein, in competitive binding measurements. Preferential binding to the light-isotope-labeled substrate results in a normal BIE (>1.0) and indicates that the isotope-substituted bond is weaker in the bound complex, while binding to the heavy isotope results in an inverse BIE (<1.0), indicating a strengthening of the substituted bond in the bound state. Thus, measuring equilibrium BIEs allows us to distinguish between changes in bonding occurring in the enzyme−substrate complex and changes in bonding at the TS.
Here, tritium BIEs for [Me-3H3]SAM and [5′−3H2]SAM binding to NSD2 were measured via internal competition with [8-14C]SAM, which contains a remote 14C to track light 1H isotopes at the Me and C5′ positions (Figure 1b). Equilibrium BIE values were measured for the NSD2·SAM binary complex and an NSD2·SAM·Nucleosome pseudo-ternary complex via rapid equilibrium dialysis, as previously used to measure BIEs for dihydropteroate synthase (DHPS).14 For this method, BIEs are determined from the ratio of light-to-heavy isotope-labeled substrate bound to NSD2 compared to the ratio of isotopes in the unbound substrate.
A large inverse equilibrium BIE of 0.65 ± 0.02 (35% inverse; Table 1) was observed for the binary complex of NSD2 with [Me-3H3]SAM. By contrast, the binary complex of NSD2 with [5′−3H2]SAM resulted in a smaller inverse BIE of 0.973 ± 0.009 (2.7% inverse). In both cases, the equilibrium BIE values are more inverse than V/K KIEs, i.e., 0.77 ± 0.03 (23% inverse) and 1.05 ± 0.01 (5% normal),12 measured for the methylation of HeLa cell nucleosome using the same isotope substitutions. These BIEs indicate that the bonding environment experienced by SAM differs substantially in the enzyme’s substrate-bound complex compared to the enzyme’s transition-state complex or substrate free in solution.
Table 1.
Equilibrium Binding Isotope Effects for NSD2 Binary and Pseudo-ternary Complexes
| isotope label | binary complex BIE | pseudo-ternary complex BIE | V/K KIEa |
|---|---|---|---|
| [Me-3H3] SAM | 0.65 ± 0.02 (5) | 0.990 ± 0.005 (6) | 0.77 ± 0.01 (9) |
| [5′−3H2] SAM | 0.973 ± 0.009 (9) | 0.982 ± 0.008 (6) | 1.047 ± 0.004 (6) |
V/K KIEs were reported previously.12 BIEs and V/K KIEs are reported with ± standard error with the number of replicates presented in parentheses. BIE, binding isotope effect; KIE, kinetic isotope effect.
It is important to note that BIE measurements for the binary complex may not reflect changes in bonding directly related to enzyme catalysis. Any substrate-binding interactions that contribute to catalysis should be maintained in the ternary complex and result in similar magnitude BIEs. To mimic an NSD2·SAM·Nucleosome ternary complex, we have used recombinant polynucleosome in which the substrate histone H3 lysine 36 residues are mutated to methionine (H3K36-MNuc). The substitution of lysine with methionine has previously been used in crystal structures of lysine-specific demethylase 1 (LSD1), and resulted in increased binding affinity compared to that of the native lysine-containing substrate.15 H3K36MNuc binds NSD2 competitively (Ki = 0.63 nM) and with increased affinity compared to native H3K36 nucleosome substrate (Km = 14.3 nM) but does not react during the course of equilibrium BIE measurements (Figures S1 and S2).
Intriguingly, BIE measurements for this pseudo-ternary complex of NSD2 with SAM and H3K36MNuc resulted in a [Me-3H3]SAM BIE of 0.990 ± 0.005 (1.0% inverse), significantly less inverse than that of the equivalent binary complex BIE, and a [5′−3H2]SAM pseudo-ternary complex BIE of 0.982 ± 0.008 (1.8% inverse) that was not significantly different with H3K36MNuc added. It is important to note that the total fraction of bound SAM did not change upon addition of H3K36MNuc, indicating that the saturation of SAM remains constant (Km = 203 ± 13 nM). This indicates that nucleosome binding alters the chemical bonding environment of the SAM methyl group C−H bonds, while leaving the C5′−H bonding environment relatively unchanged.
