Abstract
Objective:
The purpose of this study was to detect the incidence of multi-drug resistant (MDR) and the spread of tet genes that encode tetracycline (TE) resistance in E. coli in pig farms in the city of Kupang, Indonesia.
Materials and Methods:
Samples of pig feces have been obtained from 96 pig farms in Kupang city, Indonesia. Escherichia coli bacteria were isolated and identified morphologically and biochemically, and finally confirmed by the API test. The disk diffusion method has been used to observe the antibiotic sensitivity effects and has been followed by observing resistant genes encoding TE resistance using the multiplex polymerase chain reaction (m-PCR) method to detect the presence of tet genes such as tet (A), tet (B), tet (C), tet (D), and tet (E), respectively.
Results:
A total of 82 (85.4%) of E. coli isolates have been found in all pig feces samples obtained from 96 pig farms in Kupang city. This study has shown a high level of antibiotic resistance dominated by erythromycin (85.4%) and cephalothin (58.5%) and followed by several other antibiotics with a percentage below 34.1%. The prevalence of MDR E. coli was 57.3% by showing 39 different patterns. The most common pattern was showed by the Cephalothin-Colistin-Erythromycin pattern. The resistance of E. coli to TE appears to be related to the presence of tet (A) and tet (E) genes.
Conclusion:
This study has encouraged the need for public awareness (farmers) of the wise use of antibiotics in preventing the spread of resistant bacteria that can cause health problems in animals and humans.
Keywords: Antibiotics, E. coli, tet genes, multi-drug resistance, pig farms
Introduction
Antibiotic resistance is a problem that arises throughout the world with a threat to the health and welfare of humans and animals [1]. O’Neill [2] in his report has shown that the estimated threat of the emergence of antibiotic resistance if not controlled can lead to an increase in mortality rates of up to 10 million people each year due to antibiotic-resistant bacteria in 2050. Pathogenic bacteria that have been resistant can develop in humans, animals, and the environment, disrupting public health due to unwise use of antibiotics (misuse and overuse) in various sectors (livestock and human environment). Livestock (the agricultural sector) as a place of animal production is one of the places where the development of antibiotic-resistant bacteria due to the use of antibiotics in large quantities is not only for therapeutic purposes but also as a growth promoter to increase livestock production [3]. The same opinion has been explained by Hu et al. [4] which inform that livestock was a reservoir of genes responsible for antibiotic resistance. Several studies have shown resistance in some antibiotic classes such as penicillin [5], cephalosporins [6], tetracycline (TE) [7,8], polymyxin [9], sulfonamides [8], and macrolides [5].
Besides the occurrence of antibiotic resistance in pathogenic bacteria, it has also been found in commensal bacteria such as E. coli. Similar opinion was also presented by Xia et al. [10] which states that E. coli as one of the commensal bacteria have become resistant to more than one type of antibiotics. Skočková et al. [11] also show that E. coli has become a reservoir for antibiotic-resistant genes and has the ability to transfer genes that encode the nature of antibiotic resistance to other bacterial species including pathogenic bacteria [2,12]. The presence of E. coli has become important because E. coli is the most dominant microflora in the digestive tract of humans and animals, has been found in large amounts in feces [13], and has become the most common contaminant in meat. This situation caused World Organization for Animal Health (OIE) to choose E. coli as one of the bacteria used as an indicator in the antibiotic resistance monitoring program [14]. The OIE has recommended that each country conduct an antibiotic resistance control program to prevent the spread of antibiotic-resistant bacteria [15].
The level of resistance has become increasingly dangerous with the emergence of bacteria that have become resistant to three or more antibiotic classes known as multi-drug resistant (MDR) [16]. Research conducted by Petternel et al. [17] showed the presence of E. coli bacteria that have become MDRs in animal products, as well as the results [5] of which shows the presence of MDR E. coli in livestock. This condition has become increasingly worrying regarding the existence and spread of MDR bacteria because it can endanger public health.
Antibiotic resistance gene is a gene that has been resistant to bacteria so that if bacteria have this gene it will cause the emergence of resistance in the bacteria. The antibiotic resistance gene can be spread horizontally by E. coli to other members of the Enterobacteriaceae family via the plasmid through a conjugation process [18]. Tetracycline resistant genes that can be found in resistant E. coli bacteria include tet (A), tet (B), tet (C), tet (D), and tet (E) causing the system “efflux pump” [11,19,20]. Rapid and broad developmental ability by E. coli bacteria demonstrate the need to improve our understanding of factors that influence the spread of antibiotic resistance between humans and animals so that appropriate interventions can be taken to deal with it. OIE [15] suggests the need for action to evaluate the role of bacteria contained in feces (commensal) such as E. coli in spreading antibiotic-resistant genes and mechanisms for the spread of resistance in the environment.
The city of Kupang has been known as a region with a high pig population [21] as a result of increasing demand for pork from year to year. To maintain the level of pig production has been done by maintaining the quality of health of livestock. The health of pigs has been obtained by using antibiotics as a treatment. Antibiotics have been found easily and in large quantities at veterinary drug stores, especially TEs at low prices and can be purchased with or without veterinary prescriptions. Improper use of antibiotics in pigs has led to the emergence of antibiotic residues in meat and on the other hand, can allow for the emergence of antibiotic-resistant bacteria. Furthermore, Gebreyes et al. [22] have found that antibiotic resistance is found in areas with high pig counts. The presence of TE in meat has been shown by Kale et al. [23] which shows the presence of TE residues in pigs in Kupang city.
In Indonesia, research that has been conducted on hygiene and sanitation related to E. coli contaminated in pig farms is still very limited, so research has been conducted with the aim of detecting MDR E. coli incidents and the spread of tet genes that encode resistance to TE antibiotics on farms pigs in Kupang City and also as an effort to monitor the spread of resistance.
