Abstract
Over the past century, mice have been selectively bred to give rise to the strains used in biomedical research today. Mouse models of cancer allow researchers to control variables of diet, environment, and genetic heterogeneity found in the human population to better dissect the role of these factors in cancer. Because of the important role of genetic background in cancer, the strain of the mouse can give confounding results in studies of mouse models if not properly controlled or can provide important new insights into cancer mechanisms. In this chapter the sources of genetic heterogeneity in mouse models and how it modifies cancer phenotypes is reviewed.
Origin of inbred mouse strains used in cancer research:
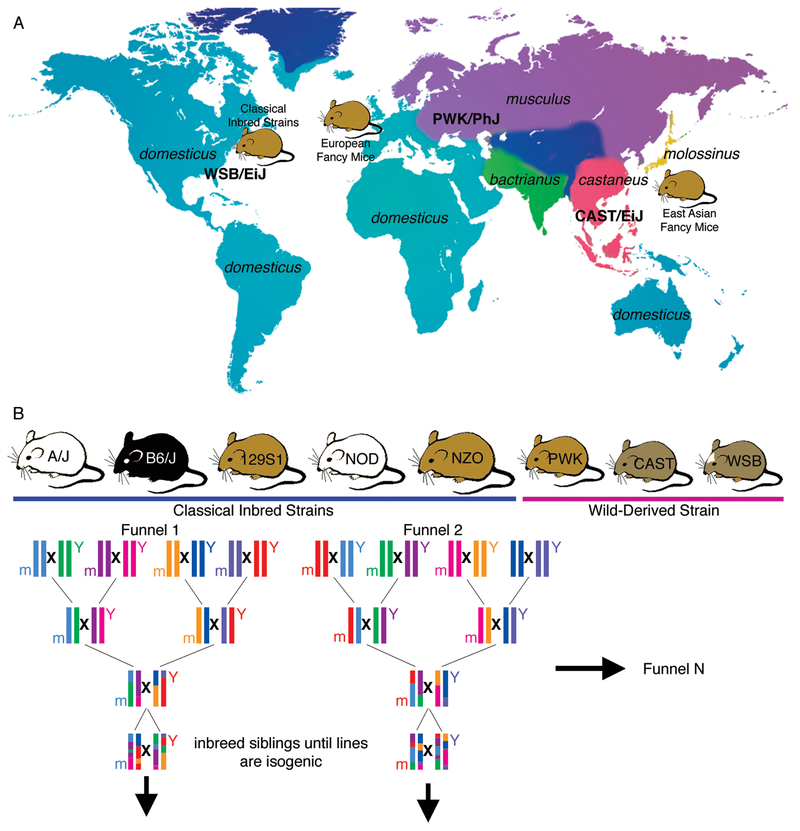
Classical inbred strains are a genetic mixture of the Mus musculus subspecies: M. m. domesticus, M. m. musculus, M. m. castaneus, and M. m. molossinus (which is itself a hybrid between M. m. musculus and M. m. casteneus) (Figure 1A) (Silver 1995). East Asian “fancy” mice were selectively bred from M. m. molossinus and M. m. musculus as pets in the eighteenth century and brought to England during the Victorian era. They were further selectively bred with M. m. domesticus, resulting in European “fancy” mice. A limited number of founders from the European “fancy” mice were brought to the US in the twentieth century and inbred to establish the current classical inbred mouse strains. Genetic analysis of these inbred strains show that they are 94% M. m. domesticus, 5% M. m. musculus, and <1% M. m. castaneus (Yang et al. 2011). In addition, many pure subspecies have been inbred to give rise to the “wild-derived” inbred strains, such as WSB/EiJ (domesticus), PWK/PhJ (musculus), MSM/Ms (molossinus), and CAST/EiJ (castaneus), which are more genetically diverse that the classical inbred strains.
