SUMMARY
Microbial exposures can define an individual’s basal immune state. Cohousing specific pathogen-free (SPF) mice with pet store mice, which harbor numerous infectious microbes, results in global changes to the immune system, including increased circulating phagocytes and elevated inflammatory cytokines. How these differences in the basal immune state influence the acute response to systemic infection is unclear. Cohoused mice exhibit enhanced protection from virulent Listeria monocytogenes (LM) infection, but increased morbidity and mortality to polymicrobial sepsis. Cohoused mice have more TLR2+ and TLR4+ phagocytes, enhancing recognition of microbes through pattern-recognition receptors. However, the response to a TLR2 ligand is muted in cohoused mice, whereas the response to aTLR4 ligand is greatly amplified, suggesting a basis for the distinct response to Listeria monocytogenes and sepsis. Our data illustrate how microbial exposure can enhance the immune response to unrelated challenges but also increase the risk of immunopathology from a severe cytokine storm.
In Brief
Cohousing of laboratory mice with pet store animals changes the immune system and alters responsiveness to future challenges. Huggins et al. demonstrate that microbial exposure results in alterations to immune cells, serum cytokines, and microbiome composition. This study shows that cohousing alters the ability to detect pathogens through pattern-recognition receptors.
Graphical Abstract
INTRODUCTION
Mice are one of the most important and heavily used tools in biomedical research, due in part to their well-decoded genetics and their ability to model complex physiological systems in humans (Müller and Grossniklaus, 2010). The availability of an extensive array of transgenic and knockout mouse strains, as well as the ease of manipulating the genome to generate novel strains as needed, has made the mouse increasingly valuable for studying functions of the immune system (Doudna and Charpentier, 2014). However, environmental pathogen exposure is an important difference between human and laboratory mouse biology that must be considered when using mouse models to evaluate the fitness of the immune system. Humans and mice are naturally exposed to both commensal and pathogenic microbes on a daily basis from birth. Laboratory mice, in contrast, are often housed under specific pathogen-free (SPF) conditions. While SPF housing has been instrumental in increasing experimental reproducibility, it has also further distanced the mouse model from humans (Masopust et al., 2017). The immune system of adult humans has been trained and shaped in the context of each infection and vaccination experienced, while the SPF mouse experiences limited microbial exposures (Foster, 1959). How these interactions between host and microbe might impact future immune responses to acute infection is unknown and prompted the current investigation.
Recent studies have highlighted notable differences in composition and function of the immune systems of SPF mice and mice with increased exposure to naturally transferred murine microbes (Beura et al., 2016; Reese et al., 2016). Natural pathogen transfer by cohousing laboratory mice with “dirty” pet store mice shifts the murine immune system to more closely resemble that seen in adult humans exposed to microbes through infection and vaccination (Beura et al., 2016). The T cell compartment of SPF laboratory mice is dominated by naive cells, whereas the majority of T cells in cohoused and pet store mice have an effector or memory phenotype and corresponding functional capacity. SPF mice have few nonlymphoid tissue-resident T cells, while they are abundant in cohoused mice. Finally, transcriptomic analysis of cells from the blood indicates the basal immune system of an SPF mouse most closely resembles a neonatal human, whereas the cohoused mouse gene expression signature correlates with that of adult humans (Beura et al., 2016).
Currently, it is poorly understood how natural microbial exposure alters the immune response to future challenges. Microbial exposure can increase resistance to bacterial and parasitic pathogens (Beura et al., 2016), but the basis for improved control, or whether it extends to other infectious challenges, is unknown. Our ability to incorporate spontaneous microbial exposure allows us to qualitatively address the cumulative effects of natural infections on future immunity. We have leveraged this mouse model where exposure to multiple naturally acquired infections generates an experienced immune system–mimicking a critical aspect of human biology–to investigate the immune response following a systemic inflammatory stimulus. Our data demonstrate substantial differences in the acute immune response after exposure to monomicrobial (using Listeria monocytogenes [LM]), polymicrobial (using the cecal ligation and puncture [CLP] model of sepsis), or sterile (using the classical model of lipopolysaccharides [LPS] endotoxin or Pam3CSK4 challenge) stimuli. Surprisingly, these differences either improved immunity for the host (Listeria monocytogenes) or caused a dramatic reduction in survival (CLP and LPS). Our investigation also identified changes in the microbiome as well as the expression and activation of two pattern-recognition receptors (PRRs), Toll-like receptor 2 (TLR2) and TLR4, following cohousing. A muted response to TLR2 ligands and an exaggerated response to TLR4 ligands indicate a role for infectious history in “training”‘ the innate immune system. Thus, our results show that an experienced immune system can differentially induce an acute inflammatory response that ultimately benefits or harms the host based on the type of challenge.
RESULTS
Normalizing Microbial Exposure Creates a More Inflammatory Host Environment
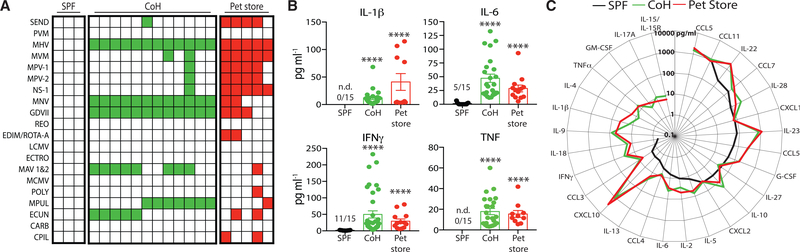
Mice from pet stores carry a number of pathogens normally absent from standard SPF laboratory strains of mice (Beura et al., 2016). After cohousing for 60 days, C57BL/6 (B6) mice acquire many of the microbes carried by the pet store mice based on serological assays (Figure 1A). Importantly, this physiological microbial exposure resulted in global changes to the immune status of the cohoused mice. Inflammatory serum cytokine and chemokine levels were elevated in long-term cohoused B6 and pet store mice, relative to SPF B6 mice. For example, the pro-inflammatory cytokines IL-1β, IL-6, IFNγ, and tumor necrosis factor (TNF) were notably higher in the serum of cohoused B6 and pet store mice after 60 days (Figure 1B). Similar increases were seen for IL-2, IL-5, IL-9, IL-10, IL-13, IL-15/IL-15R, IL-17A, IL-18, IL-22, IL-23, IL-28, G-CSF, GM-CSF, CCL3, CCL4, CCL5, and CXCL10 (Figures 1C and S1). Collectively, these data demonstrate that physiological microbial exposure establishes elevated systemic inflammation, marked by the presence of numerous cytokines and chemokines in the serum.
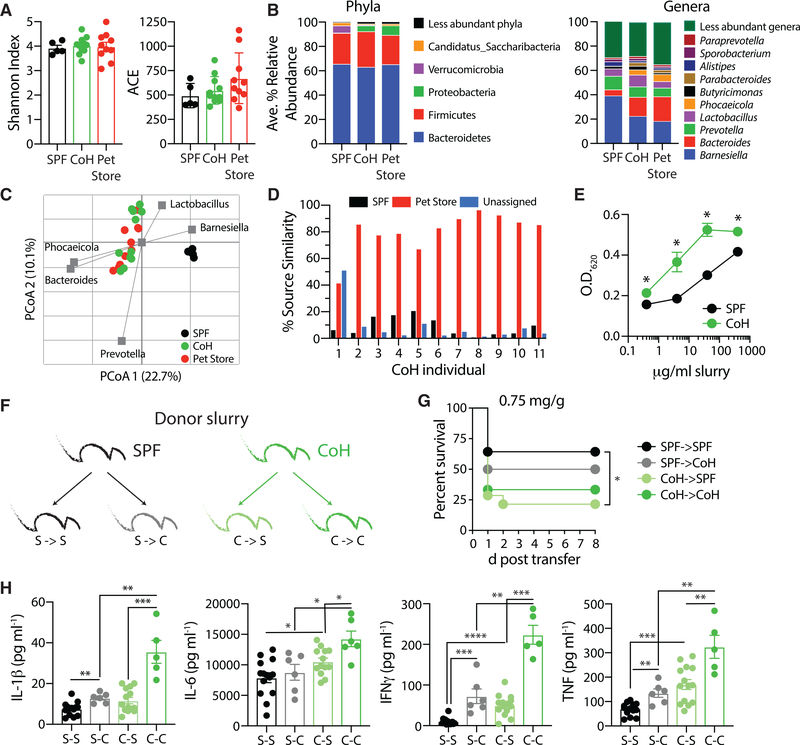
Figure 1. Physiological Microbial Exposure Creates a More Inflammatory Host Environment.
(A) Representative serology from specific pathogen free (SPF) B6, cohoused (CoH) B6, and pet store mice detecting the extent of pathogen exposure. Filled squares indicate a positive serology result for the indicated pathogen. Each column represents an individual mouse. Data are representative of at least 20 independent serology tests conducted.
(B and C) Serum was collected from SPF B6, cohoused B6, and pet store mice after 60 days. The concentration of cytokines and chemokines was determined by bioplex.
(B) The number of mice with detectable IL-1β, IL-6, IFNγ, and TNF in the serum are indicated for the SPF group.
(C) Radar plot comparing the basal serum concentrations of indicated cytokines and chemokines in SPF B6, cohoused B6, and pet store mice. For statistical comparisons, SPF mice with undetectable cytokines were given a value of “0.” 10–36 mice/group (pooled from 3 technical replicates) were analyzed in (B) and (C).
Asterisk (*) refers to statistical differences in the response of cohoused and pet store mice relative to SPF. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.005, ****p ≤ 0.001. n.d., none detected.
See also Figure S1.
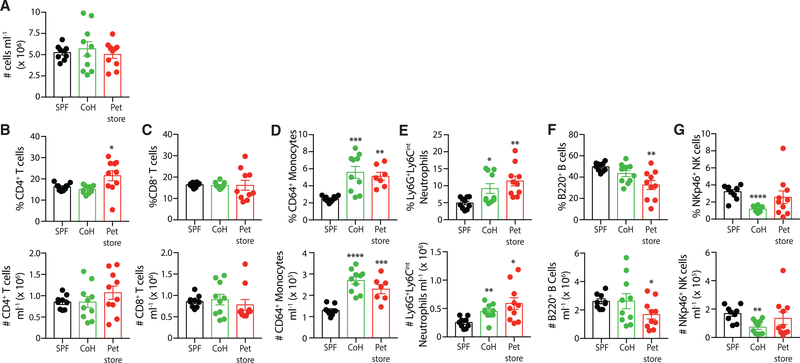
Given the increase in serum cytokines, we next defined the cellular immune composition of cohoused and pet store mice. While the total blood cell numbers were not different among SPF B6, cohoused B6, and pet store mice (Figure 2A), cohousing altered the composition of circulating immune cells. The frequency and number of circulating CD4 and CD8 T cells were not changed compared to SPF B6 mice (Figures 2B and 2C), but consistent with previous data, physiological microbial transfer resulted in increased frequencies of CD44hi and KLRG1+ and decreased frequencies of CD62L+ CD4 and CD8 T cells (collectively indicating increased frequencies of effector and effector memory T cells [Wherry et al., 2003] and long-lived effector memory T cells [Olson et al., 2013]) in the blood of cohoused B6 and pet store mice (Figure S2) (Beura et al., 2016). These data indicate a transition of naive T cells to antigen-stimulated T cells as a result of microbial exposure. The innate immune cell compartment was significantly expanded following cohousing, with increases in both the frequency and the number of circulating monocytes and neutrophils (Figures 2D and 2E). In contrast, B cells and natural killer (NK) cells showed modest reductions in the blood after microbial exposure (Figures 2F and 2G). Collectively, these data demonstrate that physiological microbial exposure establishes numerous elevated cytokines and chemokines in the serum as well as increased circulating phagocytes.
Figure 2. Cohoused Mice Have Altered Circulating Immune Cell Composition.
(A) Total circulating immune cell counts from the blood of SPF B6, cohoused B6, and pet store mice were determined by flow cytometry.
(B–G) Frequency and number (cells/ml blood) of (B) CD4 T cells, (C) CD8 T cells, (D) monocytes, (E) neutrophils, (F) B cells, and (G) NK cells. Ten mice per group were analyzed, and data are representative of 3 technical replicates.
