It was in 1959 that Russell and Burch, through their book ‘The Principles of Humane Experimental Technique’, first introduced the concept of 3Rs - Replacement, Reduction, and Refinement of animals in experimentation1. India has long since accepted and adopted the 3Rs through the mandate given by the Committee for the Purpose of Control and Supervision of Experimentation in Animals (CPCSEA). They have also inculcated a fourth R, that of Rehabilitation for the animals used in experiments2. Despite the campaign to reduce the number of animals used in research, it is estimated that more than 100 million animals are used annually, worldwide and evidence suggests that there is no decline in the use of animals in research3. One reason for the lack of decline in animal usage could be that there were, until recently, no suitable non-animal models/technologies that would effectively translate toxicology and pharmacology data to humans. In recent years, however, technology has evolved considerably with non-animal models becoming available, especially, human-derived, three-dimensional (3D) models which preserve human physiology, making extrapolation to humans possible4. Such models are rapidly evolving and replacing animal studies either to reduce or minimize animal usage in research. In addition, regulatory agencies in the western world are very receptive to the evolving novel, non-animal technologies to replace animal studies. Collaborative efforts between regulatory agencies, academia and industry in the western countries are simply revolutionizing the development of human-derived, non-animal technologies, not only for less animal dependence but also to improve the successful translational outcomes in humans. Here, we discuss the current status in India on the utility of these models to replace animal studies, identify gaps and put forward a roadmap for Indian research on non-animal technologies to support regulated industries and the life science research sector.
Though animal usage added immense knowledge to research advances made in the past several decades, the scenario is rapidly changing. The ban on the use of animals in cosmetic testing is a significant breakthrough, and recently the ban on Draize test in rabbits for drug5,6. With the development of 3D skin and eye models, industry is shifting to use these models to conduct local irritation and corrosion assays, which also have become a regulatory mandate in India7. These regulatory mandates were made, primarily because of animal ethics concerns. However, with respect to drug discovery and development (DD&D), extensive animal usage continues, all around the globe, with no strict regulatory mandates due to lack of appropriate non-animal translational models8.
Apart from ethical concerns in the use of animals for research, it is also noted that animal data are not being translated to successful clinical outcomes, as evident from failure rates of over 90 per cent between nomination for phase I clinical trials and approval of new drugs9. Within India, two decades of drug discovery R&D10 did not produce a single New Chemical Entity (NCE) to enter global markets, although a few molecules were out-licensed to pharma giants across the globe. These failures are due to unpredictable toxicity, and/or due to poor efficacy in humans, although these NCEs are efficacious in animal models with little toxicities11,12,13,14. The failure of animal testing to translate into successful clinical outcomes for many human diseases has propelled the scientific community to think about alternative methods of testing. Poor translation, along with increasing costs15,16,17 (estimated to be $2.5 billion USD or more) of DD&D has compelled the scientific community, as well as multinational companies to transition towards alternative non-animal translational technologies12,17. Despite the mounting failures in clinical trials, even today, the regulatory environment of DD&D requires at least two animal species18, rodent (mouse and rat) and non-rodent (canines and non-human primates) before human testing. The safety and efficacy are measured using pharmacokinetics (PKs), toxicokinetics, allometric scaling, PK-pharmacodynamic (PK-PD) and physiologically based PK methods across the species to predict safety- and therapeutic-margins and for selecting safe human doses for phase I clinical trials. There is a paradigm shift in the way biomedical research is being looked at. Targeted and focused library synthesis19,20 in all sectors (including agrochemical, nutraceutical and biopharmaceutical) must come up with novel, safe and efficacious chemicals for the use of humans. In technologies, such as the increased throughput in chemistry, high-throughput screening assays for pharmacology and toxicity screening, drug design using in silico and computational models as predictive pharmacology and toxicology tools, and the explosion of systems biology tools21(genomics, proteomics, transcriptomics, metabolomics and molecular diagnostics) and regenerative medicine (growing human tissue organoids from stem cells), there has been considerable improvement in the development of non-animal technologies (particularly 3D-functional organs) in the developed countries such as the US22, the UK23 and many other countries including Denmark, Brazil, Germany, Switzerland, Australia, Korea and China showing interest in developing these, through accepted forms of alternatives24.
A significant breakthrough in the recent past was the development of 3D-human-derived models8 being utilized for translation as non-animal technologies. In addition, parallel technology explosions including the advances in systems biology tools are helping to discover novel biomarkers. In silico computer-based models are also advancing rapidly to predict human pharmacology, toxicology and other biological parameters8. One of the primary reasons why these technologies have not matured yet to be of predictive value is the complexity of human physiology, biochemistry and molecular mechanisms of pharmacology and toxicology. Another reason is that data derived from animal models are constantly being extrapolated to human situations, but these extrapolations have largely proved futile in relating animal data to humans. Finally, humans have a lot of intra- and inter-individual variations including genetic and epigenetic predispositions related to pharmacology targets, drug metabolism and drug transporters, adding another dimension to the complexity of drug disposition. Therefore, future correlations for humans should be made using human-derived, non-animal technologies, utilizing dynamic flow models which mimic human physiology, biochemistry, pharmacogenetics and other relevant parameters. Optimizing these human-derived models with -omics and in silico tools should help us get more critically relevant human data that can be applied for clinical trial design and improving successful clinical outcomes. The primary goal of regenerative medicine is to provide transplantable human organs by utilizing stem cells derived from patients, but it can be utilized to develop patient-specific therapeutics in the future.