Figure 1:
(A) The distribution of Mus musculus subspecies and their contribution to modern day classical inbred strains. Classical inbred strains used in biomedical research were developed from European “fancy” mice that were descended from East Asian “fancy” mice. Classical inbred strains are mixtures of the domesticus, musculus, castaneus, and molossinus subspecies of Mus musculus. In addition, pure subspecies have been inbred to give rise to the “wild-derived” strains. WSB/EiJ is a M. m. domesticus strain originating in Maryland, USA. PWK/PhJ is a M. m. musculus strain originating near Prague, Czech Republic. CAST/EiJ is a M. m. castaneus strain originating in Thonburi, Thailand. (B) The design of the Collaborative Cross to maximize genetic diversity across a panel of recombinant inbred strains. Eight founder strains include 5 classical inbred strains and 3 wild-derived strains. By varying the position of each founder in the different breeding funnels, polymorphisms in the mitochondria (m) and the Y chromosome (Y) can be equally represented across the resulting lines.
Different inbred strains have been favored in different research fields and are differentially represented in the background of mouse cancer models. Most genetically engineered mouse models are generated in embryonic stem cells, involving the 129/Sv and/or C57BL/6 strain backgrounds, or in mouse zygotes, involving the FVB/N, C57BL/6 and/or SJL strain backgrounds. In addition, BALB/cJ and A/J are commonly used in studies of autoimmunity and DBA/2J has been important in many fields. As biomedical fields interact and overlap, mouse models on different strain backgrounds have been combined. The resulting mixed strain backgrounds have the potential to confound the interpretation of mouse models, but also allow researchers to identify interacting polymorphisms to better understand cancer pathways.
Sources of variation across different mouse strains:
Natural variation occurs in many forms and can be difficult to integrate (Scherer et al. 2007). The DNA sequence varies between different strains by single nucleotide polymorphisms (SNPs), small (<1kb) insertions and deletions (INDELs) that can give rise to restriction fragment length polymorphisms (RFLPs) or simple sequence length polymorphisms (SSLPs), and by larger (> 1kb) copy number variations (CNVs). Autosomal variants undergo germline recombination to produce additional variation in progeny of mixed strain crosses. In addition, these natural variations in DNA sequence are found on the X and Y sex chromosomes and the mitochondrial DNA. Because the sex chromosomes and mitochondrial genome do not undergo the similar recombination and inheritance as the autosomes, they can introduce bias into the mouse populations being studied.
Chromosome X:
The X chromosome undergoes germline recombination in females, but not males. On mixed strain backgrounds the X chromosome contributed by the father is fixed, but the X chromosome contributed by the mother can recombine compared to the previous generation. In female cells, one of the X chromosomes becomes inactivated to maintain the gene dosage similar to males. Whether the maternal or paternal copy of the X chromosome is inactivated is theoretically random, but is influenced by polymorphisms at the Xce locus (Chadwick et al. 2006). Loci on the X chromosome have been shown to modify ovarian granulosa cell tumors (Beamer et al. 1998; Dorward et al. 2003), mammary tumors (Koch et al. 2007), and testicular tumors (Hammond et al. 2007), so on a mixed strain background the choice of which X chromosome remains active could influence tumor phenotypes.
Chromosome Y:
The Y chromosome does not recombine, but has developed polymorphisms spontaneously between different strain backgrounds. Because the Y chromosome is inherited through the paternal line, male progeny of reciprocal F1 hybrids (AXB vs BXA) are not identical. Although there is not yet strong evidence for a role of Y chromosome polymorphisms in modifying tumorigenesis, a study of cardiac growth (Llamas et al. 2009) found expression changes in p53 pathway genes, including Pten, Cnnd1 (CyclinD1), and Cdkn1a (p21), in mice carrying the A/J Y chromosome vs the C57BL/6J Y chromosome. Therefore, it remains a formal possibility that Y chromosome polymorphisms can affect tumor phenotypes and should be controlled in genetic crosses.