Materials and Methods
Time and place of study
Sampling for this study was conducted in March 2017 until February 2018. Sampling has been carried out on intensive pig farms (household scale) in Kupang city. The laboratory where testing has been carried out in several places includes: (1) isolation and identification of bacteria has been held at the Laboratory of the Veterinary Technical Implementation Unit of the Livestock Service Office of East Nusa Tenggara Province, (2) confirmation of E.coli bacteria by API test and sensitivity test by disk diffusion method (Kirby–Bauer method) has been conducted in the Laboratory of Veterinary Public Health, Faculty of Veterinary Medicine of Bogor Agricultural University, (3) Detection of the tet gene with multiplex-PCR genes has been carried out at the Laboratory of the Veterinary Research Institute (Balai Besar Penelitian Veteriner / Indonesian Research Center for Veterinary Science) Bogor, Indonesia.
Magnitude and sampling techniques
Data have been collected through direct observation of 96 pig feces samples obtained from 96 pig farms in Kupang City, Indonesia. Samples have been taken from pigs showing healthy conditions.
Isolation and identification of E. coli
Microbiological testing has been carried out based on references from the Indonesian National Standard on microbial contamination methods [24]. This method has been carried out by isolation and identification with biochemical tests: indole, methyl red, Voges-Proskauer and citrate (IMViC) and Gram stain, KOH test, and oxidase test. The following are the steps in carrying out the isolation and identification of E. coli as follows: The first stage (sample preparation) is 25 gm of pig fecal sample dissolved in 225 ml 0.1% buffered peptone water and homogenized using a stomacher to make a 10−1 dilution. The second stage was 10 ml of homogenate transferred to a sterile reaction tube and incubated for 24 h at 37°C. The third stage was isolation and cultivation on MacConkey agar medium and incubated in aerobic conditions at 37°C for 24 h. Colonies with a round shape, 1.1–1.5 μm (width) and 2.0–6.0 μm (length) in size, red, and surrounded by turbid zones suspected as E. coli. The fourth step was identified the suspected E. coli by Gram stain test, KOH test, oxidase test, and IMViC biochemical test. Positive E. coli bacteria indicated by the characteristics in the gram staining test showed negative gram bacteria, positive KOH test, and negative oxidase test. Furthermore, all positive colonies were subcultured on Trypticase Soy Agar (TSA) media and incubated at 37°C for 24 h. Isolates are stored in Trypticase Soy Broth (TSB) containing 30% glycerol at −20°C until the subsequent execution.
Isolates that have been suspected as E. coli were confirmed to find out the species level using the API 20E test kit (Biomereaux). For refreshment, the isolate was cultivated on TSB and incubated at 37°C for 24 h, then cultivated on TSA and incubated at 37°C for 24 h. From separate coloniestwo to five colonies were taken with a sterile cotton swab and mixed into 5 ml of 0.85% sterile NaCl until turbidity was equal to 0.5 McFarland or equivalent to 1–2 × 108 CFU/ml. The bacterial suspension was then applied to the API 20E test kit and incubated at 37°C for 24 h. Finally, reading the test results by using the API 20E test kit was done through the ApiwebTM application.
Antibiotics sensitivity test method
Testing the sensitivity of E. coli bacteria to antibiotics was carried out using the Kirby–Bauer method (disc diffusion method) which refers to the method published by the Clinical and Laboratory Standards Institute [25]. The bacterial isolate was determined for its antimicrobial sensitivity by measuring the inhibitory zone. Meanwhile, the determination of susceptibility (S), intermediate (I), and resistant (R) was determined by the size of the inhibitory zone formed based on the standard interpretation of the diameter of the zone of antibiotic inhibition. Antibiotics used in this study include: amoxicillin (AML) 25 μg (CT0061B-Oxoid), cephalothin (KF) 30 μg (CT0010B-Oxoid), cefotaxime (CTX) 30 μg (CT0166B-Oxoid), TE 30 μg (CT0054B-Oxoid), doxycycline (DO) 30 μg (CT0018B-Oxoid), colistin sulfate (CT) 10 μg (CT0017B-Oxoid), trimethoprim-sulfamethoxazole (SXT) 25 μg (CT0052B-Oxoid), streptomycin (S) 10 μg (CT0047B-Oxoid), gentamicin (CN) 10 μg (CT0024B-Oxoid), erythromycin (E) 15 μg (CT0020B-Oxoid), 30 μg nalidixic acid (NA) (CT0031B-Oxoid), and 5 μg ciprofloxacin (CIP) (CT0425B-Oxoid). In each test, discs without antimicrobial ingredients (blank discs; CT0998B-Oxoid) were used as negative controls.
Detection methods of tetracycline-resisted genes on isolate E. coli
Detection of the antibiotic resistance gene in E. coli against TE antibiotics was carried out using the Multiplex-PCR method. Tests using Multiplex-PCR were carried out referring to Skočková et al. [11]. Tests have been carried out to determine the presence of resistance genes to TE antibiotics including tet (A), tet (B), tet (C), tet (D), and tet (E) genes found in E. coli and using primers according to Ng et al. [19] (Table 1).
