Abstract
Background
Circular RNA (circRNA) is a special long-chain non-coding RNA produced during the process of intracellular RNA splicing. Also, circZNF609 is abundant in human tissues, with multiple functions in human diseases, but its role in colorectal cancer remains unknown. This study aimed to evaluate the expression of circZNF609 in tumor tissue and serum samples from patients with colorectal cancer and in colorectal cancer cell lines.
Material/Methods
The expression of circZNF609 was measured by quantitative polymerase chain reaction (q-PCR) in 45 paired tissue samples from patients with colorectal cancer and 46 serum samples from patients with colorectal cancer and healthy controls, and in the normal human colorectal cell line, FHC, and human colorectal cancer cell lines, HCT116 and HT29. Protein expression of proliferating cell nuclear antigen (PCNA), c-Myc, Bax, Bcl-2, and p53 was determined by Western blot. Cell apoptosis was measured by flow cytometry.
Results
CircZNF609 was significantly down-regulated in colorectal cancer tissues compared with adjacent normal tissues and in the serum of patients with colorectal cancer compared with healthy controls, verified by receiver operating characteristic (ROC) curve analysis. There was low expression of circZNF609 in HCT116 cells, and overexpression inhibited cell proliferation but had no effect on PCNA and c-Myc protein expression. Expression of circZNF609 induced apoptosis and upregulated expression of the pro-apoptotic protein, Bax, down-regulated the expression of the anti-apoptotic protein, Bcl-2, and upregulated p53.
Conclusions
Expression of circZNF609 was down-regulated in colorectal cancer tissue and promoted apoptosis in colorectal cancer cells in vitro by upregulating p53.
MeSH Keywords: Apoptosis; Biomarkers, Pharmacological; Colorectal Neoplasms; Diagnosis; DNA, Circular
Background
Circular RNAs (circRNAs) are a novel class of non-coding RNAs that are covalently closed circular molecules mainly generated by back-splicing [1,2]. Without 5′-end structure and 3′-end poly(A) tail, circRNAs cannot be hydrolyzed by RNA enzymes and exist in a relatively stable form. Also, circRNAs are abundant in mammalian cells and have tissue and cell specificity [3,4]. Several potential functions for circRNAs have been identified, including their role as miRNA sponges, transcriptional regulation, and interactions with RNA-binding proteins [5,6]. Recent studies have shown that circRNAs are important regulatory factors for tumor gene expression and pathological networks, and have a role in tumor growth, metastasis, and drug resistance [7]. Xie et al. [8] reported that circBCRC-3 suppressed the proliferation of bladder cancer cells proliferation by promoting p27 protein expression. CircHIPK3, a sponge of miR-558, has been shown to inhibit cell migration, cell invasion, and angiogenesis associated with bladder cancer cells [9]. Therefore, studies of the association between circRNA expression and colorectal cancer is an important area of research that might lead to an understanding of the molecular mechanisms involved.
Worldwide, colorectal cancer is the fifth most common cancer and the third leading cause of cancer-related death [10]. Although the treatment of early-stage colorectal cancer can result in good clinical outcome and quality of life, patients with early-stage malignancy may not have clinical symptoms, which can delay diagnosis and treatment. Colonoscopy is the primary method for the diagnosis of colorectal cancer, and has a detection rate of up to 95% for early-stage lesions [11]. However, colonoscopy is an invasive technique with inherent risk during the endoscopic procedure, including bleeding, bowel perforation, and other complications. A minimally invasive test using blood samples that can identify tumor biomarkers is a highly attractive approach that may improve patient compliance for screening and the detection of colorectal cancer. Carcinoembryonic antigen (CEA) and carbohydrate antigen 199 (CA199) have low rates of sensitivity and specificity for the diagnosis of early-stage colorectal cancer and highlight the need for new serum biomarkers [12].
The circNA, circZNF609 (circBase ID: hsa_circ_0000615) is located at chr15: 64791491–64792365 and (relative gene symbol, ZNF609). The previous study demonstrated that circZNF609 had an essential role in the development of the central nervous system [13]. A recently published study showed that circZNF609 reduced the activity of miR-615-5p in promoting vascular endothelial dysfunction [14]. Subsequent research has also shown that circZNF609 could sponge miR-150-5p to promote the expression of AKT3 in Hirschsprung’s disease [15]. The findings from these previously published studies indicate that circZNF609 may have multiple functions in human disease. However, its biological role in colorectal cancer remains unclear.
Preliminary experimental data from our laboratory indicated that circZNF609 was down-regulated in patients with colorectal cancer. Therefore, this study aimed to evaluate the expression of circZNF609 in tumor tissue and serum samples from patients with colorectal cancer and colorectal cancer cell lines.