*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.005, ****p ≤ 0.001.
See also Figure S2.
Cohoused Mice Demonstrate Increased Resistance to Listeria Monocytogenes Infection
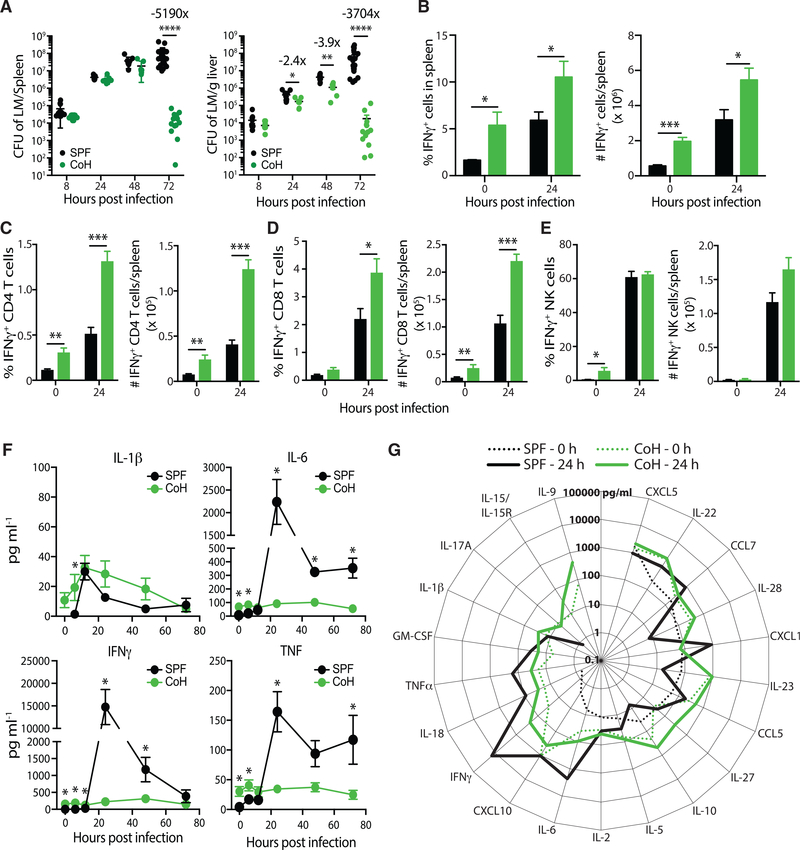
Previous data showed cohoused B6 and pet store mice display an increased ability to clear a systemic infection by virulent Listeria monocytogenes—a common bacterium used to assess immune fitness in laboratory mice—at day 3 post-infection relative to SPF B6 mice (Beura et al., 2016). While the mechanism of enhanced Listeria monocytogenes protection was undefined, it is possible that Listeria monocytogenes fails to establish infection in the cohoused mice because the increased circulating monocytes and neutrophils in these mice could mediate very early resistance, similar to that shown following murine gammaherpesvirus 68 infection (Barton et al., 2007). To more fully define Listeria monocytogenes resistance after cohousing, we measured the bacterial burden at early time points post-challenge. Both SPF and cohoused B6 mice had similar Listeria monocytogenes burdens in the spleen and liver during the first 8 h after infection, suggesting the initial establishment of bacterial infection was not impacted by cohousing (Figure 3A). While the early bacterial burden was not different, cohoused B6 mice more rapidly and effectively controlled the Listeria monocytogenes infection. The cohoused B6 mice began to show signs of controlling Listeria monocytogenes in the liver at 24 h post-infection. However, 72 h were needed to observe a significant decrease in Listeria monocytogenes burdens in the spleens of cohoused mice. By this time point, bacterial loads reached 5190- and 3704-fold lower than SPF B6 mice in the spleens and livers, respectively.
Figure 3. Cohoused Mice Are More “Immune Fit” Than SPF Mice in Response to Virulent Listeria Infection.
SPF and cohoused B6 mice were infected with virulent Listeria monocytogenes (LM; 8 × 104 CFU i.v.).
(A) Bacterial burden in the spleen and liver was determined after 8, 24, 48, and 72 h. 7–17 mice/group/time point were analyzed, pooled from >2 technical replicates.
(B–E) The frequency and number of IFNγ-producing cells in the spleen before and 24 h after Listeria monocytogenes infection were quantified by intracellular cytokine staining flow cytometry. IFNγ-producing (B) total cells, (C) CD4 T cells, (D) CD8 T cells, and (E) NKcells were identified. 4–7 mice/group/time point were analyzed, pooled from two technical replicates. *p % 0.05, **p % 0.01, ***p % 0.005.
(F) Serum samples from SPF or cohoused B6 mice were obtained at indicated hours aftervirulent Listeria monocytogenes infection. The amount of IL-1β, IL-6, IFNγ, and TNF in the serum was determined by bioplex. 4–7 mice/group/time point were analyzed. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.005 for SPF versus cohoused mice at the indicated time points.
(G) Additional serum cytokine and chemokine levels in SPF (black lines) and cohoused (green lines) B6 mice were determined prior to (dotted lines) and 24 h post-Listeria monocytogenes infection (solid lines). No line for given a cytokine indicates levels were below the limit of detection.
Data in (F) and (G) are representative from two technical replicates.
IFNγ plays an important role in the response to intracellular bacteria and effective clearing of Listeria monocytogenes (Harty and Bevan, 1995; Lertmemongkolchai et al., 2001). Because basal IFNγ levels are elevated in cohoused B6 mice (Figure 1B), we next examined the cellular source of IFNγ before and after Listeria monocytogenes infection. We saw a significant increase in the frequency and number of IFNγ-producing cells in the spleen of cohoused B6 mice before and 24 h after Listeria monocytogenes infection (Figure 3B). In particular, IFNγ-producing CD4 and CD8 T cells were increased in cohoused mice 24 h post-Listeria monocytogenes infection (Figures 3C and 3D). The increase was not ubiquitous, as IFNγ-producing NK cells, a well-established early producer of IFNγ during Listeria monocytogenes infection, were not elevated in cohoused mice after Listeria monocytogenes infection (Figure 3E). Surprisingly, despite the increased frequency of IFNγ+ cells in the spleen, the amount of IFNγ detectable in the serum of Listeria monocytogenes-infected cohoused mice remained constant throughout the 72 h after infection (Figure 3F). In contrast, there was a rapid and robust increase in serum IFNγ of Listeria monocytogenes-infected SPF mice. As a result, SPF mice had ~100 times more IFNγ in the serum 24 h after systemic virulent Listeria monocytogenes infection than was measured in cohoused mice. A similar trend was seen for TNF and IL-6 (Figure 3F). Moreover, the overall serum cytokine and chemokine milieu detected in SPF mice was markedly different 24 h after Listeria monocytogenes infection (Figure 3G). There was little change, in contrast, seen in cohoused mice when comparing serum cytokine and chemokine levels before and 24 h after Listeria monocytogenes infection. Whether these observations are influenced by increased amounts of cytokine produced in the tissues (as opposed to the circulation) in cohoused mice remains to be determined. Collectively, the data in Figure 3 show that cohoused B6 mice exhibit enhanced resistance to Listeria monocytogenes infection, compared to SPF mice with remarkably stable, yet heightened, circulating cytokine and chemokine levels.
Cohoused B6 Mice Exhibit an Exacerbated Cytokine Response and Increased Mortality to CLP-Induced Polymicrobial Sepsis
The data obtained from SPF and cohoused B6 mice after Listeria monocytogenes infection, as well as previous data following parasitic infections, suggest that a host with significant microbial exposure can deal with subsequent infections more efficiently (Beura et al., 2016). However, the duration, magnitude, and impact of an inflammatory response is pathogen dependent (Haring et al., 2006), and we reasoned the acute response in SPF and cohoused B6 mice could be similarly influenced by the infecting pathogen(s). To further explore how cohoused mice respond to newly acquired infections, we evaluated the acute response to a systemic polymicrobial infection by gut commensals generated using the CLP model of sepsis (Rittirsch et al., 2009).
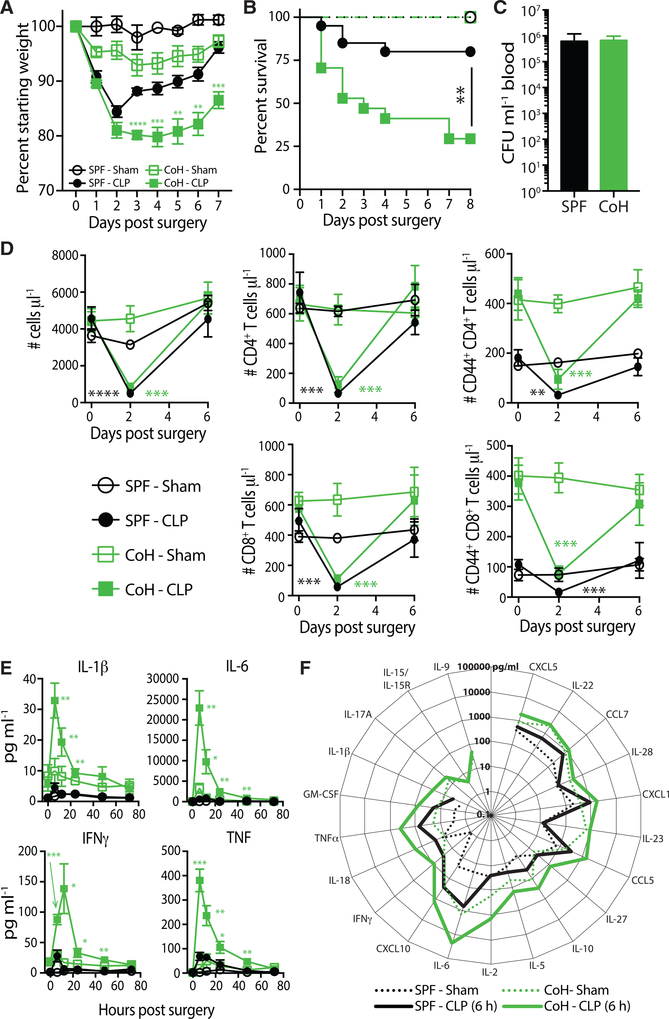
One feature of the CLP model is the ability to modulate the severity of inflammation and mortality based on the amount of cecum ligated and the number and/or size of punctures (Rittirsch et al., 2009). Our version of the CLP model induces a septic state in SPF B6 mice defined by modest weight loss that recovers by day 7 post-surgery and ~20% acute mortality (Cabrera-Perez et al., 2015; Condotta et al., 2015, 2013; Danahy et al., 2019), which is consistent with clinical rates (Figures 4A and 4B) (Levy et al., 2012). Interestingly, cohoused mice exhibited significantly increased mortality (~75%) compared to SPF mice (~20%) following CLP. Cohoused mice that survived also took longer to regain the weight lost following CLP surgery. The physiological microbial exposure during cohousing alters the cecal microbiome, and this may cause exacerbated microbial release following CLP surgery; however, there was no difference in bacteremia 24 h post-CLP (Figure 4C). Both SPF and cohoused B6 mice had similarly significant reductions in numbers of CD4 and CD8 T cells in the blood by 2 days after CLP-induced sepsis (compared to pre-CLP numbers) that recovered within 1 week (Figure 4D), suggesting the increased morbidity and mortality in the cohoused mice was not caused by a substantial deviation in the characteristic acute lymphopenia during CLP-induced sepsis (Cabrera-Perez et al., 2016, 2017, 2014, 2015; Condotta et al., 2015, 2013; Danahy et al., 2017; Duong et al., 2014; Gurung et al., 2011; Jensen et al., 2018a,b; Sjaastad et al., 2018; Strother et al., 2016; Unsinger et al., 2009).
Figure 4. Increased Morbidity and Mortality in CLP-Treated Cohoused Mice Correlates with an Exacerbated “Cytokine Storm”.
(A and B) SPF and cohoused B6 mice underwent sham or CLP surgery. Weight loss (A) and survival
(B) were monitored over time. 8–17 mice/group were analyzed, pooled from 2 technical replicates. **p ≤ 0.01, ***p ≤ 0.005, ****p ≤ 0.001.
(C) Bacterial CFU in blood at 24 h post-CLP procedure was quantified in SPF and cohoused B6 mice. 9 mice/group were analyzed, representative of 2 technical replicates.
