Abstract
Cardiovascular disease constitutes an important threat to humans after space missions beyond the Earth’s magnetosphere. Epigenetic alterations have an important role in the etiology and pathogenesis of cardiovascular disease. Previous research in animal models has shown that protons and 56Fe ions cause long-term changes in DNA methylation and expression of repetitive elements in the heart. However, astronauts will be exposed to a variety of ions, including the smaller fragmented products of heavy ions after they interact with the walls of the space craft. Here, we investigated the effects of 16O on the cardiac methylome and one-carbon metabolism in male C57BL/6J mice. Left ventricles were examined 14 and 90 days after exposure to space-relevant doses of 0.1, 0.25, or 1 Gy of 16O (600 MeV/n). At 14 days, the two higher radiation doses elicited global DNA hypomethylation in the 5’-UTR of Long Interspersed Nuclear Elements 1 (LINE-1) compared to unirradiated, sham-treated mice, whereas specific LINE-1 elements exhibited hypermethylation at day 90. The pericentromeric major satellites were affected both at the DNA methylation and expression levels at the lowest radiation dose. DNA methylation was elevated, particularly after 90 days, while expression showed first a decrease followed by an increase in transcript abundance. Metabolomics analysis revealed that metabolites involved in homocysteine remethylation, central to DNA methylation, were unaffected by radiation, but the transsulfuration pathway was impacted after 90 days, with a large increase in cystathione levels at the lowest dose. In summary, we observed dynamic changes in the cardiac epigenome and metabolome three months after exposure to a single low dose of oxygen ions.
Keywords: epigenetics, ionizing radiation, cardiotoxicity, methionine, DNA methylation, one-carbon metabolism, repetitive elements, space radiation
INTRODUCTION
The potential health hazards associated with space radiation are a major obstacle preventing deep space exploration [Chancellor et al., 2014]. Beyond the protective magnetosphere of the Earth, astronauts are exposed to radiation from galactic cosmic rays and solar flares. For even the shortest round-trip to Mars, exposure for the crew is estimated to reach 0.66 Sv (or Gy) [Zeitlin et al., 2013]. Furthermore, contrary to terrestrial radiation, current technology does not provide effective shielding against the type of high energy particles encountered in space. It is therefore crucial to learn more about the types of long-term biological changes associated with exploration beyond the orbit of the Earth.
Experimental evidence suggests cardiac remodeling, as well as apoptotic and inflammatory responses in the mouse heart as a result of exposure to high-LET radiation [Boerma et al., 2016; Ramadan et al., 2016; Tungjai et al., 2013; Boerma et al., 2015; Hughson et al., 2018]. Studies of cancer survivors indicate that cardiac injury due to exposure to ionizing radiation increases linearly with mean dose of radiation to the heart, and progresses very slowly, with onset observed decades after exposure [Darby et al., 2013]. Recent studies indicate that epigenetic and metabolic alterations may substantially contribute to pathogenesis of cardiovascular disease [Stenvinkel et al., 2007; Sharma et al., 2008]. DNA methylation is one of the key epigenetic mechanisms for the regulation of genetic information. It is responsive to environmental exposures, providing a possible mechanism linking radiation exposure and late-onset injury to the heart.
Methyl groups for DNA methylation are supplied through the methyl donor methionine, with homocysteine as a byproduct. The methionine cycle is critical for normal heart development, and folic acid supplementation during pregnancy is associated with a decreased incidence of congenital heart defects [Feng et al., 2015; Liu et al., 2016]. Beyond the development period, an elevated level of homocysteine is an independent risk factor for cardiovascular disease [Perry et al., 1995; Wald et al., 2011], highlighting the importance of the methionine cycle in cardiac health throughout life.
Studies report that radiation-induced changes in DNA methylation stem primarily from the repetitive elements rather than individual genes [Nzabarushimana et al., 2014; Miousse et al., 2017b; Koturbash et al., 2016]. Of particular interest are two subtypes of repetitive elements; LINE-1 retrotransposons and satellite DNA. LINE-1 repeats cover about 20% of the murine genome and are heavily methylated within their 5’-untranslated regions (UTRs) [Miousse and Koturbash, 2015]. Satellite DNA repeats are characteristic of the pericentromeric regions. It is a less abundant, but still heavily methylated type of repetitive element. DNA methylation is a critical regulator of aberrant transcription for both elements, and DNA hypomethylation is associated with their reactivation and insertional mutagenesis (for LINE-1 elements) and chromosomal aberrations (for satellite DNA) [Miousse and Koturbash, 2015; Miousse et al., 2015b; Koturbash et al., 2007; Miousse et al., 2015a; Qu et al., 1999; Ji et al., 1997]. Recent studies demonstrated alterations in DNA methylation during the pathogenesis of a number of heart diseases [Baccarelli et al., 2010; Kim et al., 2010; Gilsbach et al., 2014; Fiorito et al., 2014], DNA hypomethylation and accumulation of satellite DNA transcripts has also been observed in humans diagnosed with heart failure [Haider et al., 2012]. Effects of high-LET irradiation on DNA methylation of those two types of repetitive elements in the mouse heart, as well as alterations to one-carbon metabolism, were reported [Koturbash et al., 2016].
In this study, we investigate the short- and long-term (14 and 90 days post exposure, respectively) effects of exposure to one of the under-investigated heavy ions yet relevant for the radiation environment inside a space craft, oxygen (16O), on the mouse cardiac methionine cycle using epigenetic and metabolomic approaches. The most biologically relevant radiation source in free space is 56Fe [Cucinotta et al., 2003], and we previously reported epigenetic changes in the heart associated with this type of radiation [Koturbash et al., 2016]. However, interaction with the hull of the spacecraft and space suit materials also creates ions of smaller masses, such as 16O [Walker et al., 2013]. Previous results from our group and others indicate a distinctly large cardiovascular toxicity associated with exposure to 16O, which prompted us to take a closer look at the epigenetic signature of 16O in the heart [Ramadan et al., 2016; Yan et al., 2014; Seawright et al., 2019]. Levels of radiation exposure have been estimated for different types of charged particles in outer space and on the surface of Mars [Nelson, 2016; Cucinotta et al., 2003; Zeitlin et al., 2013]. Several Russian, European and Canadian space agencies have proposed 1 Sv (or 1 Gy) as the astronaut career exposure limit, a dose only slightly above the high estimates for the cumulative exposure associated with a round trip mission to Mars. Based on these estimates, we analyzed doses of 0.1, 0.25, and 1 Gy of 16O radiation (600 MeV/n) in an effort to replicate the most current estimation of exposures associated with a round trip to Mars, but with all of the dose coming from a single particle species.