Material and Methods
Clinical specimens
The study included 45 patients with a diagnosis of colorectal cancer who underwent surgical tumor resection with the availability of tumor tissue and adjacent normal colon tissue. The tissue samples were obtained from the Fourth Hospital of Hebei Medical University Second Department of General Surgery, China between January 2017 and February 2018. Adjacent normal tissues were taken 5 cm away from the tumor edge. The tissue samples were placed in liquid nitrogen immediately after surgical removal and stored at −80°C. The 46 patients with a diagnosis of colorectal cancer had peripheral blood samples collected before surgery. Peripheral blood of 46 healthy people was collected as the control samples. Serum was isolated and stored in Eppendorf tubes at 330 microliters per tube at −80°C.
Clinical data from the patients were recorded. Patients who had received radiotherapy, chemotherapy, or preoperative imaging or metastasis confirmed by pathology, were excluded from this study. Tumor, node, metastasis (TNM) staging was used to assess tumor invasion and depth, lymph node metastasis, and distant metastasis. The 7th edition of the American Joint Committee on Cancer (AJCC) staging system was used for tumor classification [16]. Patients who were enrolled in the study signed informed consent. The study was approved by the Ethics Committee of the Fourth Hospital of Hebei Medical University.
Cell lines and cell transfection
Cell lines were purchased from the Cell Bank of the Committee for the Conservation of Typical Cultures of the Chinese Academy of Sciences. The human normal colorectal mucosa cell line, FHC, was cultured in Dulbecco’s modified Eagle’s medium (DMEM) and F12 medium (Gibco, Thermofisher Scientific, Waltham, MA, USA). Human colorectal cancer cell lines HCT116 and HT29 were cultured in McCoy’s 5A medium (Gibco, Thermofisher Scientific, Waltham, MA, USA). Medium was supplemented with 10% fetal bovine serum (FBS) (Gibco, Thermofisher Scientific, Waltham, MA, USA), 100 U/ml penicillin and 100 μg/ml streptomycin (Huayao CNC Technology Co., Ltd., Wenzhou, China). All cell lines were cultured in an incubator containing 5% CO2 at 37°C. Cells were selected and inoculated into a 6-well plate for transfection. Full circZNF609 cDNA in an EcoRII–EcoRV fragment was inserted into the pCDNA3.1 vector. Negative control cells were transfected with the empty pCDNA3.1 vector. Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) was used for transfection.
Total RNA extraction and reverse transcription
TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was used to extract RNA from colorectal cancer tissue specimens and patient serum. The RNA concentration and purity were measured by the Nanodrop 1000™ (Thermofisher Scientific, Waltham, MA, USA) and cDNA was synthesized using M-MLV (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Divergent primers were designed with Primer 5 (Sangon Biotech, Beijing, China). The primer sequences were as follows:
Forward primer, circZNF609:
5′-CCTCCAGCCAGTTCCCTTG-3′;
Reverse primer, circZNF609:
5′-GTTCTCAGACCTGCCACATTG-3′;
Forward primer, ZNF609 mRNA:
5′-ACCTGAGAGCATAGAAGGGAAAG-3′;
Reverse primer, ZNF609 mRNA:
5′-CATAGGGGGGTACATAGGCATA-3′;
Forward primer, GAPDH (control):
5′-ATCTTCCAGGAGCGAGATCCC-3′;
Reverse primer, GAPDH (control):
5′-TGAGTCCTTCCAAGATACCAA-3′.
The expression levels of circZNF609 and GAPDH in tissue specimens and peripheral blood were analyzed by the 2−ΔΔCt calculation method [17], with a higher number representing a lower expression level of circZNF609. The cell RNA expression level was expressed by 2−ΔΔCt. All experimental data were expressed as the mean ± standard deviation (SD) of three independent experiments.
Cell proliferation assay
Cells were inoculated into 96-well plate after transfection. Each well contained 5000 cells. After inoculation for 24 h, 48 h, 72 h, and 96 h, 10 μl of cell counting kit-8 (CCK-8) reagent (Abmole, Shanghai, China) was added to each well and incubated for 2 h at 37°C. The absorbance was measured at 450 nm. The cells were counted after transfection and 500 cells were inoculated into a six-well plate. The colony formation was observed at between 7–10 days later and stained with crystal violet. Cell colony counts were then performed.