(D) The number of total immune cells, CD4+ T cells, CD8+ T cells, CD44+ CD4+ T cells, and CD44+ CD8+ T cells per μl blood were determined before (day 0) and after (days 2 and 6) sham or CLP surgery. 5–7 mice/group were analyzed, representative of 3 technical replicates.
(E) Serum samples from sham- or CLP-treated SPF or cohoused B6 mice were obtained at indicated hours after surgery. The amount of IL-1β, IL-6, IFNγ, and TNF in samples was determined by bioplex.
(F) Additional cytokines and chemokines in the serum 6 h post-surgery were quantified by bioplex. Data in (E) and (F) consist of 4–7 mice/group/time point and are representative of 3 technical replicates. **p ≤ 0.01, ***p ≤ 0.005, ****p ≤ 0.001 for SPF-CLP versus cohoused-CLP at the indicated time points.
The acute hyperinflammatory response during sepsis induces a variety of pathophysiological events (e.g., fever, shock, vasodilation, and organ failure) stemming from the rapid and robust production of multiple pro-inflammatory cytokines that must be controlled to prevent an unbridled response and death (Hotchkiss and Karl, 2003). The early cytokine storm is likely due to an innate response by a variety of cells within the immune system to multiple pathogen-associated molecular patterns (PAMPs) expressed by the invading microbe(s), as well as the damage-associated molecular patterns (DAMPs) released from dying cells (Chousterman et al., 2017). Consistent with our recent data (Danahy et al., 2017), a transient but relatively modest spike in serum IFNγ was detected in SPF B6 mice after CLP surgery (Figure 4E). This was also observed for IL-1β, IL-6, and TNF—cytokines of crucial importance to sepsis pathophysiology (Chousterman et al., 2017)—as well as several other cytokines and chemokines (Figure 4F). In striking contrast to what we saw in the Listeria monocytogenes-infected cohoused mice, the magnitude of this response was markedly increased in cohoused mice subjected to CLP surgery (Figures 4E and 4F). The exacerbated cytokine storm observed in cohoused mice following CLP surgery, which correlated with the increased morbidity and acute mortality, contrasted to a muted cytokine response and improved bacterial clearance in Listeria monocytogenes-infected cohoused mice (Figure 3).
Cohousing of Mice Alters Gut Microbiome Composition
Given the established role for the composition of the gut microbiota in human health and the host resistance to infection (Thiemann et al., 2017), we sought to characterize the changes to the microbiome that occur during the cohousing process. Across SPF, cohoused, and pet store mice, a mean Good’s coverage of 99.1% ± 0.1% was observed, with a range from 224 to 660 operational taxonomic units (OTUs) identified in individual samples. No differences in alpha diversity parameters were observed due to origin or cohousing (Figure 5A; p = 0.905 and 0.196 for Shannon and Abundance-based Coverage [ACE] indices, respectively). The predominant phyla in all fecal communities were Bacteroidetes and Firmicutes, and the relative ratios between these did not differ among groups (Figure 5B). The relative abundances of Proteobacteria were, however, significantly greater in pet store and cohoused communities compared to SPF (post hoc p ≤ 0.006), while SPF communities had greater abundances of Verrucomicrobia (p ≤ 0.014). Some studies suggest Proteobacteria may increase the risk of bacterial translocation and sepsis (Mai et al., 2013). At the genus level, communities predominantly comprised Barnesiella, Bacteroides, Prevotella, and Lactobacillus (Figure 5B). Communities from SPF mice had significantly greater relative abundances of Barnesiella and Alistipes than cohoused or pet store mice (post hoc p = 0.019 and p ≤ 0.006, respectively). In contrast, cohoused and pet store mice communities had significantly greater abundances of Bacteroides, Phocaeicola (phylum Bacteroidetes), and Parabacteroides (p ≤ 0.038, p ≤ 0.026, and p ≤ 0.039, respectively). Overall, bacterial communities were significantly different in SPF mice compared to cohoused and pet store mice (Figure 5C; ANOSIM R = 0.883 and 0.952, p ≤ 0.001), but cohoused and pet store mouse communities were similar (ANOSIM R = 0.135, p = 0.033, at Bonferroni-correct α = 0.017). SourceTracker analysis revealed a significantly greater similarity to pet store mouse communities than SPF communities (p < 0.0001) among cohoused mice (Figure 5D). Lastly, cecal contents from SPF and cohoused mice were applied to a TLR4 reporter cell line (HEK-Blue hTLR4) to assay the ability of the differing microbial communities to stimulate this innate pattern-recognition receptor. While there was no increase in abundance of gram-negative bacteria in the cohoused mice compared to SPF (p = 0.441), the cecal contents from cohoused mice did display a significantly higher ability to stimulate TLR4, compared to the SPF cecal contents (Figure 5E), suggesting an increased endotoxin content in the cohoused cecal material. Thus, the composition of the microbiome following cohousing was differentially able to activate the immune system.
Figure 5. Characterization of the Microbiome in Cohoused Mice.
(A) Mean alpha diversity indices (±SE) in mouse fecal samples. Shannon and ACE indices are shown on the left and right, respectively. n = 5 SPF, 11 cohoused, and 10 pet store samples.
(B) Distributions of abundant phyla and genera in mouse fecal samples. Less-abundant genera accounted for a mean of ≤2.2% of the community, among all samples.
(C) Principal coordinates analysis (PCoA) of Bray-Curtis distances (r2 = 0.47). The five most abundant genera were correlated with axes position, where positioning near sample groups indicates a greater relative abundance of that genus.
(D) Similarity of cohoused B6 mice to SPF B6 and pet store mice, determined by SourceTracker.
(E) Cecal contents from SPF or cohoused B6 mice (n = 5of each) were cultured with the TLR4 reporter cell line, HEK-Blue hTLR4. TLR4 activation was quantified after 24 h with indicated concentration of cecal material. Data are representative of 3 technical replicates.
(F) Cecal contents were harvested from SPF or cohoused B6 donor mice to challenge recipient mice in the following combinations: S-S, SPF cecal slurry transferred to SPF recipients (n = 14); S-C, SPF cecal slurry transferred to cohoused recipients (n = 6); C-S, cohoused cecal slurry transferred to SPF recipients (n = 14); and C-C, cohoused cecal slurry transferred to cohoused recipients (n = 6).
(G) Survival curve of mice after i.p. injection with 0.75 mg/g body weight of cecal slurry as indicated in (F).
(H) Serum cytokine levels 6 h post-cecal slurry injection was determined by bioplex.
Data in (G) and (H) are pooled from 2 technical replicates. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.005, ****p ≤ 0.001.
See also Figure S3.
Our data show cohousing SPF laboratory mice with pet store mice skews the composition and maturational status of the host immune system, but it also leads to a shifting of microbial communities in the gut. To assay the relative contribution of these changes toward the heightened sensitivity of cohoused mice to sepsis, we isolated the cecal contents from either SPF or co-housed mice and directly injected them into new hosts to induce an intraperitoneal polymicrobial infection (Figure 5F). Injection of cohoused cecal contents resulted in higher mortality compared to SPF cecal contents when transferred to both SPF and co-housed recipients (Figure 5G), suggesting the composition of cohoused mice microbiota induces a more severe septic shock. However, the microbial composition does not solely explain the differences between SPF and cohoused mice. Injection of SPF cecal slurry into cohoused recipients resulted in elevated IL-1β, IFNγ, and TNF 6 h post-transfer, compared to SPF cecal slurry into SPF recipients (Figures 5H and S3). Our earlier results illustrate that cohoused mice have elevated serum cytokine levels prior to challenge (Figure 1). Importantly, following the cecal slurry transfer, IL-1β, IL-6, IFNγ, and TNF were each increased significantly over their baseline levels in both cohoused and SPF recipients (Figure S3A). Thus, the differences in cytokine levels following the cecal slurry injection between SPF and cohoused hosts cannot simply be attributed to elevated baseline levels. Similar results were observed when injecting a cohoused cecal slurry into SPF and cohoused recipients, with increased cytokine production in cohoused recipients compared to SPF recipients.
Thus, microbial transfer through cohousing results in an increased sensitivity to sepsis even when the cecal transfer is normalized between the two groups. These results suggest that while the makeup of the gut microbiome does contribute to the severity of sepsis, maturation of the immune system also plays a major role in determining the cytokine response and disease outcome.
Increased Prevalence of TLR2+ and TLR4+ Immune Cells after Normalizing Microbial Exposure
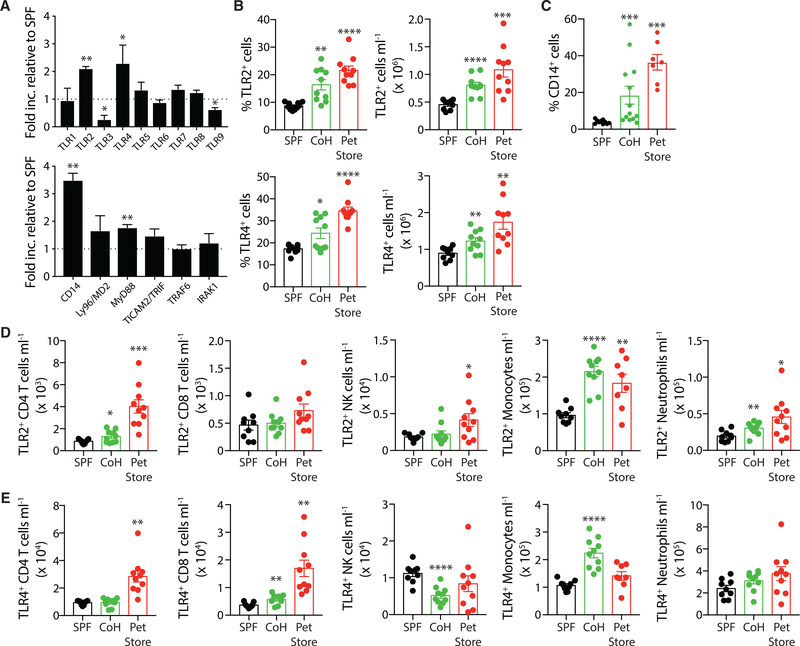
The findings presented in Figure 5 suggest there were contributions of both the cecal microbiota and the immunological experience of the mice in dictating susceptibility to sepsis, complicating interpretation. Hence, we moved to models that measured the response to isolated TLR agonists. TLRs are important pattern-recognition receptors for recognizing bacterial and viral pathogens to mobilize the innate immune response during infection (Beutler, 2009; Kawasaki and Kawai, 2014). Triacylated and diacylated lipopeptides found in gram-positive bacteria such as Listeria monocytogenes are recognized by TLR2/TLR1 and TLR2/TLR6 heterodimers, respectively (Akira et al., 2001; Takeda and Akira, 2001a,b; Takeda et al., 2003). TLR4 recognizes LPS present in the cell walls of gram-negative bacteria, and both TLR2 and TLR4 have been implicated in inducing septic shock (Heipertz et al., 2018; Lima et al., 2015; Oliveira-Nascimento et al., 2012). These surface receptors are primarily expressed by circulating innate immune cells (especially phagocytes) and serve to activate the primary immune response (Kawai and Akira, 2011). A qPCR analysis of splenic adherent myeloid cells (Inaba et al., 2009; Steinman et al., 1979) revealed elevated transcription for multiple components of both the TLR2 and TLR4 signaling pathways in cohoused mice relative to SPF mice. Both Tlr2 and Tlr4 transcripts, but not other TLRs, were upregulated in cohoused mice compared to SPF mice (Figure 6A). Additionally, mRNAfor CD14, a co-receptor for TLR4 binding to LPS, as well as the downstream signaling adaptor MyD88, were elevated in the splenic phagocytes following cohousing. These data provide more evidence to suggest cohousing alters the ability of the immune system to recognize and respond to pathogens.
Figure 6. TLR2 and TLR4 Expression Is Increased after Physiological Microbial Exposure.
(A) The qPCR analysis of TLRs and TLR signaling pathway components in the adherent myeloid cells isolated from spleens of SPF (n = 5) and cohoused (n = 5) B6 mice. Data are representative of 2 technical replicates.