MATERIALS AND METHODS
Animals and whole body irradiation
Four to 8 week-old male C57BL/6J mice purchased from the Jackson Laboratory (Bar Harbor, ME) were housed 5 per cage at the University of Arkansas for Medical Sciences (UAMS). At 6 months of age, mice were shipped to the NASA Space Radiation Laboratory (NSRL) at Brookhaven National Laboratories (BNL) in Upton, NY. Two cages, each with 5 mice, were assigned to one of the 8 groups (n=10/group) for a total of 80 mice in this study. After a one-week acclimation period, the mice received a single dose of 0.1 Gy, 0.25 Gy, or 1.0 Gy 16O (600 MeV/n) at a dose rate of 0.25–0.26 Gy/min during NSRL run 15A. We selected 600 MeV/n as a representative single energy value for the most abundant subset of ions in galactic cosmic rays (100-1000 MeV/n), as reported previously [Nelson, 2016], For each exposure, animals were individually placed into clear Lucite cubes (3 in × 1½ in × 1½ in) with breathing holes, and 4 – 5 unanesthetized mice were placed in the radiation beam. The total field of exposure is 15 ×l5 cm2, with typical beam uniformities of ±2%. The Lucite cubes represent the minimal amount of shielding for any biological sample irradiated at the NSRL beam line, amounting to approximately 1:10 (0.1 g/cm2) of the amount of matter provided by air (1.12 g/cm2) in the ion path between the accelerator vacuum environment and the animal (see dosimetry below). Unirradiated control mice were placed into the same enclosures for the same amount of time, but were not exposed to radiation. One day after irradiation, the mice were shipped to the University of Arkansas for Medical Sciences (UAMS), where they were housed under a constant 12 h light:dark cycle. A standard soy-protein free rodent diet (2020X, Harlan Teklad, Indianapolis, IN) and water were provided ad libitum. During the entire experiment, control mice were not housed together in the same cage with irradiated mice. Tissue harvest was performed as previously described by Seawright et al.[Seawright et al., 2019]. Briefly, mice were anesthetized and blood collected through the vena cava. The left ventricle was cut into four sections and flash-frozen. Unirradiated controls were sacrificed at both time points. The time points were chosen to represent short term (2 weeks) and long term (90 days) effects. All procedures in this study were approved by the Institutional Animal Care and Use Committees of the University of Arkansas for Medical Sciences and Brookhaven National Laboratory.
Dosimetry was performed by the NASA Space Radiation Laboratory physics dosimetry group at BNL to ensure the quality of exposure. Briefly, The NASA Space Radiation Laboratory beamline at BNL recorded the charge delivered to a transmission ion chamber placed just in front of the animals. The transmission ion chamber was calibrated against a National Institute of Standards and Technology (NIST)-traceable 1.0 cm3 thimble ion chamber (EG&G, Inc.) which was placed at the target position. The doses indicated in the present study were thus the absorbed doses by the animals in the ion chamber at the target surface location on the beamline. Based on calculations for the NSRL beamline performed with the GCR Event-Based Risk Model (GERMCode) Code software (https://software.nasa.gov/software/MSC-24760-1), the total particle range is in the order of 30 cm, ensuring a uniform exposure of the entire animal during irradiation [Cucinotta et al., 2011].
Analysis of LINE-1 and major satellites DNA methylation
DNA and RNA were extracted simultaneously from flash-frozen left ventricular tissue specimens using the AllPrep extraction kit (Qiagen, Germantown, MD). Nucleic acid quality and concentration was assessed by spectrophotometry (Nanodrop 2000, ThermoFisher Scientific, Waltham, MA). Analysis of the DNA methylation was performed from 250 ng of genomic DNA digested with a 5-enzyme digestion protocol using 1 U each of SmaI, HpaII, HhaI, AciI, and BstUI (New England Biolabs, Ipswitch, MA) as described before [Miousse et al., 2015b]. Digested DNA was then analyzed with intercalating dye (SYBR Select, ThermoFisher Scientific) by qRT-PCR on a ViiA 7 Real-Time PCR System (Applied Biosystems, Foster City, CA). Assays for determination of 5’-UTR LINE-1 and satellite DNA methylation are provided in Supplementary Table S1. Fold changes were calculated using the ΔΔCt method and normalized towards readings from a LINE-1 element ORF1 region that lacks CpG sites.
Gene expression analysis
cDNA was synthesized from 500 ng RNA using the Superscript ViLO reverse transcriptase kit (Life Technologies) according to the manufacturer’s protocol. Quantitative realtime PCR (qRT-PCR) was performed with Taqman Universal Master Mix (ThermoFisher Scientific) according to the manufacturer’s protocol. Expression of Dnmt1, Dnmt3a, Uhrf1, and repetitive elements was normalized to the internal control gene Hprt and expressed as fold change according to the ΔΔCT method. Primers are listed in Supplementary Table S1.
Analysis of one-carbon metabolism pathway
The tissue samples were homogenized in 200 μl of extraction buffer (methanol: Water [70:30]) containing the internal standards (metabolite standards labeled with stable isotope). Subsequently, 500 μL of ACN: MEOH (50:50) was added to the mixture to facilitate protein precipitation. The tubes were vortexed and incubated for 20 min on ice followed by centrifugation at 13,000 rpm for 20 min at 4°C.
The supernatant was transferred to another Eppendorf tube and are dried using speed vac and reconstituted in 100 μL of Methanol: Water (50:50) for MS analysis. Targeted quantification of metabolites participating in single carbon metabolism pathway was performed using stable-isotope labeled multiple reaction monitoring mass spectrometry (SID-MRM-MS). The samples were resolved on an Acquity UPLC BEH Amide 1.7μm, 2.1 × 100 mm column online with a triple quadrupole mass spectrometer (Xevo-TQ-S, Waters Corporation, USA) operating in the multiple reaction monitoring (MRM) mode. The sample cone voltage and collision energies were optimized for each metabolite to obtain maximum ion intensity for parent and daughter ions using “IntelliStart” feature of MassLynx software (Waters Corporation, USA). The instrument parameters were optimized to gain maximum specificity and sensitivity of ionization for the parent and daughter ions. Signal intensities from all MRM Q1/Q3 ion pairs were ranked to ensure selection of the most intense precursor and fragment ion pair for MRM-based quantitation. This approach resulted in selection of cone voltages and collision energies that maximized the generation of each fragment ion species.
