Abstract
In a screen of 1,000 consecutively ascertained families, we recently found that mutations in the gene RPGR are the third most common cause of all inherited retinal disease. As the two most frequent disease-causing genes, ABCA4 and USH2A, are far too large to fit into clinically relevant adeno-associated virus (AAV) vectors, RPGR is an obvious early target for AAV-based ocular gene therapy. In generating plasmids for this application, we discovered that those containing wild-type RPGR sequence, which includes the highly repetitive low complexity region ORF15, were extremely unstable (i.e., they showed consistent accumulation of genomic changes during plasmid propagation). To develop a stable RPGR gene transfer vector, we used a bioinformatics approach to identify predicted regions of genomic instability within ORF15 (i.e., potential non-B DNA conformations). Synonymous substitutions were made in these regions to reduce the repetitiveness and increase the molecular stability while leaving the encoded amino acid sequence unchanged. The resulting construct was subsequently packaged into AAV serotype 5, and the ability to drive transcript expression and functional protein production was demonstrated via subretinal injection in rat and pull-down assays, respectively. By making synonymous substitutions within the repetitive region of RPGR, we were able to stabilize the plasmid and subsequently generate a clinical-grade gene transfer vector (IA-RPGR). Following subretinal injection in rat, we demonstrated that the augmented transcript was expressed at levels similar to wild-type constructs. By performing in vitro pull-down experiments, we were able to show that IA-RPGR protein product retained normal protein binding properties (i.e., analysis revealed normal binding to PDE6D, INPP5E, and RPGRIP1L). In summary, we have generated a stable RPGR gene transfer vector capable of producing functional RPGR protein, which will facilitate safety and toxicity studies required for progression to an Investigational New Drug application.
Keywords: gene augmentation, X-linked retinitis pigmentosa, RPGR
Introduction
Vision loss from inherited retinal disease affects ∼140,000 people in the United States with about 1,700 new cases diagnosed annually.1 While more than 100 different disease-causing genes have been identified, mutations in ABCA4, USH2A, RPGR, and RHO account for more than 33% of all cases.1 ABCA4 and USH2A exceed the carrying capacity of clinically approved adeno-associated virus (AAV) vectors, and RHO-associated retinitis pigmentosa (RP) is predominantly a dominant gain of function disorder. As a result, among these four genes, only RPGR is expected to be amenable to traditional AAV-based gene augmentation therapy.
When present in the hemizygous state in males, most mutations in the X-linked gene RPGR cause a degenerative disease of rod photoreceptors known as RP. RPGR-associated RP usually comes to medical attention before the 20th birthday and in some cases can cause profound vision loss within the first decade of life.
When designing a gene augmentation treatment, it is important to consider all of the naturally occurring genetic isoforms of the disease-causing gene. For RPGR, two major transcripts exist: (1) a ubiquitous transcript that contains 19 exons and (2) a transcript that is dramatically enriched in the retina, which contains a purine-rich low complexity region encoded by an alternative exon known as ORF15 followed by an early stop codon.2–5 The fact that the overall length and charge of this low complexity domain have been evolutionarily conserved among vertebrates suggests that it has some functional importance in the retina.6 However, surprisingly, little is known about the precise biological function of RPGR beyond its localization to the connecting cilium of photoreceptor cells5 and the plethora of reported binding partners.7–13
Rescue experiments in RPGR-knockout animals show that delivery of the RPGR ORF15 isoform is sufficient to rescue photoreceptor cell function.14–17 However, the generation of RPGR ORF15 expression cassettes capable of delivering this isoform to tissues has proven to be difficult. Multiple codon-optimized gene transfer constructs have been designed by different research groups including constructs with large deletions or spontaneous mutations within the low complexity region16,18 and others with synonymous sequence changes distributed throughout the entire coding sequence.17
Due to its purine-rich sequence, the low complexity region of RPGR has been postulated to adopt non-B-DNA conformations such as triplex DNA.4 Although these alternate structures have the potential to be mutagenic,19 they also play important roles in gene regulation.20 As a result, when modifying these sequences for therapeutic purposes, one must ensure that the resulting transcript is expressed at the desired level and retains the ability to interact with downstream binding partners. To mitigate the instability previously observed when cloning canonical RPGR ORF15 cDNA,18,21 we started with the shortest naturally occurring allele, which we identified by screening RPGR ORF15 in 50 unaffected males. We subsequently made synonymous codon changes in all highly predicted triplex-DNA-forming regions and packaged the resulting RPGR ORF15 plasmid into an AAV5 expression vector (IA-RPGR). Following subretinal injection in wild-type rat, IA-RPGR-encoded transcripts were expressed at a level similar to a nonoptimized ORF15 construct. Likewise, the resulting IA-RPGR protein product was shown to retain normal protein binding properties (i.e., pull-down analysis revealed normal binding to PDE6D, INPP5E, and RPGRIP1L).