Cell apoptosis measured by fluorescence flow cytometry
A single suspension of cells was obtained by digestion with 0.25% trypsin. Cells were washed twice with cold PBS, and 5×105 cells were evaluated by flow cytometry. The manufacturer’s instructions were followed for the Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) staining and flow cytometry analysis. The experiment was performed in triplicate.
Western blot
Cells were collected and lysate was added that contained 50 mM Tris (pH 7.5), 150 mM NaCl, 1 mM phenylmethanesulfonyl fluoride, 1% NP-40, and 0.5% sodium deoxycholate. The Lowry method was used to quantify total protein concentration. Two groups of protein were separated by a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) membrane and blocked with dried skimmed milk powder at 37°C. The membrane was incubated with primary antibodies at 4°C overnight, and according to the type of primary antibody, the corresponding secondary antibody was selected and incubated on the membranes at 37°C for 1 hour. A chemiluminescence imaging analyzer was used for image scanning.
Statistical analysis
Data were presented by the mean ± standard deviation (SD). The t-test was used for comparisons between groups and one-way analysis of variance (ANOVA) was applied for multiple groups. The receiver operating characteristic (ROC) curves were calculated to evaluate the diagnostic clinical value. Data were analyzed by using SPSS software version 17.0 (IBM, Chicago, IL, USA) and GraphPad Prism version 5 (GraphPad Software, La Jolla, CA, USA). A P-value <0.05 was considered to be statistically significant.
Results
The expression and characteristics of circZNF609 in colorectal cancer tissues
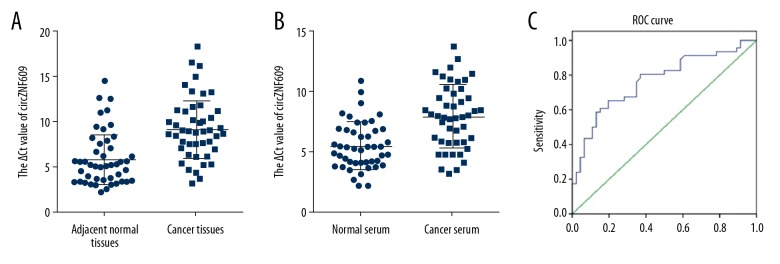
In this study, the expression level of circZNF609 in colorectal cancer tissues was significantly lower than that in adjacent normal colonic tissues (Figure 1A). Also, circZNF609 levels in the serum samples from patients with colorectal cancer were down-regulated compared with healthy controls (Figure 1B). To evaluate the potential clinical diagnostic value of circZNF609 in the serum of patients with colorectal cancer, the receiver operating characteristic (ROC) curves were created and analyzed (Figure 1C). The sensitivity and specificity were 0.652 and 0.804, respectively. The area under the curve (AUC) was 0.767 (95% CI, 0.670–0.864), and the cutoff value was 6.915. The Youden index was 0.457. The diagnostic value was used to evaluate the efficacy of circZNF609 diagnosis in 46 patients with colorectal cancer, the diagnostic value was 65.217%.
Figure 1.
The expression levels of circZNF609 in clinical specimens of colorectal cancer. The expression levels of circZNF609 in colorectal cancer tissues (A) (n=45) (P<0.05). The expression levels of circZNF609 in serum from patients with colorectal cancer (B) (n=46) (P<0.05). The higher ΔCt number indicates the lower expression level of circZNF609. Receiver operating characteristic (ROC) curve analysis was used to verify the diagnostic value of circZNF609 as a serum biomarker (C).
Because circZNF609 expression was lower in the tumor tissue and serum from patients with colorectal cancer, in 39 cases the patient clinical data and the relationship between circZNF609 expression and clinicopathological factors were analyzed (Table 1). The circZNF609 expression was significantly associated with the tumor diameter (P<0.05). The larger the tumor diameter, the lower the circZNF609 expression level. However, there were no relationships between circZNF609 expression levels and other clinicopathologic factors, including patient gender, age, tumor differentiation, TNM stage, tumor location, tumor shape, lymphatic invasion, vascular and neural invasion.
Table 1.