Table 1. Details of the primers used for the detection of tet (A), tet (B), tet (C), tet (D), and tet (E) genes.
| Gene | Primer | Primer sequence (5'–3') | Size (bp) | Reference |
|---|---|---|---|---|
| tet A |
tet A–F tet A–R |
GCT ACA TCC TGC TTG CCT TC CAT AGA TCG CCG TGA AGA GG |
210 | Ng et al. [25] |
| tet B |
tet B–F tet B–R |
TTG GTT AGG GGC AAG TTT TG GTA ATG GGC CAA TAA CAC CG |
659 | Ng et al. [25] |
| tet C |
tet C–F tet C–R |
CTT GAG AGC CTT CAA CCC AG ATG GTC GTC ATC TAC CTG CC |
418 | Ng et al. [25] |
| tet D |
tet D–F tet D–R |
AAA CCA TTA CGG CAT TCT GC GAC CGG ATA CAC CAT CCA TC |
787 | Ng et al. [25] |
| tet E |
tet E–F tet E–R |
AAA CCA CAT CCT CCA TAC GC AAA TAG GCC ACA ACC GTC AG |
278 | Ng et al. [25] |
Before the test was performed, bacterial isolates were first collected and cultivated in blood agar and incubated at 37°C for 24 h and bacterial DNA was extracted from a single colony using QIAamp DNA Qiagen Minikit (Qiagen). This test was carried out using PCR (LabCycler) apparatus with a total volume of reagents and a material used from 20 μl where the main pair each contained 0.5 μl tet (A) (12.5 pmol/μl), 0.6 μl tet (B) (15 pmol/μl), 0.6 μl tet (C) (15 pmol/μl), 0.8 μl tet (D) (20 pmol/μl), and 0.4 μl tet (E) (10 pmol/μl), 10 μl mastermix (2× KAPA 2G Readymix PCR Kit Fast Hotstart Kit), 2 μl dH2O, and 2 μl DNA templates from the sample tested.
The test cycle has been carried out including denaturation at 95°C for 3 min and followed by 35 multiplication cycles of DNA strands. The cycle consists of the denaturation phase at 95°C for 15 sec, annealing at 53°C for 15 sec and amplification (stretching) of the DNA strand at 72°C for 30 min. The DNA strand multiplication cycle ends with an amplification phase at 72°C for 10 min. The PCR results in the form of a DNA strand has been duplicated then read through the electrophoresis stage on 1.5% agarose medium by giving ethidium bromide dye (0.5 mg/ml) and visualizing using ultraviolet light. The reference bacteria used in this resistance test was based on E. coli isolates from the American Type Culture Collection type 25922.
Data analysis
The data that has been obtained were displayed in the form of tables and graphs that are used to describe the incidence of E. coli contamination in pig feces in Kupang City and analyzed descriptively related to the presence of MDR E. coli contamination in pig feces and spreading of tet gene in pig farms in Kupang city.
Results and Discussion
Prevalence of Escherichia coli in sample population
This study was conducted to determine the resistance profile of E. coli in pig farms in Kupang city and found 82 (85.40%) E. coli isolates from 96 samples. This prevalence is almost the same as that found by Urumova [20]. The high prevalence rates on farms were caused by E. coli as a commensal and pathogenic bacterium and have been found in pig farms and the surrounding environment [26]. Most pig farms in Kupang city were traditional or household-scale farms, their pens were located not far from the home environment and have low hygiene and sanitation [27]. This condition has caused the spread of E. coli bacteria from the cage to the environment (humans, other animals, and the enclosure environment) and vice versa is very possible.The presence and endurance of E. coli in pig feces have been affected by the temperature of the environment, hygiene, and sanitation of the cage (which is done on the farm). Several studies in the city of Kupang have shown the presence of E. coli contamination in pork meat (processed meat of pig origin) [28–30], refill drinking water [31], and groundwater [32]. The high number of E. coli has given side effects related to the spread of antibiotic-resistant genes in animals, animal products, and the environment [11].
Escherichia coli could be a reservoir for antibiotic-resistant genes and can transfer genes horizontally to similar bacteria or other types through conjugation (transfer of resistant genes via plasmids or other genetic material such as transposons and integrons) [33]. Furthermore, because of the important information about these bacteria in studying resistance levels in the environment, OIE has determined E. coli (commensal) and Salmonella sp. (pathogen) as an indicator bacteria for conditions of antibiotic resistance in animals and the environment in monitoring and surveillance programs [14].
Prevalence of Escherichia coli resistant
A total of 82 E. coli isolates were examined by agar diffusion method (Kirby–Bauer) to determine the resistant profile of E. coli against 12 antibiotics (eight classes of antibiotics). The results of antibiotic resistance in Table 2 have shown that a high level of resistance was found in antibiotics of erythromycin (85.4%) and KF (58.5%). Some antibiotics have also been shown to have a large tendency to increase which can be found in some antibiotics such as colistin sulfate (34.1%), streptomycin (31.7%), TE (29.3%), and AML (28%), this has become the next threat to be noticed. Whereas for a number of other antibiotics have been found at low levels of resistance (SXT 14.6%, CTX 12.2%, DO 12.2%, CN 4.9%, NA 2.4%, and CIP 1.2%). Another important thing was also to have found the potential to be resistant to some antibiotics that have intermediate levels such as CTX (52.7%), AML (42.7%), KF (32.9%), streptomycin (20.7% ), and NA (20.7%). This needs to be watched out because it could be a big threat if the control of antibiotic used was not considered properly in the future. The observed E. coli isolates have shown that 15% (12/82) have been resistant to one antibiotic while 82% (67/82) have been resistant to more than one antibiotic. E. coli strains have shown high variations in the emerging resistance patterns. The total number of isolates has shown that resistance to three types of antibiotics (23%) was the highest and followed by four types of antibiotics (22%). The number of antibiotics tested from E. coli isolates has shown various variations from 0 to a maximum of eight antibiotics that are resistant to one isolate and resistant to antibiotics Erythromycin (macrolide) was the most commonly found (Table 2).