(B) Frequency and number of TLR2+ or TLR4+ cells in the blood of SPF B6, cohoused B6, and pet store mice.
(C) Frequency of CD14+ cells in blood of SPF B6, cohoused B6, and pet store mice.
(D and E) Immune subset quantification of TLR2 (D) and TLR4 (E) expression by CD4 T cells, CD8 T cells, NK cells, monocytes, and neutrophils.
Data in (B)-(E) consist of 5–10 mice/group and are representative of 3 technical replicates. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.005.
See also Figure S4.
Given the increased frequency and number of circulating monocytes and neutrophils after cohousing (Figure 2B), we examined blood cells from SPF and cohoused mice for TLR2 and TLR4 expression as a potential mediator of increased pathogen responsiveness in the cohoused mice. Indeed, cohoused B6 and pet store mice have an increased frequency and total number of TLR2+ and TLR4+ cells circulating in the blood, compared to SPF B6 mice (Figure 6B), but no change in the expression levels of TLR2 or TLR4 as measured by mean fluorescence intensity (MFI; Figures 6B and S4). The frequency of circulating CD14+ cells is also elevated in cohoused B6 and pet store mice (Figure 6C). Following cohousing, monocytes contribute the largest increase in TLR2+ (Figure 6D) and TLR4+ cells (Figure 6E), as well as a slight, but significant, increase in the number of TLR2+ neutrophils. Both of these cell populations were also numerically expanded in cohoused B6 and pet store mice (see Figures 2D and 2E). Interestingly, we also noted a modest but significant increase in the number of CD8 T cells expressing TLR4 in the cohoused B6 mice (Figure 6E). Thus, these data suggest microbial exposure augments the capacity of the immune system to recognize infection through increased expression of pattern-recognition receptors.
Physiological Microbial Exposure Sensitizes Mice to TLR4, but Not to TLR2, Agonist-Induced Sterile Inflammation
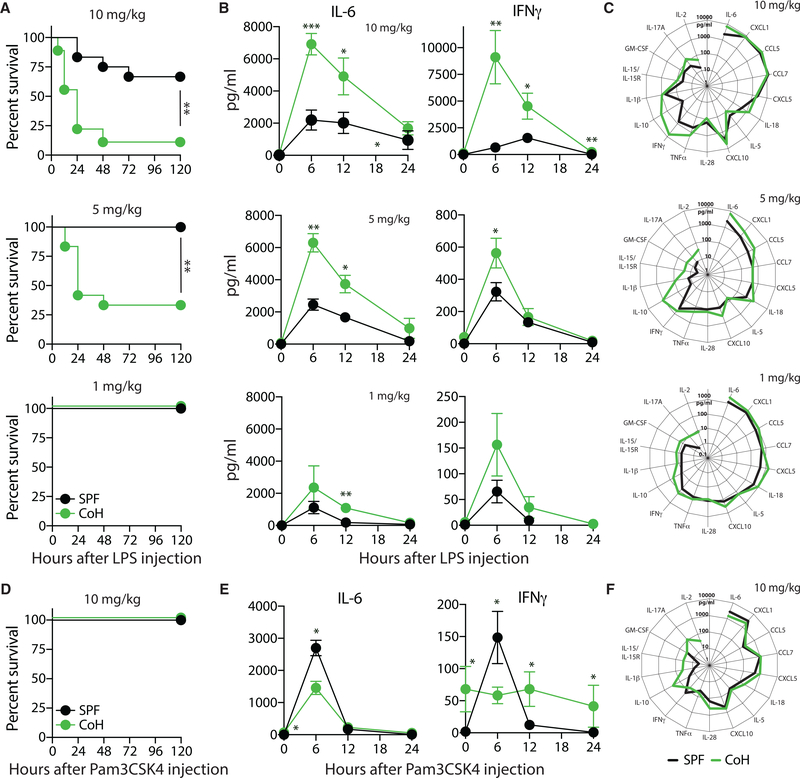
Direct injection of live, virulent Listeria monocytogenes establishes an active infection. Similarly, the CLP model of sepsis relies on the disruption of the protective barrier maintained by the intestinal epithelium, releasing a mixture of live microbes into the peritoneum. Injection of the endotoxin LPS is another common model of sepsis, which instead induces a sterile inflammation that mimics the activation of the innate immune system during a septic event (Remick et al., 2000). In contrast to plentiful data obtained from LPS endotoxemia models, there is little information regarding the sterile inflammatory response to a purified TLR2 agonist. With the increase in TLR2+ and TLR4+ immune cells (especially monocytes and neutrophils) in mice after physiological microbial exposure, we were interested to see the effect of purified TLR2 or TLR4 agonist challenge in SPF and cohoused B6 mice. Similar to the data obtained with CLP-induced polymicrobial sepsis, cohoused B6 mice demonstrated an increased susceptibility to LPS challenge and succumbed rapidly to doses (10 and 5 mg/kg intravenous [i.v.]) that were tolerated by SPF mice (Figure 7A). Examination of the serum after LPS administration found a rapid increase in IL-6 and IFNγ in both SPF and cohoused B6 mice, but the amount was significantly higher in cohoused B6 mice (Figure 7B). This effect was dose dependent, and titration of LPS to 5 mg/kg and 1 mg/kg i.v. reduced cytokine production after challenge. Interestingly, while all SPF and co-housed mice survived after a 1-mg/kg i.v. LPS challenge, there was still an increase in cytokine production at 6 h in cohoused B6 mice relative to B6 SPF (Figure 7C). These data show that similar to the live polymicrobial infection seen during CLP, co-housed mice robustly respond to the LPS challenge with rapid pro-inflammatory cytokine production and increased mortality. In contrast to the LPS challenge, injection of the TLR2/TLR1 agonist Pam3CSK4 (10 mg/kg i.v.) was well tolerated in both SPF and cohoused B6 mice (Figure 7D). Analysis of the global inflammatory response 6 h after the Pam3CSK4 challenge did reveal some cytokines (e.g., IL-1β, IL-10, IL-15/IL-15R, IL-17A, IL-18, and IL-28) and chemokines (e.g., CCL5, CCL7, CXCL5, and CXCL10) that were elevated in the cohoused mice (Figure 7F), but SPF mice had higher amounts of IL-6 and IFNγ in the serum than did cohoused mice (Figure 7E), much like what we saw in the Listeria monocytogenes-infected mice (see Figure 3F). We also find that the TLR3 agonist poly I:C (10 mg/kg i.v.) induces elevated IL-6 and IFNγ in cohoused mice, but the adjuvant was well tolerated by both SPF and cohoused hosts (data not shown). Collectively, these data (when combined with the data in Figure 6) suggest the increased presence of TLR-expressing immune cells following physiological microbial exposure affects the composition and magnitude of an inflammatory response to systemically administered TLR agonists. However, there is also differential responsiveness, such that the TLR2 ligands induce somewhat muted cytokine responses, whereas TLR4 ligands induce dramatic increases in systemic pro-inflammatory cytokines. These distinct responses to TLR stimulation may explain the different responses of cohoused mice to Listeria monocytogenes and sepsis, which are major drivers of TLR2 and TLR4, respectively.
Figure 7. Increased Cytokine Production in Cohoused Mice after LPS Injection Correlates with Increased Mortality.
SPF and cohoused B6 mice were injected with LPS (10, 5, or 1 mg/kg i.v.).
(A) Survival after LPS injection.
(B) Serum samples were obtained at indicated hours after LPS injection, and the amount of IL-6 and IFNγ was determined by bioplex.
(C) Additional cytokines and chemokines in the serum 6 h post-LPS injection were quantified by bioplex.
(D–F) SPF and cohoused B6 mice were injected with Pam3CSK4 (10 mg/kg i.v.).
(D and E) Survival was monitored (D) and the amount of IL-6 and IFNγ in the serum at the indicated time points was determined by bioplex (E).
(F) Additional cytokines and chemokines in the serum 6 h post-Pam3CKS4 injection were quantified by bioplex.
Survival data (A and D) consist of 12–21 mice/group and are pooled from3technical replicates. Cytokine data(B, C, E, and F) consist of 4–7 mice/group/time point and are representative of 3 technical replicates. *p ≤ 0.05, **p ≤ 0.01 for SPF versus cohoused at the indicated time points.
DISCUSSION
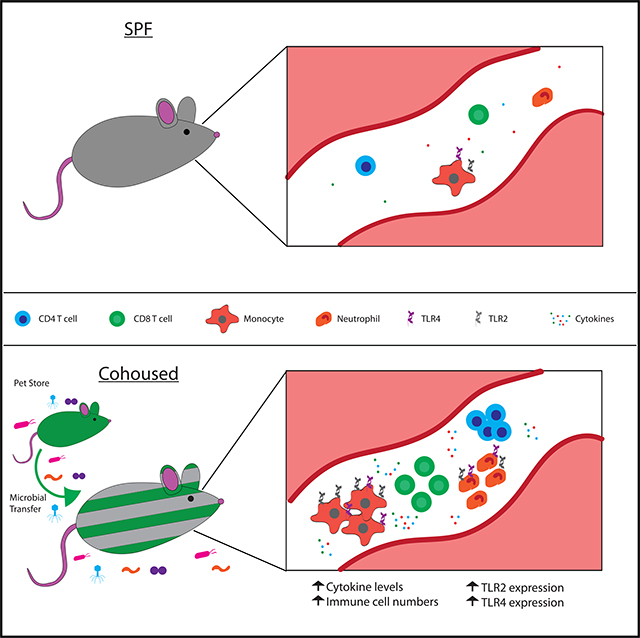
Animals (including humans) are exposed to commensal and pathogenic microbes throughout their lives, and this physiological microbial exposure has a profound impact on immune competency and overall health (Huggins et al., 2019). The use of laboratory mice housed under SPF conditions in the vast majority of immunology research has been an important step to improve experimental consistency, but these unusually clean living conditions have inadvertently left the laboratory mice with an underdeveloped immune system—far different from the immune system found in adult humans. The recent description of a mouse model of physiological microbial exposure introduced the important effects of the environment on the basal immune state (Beura et al., 2016). The data presented herein have extended those initial findings to demonstrate that microbial exposure alters the inflammatory state of the immune system. Former SPF laboratory mice develop elevated profiles of numerous inflammatory cytokines after microbial transfer through cohousing with outbred pet store mice. In addition, the cellular composition of the immune system is altered after cohousing to favor increased circulating monocytes and neutrophils expressing TLRs. These changes dramatically altered the responsiveness to future unrelated immune challenges. Intriguingly, the nature of the specific challenge determines whether previous microbial exposure enhances or inhibits effective immune function, pathogen clearance, and ultimately survival of the host. Specifically, cohoused mice demonstrate enhanced bacterial clearance after Listeria monocytogenes infection but are more susceptible to death from polymicrobial sepsis and LPS.
Recent studies have drawn attention to the idea that the innate immune system is capable of forming long-term memory in response to stimulation. Mechanisms of this altered responsiveness, or “trained immunity,” include changes in expression of pathogen-sensing receptors, signaling proteins, and innate cell numbers (Foster et al., 2007; Netea et al., 2016). In the cohoused mouse model, laboratory mice are exposed to a variety of viral, bacterial, and parasitic microbes, resulting in the activation of innate immune cells through pattern-recognition receptors. Signaling through pattern-recognition receptors induces stable changes in histone methylation (Quintin et al., 2012). Here, we demonstrate that following cohousing, the “trained” immune system contains increased numbers of TLR2- and TLR4-expressing cells. Furthermore, transcriptional studies by Beura et al. (2016) identified elevated Myd88 mRNA transcripts in cohoused mice, a critical downstream signaling component utilized by most TLRs, including TLR2 and TLR4 (Beutler, 2009; Kawai and Akira, 2011; Kawasaki and Kawai, 2014; Netea et al., 2016). We confirm that Myd88 is elevated in cohoused mice (Figure 6A) and also show elevated CD14 expression (Figure 6C). Thus, innate stimulation through pattern-recognition receptors can be regulated through prior microbial experience to unrelated organisms. The benefit of these changes is illustrated through enhanced host protection to Listeria monocytogenes, but at the cost of increased sensitivity to cytokine storms during polymicrobial sepsis, leading to poorer survival rates. Further studies are required to understand the full landscape of innate changes induced through cohousing and how they potentially synergize to shape the responsiveness to future challenges.
