Abstract
This study proposes and validates an automated method for counting neurons in spinal cord injury (SCI) and then uses it to examine and compare the surviving cells in common types of SCI mechanisms. Moderate contusion, dislocation, and distraction SCIs were surgically induced in Sprague Dawley male rats (n = 6 for each type of injury). Their spinal cords were harvested 8 weeks post injury with 5 normal weight-matched rats. The spinal cords were cut, stained with anti-NeuN antibody and fluorescent Nissl, and imaged in the dorsal and ventral horns at various distances to the epicenter. Neurons in the images were automatically counted using an algorithm that was designed to filter non-soma-like objects based on morphological characteristics (size, solidity, circular pattern) and check the remaining objects for the double-stained nucleus/cell body features (brightness variation, brightness distribution, color). To validate the automated method, some of the images were randomly selected for manual counting. The number of surviving cells that were automatically measured by the algorithm was found to be correlated with the values that were manually measured by 2 observers (P < .001) with similar differences (P > .05). Neurons in the dorsal and ventral horns were reduced after the SCIs (P < .05). Dislocation and distraction, respectively, caused the most severe damage to the ventral horn neurons especially near the epicenter and the most extensive and uniform damage to the dorsal horn neurons (P < .05). Our method was proved to be reliable, which is suitable for studying different types of SCI.
Keywords: Cell count, automatic, NeuN, Nissl, histology, rat
Introduction
Neurons play an essential role in the function and structure of the spinal cord, and cell counting is very common in studies of spinal cord injury (SCI). When the spinal cord is initially injured, most often due to physical trauma, the spinal neurons can be acutely damaged, which is followed by a cascade of secondary injury events that can potentially lead to further neuronal damage.1 The resulting damage depends on the level, type, severity, and time of injury and possibly other factors.2-4 Measurement of the surviving cells is usually performed with histology to analyze the injury and recovery mechanisms.5 However, manual cell counting is generally time-consuming and error-prone. A reliable method to count cells automatically would greatly benefit the field.
Numerous automated methods have been developed for counting neurons in various research areas (eg, brain, retina, dorsal root ganglion, cell culture, and graft).6-24 Many automated methods have also been proposed for counting neurons in the spinal cord and other types of cells after SCI (eg, macrophages, microglia, lymphocytes, and proliferating cells).4,25-44 However, as far as we know, these methods of counting spinal neurons are only semi-automatic (eg, manual brightness threshold settings) and/or have not been validated. Furthermore, it is not known whether they are capable of detecting the differences among different types of injury as it has not been done, which would be important to analyze the SCI mechanisms.
This study has 2 aims. The first is to propose and validate an automated method for counting neurons in SCI. The second is to use the method to examine and compare the surviving cells in the common types of SCI mechanisms.
Materials and Methods
Animals
The study was approved by the UBC Committee on Animal Care in accordance with the Guide to the Care and Use of Experimental Animals by the Canadian Council on Animal Care, and the raw data were generously provided by ICORD. The study was also approved by the Ethics Committee of the School of Biological Science and Medical Engineering of the BUAA. A total of 23 Sprague Dawley male rats were acquired for the experiment; 18 rats in 3 groups, respectively, received 3 types of SCI (n = 6 for contusion, dislocation, and distraction; 280 g before surgery and 340 g before euthanasia). One group of 5 rats was uninjured (n = 5 for control; 340 g before euthanasia). They were provided with food and water ad libitum.
Surgery
Standard surgical procedure for inducing moderate bilateral cervical contusion, dislocation, and distraction SCIs was performed in the 3 injury groups, with the injury thresholds that were previously determined from the resulting behavioral deficits.2 A dorsal midline incision was made from approximately C2 to C7. For contusion, the spinal cord between C5 and C6 was exposed by laminectomy. Custom clamps held the transverse processes at C4 to C7 on a stereotaxic surgical frame. A linear actuator applied a small preload (0.03 N) to the surface of the dura mater with a spherical head impactor between C5 and C6, retracted 6 mm above the dura mater, and accelerated into the spinal cord to 1.6 mm with a peak velocity of 1.2 m/s. For dislocation and distraction, the posterior ligaments between C5 and C6 were transected, and a C5/C6 facetectomy was performed. A pair of custom clamps, respectively, held the transverse processes at C4 to C5 on the stereotaxic surgical frame and the transverse processes at C6 to C7 on the linear actuator. The linear actuator applied a small preload (2 N) to the spinal column and acutely translated C6 to C7 relative to C4 to C5, either dorsally in dislocation to 1.8 mm with a peak velocity of 0.9 m/s or caudally in distraction to 5.6 mm with a peak velocity of 1.3 m/s. A custom implant was used to hold the transverse processes at C5 to C6 together after dislocation and distraction.
Histology
The injury groups were euthanized 8 weeks post injury together with the noninjury group. They were perfused with fixative (4% paraformaldehyde in phosphate buffered saline, pH 7.4). The spinal cords were harvested, post-fixed overnight in fixative, washed in several changes of sucrose (12%, 18%, and 24%) every 24 hours, frozen in optimal cutting temperature compound, and cut in cross section at 20 µm thickness.