Materials and Methods
Ethics statement
This study was approved by the Institutional Review Board of the University of Iowa and adhered to the tenets set forth in the Declaration of Helsinki. All patients were seen by a retina specialist in the Retina Clinic of the University of Iowa Department of Ophthalmology and Visual Sciences. All rat experiments were conducted with the approval of the University of Iowa Animal Care and Use Committee and were consistent with the Association for Research in Vision and Ophthalmology Statement for the Use of Animals in Ophthalmic and Vision Research.
Identification of shortest naturally occurring wild-type human RPGR ORF15 allele
RPGR ORF15 alleles from 50 normal males were cloned, and sequencing was performed as previously published.1 In short, the low complexity region of RPGR was amplified, and the resulting PCR products were TA cloned and transformed using the TOPO® TA Cloning® Kit (cat. no. K450040; Invitrogen). Transformed cells were grown on Ampicillin, IPTG, X-gal (AIX) plates for blue-white colony screening. White colonies were picked, expanded in lysogeny broth, purified via mini prep and Sanger sequenced.
Construction of AAV vectors
pFB-CMVp-hRPGR ORF15 was constructed in a stepwise manner. The 5′ end of the gene was amplified from RPGR cDNA purchased from Origene (cat. no. RG212243), whereas the terminal exon ORF15 was amplified from normal control gDNA. The ends were ligated via a FauI restriction site. To generate IA-RPGR, triplex DNA predictions were made using Triplex, a R/Bioconductor package.22 The cDNA for IA-RPGR was constructed by GenScript. Both full-length cDNAs were then cloned into the pFBAAVCMVmcs plasmid (cat. no. G0347; University of Iowa Gene Transfer Vector Core).
Sequencing reactions
Sanger sequencing was performed using an ABI 3730xl instrument. For highly repetitive regions, optimized sequencing chemistry was used, as previously published.1
AAV virus production and quality control
Virus production
pFB-CMVp-hRPGR ORF15 and pFB-CMVp-IA-RPGR vectors were packaged in AAV serotype 5 (AAV2/5) via calcium phosphate transfections of HEK293T (National Gene Vector Biorepository, Indiana University) cells, as previously published.23
Silver staining
Viral samples were mixed with lithium dodecyl sulfate (cat. no. NP0007; Invitrogen) and reducing agent (cat. no. NP0009; Invitrogen) and boiled for 5 min. Samples and ladder (cat. no. 10747-012; Invitrogen) were then electrophoresed on a Bis-Tris gel at 90 V (cat. no. NP022BOX; Invitrogen). A SilverQuest Staining Kit (cat. no. LC6070; Invitrogen) was used to visualize the samples, and images were recorded using the VersaDoc Gel Imager (cat. no. 1708640; Bio-Rad).
Vector integrity
Recombinant AAV particles were amplified using the Herculase II Fusion DNA Polymerase Kit (cat. no. 600679; Agilent) and the pFBAAVforward primer (5′-TGCTACTTATCTACGTAGCCATGCTCTAGA-3′) and the pFBAAVreverse primer (5′-ATGCTACTTATCTACGTAGCCATGCTCTAGT-3′).
The following conditions were used:
Stage 1: at 98°C for 10 min
Stage 2: × 34
Step 1 at 98°C for 15 s
Step 2 at 55°C for 20 s
Step 3 at 68°C for 3 min
Stage 3: at 68°C for 4 min
Stage 4: Hold at 12°C
Subsequent PCR products were purified using the MinElute PCR Purification Kit (no. 28006; Qiagen), and gene cassette identity was confirmed via Sanger sequencing.
FLAG-immunoprecipitation
Both RPGR ORF15 and IA-RPGR were N-terminal FLAG-S-tagged and transfected into HEK293T cells. After 48 h, the cells were treated with lysis buffer (1 × phosphate-buffered saline, 1% Triton X-100, and protease inhibitor (cat. no. 11 836 153 001; Roche) and pelleted at 20,000 g for 15 min at 40°C. The resulting supernatant was incubated with anti-FLAG beads (cat. no. B26102; Biotool) for 4 h. The beads were washed four times with lysis buffer, and the interactions were detected by Western blotting.