To better understand the changes in bonding environment contributing to observed BIEs, density functional theory (DFT) calculations were employed to model environmental and bonding changes experienced by SAM in binary and pseudo-ternary complexes with NSD2. Specifically, we considered the effects of (1) SAM conformation, (2) protein dielectric environment, (3) interactions with active-site amino acids, and (4) restriction C−H bond lengths on the vibrational frequencies of the Me C−H bonds. In each case, equilibrium BIEs were calculated from scaled vibrational frequencies of optimized structures (Gaussian 09,16 M062X/6–31g(d) theory) of SAM in the “free” and predicted “bound” conformations. Initial coordinates for SAM from the crystal structure of NSD1 (PDB code: 3OOI)9 were used for calculations of the NSD2 “bound” state, and the low-energy conformation of SAM from PubChem 3D17 (PubChem CID 34756) was used to model the “free”-state solution conformation of SAM, including water as an implicit solvent model. This conformation is consistent with nuclear magnetic resonance (NMR) measurements for SAM in aqueous solution,18 which predict an extended SAM conformation with little restriction of the methionine group conformation,19 and it has previously been used to calculate SAM KIEs for the methylation of H3K36 catalyzed by NSD2,12 and the methylation of DNA by DNA methyltransferase 1 (DNMT1).20
SAM in crystal structures of SET domain PKMTs adopts a “folded” conformation, restricting the position of the adenosine ribose and methionine groups (Figure 1a).3 When we investigated the impact of this conformational change on SAM equilibrium, BIEs predicted small inverse [Me-3H3]SAM and [5′−3H2]SAM BIEs of 0.994 (0.6% inverse) and 0.964(3.6% inverse), respectively, that agree within error with observed pseudo-ternary complex BIEs. This change in SAM conformation upon NSD2 binding is sufficient to explain the pseudo-ternary complex BIEs observed in the presence of H3K36MNuc without any other changes in bonding; however, it is unable to explain the large inverse [Me-3H3]SAM BIE observed in the binary complex.
The change in dielectric environment between water and the protein active site has been shown to influence the value of predicted BIEs.14 As a result, the influence of protein dielectric environment on [Me-3H3]SAM BIEs was examined using DFT calculations of the SAM “bound” state using implicit solvent models (polarizable continuum model, PCM) shown in Figure S3 to mimic the NSD2 active-site dielectric. Changing the solvent dielectric between water (ε = 78.3) and chlorobenzene (ε = 5.7) did not significantly alter the magnitude of the predicted [Me-3H3]SAM BIE (0.984 and 0.999 in water and chlorobenzene, respectively; Table S1). These models indicate that the protein dielectric environment cannot sufficiently explain the large [Me-3H3]SAM BIEs for the binary complex.