The mitochondrial genome:
The mitochondrial genome is inherited through the maternal line and, like the Y chromosome, does not recombine, but accumulates polymorphisms spontaneously. Although the role of mitochondrial variation in tumorigenesis is only beginning to be appreciated, it clearly affects tumor related phenotypes of apoptosis, proliferation, and invasion (Jandova et al. 2012a; Jandova et al. 2012b). Mitochondrial polymorphisms also affect phenotypes related to diabetes (Chen et al. 2011; Weiss et al. 2012), autoimmune disease (Jonsen et al. 2009; Yu et al. 2009a; Yu et al. 2009b), and cell metabolism (Moreno-Loshuertos et al. 2006).
Controlling variation in genetic crosses of mouse cancer models:
Given the effects of strain on mouse phenotypes, it is critical to control for genetic background in experiments testing hypotheses of how genes, carcinogens, or therapies affect tumorigenesis (see Protocol 1). This can be achieved using well-controlled inbred backgrounds or by using appropriate sibling controls in mixed backgrounds, so that the extent of variation in the control group matches the variation in the experimental group. In this case, it is important to consider how the crosses are set up to ensure that variations in sex chromosomes and the mitochondrial genome are equally represented in all groups.
Once mutant mouse models are generated they can be switched to a different strain background by 10 or more generations of backcrossing; however, it is very difficult to completely remove the original strain polymorphisms in the region of the gene mutation. This leads to a window of strain contamination around the gene of interest that can modify the phenotype of the gene of interest (Bolivar et al. 2001; Reilly et al. 2004). It is particularly difficult to control this type of variation, particularly in heterozygous mutant models where mutants are compared with wild-type siblings. The wild-type siblings will not inherit the window of strain contamination, whereas the mutants will, leading to bias between the groups that is independent of the gene mutation. It is important to consider this caveat when interpreting results from this type of model system.
Modification of cancer genes by genetic background:
Genes that play important roles in tumorigenesis can be modified by strain background and can themselves by be polymorphic between different strains (Table 1). A better understanding of how strain-specific polymorphisms modify the cancer phenotypes can improve the understanding of cancer pathways. The role of a mutant gene in cancer, i.e. whether it is an oncogene, a tumor suppressor gene, or has no apparent phenotype, can be dramatically affected by the genetic background. For example, p53−/+ mice develop mammary tumors on the BALB/c background, but not the C57BL/6J background (Kuperwasser et al. 2000; Blackburn et al. 2007; Koch et al. 2007). In another example, the expression level of Nf1 varies between the C57BL/6J and 129S4/SvJae strains to the same extent as knocking out one copy of the gene in C57BL/6J (Hawes et al. 2007). Both Mtor and Cdkn2a have been found to carry polymorphisms between the BALB/c and DBA/2 strains that modify tumorigenesis (Zhang et al. 1998; Bliskovsky et al. 2003). Studies of modifier effects on many cancer genes (Table 1) all contribute to the idea that any pathway relevant to cancer will likely be influenced by the genetic background of mouse models.