An RPGR ORF15 missense variant (RPGR ORF15G275S) was generated by site-directed mutagenesis (cat. no. 200523; Stratagene) using N-terminal FLAG-S-tagged RPGR ORF15 as template, and mutations were verified by Sanger sequencing.
Antibodies and reagents
The following antibodies were used: anti-PDE6D rabbit polyclonal antibody (cat. no. MBS7005086; MyBioSource), anti-INPP5E rabbit polyclonal antibody (cat. no. 17797-1-AP; Proteintech), anti-RPGRIP1L (cat. no. HPA039405; Sigma), horseradish peroxidaseperoxidase-conjugated anti-FLAG mouse monoclonal antibody (cat. no. A8592; Sigma), and anti-glyceraldehyde 3-phosphate dehydrogenase (cat. no. ma5-15738; Invitrogen).
AAV characterization from injected rat eyes
To evaluate transcript expression in vivo, AAV 2/5-CMV-RPGR ORF15, AAV 2/5-CMV-IA-RPGR, or AAV storage and injection buffer (vehicle control) were injected into the subretinal space of wild-type Sprague Dawley rats. Subretinal injections were made by inserting a 33-gauge blunt needle on a Hamilton syringe (Hamilton Company, Reno, Nevada) through the sclera into the temporal midperipheral subretinal space and slowly depressing the plunger until a 10 μL subretinal bleb containing each AAV (1 × 1012 vector genome/mL) was injected. To prevent efflux, the needle was left in place for several seconds postinjection. The resulting retinal bleb was visualized through the operating microscope for all eyes and as needed with fundus photography and optical coherence tomography (Micron IV with OCT2; Phoenix Research Laboratories, CA) to confirm location. Animals were allowed to fully recover (i.e., until mobile) under continuous surveillance before being returned to their husbandry room. Seventy-two hours after transduction, the injected eyes were enucleated, and retinas were processed for RNA analysis. RNA extractions were performed using the NucleoSpin RNA Kit (cat. no. 740955; Macherey-Nagel), and cDNA was generated using the SuperScript™ VILO™ cDNA Synthesis Kit (cat. no. 11754-250; Thermo). Quantitative reverse transcription (RT)-PCR was performed using Power SYBR Green Master Mix (cat. no. 4367659; Applied Biosystems™). To obtain vector-driven expression levels, residual viral gDNA was quantified and subtracted from the copies of the gene of interest. For this analysis, the following primers were used: CMV forward-CATGACCTTATGGGACTTTCCT and CMV reverse-CTATCCACGCCCATTGATGTA, and RPGR 14F-AGGGATTTTCATGACGCAGC and RPGR ORF15 R-AGGTTCCATCCCCTCTACCT.24