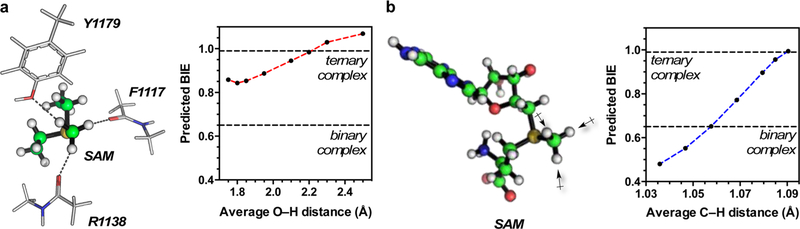
DFT calculations from our laboratory used to predict human brain hexokinase BIEs found that interactions with either hydrophobic side-chain residues or carbonyl oxygen of amino acid backbones predict inverse BIEs.21 It is therefore possible that the inverse [Me-3H3]SAM BIE for the SAM·NSD2 binary complex results from specific active-site interactions. Specifically, the SET domain of PKMT enzymes contains a structurally conserved “methyltransfer pore” composed of amino acid residues in direct contact with the SAM methyl group and lysine ε-amino group.3 NMR studies of SET7/9 indicate the presence of nonclassical C−H⋯O hydrogen-bonding interactions between the oxygen atoms of two backbone carbonyls and one tyrosine side chain in the “methyltransfer pore” and the SAM methyl group hydrogens.22,23 We observed no similar 1H chemical shift change in heteronuclear single quantum coherence NMR spectroscopy measurements of the bound complex (Figure S4); however, we did observe a significant decrease in SAM Me C−H signal intensity with increasing concentration of NSD2. Since no new signal was observed, it is possible that the SAM binding/release is in slow to intermediate exchange on the NMR time scale or that intermediate exchange between multiple bound conformations suppresses the SAM methyl group signal. Therefore, we investigated the impact of C−H⋯O interactions on predicted BIEs computationally by including two N-methylacetamide and p-hydroxytoluene in DFT calculations to mimic the conserved backbone carbonyls of F1117 and R1138 and side chain of Y1179, respectively. The crystallographic position of these residues, determined from the structure of NSD1 (PDB code: 3OOI), was used as the starting point for DFT calculations (Figure 2a). Without constraints on the H⋯O distance, this model predicts a 1.021 (2.1% normal) [Me-3H3]-SAM BIE that is inconsistent with the inverse BIEs observed for both binary and pseudo-ternary complexes. For SET7/9, it was shown that the “methyltransfer pore” region can contract as the result of substrate binding.23 Thus, we looked at the relationship between average H⋯O distance and predicted [Me-3H3]SAM BIE shown in Figure 2a. Shortening the H⋯O distance between the SAM methyl group hydrogen and the “methyltransfer pore” residues results in increasingly more inverse [Me-3H3]SAM BIE values (<1.0). Regardless, the magnitudes of the predicted BIEs remain less inverse than those of the observed binary complex BIE, even at a H⋯O distance of 1.75 Å. These vibrational frequencies used for BIE measurements were all calculated using the harmonic approximation; it is possible that anharmonicity could alter the magnitude of these BIEs, but we would expect the trend to remain the same. Given these results, it is unlikely that C−H⋯O hydrogen-bonding interactions are solely responsible for the magnitude of observed binary complex BIEs.
Figure 2.
Influence of C−H⋯O hydrogen-bonding interactions and geometric constraint on predicted [Me-3H3]SAM BIEs. (a) Equilibrium BIEs were calculated at fixed H⋯O distance between the methyl group hydrogen and backbone carbonyl oxygen of F1117 and R1138, and the side-chain oxygen of Y1179. (b) Calculated BIEs for compression of the average Me group C−H bond length. The value of experimentaly observed BIEs for the pseudo-ternary complex and binary complex are displayed as dashed lines.