The relationship between circZNF609 expression levels in colorectal cancer tissues and patient clinicopathological factors.
| Clinicopathological factors | No. of patients | Mean ± SD | t-value | P-value |
|---|---|---|---|---|
| Gender | ||||
| Male | 24 | 8.72±3.24 | 0.65 | 0.52 |
| Female | 21 | 9.34±3.13 | ||
| Age | ||||
| <60 | 15 | 9.74±3.01 | 1.10 | 0.28 |
| ≥60 | 30 | 8.64±3.23 | ||
| Tumor diameter | ||||
| <5 cm | 22 | 7.98±3.09 | 2.22 | 0.03* |
| ≥5 cm | 23 | 9.99±2.98 | ||
| Tumor differentiation | ||||
| II | 39 | 9.11±3.21 | 0.57 | 0.58 |
| III | 6 | 8.32±3.01 | ||
| TNM stage | ||||
| II | 30 | 8.85±2.98 | 0.48 | 0.64 |
| III | 15 | 9.32±3.58 | ||
| Tumor location | ||||
| Colon | 25 | 8.83±3.27 | 0.41 | 0.68 |
| Rectum | 20 | 9.23±3.10 | ||
| Tumor shape | ||||
| Ulcer type | 37 | 8.89±3.07 | 0.51 | 0.62 |
| Swelling type | 8 | 9.52±3.75 | ||
| lymphatic metastasis | ||||
| Negative | 29 | 8.85±3.04 | 0.44 | 0.66 |
| Positive | 16 | 9.29±3.47 | ||
| Vascular tumors bolt | ||||
| Negative | 39 | 8.93±3.30 | 0.40 | 0.69 |
| Positive | 6 | 9.49±2.31 | ||
| Nerves invasion | ||||
| Negative | 25 | 8.48±2.78 | 1.26 | 0.22 |
| Positive | 20 | 9.67±3.56 | ||
P<0.05.
The circZNF609 overexpression vector was constructed and verified
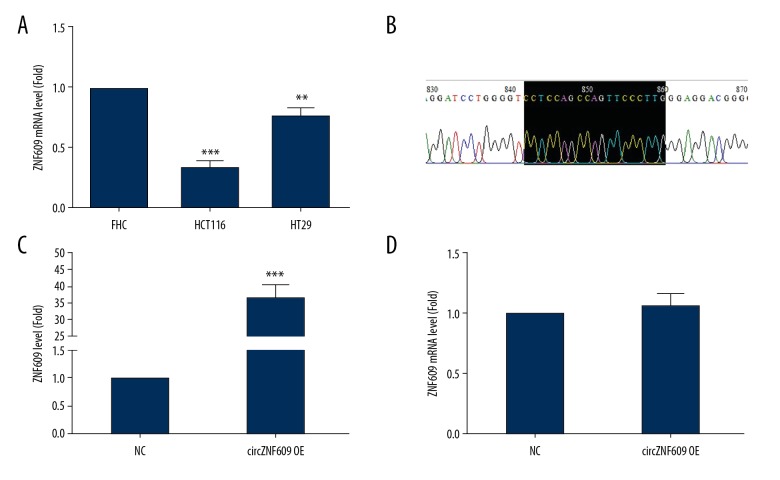
Compared with the normal human colonic mucosal FHC cell line, circZNF609 expression was significantly reduced in the HCT116 and HT29 cells (P<0.01) (Figure 2A). Because the decrease in the HCT116 cells was the most significant, the HCT cell line was selected for the subsequent experiments. The circZNF609 overexpression vector was constructed and the sequencing results indicated that the vector was successfully constructed (Figure 2B). The cells were transfected with Lipofectamine 2000. Compared with the empty vector, the circZNF609 expression level was significantly increased in the circZNF609 overexpression group (P<0.01) (Figure 2C), while the ZNF609mRNA level showed no significant change (P>0.05) (Figure 2D).
Figure 2.
The expression levels of circZNF609 in colorectal cancer cell lines. The expression levels of circZNF609 in the human colorectal cancer cell lines, FHC, HCT116, and HT29 (A) (** P<0.01, *** P<0.001). The sequencing result of circZNF609 overexpression vector (B). HCT116 cells were transfected with circZNF609 vector and vector (C) (*** P<0.001). (D) Overexpression of circZNF609 had no effect on the ZNF609 mRNA level (D) (P>0.05).
Overexpression of circZNF609 inhibited the activity of HCT116 cells
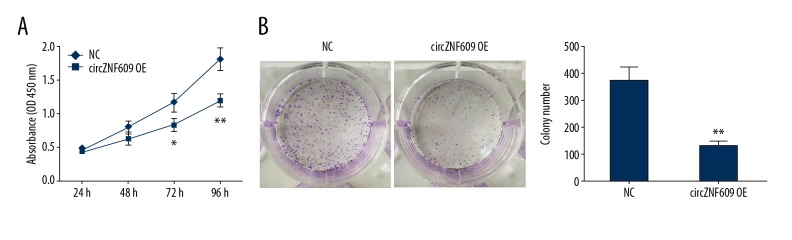
Cell activity of HCT116 cells following circZNF609 overexpression was detected by the cell counting kit-8 assay (CCK-8). As shown in Figure 3A, the cell activity of HCT116 was significantly inhibited after circZNF609 overexpression (P<0.05). Similar results were obtained from the colony formation experiment (P<0.01) (Figure 3B).
