Abstract
Mouse trophoblast stem cells (TSCs) proliferate indefinitely in vitro, despite their highly heterogeneous nature. In this study, we sought to characterize TSC colony types by using methods based on cell biology and biochemistry for a better understanding of how TSCs are maintained over multiple passages. Colonies of TSCs could be classified into four major types: type 1 is compact and dome-shaped, type 4 is flattened but with a large multilayered cell cluster, and types 2 and 3 are their intermediates. A time-lapse analysis indicated that type 1 colonies predominantly appeared after passaging, and a single type 1 colony gave rise to all other types. These colony transitions were irreversible, but at least some type 1 colonies persisted throughout culture. The typical cells comprising type 1 colonies were small and highly motile, and they aggregated together to form primary colonies. A hierarchical clustering based on global gene expression profiles suggested that a TSC line containing more type 1 colony cells was similar to in vivo extraembryonic tissues. Among the known TSC genes examined, Elf5 showed a differential expression pattern according to colony type, indicating that this gene might be a reliable marker of undifferentiated TSCs. When aggregated with fertilized embryos, cells from types 1 and 2, but not from type 4, distributed to the polar trophectoderm in blastocysts. These findings indicate that cells typically found in type 1 colonies can persist indefinitely as stem cells and are responsible for the maintenance of TSC lines. They may provide key information for future improvements in the quality of TSC lines.
Keywords: mouse, placenta, trophoblast stem cell
Introduction
During early development in mammals, the extraembryonic lineage, the trophectoderm (TE), is segregated away from the inner cell mass (ICM) of blastocysts. The TE is committed to the formation of the outer monolayer of blastocysts, and these develop into the placental tissues required for supporting fetal development through the physical and physiological maternal-fetal interface [1]. Following implantation, the TE overlying the ICM (polar TE) continues to form the extraembryonic ectoderm (ExE) and, more apically, the ectoplacental cone (EPC). The polar TE cells give rise to all trophoblast lineages in the placenta, whereas the mural TE cells cease cell division and become trophoblast giant cells (TGCs) by endoreduplication [1].
Trophoblast stem cells (TSCs) are established from the polar TE or ExE in the presence of fibroblast growth factor 4 (FGF4), heparin, and mouse embryonic fibroblasts (MEFs) as feeder cells, or in MEF-conditioned medium [2]. FGF signaling has been shown to be crucial for the self-renewal of TSCs [3, 4], and its withdrawal promotes differentiation of TSCs into TGCs [2]. Trophoblast stem cells retain the capacity to differentiate into all lineages of the placenta in vivo, as evidenced by chimeric embryo experiments. Thus, when the TSCs are injected into murine blastocysts or aggregated with early embryos, they contribute to all of the placental layers of the subsequent conceptuses [2].
Trophoblast stem cells proliferate in vitro indefinitely while maintaining their in vivo differentiation ability. However, TSC lines are thought to consist of heterogeneous cell populations because they form colonies of different sizes and morphologies [2, 5]. Two studies have reported that TSC lines could be established efficiently using chemically defined media with or without feeder cells [5, 6]. These newly established TSCs formed relatively homologous colonies, but they still tended to change their morphology with time after passage. Thus, the TSC lines established to date contain different cell types that can change their characteristics dynamically during culture, although some TSCs should retain their undifferentiated status over several passages. However, there is little information on the identity of such stable cell type(s) and how they contribute to the persistence of TSC lines. The present study was undertaken to obtain clues to solving these key questions about the nature of TSCs. For this purpose, we classified TSC colonies based on their morphology and identified their colony type-specific characteristics using approaches based on cell biology and biochemistry.
Materials and Methods
Animals
C57BL/6N (B6), DBA/2, and (B6 × DBA/2) F(1) (BDF1) strains of mice were purchased from Japan SLC Inc. (Shizuoka, Japan), and ICR strain mice were purchased from CLEA Japan Inc. (Tokyo, Japan). Experiments were approved by the Animal Experimentation Committees of the RIKEN Tsukuba Institute and were performed in accordance with the committees' guiding principles.
Cell Lines
Most cell culture experiments in this study were performed with TSC lines established by the conventional method developed by Tanaka et al. [2] (“conventional TSC lines” hereafter). Additional experiments were performed using a TSC line established with a chemically defined culture condition (“defined TSC line” hereafter) [6]. The cell lines used are listed in 7]. Conventional TSCs were cultured as described previously [2]. In brief, cells were cultured on mitomycin-C-treated (Sigma-Aldrich, St. Louis, MO) primary MEFs in RPMI1640 medium (11875-093; Thermo Fisher Scientific, San Jose, CA) with 20% fetal bovine serum (Thermo Fisher Scientific), 25 ng/ml human recombinant FGF4 (062-04341; Wako, Osaka, Japan), 1 μg/ml heparin (H3149; Sigma-Aldrich), 100 μM 2-mercaptoethanol (Sigma-Aldrich), 1% Glutamax (Thermo Fisher Scientific), and 1 mM sodium pyruvate (Thermo Fisher Scientific). Trophoblast stem cells established with chemically defined medium were cultured under feeder-free, serum-free conditions as described previously [6].
Immunofluorescence
Trophoblast stem cells were fixed in 4% paraformaldehyde in PBS for 30 min and were permeabilized with 0.5% Triton X-100 in PBS for 10 min, followed by blocking with 1% bovine serum albumin in PBS for 1 h. They were treated with a primary antibody overnight at 4°C and with a secondary antibody for 1 h at room temperature. The primary antibodies used were a rabbit monoclonal anti-CDX2 (1:500; ab76541; Abcam, Cambridge, U.K.) and a rat monoclonal anti-mouse CDH1 (1:100; ECCD2; M108; TaKaRa Bio Inc., Otsu, Japan). The secondary antibodies were goat anti-rabbit immunoglobulin G (IgG) conjugated to Alexa Fluor 555 (1:500) and goat anti-rat IgG conjugated to Alexa Fluor 546 (1:500; Thermo Fisher Scientific). Samples were also stained with 4′,6-diamidino-2-phenylindole (DAPI) for localizing the nuclei. Images were acquired using a confocal scanning laser microscope (CV1000; Yokogawa Electric, Kanazawa, Japan) or a fluorescence microscope (BZ-9000; Keyence, Osaka, Japan) equipped with a Plan-Fluor ELWD 20×/0.45 objective lens (Nikon, Tokyo, Japan).
Differential Interference Contrast Imaging
Trophoblast stem cells were dissociated with a mixture of TrypLE Select (1×; 12563-011; Thermo Fisher Scientific) and 0.1 mg/ml polyvinyl alcohol (P-8136; Sigma-Aldrich) at 37°C under 5% CO2 in humidified air for 2 min, and were subsequently inactivated with an equal volume of fetal bovine serum. A drop of TSC suspension was placed on a glass slide. Differential interference contrast images were acquired using an Olympus BX53 microscope (Olympus, Tokyo, Japan) equipped with a 60×/0.90 UPlanSApo objective lens (Olympus) and a NEX-5T camera (Sony, Tokyo, Japan).