Previous work on human brain hexokinase21 and DHPS14 found that restriction of stretching vibrational modes of C−H bonds as the result of steric impingement contributed to inverse tritium BIEs. We modeled steric imposition by restricting the methyl group C−H bond lengths in calculation of the SAM “bound”-state conformation, as previously described for DHPS BIE calculations.14 Although these constrained geometries are not true stationary points on the energy surface for the free substrate, they mimic the effect of restrictions imposed by the enzyme. The effects of constrained geometry on quantum mechanics analysis of isotope effects have been analyzed in great detail and found to be within experimental errors of isotope effect measurements.24 The results from these calculations shown in Figure 2B indicate a symmetric 3% compression of the methyl group C−H bond lengths (relative to the equilibrium bond lengths), or 10% compression of a single C−H bond length (Table S4), consistent with the [Me-3H3]SAM BIE of 0.65 (35% inverse) observed for the binary complex. Thus, steric impingement of the SAM methyl group is consistent with the inverse BIEs observed in the SAM NSD2 binary complex. It is possible that this steric impingement results from interaction with the “methyltransfer pore” residues, but we would expect that these contributions would also be maintained in the ternary complex. Instead, it is possible that the steric impingement results from interactions with the NSD2 autoinhibitory loop that are unrelated to catalysis, and that these interactions are displaced by binding of H3K36MNuc. This would be consistent with previous molecular dynamics simulations of NSD1 that show that the autoinhibitory loop is displaced during nucleosome binding.25
In conclusion, the evidence provided here from the measurement of equilibrium BIEs indicates that the methyl group of bound SAM in a binary complex with NSD2 experiences a sterically constrained bonding environment compared to that in the SAM·NSD2·H3K36MNuc pseudo-ternary complex. DFT calculations indicate that the large inverse [Me-3H3]SAM BIE (35% inverse) in the binary complex is consistent with a restriction of the Me group C−H bond length due to steric impingement. By contrast, the small inverse BIEs observed for [Me-3H3]SAM and [5′−3H2]SAM in the pseudo-ternary complex (1.0% and 1.8%, respectively) can be adequately explained by differences in SAM conformation between the free and bound states without requiring any additional changes in bonding. The structural similarity between H3K36MNuc used in this study and native nucleosome substrates of NSD2 gives us confidence that the pseudo-ternary complex BIE measurements more accurately reflect the bonding environment experienced by SAM in the catalytically active Michaelis complex. This indicates that the changes in bonding for bound SAM will not significantly contribute to previously observed V/K KIEs,12 and that V/K KIEs report on changes in bonding in the transition-state structure, not in the bound state. This study clearly demonstrates the utility of equilibrium BIE measurements for studying changes in the bonding environment of enzyme-bound substrates, and shows that nucleosome binding to NSD2 alters the chemical environment of the protein-bound SAM.
Supplementary Material
ACKNOWLEDGMENTS
We thank C. Stratton, Q. Du, and Z. Wang (Albert Einstein College of Medicine) for helpful discussion. This work was supported by a collaborative research agreement between GlaxoSmithKline Pharmaceuticals, the Albert Einstein College of Medicine, and NIH research grant GM041916. M.B.P. is the recipient of a Postdoctoral Fellowship from the Natural Sciences and Engineering Research Council of Canada.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.6b01612.
Figures S1−S3 and Tables S1−S3; experimental methods for reagent preparation; equilibrium BIE measurements; and DFT calculations (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Jenuwein T; Allis CD Science 2001, 293, 1074. [DOI] [PubMed] [Google Scholar]