Table 2. Percentage of antibiotic resistance, number of resistant antibiotics, and MDR incidence in pig farm Kupang city, Indonesia.
| Types of antibiotics | Antibiotic class | Breakpoints [8] | Number and percentage of resistance | Number and percentage of intermediates | Number and percentage of susceptible | |||
|---|---|---|---|---|---|---|---|---|
| R/I/S (mm) | Σ | % | Σ | % | Σ | % | ||
| AML | Penicillin | ≤13/14–17/≥18 | 23 | 29.1 | 35 | 44.3 | 24 | 30.4 |
| KF | Cephalosporins | ≤14/15–17/≥18 | 48 | 60.8 | 27 | 34.2 | 7 | 8.9 |
| CTX | ≤22/23–25/≥26 | 10 | 12.7 | 44 | 55.7 | 28 | 35.4 | |
| TE | Tetracycline | ≤11/12–14/≥15 | 24 | 30.4 | 2 | 2.5 | 56 | 70.9 |
| DO | ≤10/11–13/≥14 | 10 | 12.7 | 14 | 17.7 | 58 | 73.4 | |
| CT | Polymyxin | ≤10/–/≥11 | 28 | 35.4 | 0 | 0.0 | 54 | 68.4 |
| SXT | Diaminopyrimidine-Sulfonamide | ≤10/11–15/≥16 | 12 | 15.2 | 0 | 0.0 | 70 | 88.6 |
| S | Aminoglycosides | ≤11/12–14/≥15 | 26 | 32.9 | 17 | 21.5 | 39 | 49.4 |
| CN | ≤12/13–14/≥15 | 4 | 5.1 | 3 | 3.8 | 75 | 94.9 | |
| E | Makrolide | ≤13/14–22/≥23 | 70 | 88.6 | 12 | 15.2 | 0 | 0.0 |
| CIP | Fuorokuinolon | ≤15/16–20/≥21 | 1 | 1.3 | 4 | 5.1 | 77 | 97.5 |
| NA | ≤13/14–18/≥19 | 2 | 2.5 | 17 | 21.5 | 63 | 79.7 | |
CT = colistin sulphate.
The uncontrolled use of antibiotics for erythromycin in pig farms has been one of the causes of resistance to erythromycin [34]. This resistance has been caused by protein synthesis of erythromycin inhibitors, more specifically due to the emergence of methyl groups produced by E. coli which have become inhibitors of erythromycin in binding to the 50S ribosome subunit [35]. Erythromycin ribosome methylation (erm) genes have been responsible for the occurrence of erythromycin resistance that occurs in the environment, as shown by Koike et al. [36] related to the presence of genes such as ermB, ermC, and ermF found in pig farms areas. Some countries have banned the use of antibiotics as growth promoters, but Schipp [37] informs that erythromycin was still used as a growth promoter in Australia.
Wasiński et al. [38] have revealed the number of cephalosporin resistance occurring on farms. The incidence of resistance to cephalosporins has been widely reported and has a lot to do with pig farming [39]. The use of cephalotine antibiotics in pig farms in Indonesia has rarely been done with regard to the clinical management of animals but these antibiotics are more commonly used in poultry farms. The incidence of cephalosporin resistance has been widely reported in poultry farms, which has raised fears of cross-contamination from poultry farms to other livestock, humans, and the environment. At present, the extended-spectrum β-lactamase (ESBL) has been the cause of resistance to class 1 and 3 antibiotic cephalosporins such as KF, CTX that occur in humans and animals and in the environment. ESBL E. coli has been widely reported to be found around the enclosure of pig farms, as reported by Von Salviati et al. [40] in Germany.
Attention must also be directed to colistin sulfate antibiotics because colistin sulfate has been considered as one of the last drugs to treat MDR bacteria has shown a threat to the ability of this drug because of the development of antibiotic-resistant bacteria and this has also been supported by Shen et al. [41]. The spread of the coding resistance gene was resistant to colistin by mcr-1, mcr-2, and mcr-3 have been responsible for this [42].
Prevalence and pattern of MDR Escherichia coli
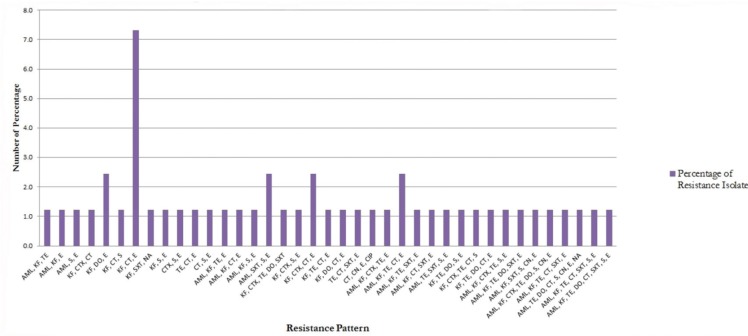
The prevalence of MDR E. coli in pig farms in the city of Kupang has shown a high percentage.From the data that has been obtained, overall around 57.3% (47/82) have shown MDR E. coli, which has been resistant to three or more classes of antibiotics. Escherichia coli isolates from six sub-districts in Kupang city had different resistance patterns, both in MDR E. coli and non-MDR E. coli isolates. The most common resistance pattern in non-MDR E. coli isolates was the E (Erythromycin) pattern and was followed by the KF-E pattern (Cephalothin-Erythromycin) and S–E (Streptomycin–Erythromycin). The most common resistance pattern in MDR E. coli isolates is the KF-CT-E pattern (Cephalothin-Colistin-Erythromycin) (Fig. 1). In general, resistance to antibiotics of erythromycin and KF has dominated the emerging resistance patterns.
Figure 1. Percentage of antibiotic resistance pattern on MDR E. coli. AML = Amoxicillin; KF = cephalothin; CTX = cefotaxime; TE = tetracycline; DO = doxycycline; CT = colistin sulphate; SXT = trimethoprim-sulfamethoxazole; S = streptomycin; CN = gentamicin; E = erythromycin; NA = nalidixic acid; CP = ciprofloxacin; and MDR = multi-drug resistance.
A high pattern of observations of multi-drug resistance has shown that pigs are a reservoir for antibiotic-resistant genes. Zhang et al. [8] has also shown that E. coli with high MDR properties has experienced an increase in numbers in pig farms, as well as research conducted by Park et al. [26] have shown the consequences of high antibiotic use which cause high levels of MDR in E. coli bacteria released into the environment (water environment) around pig farms by being associated with the number of integrons as carriers of resistant genes to the environment. MDR events often occur in ESBL and methicillin resistant Staphylococcus aureus-producing bacteria [17], this has caused concerns about the use of various antibiotics and has been at risk for the selective bacterial pressure that encourages an increase in the number of resistant bacteria. Van Breda et al. [43] has emphasized that emerging multi-drug resistance has made effective treatment difficult, and requires the use of other drugs that are more effective to anticipate it.