Time-Lapse Live Imaging
Trophoblast stem cell colonies in culture were monitored using an Axio Observer Z1 fluorescence microscope equipped with a 10×/0.30 Plan-Neofluar Ph1 objective lens, a Colibri LED light source, and an AxioCam MRm camera (all from Carl Zeiss Microimaging GmbH, Jena, Germany). The culturing environment was maintained with a Heating Insert P S and the incubation system S (Carl Zeiss) at 37°C under 5% CO2 in air with 100% humidity. The fields of view were acquired every 30 min for 48 h to monitor any conversion of type 1 colonies. Images were acquired, merged, and processed using AxioVision 4.8 software (Carl Zeiss).
Quantification of Single Cell Sizes
The EGFP fluorescent signals were detected and analyzed quantitatively using a fluorescence microscope (BZ-9000; Keyence) equipped with a Plan-Fluor ELWD 40×/0.60 objective lens (Nikon). Images were recorded with the BZ-II viewer software, and the diameters of single cells were measured using BZ-II analyzer software (Keyence). The nuclear shape was observed after staining with Hoechst 33342.
RNA Isolation, Amplification, and Hybridization
Separation of EPC and ExE lines was performed as described previously [8]. Each colony of TSCs was picked up with a pulled-glass capillary pipette. Total RNA was extracted with TRIzol reagent (15596018; Thermo Fisher Scientific) from 30–50 TSC colonies or from the extraembryonic tissues of Embryonic Day 6.5 (E 6.5). RNA samples (500 pg) were amplified with a two-round RNA amplification procedure performed with the TargetAmp 2-Round Aminoallyl-aRNA Amplification Kit 1.0 (TAA2R4924; Epicenter Biotechnologies, Madison, WI) according to the manufacturer's instructions. Amplified RNA was labeled with Cy3 dye (GE Healthcare, Little Chalfont, U.K.) and hybridized to a Whole Mouse Genome oligo DNA microarray (8 × 60K; G4858A; Agilent Technologies, Palo Alto, CA) for 16 h at 65°C according to the manufacturer's instructions.
Microarray Analysis
Hybridized microarray slides were exposed to a DNA microarray scanner (Agilent Technologies) at 3-μm resolution. Fluorescence intensities on scanned images were quantified, corrected for background, and normalized using Feature Extraction software (Agilent Technologies). The raw signals were imported into Gene Spring GX 12.5 software (Agilent Technologies) and normalized using the default settings. Data sets for each array were filtered by expression levels to remove unreliable raw intensity values that were less than 100 (27 874 probes). The normalized and filtered data were analyzed to characterize the global relationships of individual samples by two-way hierarchical clustering and principal component analysis (PCA). To extract differentially expressed genes, one-way ANOVA and Tukey post hoc statistical analysis at a 5% significance level were performed using the Benjamini and Hochberg false-discovery rate for correcting multiple tests. Additionally, 2-fold change selection was performed to identify significantly differentially expressed genes compared with type 4 colonies. Gene Ontology (GO) analysis was performed with FuncAssociate 2.1 (http://llama.mshri.on.ca/funcassociate/)[9]. Statistical analysis was done using a modified Fisher exact test at a 5% significance level for enrichment terms and resampling-based multiple testing.
RT-Quantitative PCR
Total RNA for RT-quantitative PCR (RT-qPCR) was extracted with RNeasy Micro kits (74004; Qiagen, Venlo, the Netherlands) from 60–80 TSC colonies. Following extraction, the first-strand cDNA was synthesized with a SuperScript III reverse transcriptase reagent set (18080-044; Thermo Fisher Scientific). Gene expression was assessed by qPCR on a StepOnePlus instrument (Thermo Fisher Scientific) using Quantitect SYBR Green PCR kits (204145; Qiagen) according to the manufacturer's instructions. The Actb gene in each sample was used as an endogenous reference [10]. Data analysis was performed using StepOne software v. 2.1 (Thermo Fisher Scientific). The primer sets used for quantification were as follows: Cdx2, 5′-GCAGTCCCTAGGAAGCCAAG-3′ and 5′-GCAGCCAGCTCACTTTTCCT-3′; Hand1, 5′-AGCAAGCGGAAAAGGGAGT-3′ and 5′-GTGCGCCCTTTAATCCTCTT-3′; Elf5, 5′-GTGGCATCCTGGAATGGGAA-3′ and 5′-CACTAACCTCCGGTCAACCC-3′; and Actb, 5′-CTGTCGAGTCGCGTCCA-3′ and 5′-ACCCATTCCCACCATCACAC-3′.
Aggregation Chimeras
BDF1 strain female mice were induced to superovulate by injections of 7.5 IU of pregnant mare serum gonadotropin (Sankyo, Tokyo, Japan), followed by an injection of human chorionic gonadotropin (Sankyo) 48–50 h later. Mature metaphase II oocytes were collected from oviducts and ovarian follicles, and epididymal spermatozoa were collected from male C57BL/6N (B6) mice. In vitro fertilization was performed as described previously [11]. Aggregation of eight-cell-stage embryos with TSCs was used as the protocol for generating embryonic stem cells (ESCs) [12]. Clumps of 10–15 TSCs after dissociation were chosen and transferred into drops containing zona-free eight-cell embryos. However, type 4 colonies (see below) have low adhesion, so the TSCs of such colonies were attached to the surfaces of an eight-cell embryo. Aggregates were cultured for 40–48 h at 37°C under 5% CO2 in humidified air until the blastocyst stage. Images were acquired with an Olympus IX-70 inverted microscope equipped with both Hoffman modulation and fluorescence optics. Chimeric blastocysts were stained with 5 μg/ml Hoechst 33342 (Dojindo, Kumamoto, Japan) for 10 min at 37°C under 5% CO2 in humidified air and were subsequently observed using an HS all-in-one fluorescence microscope (BZ-9000) equipped with a Plan-Fluor ELWD 20×/0.45 objective lens (Nikon). In some experiments, chimeric blastocysts were transferred into the uteri of E 2.5 pseudopregnant females, which had been mated with vasectomized males. The placentas were removed from the uteri at E 8.5, E 9.5, E 12.5, or E13.5 for determining the presence of TSC-derived placental cells. The primer set used for the detection of EGFP transgene was: 5′-GGCAAGCTGACCCTGAAGTT-3′ and 5′-TCAGCTCGATGCGGTTCAC-3′.
Statistical Analysis
For comparing gene expression levels between colony types within the TSC lines, a multiple comparison test (Shirley-Williams test) was performed and P < 0.05 was considered statistically significant.