TLR expression on immune cells present in neonatal cord blood is reduced relative to adult blood (Kollmann et al., 2009; Pedraza-Sánchez et al., 2013). Furthermore, neonatal T cells and monocytes have a reduced functional capacity to produce cytokines following stimulation (Chipeta et al., 1998; Gasparoni et al., 2003), and the ability of immune cells obtained from infants to produce IFNγ, TNFα, IL-2, and IL-4 is significantly reduced compared to cells from adults (Härtel et al., 2005). Neonatal blood monocytes are reduced in number compared to adults, and they are less responsive to TLR4 stimulation. This work goes on to demonstrate that neonatal cord blood exposed to malaria in utero causes an elevation in circulating monocytes and responsiveness to TLR4 agonist stimulation. These differences in TLR expression between neonatal and adult humans are consistent with our findings, as we noted substantial increases in the frequency of circulating TLR2+ and TLR4+ cells in cohoused mice (see Figure 6) but, intriguingly, not other TLRs (Figure 6A). Thus, our study provides additional rationale for using the cohoused murine system to identify the extent that microbial exposure contributes to age-associated differences in the immune system. One hypothesis for the correlation among increasing age, elevation in serum cytokines, and TLR expression (especially in humans) is the increase in microbial exposure through vaccination and natural infection. The cohousing model offers an appealing and unique opportunity to study the impact of physiological microbial exposure on the immune system in a manner that complements studies using neonatal cord blood and adult human blood.
Cohoused mice are able to more rapidly clear Listeria monocytogenes infection than SPF mice (Figure 3A) (Beura et al., 2016). Our data show SPF and cohoused mice had similar bacterial burdens in both the liver and spleen at 8 h after Listeria monocytogenes infection. It was not until 48–72 h post-infection that a reduction in pathogen burden became apparent in the cohoused mice. We observed increased IFNγ production, particularly from CD4+ and CD8+ T cells, after cohousing. Interestingly, the number of NK cells did not increase in cohoused mice (Figure 2G), and they were not elevated in their production of IFNγ (Figure 3E) as might be expected following immune “training” (Netea et al., 2016). Whether this is an indication that specific pathogens are required to train NK cells will require further study. However, since memory T cells can make IFNγ in response to early inflammatory cues in an antigen-independent manner, it is possible that the elevated numbers of antigen-experienced T cells in cohoused mice could lead to improved clearance of Listeria monocytogenes and other intracellular bacteria (Berg et al., 2003, 2005; Lertmemongkolchai et al., 2001). These data indicate that the enhanced adaptive immune response may be stimulated via increased TLR expression following microbial exposure. TLR stimulation alerts the host to respond to infectious challenges and recruits T cells to the site of the infection through inflammatory signals (Akira et al., 2006). In humans and mice, prior infectious experience can regulate TLR expression and overall immune responsiveness. Humans infected with Plasmodium have increased responsiveness to TLR agonists (McCall et al., 2007). TLR expression increases in a feedback loop where IFNγ production enhances TLR expression and allows the host to more effectively respond to future infectious challenges (Franklin et al., 2009). This increased responsiveness reveals a “double-edged sword” seen in our cohoused model system, where the host can respond more effectively to TLR2-triggering bacterial infections but is also at a higher risk of pathology from hyper-reactive cytokine responses to TLR4 ligands. We found increased IFNγ-producing cells in the blood 24 h after virulent Listeria monocytogenes infection of cohoused mice. Unexpectedly, the amount of IFNγ and a number of other inflammatory cytokines in the serum was relatively unchanged following Listeria monocytogenes infection of cohoused mice, though they were markedly elevated in SPF mice. One potential explanation for the reduced numbers of circulating cytokines in the Listeria monocytogenes-infected cohoused mice is that memory T cells and “trained” innate cells may be rapidly absorbing the cytokines as they are being produced to facilitate their increased function.
In striking contrast to Listeria monocytogenes infection, CLP-induced polymicrobial sepsis results in a dramatic increase in pro-inflammatory cytokines and chemokines in cohoused mice relative to their SPF counterparts. For example, there were exceedingly high amounts of IL-6 (>20,000 pg/ml at 6 h post-CLP) in the serum of cohoused mice after CLP surgery. Previous reports have highlighted the predictive nature of circulating IL-6 levels (i.e., increased mortality with increased IL-6) in regard to mortality in preclinical and clinical settings (Hack et al., 1989; Remick et al., 2002; Waage et al., 1989). Similar correlations between the amount of IL-1β and TNF and survival have also been made, prompting the clinical testing of various strategies for neutralizing IL-1β and TNF activity (Zanotti et al., 2002). Use of the CLP model has led to a vast expansion of knowledge regarding the intricate changes within the immune system following a septic event. Recent clinical data using agents (e.g., IL-7 and anti-PD-1 mAb) (Shindo et al., 2017, 2015; Spec et al., 2016; Venet et al., 2017, 2012; West et al., 2013) to manipulate the number and responsiveness of immune cells in sepsis survivors highlight the importance of preclinical experimentation, but the success of these new approaches is atypical. Over 100 agents (many targeting cytokines) with preclinical efficacy in mouse models of sepsis have been unsuccessful in humans (Fink, 2014); it is tempting to speculate that the exclusive use of SPF mice in previous preclinical studies may have underestimated the severity of the cytokine response following CLP surgery.
Laboratory mice cohoused with pet store mice not only encounter numerous different pathogenic infections, but also develop changes to the microbiome. The link between microbiome composition and immune status has been increasingly appreciated in the past decade. Immune cell interactions with the diverse components of the microbiome are critical for the development of a healthy immune system (Belkaid and Hand, 2014). Dysbiosis of a healthy microbiome results in aberrant immune responses and susceptibility to both autoimmunity and inflammatory diseases (Kamada et al., 2013). Our cohousing experiments demonstrate that the gut microbial contents contribute to the heightened immune response of cohoused mice during sepsis. Cohoused cecal contents injected into the peritoneum of SPF mice induced a more robust cytokine storm, compared to that seen in SPF mice injected with SPF cecal contents. However, our data suggest the gut contents do not fully explain the phenotype, as cohoused recipients displayed an exacerbated cytokine response after injection of either SPF or cohoused cecal slurries. Additional studies are needed to evaluate how pathogen exposure and variations in commensal microbes influence the maturation and function of the immune system to future infections.
Additionally, we noted similar increases in pro-inflammatory cytokines and increased mortality after exposing cohoused mice to LPS—a potent TLR4 stimulant that induces sterile inflammation. Bolus injection of endotoxin (LPS) is considered a model of sepsis, mostly because of the rapid activation of the innate immune system (Stortz et al., 2017). Despite having a number of similarities to CLP-induced sepsis (e.g., lethargy, piloerection, and acute leukopenia), there are also a number of important differences, such as the kinetics and magnitude of proinflammatory cytokine (IL-6, TNF) production (Remick et al., 2000). Another criticism of LPS endotoxemia as a model of sepsis was based on data suggesting humans to be significantly more sensitive to the effects of the LPS challenge compared to mice (Warren et al., 2010). As expected, this conclusion came (in part) from experiments comparing the responsiveness of cells from the blood of adult humans and SPF mice. As now evidenced by data presented herein and previously (Beura et al., 2016), there are distinct phenotypic and transcriptomic changes within the cells of the immune systems of SPF and cohoused mice that indicate their resemblance to the immune systems of neonate and adult humans, respectively. It stands to reason that the conclusions drawn by Warren et al. (2010) may be the result of comparing the response of the naive, neonate-like immune system of SPF mice to that of the matured, microbially experienced immune system of adult humans. We posit that the differences noted between mice and humans may be more complex than simply a “species-specific” difference, and the underappreciated extent of physiological microbial exposure may have played a more significant role.
In conclusion, we find that mice with normal physiologic exposure to microbes exhibit increased serum cytokine levels and circulating phagocytes, in addition to phenotypic conversion of the CD4 and CD8 T cell compartments. These changes in the basal state of the immune system, however, do not lead to a “blanket” improvement in immunity. Instead, depending on the nature of the immune insult, previous immune history is either beneficial (as in the case of the Listeria monocytogenes challenge) or detrimental (as in the case of CLP-induced sepsis). This conclusion differs slightly from previous studies, which have thus far shown improved immune responses after diverse microbial exposure (Beura et al., 2016; Reese et al., 2016; Rosshart et al., 2017). The data presented herein suggest that the cumulative microbial exposure of the host defines the expression and differing activation state of individual pattern-recognition receptors. Thus, we cannot exclude the possibility that the more robust immunity acquired via exposure to numerous pathogens could decrease the incidence of sepsis by rapidly controlling pathogens acquired by normal routes of infection-even though our data with cohoused mice clearly develop increased severity of experimental sepsis in the CLP and LPS models. Using the cohousing system, future studies can be designed to differentiate the specific traits influenced by microbial exposure and show how to generate an optimal immune response that minimizes the chances for immunopathology. Applying this cohousing system to diseases that, to date, have been poorly modeled in mice may open new opportunities in preclinical research.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for reagents may be directed to and will be fulfilled by the Lead Contact, Sara Hamilton (hamil062@umn.edu). This study did not generate unique reagents.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Female C57BL/6N (B6; 8–10 weeks of age) mice were purchased from Charles River (Wilmington, MA). Female pet store mice were purchased from local pet stores in the Minneapolis-St. Paul, MN metropolitan area. All mice were housed in AALAC-approved animal facilities at the University of Minnesota (BSL-1/BSL-2 for SPF B6 mice, and BSL-3 for cohoused B6 and pet store mice). SPF B6 and pet store mice were cohoused at a ratio of 8:1 in large rat cages for 60 days to facilitate microbe transfer. Littermates were randomly assigned to experimental groups. Experimental procedures were approved by the University of Minneapolis Animal Care and Use Committee and performed following the Office of Laboratory Animal Welfare guidelines and PHS Policy on Human Cancer and Use of Laboratory Animals.
METHOD DETAILS
Serology and immune cell phenotyping
Serum was collected prior to and after 60 days of co-housing for serology and cytokine analysis. Serum was screened using EZ-spot and PCR Rodent Infectious Agent (PRIA) array methods (Charles River Laboratories) for sendai virus (SEND), pneumonia virus of mice (PVM), mouse hepatitis virus (MHV), Minute virus of mice (MVM), mouse parvovirus type 1 (MPV-1), mouse parvovirus type 2 (MPV-2), mouse parvovirus-NS1 (NS-1), murine norovirus (MNV), Theiler’s murine encephalomyelitis virus (GDVII), reovirus (REO), Rotavirus EDIM (EDIM/ROTA-A), lymphocytic choriomeningitis virus (LCMV), ectromelia virus (ECTRO), mouse adenovirus 1 and 2 (MAV1&2), mouse cytomegalovirus (MCMV), polyoma virus (POLY), mycoplasma pulmonis (MPUL), enchephalitozoon cuniculi (ECUN), cilia-associated respiratory bacillus (CARB), and clostridium piliforme (CPIL). Blood was also collected before and after co-housing to determine the composition and phenotype of circulating immune cells.
Flow cytometry
The following fluorochrome-conjugated mAb were purchased from Biolegend: Ly6C (HK1.4), Nkp46 (29A1.4), MHC II (I:A/I:E) (M5/114.15.2), CD62L (MEL-14), CD45.2 (104), CD44 (IM7), Ly6G (1A8), TLR2 (QA16A01), TLR4-MD2 (MTS510), CD64 (X54–5/7.1, CD14 (Sa14–2) and Tonbo Biosciences: CD8a (53–6.7), CD4 (RM4–5), IFNγ (XMG1.2), Ghost 780 (live/dead).
Measurement of serum cytokines and chemokines
Serum cytokines and chemokines were quantitated with the ProcartaPlex Cytokine & Chemokine 36-plex mouse panel or custom 17-plex panel (CCL5, CCL7, CXCL1, CXCL5, CXCL10, GM-CSF, IL-1β, IL-2, IL-5, IL-6, IL-10, IL-15/IL-15R, IL-17A, IL-18, IL-28, IFNγ, and TNF; Invitrogen) using a Luminex 200 with Bio-plex Manager Software 5.0.
