Abstract
Objective:
This study aimed to design specific primers derived from mitochondrial cytb of Sus Scrofa (1F1R primer) used in the pork meatball analysis using real time polymerase chain reaction (RT-PCR) method.
Materials and Methods:
Such designed primers were validated and these included specificity of primer, linearity, and sensitivity of the method as well as the repeatability test. The primers were specifically affirmed in the fresh tissue of chickens, cows, pigs, and goats. The linearity and sensitivity of the method was conducted by measuring the amplification curve from a series of dilution (0, 1, 1, 10, 100, 1,000, and 10,000 pg/μl of DNA) extracted from 100% pork meatball formulation. The repeatability test was conducted by determining the cycle threshold (Ct) values of RT-PCR amplification from 100% pork meatball formulation as many as six times.
Results:
Primer of 1F1R (forward: 5′-ACG CGA TAT AAG CAG GTA AA-3′; reverse: 5′-CTG CTT TCG TAG CAC GTA TT-3′) was specific in analyzing the presence of pork in meatball formulation at 47.1°C, which was optimum annealing temperature. The DNA identification was able to use the primers by RT-PCR with 1 pg as the limit of detection, efficiency value was 242.58%, and the coefficient of determination value (R2) was 0.956. The coefficient of variance was 4.13%. The developed method was also fruitfully applied to analyze commercial meatballs.
Conclusion:
RT-PCR method using specific primers targeting on mitochondrial gene (1F1R primer) could be used as the standard method for identification of pork in food samples intended for halal authentication studies.
Keywords: Pork meat, meatball, mitochondrial cyt-b, real-time PCR
Introduction
Meat-based food products, such as meatball, are well known as protein sources needed by human body and become an important diet due to its acceptability in terms of taste and for almost people [1]. FAO-UN has reported that the need on meat and meat-based food has grown rapidly [2]. Caused by price difference between halal beef and non-halal meat, such as pork and motivated by economical gain, some meatball producers try to blend or substitute halal beef with pork [3]. This substitution action is prohibited either from economical aspect or religion reason. The National Sanitation Foundation, from economic point of view, has officially reported that practices of industrial food adulteration were above 49 billion dollars annually worldwide. In addition, from religion point of view, pork is considered as non-halal for Muslim and non-kosher for Jewish; therefore, the accurate labeling in meat types and meat-based products is needed in order to allow customers to know what they eat [4,5].
The authenticity and traceability of meat-based products in our modern society is the most important issues in the food industry sectors, for all the parties, including regulators, producers, and consumers. A specific and harmonized definition of food adulteration is still not available. The European commission (EC) has described food adulteration, including processed meat, as an intentional violation of the food laws to benefit economical profits causing in consumer’s deception and health risk [5]. In addition, traceability of food according to EC 2002 is “the ability to trace and follow a food, feed, food producing animal or ingredients, through all the stages of production and distribution” [6]. Authentication of meat-based food products describes verifying the labels of meat-based food product, detecting where the meat comes from, and assuring the presence of non-claimed meat proposed to be in their product formulation [7], and the mixing of beef and pork in meatball formulation can be categorized as an adulteration practice [8]. Adulteration of meat species in comminuted and ground products has been a widespread issue in retail markets in the last years [9]. Therefore, the detection of authenticity of food products becomes very important to protect consumers.
The analytical methods capable of identifying non-halal meats with high accuracy, precision and sensitivity, inexpensive, and easily accessible greatly assist the process of authentication supervision [10]. Spectroscopic-based methods, such as Fourier infrared spectroscopy [11] and chromatographic methods [12], especially liquid chromatography combined with an advanced analytical method, mass spectrometry or LC-MS/MS [13,14], and GC-MS combined with powerful chemometrics [15], have been widely applied for identification of non-halal meats; however, spectroscopic method is lack in selectivity and LC-MS/MS need sophisticated instrument, as a consequence, methods based on DNA, especially polymerase chain reaction (PCR) were typically used in the analysis of pork in the meat-based foods.