The cut sections were blocked with normal donkey serum (1:10; Jackson ImmunoResearch Laboratories, West Grove, PA) and stained with primary antibodies against NeuN (mouse host, 1:200; Millipore, Billerica, MA) in combination with secondary antibodies conjugated with DyLight 594 (donkey host, mouse antigen, 1:200; Jackson ImmunoResearch Laboratories) and green fluorescent Nissl (FluoroNissl; 1:200; Molecular Probes, Eugene, OR). All the sections were stained at the same time and under the same conditions using a standard staining procedure.2
The NeuN/FluoroNissl-stained sections were examined under a fluorescence microscope (AxioObserver Z1 inverted confocal; Carl Zeiss, Jena, Germany). Epicenter was identified based on the lesion size, and images at the epicenter and 0.2, 0.6, 1, 1.6, 2.2, and 3 mm rostral and caudal to the epicenter were taken. The dorsal and ventral horns were photographed (dorsal: Plan-Apochromat 20×, NA 0.8, 1040 × 1040 pixels, 335.40 µm × 335.40 µm; ventral: Plan-Apochromat 10×, NA 0.45, 1040 × 1040 pixels, 670.80 µm × 670.80 µm) by aligning the lateral and dorsal/ventral edges of the horns with the borders of the images. Brightness and contrast were kept constant for all the images.
Automated Counting
Neurons in the dorsal and ventral horns were counted automatically using a custom MATLAB (MathWorks, Natick, MA) script (Figure 1). The script was designed to find neurons based on the double-stained features of the nucleus and the rest of the cell body (Figure 2). The number of surviving cells in the dorsal horn, the number of surviving cells in the ventral horn, and the number of surviving large cells (soma area >591.5 µm2 or 1400 pixels) in the ventral horn were measured.
Figure 1.
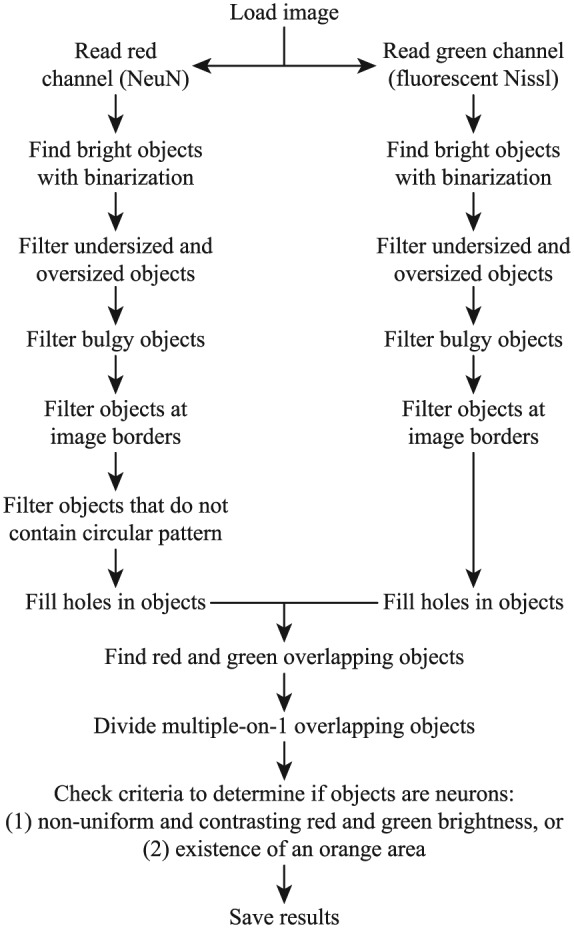
Flowchart of the automated algorithm for counting neurons. The red (for NeuN) and green (for FluoroNissl) channels of the input image were separated into 2 images that were processed to find soma-like objects. The red and green images were double-binarized using Otsu method and an adaptive method (based on local first-order image statistics) to identify bright objects from the global and local dark background automatically. Objects that were too small and too large, had low solidity (area ÷ convex area <0.55), and touched the image borders were neglected. In addition, the red image was processed using a Circular Hough Transform–based algorithm to neglect objects containing no circular pattern (for the nucleus). Holes in the objects were filled. When a red soma-like object and a green soma-like object clearly overlapped with each other, it was considered as a candidate of neuron. When multiple red soma-like objects clearly overlapped with a single green soma-like object (it sometimes happened when neurons were close to each other), the green soma-like object was artificially divided in the midline between the red soma-like objects, and they were considered as multiple candidates of neuron. Neuronal candidates that met 1 of the 2 criteria were confirmed to be neurons: (1) red and green brightness was nonuniform (brightness coefficient of variation >0.25) and in contrast to each other (ratio of pixels in bright red/medium green and medium red/bright green >0.25), and (2) an orange area existed (number of pixels in bright red and medium green >16). These criteria were designed to capture the features of the stained neurons that were not observed in other undesirably stained objects. Although both stains can highlight the soma, NeuN-stained soma generally appears to be darker in the nucleolus, brighter in the rest of the nucleus, and darker in the rest of the body, and the opposite is generally true in FluoroNissl-stained soma. The first criterion was more useful to detect larger neurons, which had more explicit brightness variation and contrast. The second criterion was more useful to detect smaller neurons, which exhibited more consistent nucleus in orange (small neurons generally have less FluoroNissl stain).