Results
TSC Colonies Are Heterogeneous in Shape and Contain Two Different Cell Types
Trophoblast stem cell colonies could be classified into four major types by their morphology (Fig. 1A): type 1 was small, compact, and dome shaped; type 2 was compact and flattened; type 3 was similar to type 2, but the colonies had loose and multilayered cell clusters in their centers; and type 4 was similar to type 3 but with an extensive multilayered area. There was also an additional type 5, with a sparse monolayered appearance that was observed only rarely but expanded rapidly once it appeared. Immunostaining revealed that all colony types were positive for CDX2, a marker for undifferentiated trophoblastic cells, but the intensity of the CDX2-positive cells was lower in type 5 colonies than in the others (Fig. 1A). Therefore, we did not perform further detailed analyses for type 5 in this study. Besides these TSC colonies, TGCs appeared in the vicinity of types 2 and 3 colonies. Trophoblast giant cells were negative for CDX2 (Fig. 1A). The defined TSC line, CD1-TS, formed type 1-like and type 2-like colonies of different sizes, but not type 3-like or type 4-like colonies (Fig. 1B). Unlike conventional TSC colonies, the defined TSC line showed a relatively homogeneous colony transition; most type 1-like colonies transformed synchronously into large type 2-like colonies within 4 days (Fig. 1B).
Fig. 1.
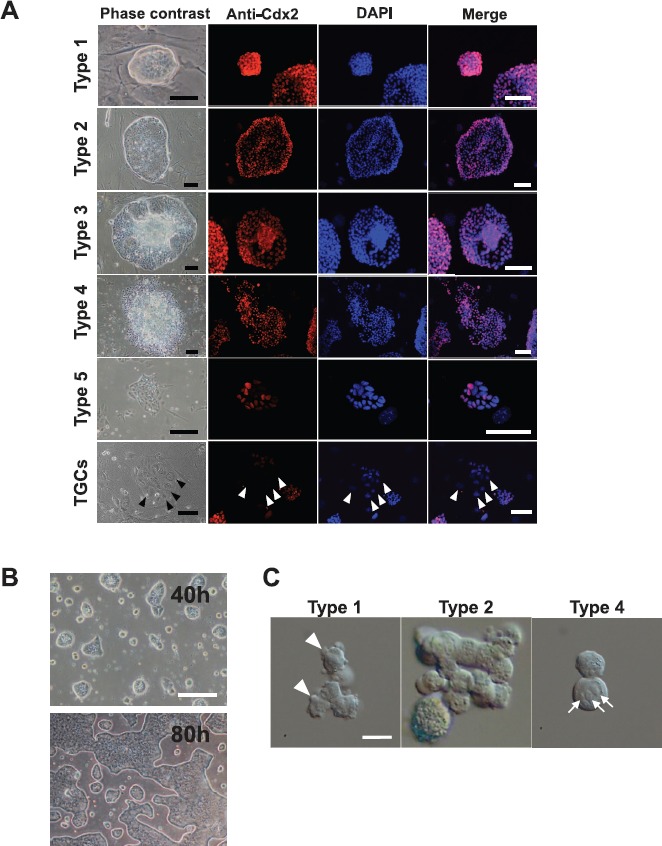
Colony types in conventional TSC lines. A) Morphology of TSC colonies in the B6TS4 line. They were immunostained for CDX2 (red); nuclei are stained with DAPI (blue). Most cells in the type 5 colonies were negative for CDX2 (see merged image). Trophoblastic giant cells were also CDX2 negative (arrowheads). Bar = 100 μm. B) Colonies in defined TSCs. They are similar to types 1 and 2 colonies in conventional TSCs. During culture, most type 1 colonies were transformed synchronously into large type 2 colonies, maintaining a relatively homogeneous colony appearance (see the transition from 40 to 80 h). Bar = 100 μm. C) Differential interference contrast microscope images of single cells from types 1–2, and type 4 colonies in the B6TS4 line. Cells in type 1 colonies were small and had cytoplasmic protrusions (pseudopods; arrowheads). Cells in type 4 colonies were large and had a clear, round nucleus containing a few large nucleoli (arrows). Type 2 colonies had a mixture of cells of both types. Bar = 20 μm.
We next observed the morphology of individual TSCs by dissociating colonies of different types into single-cell suspensions. The cell types were roughly divided into two categories (Fig. 1C and Supplemental Fig. S1A; Supplemental Data are available online at www.biolreprod.org): a smaller cell type 6–18 μm in diameter with an unclear nuclear membrane, and having an irregular contour because of small cytoplasmic protrusions (pseudopods) on the surface; and a larger round cell type 10–26 μm in diameter with a clear, round nucleus containing a few large nucleoli. The ratios of these two cell types varied according to the colony type and the TSC line. In the B6TS4 line, type 1 colonies were predominantly composed of the smaller cells (70%–80%), whereas types 2 and 3 colonies had a mixture of cells of both types, and type 4 colonies contained only the larger cells (Fig. 1B). This caused differences in the cell size populations among the colony types. As expected, types 1 and 2 colonies contained significantly smaller cells than types 3 and 4 (Supplemental Fig. S1B and Supplemental Table S1). In the EGFP-TS3.5 and XGFP lines derived from ICR cells (see below), type 1 colonies contained smaller populations of the smaller cells (20%–40%), and types 2, 3, and 4 colonies were composed of only the larger cells. Notably, in the defined TSC line, only the larger cells were observed (data not shown).
Type 1 Is the Primary Colony Category
To evaluate how this heterogeneity of TSC colonies was established in culture, we undertook a time-lapse live-imaging analysis after passaging a TSC line. Trophoblast stem cells from the B6TS4 line were dissociated into single-cell suspensions and then seeded onto a feeder cell layer. Among the single cells traced, the smaller cells typically found in type 1 colonies moved actively on the feeder cell layer and occasionally collided, forming small cell masses (Fig. 2A and Supplemental Movie S1). These cell masses increased with time and formed small type 1 colonies within 24 h after seeding (Fig. 2A and Supplemental Movie S1). Thus, type 1 was the category that first appeared after passage. However, TSC colonies were never formed from single cells in low-density cultures. Trophoblast stem cells from type 1 or type 4 colonies could not divide under single-cell culture conditions and finally underwent apoptosis (Fig. 3 and Supplemental Movies S2 and S3). Next, we traced the transitions of a single type 1 colony, which reached its maximal size at 48 h after seeding (the end of Day 2; Fig. 4A). The results are summarized in a dendrogram with the numbers of each colony type every 12 h (Fig. 4B). All colony types were derived from type 1, and these conversions were mostly irreversible (Fig. 4B); only one case among all 38 conversions was a reversion into type 1 from type 2 at 24 h. The proportion of type 1 colonies decreased with time, but it remained at about 20% at 48 h (Fig. 4C). Type 4 was the colony category that emerged from type 1, 2, or 3 at the final stage of culture (48 h). Only one type 5 colony was found in this experiment. These results indicate that type 1 is the primary colony category that can give rise to all types of TSC colonies in conventional TSC lines.
Fig. 2.