Analysis using PCR primers, specifically for species, has been applied for specific and sensitive detection of various species of animals derived from products highly processed [16,17]. Additionally, another method of analysis known as quantitative PCR or referred to as qPCR or real-time PCR (RT-PCR) has also been increasingly applied for rapid, specific, and sensitive detection of DNA specifically for species in a various processed products [18]. PCR and its variation including RT-PCR [19], Restriction Fragment Length Polymorphism analysis [20], multiplex-based PCR [21], and PCR specifically for species or known as species-specific PCR [22]. Primer specificity is an essential thing to note on the PCR technique, thus the design of specific primers describes the success of PCR analysis [23]. The purpose of this study is to design new primer specific to pig DNA targeting on mitochondrial gene and the primer as well as to validate RT-PCR in identifying DNA of pig in meatball-based products.
Materials and Methods
Samples, including pork and other animal meats, were obtained from different slaughter houses around Yogyakarta. Meatball samples were prepared in the laboratory, according to Purnomo and Rahardiyan [24] with slight modifications. Laboratory meatball samples were formulated by mixing 90% of beef and pork, which previously were finely ground, with 10% of tapioca flour and added salt and other spices commonly used in making meatballs. The laboratory meatball samples were prepared with different levels of pork. The samples of commercial meatball were bought from local markets randomly in the Province of Yogyakarta, Indonesia.
Design of primers
Pig DNA primers were designed with NCBI-Primer BLAST software in accordance with the desired criteria. In silico, the designed primer was subjected to BLAST or primer-basic local alignment search tool using tools available in website at http://blast.ncbi.nlm.
DNA isolation and purity assessment
The DNA isolation was carried out using Pure Link Genomic DNA Mini Kit, USA (Cat No. K1820-01; Lot No. 1670947) according to the manufacture instruction. The analysis of purity of isolated DNA was conducted by measuring solution containing DNA (20 μl in 980 μl of sterile aquabidest) at a wavelength of 260 and 280 nm. In addition, the concentration of the isolated DNA was measured from the absorbance values on a wavelength of 260 nm (A260) multiplied by the dilution factor and the absorption constant (50 μl/ml) [25].
Optimizing annealing temperature of primer
The optimization of annealing temperature of primer that has been designed for analysis pig DNA was carried out in the temperature ranged from 46°C to 52°C based on the prediction of melting temperature of designed primer. The temperature used for optimization was 46.6°C, 47.1°C, 48.0°C, 49.4°C, and 51.1°C.
Real-time PCR analysis
The analysis of RT-PCR used reaction mixture (20 μl) consisting of 10 μl universal PCR master mix of SYBR Green®, 1 μl of reverse and forward primer, 2 μl of 50 ng template DNA, and 6 μl of water-free nuclease. The temperature program used 94°C for 30 sec, which was followed by 30 cycles at 94°C for 3 sec for denaturation stage. The next stage was the annealing of primer in the annealing temperature 47.1°C that has been optimized for 3 sec in addition to 72°C for 3 sec for the elongation stage. Melting curve was carried out at a temperature of 65°C–95°C with a slope of 0.5°C/5 sec.
Validation of real-time PCR
The primer previously designed was then subjected to method validation according to Codex Alimentarius Comission/GL 74 [26] which included primer specificity, construction of calibration curve for determining efficiency and sensitivity, and determination of repeatability. Primer specificity was confirmed by amplifying 50 ng/μl of pig, cow, chicken, and goat DNAs as well as a negative control (without DNA) called No Template Control (NTC). Preparation of standard curve was carried out by plotting the levels of DNA extracted from prepared laboratory meatballs with concentrations of pork of 0%, 10%, 25%, 50%, 75%, and 100%, respectively (x-axis) and Cycle threshold (Ct) value (y-axis). The equation of linear regression obtained was then used for determination of efficiency value (E). Determining the limit of detection (LoD) was based on the results of amplification on 100% concentration of pork meatballs by serial dilution in order to obtain a concentration of DNA at levels of 0.1, 1, 10, 100, 1,000, and 10,000 pg. The LoD was the smallest concentration that could still give amplification results. Repeatability of RT-PCR method was evaluated by calculating coefficient variation (CV) from six replication times of DNA amplification from meatballs with 100% pork. The validated RT-PCR methods were applied in the analysis of commercial samples available in the market.