Development and use of non-animal technologies
In India, major initiatives are yet to be taken for the development of many of the new technologies and infrastructures. While regulatory acceptance of alternatives is increasing with the (i) prohibition of animal tests for cosmetics5, (ii) prohibition of Draize test in the testing of drugs and other chemicals6, and (iii) amendment in safety testing requirement for pesticides25 over the past few years, laboratories dedicated to develop these non-animal technologies, at both public and private sectors, have been missing or are very limited. The diversity in the Indian populations is an added dimension that is to be kept in mind for translation of PK-PD carried out in animals to humans.
With all the above mentioned issues in mind, the Indian Council of Medical Research (ICMR), New Delhi, constituted an expert committee in 2017 and did the brainstorming. With this initiative, it is hoped that the country will look towards a new era where basic science research, as well as drug testing, will not only be more relevant to human diseases but also time and cost-effective in addition to being humane. This article focuses on DD&D as a major beneficiary of advanced non-animal technologies, but these technologies will also benefit agrochemical, pesticide and cosmetic industries as well.
Evolving strategies for alternatives to animals and current state of the art
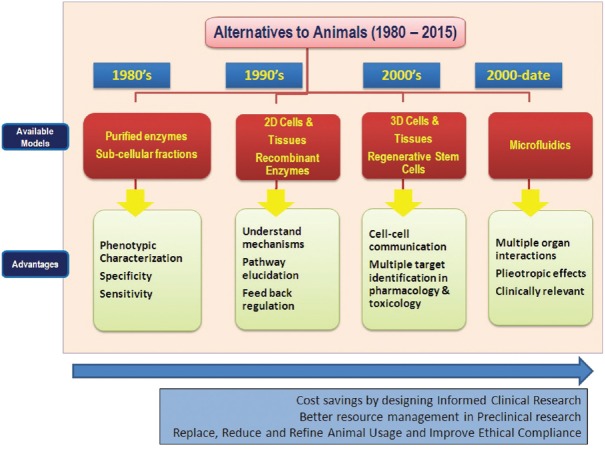
In parallel with the campaign of 3Rs in the 1950s, scientific communities around the world recognized the need for mechanistic translational human-derived models to complement, or supersede, animal data. Figure 1 provides the evolution of these technologies along with the advantages and disadvantages of all these models with a focus on the current state-of-the-art, non-animal and translational models.
Fig. 1.
Evolution of alternatives to animal research in the last five decades. Source: www.reagenebiosciences.com/about/why-alternatives-to-animals. Reproduced with permission.
In vitro models
In the early 1950s, it was widely recognized that hepatic drug metabolism, primarily mediated by cytochrome p450 enzymes, plays a central role in the disposition of xenobiotic chemicals26. In the 1980s and 1990s, variations in drug metabolism across the species, including humans, were recognized as a hurdle to translate the animal data to human relevance. Thus, human-derived in vitro subcellular fractions, especially liver microsomes and hepatocytes, served as useful tools of mechanistic dissemination of drug disposition allowing investigative toxicology to be set in motion17. Further advances in molecular cloning approaches and recombinant technologies allowed testing and screening of chemicals using in vitro recombinant human enzymes and human cell-based assays for target specific pharmacology and selecting the potential drug candidates for testing in animals and clinical trials. In the early 1990s, pharmacogenetics research unraveled ethnic variations in human drug metabolism that could impact human safety and efficacy of drugs27. In the 2000s, it was recognized that drug transporters also modulated xenobiotic disposition and together drug transporters and drug metabolism could account for fatal drug-drug interactions and genetically variable drug disposition across various human populations and sub-populations28.
A series of regulatory guidance documents were issued by the US Food and Drug Administration (FDA) and International Conference on Harmonization (ICH) as a mandate to understand human drug metabolism relative to animal drug metabolism including identification of various metabolites in all the species as well as the potential drug-drug interactions and cardiovascular toxicities29,30,31,32. If a metabolite seen in in vitro human models is not seen in animal models or is present in higher concentration relative to animals, regulatory agencies require more animal safety studies using unique human metabolite(s). A rapid advancement of human clinical trials based on pharmacogenetic principles shifted the drug discovery paradigm to personalized and precision medicine33, further weakening the relevance of animal studies in conducting appropriate clinical trials. In the late 1990s, many marketed drugs were withdrawn due to unanticipated deaths in humans primarily attributed to drug-drug interactions and idiosyncratic drug toxicities including hepatotoxicities and cardiotoxicities34. Recent analysis indicated that close to 90 per cent of the drugs that failed during clinical trials, led the pharmaceutical and biotech industry to further explore non-animal technologies for human drug development9. Another point to be considered here is that Indian populations are ethnically different from western populations and data on pharmacogenetic differences in Indian populations are lacking.