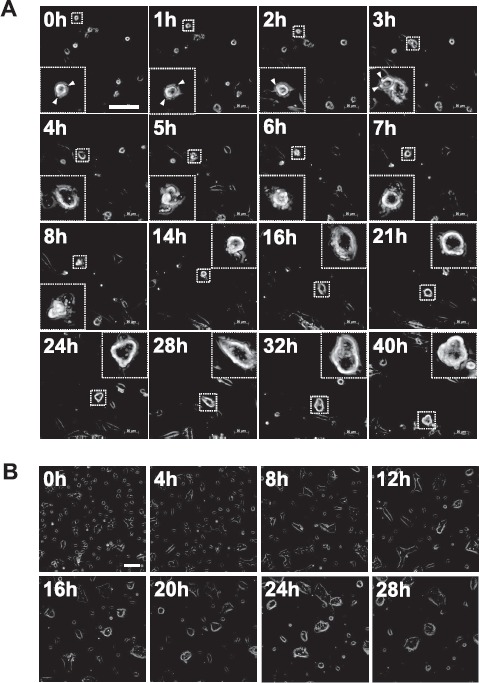
Time-lapse live imaging for tracing the formation of primary colonies after seeding single TSC cells. See also Supplemental Movies S1 and S4. A) B6TS4 line. Small cells with pseudopods (arrowheads) moved actively and aggregated to form small colonies. B) CD1-TS line. Round, larger TSCs were transformed into fibroblast-like cells upon attachment to the bottom of the dish, and started random movements. Then, adjacent cells formed small colonies while they continued cell division. Subsequently, groups of many small colonies aggregated into a smaller number of large ones. Bar = 100 μm.
Fig. 3.
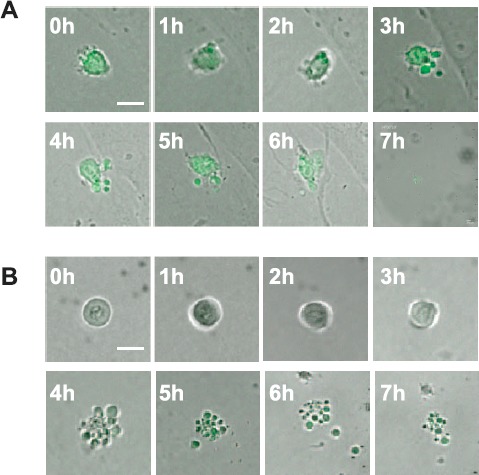
Time-lapse live imaging to trace the behavior of single B6TS4 cells after low-density culture. Typical TSCs from type 1 (A; small, amorphous cell) and type 4 (B; large, round cell) colonies did not divide, and they underwent apoptosis. See also Supplemental Movies S2 and S3. Bar = 20 μm.
Fig. 4.
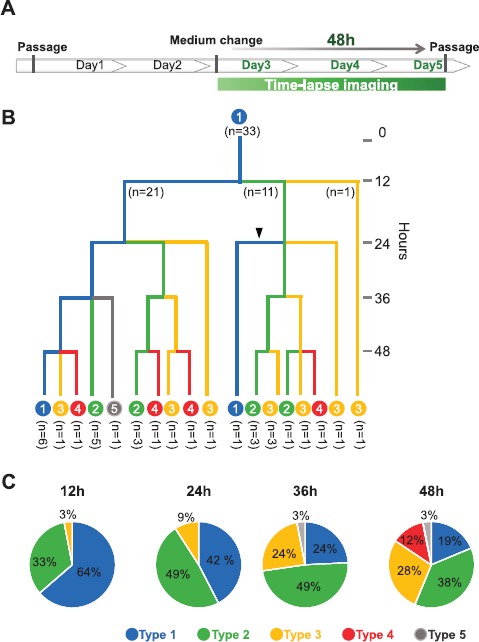
A representative pattern of the transition of colony types from a single type 1 colony in the B6TS4 line. A) A fully developed type 1 colony at 48 h (the end of Day 2) was selected and traced by time-lapse live imaging. B) A schematic diagram of the colony transition from a type 1 colony at every 12 h. All colony types appeared from a single type 1 colony and the conversion was irreversible, except for one case in which a type 1 colony was formed from a type 2 colony (arrowhead). C) A pie chart indicating the percentages of colony types counted every 12 h.
In the defined TSC line, which did not contain the smaller cell type, primary colonies were formed in a different way. Round cells of the larger type were transformed into fibroblast-like cells upon attachment to the bottom of the dish, and they started random movements. Then, adjacent cells formed small colonies while they continued cell division. Subsequently, groups of many small colonies aggregated into a smaller number of large ones (Fig. 2B and Supplemental Movie S4).
Global Gene Expression Profiles of TSC Colonies Are Line and Genotype Dependent
To characterize the colony types by their gene expression patterns, we performed microarray analysis using pooled colony samples from each cell line, together with in vivo-derived E 6.5 extraembryonic tissues. We analyzed types 1 and 2, the forms predominating in early days after passage, and type 4, the form that appeared in later days. The samples analyzed are listed in Table 1. To visualize the relationship between the samples, we analyzed the data by hierarchical clustering and PCA. Both analytical methods suggested that the samples could be grouped principally by their cell line rather than by their colony type (Fig. 5). Intriguingly, samples from the B6 cell line (B6TS4) proved closer to in vivo-derived samples (ExE and EPC) than other cell lines derived from ICR cells (EGFP-TS3.5, XGFP, and CD1-TS). As a result, the defined TSC line (CD1-TS) was classified as one of the ICR-derived cell lines. These results indicate that the genetic background of the mice (B6 or ICR) was a primary determining factor that distinguished the TSC colonies in their gene expression profiles.
Table 1.
List of TSC lines and samples used for microarray analysis.
| TSC line (sex) | Genetic background | Culture medium | Samples for microarray analysis | |
|---|---|---|---|---|
| Colony/tissue typea | No. of samples | |||
| B6TS4 (male) | C57BL/6 (B6) | Conventional | Type 1 | 4 |
| Type 2 | 2 | |||
| Type 4 | 3 | |||
| EGFP-TS3.5 (female) | ICR | Conventional | Type 1 | 3 |
| Type 2 | 3 | |||
| Type 4 | 4 | |||
| XGFP (female) | ICR | Conventional | Type 1 | 3 |
| Type 2 | 4 | |||
| Type 4 | 4 | |||
| CD1-TS-Tg(CAG-GFP) #3Fb (female) | ICR (CD1) | Chemically defined | 3 | |
| E6.5 extraembryonic tissues (male and female) | C57BL/6 (B6) | ExE | 3 | |
| EPC | 3 | |||
| ICR | ExE | 3 | ||
| EPC | 3 | |||
ExE, extraembryonic ectoderm; EPC, ectoplacental cone.
Referred to as “CD1-TS” in the text.
Fig. 5.
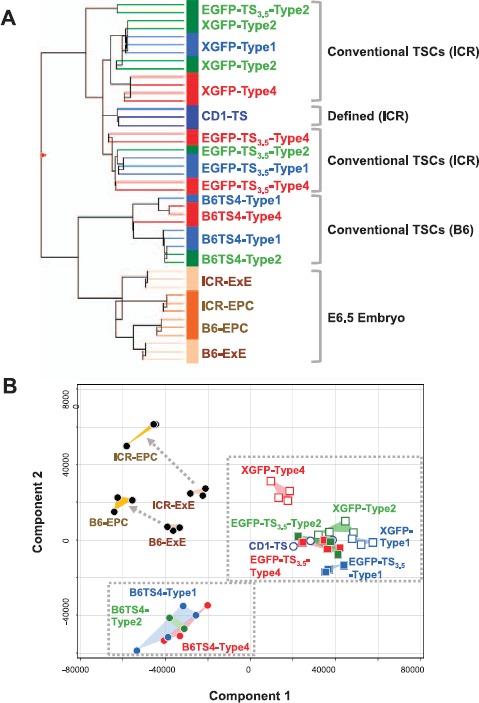
Microarray analysis for global gene expression profiles in TSC colonies and in vivo-derived extraembryonic tissues. A) Hierarchical clustering analysis. Trophoblast stem cell samples were clustered by their genetic background (B6 vs. ICR strain) rather than by their colony type. B) Principal component analysis. The TSC samples were grouped by their genetic backgrounds. For the numbers of samples, see Table 1.