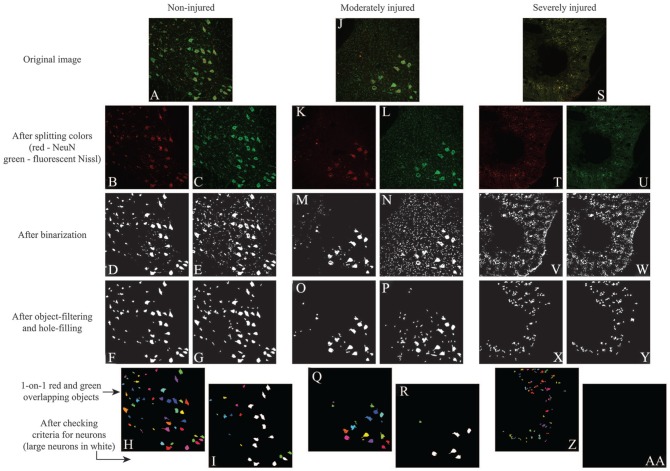
Figure 2.
Demonstration of the automated algorithm for counting neurons. In the (A) normal spinal cords, NeuN staining highlighted the (B) somas and (D) some non-soma-like objects, while FluoroNissl staining highlighted the (C) somas and (E) many non-soma-like objects. (F-H) Simple object-filtering was able to remove most of the non-soma-like objects in both stains. (I) The neuronal criteria helped to detect the objects that exhibited a clear nucleus and soma. In (J) moderately injured cord sites, both staining highlighted (K-N) the somas but also more non-soma-like objects comparing to normal. (O and P) Simple object filtering was able to remove most of the non-soma-like objects in both stains, but there were still a considerable number of them left. (Q) Checking double-stained objects helped to remove most of the remaining non-soma-like objects. (R) The neuronal criteria helped to remove the objects that did not exhibit a clear nucleus and soma. In (S) severely injured cord sites, both staining highlighted (T-W) many non-soma-like objects. Simple object-filtering was able to remove most of the non-soma-like objects in both stains, but there were still (X and Y) many non-soma-like objects left, and most of them were (Z) double-stained objects. The neuronal criteria helped to remove the remaining non-soma-like objects (aa).
Manual Counting
One rat and 1 side of the spinal cords (left or right) were randomly selected from each group (4 groups: normal, contusion, dislocation, and distraction) for manual counting. Neurons in the dorsal and ventral horns were counted by 2 independent experts in histology who were blinded to the study design. Only neurons with a clear cell body and nucleus were considered. Neurons at the image borders were neglected. The number of distinctively large neurons in the ventral horn was also recorded.
Statistical Analysis
The correlations between automatic neuron counts by the algorithm and manual neuron counts by the 2 observers were assessed using Pearson correlation coefficient (r; 2-tailed, P < .001). Their differences were analyzed by 1-way analysis of variance (ANOVA) followed by Tukey test (P < .05). The number of surviving cells between normal and contusion, dislocation, and distraction injury mechanisms that were measured by the algorithm were compared using the Kruskal-Wallis test followed by the Mann-Whitney U test (2-tailed, P < .05). Data from the left and right sides were averaged for the comparison. Analysis was performed using SPSS (IBM, Chicago, IL).
Results
Automated Versus Manual Counting
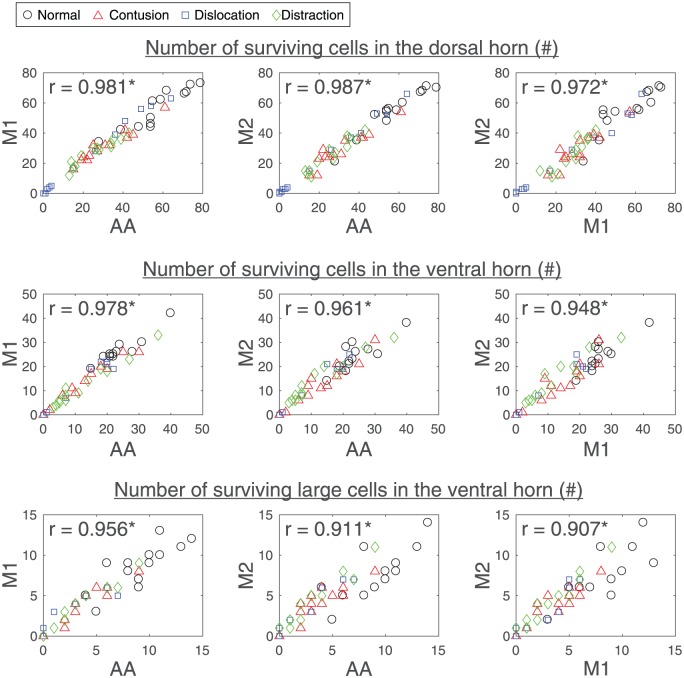
The number of surviving cells in the dorsal horn, the number of surviving cells in the ventral horn, and the number of surviving large cells in the ventral horn that were automatically measured by the algorithm were highly correlated with the values that were manually measured by the 2 observers, and the values between the 2 observers were also highly correlated (Figure 3). Their differences were found to be similar (Table 1). Interestingly, 1 observer tended to have less strict cell counting criteria than the algorithm (M1 in Figure 3), and the other observer tended to have stricter cell counting criteria than the algorithm (M2 in Figure 3), making the differences between the 2 observers greater than their differences to the algorithm, although none of these differences were significant (Table 1).