Results
Generation of a stable RPGR ORF15 AAV plasmid
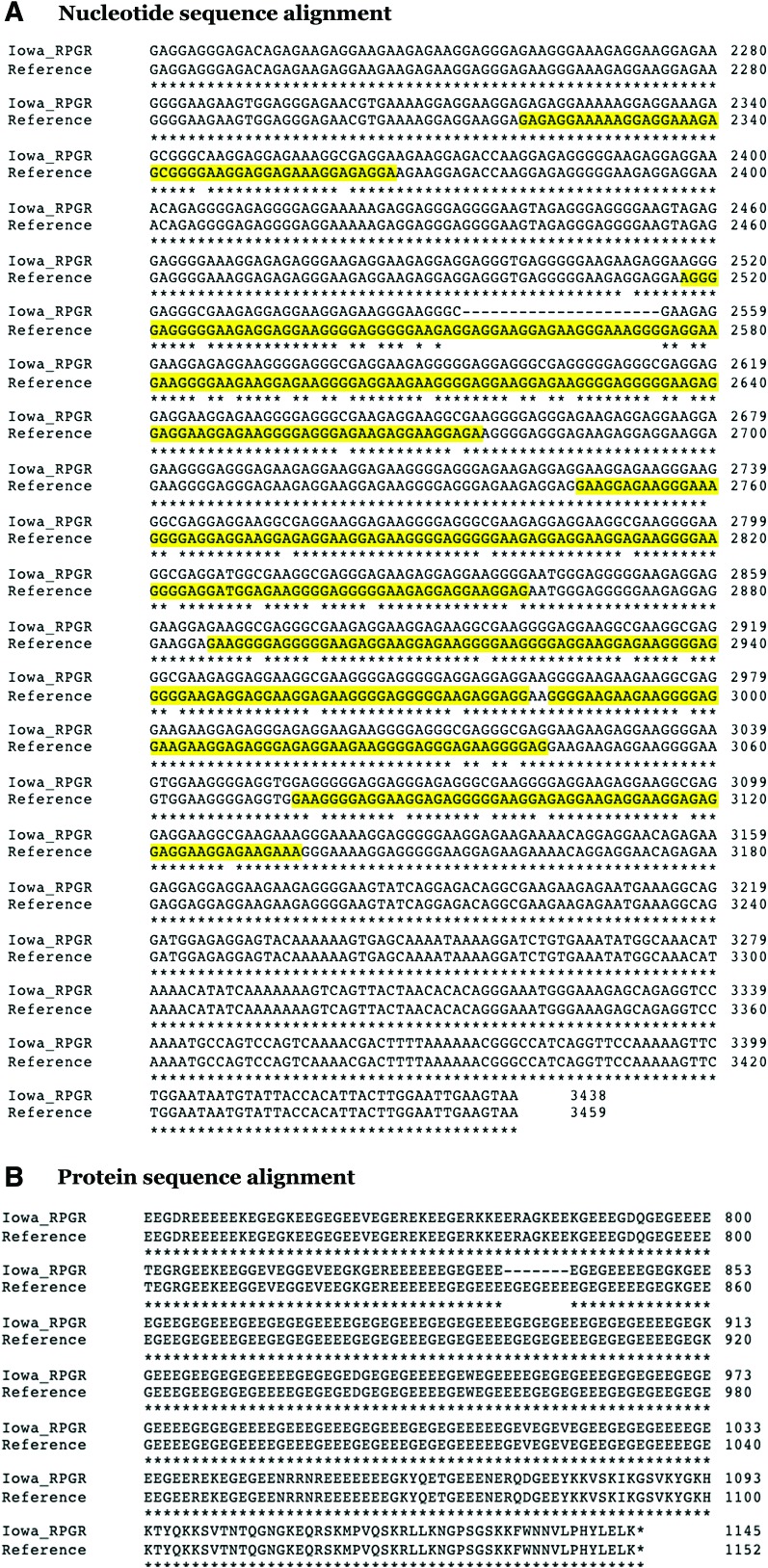
To inform our gene transfer vector design strategy, we began by identifying the natural nondisease-causing variations that occur within the low complexity region of RPGR ORF15 in 50 normal males. As shown in Table 1, eight different nondisease-causing alleles were identified among these normal males. The allele containing the largest nondisease-causing in-frame deletion (Fig. 1A) was chosen as the starting point for generating a codon-optimized RPGR ORF15 construct. Next, in an attempt to identify regions that may be contributing to plasmid instability (which we define as the ability of the ORF15 sequence to be replicated and maintained in Escherichia coli), triplex DNA predictions were made using the bioinformatic prediction package Triplex.22 As shown in Table 2, 12 sites ranging from 47 to 61 bp in length scored highly in this analysis, indicating a potential for triplex DNA formation at these sites. Synonymous substitutions were subsequently created within these high scoring sequences (Fig. 1A, yellow highlighted regions), and the Triplex analysis was repeated. As shown in Table 3, the synonymous changes proposed were predicted to mitigate the formation of triplex DNA without altering the protein sequence (Fig. 1B). To determine if the resulting RPGR AAV expression plasmid (termed IA-RPGR) was more stable than the RPGR ORF15 containing construct, both plasmids were transformed in SURE2 competent cells and subjected to diagnostic digest to confirm identity, as described previously. For the RPGR ORF15 cDNA, only 1 of 24 clones contained the correct insert. In contrast, 100% of the IA-RPGR clones retained the correct sequence, suggesting that the synonymous changes were sufficient to stabilize the plasmid.
Table 1.
Normal variation in RPGR ORF15 observed in 50 normal males
| DNA Variants |
|---|
| Ala781Thr GCG>ACG hemi |
| Glu881Gly GAG>GGG hemi |
| Gly834 Del21ggtGAGGGGGAAGAGGAGGAAGGG hemi |
| Glu869 del15gagGAAGAAGGGGAGGAA hemi |
| Glu889 del3gaaGAG Hemi |
| Glu937 ins21gaaGGAGAAGGGGAAGGGGAGGAT hemi |
| Glu1017 del12gaGGAAGGGGAAGT hemi |
| Lys1048Arg AAG>AGG hemi |
Bold, allele selected as the starting sequence for IA-RPGR.