Elf5 Might Be a Reliable Marker for Distinguishing the Status of Differentiation in TSC Colonies
We next observed the expression levels of marker genes for undifferentiated or differentiated trophoblastic cells using a microarray analysis. Among the undifferentiated state markers (Cdx2, Esrrb, Sox2, and Elf5), Elf5 was specifically decreased in type 4 colonies in both B6 (B6TS4) and ICR (EGFP-TS3.5) TSC lines (Fig. 6). These four genes were downregulated in the EPC compared with the ExE, which is consistent with the fact that the EPC differentiates from the ExE (Fig. 6). A spongiotrophoblast cell marker, Ascl2, was expressed consistently in the B6ST4 and EGFP-TS3.5 lines but was downregulated in type 1 colonies in the XGFP line. A TGC marker gene, Hand1, was consistently expressed in these three conventional TSC lines but was downregulated in the defined TSC line (CD1-TS). These findings suggest that Elf5 is a reliable marker for distinguishing the status of differentiation in TSC colonies, and that TSC lines established by a conventional method inherently contained differentiating TSCs, whereas the TSC line established by a chemically defined method was at a relatively undifferentiated state. Some of the other trophoblast marker genes—Eomes, Fgfr2, Ets2, Tfap2c, Prl3d1/Pl-1, and Krt7I [13–15]—also showed expression levels in a colony type-dependent manner (Supplemental Fig. S2). Genes specific for endoderm, mesoderm, and ectoderm lineages showed no expressions, indicating that these TSCs did not undergo transdifferentiation during culture (Supplemental Fig. S3).
Fig. 6.
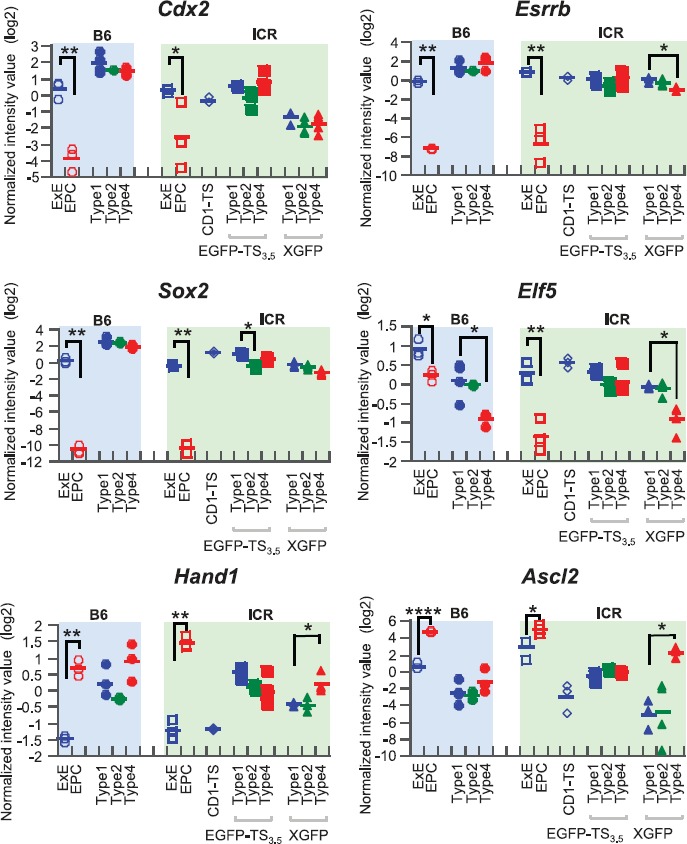
Expression levels of TSC marker genes. Among the four undifferentiated markers (Cdx2, Esrrb, Sox2, and Elf5), only Efl5 showed a colony type-specific expression pattern in both B6 (B6TS4) and ICR (XGFP) lines. In the in vivo-derived samples, EPC consistently showed lower expression levels than ExE for these four genes. A spongiotrophoblast cell marker, Ascl2, and a TGC marker gene, Hand1, were consistently expressed in the three conventional lines, with slight variations, but Hand1 was downregulated in the defined line. Bars represent the means of each group. *P < 0.05; **P < 0.01; ***P < 0.001.
Type 4 Colonies Decrease the Expression of “Cell Junction” Categories of Genes
The results shown above indicate that type 4 might represent a differentiated colony type. Therefore, we sought to identify those genes with significantly different expression levels between type 4 and types 1 and 2 in different TSC lines. However, when the data from all TSC lines were analyzed together, very few common differentially expressed genes were extracted, probably because of variations in the gene expression levels among the TSC lines. Therefore, we used the data from B6TS4 and EGFP-TS3.5 cells as representatives of the B6 and ICR lines, respectively, because the EGFP-TS3.5 line has been used extensively elsewhere [3, 16]. We identified 71 commonly upregulated and 116 commonly downregulated genes in these two cell lines (Fig. 7A). The GO analysis revealed that the two major categories for upregulated genes in type 4 colonies were angiogenesis-related GO terms (Fig. 7B). This is in accordance with differentiation of the trophoblast lineage in the hemochorial placenta of rodents, because in such placental types trophoblastic cells make direct contact with the maternal blood to form vessels for the maternal blood supply [17, 18]. The upregulated angiogenesis-related genes included Nos3 (nitric oxide synthase 3), Robo4 (roundabout homolog 4), and Ramp2 (receptor [calcitonin] activity-modifying protein 2), which are known to be produced by trophoblastic lineage cells and have direct or indirect effects on vasodilatation and angiogenesis [19–21]. The genes downregulated in type 4 colonies were enriched in cell junction-related GO terms. This might reflect the loosened cell-cell contacts in type 4 colonies. Of these GO terms, “tight junction” and “cell-cell junction” included Cdh1, the gene encoding E-cadherin (CDH1). The decreased expression levels of Cdh1 are shown in Figure 7C. CDH1 is known to be one of the key molecules necessary for establishing cell polarity and the structural integrity of several tissues [22]. We then investigated the localization of CDH1 in TSC colonies by immunostaining. Fluorescence microscopy images for CDH1 showed meshwork patterns in types 1 and 2 colonies, indicating that this protein was localized in entire cell-to-cell adjacent areas (Fig. 7D). By contrast, the localization of CDH1 was coarse and patchy in type 4 colonies, reflecting their weaker cell contacts (Fig. 7D).
Fig. 7.