Statistical analysis
For both DNA methylation and expression, values for each LINE-1 element family were expressed as fold-change from the mean of control animals. Each animal provided a value for each of the 18 LINE-1 elements. The values from a single animal are like repeated measures for longitudinal data, where evolutionary age of the LINE-1 element is the “longitudinal” factor. We thus used a repeated measures ANOVA having dose, LINE-1 element, and their interaction as factors. The dose factor was among-animal, the other two were within-animal. We analyzed values taken at 14 days separate from values taken at 90 days because the variances differed. We used the Bayesian Information Criterion to choose the best-fitting (within-animal) covariance structure. The structures we considered were exchangeable, 1st order autoregressive, Toeplitz, and general. We then compared each non-0 dose to control averaged over all LINE-1 elements and within a LINE-1 element, and present 95% confidence intervals (CIs) for a difference from control. In so doing, we made 57 comparisons within a repeated measures ANOVA. Since each LINE-1 element is of interest, we do not adjust the significance level for multiple comparisons, but rather provide the positive false discovery rates (pFDR) estimated with Storey’s 3rd algorithm [Storey, 2002]. The positive false discovery rate is the proportion of Type I errors (i.e., false discoveries), given at least one significant result. We used SAS/STAT software, version 9.4, SAS System for Windows (SAS Institute, Cary, NC) for the repeated measures ANOVAs; specifically, using the MIXED procedure, employing the Kenward-Roger method for estimating error degrees of freedom. For determination of pFDR, we used internally created code in R version 3.2.2; available upon request to RDL.
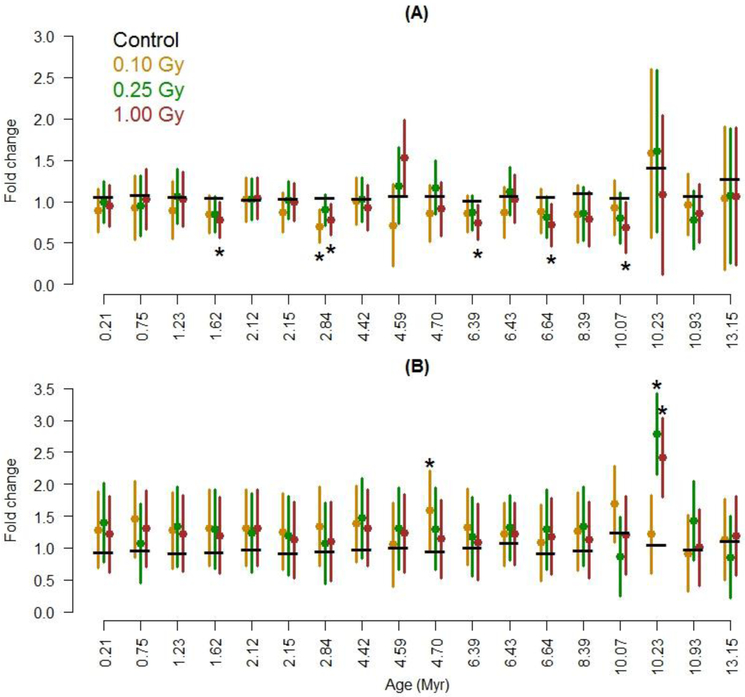
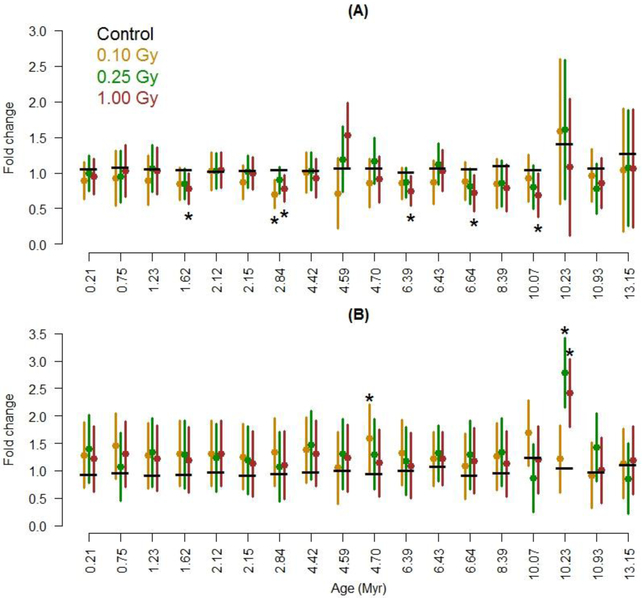
In Figures 1 and 2, we present error bars which are half the width of 95% confidence intervals for a difference from the control (for the given LINE-1 repetitive element). These error bars include variability from both the unirradiated control and the radiation group to which the control is compared. To obtain a 95% confidence interval for a difference, subtract the control’s mean from the lower and upper bound of the error bar. For example, if the control’s mean is 1.0 and the error bars start and end at 0.8 and 1.6, the 95% CI for a difference is (−0.2, 0.6).
Figure 1:
Effects of 16O exposure on methylation of different families of LINE-1 repetitive elements, ordered by evolutionary age. Measured (A) 14 days and (B) 90 days after irradiation. The means for control animals are indicated with a horizontal black bar; means for non-0 radiation groups are in color. Error bars that do not cover the control mean indicate a statistical difference at p ≤ 0.05 (see Statistical Analysis for more details), as do asterisks. Each radiation group had 10 mice (40 total).
Figure 2:
Effects of 16O exposure on expression of different families of LINE-1 repetitive elements, ordered by evolutionary age. Measured (A) 14 days and (B) 90 days after irradiation. The means for control animals are indicated with a horizontal black bar; means for non-0 radiation groups are in color. Error bars that do not cover the control mean indicate a statistical difference at p ≤ 0.05 (see Statistical Analysis for more details), as do asterisks. Each radiation group had 10 mice (40 total).
Sample size considerations
The sample sizes for dose×day groups were based on primary outcomes from a larger study not reported here [Wang et al., 2017]. Based on these sample sizes and variances estimates from a prior experiment (described below), we computed effect sizes detectable with 0.80 power using 0.05 significance level t-tests conducted within the aforementioned repeated measures ANOVA framework. We first estimated error variances and within-mouse correlations (in an exchangeable covariance structure) with methylation values reported in Miousse et al. [Miousse et al., 2017c]. Variances ranged from 0.06 to 0.13 (SDs from 0.24 to 0.36) and within-mouse correlations ranged from 0.24 to 0.58. We assumed a correlation of 0.40. With 10 mice per dose×day group, 1.62 SD was the detectable difference in means between control and a non-0 radiation dose for any given LINE-1 element. This 1.62 SDs translates to mean differences between 0.39 and 0.58 units depending on the SD. For example, if a non-0 radiation dose truly causes methylation for a given LINE-1 element to be 1.39 compared to control’s standardized 1.0, then this difference would be detectable at 0.80 power. We wrote code particular to these calculations in SAS/STAT® (available upon request to RDL).