Figure 3.
Overexpression of circZNF609 inhibited cell activity of HCT116 cells. Cell counting kit-8 (CCK-8) assay showed that the cell activity of HCT116 cells was significantly inhibited (A) (P<0.05). The results of plate cloning experiments showed that the number of cell colony formation was significantly reduced in the circZNF609 overexpression group (B) (P<0.01).
Overexpression of circZNF609 did not affect the expression of PCNA and c-Myc in HCT116 cells
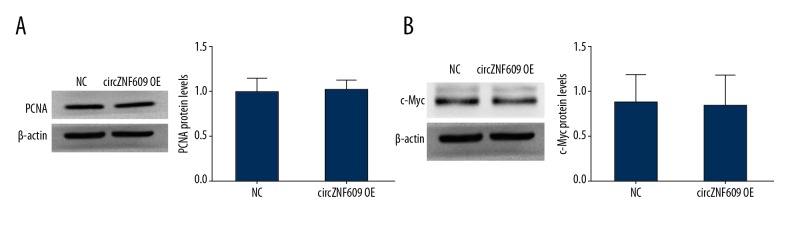
To explore the molecular mechanism of circZNF609 inhibition of HCT116 cell proliferation, the expression of PCNA and c-Myc were detected by Western blot. PCNA is an acidic non-histone nucleoprotein that regulates cell proliferation and can be used to assess tumor cell proliferation and is a prognostic marker in malignancy [17]. The c-MYC gene, an important member of MYC gene family, is an oncogene, and its translation product, the c-Myc protein, has the function of promoting cell division, regulating cell proliferation, and differentiation [18]. The results showed that there was no difference in expression between the two groups for PCNA (P>0.05) (Figure 4A) or c-Myc (P>0.05) (Figure 4B). Therefore, it was speculated that the decreased proliferation activity and cell number of HCT116 cells might not be inhibited by cell proliferation.
Figure 4.
The expression levels of proliferation-associated proteins in HCT116 cells. There was no difference in the protein expression levels of PCNA (A) and c-Myc (B) between the circZNF609 overexpression group and the control group (P>0.05).
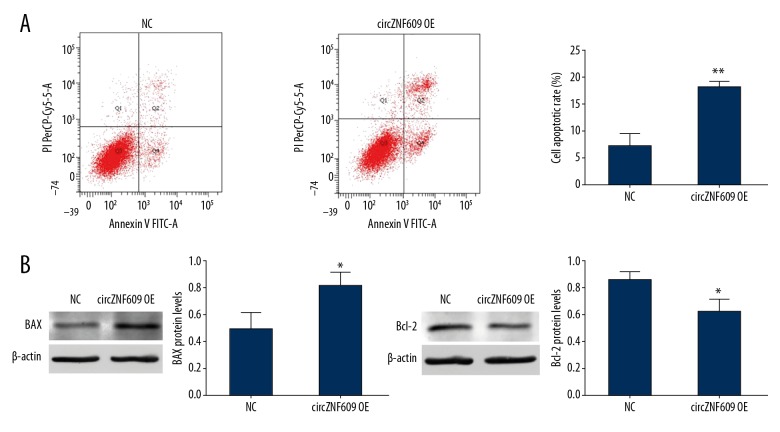
Overexpression of circZNF609 induced apoptosis of HCT116 cells
Flow cytometry showed that the cell apoptosis rate, calculated by Q2+Q4, in the circZNF609 overexpression group was 17.87±1.33% greater than in the control group (8.37±1.81%) (P<0.01) (Figure 5A). Q2 and Q4 represented late and early apoptosis, respectively. The early-stage Q4 was 9.30±0.30% in the circZNF609 overexpression group, which was higher 4.13±2.40% than in the control group (P<0.05). Also, the circZNF609 overexpression group was 8.57±1.04% higher than the control group, 3.90±2.52%, in late-stage Q2 (P<0.05).
Figure 5.