In silico models
Advancements in bio-informatics and other in silico computational tools as alternatives to animals have started appearing since the early 1990s35,36. These models have raised some regulatory interest in pharmaceutical and non-pharmaceutical sectors including the US FDA and Environmental Protection Agency, European Medicines Agency and EU ‘REACH’ chemicals legislation and the Organization for Economic Cooperation and Development. For example, the FDA encourages the use of in silico tools as a Critical Path Initiative37. As part of this initiative, they encourage industries to conduct in silico modelling of cardiotoxicity through QT-prolongation, phospholipidosis, hepatotoxicity, ecotoxicology of pharmaceuticals, human-specific drug metabolite prediction and applications in nanotoxicology. Most of the in silico models work on the principles of (Quantitative) structure-activity correlations where several chemicals which are toxic and non-toxic are populated in databases and serve as guiding principles for the synthesis of NCEs. Many modules were developed including genotoxicity, carcinogenicity, developmental toxicity, skin sensitization, general systemic toxicity and absorption, distribution, metabolism and excretion among various others. These tools have been primarily used for screening purposes to eliminate potential structures with positive signals of toxicity and avoid further testing. However, their predictive value is not realized yet due to requirement of more experimental data from in vitro or in vivo animal models before these models could accurately predict human outcomes. There are also several in silico and computational models of pharmacology17, but such models are still not mandated by regulatory agencies, because of poor predictions of in silico data with biological data but are encouraged by industries to utilize these models. These tools primarily serve as research tools for mechanistic understanding and designing potentially active pharmaceuticals.
Non-vertebrate models
Drosophila melanogaster38, Caenorhabditis elegans39 and zebra fish40,41 are touted as species to use as efficacy and toxicity models with human relevance. The possibility of depending on these models is the fact that many signaling pathways are conserved and similar to those in humans. Furthermore, genomic, transcriptomic and proteomic analyses have shown that the extent of homology in drug targets is comparable in these small organisms with that in rodents and other small mammals. However, very little comparative data are available in these models as to whether they are better/equal/worse than rats, dogs and monkeys, which are widely used for both toxicity and efficacy. In several studies these models have been successfully utilized for extensive pre-clinical testing and have led to the discovery of novel drugs42,43. Newer non-vertebrate models do not have such historical databases, and the available data are very scanty; and thus, many companies are not including these models for their pharmacology and toxicology studies. These models are simply learning tools at this stage and relevance to human outcomes is not known. Thus, these models are used in exploratory studies, where necessary, but have not been included as a regulatory mandate. Considering the low cost of maintaining these non-mammalian animal models, initiative can be taken in India for comparative studies that can eventually help acceptance by regulatory agencies.
Current state of the art
A disadvantage of classical in vitro cell-based assays is that these are static models and devoid of the niche, cell-to-cell and organ-to-organ communications which is the reality in in vivo situations.
The two important advancements are (i) Organs (animal/human)-on-a-chip models, which employ multi-channel 3D-microfluidic cell culture chips to simulate the activity, mechanics and physiological responses of entire organs and organ systems, acting as a type of artificial organ (Fig. 2). In this model, a tandem connection to other organs using flow dynamics can be used to create more relevant in vivo systems41; (ii) Disease-in-a-dish models, where patient-derived cells are grown into organoids for prediction of individual drug responses, and successful phase III trial outcomes, and hopefully making personalized medicine a future reality44,45. The ability to grow cell populations in 3D in controlled environments has aided the development of organ cultures that show mechanical and functional properties of organs in the human body. These organoids are useful tools for drug testing, disease research and regenerative medicine, as these replicate human physiology, diseases, and drug responses reasonably more reliably. 3D-culture systems provide cost-effective in vivo information plus a better translational model compared to cost-intensive experimental animal testing. Furthermore, GLP animal testing requires a large quantity (in kg) of drug materials, whereas organ-on-a-chip and disease-in-a-dish models with microfluidics require only a few hundred grams of drug material. Thus, fluid dynamic models incorporating human-derived organoid models are believed to be more closely relevant to human physiology, including chemotherapeutic response of cells, tumour modelling, cellular adaptation, differentiation, biochemical events relevant to cell functional activities, tissue remodelling/engineering and co-culture response46. At present, organoid development has been taken up on priority in many countries to aid drug discovery. In fact, some countries have institutes dedicated for this purpose. There is an urgent need for the Government in India to invest in organoid development specifically for drug development, keeping in mind the wide heterogeneity of Indian subjects.
Fig. 2.