Figure 3.
Scatterplots showing the relation between automatic neuron counts by the algorithm (AA) and manual neuron counts by the 2 observers (M1 and M2) that were evaluated by Pearson correlation coefficient (r). Images of the NeuN/FluoroNissl-stained sections at various locations with respect to the epicenter of 1 side of a spinal cord for each of the different SCI mechanisms and the control were randomly selected for the validation analysis (1 rat × 1 cord side × 13 spinal cord locations × 4 study groups = 52 data points in each scatterplot). SCI indicates spinal cord injury.
*Statistically correlated with each other (P < .001).
Table 1.
Absolute differences between automatic neuron counts by the algorithm (AA) and manual neuron counts by the 2 observers (M1 and M2).
| Absolute differences | AA-M1 | AA-M2 | M1-M2 |
|---|---|---|---|
| Number of surviving cells in the dorsal horn | 3.3 ± 2.4 | 2.9 ± 2.2 | 3.8 ± 3.2 |
| Number of surviving cells in the ventral horn | 1.8 ± 1.5 | 2.1 ± 2.0 | 2.7 ± 2.1 |
| Number of surviving large cells in the ventral horn | 0.8 ± 0.8 | 1.2 ± 1.1 | 1.0 ± 1.1 |
The values were calculated using the data points in Figure 3 and are presented as mean ± standard deviation. No significant differences were found among the absolute differences of AA, M1, and M2 in the number of surviving cells in the dorsal horn, the number of surviving cells in the ventral horn, and the number of surviving large cells in the ventral horn (P > .05).
Dorsal Horn Neurons After SCI
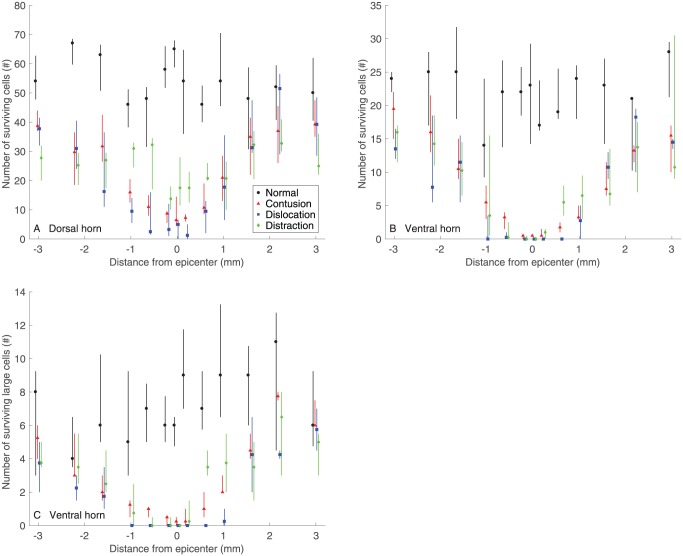
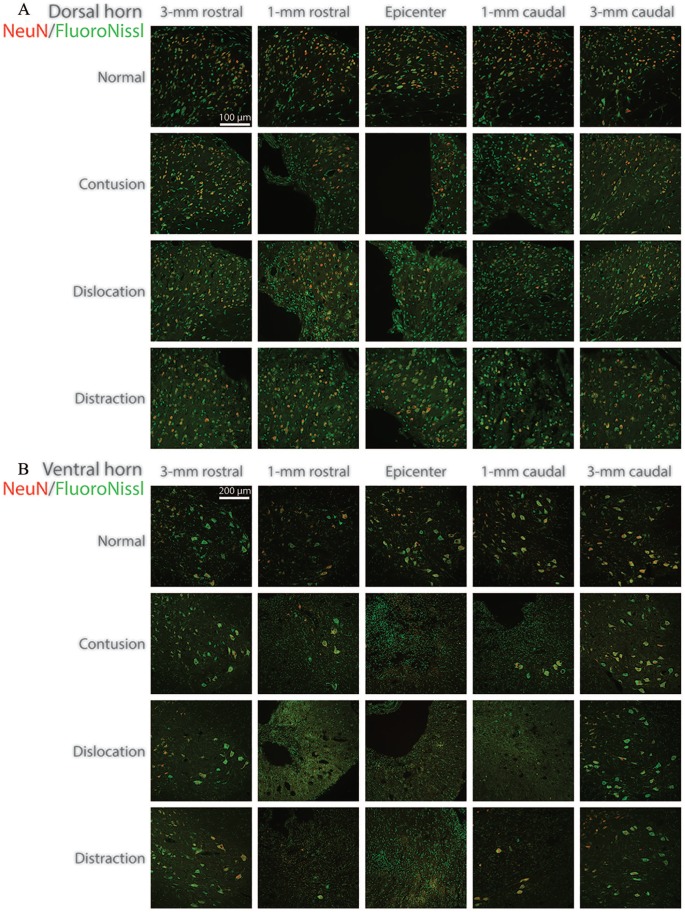
The number of surviving cells in the dorsal horn after contusion, dislocation, and distraction SCIs were significantly different from normal (Figure 4A). Neuronal reduction was found between 2.2 mm rostral and 1 mm caudal to the epicenter in contusion and dislocation, while it was found between 3 mm rostral and 3 mm caudal to the epicenter in distraction, which was more extensive longitudinally. Lesions (including the cracks in distraction) visibly reduced the amount of tissue in the dorsal horn near the epicenter (eg, at 1 mm), which presumably contributed to the neuronal reduction (Figure 5A). The extensive damage to the dorsal horn neurons in distraction was reflected in the visibly low density of surviving cells far away from the epicenter (eg, at 3 mm) when comparing to normal.