Furthermore, Strom et al. [44] in his research has concluded that the incidence of MDR is more common in medium to small-scale farms and is more common on farms with many livestock. The results of this study (MDR level) are in line with the results of research that has been shown by Moredo et al. [45] of 52.5%, but very different from what was shown by Luque et al. [5] of 30.77%. MDR resistant patterns that have been shown in this study indicate the emergence of 39 patterns (Fig. 1) and these results are similar to those shown by Tang et al. [46].
Moredo et al. [45] have shown that around 17.5% of E. coli isolates from pig farms carry an integron that has been responsible as one of the carriers of resistance to the environment. Besides that, Tang et al. [46] have also explained that first class integrons in 27.3% of isolates observed from pigs have influenced resistance to trimethoprim and aminoglycosides. Cho et al. [47] in their study have shown that resistant E. coli will occur more frequently in individual agricultural workers (pigs and poultry) than workers who do not work on farms. Bacterial strains that already have several genes that encode resistance to some antibiotics often become multi-resistant compared to others [46].
Prevalence of tet genes
The prevalence of TE-resistant coding genes from E. coli isolates has been shown in Table 3. In studies that have been conducted to detect this resistant gene, it only focuses on knowing the presence of genes tet (A), tet (B), tet (C), tet (D), and tet (E) that encode the gene to TE resistance by causing pumping TE antibiotic molecules from bacterial cells (Efflux pump). Twenty-six samples of E. coli isolates consisting of 24 resistant isolates and two isolates were intermediates to TE antibiotics, have shown the presence of tet (A) gene from six isolates (23%), tet (E) genes from six isolates (23%), and gene combination of tet (A) + tet (E) from 12 isolates (46%) (Table 3). Several determinants of resistance to TE antibiotics have been found in E. coli isolates from pigs, namely genes tet (A) and genes tet (E), whereas genes tet (B), tet (C), and tet (D) were not found in this study (Fig. 2). This research has been slightly different from the research conducted by Urumova [20] which found the prevalence of genes tet (A) and tet (B) in E. coli isolates that have been studied. Tet gene was responsible for the emergence of bacterial resistance to TE antibiotics.
Table 3. Percentage of tet gene types appearing in the observed sample.
| Description | Total number of samples | Percentage of the total number of samples | |
|---|---|---|---|
| Pattern of tet gene | A | 6 | 23 |
| A, E | 12 | 46 | |
| E | 6 | 23 | |
| Not detection | 2 | 8 | |
| Total | 26 | 100 | |
Figure 2. Results Amplification of tet A and tet E gene encoding TE resistance in E. coli at Kupang city pig farm. Lane M: marker 100 bp DNA ladder, Lane 64–96: E. coli isolates from a pig farm that resistance with TE.
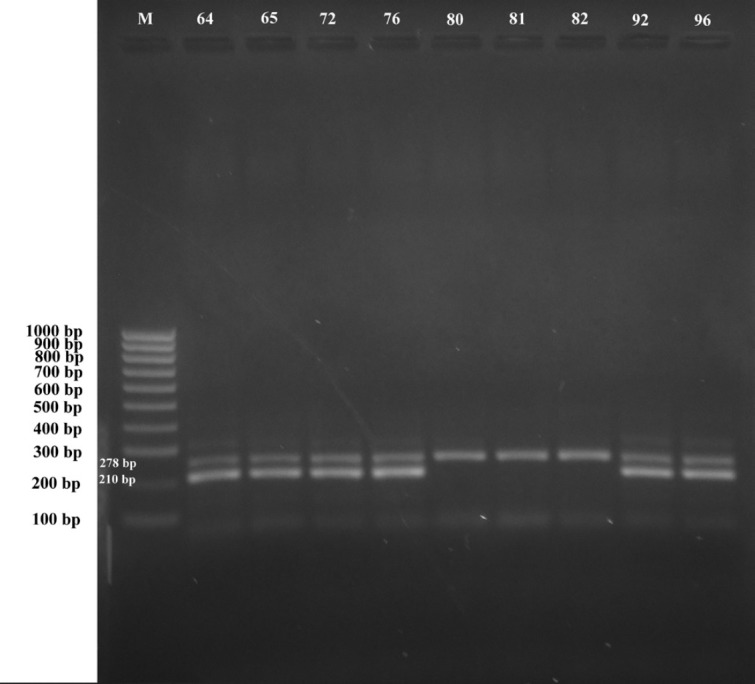
Tetracycline resistant genes have the highest prevalence in pig fecal samples and are responsible for the efflux pump process [35,46]. The incidence rate of tet genes in this study has shown a high prevalence of resistance to TE antibiotics in samples from pig feces. Several types of tet genes have been reported to cause resistance to TE antibiotics such as tet (A), tet (B), tet (C), tet (D), tet (E), and tet (G) [7,11,19,46]. The diversity of distribution of TE resistant genes has been dependent on certain environmental conditions such as waste, soil, and underground water and Horizontal gene transfer has been known as the main mechanism in the rapid spread of TE resistant genes to other types of bacteria in the environment [20]. The tet (A) gene has been associated with conjugated plasmids, whereas the tet (E) gene was associated with non-conjugated plasmids [19], so the spread of the tet (A) gene should be wider or faster than tet (E), but in this study showed different results, which also showed an increase in the number of spreads of the tet (E) gene in TE resistant E. coli. Antimicrobial resistance (AMR) that has spread from pigs needs to be monitored. Birkegard et al. [48] have also emphasized that pigs are a potential reservoir for the AMR gene that can be transferred to pathogenic bacteria in humans (through direct contact with pigs or releasing pig manure into the environment).