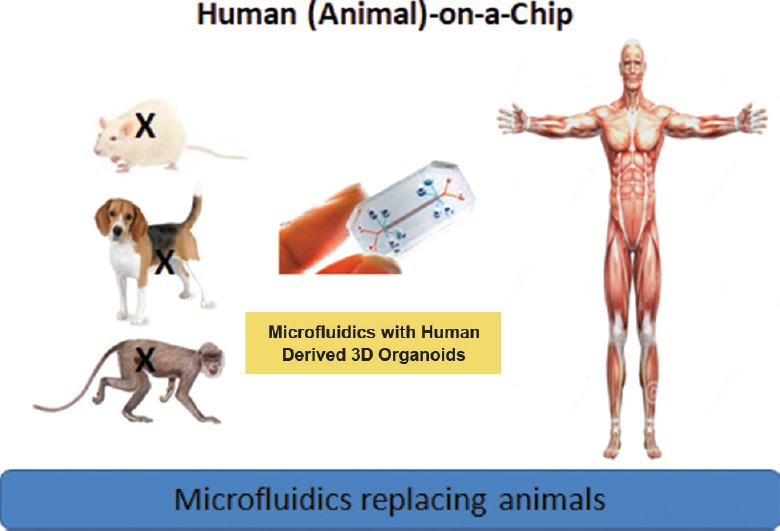
The concept of organ (human/animal)-on-a-chipa,b,c. a=Multi-organ chip using biosensors to mimic human organs and dynamic flow are in development. b=Organoids derived from stem cells are also being considered. c=Patient-derived diseased organs are also being used. Source: www.reagenebiosciences.com. Reproduced with permission.
Organ-on-a-chip and disease-in-a-dish models
In the early 2000s, HμREL, a startup biotech company in collaboration with Johnson and Johnson developed the first prototype of organ-on-a-chip models for drug metabolism studies47. These dynamic, microfluidic models showed several advantages compared to static cell-based assays, including mimicking the activities of human drug metabolizing enzymes and drug transporters. Since then, a dozen companies in the US and Europe ventured to develop such models (Table). For example, Organovo, founded in 2007 with Angel financing of $3 million USD, went public in 2013 with $47 million USD financing48. Together with Massachusetts Institute of Technology (MIT), the organ-chip company CN Bio was awarded $26 million funding from Defense Advanced Research Projects Agency (DARPA) and recently announced the successful engineering of a micophysiological system that allows connection of up to seven tissue-engineered human organs49,50. In addition, a significant regulatory advancement and welcome approach was made by US FDA by inking a collaboration agreement with Emulate Bio (spun out from the Wyss Institute, Boston, MA, USA51,52), an organ-on-a-chip company. This agreement is aimed to evaluate whether chip technology can be used to detect potential chemical hazards found in foods, cosmetics and/or dietary supplements, focusing initially on liver chips, but with an eye to expand and test kidney, lung and intestine models.
Table.
Some organ-on-a-chip startups founded in the last decade
| Company | Year founded | Founded/incubated by | Funding received (USD-millions) | Technology |
|---|---|---|---|---|
| HµREL Corporation | 2005 | J and J, Merck, Humane Society of United States | >10 | Liver |
| Organovo | 2007 | Angel financing | ~3 Subsequently received $15 million in 2012 and went public in 2013 with $47 million | Liver and kidney |
| Hepregen | 2008 | Massachusetts Institute of Technology (MIT), MA, USA | ~5 | Liver |
| InSphero | 2009 | Swiss-Federal Institute of Technology-Zurich, Switzerland | ~2.5 | Liver, Islet, tumour of various species |
| TissUse | 2010 | Technische Universitat, Berlin, Germany | ~2.65 | Kidney |
| NORTIS | 2011 | University Washington, Seattle, Bill and Melinda Gates Foundation, WA, USA | ~2.65 | 10 organs on a chip announced in 2018 |
| MIT* | 2012 | Defense Advanced Research Projects Agency (DARPA), VA, USA | $32 | Body on a chip |
| Emulate Bio | 2013 | Wyss Institute (Harvard University), MA, USA | $12 | Liver |
| AxoSim | 2014 | Tulane University, New Orleans, USA | $1 | Nerve |
| TARA Biosystems | 2015 | Columbia University, New York, USA | $11.3 | Heart |
| ReaGene Biosciences | 2016 | Private organization, Bengaluru, India | - | Liver Heart Skin |
On the efficacy side, as personalized medicine concepts are rapidly advancing, there is the advancement of disease-in-a-dish models53,54 which utilize patient-derived stem cells and diseased cells and are helping to discover and/or develop drugs based on patient pharmacogenomics, drug metabolism and other variables. One down-side, however, is to obtain the patient-derived cells in advance to screen and recommend appropriate medicines to the patient. A collaboration of research companies with medical hospitals should be established. These tools are future models to rapidly advance medicines from bench side to the bedside.