RESULTS
Repetitive element DNA methylation
To investigate the effect of 16O radiation on the cardiac epigenome, DNA methylation at LINE-1 elements and satellite DNA was first measured. DNA methylation status of 18 most represented LINE-1 elements that evolved in the murine genome during the last 13 million years (Myr) was measured. At the 14 day time point, exposure to two doses of 16O (0.25 and 1 Gy) led to significant global DNA hypomethylation of LINE-1 elements within their 5’-UTRs, independently of evolutionary age (Fig.1A). Comparing the irradiated groups to control in each of the 18 LINE-1 families showed either an estimated decrease or a significant decrease in DNA methylation with radiation at the 0.25 and 1 Gy doses. Among the 57 comparisons made within the 14 day time point, 7 were significant. The positive false discovery rate (pFDR) was 16%, and its 95% upper confidence bound was 50%. This means the estimated number of false discoveries was likely 1 of 7, but could have been as high as 4 out of 7.
At the 90 day time point, the variability was greater than that observed in the 14 day values. Across the LINE-1 families, DNA methylation experienced in irradiated animals differed little from that in control animals (Fig 1B). Comparing the exposed groups to control in each of the 18 LINE-1 families, the L1MdA_V family (2.84 Myr) experienced significant DNA hypermethylation after exposure to the lowest dose, 0.1 Gy (Figure S1). This was the only significant comparison among 57; the pFDR was over 100% indicating this significant result could be a false discovery.
Repetitive element expression
Along with DNA methylation, the levels of transcript coding for LINE-1 elements and major satellite were also assessed. Exposure to 16O tended to decrease overall expression of the LINE-1 families at the 14 day time point, but not significantly so for the 3 dose groups (p-values of 0.14, 0.41, and 0.14 for doses 0.10, 0.25, and 1.0 Gy, respectively). Among the 57 comparisons, 2 were significant. The pFDR was 91%, indicating both could be false discoveries.
For the 90 day time point, overall expression was estimated to be increased after 16O exposure (Fig. 2A). However, these increases in the 90 day time points were not significant (p-values of 0.13, 0.10, 0.16 for doses 0.10, 0.25, and 1.0 Gy, respectively). Comparing the non-0 radiation dose groups to control for each of the 18 LINE-1 families, the L1Lx_III family (10.23 Myr) was significantly overexpressed for the 0.25 and 1.0 Gy groups. Expression of the L1MdA_VI family (4.7 Myr) was also higher than that for the control group in animals exposed to 0.1 Gy. Expression in other LINE-1 families was little affected. The pFDR for the 3 significant comparisons among the 57 was 36%. This means that we expect one false discovery among these 3. We did not observe any correlation between methylation and expression in LINE-1 elements.
Pericentromeric satellite
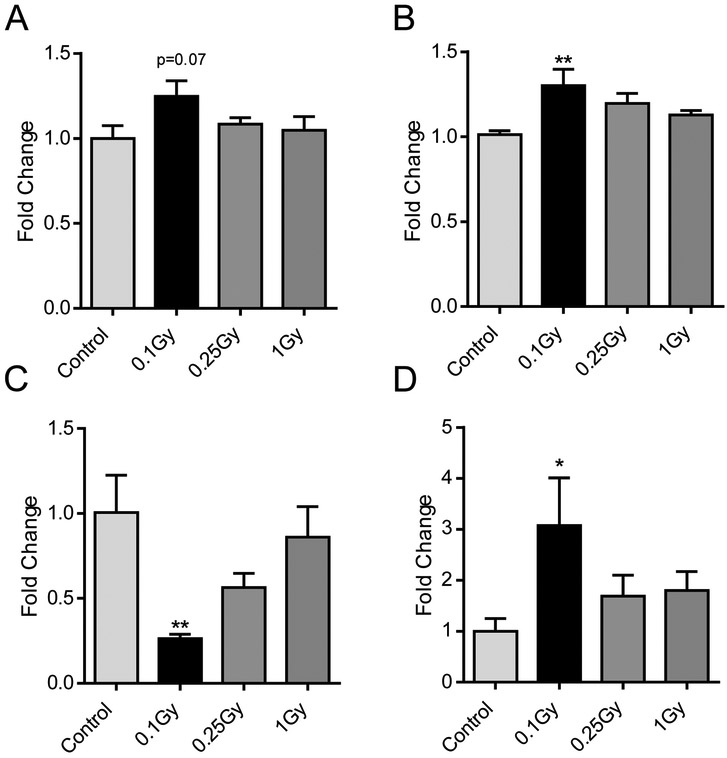
DNA methylation was then measured at the pericentromeric major satellite DNA. Contrary to LINE-1, we did not observe hypomethylation of major satellites 14 days after irradiation (Fig.3A). Interestingly, an upward, near-significant trend towards hypermethylation was identified at the lowest dose (0.1 Gy, p=0.07). The same trend was found to be significant at 90 days (Fig.3B, p= 0.009), similar to what was identified in the LINE-1 families at the same low dose and late time point. Of particular interest are the effects of 16O on the expression at these regions. In this study, a four-fold decrease was observed in major satellite expression 14 days after exposure to 0.1 Gy of 16O (p= 0.006) (Fig. 3C). This was followed at the 90 day time point by a four-fold increase (p= 0.04) of major satellites, once again limited to the lowest radiation dose (Fig.3D).
Figure 3:
Effects of radiation on methylation (A,B) and expression (C,D) of the pericentromeric major satellite. Values for 14 (A,C) and 90 (B, D) days after IR. Bars indicate average and SEM, N=10. *: Dunnett's multiple comparisons test, p< 0.05, **: Dunnett's multiple comparisons test, p< 0.01
Gene expression in the DNA methylation machinery
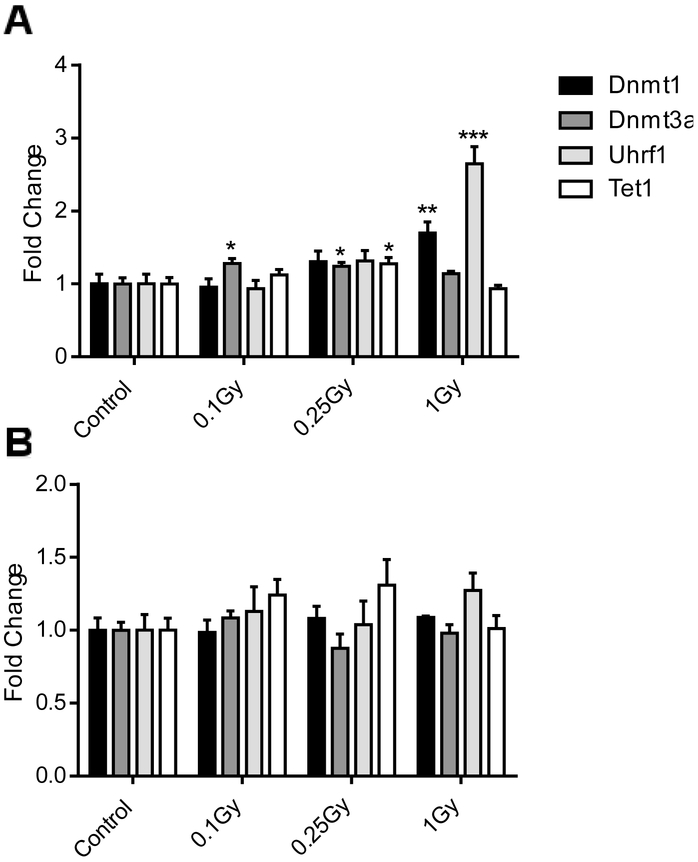
Exposure to 16O also significantly affected the DNA methylation machinery. The expression of major maintenance DNA (cytosine-5)-methyltransferase 1 (Dnmt1) showed a 2-fold increase (p= 0.004) in response to 1 Gy 14 days after irradiation (Fig. 4A). Similarly, the accessory protein E3 ubiquitin-protein ligase (Uhrf1) mimicked the patterns of Dnmt1 expression, with a nearly three-fold increase after 1 Gy of irradiation (p<0.001). At the same time, expression of the de novo methyltransferase Dnmt3a was affected primarily by lower doses (0.1 and 0.25 Gy; p=0.01 and 0.02, respectively). Elevation of the expression of the DNA demethylase, methylcytosine dioxygenase (Tet1) 14 days after 0.25 Gy of 16O was also found (1.25 fold, p=0.03). The observed effects were limited to an early time point only and were completely resolved by the 90 day time point (Fig. 4B).