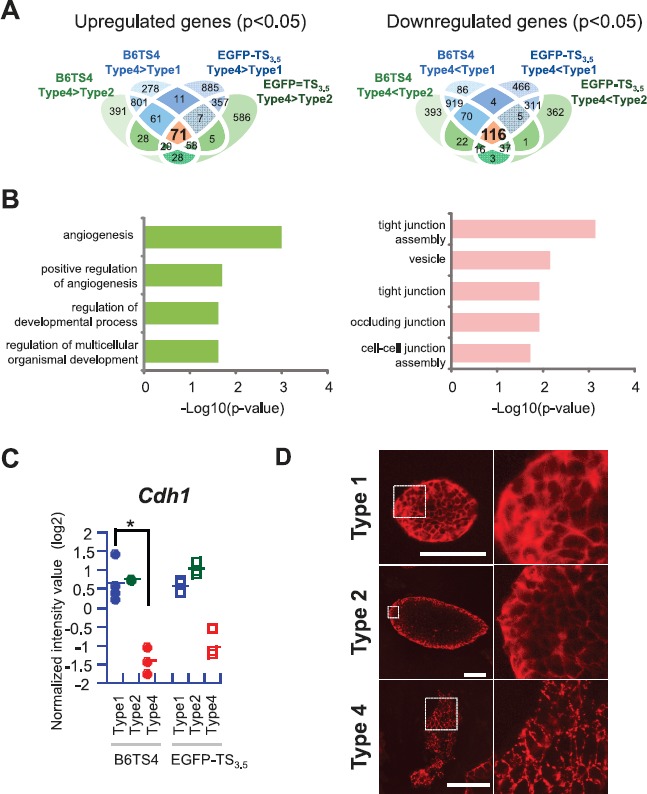
Extraction of differentially expressed genes (DEGs) between type 4 and types 1 and 2 colonies and their GO analysis. A) A Venn diagram showing the overlaps of DEGs in two TSC lines. In these lines, 71 commonly upregulated and 116 commonly downregulated genes were identified. B) The GO analyses showing functional annotation of the overlapping genes extracted in the Venn diagram. C) Expression levels of Cdh1 showing lower expressions in type 4 colonies in both lines, although there was no statistical significance for the ICR line. *P < 0.05. D) Immunofluorescence staining for CDH1. Types 1 and 2 colonies showed intense meshwork patterns, whereas type 4 showed a patchy staining pattern. Bars = 100 μm.
Quantitative Gene Expression Analysis of Trophoblast Marker Genes
To confirm the results obtained by the microarray analysis, the expression levels of some marker genes were analyzed by RT-qPCR (Fig. 8). We used B6TS4 and EGFP-TS3.5 cells, the representative B6- and ICR-derived conventional lines (see above). In both cell lines, Cdx2 was repressed in type 5 colonies, in agreement with the results from immunostaining experiments (Fig. 1A). Elf5 was repressed specifically in types 3 and 4 colonies. A marker for TGC, Hand1, did not show any colony type-dependent pattern. Thus, our RT-qPCR analysis further supported the findings obtained by the microarray analysis: Elf5 could be a better marker for the undifferentiated state than Cdx2, and Hand1 was consistently expressed in TSCs regardless of the undifferentiated/differentiated state of TSCs. Type 3 colonies, which were not analyzed by microarray, seemed to have characteristics intermediate to those of types 2 and 4.
Fig. 8.
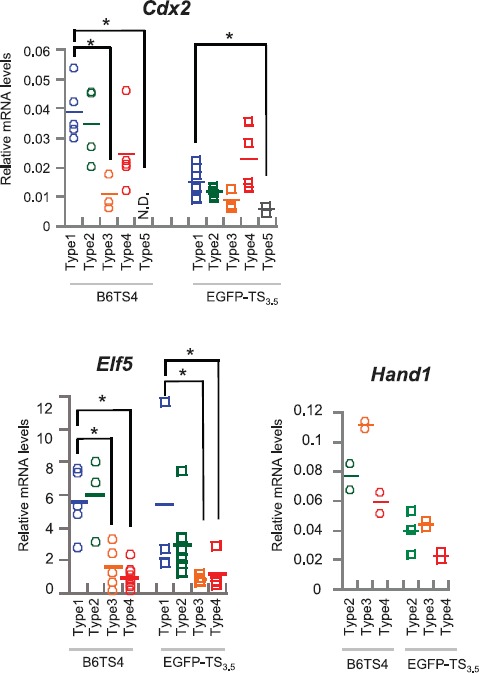
Analysis for TSC marker genes by RT-qPCR. Cdx2 was repressed in type 5 colonies in both TSC lines, consistent with the immunofluorescence analysis shown in Figure 1A. Elf5 showed different expression patterns according to the colony type. The expression levels of Hand1 were not significantly different among the samples. The relative expression levels were normalized against that of Actb. *P < 0.05.
TSCs from Types 1 and 2 Colonies, but Not from Type 4, Contribute to the TE in Chimeric Blastocysts
Finally, we examined the ability of TSCs from types 1, 2, and 4 colonies to contribute to polar TE formation after aggregation with normal embryos. The distributions of TSC-derived cells were identified from the EGFP expression levels of the original TSCs (B6TS4). At the blastocyst stage, TSCs from types 1, 2, and 4 colonies were localized to the polar TE of blastocysts in 27 of 35 (77%), 18 of 25 (72%), and 4 of 41 (9%), respectively (Fig. 9). Only TSCs from type 4 colonies contributed to the mural TE, but the proportion was low at 2 of 41 (5%). When the ability of TSCs to contribute to the placental tissues was examined by embryo transfer experiments, TSCs from types 1 and 2 were distributed to the placentas, but those from type 4 were not detected even with genomic PCR analysis (Supplemental Fig. S4A). We did not observe EGFP-specific fluorescence in these placentas, whereas those derived from chimeric blastocysts using TSCs from type 1 colonies of EGFP-TS3.5 line did show it (Supplemental Fig. S4B). Although we do not know why B6TSC4-derived chimeric placentas showed no fluorescence, these findings indicate that not only TSCs in type 1 colonies but also those in type 2 colonies maintained their ability to contribute to a placenta. However, TSCs in type 4 colonies had lost this ability.
Fig. 9.
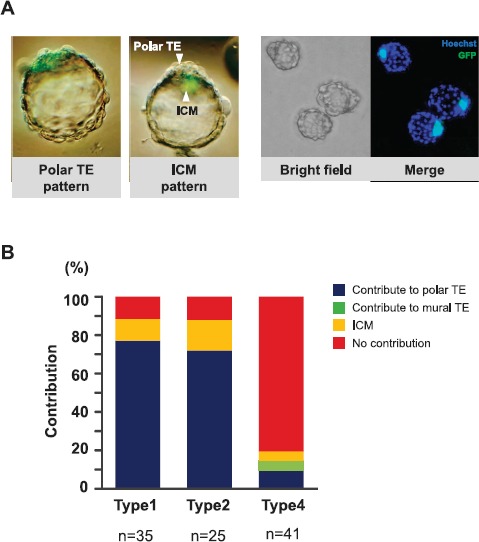
Assessment of the chimera formation ability of TSCs by aggregation with eight-cell embryos. A) Images of chimeric blastocysts at 40–48 h after aggregation. Most EGFP-positive TSC-derived cells, if present, were distributed to the polar TE or ICM. Right side: representative images of blastocysts containing cells derived from type 1 colonies. B) The proportions of blastocysts classified by the distribution pattern of TSC-derived cells.