Result and Discussion
Primer design and DNA extraction
Primer design was the initial step for DNA analysis using RT-PCR method. Primer design was aimed to obtain specific primer of pig (Sus Scrofa) mitochondrial cytochrome-b (Cyt-b) gene. The selection of this gene was based on degree variability of intra- and inter-species in cyt-b. This gene also offers high number of copies for each cell which increases the sensitivity of real-time assay significantly and contributes to the survival of few copies of DNA when tissue has been subjected to extreme processing condition, for example sterilization and boiling processes [26,27]. There were three pairs of primer candidates from cyt-b gene of pig, called as 1F1R, 2F2R, and 3F3R primers, as shown in Table 1.
Table 1. The primer candidates targeting on mitochondrial cytochrome-b (Cyt-b) for identification of pig (Sus Srofa) DNA.
| Primers* | Sekuen 5′–3′ | Tm (°C) | %GC |
|---|---|---|---|
| 1F1R | F: 5′-ACG CGA TAT AAG CAG GTA AA-3′ | 51.4 | 40.0 |
| R: 5′-CTG CTT TCG TAG CAC GTA TT-3′ | 52.9 | 45.0 | |
| 2F2R | 5′-TCC TAT TCC TGC ACG AAA-3′ | 54.8 | 50.0 |
| 5′-TGC TGG GGT GTA GTT GTC TG-3′ | 57.0 | 55.0 | |
| 3F3R | 5′ GCC CTA GTA GCC TCC ATC CT-3′ | 58.2 | 60.0 |
| 5′ TAG TTG GCC GAT GAT GAT GA-3′ | 53.5 | 45.0 |
Only primer 1F1R used for next study.
Specific primer can be considered as a primer which only amplifies DNA target (porcine DNA) and did not amplify DNAs from other species. The result of primer BLAST showed that only primer pair of 1F1R that attached to pig DNA (Sus Scrofa), and it did not attach to other organisms, such as cow, chicken, and goat. Therefore, from those three primer pairs, primer 1F1R was chosen to be used for the next study. The designed primer pair of 1F1R had primer parameters close to ideal primer, such as the number of base pair of 19–23 pairs, adjacent to melting temperature, and there was no recurrence on G or C on 3′ end that might cause GC clamp. In addition, the length of amplicon less than 250 base pair (bp) could increase the efficiency of PCR amplification [28].
DNA isolation was carried out by PureLink Genomic DNA Mini Kit. Isolation of DNA was aimed to separate DNA from other cell matrices in the cell and was conducted on both fresh meat and meatball, either in the laboratory meatball or commercial meatballs. The process of DNA isolation was carried out several stages, including lysis or destruction of cell membranes, DNA extraction process using organic solvents, refining process, precipitating process, and concentration. DNA isolate from the extraction results was analyzed quantitatively using spectrophotometer on wavelengths of 260 nm (A260) and 280 nm (A280) to obtain DNA concentration and DNA purity. The result of A260/A280 ratio was around 1.80, indicating that the isolated DNA was relatively pure.
Validation of real-time PCR
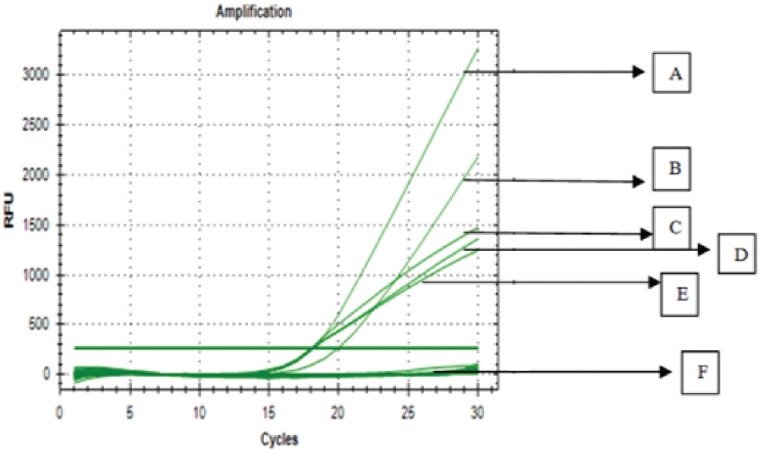
Before validating RT-PCR method, the optimization of temperatures, especially annealing temperature, capable of offering good PCR results must be done. The annealing temperature was carried out in the range of 46.6°C–53.6°C based on the predicted designed primer (F1R1). The temperature used for optimizing was exactly set at 46.6°C, 47.1°C, 48.0°C, 49.4°C, and 51.1°C, respectively. Based on the amplification result of primer F1R1 on porcine DNA, the temperature of 47.1°C was selected to be used due to its capability to provide the highest response of amplification (relative fluorescence unit) with the smallest Cq, as shown in Figure 1.