Figure 4.
(A) Number of surviving cells in the dorsal horn, (B) number of surviving cells in the ventral horn (B), and (C) number of surviving large cells in the ventral horn for the different SCI mechanisms (n = 6) and the control (n = 5). Data are presented as median with quartiles and offset horizontally for clarity. The position along the spinal cord extended from −3 mm (rostral) through +3 mm (caudal) to the epicenter (0 mm). Statistical differences (P < .05) in (A) are as follows: (1) differences from Normal: contusion −2.2, –1.6, –1, –0.6, –0.2, 0, +0.2, +0.6, +1 mm; distraction −3.2, –1.6, –1, –0.6, –0.2, 0, 0.2, 0.6, 1 mm; distraction −3, –2.2, –1.6, –1, –0.2, 0, +0.2, +0.6, +1, +2.2, +3 mm; (2) differences between contusion and dislocation: none; (3) differences between contusion and distraction: –3, –0.6, 0, +0.2, +0.6 mm; (4) differences between dislocation and distraction: –1, –0.6, 0, +0.2, +0.6, +3 mm. Statistical differences (P < .05) in (B) are as follows: (1) differences from Normal: contusion −1.6, –1, –0.6, –0.2, 0, +0.2, +0.6, +1, +1.6, +3 mm; distraction −3, –2.2, –1.6, –1, –0.6, –0.2, 0, +0.2, +0.6, +1, +1.6, +3 mm; distraction −3, –1.6, –0.6, –0.2, 0, +0.2, +0.6, +1, +1.6 mm; (2) differences between contusion and dislocation: –0.2, 0, +0.2 mm; (3) differences between contusion and distraction: 0, +0.6 mm; (4) differences between dislocation and distraction: +0.2, +0.6 mm. Statistical differences (P < .05) in (C) are as follows: (1) differences from normal: contusion −1.6, –1, –0.6, –0.2, 0, +0.2, +0.6, +1 mm; distraction −3.2, –1.6, –1, –0.6, –0.2, 0, +0.2, +0.6, +1, +1.6 mm; contusion −1.6, –1, –0.6, –0.2, 0, +0.2, +0.6, +1.6 mm; (2) differences between contusion and dislocation: –1, –0.6, –0.2, 0, +0.2, +0.6, +1, +2.2 mm; (3) differences between contusion and distraction: +0.6 mm; and (4) differences between dislocation and distraction: +0.2, +0.6, and +1 mm. SCI indicates spinal cord injury.
Figure 5.
Representative images of the NeuN/FluoroNissl-stained sections in the (A) dorsal horn and the (B) ventral horn for the different SCI mechanisms and the control. SCI indicates spinal cord injury.
The number of surviving cells in the dorsal horn was significantly different among the different types of SCI (Figure 4A). Particularly, although the values were similar between contusion and dislocation, they were different from distraction. In comparison with distraction, damage to the dorsal horn neurons was more severe near the epicenter in contusion (between 0.6 mm rostral and 0.6 mm caudal) and dislocation (between 1 mm rostral and 0.6 mm caudal), but it was less severe far away from the epicenter in contusion (at 3 mm rostral) and dislocation (at 3 mm caudal). Neuronal reduction was visibly more uniform rostrocaudally in distraction than in contusion and dislocation (Figure 5A).
Ventral Horn Neurons After SCI
The number of surviving cells in the ventral horn after contusion, dislocation, and distraction SCIs was significantly different from normal (Figure 4B). Neuronal reduction was found between 1.6 mm rostral and 3 mm caudal, between 3 mm rostral and 3 mm caudal, and between 3 mm rostral and 1.6 mm caudal to the epicenter in contusion, dislocation, and distraction, respectively. Specifically, the reduction of large neurons was, respectively, retained between 1.6 mm rostral and 1 mm caudal, between 2.2 mm rostral and 1.6 mm caudal, and between 1.6 mm rostral and 1.6 mm caudal to the epicenter in contusion, dislocation, and distraction (Figure 4C). This implied that damage to the small neurons was more extensive than damage to the large neurons in the ventral horn for the 3 types of SCI mechanisms. The phenomenon was likely associated with the observation that the central lesion generally caused more severe tissue loss in the ventral horn where it was closer to the spinal cord center with more small neurons than where it was farther from the spinal cord center with more large neurons (Figure 5B).