Listeria infection
Wild-type, virulent Listeria monocytogenes (LM) strain 10403s (provided by John Harty, University of Iowa) was grown to log phase in tryptic soy broth containing streptomycin. Mice were infected i.v. with 8 × 104 CFU. Bacterial load in the spleen and liver was determined at the indicated days post-challenge (Hamilton et al., 2006).
Cecal ligation and puncture
Sepsis was induced by CLP (Rittirsch et al., 2009). Briefly, mice were anesthetized using isoflurane (2.5% gas via inhalation) or Ketamine/xylazine (87.5mg/kg and 12.5mg/kg, respectively, i.p.). The abdomen was shaved and disinfected with 5% povidone-iodine antiseptic. Bupivicaine (6 mg/kg s.c.) was then administered at the site where a midline incision was made. The distal third of the cecum was ligated with 4–0 silk suture and punctured once with a 25-g needle to extrude a small amount of cecal content. The cecum was returned to the abdomen, the peritoneum was closed via continuous suture, and the skin was sealed using surgical glue (Vetbond; 3M, St. Paul, MN). Meloxicam (2 mg/kg) in 1 mL saline was administered at the time of surgery and for 3 additional days for postoperative analgesia and fluid resuscitation. Mice were monitored daily for weight loss and pain for at least 5 days post-surgery. Sham-treated mice underwent the same laparotomy procedure excluding ligation and puncture.
Bacteremia quantitation
Bacterial burden in the blood was assessed at 24 hours after CLP surgery. Heparinized blood was collected via cardiac puncture using aseptic techniques, serially diluted in sterile PBS, and plated on TSA plates. After overnight incubation at 37°C, colonies were counted.
DNA extraction and sequencing
DNA was extracted from single mouse fecal pellets using the DNeasy PowerSoil kit (QIAGEN, Hilden, Germany). The V5-V6 hypervariable regions of the 16S rRNA gene were amplified and paired-end sequenced using the BSF784/1064R primer set (Claesson et al., 2010) by the University of Minnesota Genomics center (UMGC; Minneapolis, MN). Sequencing was done using the Illumina MiSeq platform (Illumina, Inc., San Diego, CA) at a read length of 300 nucleotides (nt). Raw data are deposited in the Sequence Read Archive (Leinonen et al., 2011) under BioProject accession number SRP193509.
TLR4 Reporter Cell Assay
Cecal contents were collected and prepared according to previously published protocols (Hilbert et al., 2017). Briefly, SPF and CoH mice were euthanized and cecal contents harvested and weighed. Samples from individual mice were initially mixed with a 5% dextrose solution to create a 40 mg/ml slurry. The cecal slurry was then passed through 70 mm filter to remove large particulates. Slurries were then added at the indicated concentrations to triplicate wells of 96-well flat-bottom plates seeded with 4 × 104 TLR4 reporter cells (HEK-Blue hTLR4; InvivoGen [Simons et al., 2007]). HEK-Blue hTLR4 were maintained according to manufacturer’s recommendations. After 24 h, conditioned media was collected to measure SEAP activity according to the manufacturer’s protocol (QUANTI-Blue Solution, InvivoGen).
Intraperitoneal injection of cecal slurry
Cecal contents from 3–4 donor mice were pooled and mixed with 5% dextrose solution to create a 40 mg/ml cecal slurry. The cecal slurry was then passed through 70 μm filter and then injected i.p. into recipient mice (0.75 or 0.5 mg slurry/g mouse in 300 μl) using a 21G needle. Meloxicam (2 mg/kg) in 1 mL saline was administered at the time of slurry injection and for 3 additional days for postoperative analgesia and fluid resuscitation. Mice were monitored daily for weight loss and pain for at least 5 days post-injection.
Quantitative real-time PCR (qPCR)
Myeloid cells were isolated from the spleens of SPF and CoH mice by plastic adherence (Inaba et al., 2009; Steinman et al., 1979). Spleens (5 SPF and 5 CoH) were prepared into a single cell suspension in 2 mL HBSS, washed, and resuspended in 10 mL of complete RPMI. Each cell suspension was added to two 10 cm culture dishes and incubated at37°Cfor90 min. Plates were washed twice with 5 mL PBS to remove nonadherent cells. Total RNA was isolated from the remaining adherent myeloid cells using TRIzol reagent (Invitrogen, Carlsbad, CA), and 1 mg was reverse-transcribed using Superscript III (Invitrogen). Resulting cDNA was used as a template for qPCR using TaqMan primer/probe sets for Tlr1, Tlr2, Tlr3, Tlr4, Tlr5, Tlr6, Tlr7, Tlr8, Tlr9, Cd14, Ly96, Myd88, Ticam2, Traf6, Iraki, and 18 s rRNA (Applied Biosystems).
Bioinformatics
Sequence processing was done using mothur (version 1.33.3) (Schloss et al., 2009), as previously described (Staley et al., 2018). Briefly, sequences were paired-end join, trimmed for quality, and aligned against the SILVA database (version 123) for clustering. A 2% pre-cluster was used to remove sequences likely to contain errors (Huse et al., 2010). Chimeras were identified and removed using UCHIME (version 4.2.40) (Edgar et al., 2011). Operational taxonomic units (OTUs) were identified at 97% similarity using furthest-neighbor clustering and classified against the Ribosomal Database Project release (version 14). Alpha diversity (within-sample diversity), measured as Good’s coverage, Shannon index, and abundance-based coverage estimate (ACE), was calculated in mothur. Differences in beta diversity (between-sample diversity) were calculated using Bray-Curtis dissimilarities (Bray and Curtis, 1957). To determine engraftment among CoH mice, SourceTracker (version 0.9.8) was used with default parameters (Knights et al., 2011). This program uses a Bayesian algorithm to determine the percent of the community in user-defined sink (co-housed mice) samples that can be attributed to user defined source (SPF and pet store mice) samples.
LPS/Pam3CSK4 challenge
A rodent model of endotoxemia was induced by a single injection of LPS-EBfrom E. coliO111:B4 (10, 5, or 1 mg/kg body weight i.v.; InvivoGen) (Markwart et al., 2014). Similarly, some mice were challenged with a single dose (10 mg/kg body weight i.v.) of Pam3CSK4 VacciGrade (InvivoGen).
QUANTIFICATION AND STATISTICAL ANALYSIS
Data shown are presented as mean values ± SEM. GraphPad Prism 8 was used for statistical analysis, where statistical significance was determined using two-tailed Unpaired, nonparametric Mann-Whitney U test (for 2 individual groups) or group-wise, one-way ANOVA analyses followed by multiple-testing correction using the Holm-Sidak method, with α = 0.05. * p < 0.05, ** p < 0.01, *** p < 0.005, **** p < 0.001. Differences in alpha diversity were determined using ANOVA and differences in relative abundances of genera were determine by Kruskal-Wallis test using the Steel-Dwass-Critchlow-Fligner procedure for pairwise comparisons. Analysis of similarity (ANOSIM) (Clarke, 1993) was used to evaluate differences in community composition, and ordination was done using principal coordinates analysis (PCoA) (Anderson and Willis, 2003). Correlation of phyla and genera abundances with axes positions were done using Spearman correlations. ANOVA and Kruskal-Wallis tests were done using XLSTAT software (version 17.06; Addin-soft, Belmont, MA). ANOSIM, PCoA, and correlation analyses were done using mothur. All statistics were evaluated at α = 0.05 with Bonferroni correction for multiple comparisons.
DATA AND CODE AVAILABILITY
The accession number for the raw 16 s rRNA sequencing data reported in this paper is SRA: SRP193509 (Leinonen et al., 2011).
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-mouse CD8a (53–6.7) BUV395 | 3D Bioscience | Cat #: 563786; RRID:AB_2732919 |
| Anti-mouse CD4 (RM4–5) APC | eBioscience | Cat #: 17–0041-82; RRID:AB_469320 |
| Anti-mouse CD45 (30 F11) BV711 | BD Bioscience | Cat #: 563709; RRID:AB_2687455 |
| Anti-mouse CD45.2 (104) PerCP-Cy5.5 | Tonbo Biosciences | Cat #: 65–0454-U100; RRID:AB_2621894 |
| Anti-mouse CD44 (IM7) BUV737 | BD Bioscience | Cat #: 564392; RRID:AB_2738785 |
| Anti-mouse CD64 (X54–5/7.1) BV605 | Biolegend | Cat #: 139323; RRID:AB_2629778 |
| Anti-mouse Ly6C (HK1.4) APC-eF780 | eBioscience | Cat #: 47–5932-80; RRID:AB_2573991 |
| Anti-mouse Nkp46 (29A1.4) BV421 | Biolegend | Cat #: 137612; RRID:AB_2563104 |
| Anti-mouse MHC II (I:A/I:E) BV510 | Biolegend | Cat #: 107635; RRID:AB_2561397 |
| Anti-mouse Ly6G (1A8) BV785 | Biolegend | Cat #: 127645; RRID:AB_2566317 |
| Anti-mouse TLR2 (QA16A01) | Biolegend | Cat #: 153006; RRID:AB_2728206 |
| Anti-mouse TLR4-MD2 (MTS510) | Biolegend | Cat #: 117610; RRID:AB_2044020 |
| Anti-mouse IFNγ (XMG1.2) PE-Cy7 | Tonbo Biosciences | Cat #: 60–7311-U100; RRID:AB_2621871 |
| Anti-mouse CD62L (MEL-14) BV786 | BD Bioscience | Cat #: 564109; RRID:AB_2738598 |
| Anti-mouse CD14 (Sa14–2) BV421 | Biolegend | Cat #: 123329; RRID:AB_2721526 |
| Ghost 780 Viability Dye | Tonbo Biosciences | Cat#: 13–0865-T100 |
| Bacterial and Virus Strains | ||
| Listeria Monocytogenes Strain 10403s | John T. Harty, University of Iowa; Hamilton et al., 2006 | NCBITaxon: 393133 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| LPS (from E. coli O111:B4) | InvivoGen | tlrl-eblps |
| Pam3CSK4 | InvivoGen | tlrl-pms |
| Critical Commercial Assays | ||
| ProcartaPlex Cytokine & Chemokine 36-plex mouse panel | Thermo Fisher Scientific | EPX360–26092-901; RRID:AB_2576123 |
| Mouse Custom ProcartaPlex 17-plex (CCL5, CCL7, CXCL1, CXCL5, CXCL10, GM-CSF, IFNγ, IL-1P, IL-2, IL-5, IL-6, IL-10, IL-15/IL-15R, IL-17A, IL-18, IL-28, TNFα) | Thermo Fisher Scientific | PPX-17-MXKA3NP |
| Mouse PCR Rodent Infectious Agent Array | Charles River Laboratory | N/A |
| Deposited Data | ||
| Raw data (16S rRNA sequencing) | Sequence Read Archive | BioProject accession #: SRP193509 |
| Experimental Models: Cell Lines | ||
| HEK-Blue hTLR4 | InvivoGen | Cat #: hkb-htlr4; RRID:CVCL_IM82 |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6Ncr | NCI Charles River | #556 |
| Oligonucleotides | ||
| 16S rRNA primer set | Claesson et al., 2010 | BSF784/1064R |
| TaqManGene Expression Assay (FAM) -Assay ID: Mm00446095_m1; Gene Symbol: Tlr1 | Life Technologies | Cat #: 4453320 |
| TaqManGene Expression Assay (FAM) -Assay ID: Mm00442346_m1; Gene Symbol: Tlr2 | Life Technologies | Cat #: 4453320 |
| TaqManGene Expression Assay (FAM) - Assay ID: Mm01207404_m1; Gene Symbol: Tlr3 | Life Technologies | Cat #: 4453320 |
| TaqManGene Expression Assay (FAM) - Assay ID: Mm00445273_m1; Gene Symbol: Tlr4 | Life Technologies | Cat #: 4448892 |
| TaqManGene Expression Assay (FAM) - Assay ID: Mm00546288_s1; Gene Symbol: Tlr5 | Life Technologies | Cat #: 4453320 |
| TaqManGene Expression Assay (FAM) - Assay ID: Mm02529782_s1; Gene Symbol: Tlr6 | Life Technologies | Cat #: 4453320 |
| TaqManGene Expression Assay (FAM) - Assay ID: Mm00446590_m1; Gene Symbol: Tlr7 | Life Technologies | Cat #: 4453320 |
| TaqManGene Expression Assay (FAM) - Assay ID: Mm01157262_m1; Gene Symbol: Tlr8 | Life Technologies | Cat #: 4453320 |
| TaqManGene Expression Assay (FAM) - Assay ID: Mm00446193_m1; Gene Symbol: Tlr9 | Life Technologies | Cat #: 4453320 |
| TaqManGene Expression Assay (FAM) - Assay ID: Mm00438094_g1; Gene Symbol: Cd14 | Life Technologies | Cat #: 4453320 |
| TaqManGene Expression Assay (FAM) - Assay ID: Mm01227593_m1; Gene Symbol: Ly96 | Life Technologies | Cat #: 4448892 |
| TaqManGene Expression Assay (FAM) - Assay ID: Mm00440338_m1; Gene Symbol: Myd88 | Life Technologies | Cat #: 4453320 |
| TaqManGene Expression Assay (FAM) - Assay ID: Mm01260003_m1; Gene Symbol: Ticam2 | Life Technologies | Cat #: 4448892 |
| TaqManGene Expression Assay (FAM) - Assay ID: Mm00493836_m1; Gene Symbol: Traf6 | Life Technologies | Cat #: 4453320 |
| TaqManGene Expression Assay (FAM) - Assay ID: Mm01193538_m1; Gene Symbol: Irak1 | Life Technologies | Cat #: 4448892 |
| Software and Algorithms | ||
| Prism 8.0 | Graphpad | https://www.graphpad.com/scientific-software/prism/ |
| FlowJo | BD Biosciences | www.flowjo.com |
| FacsDiva | BD Biosciences | N/A |
| Mothur (v1.33.3) | Schloss et al., 2009 | https://www.mothur.org/ |
| UCHIME (version 4.2.40) | Edgar et al., 2011 | https://www.drive5.com/uchime/ |
| Ribosomal Database Project (version 14) | https://rdp.cme.msu.edu/ | |
| SourceTracker (version 0.9.8) | Knights et al., 2011 | https://github.com/danknights/sourcetracker/releases |
Highlights.