Conclusion
In this study, it has been shown that E. coli in pig farms in the city of Kupang has experienced high resistance to erythromycin and KF and several other antibiotics that have been increased such as colistin sulfate, streptomycin, TE, and AML. Some antibiotics show low resistance such as SXT, CTX, DO, CN, NA, and CIP, respectively. This increase in resistance was likely to be attributed to excessive antibiotic use in animals for production purposes and the close relationship between humans and livestock that can cause gene transfer that encodes antibiotic resistance from humans to animals and vice versa. This study also showed that E. coli isolates had an MDR prevalence of 57.3% (47/82) with 39 resistant patterns and could pose a threat to human health due to high resistance to some commonly used antibiotics. Tet (A) and tet (E) genes become resistance-coding genes for TE antibiotics commonly found in E. coli in pig farms in Kupang City. Public awareness due to the proper use of antibiotics in pig farms in the city of Kupang, Indonesia may be an important issue. Thus, the management of antibiotic use by farmers in the need to be increased in reducing the emergence of antibiotic resistance events on farms is at risk of posing a threat to the health of humans and animals around them.
Acknowledgments
The author would like to express the appreciation to Fenny Billy, SKH and Dr. Rahmat Setya Adji, M.Si who has helped provide suggestions and inputs during the author doing research in the laboratory. Acknowledgments are also conveyed to the Ministry of Technology and Higher Education Research which has supported funds in carrying out the research program (Research Contract number 37/UN15.19/LT/2018.).
Conflict of interest
No conflict of interest.
Authors’ contribution
Novalino Harold Geoffrey Kallau conducted the research work, did the necessary analysis, and made the primary draft of this manuscript. I Wayan Teguh Wibawan, Denny Widaya Lukman, and Mirnawati Bachrum Sudarwanto critically reviewed and improved the manuscript.
References
- 1.Zawack K, Li M, Booth JG, Love W, Lanzas C, Grohn YT. Monitoring antimicrobial resistance in the food supply chain and its implications for FDA policy initiatives. Antimicrob Agents Chemother. 2016;60(9):5302–11. doi: 10.1128/AAC.00688-16. https://doi.org/10.1128/AAC.00688-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Neill J. Wellcome trust and HM Government. London, UK: 2016. Tackling drug-resistant infections globally: final report and recommendations. [Google Scholar]
- 3.Molton JS, Tambyah PA, Ang BS, Ling ML, Fisher DA. The global spread of healthcare-associated multidrug-resistant bacteria: a perspective from Asia. Clin Infect Dis. 2013;56(9):1310–8. doi: 10.1093/cid/cit020. https://doi.org/10.1093/cid/cit020. [DOI] [PubMed] [Google Scholar]
- 4.Hu Y, Yang X, Li J, Lv N, Liu F, Wu J, et al. The bacterial mobile resistome transfer network connecting the animal and human microbiomes. Appl Environ Microbiol. 2016;82(22):6672–81. doi: 10.1128/AEM.01802-16. https://doi.org/10.1128/AEM.01802-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luque AT, Moreno CG, Pasteris SE, Orden JA, de la Fuente R, Otero MC. Antimicrobial resistant Escherichia coli in the reproductive tract microbiota of cows and sows. Comp Immunol Microbiol Infect Dis. 2017;55:13–9. doi: 10.1016/j.cimid.2017.09.002. https://doi.org/10.1016/j.cimid.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 6.Gao L, Tan Y, Zhang X, Hu J, Miao Z, Wei L, Chai T. Emissions of Escherichia coli carrying extended-spectrum beta-lactamase resistance from pig farms to the surrounding environment. Int J Environ Res Public Health. 2015;12(4):4203–13. doi: 10.3390/ijerph120404203. https://doi.org/10.3390/ijerph120404203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J, Ben W, Yang M, Zhang Y, Qiang Z. Dissemination of veterinary antibiotics and corresponding resistance genes from a concentrated swine feedlot along the waste treatment paths. Environ Int. 2016;92–93:317–23. doi: 10.1016/j.envint.2016.04.020. https://doi.org/10.1016/j.envint.2016.04.020. [DOI] [PubMed] [Google Scholar]
- 8.Zhang P, Shen Z, Zhang C, Song L, Wang B, Shang J, et al. Surveillance of antimicrobial resistance among Escherichia coli from chicken and swine, China, 2008–2015. Vet Microbiol. 2017;203:49–55. doi: 10.1016/j.vetmic.2017.02.008. https://doi.org/10.1016/j.vetmic.2017.02.008. [DOI] [PubMed] [Google Scholar]
- 9.Li XS, Liu BG, Dong P, Li FL, Yuan L, Hu GZ. The prevalence of mcr-1 and resistance characteristics of Escherichia coli isolates from diseased and healthy pigs. Diagn Microbiol Infect Dis. 2017;91(1):63–5. doi: 10.1016/j.diagmicrobio.2017.12.014. https://doi.org/10.1016/j.diagmicrobio.2017.12.014. [DOI] [PubMed] [Google Scholar]
- 10.Xia J, Sun J, Cheng K, Li L, Fang LX, Zou MT, et al. Persistent spread of the rmtB 16S rRNA methyltransferase gene among Escherichia coli isolates from diseased food-producing animals in China. J Vet Mic. 2016;188:41–6. doi: 10.1016/j.vetmic.2016.03.018. https://doi.org/10.1016/j.vetmic.2016.03.018. [DOI] [PubMed] [Google Scholar]