- (2).Kouzarides T Cell 2007, 128, 693. [DOI] [PubMed] [Google Scholar]
- (3).Dillon SC; Zhang X; Trievel RC; Cheng X Genome Biol. 2005, 6, 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Couture JF; Dirk LM; Brunzelle JS; Houtz RL; Trievel RC Proc. Natl. Acad. Sci. U. S. A 2008, 105, 20659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Del Rizzo PA; Trievel RC Epigenetics 2011, 6, 1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Li Y; Trojer P; Xu CF; Cheung P; Kuo A; Drury WJ; Qiao Q; Neubert TA; Xu RM; Gozani O; Reinberg DJ Biol. Chem 2009, 284, 34283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Kuo AJ; Cheung P; Chen K; Zee BM; Kioi M; Lauring J; Xi Y; Park BH; Shi X; Garcia BA; Li W; Gozani O Mol. Cell 2011, 44, 609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Allali-Hassani A; Kuznetsova E; Hajian T; Wu H; Dombrovski L; Li Y; Graslund S; Arrowsmith CH; Schapira M; Vedadi MJ Biomol. Screening 2014, 19, 928. [DOI] [PubMed] [Google Scholar]
- (9).Qiao Q; Li Y; Chen Z; Wang M; Reinberg D; Xu RM J. Biol. Chem 2011, 286, 8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Hudlebusch HR; Santoni-Rugiu E; Simon R; Ralfkiaer E; Rossing HH; Johansen JV; Jorgensen M; Sauter G; Helin K Clin. Cancer Res 2011, 17, 2919. [DOI] [PubMed] [Google Scholar]
- (11).Morishita M; di Luccio E Biochim. Biophys. Acta, Rev. Cancer 2011, 1816, 158. [DOI] [PubMed] [Google Scholar]
- (12).Poulin MB; Schneck JL; Matico RE; McDevitt PJ; Huddleston MJ; Hou W; Johnson NW; Thrall SH; Meek TD; Schramm VL Proc. Natl. Acad. Sci. U. S. A 2016, 113, 1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Schramm VL Curr. Opin. Chem. Biol 2007, 11, 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Stratton CF; Namanja-Magliano HA; Cameron SA; Schramm VL ACS Chem. Biol 2015, 10, 2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Forneris F; Binda C; Adamo A; Battaglioli E; Mattevi AJ Biol. Chem 2007, 282, 20070. [DOI] [PubMed] [Google Scholar]
- (16).Frisch MJ; Trucks GW; Schlegel HB; Scuseria GE; Robb MA; Cheeseman JR; Scalmani G; Barone V; Mennucci B; Petersson GA; Nakatsuji H; Caricato M; Li X; Hratchian HP; Izmaylov AF; Bloino J; Zheng G; Sonnenberg JL; Hada M; Ehara M; Toyota K; Fukuda R; Hasegawa J; Ishida M; Nakajima T; Honda Y; Kitao O; Nakai H; Vreven T; Montgomery JA Jr.; Peralta JE; Ogliaro F; Bearpark MJ; Heyd J; Brothers EN; Kudin KN; Staroverov VN; Kobayashi R; Normand J; Raghavachari K; Rendell AP; Burant JC; Iyengar SS; Tomasi J; Cossi M; Rega N; Millam NJ; Klene M; Knox JE; Cross JB; Bakken V; Adamo C; Jaramillo J; Gomperts R; Stratmann RE; Yazyev O; Austin AJ; Cammi R; Pomelli C; Ochterski JW; Martin RL; Morokuma K; Zakrzewski VG; Voth GA; Salvador P; Dannenberg JJ; Dapprich S; Daniels AD; Farkas Ö ; Foresman JB; Ortiz JV; Cioslowski J; Fox DJ Gaussian 09; Gaussian, Inc: Wallingford, CT, 2009. [Google Scholar]
- (17).Kim S; Bolton EE; Bryant SH J. Cheminf 2013, 5, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Markham GD; Norrby P-O; Bock CW Biochemistry 2002, 41, 7636. [DOI] [PubMed] [Google Scholar]
- (19).Stolowitz ML; Minch MJ J. Am. Chem. Soc 1981, 103, 6015. [Google Scholar]
- (20).Du Q; Wang Z; Schramm VL Proc. Natl. Acad. Sci. U. S. A 2016, 113, 2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Lewis BE; Schramm VL J. Am. Chem. Soc 2003, 125, 4785. [DOI] [PubMed] [Google Scholar]
- (22).Horowitz S; Yesselman JD; Al-Hashimi HM; Trievel RC J. Biol. Chem 2011, 286, 18658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Horowitz S; Dirk LM; Yesselman JD; Nimtz JS; Adhikari U; Mehl RA; Scheiner S; Houtz RL; Al-Hashimi HM; Trievel RC J. Am. Chem. Soc 2013, 135, 15536. [DOI] [PubMed] [Google Scholar]
- (24).Hirschi JS; Takeya T; Hang C; Singleton DA J. Am. Chem. Soc 2009, 131, 2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Morishita M; di Luccio E Biochem. Biophys. Res. Commun 2011, 412, 214. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.