Apoptosis associated with circZNF609 in HCT116 cells. The results of flow cytometry showed that apoptosis in HCT116 cells was promoted after transfection with circZNF609 compared with normal control (NC) group (A) (** P<0.01). Compared with the NC group, the expression levels of Bax protein was significantly increased (B) (* P<0.05). Protein expression of Bcl-2 was significantly decreased in the circZNF609 overexpression group (C) (* P<0.05)
To further investigate the molecular mechanism of circZNF609 in the induction of apoptosis in HCT116 cells, the expressions of apoptosis-related proteins Bax and Bcl-2 in the two groups were detected. The expression and regulation of the Bcl-2 family are key factors affecting apoptosis and BAX and BCL2 are the most representative genes in the Bcl-2 family that promote and inhibit apoptosis, respectively [19,20]. The Western blot results showed that, compared with the control group, Bax expression was upregulated (P<0.05) (Figure 5B), while Bcl-2 expression was decreased in the circZNF609 overexpression group (P<0.05) (Figure 5C).
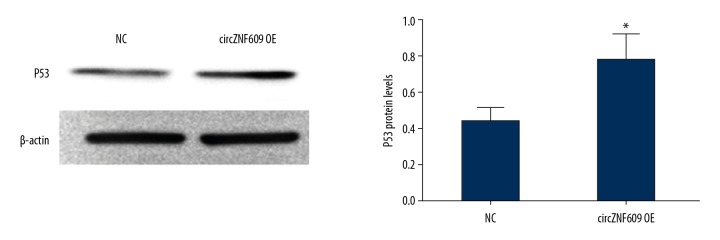
Overexpression of circZNF609 promoted the expression of p53 protein
To further dissect the molecular mechanism of how circZNF609 triggered HCT116 apoptosis, the expression of p53 protein was investigated. It has previously been reported that Bax can be regulated by p53 [21]. The Western blot results showed that, when compared with the control group, p53 expression was upregulated in the circZNF609 overexpression group (P<0.05) (Figure 6).
Figure 6.
The expression levels of the p53 protein in the circZNF609 overexpression group. Western blot results showed that, compared with the normal control (NC) group, p53 expression was upregulated in the circZNF609 overexpression group (* P<0.05).
Discussion
With the development of high-throughput gene sequencing technology, a large amount of non-coding RNA has been identified in humans, which performs a variety of biological functions and is involved in the occurrence and development of diseases, including malignant tumors [22]. Recently, an increasing number of studies have shown that circular RNAs (circRNAs), special long-chain non-coding RNAs, are produced during the process of intracellular RNA splicing and involve in the regulation of gene expression following transcription [4,5,23]. The circRNAs are widely expressed in human cells and can be even more highly expressed than linear isoforms [24]. Also, high conservation and long half-life are two other important characteristics of circRNAs [25]. Disease-related clinical research had identified a resource of circRNA expression from 20 human tissues [26]. Current studies have shown that circRNAs might be novel biomarkers for the diagnosis of several diseases and potential therapeutic targets.
The findings of the present study showed that circZNF609 was significantly down-regulated in colorectal cancer tissues compared with adjacent normal tissues (Figure 1A), which indicated that it might have a role in the occurrence and development of colorectal cancer [27]. In the present study, low expression levels of circZNF609 were associated with tumor diameter (Table 1), as the greater the tumor diameter, the lower the expression level of circZNF609. However, the expression level of circZNF609 was independent of other clinicopathologic factors, such as TNM stage and metastasis. These preliminary findings support that further studies are needed to determine whether circZNF609 is an early biomarker for colorectal cancer.
Further studies on the molecular associations with colorectal cancer are committed to the discovery of new tumor markers. Yang et al. showed that increased expression levels of the aldehyde dehydrogenase 1 family member A1 (ALDH1A1) protein in colorectal cancer tissues was associated with poor prognosis [28]. Currently, the main method of detection of circRNA in tumors is from tissue samples, but this method requires surgery, which is invasive and not suitable for the diagnosis of early tumors. Compared with tissue samples, other clinical samples such as serum, urine, and body fluids are more easily available and non-traumatic. For example, Dong et al. found that high expression levels of miR-429 in the serum of patients with colorectal cancer was correlated with poor prognosis [29]. Studies have shown that circRNA is widely found in blood, urine, cerebrospinal fluid, and other body fluids. Also, cells can secrete blood circulating RNA into the blood through exosomes, suggesting that serum circRNA may play an important role in cell communication [30,31]. Therefore, in this study, circZNF609 expression levels were detected in the serum of patients with colorectal cancer and were down-regulated compared with normal controls (Figure 1B). To further determine the diagnostic value of serum levels of circZNF609, receiver operating characteristic (ROC) curve analysis was performed (Figure 1C). The sensitivity and specificity were 0.64 and 0.78, respectively. The area under the curve (AUC) was 0.762 (95% CI, 0.652–0.873), supporting its potential value as a diagnostic biomarker for colorectal cancer.