Figure 1. The amplification curve of pig DNA using primer 1F1R at various annealing temperatures, namely at 47.1°C (A); 46.6°C (B); 51.1°C (C); 48.0°C (D); 49.4°C (E); and NTC (F).
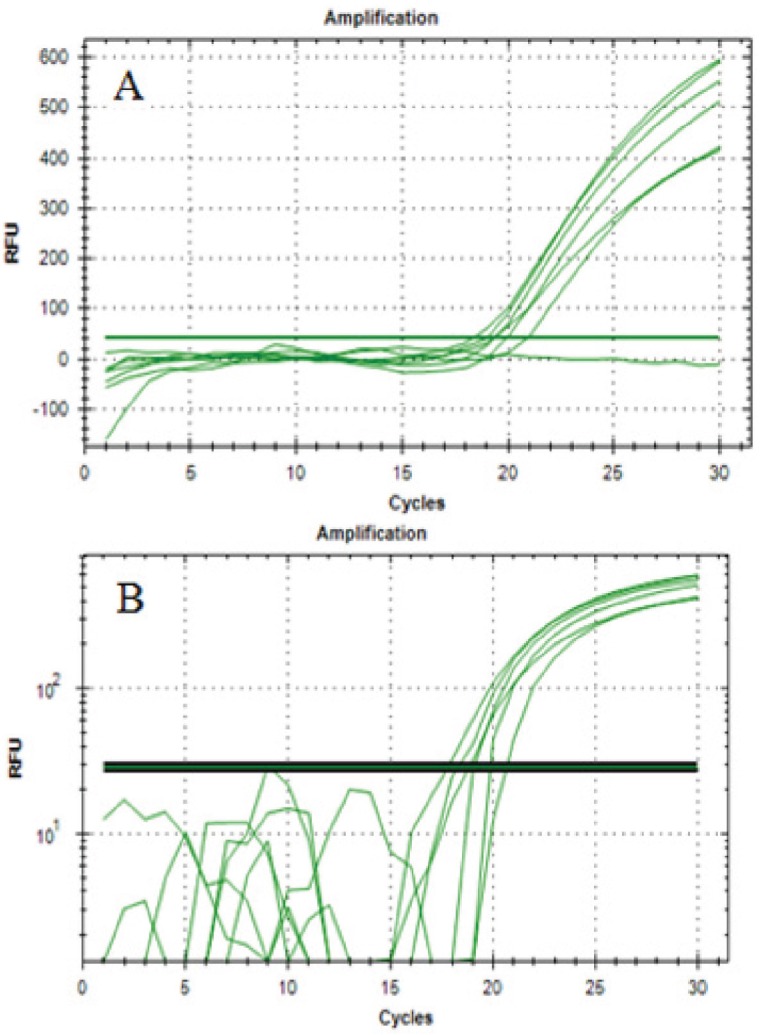
The specificity test of 1F1R primer was conducted using DNA template, isolated from fresh pig, beef, chicken, and goat using optimum annealing temperature which was 47.1°C. The result of amplification showed that only pig DNA could be amplified, while the DNA on beef, chicken, and goat were not amplified (Fig. 2). It means that 1F1R primer was specific on pig DNA supporting BLAST results which had been previously conducted. Repeatability of PCR methods was conducted using meatballs with 100% pork and was evaluated by calculating CV values of quantification cycle (Cq) from six replicates of DNA amplification (Fig. 3). Codex Alimentarius Comission [26] stated that the acceptable CV values for PCR was <25%. The obtained CV values were 4.13%, indicating that RT-PCR was precise for prediction of DNA in meatball samples.
Figure 2. The amplification curve using DNA template, extracted from pig (A), beef (B), chicken (C), goat (D), and NTC (E).