Table 1:
Genetic Effects on Cancer Genes in Mouse Models
| Gene/Protein | Modifier Effect | Strains | References |
|---|---|---|---|
| Alk | modifier | C57BL/6J vs C3H | Chun et al. 2010 |
| Apc | modified | C57BL/6J vs AKR/J,MA/MyJ, CAST, SWR/J, DBA/2J, BALB/cByJ; BTBR/Pas, A/J | Dietrich et al. 1993; Gould et al. 1996a; Gould et al. 1996b; Cormier et al. 1997; Shoemaker et al. 1998; Cormier et al. 2000; Kwong et al. 2007; Halberg et al. 2009 |
| Aurka | modifier | NIH/Ola vs M. spretus, SEG/Pas, SPRET/EiJ | Ewart-Toland et al. 2003 |
| Cdkn2a/p16INK4a | modifier | BALB/c vs DBA/2 | Zhang et al. 1998 |
| Egfr | modified | CF1 vs 129/Sv vs CD1; FVB/NJ, ICR/HaROS vs 129S6/SvEvTAC, BALC/cJ, NON/LtJ, NOD/LtJ, C57BL/6J, SJL/J, DBA2/J vs AKR/J, C3H/HeJ, SWR/J, ALR/LtJ, ALS/LtJ, APN, APS; C57BL/6J vs A/J | Threadgill et al. 1995; Strunk et al. 2004; Rinella and Threadgill 2012 |
| Fbxw7 | modifier | 129/Sv, M. spretus vs C57BL/6J, FVB/N, NIH/Ola | Kwon et al. 2012; Perez-Losada et al. 2012 |
| Hras | modified | NIH/Ola vs M. spretus; C57BL/6J, BALC/c vs C3H/HeJ, CBA, CF1; C57BL/6J vs FVB/N | Buchmann et al. 1991; Nagase et al. 2003; Wakabayashi et al. 2007 |
| Kras | modifier | C57BL/6J vs A/J | Lin et al. 1998 |
| Met | modified | C57BL/6J vs FVB/N | Graveel et al. 2010 |
| Mtor (Frap)/mTOR | modifier | BALB/c vs DBA/2 | Bliskovsky et al. 2003 |
| Nf1 | modified | 129S4/SvJae vs C57BL/6J | Hawes et al. 2007 |
| Nf1;Trp53 double mutant | modified | C57BL/6J vs 129/Sv, A/J, CAST/EiJ, SJL/J, CBA/J | Reilly et al. 2000; Reilly et al. 2004; Reilly et al. 2006; Hawes et al. 2007; Walrath et al. 2009; Amlin-Van Schaick et al. 2012a; Amlin-Van Schaick et al. 2012b |
| Ptch1 | modified | C57BL/6J vs BALB/c | Hahn et al. 2004 |
| Ptch1 | modifier | C57BL/6J vs FVB/N | Wakabayashi et al. 2007 |
| Tgfb1 | modified | C57BL/6J/Ola vs NIH/Ola | Bonyadi et al. 1997 |
| Tgfb1 | modifier | NIH/Ola vs M. spretus, SEG/Pas, SPRET/EiJ | Mao et al. 2006 |
| Trp53/p53 | modified | BALB/c vs MSM; CE/J vs 129/Sv; BALB/c vs C57BL/6J; FVB/N vs MSM/Ms, 129/Sv vs C57BL/6J | Harvey et al. 1993; Donehower et al. 1995; Kuperwasser et al. 2000; Biggs et al. 2003; Ochiai et al. 2003; Evans et al. 2004; Blackburn et al. 2007; Koch et al. 2007; Liang et al. 2008; Okumura et al. 2012; Bohringer et al. 2013 |
The Collaborative Cross Mouse Resource:
Although variation in genetic background can be a confounder in experiments with mouse models of cancer, understanding how this variation alters cancer phenotypes is critical for understanding cancer pathways and modeling the genetic heterogeneity found in human populations. Comparisons of different inbred strains have yielded some results (Table 1), but are limited in scope. Over the past decade a new resource, the Collaborative Cross, has been developed to more robustly study genetic variation in mice (Threadgill et al. 2002; Churchill et al. 2004; Chesler et al. 2008; Iraqi et al. 2008; Morahan et al. 2008; Collaborative Cross Consortium 2012; Threadgill and Churchill 2012). Eight founder strains (Figure 1B) were chosen to represent the diversity of the M. musculus species and the most commonly used classical inbred strains in biomedical research. The eight strains were mated pairwise to combine the eight genomes and then inbred to homozygosity to generate approximately 350 recombinant inbred lines. Because the lines are inbred, they are genetically stable and reproducible. The lines capture 90% of the genetic variation found across M. musculus and, unlike the classical inbred strains, the variation is evenly distributed across the genome, such that there are no “blind spots” for understanding the role of genetic background in disease (Roberts et al. 2007; Yang et al. 2011; Collaborative Cross Consortium 2012). It has been estimated that the Collaborative Cross panel carries 4–5 times the number of variants found in the human population. The Collaborative Cross will be a powerful tool in future research to improve mouse cancer models to represent the heterogeneity of the human population (see Protocol 2).