Figure 4:
Expression of genes involved in DNA methylation, 14 (A) and 90 (B) days after radiation. Bars indicate average and SEM, N=10. *: Dunnett's multiple comparisons test, p< 0.05, **: Dunnett's multiple comparisons test, p< 0.01, ***: Dunnett's multiple comparisons test, p< 0.001
Metabolites in the one-carbon pathway
Given the changes observed in DNA methylation, the levels of six different metabolites in the one-carbon metabolism were analyzed in the hearts of mice exposed to 16O radiation. The specific targets were the methyl donors methionine and betaine, the by-products of DNA methylation S-adenosylhomocysteine (SAH) and homocysteine, and the metabolic product of homocysteine through the transsulfuration pathway, cystathione. S-adenosylmethionine was below the detection limit in these samples. Further down the transsulfuration pathway, glutathione (GSH), an antioxidant, was also measured, as well as its oxidized form GSSG. Fourteen days after exposure, there was no significant change in any of the metabolites measured in left ventricular tissue specimens of mice exposed to either 0.1, 0.25, or 1 Gy of 16O. Interestingly, it is at the later time point, 90 days post exposure, that effects were visible. An increase in cystathione levels was measured compared to control for the 0.1 and 0.25 Gy doses (Table 1 and Supplementary Table S2). The fold change compared to control was the largest after exposure to the lowest dose of 0.1 Gy (12 fold, p< 0.0001), and was still significant although lower for the 0.25 Gy dose (6 fold, p=0.06). We also saw a decrease in GSH for the 0.25 Gy dose. Our observations indicate that there is a non-linear response in the levels of metabolites in the transsulfuration branch of one-carbon metabolism.
TABLE 1:
Heart Metabolites
| METABOLITES | 16O- 14 DAYS | |||||
|---|---|---|---|---|---|---|
| 0.1 Gy | 0.25 Gy | 1 Gy | ||||
| FC | SD | FC | SD | FC | SD | |
| BETAINE | 1.54 | 1.05 | 2.38 | 2.51 | 1.26 | 0.34 |
| L-METHIONINE | 1.26 | 0.29 | 1.18 | 0.27 | 1.13 | 0.17 |
| SAH | 0.97 | 0.07 | 0.97 | 0.11 | 0.97 | 0.06 |
| L-HOMOCYSTEINE | 1.88 | 1.70 | 1.59 | 1.17 | 1.31 | 1.02 |
| CYSTATHIONE | 1.28 | 1.06 | 1.42 | 1.52 | 1.00 | 0.88 |
| GSH | 0.92 | 0.18 | 0.95 | 0.27 | 1.08 | 0.46 |
| GSSG | 1.00 | 1.29 | 0.87 | 0.85 | 1.05 | 0.60 |
| GSH/GSSG | 1.00 | 0.59 | 1.41 | 0.92 | 1.07 | 0.61 |
| METABOLITES | 16O- 90 DAYS | |||||
| 0.1 Gy | 0.25 Gy | 1 Gy | ||||
| FC | SD | FC | SD | FC | SD | |
| BETAINE | 3.23 | 2.93 | 0.89 | 0.50 | 0.82 | 0.37 |
| L-METHIONINE | 1.01 | 0.42 | 0.79 | 0.41 | 0.68 | 0.28 |
| SAH | 1.05 | 0.06 | 1.02 | 0.07 | 1.04 | 0.06 |
| L-HOMOCYSTEINE | 0.60 | 0.24 | 1.12 | 0.60 | 0.83 | 0.35 |
| CYSTATHIONE | 12.97### | 5.33 | 6.37## | 4.88 | 1.25 | 0.93 |
| GSH | 0.87 | 0.38 | 0.56** | 0.18 | 0.77 | 0.33 |
| GSSG | 0.49 | 0.24 | 1.20 | 0.86 | 0.56 | 0.41 |
| GSH/GSSG | 1.41 | 1.23 | 0.52 | 0.54 | 1.41 | 1.08 |
FC: Fold change compared to control
Dunn's multiple comparisons test, p< 0.001
Dunn's multiple comparisons test, p< 0.01
Dunnett's multiple comparisons test, p< 0.01
DISCUSSION
The present study investigated the short and long term effects of exposure to space-relevant doses of high energy 16O ions on DNA methylation and methyl group metabolism in the heart. Previous studies indicated changes in DNA methylation, repetitive element repression, and expression of methylation-related genes in this organ caused by exposure to two other types of charged particles, protons and 56Fe [Koturbash et al., 2016]. This new study confirms and expands these findings. In addition, the epigenetic changes described here are associated with functional alterations suggesting a reduced systolic function, with no indication of fibrosis, in these same animals [Seawright et al., 2019].
A limitation of our study is the use of a single exposure to a single type of radiation. By comparison, a more constant rate of exposure to many types of heavy ion with short bursts of high energy particles is expected for travel beyond the orbit of the Earth. While models are emerging for continuous exposure to low-LET radiation [Mao et al., 2016], the reliance on synchrotron-generated particles for the study of high-LET radiation remains a challenge. Therefore, we utilized the best available yet imperfect exposure model provided by the NASA Space Radiation Laboratory at the Brookhaven National Laboratory at the time of the study. Future experiments may take advantage of the implementation of galactic cosmic ray simulation, that is now enabling researchers to study multiple ion beams simultaneously. The analysis was also performed on whole left ventricles, composed of a heterogeneous mix of cell types. We expect that the composition of the tissue is predominantly of muscular origin, with a smaller vascular contribution. Future studies may address potential differences in the response of these different tissue types within the heart.