To further explore the mechanism for circZNF609 in the occurrence and development of colorectal cancer, functional studies were performed at the cellular level. Since the expression level was related to tumor size, its function might affect cell proliferation activity. The cell counting kit-8 (CCK-8) and the colony formation assays showed that circZNF609 inhibited the proliferation activity of HCT116 cells. Evaluation of the proliferation protein markers, PCNA and c-Myc protein showed no significant changes in their expression between the two groups.
Based on the above results, it might be proposed that the mechanism of the effects of circZNF609 to reduce the proliferation activity and cell number of HCT116 cells might not be through inhibition of cell proliferation, but the induction of apoptosis. Flow cytometry was used to detect apoptosis and Western blot was used to detect the expression of apoptosis-related proteins. Flow cytometry showed that circZNF609 promoted HCT116 cell apoptosis. The expression of the pro-apoptotic protein, Bax, was upregulated, while the expression of the anti-apoptotic protein, bcl-2, was down-regulated.
The p53 protein is produced by the expression of the P53 gene on the short arm of chromosome 17 and is involved in the processes of apoptosis, aging, and DNA damage repair [32]. The role of the p53 protein in cell apoptosis has been established, and the results of the present study showed that the expression of the p53 protein increased in the circZNF609 overexpression group compared with the control group. Therefore, circZNF609 might induce apoptosis by promoting the expression of p53, thereby inhibiting cell proliferation in colorectal cancer.
The expression of circZNF609 has been previously shown to have different effects in different diseases. Recent studies have shown that circZNF609 silencing protected human umbilical vein endothelial cells (HUVECs) from oxidative stress or hypoxia-induced apoptosis [14]. However, a study by Wang et al. [33] showed that circZNF609 promoted cell growth, cell migration, and cell migration breast cancer cells, which might be due to the tissue specificity of circRNAs. Also, circRNAs generated by DOCK1 gene expression are highly expressed in the MCF-7 breast cancer cell line but were minimally expressed in the A549 lung cancer cell line [34]. The preliminary findings of the present study showed that circZNF609 promoted apoptosis of HCT116 colorectal cancer cells by upregulating p53. However, the molecular mechanisms underlying the effects of circZNF609 on the upregulation of p53 remain unknown. The molecular mechanisms require further study to identify new molecular biomarkers or the clinical diagnosis and new targets for the treatment of colorectal cancer.
Conclusions
This study aimed to evaluate the expression of circZNF609 in tumor tissue and serum samples from patients with colorectal cancer and in colorectal cancer cell lines. Expression of circZNF609 was down-regulated in colorectal cancer tissue and promoted apoptosis in colorectal cancer cells in vitro by upregulating p53.
Footnotes
Source of support: This study was supported by the National Natural Science Foundation of China (Grant No. 81372150/91739301/91849102)
Conflict of interest
None declared.
References
- 1.Han B, Chao J, Yao H. Circular RNA and its mechanisms in disease: From the bench to the clinic. Pharmacol Ther. 2018;187:31–44. doi: 10.1016/j.pharmthera.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 2.Hentze MW, Preiss T. Circular RNAs: Splicing’s enigma variations. EMBO J. 2013;32(7):923–25. doi: 10.1038/emboj.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrett SP, Salzman J. Circular RNAs: Analysis, expression and potential functions. Development. 2016;143(11):1838–47. doi: 10.1242/dev.128074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–57. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–38. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Zhang XO, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51(6):792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 7.Guarnerio J, Bezzi M, Jeong JC, et al. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell. 2016;166(4):1055–56. doi: 10.1016/j.cell.2016.07.035. [DOI] [PubMed] [Google Scholar]
- 8.Xie F, Li Y, Wang M, et al. Circular RNA BCRC-3 suppresses bladder cancer proliferation through miR-182-5p/p27 axis. Mol Cancer. 2018;17(1):144. doi: 10.1186/s12943-018-0892-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, Zheng F, Xiao X, et al. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017;18(9):1646–59. doi: 10.15252/embr.201643581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. Cancer J Clin. 2017;67(3):177–93. doi: 10.3322/caac.21395. [DOI] [PubMed] [Google Scholar]