Summary:
Different mouse strains carry variants in cancer genes or in modifiers of cancer genes. If not properly controlled, experimental or control groups in cancer studies can carry biased representation of these variants, which can affect the phenotype as much as the experimental variable being studied. It is therefore important to take genetic background effects into consideration in designing mouse crosses and control groups. A new mouse resource, the Collaborative Cross, is making it more feasible to identify the genes and pathways underlying these genetic effects.
Acknowledgements:
This work was supported by the Intramural Research Program of the NIH, NCI.
References:
- Amlin-Van Schaick J, Kim S, Broman KW, Reilly KM. 2012a. Scram1 is a modifier of spinal cord resistance for astrocytoma on mouse Chr 5. Mamm Genome 23: 277–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amlin-Van Schaick JC, Kim S, DiFabio C, Lee MH, Broman KW, Reilly KM. 2012b. Arlm1 is a male-specific modifier of astrocytoma resistance on mouse Chr 12. Neuro Oncol 14: 160–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beamer WG, Shultz KL, Tennent BJ, Nadeau JH, Churchill GA, Eicher EM. 1998. Multigenic and imprinting control of ovarian granulosa cell tumorigenesis in mice. Cancer Res 58: 3694–3699. [PubMed] [Google Scholar]
- Blackburn AC, Hill LZ, Roberts AL, Wang J, Aud D, Jung J, Nikolcheva T, Allard J, Peltz G, Otis CN et al. 2007. Genetic mapping in mice identifies DMBT1 as a candidate modifier of mammary tumors and breast cancer risk. Am J Pathol 170: 2030–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliskovsky V, Ramsay ES, Scott J, DuBois W, Shi W, Zhang S, Qian X, Lowy DR, Mock BA. 2003. Frap, FKBP12 rapamycin-associated protein, is a candidate gene for the plasmacytoma resistance locus Pctr2 and can act as a tumor suppressor gene. Proc Natl Acad Sci U S A 100: 14982–14987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar VJ, Cook MN, Flaherty L. 2001. Mapping of quantitative trait loci with knockout/congenic strains. Genome Res 11: 1549–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchmann A, Bauer-Hofmann R, Mahr J, Drinkwater NR, Luz A, Schwarz M. 1991. Mutational activation of the c-Ha-ras gene in liver tumors of different rodent strains: correlation with susceptibility to hepatocarcinogenesis. Proc Natl Acad Sci U S A 88: 911–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick LH, Pertz LM, Broman KW, Bartolomei MS, Willard HF. 2006. Genetic control of X chromosome inactivation in mice: definition of the Xce candidate interval. Genetics 173: 2103–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Gusdon AM, Piganelli J, Leiter EH, Mathews CE. 2011. mt-Nd2(a) Modifies resistance against autoimmune type 1 diabetes in NOD mice at the level of the pancreatic beta-cell. Diabetes 60: 355–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesler EJ, Miller DR, Branstetter LR, Galloway LD, Jackson BL, Philip VM, Voy BH, Culiat CT, Threadgill DW, Williams RW et al. 2008. The Collaborative Cross at Oak Ridge National Laboratory: developing a powerful resource for systems genetics. Mamm Genome 19: 382–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun MG, Mao JH, Chiu CW, Balmain A, Hanahan D. 2010. Polymorphic genetic control of tumor invasion in a mouse model of pancreatic neuroendocrine carcinogenesis. Proc Natl Acad Sci U S A 107: 17268–17273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA Airey DC Allayee H Angel JM Attie AD Beatty J Beavis WD Belknap JK Bennett B Berrettini W et al. 2004. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat Genet 36: 1133–1137. [DOI] [PubMed] [Google Scholar]