In concordance with many others studying DNA methylation and radiation exposure [Koturbash et al., 2016; Miousse et al., 2017a; Aypar et al., 2011], we observed overall DNA hypomethylation in the 5’UTR of the retrotransposon LINE-1 14 days after exposure. This region of LINE-1 is particularly important as it contains the regulatory elements involved in transcription. An inverse relationship between DNA methylation within the CpG-rich 5’-UTR and LINE-1 expression is typically expected. However, in this experiment, both DNA methylation and expression of LINE-1 decreased. Only one family, L1MdA_II, showed changes in both DNA methylation and expression.
A different pattern emerged in samples collected 90 days after exposure. At this later time point, there was no global change in LINE-1 DNA methylation, and two specific LINE-1 families showed a significant DNA hypermethylation. This effect was restricted to the lowest, 0.1 Gy dose. This same hypermethylation was observed after the lowest dose and at the late time point in major satellite. A low-dose effect was also observed at both 14 and 90 days for the expression of major satellite. In our previous study on protons and 56Fe irradiation, a decrease in satellites expression was observed 1 week after irradiation, followed by the accumulation of their mRNA transcripts at day 90 post exposure [Koturbash et al., 2016]. This pattern of early decrease followed by an increase was thus replicated in the current study. Interestingly, an overall increase in LINE-1 expression was observed at the 90 day time point, although for this element, the increase was found evenly throughout the three doses tested.
The disconnect observed between DNA methylation and expected responses in transcript levels may be indicative of additional and possible compensatory mechanisms of expression regulation, histone modifications for example. For instance, in our previous study, we reported histone-mediated reactivation of LINE-1 after exposure to protons and 56Fe despite DNA hypermethylation observed at the 5’-UTRs of the elements [Prior et al., 2016], It is also possible that the expression of Dnmt1 and Urhf1 is a compensatory mechanism/response activated under the highest radiation dose at the early time point. This activation may contribute to successfully prevent loss of DNA methylation, providing an explanation for the lack of a dose-response relationship. Some studies support a role for Dnmt1 and Uhrf1 in the silencing of LINE-1 [Sharif et al., 2007] and of Uhrf1 in the silencing of major satellite [Papait et al., 2007]. Alternatively, the increase in Dnmt1 and Uhrf1 expression may be related to their role in DNA repair [Ha et al., 2011; Tian et al., 2015], independent of their effect on DNA methylation. Indeed, increases in DNA damage was noted in hematopoietic stem cells isolated from these same animals [Chang et al., 2016]. The expression of the de novo methyltransferase Dnmt3a and the methylcytosine dioxygenase Tet1, neither of which is known to have a direct role in DNA repair, showed some increase at the low and/or middle dose at the early time point, but none of these effects on the DNA methylation machinery genes were found by the later time point. These early changes in gene expression parallel the decrease in LINE-1 DNA methylation identified at the 14 day time point.
Lastly, the levels of the different metabolites in the one-carbon metabolism that provides and recycles methyl groups to transfer to DNA were measured. Methionine, in addition to its role in protein synthesis, is an important source of methyl groups, and contributes to the maintenance of the normal redox status via synthesis of cysteine and glutathione. Other groups have identified alterations in the stability and absorption of one-carbon intermediates and cofactors induced by radiation [Batra et al., 2004; Guren et al., 2004; Kesavan et al., 2003]. Furthermore, multifaceted interactions between ionizing radiation and one-carbon metabolism is proposed as a pivotal trigger in health effects from radiation exposure [Miousse et al., 2017d]. Our results also indicate that effects of radiation on these metabolites, instead of subsiding, rather amplify over time. This effect was restricted to lower doses of 16O, and affected the transsulfuration pathway. Cystathione and glutathione bridge the gap between one-carbon metabolism and oxidative stress, with glutathione being a major cellular antioxidant. This accumulation of cystathione, to a certain degree to the expense of GSH, indicates a possible long-term impact on the redox state of the heart and warrants further investigations. Our results further indicate that effects on metabolites in the remethylation pathway and DNA methylation may primarily occur within a few weeks after exposure, revealing a need for additional time points to investigate that early period. In humans, a common polymorphism in a gene involved in the remethylation of homocysteine to methionine, methylene tetrahydrofolate reductase (MTHFR) C677T, has been associated with an increased risk of ischemic stroke [Cronin Simon et al., 2005]. In the light of our results, this polymorphism may also be associated with a greater sensitivity to cardiac effects of radiation.
In conclusion, our findings suggest that exposure to higher doses of 16O result in profound epigenetic short-term reprogramming. Importantly, long-term consequences of exposure were also observed three months after exposure. Of particular concern, the long term response displayed a non-linear behavior, with effects on the DNA methylation and the methylation cycle primarily associated with the lowest dose (0.1 Gy) of 16O exposure. This suggests that unexpected adverse consequences may arise at doses previously considered safe, warranting more research on the effects of low doses of radiation during mission beyond the Earth’s protective magnetosphere.
Supplementary Material
Highlights:
Space-relevant doses of radiation lead to long-lasting changes in DNA methylation.
Major satellites are particularly affected, at the methylation and expression levels.
Transsulfuration of methionine is impacted at low dose.