The number of surviving cells in the ventral horn was significantly different between the different types of SCI (Figure 4B). Dislocation caused the most severe damage to the ventral horn neurons, as its values were lower than those in contusion and distraction between 0.2 mm rostral and 0.2 mm caudal and between 0.2 mm and 0.6 mm caudal to the epicenter, respectively. The differences were more evident in the large neurons; the number of surviving large cells was found to be, respectively, lower between 1 mm rostral and 2.2 mm caudal and between 0.2 mm and 1 mm caudal to the epicenter in dislocation than in contusion and distraction (Figure 4 C). These injury differences were noticeable around the epicenter (eg, at 1 mm; Figure 5B). Furthermore, damage to the ventral horn neurons was more severe in distraction than in contusion at the epicenter (Figure 4B), but it was more severe in contusion than in distraction at 0.6 mm caudal to the epicenter (Figure 4B and C).
Discussion
This study proposed an automated method for counting neurons in SCI based on their histochemically double-stained characteristics. The method was validated against the manual approach with 2 histologists, and it was proved to be as reliable as the currently most accepted method. Moreover, the method was very efficient; while it typically took minutes to process an image manually by an expert, it took only seconds to process an image automatically using our method with a modern computer.
In the previous studies that used an automatic method for counting neurons in uninjured and injured spinal cords, many of these methods were not described in detail, and the methods that were clearly explained all relied on setting brightness threshold to filter the background and then count the remaining objects.4,25-27,29,30,32,34,37,40,44 Based on our results, a simple threshold method (ie, binarization) with a marker that is highly specific (eg, NeuN) could be suitable for counting neurons in normal spinal cords; however, it would still not be appropriate for counting neurons in injured spinal cords (Figure 2). In our experience and the experience of others, no marker is specific enough to only highlight the neurons in SCI without also highlighting a considerable number of non-neuron-like objects and artifacts. There may be an automated way to intelligently recognize the neurons in SCI with 1 marker, but the algorithm, hardware, and work needed for such method are probably too demanding for general use. Our method is relatively simple; the program can be easily written with MATLAB built-in functions, and it efficiently identifies neurons by taking advantages of their unique features using NeuN/FluoroNissl double-staining. Note that the method should be insensitive to the staining procedure and imaging settings as long as the features are detectable.
The surviving cells in the common types of SCI mechanisms were examined and compared with each other using the automated method. Distraction was found to cause the most extensive and uniform damage to the dorsal horn neurons, which coincides with previous findings; distraction was previously reported to cause the most extensive central lesion particularly in the dorsal region (up to 5 mm rostral and caudal) and deformation in the node of Ranvier (up to 4 mm rostral) and the least concentrated white matter damage and gray matter hemorrhage comparing to contusion and dislocation.2,3,45 Dislocation was found to cause the most severe damage to the ventral horn neurons especially near the epicenter, which is also in agreement with previous findings; dislocation was previously shown to cause the greatest overall tissue loss near the epicenter (between 1 mm rostral and caudal) and deformation in the node of Ranvier at the epicenter comparing to contusion and distraction.2,45 Obviously, the automated method was able to detect the differences among different types of SCI.
The present results share many similarities with our previous results that used the same experimental setup to study the common types of SCI mechanisms, but there are also many differences.2 The differences were mostly due to the different ways of processing and analyzing the data. In this study, data from the left and right sides of the spinal cord were averaged for the analysis, but in the previous study, only data from 1 side of the spinal cord were processed and analyzed. Although these were bilateral injuries, differences will always exist between the left and right sides even with precise injury induction; therefore, averaging the data from both sides will provide a more accurate assessment of the injury outcomes. In addition, in this study, a neuron was counted when the nucleus was identifiable, but in the previous study, a neuron was counted only if its nucleus and nucleolus were both visible to the observer, which greatly limited the number of surviving cells that could be found. As a result, the variation in the data was generally much higher in the previous study than in this study. Therefore, the exact way of processing and analyzing the cell count data is very important, which should be clearly stated in the study.
We believe that the present method will greatly benefit the field, and the present findings will help to better understand the SCI mechanisms.
Footnotes
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Natural Science Foundation of China (project no. 81771347). The raw data was provided by ICORD, UBC.
Declaration of conflicting interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Equal contribution first authors
Author Contributions: JW, MZ, and KC conceived and led the project. KC performed the experiments. All authors processed the data. JW, MZ, and KC analyzed the data. All authors wrote the paper.