- Collaborative Cross Consortium. 2012. The genome architecture of the Collaborative Cross mouse genetic reference population. Genetics 190: 389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier R, Hong K, Halberg R, Hawkins T, Richardson P, Mulherkar R, Dove W, Lander E. 1997. Secretory phospholipase Pla2g2a confers resistance to intestinal tumorigenesis. Nature Genetics 17: 88–91. [DOI] [PubMed] [Google Scholar]
- Cormier RT, Bilger A, Lillich AJ, Halberg RB, Hong KH, Gould KA, Borenstein N, Lander ES, Dove WF. 2000. The Mom1AKR intestinal tumor resistance region consists of Pla2g2a and a locus distal to D4Mit64. Oncogene 19: 3182–3192. [DOI] [PubMed] [Google Scholar]
- Dietrich W, Lander E, Smith J, Moser A, Gould K, Luongo C, Borenstein N, Dove W. 1993. Genetic identification of Mom-1, a major modifier locus affecting Min-induced intestinal neoplasia in the mouse. Cell 75: 631–639. [DOI] [PubMed] [Google Scholar]
- Dorward AM, Shultz KL, Ackert-Bicknell CL, Eicher EM, Beamer WG. 2003. High-resolution genetic map of X-linked juvenile-type granulosa cell tumor susceptibility genes in mouse. Cancer Res 63: 8197–8202. [PubMed] [Google Scholar]
- Ewart-Toland A, Briassouli P, de Koning JP, Mao JH, Yuan J, Chan F, MacCarthy-Morrogh L, Ponder BA, Nagase H, Burn J et al. 2003. Identification of Stk6/STK15 as a candidate low-penetrance tumor-susceptibility gene in mouse and human. Nat Genet 34: 403–412. [DOI] [PubMed] [Google Scholar]
- Gould K, Dietrich W, Borenstein N, Lander E, Dove W. 1996a. Mom1 is a semi-dominant modifier of intestinal adenoma size and multiplicity in Min/+ mice. Genetics 144: 1769–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould K, Luongo C, Moser A, McNeley M, Borenstein N, Shedlovsky A, Dove W, Hong K, Dietrich W, Lander E. 1996b. Genetic evaluation of candidate genes for the Mom1 modifier of intestinal neoplasia in mice. Genetics 144: 1777–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveel CR, DeGroot JD, Sigler RE, Vande Woude GF. 2010. Germline met mutations in mice reveal mutation- and background-associated differences in tumor profiles. PLoS ONE 5: e13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halberg RB, Waggoner J, Rasmussen K, White A, Clipson L, Prunuske AJ, Bacher JW, Sullivan R, Washington MK, Pitot HC et al. 2009. Long-lived Min mice develop advanced intestinal cancers through a genetically conservative pathway. Cancer Res 69: 5768–5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond S, Zhu R, Youngren KK, Lam J, Anderson P, Matin A. 2007. Chromosome X modulates incidence of testicular germ cell tumors in Ter mice. Mamm Genome 18: 832–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawes JJ, Tuskan RG, Reilly KM. 2007. Nf1 expression is dependent on strain background: implications for tumor suppressor haploinsufficiency studies. Neurogenetics 8: 121–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iraqi FA, Churchill G, Mott R. 2008. The Collaborative Cross, developing a resource for mammalian systems genetics: A status report of the Wellcome Trust cohort. Mamm Genome 9: 379–381. [DOI] [PubMed] [Google Scholar]
- Jandova J, Janda J, Sligh JE. 2012a. Changes in mitochondrial DNA alter expression of nuclear encoded genes associated with tumorigenesis. Exp Cell Res 318: 2215–2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jandova J, Shi M, Norman KG, Stricklin GP, Sligh JE. 2012b. Somatic alterations in mitochondrial DNA produce changes in cell growth and metabolism supporting a tumorigenic phenotype. Biochim Biophys Acta 1822: 293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsen A, Yu X, Truedsson L, Nived O, Sturfelt G, Ibrahim S, Bengtsson A. 2009. Mitochondrial DNA polymorphisms are associated with susceptibility and phenotype of systemic lupus erythematosus. Lupus 18: 309–312. [DOI] [PubMed] [Google Scholar]