Cohousing elevates basal cytokine and chemokine levels
Cohousing alters immune responsiveness to new challenges
Cohoused mice have an altered microbiome composition
TLR4 expression and LPS sensitivity are increased after microbial exposure
ACKNOWLEDGMENTS
This study was supported by NIH grants R01GM115462 (to T.S.G.), R01AI116678 (to S.E.H.), R01AIGM113961 (to V.P.B.), T32CA009138 and T32AI007313 (to F.V.S.), T32AI007485 (to D.B.D. and I.J.J.), and T32AI007511 (to I.J.J.); an American Association of Immunologists Careers in Immunology Fellowship (to S.E.H. and M.A.H.); and aVeterans Administration Merit Review Award (I01BX001324; to T.S.G.). This work was also supported in part by NIH P30 CA77598 utilizing the Masonic Cancer Center, University of Minnesota, Flow Cytometry shared resource. Finally, we thank Drs. Kristin Hogquist and Daniel Mueller for critical review of the manuscript and helpful suggestions.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.07.028.
REFERENCES
- Akira S, Takeda K, and Kaisho T (2001). Toll-like receptors: critical proteins linking innate and acquired immunity. Nat. Immunol 2, 675–680. [DOI] [PubMed] [Google Scholar]
- Akira S, Uematsu S, and Takeuchi O (2006). Pathogen recognition and innate immunity. Cell 724, 783–801. [DOI] [PubMed] [Google Scholar]
- Anderson MJ, and Willis TJ (2003). Canonical analysis of principal coordinates: A useful method of constrained ordination for ecology. Ecology 84, 511–525. [Google Scholar]
- Barton ES, White DW, Cathelyn JS, Brett-McClellan KA, Engle M, Diamond MS, Miller VL, and Virgin HW 4th. (2007). Herpesvirus latency confers symbiotic protection from bacterial infection. Nature 447, 326–329. [DOI] [PubMed] [Google Scholar]
- Belkaid Y, and Hand TW (2014). Role of the microbiota in immunity and inflammation. Cell 157, 121–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg RE, Crossley E, Murray S, and Forman J (2003). Memory CD8+ T cells provide innate immune protection against Listeria monocytogenes in the absence of cognate antigen. J. Exp. Med 198, 1583–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg RE, Crossley E, Murray S, and Forman J (2005). Relative contributions of NK and CD8Tcellsto IFN-gamma mediated innate immune protection against Listeria monocytogenes. J. Immunol 175, 1751–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beura LK, Hamilton SE, Bi K, Schenkel JM, Odumade OA, Casey KA, Thompson EA, Fraser KA, Rosato PC, Filali-Mouhim A, et al. (2016). Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature 532, 512–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutler BA (2009). TLRs and innate immunity. Blood 113, 1399–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray JR, and Curtis JT (1957). An ordination of the upland forest communities of southern Wisconsin. Ecol. Monogr. 27, 325–349. [Google Scholar]
- Cabrera-Perez J, Condotta SA, Badovinac VP, and Griffith TS (2014). Impact of sepsis on CD4 T cell immunity. J. Leukoc. Biol 96, 767–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera-Perez J, Condotta SA, James BR, Kashem SW, Brincks EL, Rai D, Kucaba TA, Badovinac VP, and Griffith TS (2015). Alterations in antigen-specific naive CD4 T cell precursors after sepsis impairs their responsiveness to pathogen challenge. J. Immunol 194, 1609–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera-Perez J, Babcock JC, Dileepan T, Murphy KA, Kucaba TA, Badovinac VP, and Griffith TS (2016). Gut Microbial Membership Modulates CD4 T Cell Reconstitution and Function after Sepsis. J. Immunol 197, 1692–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera-Perez J, Badovinac VP, and Griffith TS (2017). Enteric immunity, the gut microbiome, and sepsis: Rethinking the germ theory of disease. Exp. Biol. Med. (Maywood) 242, 127–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipeta J, Komada Y, Zhang XL, Deguchi T, Sugiyama K, Azuma E, and Sakurai M (1998). CD4+ and CD8+ cell cytokine profiles in neonates, older children, and adults: increasing T helper type 1 and T cytotoxic type 1 cell populations with age. Cell. Immunol 183, 149–156. [DOI] [PubMed] [Google Scholar]
- Chousterman BG, Swirski FK, and Weber GF (2017). Cytokine storm and sepsis disease pathogenesis. Semin. Immunopathol. 39, 517–528. [DOI] [PubMed] [Google Scholar]
- Claesson MJ, Wang Q, O’Sullivan O, Greene-Diniz R, Cole JR, Ross RP, and O’Toole PW (2010). Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 38, e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke KR (1993). Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol 18, 117–143. [Google Scholar]
- Condotta SA, Rai D, James BR, Griffith TS, and Badovinac VP (2013). Sustained and incomplete recovery of naive CD8+ T cell precursors after sepsis contributes to impaired CD8+ T cell responses to infection. J. Immunol 190, 1991–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condotta SA, Khan SH, Rai D, Griffith TS, and Badovinac VP (2015). Polymicrobial sepsis increases susceptibility to chronic viral infection and exacerbates CD8+ T cell exhaustion. J. Immunol 195, 116–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danahy DB, Anthony SM, Jensen IJ, Hartwig SM, Shan Q, Xue HH, Harty JT, Griffith TS, and Badovinac VP (2017). Polymicrobial sepsis impairs bystander recruitment of effector cells to infected skin despite optimal sensing and alarming function of skin resident memory CD8 T cells. PLoS Pathog. 13, e1006569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danahy DB, Jensen IJ, Griffith TS, and Badovinac VP (2019). Cutting Edge: Polymicrobial Sepsis Has the Capacity to Reinvigorate Tumor-Infiltrating CD8 T Cells and Prolong Host Survival. J. Immunol 202, 2843–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, and Charpentier E (2014). Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096. [DOI] [PubMed] [Google Scholar]
- Duong S, Condotta SA, Rai D, Martin MD, Griffith TS, and Badovinac VP (2014). Polymicrobial sepsis alters antigen-dependent and -independent memory CD8 T cell functions. J. Immunol 192, 3618–3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, and Knight R (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink MP (2014). Animal models of sepsis. Virulence 5, 143–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster HL (1959). Housing of disease-free vertebrates. Ann. N Y Acad. Sci 78, 80–88. [DOI] [PubMed] [Google Scholar]
- Foster SL, Hargreaves DC, and Medzhitov R (2007). Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature 447, 972–978. [DOI] [PubMed] [Google Scholar]
- Franklin BS, Parroche P, Ataide MA, Lauw F, Ropert C, de Oliveira RB, Pereira D, Tada MS, Nogueira P, da Silva LH, et al. (2009). Malaria primes the innate immune response due to interferon-gamma induced enhancement of toll-like receptor expression and function. Proc. Natl. Acad. Sci. USA 106, 5789–5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparoni A, Ciardelli L, Avanzini A, Castellazzi AM, Carini R, Rondini G, and Chirico G (2003). Age-related changes in intracellular TH1/TH2 cytokine production, immunoproliferative T lymphocyte response and natural killer cell activity in newborns, children and adults. Biol. Neonate 84, 297–303. [DOI] [PubMed] [Google Scholar]
- Gurung P, Rai D, Condotta SA, Babcock JC, Badovinac VP, and Griffith TS (2011). Immune unresponsiveness to secondary heterologous bacterial infection after sepsis induction is TRAIL dependent. J. Immunol 187, 2148–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hack CE, DeGroot ER, Felt-Bersma RJ, Nuijens JH, StrackVan Schijndel RJ, Eerenberg-Belmer AJ, Thijs LG, and Aarden LA (1989). Increased plasma levels of interleukin-6 in sepsis. Blood 74, 1704–1710. [PubMed] [Google Scholar]
- Hamilton SE, Wolkers MC, Schoenberger SP, and Jameson SC (2006). The generation of protective memory-like CD8+ T cells during homeostatic proliferation requires CD4+ T cells. Nat. Immunol 7, 475–481. [DOI] [PubMed] [Google Scholar]
- Haring JS, Badovinac VP, and Harty JT (2006). Inflaming the CD8+ T cell response. Immunity 25, 19–29. [DOI] [PubMed] [Google Scholar]
- Härtel C, Adam N, Strunk T, Temming P, Müller-Steinhardt M, and Schultz C (2005). Cytokine responses correlate differentially with age in infancy and early childhood. Clin. Exp. Immunol 142, 446–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harty JT, and Bevan MJ (1995). Specific immunity to Listeria monocytogenes in the absence of IFN gamma. Immunity 3, 109–117. [DOI] [PubMed] [Google Scholar]
- Heipertz EL, Harper J, Lopez CA, Fikrig E, Hughes ME, and Walker WE (2018). Circadian Rhythms Influence the Severity of Sepsis in Mice via a TLR2-Dependent, Leukocyte-Intrinsic Mechanism. J. Immunol 201, 193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilbert T, Steinhagen F, Senzig S, Cramer N, Bekeredjian-Ding I, Parcina M, Baumgarten G, Hoeft A, Frede S, Boehm O, and Klaschik S (2017). Vendor effects on murine gut microbiota influence experimental abdominal sepsis. J. Surg. Res 211, 126–136. [DOI] [PubMed] [Google Scholar]
- Hotchkiss RS, and Karl IE (2003). The pathophysiology and treatment of sepsis. N. Engl. J. Med 348, 138–150. [DOI] [PubMed] [Google Scholar]
- Huggins MA, Jameson SC, and Hamilton SE (2019). Embracing microbial exposure in mouse research. J. Leukoc. Biol 105, 73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse SM, Welch DM, Morrison HG, and Sogin ML (2010). Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ. Microbiol 12, 1889–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba K, Swiggard WJ, Steinman RM, Romani N, Schuler G, and Brinster C (2009). Isolation of dendritic cells. Curr Protoc Immunol. Chapter 3, Unit 3.7. [DOI] [PubMed] [Google Scholar]
- Jensen IJ, Sjaastad FV, Griffith TS, and Badovinac VP (2018a). Sepsis-Induced T Cell Immunoparalysis: The Ins and Outs of Impaired T Cell Immunity. J. Immunol 200, 1543–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen IJ, Winborn CS, Fosdick MG, Shao P, Tremblay MM, Shan Q, Tripathy SK, Snyder CM, Xue HH, Griffith TS, et al. (2018b). Polymicrobial sepsis influences NK-cell-mediated immunity by diminishing NK-cell-intrinsic receptor-mediated effector responses to viral ligands or infections. PLoS Pathog. 14, e1007405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N, Seo SU, Chen GY, and Núñez G (2013). Role of the gut microbiota in immunity and inflammatory disease. Nat. Rev. Immunol 13, 321–335. [DOI] [PubMed] [Google Scholar]
- Kawai T, and Akira S (2011).Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 34, 637–650. [DOI] [PubMed] [Google Scholar]