Discussion
Although TSCs are characterized by their heterogeneous nature and inherent tendency for autonomous differentiation, they can be maintained through multiple passages without losing their stem cell features. The present study was undertaken to determine how different types of colonies appeared in TSC cultures and how their stemness was maintained in vitro through multiple passages. Our systematic analyses of gene expression, live imaging, and chimera formation experiments cumulatively suggest that the cell type typically found in type 1 colonies was responsible for maintaining the stemness of these TSC lines (Fig. 10). During culture, some type 1 colonies were converted into other forms in an irreversible manner, but the remaining type 1 colonies could contribute to the formation of new primary colonies at the next passage (Fig. 10). The cells in type 1 colonies were characterized by their small, amorphous shape with surface pseudopods. They were highly motile and aggregated to form these colonies, as revealed by a time-lapse live-imaging analysis. Importantly, type 1 colonies gave rise to all the other colony types, and at least some of them maintained their original morphology throughout a single passage (about 4 days). As far as we observed, type 1 colonies from the B6TS4 cell line contained the largest population of these cells, indicating their relatively undifferentiated state compared with other TSC lines. Indeed, our hierarchical clustering analysis indicated a closer relationship of B6TS4 with in vivo-derived extraembryonic tissues than with other TSC lines (Fig. 5A). Intriguingly, although the defined TSCs seemed to be more homogeneous, and consistently expressed TSC markers, they rarely contained type 1 colony cells and were distant from ExE tissues in their global gene expression pattern. Possibly the ICR strain genetic background might have impeded the formation of a highly undifferentiated status, as suggested by our hierarchical clustering analysis. A predominant effect of the genetic background on the transcriptional profiles was also reported for ESCs derived from blastocysts with different origins (normally fertilized embryos or those produced by somatic cell nuclear cloning) [23]. Nonetheless, we propose that the consistent appearance of type 1 colonies and their typical small cell type might be important indicators for future improvements in the quality of TSCs.
Fig. 10.
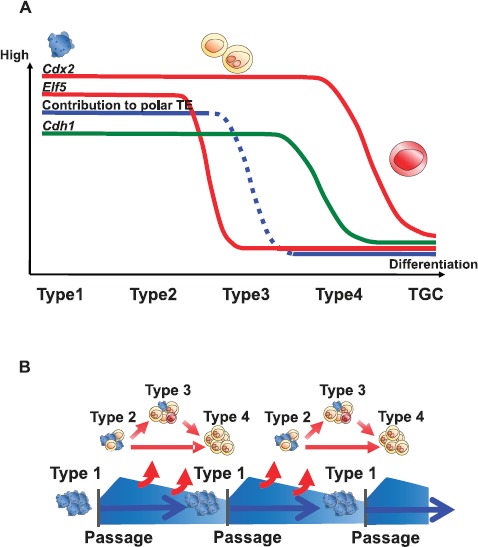
Schemes of cellular dynamics in mouse TSC colonies. A) Changes in the levels of TSC markers based on colony transitions. B) Presumed mechanism ensuring the stemness of the TSC line. Although type 1 colonies converted to other colony types with time in an irreversible manner, the small undifferentiated-type TSCs could contribute to the formation of primary colonies after the next passage.
Among the TSC marker genes we examined, Elf5 is a promising candidate for better understanding the maintenance of stemness among TSC lines. The expression of Elf5 was downregulated in types 3 and 4 colonies; importantly, cells from type 4 colonies had lost the ability to contribute to the TE in experiments to generate aggregation chimeras (Fig. 9). By contrast, other TSC marker genes—Cdx2, Essrb, and Sox2—were consistently expressed in all colony types examined (except for type 5 analyzed by RT-qPCR), with slight variations among samples. Elf5 encodes the Elf5 protein, a second ETS-domain transcription factor, which is known to be essential for the establishment of TSCs through sustaining the self-renewal of mouse ExE cells [24]. This is also the case for early human placentas because cytotrophoblast cells positive for both ELF5 and CDX2 were identified as a putative TSC population [25]. Interestingly, a recent study on TSC-specific transcriptional networks proposed that Essrb is located downstream of the FGF signaling pathway and might regulate the key TSC genes, including Elf5, directly to maintain self-renewal ability [4]. However, our differential expression analysis using separated colony types clearly showed that the expression levels of Essrb and Elf5 were not always correlated, as shown in type 4 colonies in the B6TS4 and XGFP lines (Fig. 5). Identification of another pathway that regulates the expression of Elf5 might provide new information on the mechanisms for the self-renewal of TSCs.
It is known that the Elf5 protein inhibits the epithelial-mesenchymal transition (EMT) in several epithelial tissues, including mammary glands. During trophoblast differentiation, an EMT-like change also occurs in association with the loss of CDH1 and the gain of trophoblast invasiveness [26]. We also noted that the GO terms of genes downregulated in type 4 colonies were enriched with cell junction categories, including the gene encoding CDH1. Therefore, the decrease of Elf5 in type 4 colonies might be responsible for their loosened colony shape through downregulation of CDH1-mediated cell-cell connections.
Unlike TSCs, their embryonic counterparts, ESCs, are highly homogenous and exhibit a potentiated ability to contribute to the embryonic tissues proper. High-quality ESCs enable the generation of ESC-only-derived mice by combining with tetraploid embryos that contribute predominantly to the placental tissue. This superior feature of ESCs was achieved by inhibiting differentiation with inhibitors for Erk1/2 and Gsk3β signaling [27]. If the same idea could be applied to TSCs, their quality as stem cells would also be improved. Comprehensive epigenetic and biochemical analyses using isolated colony types, as in the present study, would help in identifying the signaling cascades that need to be inhibited to maintain an undifferentiated status. The same strategy could be applied to the rat choriocarcinoma cell lines (RCHO or Rcho-1), which are prone to differentiation into trophoblast giant cells [28]. There are still many undefined epigenetic mechanisms during trophoblast differentiation, such as the imprinting-independent paternal inheritance of the memory for X chromosome inactivation [29, 30] and precocious differentiation of ExE in somatically cloned embryos [31]. Future studies using such high-quality TSCs would help elucidate their underlying mechanisms.
Supplementary Material
Acknowledgment
We thank Dr. Satoshi Tanaka for providing the B6TS4 and EGFP-TS3.5 TSC lines. The CAG-EGFP mouse line (green mice, RBRC00267) used for the positive control (Supplemental Fig. S4B) was originally generated by Professor M. Okabe (Osaka University) and was provided by the RIKEN BioResource Center, with the support of the National BioResources Project of MEXT, Japan.