ACKNOWLEDGEMENTS
We would like to thank Dr. Gregory Nelson for his assistance with the revision of this paper. The study was supported in part by the National Space Biomedical Research Institute through NCC 9–58 (MB), and Centers of Biomedical Research Excellence (P20, GM109005, MHJ and IK)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aypar U, Morgan WF, Baulch JE. 2011. Radiation-induced epigenetic alterations after low and high LET irradiations. Mutat. Res 707: 24–33. [DOI] [PubMed] [Google Scholar]
- Baccarelli A, Wright R, Bollati V, Litonjua A, Zanobetti A, Tarantini L, Sparrow D, Vokonas P, Schwartz J 2010. Ischemic heart disease and stroke in relation to blood DNA methylation. Epidemiol. Camb. Mass 21: 819–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batra V, Kesavan V, Mishra KP. 2004. Modulation of enzymes involved in folate dependent one-carbon metabolism by gamma-radiation stress in mice. J. Radiat. Res. (Tokyo) 45: 527–533. [DOI] [PubMed] [Google Scholar]
- Boerma M, Nelson GA, Sridharan V, Mao X-W, Koturbash I, Hauer-Jensen M. 2015. Space radiation and cardiovascular disease risk. World J. Cardiol 7: 882–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerma M, Sridharan V, Mao X-W, Nelson GA, Cheema AK, Koturbash I, Singh SP, Tackett AJ, Hauer-Jensen M. 2016. Effects of ionizing radiation on the heart. Mutat. Res 770: 319–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chancellor JC, Scott GBI, Sutton JP. 2014. Space Radiation: The Number One Risk to Astronaut Health beyond Low Earth Orbit. Life 4: 491–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Luo Y, Wang Y, Pathak R, Sridharan V, Jones T, Mao XW, Nelson G, Boerma M, Hauer-Jensen M, Zhou D, Shao L. 2016. Low Doses of Oxygen Ion Irradiation Cause Acute Damage to Hematopoietic Cells in Mice. PloS One 11: e0158097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon Cronin, Furie Karen L., Kelly Peter J. 2005. Dose-Related Association of MTHFR 677T Allele With Risk of Ischemic Stroke. Stroke 36: 1581–1587. [DOI] [PubMed] [Google Scholar]
- Cucinotta FA, Plante I, Ponomarev AL, Kim M-HY. 2011. Nuclear interactions in heavy ion transport and event-based risk models. Radiat. Prot. Dosimetry 143: 384–390. [DOI] [PubMed] [Google Scholar]
- Cucinotta FA, Wu H, Shavers MR, George K. 2003. Radiation dosimetry and biophysical models of space radiation effects. Gravitational Space Biol. Bull. Publ. Am. Soc. Gravitational Space Biol 16: 11–18. [PubMed] [Google Scholar]
- Darby SC, Ewertz M, McGale P, Bennet AM, Blom-Goldman U, Brønnum D, Correa C, Cutter D, Gagliardi G, Gigante B, Jensen M-B, Nisbet A, Peto R, Rahimi K, Taylor C, Hall P. 2013. Risk of ischemic heart disease in women after radiotherapy for breast cancer. N. Engl. J. Med 368: 987–998. [DOI] [PubMed] [Google Scholar]
- Feng Y, Wang S, Chen R, Tong X, Wu Z, Mo X. 2015. Maternal Folic Acid Supplementation and the Risk of Congenital Heart Defects in Offspring: A Meta-Analysis of Epidemiological Observational Studies. Sci. Rep 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorito G, Guarrera S, Valle C, Ricceri F, Russo A, Grioni S, Mattiello A, Di Gaetano C, Rosa F, Modica F, Iacoviello L, Frasca G, Tumino R, Krogh V, Panico S, Vineis P, Sacerdote C, Matullo G. 2014. B-vitamins intake, DNA-methylation of One Carbon Metabolism and homocysteine pathway genes and myocardial infarction risk: The EPICOR study. Nutr. Metab. Cardiovasc. Dis 24: 483–488. [DOI] [PubMed] [Google Scholar]
- Gilsbach R, Preissl S, Grüning BA, Schnick T, Burger L, Benes V, Würch A, Bönisch U, Günther S, Backofen R, Fleischmann BK, Schübeler D, Hein L. 2014. Dynamic DNA methylation orchestrates cardiomyocyte development, maturation and disease. Nat. Commun 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guren MG, Schneede J, Magne Tveit K, Magne Ueland P, Nexø E, Dueland S. 2004. Biochemical signs of impaired cobalamin status during and after radiotherapy for rectal cancer. Int. J. Radiat. Oncol 60: 807–813. [DOI] [PubMed] [Google Scholar]
- Ha K, Lee GE, Palii SS, Brown KD, Takeda Y, Liu K, Bhalla KN, Robertson KD. 2011. Rapid and transient recruitment of DNMT1 to DNA double-strand breaks is mediated by its interaction with multiple components of the DNA damage response machinery. Hum. Mol. Genet 20: 126–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haider S, Cordeddu L, Robinson E, Movassagh M, Siggens L, Vujic A, Choy M-K, Goddard M, Lio P, Foo R. 2012. The landscape of DNA repeat elements in human heart failure. Genome Biol. 13: R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughson RL, Helm A, Durante M. 2018. Heart in space: effect of the extraterrestrial environment on the cardiovascular system. Nat. Rev. Cardiol 15: 167–180. [DOI] [PubMed] [Google Scholar]
- Ji W, Hernandez R, Zhang XY, Qu GZ, Frady A, Varela M, Ehrlich M. 1997. DNA demethylation and pericentromeric rearrangements of chromosome 1. Mutat. Res 379: 33–41. [DOI] [PubMed] [Google Scholar]
- Kesavan V, Pote MS, Batra V, Viswanathan G. 2003. Increased folate catabolism following total body gamma-irradiation in mice. J. Radiat. Res. (Tokyo) 44: 141–144. [DOI] [PubMed] [Google Scholar]
- Kim M, Long TI, Arakawa K, Wang R, Yu MC, Laird PW. 2010. DNA methylation as a biomarker for cardiovascular disease risk. PLoS One 5: e9692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koturbash I, Boyko A, Rodriguez-Juarez R, McDonald RJ, Tryndyak VP, Kovalchuk I, Pogribny IP, Kovalchuk O. 2007. Role of epigenetic effectors in maintenance of the long-term persistent bystander effect in spleen in vivo. Carcinogenesis 28: 1831–1838. [DOI] [PubMed] [Google Scholar]
- Koturbash I, Miousse IR, Sridharan V, Nzabarushimana E, Skinner CM, Melnyk SB, Pavliv O, Hauer-Jensen M, Nelson GA, Boerma M. 2016. Radiation-induced changes in DNA methylation of repetitive elements in the mouse heart. Mutat. Res 787: 43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Joseph KS, Luo W, León JA, Lisonkova S, Hof MV den, Evans J, Lim K, Little J, Sauve R, Kramer MS. 2016. Effect of Folic Acid Food Fortification in Canada on Congenital Heart Disease Subtypes. Circulation 134: 647–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao XW, Nishiyama NC, Pecaut MJ, Campbell-Beachler M, Gifford P, Haynes KE, Becronis C, Gridley DS. 2016. Simulated Microgravity and Low-Dose/Low-Dose-Rate Radiation Induces Oxidative Damage in the Mouse Brain. Radiat. Res 185: 647–657. [DOI] [PubMed] [Google Scholar]