As a cautionary note, if personalized, precision medicine is the future reality, these models need integrations with systems biology (genomics, transcriptomics, proteomics and metabolomics), for successful clinical outcomes. However, -omics in precision is rapidly evolving with combined computational bioinformatics tools where big data analytics are replacing conventional human eye-balling data analysis. The conventional clinical diagnostics tools, with limited measures of metabolites such as plasma creatinine, cholesterol and glucose are being expanded to several hundred by utilizing -omics tools54.
Big data analytics of -omics-machine learning, artificial intelligence and their applications for predictive toxicology and pharmacology
The explosion of -omics and other systems biology research now generates several thousands of terabytes of data/day, all around the world55. The evaluation of such big data is humanly impossible to review and make informed decisions for future healthcare applications. Much of these data are being stored in cloud and are retrieved. Adverse outcome pathways (AOPs) are also used as a framework for collecting, organizing and evaluating the existing knowledge obtained from high throughput, -omics, guideline studies, clinical, epidemiology and eco-field studies, linking a molecular-level perturbation of a biological system to an adverse outcome. These are designed to offer a distinct mechanistic representation of the critical adverse (toxicological/disease) effects that traverse various layers of the biological organization. An AOP describes the progression of relationships of biological perturbations from the lower to higher levels of biological organization, concluding an adverse outcome which has regulatory relevance. This benefits the process of safety assessment without the use of non-human animals if the information is used and interpreted pragmatically56.
Way forward
The educational institutes lack a comprehensive DD&D curriculum and research programme that can help students that graduate to be ready with appropriate skills to conduct independent and innovative research. It is important that the Government encourages the creation of ‘Centers of Excellence (COE)’ where ‘Alternatives to Animals’ research in India can be compared and compete with the elite COE in western countries. Greater emphasis on human relevance will bring about a true paradigm shift. This will include top-down funding decisions, data generation, building databases and/or knowledge management tools. International and interagency collaborations should be established between major organizations and funding bodies. Funding for research focusing on human-based biology, rather than ‘improved’ animal models, should be prioritized. The data should be collected in collaborative, open access and high-quality databases. There is also an immediate need for creating case studies to indicate the applications and benefits of these predictive and mechanism-based approaches with respect to translation and biology of human disease and to identify new therapeutics57. Eventually, these COEs should enable the regulatory agencies to make necessary policy changes that will minimize animal testing in all sectors where animals are used.
Although India made some advances in regenerative medicine technology (primarily by academic and government institutions) using stem cells derived from induced pluripotent cells and mesenchymal stem cells, such research has not been integrated or applied for DD&D research of private sector58,59. If India is to emerge as the innovation centre for DD&D research, the current state of the art should be embraced by both public and private sectors. Government, academia, and industry collaborations need to be encouraged. In addition, regulatory bodies should be in closed loop with both public and private sectors for any policy changes that may be required in the future in support of non-animal technologies. Government funding agencies should fund private sectors with these kinds of innovations in alternatives to animal technology startups to give the necessary boost for many of the concepts to grow to a prototype stage. India should become self-reliant in such technologies that support research activities, rather than depending on costly imported technologies.
ICMR Expert Committee: Drs V.P. Kamboj, Central Drug Research Institute, Lucknow; A. Ray, Hamdard Institute of Medical Sciences & Research, New Delhi; Alokparna Sengupta, Humane Society International, Hyderabad; V. Radha, Centre for Cellular & Molecular Biology, Hyderabad; Mani Subrahmanyam Vangala, ReaGene Biosciences Pvt. Ltd., Bengaluru; O.P. Agarwal, Council of Scientific & Industrial Research, New Delhi.
Acknowledgment
The contributions of the following scientists are acknowledged: Drs Manoj Kumar Bhat, National Centre for Cell Science, Pune; A.B. Pant, Industrial Toxicology Research Centre, Lucknow; S. Majumdar, Postgraduate Institute of Medical Education & Research, Chandigarh; Deepa Bhartiya and Sumita D. Madhale, National Institute for Research in Reproductive Health (NIRRH), Mumbai; Renu Tripathi, Central Drug Research Institute, Lucknow; Dinesh Kumar Badyal, Christian Medical College, Ludhiana; Shri Raghuram Rao, National Institute of Pharmaceutical Education and Research, Mohali; Pankaj Seth, National Brain Research Centre, Gurugram; Suresh Kumar, National Institute of Biologicals, Noida; R.S. Sharma, Monika Pahuja, and Heena Tabazsum, Indian Council of Medical Research (ICMR), Headquarters, New Delhi; Ms Michelle Thew and Shri Jarrod Bailey, Cruelty Free International, UK.
Footnotes
Conflicts of Interest: None.
References
- 1.Russell WMS, Burch RL. The principles of humane experimental technique. Wheathampstead (UK): Universities Federation for Animal Welfare; 1959. [Google Scholar]
- 2.Pereira S, Tettamanti M. Ahimsa and alternatives – The concept of the 4th R. The CPCSEA in India. ALTEX. 2005;22:3–6. [PubMed] [Google Scholar]
- 3.Ferdowsian HR, Beck N. Ethical and scientific considerations regarding animal testing and research. PLoS One. 2011;6:e24059. doi: 10.1371/journal.pone.0024059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.National Research Council of the National Academies. Toxicity testing in the 21st century: A vision and a strategy. 2007. [accessed on April 3, 2019]. Available from: https://www.nap.edu/read/11970/chapter/3 .