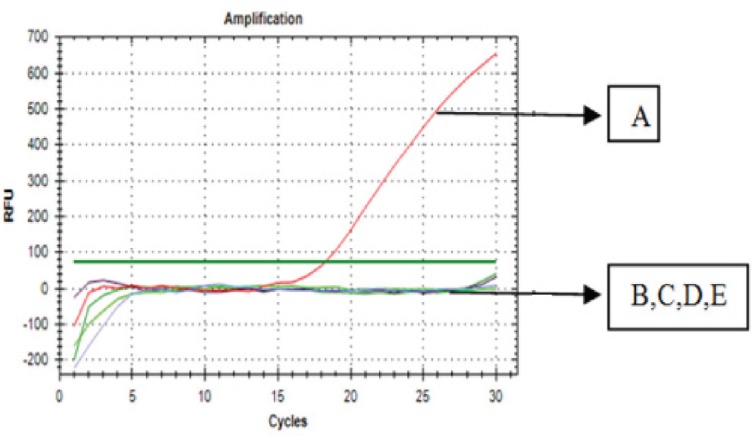
Figure 3. Amplification curve (A) and logarithmic amplification (B) of meatball formulation with concentration 100% pork meat (number of replication was 6).
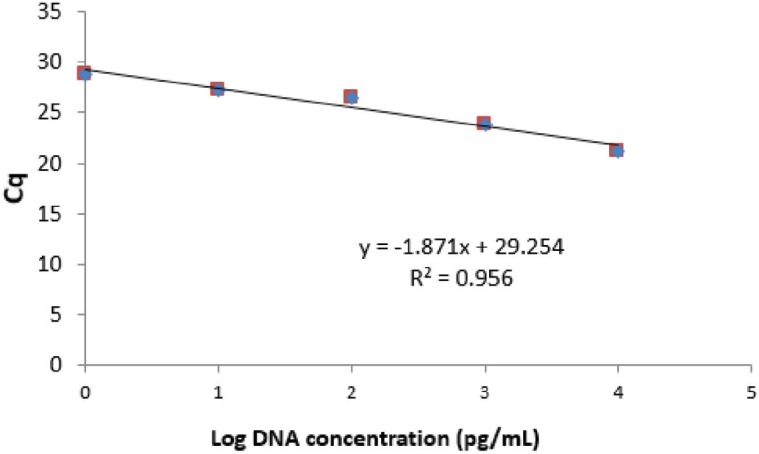
Determination of LoD was conducted on pork meatball 100% which was diluted serially to obtain DNA with final concentrations of concentration of 0.1, 1, 10, 100, 1,000, and 10,000 pg, respectively. Figure 4 showed that at concentration of 1 pg, DNA could be amplified with Cq value of 28.79. In addition, at lower concentration of DNA (<1 pg), there was no amplification observed until the 30th cycles. Based on this result, it could be concluded that the LoD value of pig DNA using primer 1F1R in combination with RT-PCR was 1 pg. Standard curve describing the correlation between logarithm of pork DNA concentration (x-axis) with Cq value (y-axis) was shown in Figure 5. The efficiency value (E) of PCR amplification was 242.58%, calculated from the slope of −1.871 exceeding acceptance criteria from Codex [26], which is 90%–110% with coefficient of the determination value (R2) of 0.956 [29]. The E-value out of acceptance criteria may be due to less pipetting precision as well as methods of DNA extraction [30].
Figure 4. The amplification curve of pig DNA extracted from pork meatball 100% with DNA concentrations of 0.1 pg (F), 1 pg (E), 10 pg (D), 100 pg (C), 1,000 pg (B), and 10,000 pg (A).
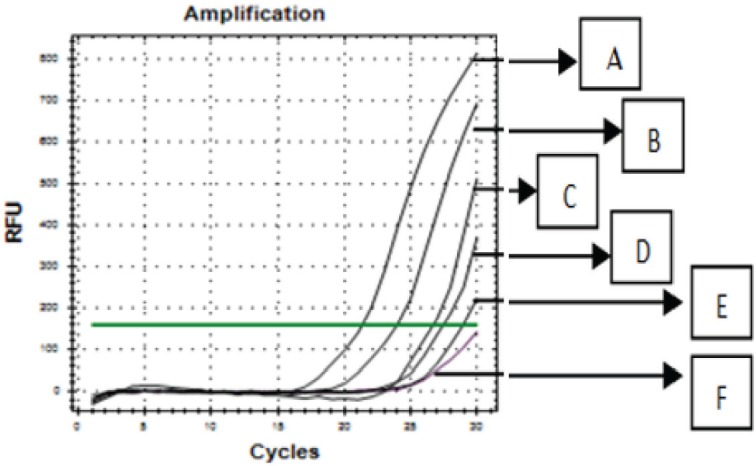
Figure 5. Standard curve for the correlation between logarithm of pork DNA concentration (x-axis) with Cq value (y-axis).