ORCID iD: Kinon Chen  https://orcid.org/0000-0003-4539-3285
https://orcid.org/0000-0003-4539-3285
References
- 1. Norenberg MD, Smith J, Marcillo A. The pathology of human spinal cord injury: defining the problems. J Neurotrauma. 2004;21:429-440. [DOI] [PubMed] [Google Scholar]
- 2. Chen K, Liu J, Assinck Pet al. Differential histopathological and behavioral outcomes eight weeks after rat spinal cord injury by contusion, dislocation, and distraction mechanisms. J Neurotrauma. 2016;33:1667-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Choo AM, Liu J, Lam CK, Dvorak M, Tetzlaff W, Oxland TR. Contusion, dislocation, and distraction: primary hemorrhage and membrane permeability in distinct mechanisms of spinal cord injury. J Neurosurg Spine. 2007;6:255-266. [DOI] [PubMed] [Google Scholar]
- 4. Zuchner M, Kondratskaya E, Sylte CB, Glover JC, Boulland JL. Rapid recovery and altered neurochemical dependence of locomotor central pattern generation following lumbar neonatal spinal cord injury. J Physiol. 2018;596:281-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sharif-Alhoseini M, Khormali M, Rezaei Met al. Animal models of spinal cord injury: a systematic review. Spinal Cord. 2017;55:714-721. [DOI] [PubMed] [Google Scholar]
- 6. Acciai L, Soda P, Iannello G. Automated neuron tracing methods: an updated account. Neuroinformatics. 2016;14:353-367. [DOI] [PubMed] [Google Scholar]
- 7. Akbar MT, Lundberg AM, Liu Ket al. The neuroprotective effects of heat shock protein 27 overexpression in transgenic animals against kainate-induced seizures and hippocampal cell death. J Biol Chem. 2003;278:19956-19965. [DOI] [PubMed] [Google Scholar]
- 8. Basso M, Pozzi S, Tortarolo Met al. Mutant copper-zinc superoxide dismutase (SOD1) induces protein secretion pathway alterations and exosome release in astrocytes: implications for disease spreading and motor neuron pathology in amyotrophic lateral sclerosis. J Biol Chem. 2013;288:15699-15711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dobremez E, Bouali-Benazzouz R, Fossat Pet al. Distribution and regulation of L-type calcium channels in deep dorsal horn neurons after sciatic nerve injury in rats. Eur J Neurosci. 2005;21:3321-3333. [DOI] [PubMed] [Google Scholar]
- 10. Donohue DE, Ascoli GA. Automated reconstruction of neuronal morphology: an overview. Brain Res Rev. 2011;67: 94-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang Y, Sun X, Hu G. An automatic integrated approach for stained neuron detection in studying neuron migration. Microsc Res Tech. 2010;73:109-118. [DOI] [PubMed] [Google Scholar]
- 12. Kaynig V, Vazquez-Reina A, Knowles-Barley Set al. Large-scale automatic reconstruction of neuronal processes from electron microscopy images. Med Image Anal. 2015;22:77-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kerekes RA, Martins RA, Davis D, Karakaya M, Gleason S, Dyer MA. Automated tracing of horizontal neuron processes during retinal development. Neurochem Res. 2011;36:583-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim KM, Son K, Palmore GT. Neuron image analyzer: automated and accurate extraction of neuronal data from low quality images. Sci Rep. 2015;5:17062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu T, Li G, Nie Jet al. An automated method for cell detection in zebrafish. Neuroinformatics. 2008;6:5-21. [DOI] [PubMed] [Google Scholar]
- 16. Narro ML, Yang F, Kraft R, Wenk C, Efrat A, Restifo LL. NeuronMetrics: software for semi-automated processing of cultured neuron images. Brain Res. 2007;1138:57-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Oberlaender M, Dercksen VJ, Egger R, Gensel M, Sakmann B, Hege HC. Automated three-dimensional detection and counting of neuron somata. J Neurosci Methods. 2009;180:147-160. [DOI] [PubMed] [Google Scholar]
- 18. Pleticha J, Jeng-Singh C, Rezek R, Zaibak M, Beutler AS. Intraneural convection enhanced delivery of AAVrh20 for targeting primary sensory neurons. Mol Cell Neurosci. 2014;60:72-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Quan T, Zhou H, Li Jet al. NeuroGPS-Tree: automatic reconstruction of large-scale neuronal populations with dense neurites. Nat Methods. 2016;13:51-54. [DOI] [PubMed] [Google Scholar]
- 20. Rennaker RL, Miller J, Tang H, Wilson DA. Minocycline increases quality and longevity of chronic neural recordings. J Neural Eng. 2007;4:L1-L5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Robinson J, Lu P. Optimization of trophic support for neural stem cell grafts in sites of spinal cord injury. Exp Neurol. 2017;291:87-97. [DOI] [PubMed] [Google Scholar]
- 22. Stegmaier J, Shahid M, Takamiya Met al. Automated prior knowledge-based quantification of neuronal patterns in the spinal cord of zebrafish. Bioinformatics. 2014;30:726-733. [DOI] [PubMed] [Google Scholar]
- 23. Wu J, Xue J, Huang R, Zheng C, Cui Y, Rao S. A rabbit model of lumbar distraction spinal cord injury. Spine J. 2016;16:643-658. [DOI] [PubMed] [Google Scholar]