- 5.Ministry of Health and Family Welfare. Guidelines for Implementation of GSR 346 (E), F. No. X-11014/7/2013- DFQC, Issued by Ministry of Health and Family Welfare. New Delhi: Department of Health and Family Welfare; 2014. May 21, [accessed on April 3, 2019]. Available from: http://ficci.in/sector/73/add_Docs/AniTestGuidelinesFICCIFINAL16092014.pdf . [Google Scholar]
- 6.India bans cruel eye and skin test on rabbits for drug testing. 2016. Nov 7, [accessed on April 3, 2019]. Available from: https://www.hsi.org/news-media/draize-test-banned-india-110716/
- 7.Bureau of Indian Standards. Methods of test for safety evaluation of cosmetics. [accessed on April 3, 2019]. Available from: http://bwcindia.org/Web/Info&Action/Legislation/TestingofCosmetics.pdf .
- 8.Langley G, Austin CP, Balapure AK, Birnbaum LS, Bucher JR, Fentem J, et al. Lessons from toxicology: Developing a 21st-century paradigm for medical research. Environ Health Perspect. 2015;123:A268–72. doi: 10.1289/ehp.1510345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plenge RM, Scolnick EM, Altshuler D. Validating therapeutic targets through human genetics. Nat Rev Drug Discov. 2013;12:581–94. doi: 10.1038/nrd4051. [DOI] [PubMed] [Google Scholar]
- 10.Differding E. The drug discovery and development industry in India-two decades of proprietary small-molecule R&D. ChemMedChem. 2017;12:786–818. doi: 10.1002/cmdc.201700043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta R. Clinical success versus attrition of investigational pharmaceuticals: A vignette. Crit Rev Ther Drug Carrier Syst. 2017;34:527–49. doi: 10.1615/CritRevTherDrugCarrierSyst.2017018747. [DOI] [PubMed] [Google Scholar]
- 12.Subrahmanyam V, Pinjari J, Patole P, Ravindran S, Gangal R, Wangikar P, et al. Translational drug discovery research: Integration of medicinal chemistry, computational modeling, pharmacology, ADME, and toxicology. In: Lyubinov AV, editor. Encyclopedia of drug metabolism and interactions. Hoboken, NJ: Wiley Inc., JWS; 2012. [Google Scholar]
- 13.Subrahmanyam V, Kolachana P. Endogenous toxins as disease initiating events: Future targets of drug discovery research. J Anal Pharm Res. 2016;2:00036. [Google Scholar]
- 14.Thomas DW, Burns J, Audette J, Carroll A, Hygelund CD, Hay M. Clinical development success rates; 2006-2015. BIO Industry Analysis. Biotechnology Innovation Organization (BIO), Biomedtracker, Amplion Inco. 2016. [accessed on April 4, 2019]. pp. 1–16. Available from: https://www.bio.org/sites/default/files/Clinical%20Development%20Success%20Rates%202006-2015%20-%20BIO,%20Biomedtracker,%20Amplion%202016.pdf .
- 15.Herper M. The truly staggering costs of inventing new drugs. Forbes. 2012;189:38–9. [Google Scholar]
- 16.Dickson Mgagnon JP. The cost of new drug discovery and development. Discov Med. 2004;4:172–9. [PubMed] [Google Scholar]
- 17.Prasad V, Mailankody S. Research and development spending to bring a single cancer drug to market and revenues after approval. JAMA Intern Med. 2017;177:1569–75. doi: 10.1001/jamainternmed.2017.3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Preclinical safety evaluation of biotechnology derived pharmaceuticals - S6(R1) - ICH Harmonised Tripartite Guideline. 1997. Jul, [accessed on April 4, 2019]. Available from: http://www.ich.org/fileadmin/Public_Web_Site/ICH_Products/Guidelines/Safety/S6_R1/Step4/S6_R1_Guideline.pdf .
- 19.Segler MHS, Kogej T, Tyrchan C, Waller MP. Generating focused molecule libraries for drug discovery with recurrent neural networks. ACS Cent Sci. 2018;4:120–31. doi: 10.1021/acscentsci.7b00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X, Betzi S, Morelli X, Roche P. Focused chemical libraries – Design and enrichment: An example of protein-protein interaction chemical space. Future Med Chem. 2014;6:1291–307. doi: 10.4155/fmc.14.57. [DOI] [PubMed] [Google Scholar]
- 21.Benson N. Network-based discovery through mechanistic systems biology. Implications for applications – SMEs and drug discovery: Where the action is. Drug Discov Today Technol. 2015;15:41–8. doi: 10.1016/j.ddtec.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 22.Interagency coordinating committee on the validation of Alternative Methods. A strategic roadmap for establishing new approaches to evaluate the safety of chemicals and medical products in the United States. 2018. Jan, [accessed on April 4, 2019]. Available from: https://www.ntp.niehs.nih.gov/iccvam/docs/roadmap/iccvam_strategicroadmap_january2018_document_508.pdf .