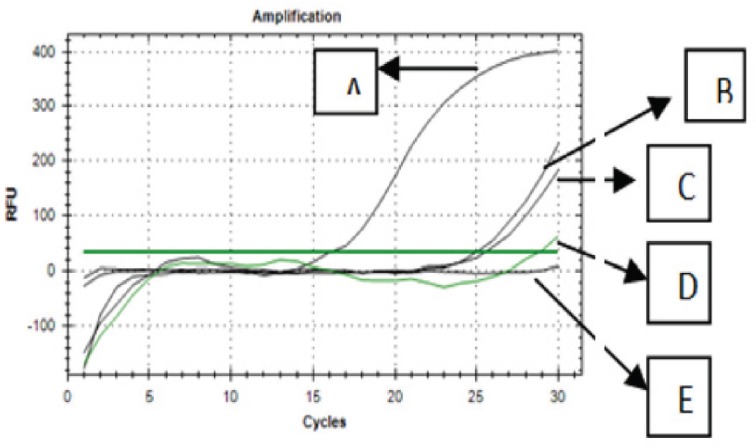
The validated RT-PCR method using primer 1F1R was used to amplify DNAs extracted from commercial samples along with positive control (containing porcine DNA) and no negative control (NTC) using annealing temperature of 47.1°C for 30 cycles. The result showed that all the samples were amplified (Fig. 6). Regarding to the result of this study, it could be confirmed that commercial samples were contaminated with pork.
Figure 6. The amplification curves of DNAs extracted from three commercial samples (B, C, and D) and positive control (A) and no template control (E).
Conclusion
Primer pairs of 1F1R 5′-ACG CGA TAT AAG CAG GTA AA-3′ (forward) and 5′-CTG CTT TCG TAG CAC GTA TT-3′ (reverse) targeting on mitochondrial cytochrome-b (Cyt-b) was specific to be used for the identification of pig DNA using RT-PCR with the optimum annealing temperature was 47.1°C. The validated RT-PCR using primer F1R1 could be proposed as a standard method for detection of pork DNA in commercial samples.
Acknowledgments
The authors would like to thank the Ministry of Research and Higher Education, Republic of Indonesia for financial support via project “Penelitian Unggulan Perguruan Tinggi (PUPT)” year 2017.
Conflict of Interest
There is no conflict of interest to declare.
Authors’ contribution
Salmah Orbayinah and Hari Widada designed and conducted research activities and prepared the manuscript. Adam Hermawan, Sismindari Sudjadi, and Abdul Rohman designed research, analyzed data, and prepared the manuscript.
References
- [1].Rohman A. The employment of Fourier transform infrared spectroscopy coupled with chemometrics techniques for traceability and authentication of meat and meat products. J Adv Vet Anim Res. 2019;6:9–17. doi: 10.5455/javar.2019.f306. http://doi.org/10.5455/javar.2019.f306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Rahmati S, Julkapli NM, Yehye WA, Basirun WJ. Identification of meat origin in food products: a review. Food Control. 2016;68:379–90. https://doi.org/10.1016/j.foodcont.2016.04.013. [Google Scholar]
- [3].Montowska M, Pospiech E. Authenticity determination of meat and meat products on the protein and DNA basis. Food Rev Int. 2011;27:84–100. https://doi.org/10.1080/87559129.2010.518297. [Google Scholar]
- [4].Murugaiah C, Noor ZM, Mastakim M, Bilung LM, Selamat J, Radu S. Meat species identification and Halal authentication analysis using mitochondrial DNA. Meat Sci. 2009;83(1):57–61. doi: 10.1016/j.meatsci.2009.03.015. https://doi.org/10.1016/j.meatsci.2009.03.015. [DOI] [PubMed] [Google Scholar]
- [5].Valdés A, Beltrán A, Mellinas C. Jiménez A, Garrigós MC. Analytical methods combined with multivariate analysis for authentication of animal and vegetable food products with high fat content. Trends Food Sci Technol. 2018;77:120–30. https://doi.org/10.1016/j.tifs.2018.05.014. [Google Scholar]
- [6].Laliotis GP, Koutsouli P, Bizelis LA. Implementation of contemporary DNA based techniques on traceability process of small ruminant species and products. J Adv Vet Anim Res. 2018;5(3):255–64. http://doi.org/10.5455/javar.2018.e274. [Google Scholar]