- 11.Skočková A, Koláčková I, Bogdanovičová K, Karpíšková R. Characteristic and antimicrobial resistance in Escherichia coli from retail meats purchased in the Czech Republic. J Food Cont. 2015;47:401–6. https://doi.org/10.1016/j.foodcont.2014.07.034. [Google Scholar]
- 12.Laube H, Friese A, von Salviati C, Guerra B, Rosler U. Transmission of ESBL/AmpC-producing Escherichia coli from broiler chicken farms to surrounding areas. J Vet Mic. 2014;172(3–4):519–27. doi: 10.1016/j.vetmic.2014.06.008. https://doi.org/10.1016/j.vetmic.2014.06.008. [DOI] [PubMed] [Google Scholar]
- 13.Hinenoya A, Shima K, Asakura M, Nishimura K, Tsukamoto T, Ooka T, et al. Molecular characterization of cytolethal distending toxin gene-positive Escherichia coli from healthy cattle and swine in Nara, Japan. BMC Microbiol. 2014;14(97):1–13. doi: 10.1186/1471-2180-14-97. https://doi.org/10.1186/1471-2180-14-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.OIE. World Organisation For Animal Health (OIE); Paris, France: 2017. Terrestrial animal health code volume I: general provisions. [Google Scholar]
- 15.OIE. OIE; 2016. The OIE strategy on antimicrobial resistance and the prudent use of antimicrobials. [Google Scholar]
- 16.Magiorakos AAS, Carey R, Carmeli Y, Falagas M, Giske C, Harbarth S, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–81. doi: 10.1111/j.1469-0691.2011.03570.x. https://doi.org/10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 17.Petternel C, Galler H, Zarfel G, Luxner J, Haas D, Grisold AJ, et al. Isolation and characterization of multidrug-resistant bacteria from minced meat in Austria. Food Microbiol. 2014;44:41–6. doi: 10.1016/j.fm.2014.04.013. https://doi.org/10.1016/j.fm.2014.04.013. [DOI] [PubMed] [Google Scholar]
- 18.Dahmen S, Metayer V, Gay E, Madec JY, Haenni M. Characterization of extended-spectrum beta-lactamase (ESBL)-carrying plasmids and clones of Enterobacteriaceae causing cattle mastitis in France. J Vet Mic. 2012;162(2–4):1–7. doi: 10.1016/j.vetmic.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 19.Ng LK, Martin I, Alfa M, Mulvey M. Multiplex PCR for the detection of tetracycline resistant genes. Mol Cell Probes. 2001;15(4):209–15. doi: 10.1006/mcpr.2001.0363. https://doi.org/10.1006/mcpr.2001.0363. [DOI] [PubMed] [Google Scholar]
- 20.Urumova V. Investigations on tetracycline resistance in commensal Escherichia coli isolates from swine. Bulgarian J Vet Med. 2016;19(3):179–88. [Google Scholar]
- 21.BPS. BPS Kota Kupang; Kupang, Indonesia: 2014. Badan Pusat Statistik Kota Kupang: Kota Kupang Dalam Angka 2014. [Google Scholar]
- 22.Gebreyes WA, Wittum T, Habing G, Alali W, Usui M, Suzuki S. Spread of antibiotic resistance in food animal production systems. Foodborne Dis. 2017:105–30. https://doi.org/10.1016/b978-0-12-385007-2.00004-8. [Google Scholar]
- 23.Kale MLF, Kallau NHG, Wuri DA. TPFKHU Seminar Nasional Ke-3 FKH Undana. Lembaga Penelitian Undana; Kupang, Indonesia: 2015. Uji Residu Antibiotik Tetrasiklin dan Aminoglikosida Pada Daging Babi Yang Beredar di Kota Kupang; pp. 105–10. [Google Scholar]
- 24.BSN. Badan Standardisasi Nasional; Indonesia: 2008. Standar Nasional Indonesia (SNI) 2897:2008 Metode pengujian cemaran mikroba dalam daging, telur dan susu, serta hasil olahannya. [Google Scholar]
- 25.CLSI. Clinical and Laboratory Standards Institute; Indonesia: 2014. M100-S24 performance standards for antimicrobial susceptibility testing; twenty-fourth informational supplement. [Google Scholar]
- 26.Park J, Gasparrini AJ, Reck MR, Symister CT, Elliott JL, Vogel JP, et al. Plasticity, dynamics, and inhibition of emerging tetracycline resistance enzymes. Nat Chem Biol. 2017;13:730–36. doi: 10.1038/nchembio.2376. https://doi.org/10.1038/nchembio.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Angi AH, Satrija F, Lukman DW, Sudarwanto M, Sudarnika E. Profil Peternakan Babi Di Kota Kupang Dan Potensi Penularan Trichinellosis. J Kajian Vet. 2014;2(2):131–41. [Google Scholar]
- 28.Liwa SR, Detta AIR, Kallau NHG. TPFKHU Seminar Nasional “One Health” Fakultas Kedokteran Hewan Universitas Nusa Cendana. FKH Undana; Kupang, Indonesia: 2014. Tingkat Cemaran Bakteri Escherichia coli dan Salmonella sp. pada Daging Babi yang Dijual di Pasar Tradisional dan Penjual Daging Eceran di Kota Kupang; pp. 152–67. [Google Scholar]
- 29.Raza EMU, Suada IS, Mahatmi H. Beban Cemaran Bakteri Escherichia coli pada Daging Asap Se’i Babi yang Dipasarkan di Kota Kupang. Indonesia Med Vet. 2012;1(4):453–70. [Google Scholar]
- 30.Susilawati NM, Ramona Y, Parwata IMOA. Pengaruh Konsentrasi Ekstrak Kasar Kulit Batang Kusambi (Schleichera oleosa (Lour) Oken) Terhadap Pertumbuhan In Vitro Bakteri E. coli. J Biol Sci. 2016;3(2):96–102. [Google Scholar]
- 31.Pakpahan RS, Picauly I, Mahayasa INW. Cemaran Mikroba Escherichia coli dan Total Bakteri Koliform pada Air Minum Isi Ulang. J Kesehatan Masyarakat Nasional. 2015;9(4):300–7. https://doi.org/10.21109/kesmas.v9i4.733. [Google Scholar]