- 24. Yan C, Li A, Zhang B, Ding W, Luo Q, Gong H. Automated and accurate detection of soma location and surface morphology in large-scale 3D neuron images. PLoS ONE. 2013;8:e62579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Adams KL, Rousso DL, Umbach JA, Novitch BG. Foxp1-mediated programming of limb-innervating motor neurons from mouse and human embryonic stem cells. Nat Commun. 2015;6:6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Beattie MS, Ferguson AR, Bresnahan JC. AMPA-receptor trafficking and injury-induced cell death. Eur J Neurosci. 2010;32:290-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen YJ, Zhu H, Zhang Net al. Temporal kinetics of macrophage polarization in the injured rat spinal cord. J Neurosci Res. 2015;93:1526-1533. [DOI] [PubMed] [Google Scholar]
- 28. Chen YJ, Chung K, Pivetti Cet al. Fetal surgical repair with placenta-derived mesenchymal stromal cell engineered patch in a rodent model of myelomeningocele [published online ahead of print October 12, 2017]. J Pediatr Surg. doi: 10.1016/j.jpedsurg.2017.10.040. [DOI] [PubMed] [Google Scholar]
- 29. Church JS, Kigerl KA, Lerch JK, Popovich PG, McTigue DM. TLR4 deficiency impairs oligodendrocyte formation in the injured spinal cord. J Neurosci. 2016;36:6352-6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ferguson AR, Christensen RN, Gensel JCet al. Cell death after spinal cord injury is exacerbated by rapid TNF alpha-induced trafficking of GluR2-lacking AMPARs to the plasma membrane. J Neurosci. 2008;28:11391-11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Han H, Xia Y, Wang S, Zhao B, Sun Z, Yuan L. Synergistic effects of galectin-1 and reactive astrocytes on functional recovery after contusive spinal cord injury. Arch Orthop Trauma Surg. 2011;131:829-839. [DOI] [PubMed] [Google Scholar]
- 32. He Z, Zhou Y, Lin Let al. Dl-3-n-butylphthalide attenuates acute inflammatory activation in rats with spinal cord injury by inhibiting microglial TLR4/NF-kappaB signalling. J Cell Mol Med. 2017;21:3010-3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jiang S, Ballerini P, Buccella Set al. Remyelination after chronic spinal cord injury is associated with proliferation of endogenous adult progenitor cells after systemic administration of guanosine. Purinergic Signal. 2008;4:61-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kalkan E, Cicek O, Unlu Aet al. The effects of prophylactic zinc and melatonin application on experimental spinal cord ischemia-reperfusion injury in rabbits: experimental study. Spinal Cord. 2007;45:722-730. [DOI] [PubMed] [Google Scholar]
- 35. Li F, Cheng B, Cheng J, Wang D, Li H, He X. CCR5 blockade promotes M2 macrophage activation and improves locomotor recovery after spinal cord injury in mice. Inflammation. 2015;38:126-133. [DOI] [PubMed] [Google Scholar]
- 36. Lim SN, Gladman SJ, Dyall SCet al. Transgenic mice with high endogenous omega-3 fatty acids are protected from spinal cord injury. Neurobiol Dis. 2013;51:104-112. [DOI] [PubMed] [Google Scholar]
- 37. Liu Y, Marin A, Ejlerskov P, Rasmussen LM, Prinz M, Issazadeh-Navikas S. Corrigendum: Neuronal IFN-beta-induced PI3K/Akt-FoxA1 signalling is essential for generation of FoxA1(+)Treg cells. Nat Commun. 2017;8:15792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Matak I, Tekus V, Bolcskei K, Lackovic Z, Helyes Z. Involvement of substance P in the antinociceptive effect of botulinum toxin type A: Evidence from knockout mice. Neuroscience. 2017;358:137-145. [DOI] [PubMed] [Google Scholar]
- 39. McTigue DM, Tripathi R, Wei P, Lash AT. The PPAR gamma agonist pioglitazone improves anatomical and locomotor recovery after rodent spinal cord injury. Exp Neurol. 2007;205:396-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vinit S, Darlot F, Stamegna JC, Sanchez P, Gauthier P, Kastner A. Long-term reorganization of respiratory pathways after partial cervical spinal cord injury. Eur J Neurosci. 2008;27:897-908. [DOI] [PubMed] [Google Scholar]
- 41. Yang R, He J, Wang Y. Activation of the niacin receptor HCA2 reduces demyelination and neurofilament loss, and promotes functional recovery after spinal cord injury in mice. Eur J Pharmacol. 2016;791:124-136. [DOI] [PubMed] [Google Scholar]
- 42. Yang CC, Jou IM. Caffeine treatment aggravates secondary degeneration after spinal cord injury. Brain Res. 2016;1634:75-82. [DOI] [PubMed] [Google Scholar]
- 43. Zhou KL, Zhou YF, Wu Ket al. Stimulation of autophagy promotes functional recovery in diabetic rats with spinal cord injury. Sci Rep. 2015;5:17130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhou KL, Chen DH, Jin HMet al. Effects of calcitriol on experimental spinal cord injury in rats. Spinal Cord. 2016;54:510-516. [DOI] [PubMed] [Google Scholar]
- 45. Choo AM, Liu J, Liu Z, Dvorak M, Tetzlaff W, Oxland TR. Modeling spinal cord contusion, dislocation, and distraction: characterization of vertebral clamps, injury severities, and node of Ranvier deformations. J Neurosci Methods. 2009;181:6-17. [DOI] [PubMed] [Google Scholar]