- [7].Fajardo V, González-Isabel I, Rojas M, García T, Martín R. A review of current PCR-based methodologies for the authentication of meats from game animal species. Trends Food Sci Technol. 2010;21:408–21. https://doi.org/10.1016/j.tifs.2010.06.002. [Google Scholar]
- [8].Ali ME, Razzak MA, Hamid SBA, Rahman MM, Amin MA, Rashid NRA, et al. Multiplex PCR assay for the detection of five meat species forbidden in Islamic foods. Food Chem. 2015;177:214–24. doi: 10.1016/j.foodchem.2014.12.098. https://doi.org/10.1016/j.foodchem.2014.12.098. [DOI] [PubMed] [Google Scholar]
- [9].Asensio L. González I, García T, Martín R. Determination of food authenticity by enzyme-linked immunosorbent assay (ELISA). Food Control. 2008;19:1–8. https://doi.org/10.1016/j.tifs.2010.06.002. [Google Scholar]
- [10].Abu-Ghoush M, Fasfous I, Al-Degs Y, Al-Holy M, Issa AA, Al-Reyahi AY, et al. Application of mid-infrared spectroscopy and PLS-Kernel calibration for quick detection of pork in higher value meat mixes. Food Meas. 2017;11:337–46. https://doi.org/10.1007/s11694-016-9402-4. [Google Scholar]
- [11].Rohman A, Putri AR. The chemometrics techniques in combination with instrumental analytical methods applied in halal authentication analysis. Indones J Chem. 2019;19:262–72. https://doi.org/10.22146/ijc.28721. [Google Scholar]
- [12].Rohman A, Sismindari Erwanto Y, Man YBC. Analysis of pork adulteration in beef meatball using fourier transform infrared (FTIR) spectroscopy. Meat Sci. 2011b;88:91–5. doi: 10.1016/j.meatsci.2010.12.007. https://doi.org/10.1016/j.meatsci.2010.12.007. [DOI] [PubMed] [Google Scholar]
- [13].von Bargen C, Brockmeyer J, Hump H-U. Meat authentication: a new HPLC-MS/MS based method for the fast and sensitive detection of horse and pork in highly processed food. J Agr Food Chem. 2014;62:9428–35. doi: 10.1021/jf503468t. https://doi.org/10.1021/jf503468t. [DOI] [PubMed] [Google Scholar]
- [14].von Bargen C Dojahn, Jr, Waidelich D, Humpf H-U, Brockmeyer J. New sensitive high-performance liquid chromatographyetandem mass spec- trometry method for the detection of horse and pork in halal beef. J Agr Food Chem. 2013;61:11986–94. doi: 10.1021/jf404121b. https://doi.org/10.1021/jf404121b. [DOI] [PubMed] [Google Scholar]
- [15].Indrasti D, Che Man YB, Mustafa S, Hashim DM. Lard detection based on fatty acids profile using comprehensive gas chromatography hyphenated with time-of-flight mass spectrometry. Food Chem. 2010;122:1273–7. https://doi.org/10.1016/j.foodchem.2010.03.082. [Google Scholar]
- [16].Dalmasso A, Fontanella E, Piatti P, Civera T, Rosati S, Bottero MT. A multiplex PCR assay for the identification of animal species in feedstuffs. Mol Cell Probes. 2004;18:81–7. doi: 10.1016/j.mcp.2003.09.006. https://doi.org/10.1016/j.mcp.2003.09.006. [DOI] [PubMed] [Google Scholar]
- [17].Fumiere O, Dubois M, Baeten V, von Holst C, Berben G. Effective PCR detection of animal species in highly processed animal byproducts and compound feeds. Analyt Bioanalyt Chem. 2006;385:1045–54. doi: 10.1007/s00216-006-0533-z. https://doi.org/10.1007/s00216-006-0533-z. [DOI] [PubMed] [Google Scholar]
- [18].Kesmen Z, Gulluce A, Sahin F, Yetim H. Identification of meat species by TaqMan-based real-time PCR assay. Meat Sci. 2009;82:444–9. doi: 10.1016/j.meatsci.2009.02.019. https://doi.org/10.1016/j.meatsci.2009.02.019. [DOI] [PubMed] [Google Scholar]