- 11.Rahier JF, Druez A, Faugeras L, et al. Circulating nucleosomes as new blood-based biomarkers for detection of colorectal cancer. Clin Epigenetics. 2017;9:53. doi: 10.1186/s13148-017-0351-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bagaria B, Sood S, Sharma R, Lalwani S. Comparative study of CEA and CA19-9 in esophageal, gastric and colon cancers individually and in combination (ROC curve analysis) Cancer Biol Med. 2013;10(3):148–57. doi: 10.7497/j.issn.2095-3941.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rybak-Wolf A, Stottmeister C, Glazar P, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58(5):870–85. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 14.Liu C, Yao MD, Li CP, et al. Silencing of circular RNA-ZNF609 ameliorates vascular endothelial dysfunction. Theranostics. 2017;7(11):2863–77. doi: 10.7150/thno.19353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng L, Chen G, Zhu Z, et al. Circular RNA ZNF609 functions as a competitive endogenous RNA to regulate AKT3 expression by sponging miR-150-5p in Hirschsprung’s disease. Oncotarget. 2017;8(1):808–18. doi: 10.18632/oncotarget.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edge SB, Compton CC. The American Joint Committee on Cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Annal Surg Oncol. 2010;17(6):1471–74. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 17.Tsai WC, Yu TY, Lin LP, et al. Platelet rich plasma releasate promotes proliferation of skeletal muscle cells in association with upregulation of PCNA, cyclins and cyclin dependent kinases. Platelets. 2017;28(5):491–97. doi: 10.1080/09537104.2016.1227061. [DOI] [PubMed] [Google Scholar]
- 18.Hung CL, Wang LY, Yu YL, et al. A long noncoding RNA connects C-MYC to tumor metabolism. Proc Natl Acad Sci USA. 2014;111(52):18697–702. doi: 10.1073/pnas.1415669112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Volkmann N, Marassi FM, Newmeyer DD, Hanein D. The rheostat in the membrane: BCL-2 family proteins and apoptosis. Cell Death Differ. 2014;21(2):206–15. doi: 10.1038/cdd.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kvansakul M, Hinds MG. Structural biology of the Bcl-2 family and its mimicry by viral proteins. Cell Death Dis. 2013;4:e909. doi: 10.1038/cddis.2013.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greijer AE, van der Wall E. The role of hypoxia inducible factor 1 (HIF-1) in hypoxia induced apoptosis. J Clin Pathol. 2004;57(10):1009–14. doi: 10.1136/jcp.2003.015032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96(4):1297–325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 23.Salzman J, Gawad C, Wang PL, et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7(2):e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen S, Li T, Zhao Q, et al. Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin Chim Acta. 2017;466:167–71. doi: 10.1016/j.cca.2017.01.025. [DOI] [PubMed] [Google Scholar]
- 25.Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–88. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 26.Maass PG, Glazar P, Memczak S, et al. A map of human circular RNAs in clinically relevant tissues. Journal Mol Med. 2017;95(11):1179–89. doi: 10.1007/s00109-017-1582-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Y, Zhang J, Chen X, et al. LncRNA FTX sponges miR-215 and inhibits phosphorylation of vimentin for promoting colorectal cancer progression. Gene Ther. 2018;25(5):321–30. doi: 10.1038/s41434-018-0026-7. [DOI] [PubMed] [Google Scholar]
- 28.Yang W, Wang Y, Wang W, et al. Expression of aldehyde dehydrogenase 1A1 (ALDH1A1) as a prognostic biomarker in colorectal cancer using immunohistochemistry. Med Sci Monit. 2018;24:2864–72. doi: 10.12659/MSM.910109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong SJ, Cai XJ, Li SJ. The clinical significance of MiR-429 as a predictive biomarker in colorectal cancer patients receiving 5-fluorouracil treatment. Med Sci Monit. 2016;22:3352–61. doi: 10.12659/MSM.900674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khan MA, Reckman YJ, Aufiero S, et al. RBM20 regulates circular RNA production from the titin gene. Circ Res. 2016;119(9):996–1003. doi: 10.1161/CIRCRESAHA.116.309568. [DOI] [PubMed] [Google Scholar]
- 31.Lasda E, Parker R. Circular RNAs co-precipitate with extracellular vesicles: A possible mechanism for circRNA clearance. PloS One. 2016;11(2):e0148407. doi: 10.1371/journal.pone.0148407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tentler JJ, Ionkina AA, Tan AC, et al. p53 family members regulate phenotypic response to aurora kinase A inhibition in triple-negative breast cancer. Mol Cancer Ther. 2015;14(5):1117–29. doi: 10.1158/1535-7163.MCT-14-0538-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang S, Xue X, Wang R, et al. CircZNF609 promotes breast cancer cell growth, migration, and invasion by elevating p70S6K1 via sponging miR-145-5p. Cancer Manag Res. 2018;10:3881–90. doi: 10.2147/CMAR.S174778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salzman J, Chen RE, Olsen MN, et al. Cell-type specific features of circular RNA expression. PLoS Genetics. 2013;9(9):e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]