- Kawasaki T, and Kawai T (2014). Toll-like receptor signaling pathways. Front. Immunol 5, 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knights D, Kuczynski J, Charlson ES, Zaneveld J, Mozer MC, Collman RG, Bushman FD, Knight R, and Kelley ST (2011). Bayesian community-wide culture-independent microbial source tracking. Nat. Methods 8, 761–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollmann TR, Crabtree J, Rein-Weston A, Blimkie D, Thommai F, Wang XY, Lavoie PM, Furlong J, Fortuno ES 3rd, Hajjar AM, et al. (2009). Neonatal innate TLR-mediated responses are distinct from those of adults. J. Immunol 183, 7150–7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinonen R, Sugawara H, and Shumway M; International Nucleotide Sequence Database Collaboration (2011). The sequence read archive. Nucleic Acids Res. 39, D19–D21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lertmemongkolchai G, Cai G, Hunter CA, and Bancroft GJ (2001). Bystander activation of CD8+ T cells contributes to the rapid production of IFN-gamma in response to bacterial pathogens. J. Immunol 166, 1097–1105. [DOI] [PubMed] [Google Scholar]
- Levy MM, Artigas A, Phillips GS, Rhodes A, Beale R, Osborn T, Vincent JL, Townsend S, Lemeshow S, and Dellinger RP (2012). Outcomes of the Surviving Sepsis Campaign in intensive care units in the USA and Europe: a prospective cohort study. Lancet Infect. Dis 12, 919–924. [DOI] [PubMed] [Google Scholar]
- Lima CX, Souza DG, Amaral FA, Fagundes CT, Rodrigues IP, Alves-Filho JC, Kosco-Vilbois M, Ferlin W, Shang L, Elson G, and Teixeira MM (2015). Therapeutic Effects of Treatment with Anti-TLR2 and Anti-TLR4 Monoclonal Antibodies in Polymicrobial Sepsis. PLoS ONE 10, e0132336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai V, Torrazza RM, Ukhanova M, Wang X, Sun Y, Li N, Shuster J, Sharma R, Hudak ML, and Neu J (2013). Distortions in development of intestinal microbiota associated with late onset sepsis in preterm infants. PLoS ONE 8, e52876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markwart R, Condotta SA, Requardt RP, Borken F, Schubert K, Wei-gel C, Bauer M, Griffith TS, Förster M, Brunkhorst FM, et al. (2014). Immunosuppression after sepsis: systemic inflammation and sepsis induce a loss of naïve T-cells but no enduring cell-autonomous defects in T-cell function. PLoS ONE 9, e115094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masopust D, Sivula CP, and Jameson SC (2017). Of Mice, Dirty Mice, and Men: Using Mice To Understand Human Immunology. J. Immunol 199, 383–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall MB, Netea MG, Hermsen CC, Jansen T, Jacobs L, Golenbock D, van der Ven AJ, and Sauerwein RW (2007). Plasmodium falciparum infection causes proinflammatory priming of human TLR responses. J. Immunol 179, 162–171. [DOI] [PubMed] [Google Scholar]
- Müller B, and Grossniklaus U (2010). Model organisms-A historical perspective. J. Proteomics 73, 2054–2063. [DOI] [PubMed] [Google Scholar]
- Netea MG, Joosten LA, Latz E, Mills KH, Natoli G, Stunnenberg HG, O’Neill LA, and Xavier RJ (2016).Trained immunity: A program of innate immune memory in health and disease. Science 352, aaf1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira-Nascimento L, Massari P, and Wetzler LM (2012). The Role of TLR2 in Infection and Immunity. Front. Immunol 3, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson JA, McDonald-Hyman C, Jameson SC, and Hamilton SE (2013). Effector-like CD8+ T cells in the memory population mediate potent protective immunity. Immunity 38, 1250–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedraza-Sánchez S, Hise AG, Ramachandra L, Arechavaleta-Velasco F, and King CL (2013). Reduced frequency of a CD14+ CD16+ monocyte subset with high Toll-like receptor 4 expression in cord blood compared to adult blood contributes to lipopolysaccharide hyporesponsiveness in newborns. Clin. Vaccine Immunol 20, 962–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintin J, Saeed S, Martens JHA, Giamarellos-Bourboulis EJ, Ifrim DC, Logie C, Jacobs L, Jansen T, Kullberg BJ, Wijmenga C, et al. (2012). Candida albicans infection affords protection against reinfection via functional reprogramming of monocytes. Cell Host Microbe 12, 223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese TA, Bi K, Kambal A, Filali-Mouhim A, Beüra LK, Bürger MC, Pulendran B, Sekaly RP, Jameson SC, Masopust D, et al. (2016). Sequential Infection with Common Pathogens Promotes Human-like Immune Gene Expression and Altered Vaccine Response. Cell Host Microbe 19, 713–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remick DG, Newcomb DE, Bolgos GL, and Call DR (2000). Comparison of the mortality and inflammatory response of two models of sepsis: lipopolysaccharide vs. cecal ligation and puncture. Shock 13, 110–116. [DOI] [PubMed] [Google Scholar]
- Remick DG, Bolgos GR, Siddiqui J, Shin J, and Nemzek JA (2002). Six at six: interleukin-6 measured 6 h after the initiation of sepsis predicts mortality over 3 days. Shock 17, 463–67. [DOI] [PubMed] [Google Scholar]
- Rittirsch D, Huber-Lang MS, Flierl MA, and Ward PA (2009). Immunodesign of experimental sepsis by cecal ligation and puncture. Nat. Protoc. 4, 31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosshart SP, Vassallo BG, Angeletti D, Hutchinson DS, Morgan AP, Takeda K, Hickman HD, McCulloch JA, Badger JH, Ajami NJ, et al. (2017). Wild Mouse Gut Microbiota Promotes Host Fitness and Improves Disease Resistance. Cell 171, 1015–1028.e1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol 75, 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo Y, Unsinger J, Burnham CA, Green JM, and Hotchkiss RS (2015). Interleukin-7 and anti-programmed cell death 1 antibody havediffering effects to reverse sepsis-induced immunosuppression. Shock 43, 334–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo Y, Fuchs AG, Davis CG, Eitas T, Unsinger J, Burnham CD, Green JM, Morre M, Bochicchio GV, and Hotchkiss RS (2017). Interleukin 7 immunotherapy improves host immunity and survival in a two-hit model of Pseudomonas aeruginosa pneumonia. J. Leukoc. Biol 101, 543–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons MP, Moore JM, Kemp TJ, and Griffith TS (2007). Identification of the mycobacterial subcomponents involved in the release of tumor necrosis factor-related apoptosis-inducing ligand from human neutrophils. Infect. Immun 75, 1265–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjaastad FV, Condotta SA, Kotov JA, Pape KA, Dail C, Danahy DB, Kucaba TA, Tygrett LT, Murphy KA, Cabrera-Perez J,et al. (2018). Polymicrobial Sepsis Chronic Immunoparalysis Is Defined by Diminished Ag-Specific T Cell-Dependent B Cell Responses. Front. Immunol 9, 2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spec A, Shindo Y, Burnham CA, Wilson S, Ablordeppey EA, Beiter ER, Chang K, Drewry AM, and Hotchkiss RS (2016). T cells from patients with Candida sepsis display a suppressive immunophenotype. Crit. Care 20, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley C, Kaiser T, Vaughn BP, Graiziger CT, Hamilton MJ, Rehman TU, Song K, Khoruts A, and Sadowsky MJ (2018). Predicting recurrence of Clostridium difficile infection following encapsulated fecal microbiota transplantation. Microbiome 6, 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinman RM, Kaplan G, Witmer MD, and Cohn ZA (1979). Identification of a novel cell type in peripheral lymphoid organs of mice. V. Purification of spleen dendritic cells, new surface markers, and maintenance in vitro. J. Exp. Med 749, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stortz JA, Raymond SL, Mira JC, Moldawer LL, Mohr AM, and Efron PA (2017). Murine Models of Sepsis and Trauma: Can We Bridge the Gap? ILARJ. 58, 90–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strother RK, Danahy DB, Kotov DI, Kucaba TA, Zacharias ZR, Griffith TS, Legge KL, and Badovinac VP (2016). Polymicrobial Sepsis Diminishes Dendritic Cell Numbers and Function Directly Contributing to Impaired Primary CD8 T Cell Responses In Vivo. J. Immunol 197, 4301–4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K, and Akira S (2001a). Regulation of innate immune responses by Toll-like receptors. Jpn. J. Infect. Dis 54, 209–219. [PubMed] [Google Scholar]
- Takeda K, and Akira S (2001b). Roles ofToll-like receptors in innate immune responses. Genes Cells 6, 733–742. [DOI] [PubMed] [Google Scholar]
- Takeda K, Kaisho T, and Akira S (2003). Toll-like receptors. Annu. Rev. Immunol 21, 335–376. [DOI] [PubMed] [Google Scholar]
- Thiemann S, Smit N, Roy U, Lesker TR, Galvez EJC, Helmecke J, Basic M, Bleich A, Goodman AL, Kalinke U, et al. (2017). Enhancement of IFNgamma Production by Distinct Commensals Ameliorates Salmonella-Induced Disease. Cell Host Microbe 21, 682–694.e685. [DOI] [PubMed] [Google Scholar]
- Unsinger J, Kazama H, McDonough JS, Hotchkiss RS, and Ferguson TA (2009). Differential lymphopenia-induced homeostatic proliferation for CD4+ and CD8+ T cells following septic injury. J. Leukoc. Biol 85, 382–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venet F, Foray A-P, Villars-Méchin A, Malcus C, Poitevin-Later F, Lepape A, and Monneret G (2012). IL-7 restores lymphocyte functions in septic patients. J. Immunol 189, 5073–5081. [DOI] [PubMed] [Google Scholar]
- Venet F, Demaret J, Blaise BJ, Rouget C, Girardot T, Idealisoa E, Rimmelé T, Mallet F, Lepape A, Textoris J, and Monneret G (2017). IL-7 Restores T Lymphocyte Immunometabolic Failure in Septic Shock Patients through mTOR Activation. J. Immunol 199, 1606–1615. [DOI] [PubMed] [Google Scholar]
- Waage A, Brandtzaeg P, Halstensen A, Kierulf P, and Espevik T (1989). The complex pattern of cytokines in serum from patients with meningococcal septic shock. Association between interleukin 6, interleukin 1, and fatal outcome. J. Exp. Med 169, 333–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren HS, Fitting C, Hoff E, Adib-Conquy M, Beasley-Topliffe L, Tesini B, Liang X, Valentine C, Hellman J, Hayden D, and Cavaillon JM (2010). Resilience to bacterial infection: difference between species could be due to proteins in serum. J. Infect. Dis 201, 223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West EE, Jin H-T, Rasheed A-U, Penaloza-Macmaster P, Ha S-J, Tan WG, Youngblood B, Freeman GJ, Smith KA, and Ahmed R (2013). PD-L1 blockade synergizes with IL-2 therapy in reinvigorating exhausted T cells. J. Clin. Invest 123, 2604–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry EJ, Teichgräber V, Becker TC, Masopust D, Kaech SM, Antia R, von Andrian UH, and Ahmed R (2003). Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat. Immunol 4, 225–234. [DOI] [PubMed] [Google Scholar]
- Zanotti S, Kumar A, and Kumar A (2002). Cytokine modulation in sepsis and septic shock. Expert Opin. Investig. Drugs 11, 1061–1075. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the raw 16 s rRNA sequencing data reported in this paper is SRA: SRP193509 (Leinonen et al., 2011).