- 23.A non-animal technologies roadmap for the UK: Advancing predictive biology. Technology Strategy Board November 2015 C15/CO082 Innovate UK. 2015. [accessed on April 4, 2019]. Available from: https://www.nc3rs.org.uk/sites/default/files/documents/NonAnimal TechCO082_RYE_4_nrfinal2.pdf .
- 24.Gordon S, Daneshian M, Bouwstra J, Caloni F, Constant S, Davies DE, et al. Non-animal models of epithelial barriers (skin, intestine and lung) in research, industrial applications and regulatory toxicology. ALTEX. 2015;32:327–78. doi: 10.14573/altex.1510051. [DOI] [PubMed] [Google Scholar]
- 25.Humane Society International-India welcomes govt's acceptance of non-animal tests for pesticide safety. 2018. Jan 12, [accessed on April 3, 2019]. Available from: https://www.downtoearth.org.in/news/governance/humane-society-international-welcomesindian-govt-s-acceptance-of-non-animal-skin-and-eyetests-for-pesticide-safety-59492 .
- 26.Estabrook RW. A passion for P450s (rememberances of the early history of research on cytochrome P450) Drug Metab Dispos. 2003;31:1461–73. doi: 10.1124/dmd.31.12.1461. [DOI] [PubMed] [Google Scholar]
- 27.Williams JA, Andersson T, Andersson TB, Blanchard R, Behm MO, Cohen N, et al. PhRMA white paper on ADME pharmacogenomics. J Clin Pharmacol. 2008;48:849–89. doi: 10.1177/0091270008319329. [DOI] [PubMed] [Google Scholar]
- 28.Mao Q, Lai Y, Wang J. Drug transporters in xenobiotic disposition and pharmacokinetic prediction. Drug Metab Dispos. 2018;46:561–6. doi: 10.1124/dmd.118.081356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.U.S. Department of Health and Human Services, Food and Drug Administration, Center for Drug Evaluation and Research (CDER) In vitro metabolism and transportermediated drug-drug interaction studies guidance for industry DRAFT GUIDANCE. 2017. Oct, [accessed on April 3, 2019]. Available from: https://www.fda.gov/media/108130/download .
- 30.U.S. Department of Health and Human Services, Food and Drug Administration, Center for Drug Evaluation and Research (CDER) Clinical Drug Interaction Studies – Study design, data analysis, and clinical implications guidance for industry: DRAFT GUIDANCE. 2017. Oct, [accessed on April 3, 2019]. Available from: https://www.fda.gov/media/82734/download .
- 31.U.S. Department of Health and Human Services, Food and Drug Administration, Center for Drug Evaluation and Research (CDER) Safety testing of drug metabolites: Guidance for industry. *Revision 1. 2016. Nov, [accessed on April 3, 2019]. Available from: https://www.fda.gov/media/72279/download .
- 32.The non-clinical evaluation of the potential for delayed ventricular repolarization (QT interval prolongation) by human pharmaceuticals – S7B. ICH Harmonised Tripartite Guideline. 2005. May 12, [accessed April 4, 2019]. Available from: http://www.ich.org/fileadmin/Public_Web_Site/ICH_Products/Guidelines/Safety/S7B/Step4/S7B_Guideline.pdf .