- 32.Novicadlitha Y. Universitas Katolik Widya Mandira; Kupang, Indonesia: 2016. Penentuan Kualitas Air Sumur Gali Secara Bakteriologi di Kelurahan Nunbaun Sabu Kecamatan Alak Kota Kupang. [Google Scholar]
- 33.Rizzo L, Manaia C, Merlin C, Schwartz T, Dagot C, Ploy MC, et al. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: a review. Sci Total Environ. 2013;447:345–60. doi: 10.1016/j.scitotenv.2013.01.032. https://doi.org/10.1016/j.scitotenv.2013.01.032. [DOI] [PubMed] [Google Scholar]
- 34.Byarugaba DK, Kisame R, Olet S. Multi-drug resistance in commensal bacteria of food of animal origin in Uganda. AJMR. 2011;5(12):1539–48. [Google Scholar]
- 35.Luby EM, Moorman TB, Soupir ML. Fate and transport of tylosin-resistant bacteria and macrolide resistance genes in artificially drained agricultural fields receiving swine manure. Sci Total Environ. 2016;550:1126–33. doi: 10.1016/j.scitotenv.2016.01.132. https://doi.org/10.1016/j.scitotenv.2016.01.132. [DOI] [PubMed] [Google Scholar]
- 36.Koike S, Aminov RI, Yannarell AC, Gans HD, Krapac IG, Chee-Sanford JC, et al. Molecular ecology of macrolide-lincosamide-streptogramin B methylases in waste lagoons and subsurface waters associated with swine production. Environ Microbiol. 2010;59(3):487–98. doi: 10.1007/s00248-009-9610-0. [DOI] [PubMed] [Google Scholar]
- 37.Schipp M. Country reports on antimicrobial use and resistance: Australia. A International Workshop on the Use of Antimicrobials in Livestock Production and Antimicrobial Resistance in the Asia-Pacific Region; Negombo, Sri Langka. 2012. pp. 6–17. APHCA FAO, FAO. [Google Scholar]
- 38.Wasiński B, Rózanska H, Osek J. Occurrence of extended spectrum ß-Lactamaseand AmpC-Producing Escherichia coli in meat samples. Bull Vet Inst Pulawy. 2013;57(4):513–7. https://doi.org/10.2478/bvip-2013-0089. [Google Scholar]
- 39.EFSA; ECDC. The European Union Summary Report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2011. EFSA J. 2013;11(5):3196. [Google Scholar]
- 40.Von Salviati C, Friese A, Roschanski N, Laube H, Guerra B, Käsbohrer A, et al. Extended-spectrum beta-lactamases (ESBL)/AmpC beta-lactamases-producing Escherichia coli in German fattening pig farms: a longitudinal study. Berl Munch Tierarztl Wochenschr. 2014;127(9–10):412–9. [PubMed] [Google Scholar]
- 41.Shen Z, Wang Y, Shen Y, Shen J, Wu C. Early emergence of mcr-1 in Escherichia coli from food-producing animals. Lancet Infect Dis. 2016;16(3):293. doi: 10.1016/S1473-3099(16)00061-X. https://doi.org/10.1016/S1473-3099(16)00061-X. [DOI] [PubMed] [Google Scholar]
- 42.Bitrus A, Chuanchuen R, Luangtongkum T. Emergence of colistin resistance in extended-spectrum beta lactamase producing Enterobacteriaceae isolated from food animals and its public health implication: a review. J Adv Vet Ani Res. 2018;5(1):1–11. https://doi.org/10.5455/javar.2018.e246. [Google Scholar]
- 43.Van Breda L, Dhungyel O, Ward M. Antibiotic resistant Escherichia coli in southeastern Australian pig herds and implications for surveillance. Zoonoses Public Health. 2018;65(1):e1–7. doi: 10.1111/zph.12402. https://doi.org/10.1111/zph.12402. [DOI] [PubMed] [Google Scholar]
- 44.Strom G, Halje M, Karlsson D, Jiwakanon J, Pringle M, Fernstrom LL, et al. Antimicrobial use and antimicrobial susceptibility in Escherichia coli on small- and medium-scale pig farms in north-eastern Thailand. Antimicrob Resist Infect Control. 2017;6:75. doi: 10.1186/s13756-017-0233-9. https://doi.org/10.1186/s13756-017-0233-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moredo FA, Pineyro PE, Marquez GC, Sanz M, Colello R, Etcheverria A, et al. Enterotoxigenic Escherichia coli subclinical infection in pigs: bacteriological and genotypic characterization and antimicrobial resistance profiles. Foodborne Pathog Dis. 2015;12(8):704–11. doi: 10.1089/fpd.2015.1959. https://doi.org/10.1089/fpd.2015.1959. [DOI] [PubMed] [Google Scholar]
- 46.Tang X, Tan C, Zhang X, Zhao Z, Xia X, Wu B, et al. Antimicrobial resistances of extraintestinal pathogenic Escherichia coli isolates from swine in China. Microb Pathog. 2011;50(5):207–12. doi: 10.1016/j.micpath.2011.01.004. https://doi.org/10.1016/j.micpath.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 47.Cho SH, Lim YS, Kang YH. Comparison of antimicrobial resistance in Escherichia coli strains isolated from healthy poultry and swine farm workers using antibiotics in Korea. Osong Public Health Res Perspect. 2012;3(3):151–5. doi: 10.1016/j.phrp.2012.07.002. https://doi.org/10.1016/j.phrp.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Birkegard AC, Halasa T, Graesboll K, Clasen J, Folkesson A, Toft N. Association between selected antimicrobial resistance genes and antimicrobial exposure in Danish pig farms. Sci Rep. 2017;7(1):9683. doi: 10.1038/s41598-017-10092-9. https://doi.org/10.1038/s41598-017-10092-9. [DOI] [PMC free article] [PubMed] [Google Scholar]