References
- 1. Cross JC, Werb Z, Fisher SJ.. Implantation and the placenta: key pieces of the development puzzle. Science 1994; 266:1508–1518. [DOI] [PubMed] [Google Scholar]
- 2. Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J.. Promotion of trophoblast stem cell proliferation by FGF4. Science 1998; 282:2072–2075. [DOI] [PubMed] [Google Scholar]
- 3. Adachi K, Nikaido I, Ohta H, Ohtsuka S, Ura H, Kadota M, Wakayama T, Ueda HR, Niwa H.. Context-dependent wiring of Sox2 regulatory networks for self-renewal of embryonic and trophoblast stem cells. Mol Cell 2013; 52:380–392. [DOI] [PubMed] [Google Scholar]
- 4. Latos PA, Goncalves A, Oxley D, Mohammed H, Turro E, Hemberger M.. Fgf and Esrrb integrate epigenetic and transcriptional networks that regulate self-renewal of trophoblast stem cells. Nat Commun 2015; 6:7776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kubaczka C, Senner C, Arauzo-Bravo MJ, Sharma N, Kuckenberg P, Becker A, Zimmer A, Brustle O, Peitz M, Hemberger M, Schorle H.. Derivation and maintenance of murine trophoblast stem cells under defined conditions. Stem Cell Reports 2014; 2:232–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ohinata Y, Tsukiyama T.. Establishment of trophoblast stem cells under defined culture conditions in mice. PLoS One 2014; 9:e107308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Honsho K, Hirose M, Hatori M, Yasmin L, Izu H, Matoba S, Togayachi S, Miyoshi H, Sankai T, Ogura A, Honda A.. Naïve-like conversion enhances the difference in innate in vitro differentiation capacity between rabbit ES cells and iPS cells. J Reprod Dev 2015; 61:13–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nagy A, Gertsenstein M, Vintersten K, Behringer R.. Manipulating the Mouse Embryo, 3rd ed. Cold Spring Harbor, NY:Cold Spring Harbor Laboratory Press; 2003:222–223. [Google Scholar]
- 9. Berriz GF, King OD, Bryant B, Sander C, Roth FP.. Characterizing gene sets with FuncAssociate. Bioinformatics 2003; 19:2502–2504. [DOI] [PubMed] [Google Scholar]
- 10. Motomura K, Inoue K, Ogura A.. Selection of accurate reference genes in mouse trophoblast stem cells for reverse transcription-quantitative polymerase chain reaction. J Reprod Dev (in press).Published online ahead of print6February2016;DOI: 10.1262/jrd.2015-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hasegawa A, Mochida K, Tomishima T, Inoue K, Ogura A.. Microdroplet in vitro fertilization can reduce the number of spermatozoa necessary for fertilizing oocytes. J Reprod Dev 2014; 60:187–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Eakin GS, Hadjantonakis AK.. Production of chimeras by aggregation of embryonic stem cells with diploid or tetraploid mouse embryos. Nat Protoc 2006; 1:1145–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wen F, Tynan JA, Cecena G, Williams R, Múnera J, Mavrothalassitis G, Oshima RG.. Ets2 is required for trophoblast stem cell self-renewal. Dev Biol 2007; 312:284–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Latos PA, Sienerth AR, Murray A, Senner CE, Muto M, Ikawa M, Oxley D, Burge S, Cox BJ, Hemberger M.. Elf5-centered transcription factor hub controls trophoblast stem cell self-renewal and differentiation through stoichiometry-sensitive shifts in target gene networks. Genes Dev 2015; 29:2435–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhou WH, Dong L, Du MR, Zhu XY, Li DJ.. Cyclosporin A improves murine pregnancy outcome in abortion-prone matings: involvement of CD80/86 and CD28/CTLA-4. Reproduction 2008; 135:385–395. [DOI] [PubMed] [Google Scholar]
- 16. Erlebacher A, Price KA, Glimcher LH.. Maintenance of mouse trophoblast stem cell proliferation by TGF-β/activin. Dev Biol 2004; 275:158–169. [DOI] [PubMed] [Google Scholar]
- 17. Sutherland A. Mechanisms of implantation in the mouse: differentiation and functional importance of trophoblast giant cell behavior. Dev Biol 2003; 258:241–251. [DOI] [PubMed] [Google Scholar]
- 18. Ogura A, Nishida T, Hayashi Y, Mochida K.. The development of the uteroplacental vascular system in the golden hamster Mesocricetus auratus. J Anat 1991; 175:65–77. [PMC free article] [PubMed] [Google Scholar]
- 19. Hemberger M, Nozaki T, Masutani M, Cross JC.. Differential expression of angiogenic and vasodilatory factors by invasive trophoblast giant cells depending on depth of invasion. Dev Dyn 2003; 227:185–191. [DOI] [PubMed] [Google Scholar]
- 20. Liao WX, Wing DA, Geng JG, Chen DB.. Perspectives of SLIT/ROBO signaling in placental angiogenesis. Histol Histopathol 2010; 25:1181–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wilson C, Nikitenko LL, Sargent IL, Rees MC.. Adrenomedullin: multiple functions in human pregnancy. Angiogenesis 2004; 7:203–212. [DOI] [PubMed] [Google Scholar]
- 22. van Roy F, Berx G.. The cell-cell adhesion molecule E-cadherin. Cell Mol Life Sci 2008; 65:3756–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brambrink T, Hochedlinger K, Bell G, Jaenisch R.. ES cells derived from cloned and fertilized blastocysts are transcriptionally and functionally indistinguishable. Proc Natl Acad Sci U S A 2006; 103:933–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Donnison M, Beaton A, Davey HW, Broadhurst R, L'Huillier P, Pfeffer PL.. Loss of the extraembryonic ectoderm in Elf5 mutants leads to defects in embryonic patterning. Development 2005; 132:2299–2308. [DOI] [PubMed] [Google Scholar]
- 25. Hemberger M, Udayashankar R, Tesar P, Moore H, Burton GJ.. ELF5-enforced transcriptional networks define an epigenetically regulated trophoblast stem cell compartment in the human placenta. Hum Mol Genet 2010; 19:2456–2467. [DOI] [PubMed] [Google Scholar]
- 26. Abell AN, Granger DA, Johnson NL, Vincent-Jordan N, Dibble CF, Johnson GL.. Trophoblast stem cell maintenance by fibroblast growth factor 4 requires MEKK4 activation of Jun N-terminal kinase. Mol Cell Biol 2009; 29:2748–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A.. The ground state of embryonic stem cell self-renewal. Nature 2008; 453:519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Peters TJ, Chapman BM, Soares MJ.. Trophoblast differentiation: an in vitro model for trophoblast giant cell development. Methods Mol Biol 2000; 137:301–311. [DOI] [PubMed] [Google Scholar]
- 29. Takagi N, Sasaki M.. Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature 1975; 256:640–642. [DOI] [PubMed] [Google Scholar]
- 30. Oikawa M, Inoue K, Shiura H, Matoba S, Kamimura S, Hirose M, Mekada K, Yoshiki A, Tanaka S, Abe K, Ishino F, Ogura A.. Understanding the X chromosome inactivation cycle in mice: a comprehensive view provided by nuclear transfer. Epigenetics 2014; 9:204–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hirasawa R, Matoba S, Inoue K, Ogura A.. Somatic donor cell type correlates with embryonic, but not extra-embryonic, gene expression in postimplantation cloned embryos. PLoS One 2013; 8:e76422. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