- Miousse IR, Chalbot M-CG, Lumen A, Ferguson A, Kavouras IG, Koturbash I. 2015a. Response of transposable elements to environmental stressors. Mutat. Res. Rev. Mutat. Res 765: 19–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miousse IR, Chalbot M-CG, Pathak R, Lu X, Nzabarushimana E, Krager K, Aykin-Burns N, Hauer-Jensen M, Demokritou P, Kavouras IG, Koturbash I. 2015b. In Vitro Toxicity and Epigenotoxicity of Different Types of Ambient Particulate Matter. Toxicol. Sci. Off. J. Soc. Toxicol 148: 473–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miousse IR, Chang J, Shao L, Pathak R, Nzabarushimana É, Kutanzi KR, Landes RD, Tackett AJ, Hauer-Jensen M, Zhou D, Koturbash I. 2017a. Inter-Strain Differences in LINE-1 DNA Methylation in the Mouse Hematopoietic System in Response to Exposure to Ionizing Radiation. Int. J. Mol. Sci 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miousse IR, Koturbash I. 2015. The Fine LINE: Methylation Drawing the Cancer Landscape. BiomedRes Int 2015: 131547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miousse IR, Kutanzi KR, Koturbash I. 2017b. Effects of ionizing radiation on DNA methylation: from experimental biology to clinical applications. Int. J. Radiat. Biol 93: 457–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miousse IR, Pathak R, Garg S, Skinner CM, Melnyk S, Pavliv O, Hendrickson H, Landes RD, Lumen A, Tackett AJ, Deutz NEP, Hauer-Jensen M, Koturbash I. 2017c. Short-term dietary methionine supplementation affects one-carbon metabolism and DNA methylation in the mouse gut and leads to altered microbiome profiles, barrier function, gene expression and histomorphology. Genes Nutr. 12: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miousse IR, Tobacyk J, Melnyk S, James SJ, Cheema AK, Boerma M, Hauer-Jensen M, Koturbash I. 2017d. One-carbon metabolism and ionizing radiation: a multifaceted interaction. Biomol. Concepts 8: 83–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson GA. 2016. Space Radiation and Human Exposures, A Primer. Radiat. Res 185: 349–358. [DOI] [PubMed] [Google Scholar]
- Nzabarushimana E, Miousse IR, Shao L, Chang J, Allen AR, Turner J, Stewart B, Raber J, Koturbash I. 2014. Long-term epigenetic effects of exposure to low doses of 56Fe in the mouse lung. J. Radiat. Res. (Tokyo) 55: 823–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papait R, Pistore C, Negri D, Pecoraro D, Cantarini L, Bonapace IM. 2007. Np95 is implicated in pericentromeric heterochromatin replication and in major satellite silencing. Mol. Biol. Cell 18: 1098–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry IJ, Refsum H, Morris RW, Ebrahim SB, Ueland PM, Shaper AG. 1995. Prospective study of serum total homocysteine concentration and risk of stroke in middle-aged British men. Lancet Lond. Engl 346: 1395–1398. [DOI] [PubMed] [Google Scholar]
- Prior S, Miousse IR, Nzabarushimana E, Pathak R, Skinner C, Kutanzi KR, Allen AR, Raber J, Tackett AJ, Hauer-Jensen M, Nelson GA, Koturbash I. 2016. Densely ionizing radiation affects DNA methylation of selective LINE-1 elements. Environ. Res 150: 470–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu G, Dubeau L, Narayan A, Yu MC, Ehrlich M. 1999. Satellite DNA hypomethylation vs. overall genomic hypomethylation in ovarian epithelial tumors of different malignant potential. Mutat. Res 423: 91–101. [DOI] [PubMed] [Google Scholar]
- Ramadan SS, Sridharan V, Koturbash I, Miousse IR, Hauer-Jensen M, Nelson GA, Boerma M. 2016. A priming dose of protons alters the early cardiac cellular and molecular response to (56)Fe irradiation. Life Sci. Space Res 8: 8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seawright JW, Sridharan V, Landes RD, Cao M, Singh P, Koturbash I, Mao X-W, Miousse IR, Singh SP, Nelson GA, Hauer-Jensen M, Boerma M. 2019. Effects of low-dose oxygen ions and protons on cardiac function and structure in male C57BL/6J mice. Life Sci. Space Res 20: 72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, Tajima S, Mitsuya K, Okano M, Koseki H. 2007. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450: 908–912. [DOI] [PubMed] [Google Scholar]
- Sharma P, Kumar J, Garg G, Kumar A, Patowary A, Karthikeyan G, Ramakrishnan L, Brahmachari V, Sengupta S. 2008. Detection of altered global DNA methylation in coronary artery disease patients. DNA Cell Biol. 27: 357–365. [DOI] [PubMed] [Google Scholar]
- Stenvinkel P, Karimi M, Johansson S, Axelsson J, Suliman M, Lindholm B, Heimbürger O, Barany P, Alvestrand A, Nordfors L, Qureshi AR, Ekström TJ, Schalling M. 2007. Impact of inflammation on epigenetic DNA methylation – a novel risk factor for cardiovascular disease? J. Intern. Med 261:488–499. [DOI] [PubMed] [Google Scholar]
- Storey JD. 2002. A direct approach to false discovery rates. J. R. Stat. Soc. Ser. B Stat. Methodol 64: 479–498. [Google Scholar]
- Tian Y, Paramasivam M, Ghosal G, Chen D, Shen X, Huang Y, Akhter S, Legerski R, Chen J, Seidman MM, Qin J, Li L. 2015. UHRF1 contributes to DNA damage repair as a lesion recognition factor and nuclease scaffold. Cell Rep. 10: 1957–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tungjai M, Whorton EB, Rithidech KN. 2013. Persistence of apoptosis and inflammatory responses in the heart and bone marrow of mice following whole-body exposure to 28Silicon (28Si) ions. Radiat. Environ. Biophys 52: 339–350. [DOI] [PubMed] [Google Scholar]
- Wald DS, Morris JK, Wald NJ. 2011. Reconciling the evidence on serum homocysteine and ischaemic heart disease: a meta-analysis. PloS One 6: e16473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker SA, Townsend LW, Norbury JW. 2013. Heavy ion contributions to organ dose equivalent for the 1977 galactic cosmic ray spectrum. Adv. Space Res 51: 1792–1799. [Google Scholar]
- Wang Y, Chang J, Li X, Pathak R, Sridharan V, Jones T, Mao XW, Nelson G, Boerma M, Hauer-Jensen M, Zhou D, Shao L. 2017. Low doses of oxygen ion irradiation cause long-term damage to bone marrow hematopoietic progenitor and stem cells in mice. PloS One 12: e0189466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Sasi SP, Gee H, Lee J, Yang Y, Mehrzad R, Onufrak J, Song J, Enderling H, Agarwal A, Rahimi L, Morgan J, Wilson PF, Carrozza J, Walsh K, Kishore R, Goukassian DA. 2014. Cardiovascular Risks Associated with Low Dose Ionizing Particle Radiation. PLOS ONE 9: e110269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlin C, Hassler DM, Cucinotta FA, Ehresmann B, Wimmer-Schweingruber RF, Brinza DE, Kang S, Weigle G, Böttcher S, Böhm E, Burmeister S, Guo J, Köhler J, Martin C, Posner A, Rafkin S, Reitz G. 2013. Measurements of Energetic Particle Radiation in Transit to Mars on the Mars Science Laboratory. Science 340: 1080–1084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.