- Koch JG, Gu X, Han Y, El-Naggar AK, Olson MV, Medina D, Jerry DJ, Blackburn AC, Peltz G, Amos CI et al. 2007. Mammary tumor modifiers in BALB/cJ mice heterozygous for p53. Mamm Genome 18: 300–309. [DOI] [PubMed] [Google Scholar]
- Kuperwasser C, Hurlbut GD, Kittrell FS, Dickinson ES, Laucirica R, Medina D, Naber SP, Jerry DJ. 2000. Development of spontaneous mammary tumors in BALB/c p53 heterozygous mice. A model for Li-Fraumeni syndrome. Am J Pathol 157: 2151–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YW, Kim IJ, Wu D, Lu J, Stock WA Jr., Liu Y, Huang Y, Kang HC, DelRosario R, Jen KY et al. 2012. Pten regulates Aurora-A and cooperates with Fbxw7 in modulating radiation-induced tumor development. Mol Cancer Res 10: 834–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong LN, Shedlovsky A, Biehl BS, Clipson L, Pasch CA, Dove WF. 2007. Identification of Mom7, a novel modifier of Apc(Min/+) on mouse chromosome 18. Genetics 176: 1237–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Festing MF, Devereux TR, Crist KA, Christiansen SC, Wang Y, Yang A, Svenson K, Paigen B, Malkinson AM et al. 1998. Additional evidence that the K-ras protooncogene is a candidate for the major mouse pulmonary adenoma susceptibility (Pas-1) gene. Exp Lung Res 24: 481–497. [DOI] [PubMed] [Google Scholar]
- Llamas B, Verdugo RA, Churchill GA, Deschepper CF. 2009. Chromosome Y variants from different inbred mouse strains are linked to differences in the morphologic and molecular responses of cardiac cells to postpubertal testosterone. BMC Genomics 10: 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morahan G, Balmer L, Monley D. 2008. Establishment of “The Gene Mine”: a resource for rapid identification of complex trait genes. Mamm Genome 19: 390–393. [DOI] [PubMed] [Google Scholar]
- Moreno-Loshuertos R, Acin-Perez R, Fernandez-Silva P, Movilla N, Perez-Martos A, Rodriguez de Cordoba S, Gallardo ME, Enriquez JA. 2006. Differences in reactive oxygen species production explain the phenotypes associated with common mouse mitochondrial DNA variants. Nat Genet 38: 1261–1268. [DOI] [PubMed] [Google Scholar]
- Nagase H, Mao JH, Balmain A. 2003. Allele-specific Hras mutations and genetic alterations at tumor susceptibility loci in skin carcinomas from interspecific hybrid mice. Cancer Res 63: 4849–4853. [PubMed] [Google Scholar]
- Perez-Losada J, Wu D, DelRosario R, Balmain A, Mao JH. 2012. Allele-specific deletions in mouse tumors identify Fbxw7 as germline modifier of tumor susceptibility. PLoS ONE 7: e31301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly KM, Broman KW, Bronson RT, Tsang S, Loisel DA, Christy ES, Sun Z, Diehl J, Munroe DJ, Tuskan RG. 2006. An imprinted locus epistatically influences Nstr1 and Nstr2 to control resistance to nerve sheath tumors in a neurofibromatosis type 1 mouse model. Cancer Res 66: 62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly KM, Loisel DA, Bronson RT, McLaughlin ME, Jacks T. 2000. Nf1;Trp53 mutant mice develop glioblastoma with evidence of strain-specific effects. Nat Genet 26: 109–113. [DOI] [PubMed] [Google Scholar]