- 33.Chang WC. Pharmacogenomics in personalized medicine and drug metabolism. Biomed Res Int. 2014;2014:897963. doi: 10.1155/2014/897963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fermini B, Coyne ST, Coyne KP. Clinical trials in a dish: A perspective on the coming revolution in drug development. SLAS Discov. 2018;23:765–76. doi: 10.1177/2472555218775028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Danchin A, Médigue C, Gascuel O, Soldano H, Hénaut A. From data banks to data bases. Res Microbiol. 1991;142:913–6. doi: 10.1016/0923-2508(91)90073-j. [DOI] [PubMed] [Google Scholar]
- 36.Sieburg HB. Physiological studies in silico. Stud Sci Complex. 1990;12:321–42. [Google Scholar]
- 37.Valerio LG., Jr In silico toxicology models and databases as FDA critical path initiative toolkits. Hum Genomics. 2011;5:200–7. doi: 10.1186/1479-7364-5-3-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marcogliese PC, Wrangler MF. Encyclopedia of Life Sciences. Chichester: John Wiley & Sons Ltd; 2018. [accessed on April 3, 2019]. Drosophila as a model for human diseases. Available from: https://www.onlinelibrary.wiley.com/doi/pdf/10.1002/9780470015902.a0005578.pub2 . [Google Scholar]
- 39.Markaki M, Tavernarakis N. Modeling human diseases in Caenorhabditis elegans. Biotechnol J. 2010;5:1261–76. doi: 10.1002/biot.201000183. [DOI] [PubMed] [Google Scholar]
- 40.Ma X, Ding Y, Wang Y, Xu X. A doxorubicin-induced cardiomyopathy model in adult zebrafish. J Vis Exp. 2018 doi: 10.3791/57567. doi:10.3791/57567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Souza Anselmo C, Sardela VF, de Sousa VP, Pereira HMG. Zebrafish (Danio rerio): A valuable tool for predicting the metabolism of xenobiotics in humans? Comp Biochem Physiol C Toxicol Pharmacol. 2018;212:34–46. doi: 10.1016/j.cbpc.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 42.Marshall LJ, Austin CP, Casey W, Fitzpatrick SC, Willett C. Recommendations toward a human pathway-based approach to disease research. Drug Discov Today. 2018;23:1824–32. doi: 10.1016/j.drudis.2018.05.038. [DOI] [PubMed] [Google Scholar]
- 43.Fang M, Guo J, Chen D, Li A, Hinton DE, Dong W. Halogenated carbazoles induce cardiotoxicity in developing zebrafish embryos (Danio rerio) Environ Toxicol Chem. 2016;35:2523–9. doi: 10.1002/etc.3416. [DOI] [PubMed] [Google Scholar]
- 44.Orwant R. Dawn of the zombies. New Sci. 2006;255:40–3. [Google Scholar]
- 45.Hall SS. Diseases in a dish. Sci Am. 2011;304:40–5. doi: 10.1038/scientificamerican0311-40. [DOI] [PubMed] [Google Scholar]
- 46.Holloway EM, Capeling MM, Spence JR. Biologically inspired approaches to enhance human organoid complexity. Development. 2019;146 doi: 10.1242/dev.166173. pii: dev166173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tiscornia G, Vivas EL, Izpisúa Belmonte JC. Diseases in a dish: Modeling human genetic disorders using induced pluripotent cells. Nat Med. 2011;17:1570–6. doi: 10.1038/nm.2504. [DOI] [PubMed] [Google Scholar]
- 48.David Zhu. Organovo. [accessed on April 4, 2019]. Available from: https://www.cs.cmu.edu/afs/cs/academic/class/15294-f15/assignments/company/writeups/davidz.pdf .
- 49.Valerio LG., Jr In silico toxicology models and databases as FDA critical path initiative toolkits. Hum Genomics. 2011;5:200–7. doi: 10.1186/1479-7364-5-3-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thilmany J. 7 human organs one one chip. 2016. Dec, [accessed on April 4, 2019]. Available from: http://www.asme.org/engineering-topics/articles/bioengineering/7-human-organs-one-chip .
- 51.Fitzpatrick S. FDA Voice. [accessed on April 25, 2017]. Available from: https://blogs.fda.gov/fdavoice/index.php/2017/04/organs-on-chips-technology-fdatesting-groundbreaking-science/
- 52.Wyss Institute. Human Organs on Chips. [accessed on April 4, 2019]. Available from: https://wyss.harvard.edu/technology/human-organs-on-chips/
- 53.Vangala S, Tonelli A. Biomarkers, metabonomics, and drug development: Can inborn errors of metabolism help in understanding drug toxicity? AAPS J. 2007;9:E284–97. doi: 10.1208/aapsj0903031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Low SK, Zembutsu H, Nakamura Y. Breast cancer: The translation of big genomic data to cancer precision medicine. Cancer Sci. 2018;109:497–506. doi: 10.1111/cas.13463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Organisation for Economic Co-operation and Development. OCED environment, health and safety publications. Revised guidance document on developing and assessing adverse outcome pathways, series on testing and assessment, No. 184 (ENV/JM/MONO(2013)6) [accessed on April 3, 2019]. Available from: http://www.oecd.org/officialdocuments/publicdisplaydocumentpdf/?cote=env/jm/mono%282013%296&doclanguage=en .
- 56.Langley G, Austin CP, Balapure AK, Birnbaum LS, Bucher JR, Fentem J, et al. Lessons from toxicology: Developing a 21st-century paradigm for medical research. Environ Health Perspect. 2015;123:A268–72. doi: 10.1289/ehp.1510345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roda A, Michelini E, Caliceti C, Guardigli M, Mirasoli M, Simoni P. Advanced bioanalytics for precision medicine. Anal Bioanal Chem. 2018;410:669–77. doi: 10.1007/s00216-017-0660-8. [DOI] [PubMed] [Google Scholar]
- 58.Lin E, Lane HY. Machine learning and systems genomics approaches for multi-omics data. Biomark Res. 2017;5:2. doi: 10.1186/s40364-017-0082-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Top 22 artificial intelligence companies in India that are changing the world of technology. [accessed on April 3, 2019]. Available from: https://www.sumhr.com/top-artificial-intelligence-companies-india/