- [19].Rodriguez MA, Garcia T, Gonzalez I, Hernandez PE, Martin R. TaqMan real-time PCR for the detection and quantitation of pork in meat mixtures. Meat Sci. 2005;70:113–20. doi: 10.1016/j.meatsci.2004.12.005. https://doi.org/10.1016/j.meatsci.2004.12.005. [DOI] [PubMed] [Google Scholar]
- [20].Chen S-Y, Liu Y-P, Yao Y-G. Species authentication of commercial beef jerky based on PCR–RFLP analysis of the mitochondrial 12S rRNA gene. J Genet Genom. 2010;37:763–9. doi: 10.1016/S1673-8527(09)60093-X. https://doi.org/10.1016/S1673-8527(09)60093-X. [DOI] [PubMed] [Google Scholar]
- [21].Ghovvati S, Nassiri MR, Mirhoseini S, Moussavi AH, Javadmanesh A. Fraud identification in industrial meat products by multiplex PCR assay. Food Control. 2009;20:696–9. https://doi.org/10.1016/j.foodcont.2008.09.002. [Google Scholar]
- [22].Man YBC, Aida AA, Raha AR, Son R. Identification of pork derivatives in food products by species–specific polymerase chain reaction (PCR) for halal. Food Control. 2007;18:885–9. https://doi.org/10.1016/j.foodcont.2006.05.004. [Google Scholar]
- [23].Martin M, García T, Fajardo V, Rojas M, Pegels N. Hernández PE. SYBR-Green real-time PCR approach for the detection and quantification of pig DNA in feedstuffs. Meat Sci. 2009;82:252–9. doi: 10.1016/j.meatsci.2009.01.023. https://doi.org/10.1016/j.meatsci.2009.01.023. [DOI] [PubMed] [Google Scholar]
- [24].Purnomo H, Rahardiyan D. Indonesian traditional meatball. Int Food Res J. 2008;15:101–8. [Google Scholar]
- [25].Maryam S, Sismindari Raharjo TJ, Sudjadi Rohman A. Analysis of porcine contamination in dendeng using mitochondrial D-Loop 686 and Cyt B gene primers by real-time polymerase chain reaction. Int J Food Prop. 2016;19(1):187–95. https://doi.org/10.1080/10942912.2015.1020434. [Google Scholar]
- [26].Codex Alimentarius Comission/GL 74. Joint FAO/WHO Food Standards Programme, FAO. Rome, Italy: 2010. Codex guidelines on performance criteria and validation of methods for detection, identification, and quantification of spesific DNA sequences and spesific proteins in foods. [Google Scholar]
- [27].Girish PS, Anjaneyulu ASR, Viswas KN, Anand M, Rajkumar N, Shivakumar BM. Sequence analysis of mitochondrial 12S rRNA gene can identify meat species. Meat Sci. 2004;66:551–6. doi: 10.1016/S0309-1740(03)00158-X. https://doi.org/10.1016/S0309-1740(03)00158-X. [DOI] [PubMed] [Google Scholar]
- [28].Widyasari YI, Sudjadi RA, Rohman A. Analysis of rat meat in meatball formulation using real time polymerase chain reaction. Asian J Anim Sci. 2015;9(6):460–5. https://doi.org/10.3923/ajas.2015.460.465. [Google Scholar]
- [29].Svec D, Tichopad A, Novosadova V, Pfaffl MW, Kubista M. How good is a PCR efficiency estimate: recommendations for precise and robust qPCR efficiency assessments. Biomol Detect Quantificat. 2015;3:9–16. doi: 10.1016/j.bdq.2015.01.005. https://doi.org/10.1016/j.bdq.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Muhammed MA, Bindu BSC, Jini R, Prashanth KVH, Bhaskar N. Evaluation of different DNA extraction methods for the detection of adulteration in raw and processed meat through polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) J Food Sci Technol. 2015;52:514–20. https://doi.org/10.1007/s13197-013-1024-9. [Google Scholar]