- Reilly KM, Tuskan RG, Christy E, Loisel DA, Ledger J, Bronson RT, Smith CD, Tsang S, Munroe DJ, Jacks T. 2004. Susceptibility to astrocytoma in mice mutant for Nf1 and Trp53 is linked to chromosome 11 and subject to epigenetic effects. Proc Natl Acad Sci U S A 101: 13008–13013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinella ES, Threadgill DW. 2012. Efficacy of EGFR inhibition is modulated by model, sex, genetic background and diet: implications for preclinical cancer prevention and therapy trials. PLoS ONE 7: e39552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Pardo-Manuel de Villena F, Wang W, McMillan L, Threadgill DW. 2007. The polymorphism architecture of mouse genetic resources elucidated using genome-wide resequencing data: implications for QTL discovery and systems genetics. Mamm Genome 18: 473–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer SW, Lee C, Birney E, Altshuler DM, Eichler EE, Carter NP, Hurles ME, Feuk L. 2007. Challenges and standards in integrating surveys of structural variation. Nat Genet 39: S7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker AR, Moser AR, Midgley CA, Clipson L, Newton MA, Dove WF. 1998. A resistant genetic background leading to incomplete penetrance of intestinal neoplasia and reduced loss of heterozygosity in ApcMin/+ mice. Proc Natl Acad Sci U S A 95: 10826–10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver L 1995. Mouse Genetics: Concepts and Applications. Oxford University Press, New York. [Google Scholar]
- Strunk KE, Amann V, Threadgill DW. 2004. Phenotypic variation resulting from a deficiency of epidermal growth factor receptor in mice is caused by extensive genetic heterogeneity that can be genetically and molecularly partitioned. Genetics 167: 1821–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threadgill DW, Churchill GA. 2012. Ten years of the Collaborative Cross. Genetics 190: 291–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threadgill DW, Dlugosz AA, Hansen LA, Tennenbaum T, Lichti U, Yee D, LaMantia C, Mourton T, Herrup K, Harris RC et al. 1995. Targeted disruption of mouse EGF receptor: effect of genetic background on mutant phenotype. Science 269: 230–234. [DOI] [PubMed] [Google Scholar]
- Threadgill DW, Hunter KW, Williams RW. 2002. Genetic dissection of complex and quantitative traits: from fantasy to reality via a community effort. Mamm Genome 13: 175–178. [DOI] [PubMed] [Google Scholar]
- Wakabayashi Y, Mao JH, Brown K, Girardi M, Balmain A. 2007. Promotion of Hras-induced squamous carcinomas by a polymorphic variant of the Patched gene in FVB mice. Nature 445: 761–765. [DOI] [PubMed] [Google Scholar]
- Walrath JC, Fox K, Truffer E, Gregory Alvord W, Quinones OA, Reilly KM. 2009. Chr 19(A/J) modifies tumor resistance in a sex- and parent-of-origin-specific manner. Mamm Genome 20: 214–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss H, Wester-Rosenloef L, Koch C, Koch F, Baltrusch S, Tiedge M, Ibrahim S. 2012. The mitochondrial Atp8 mutation induces mitochondrial ROS generation, secretory dysfunction, and beta-cell mass adaptation in conplastic B6-mtFVB mice. Endocrinology 153: 4666–4676. [DOI] [PubMed] [Google Scholar]
- Yang H, Wang JR, Didion JP, Buus RJ, Bell TA, Welsh CE, Bonhomme F, Yu AH, Nachman MW, Pialek J et al. 2011. Subspecific origin and haplotype diversity in the laboratory mouse. Nat Genet 43: 648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Gimsa U, Wester-Rosenlof L, Kanitz E, Otten W, Kunz M, Ibrahim SM. 2009a. Dissecting the effects of mtDNA variations on complex traits using mouse conplastic strains. Genome Res 19: 159–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Wester-Rosenlof L, Gimsa U, Holzhueter SA, Marques A, Jonas L, Hagenow K, Kunz M, Nizze H, Tiedge M et al. 2009b. The mtDNA nt7778 G/T polymorphism affects autoimmune diseases and reproductive performance in the mouse. Hum Mol Genet 18: 4689–4698. [DOI] [PubMed] [Google Scholar]
- Zhang S, Ramsay ES, Mock BA. 1998. Cdkn2a, the cyclin-dependent kinase inhibitor encoding p16INK4a and p19ARF, is a candidate for the plasmacytoma susceptibility locus, Pctr1. Proc Natl Acad Sci U S A 95: 2429–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]