Figure 1.
Nucleotide and protein sequence alignment demonstrating synonymous changes made in the low complexity ORF15 region of RPGR. (A) Nucleotide sequence alignment with high scoring triplex DNA regions highlighted in yellow. (B) Protein sequence alignment demonstrating amino acid conservation. Color images are available online.
Table 2.
Top 12 predicted triplex-DNA-forming regions within wild-type RPGR ORF15
| No. | cDNA Start Position | Width (bp) | Type | Score | p |
|---|---|---|---|---|---|
| 1 | 2319 | 47 | 3 | 40 | 7.00E-11 |
| 2 | 2517 | 60 | 2 | 42 | 1.30E-11 |
| 3 | 2575 | 49 | 3 | 44 | 2.40E-12 |
| 4 | 2614 | 61 | 2 | 47 | 1.90E-13 |
| 5 | 2746 | 61 | 3 | 46 | 4.50E-13 |
| 6 | 2800 | 61 | 3 | 41 | 3.00E-11 |
| 7 | 2887 | 61 | 2 | 40 | 7.00E-11 |
| 8 | 2887 | 61 | 3 | 44 | 2.40E-12 |
| 9 | 2899 | 61 | 3 | 40 | 7.00E-11 |
| 10 | 2926 | 55 | 3 | 41 | 3.00E-11 |
| 11 | 2982 | 60 | 3 | 40 | 7.00E-11 |
| 12 | 3076 | 61 | 2 | 44 | 2.40E-12 |
Table 3.
Predicted triplex-DNA-forming regions within IA-RPGR
| No. | cDNA Start Position | Width (bp) | Type | Score | p |
|---|---|---|---|---|---|
| None | — | — | — | — | — |
Viral vector packaging
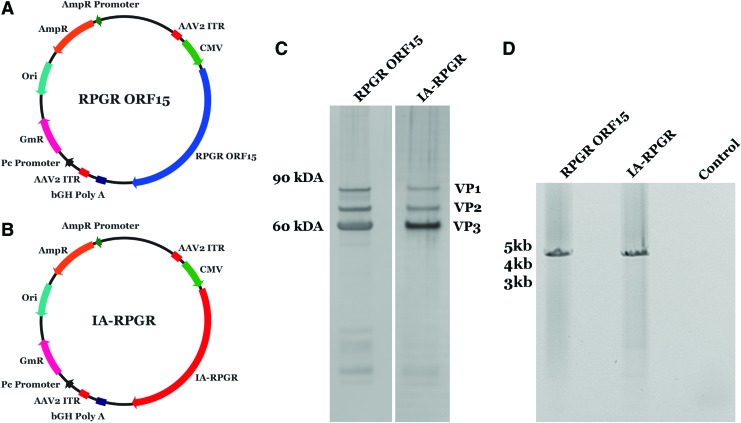
To determine whether our modification of the low complexity region within ORF15 affected viral packaging, we compared quality and quantity of viral particles after packaging the naive and optimized plasmids. To evaluate virus purity and insert identity, viral particles were subjected to a silver stain assay followed by PCR and Sanger sequencing. Clear presence of viral proteins VP1, VP2, and VP3, with little contaminating protein product, was identified for both RPGR ORF15 and IA-ORF15 (Fig. 2). Amplification and Sanger sequencing of viral gDNA confirmed sequence integrity for both RPGR ORF15 and IA-RPGR. These results suggest that the synonymous sequence changes in IA-ORF15 do not affect viral packaging and that the majority of the instability of wild-type ORF15 occurs at the level of cDNA replication rather than viral packaging.
Figure 2.
IA-RPGR AAV vector analysis. (A, B) Vector maps for RPGR-ORF15 and sequence optimized IA-RPGR. (C) Silver stain gel analysis of RPGR ORF15 and IA-RPGR viral particles indicating the presence of the AAV proteins VP1, VP2, and VP3. (D) Isolation and amplification of viral gDNA from RPGR ORF15 and IA-RPGR for confirmation of sequence identity. AAV, adeno associated virus. Color images are available online.
Transgene expression and functional analysis
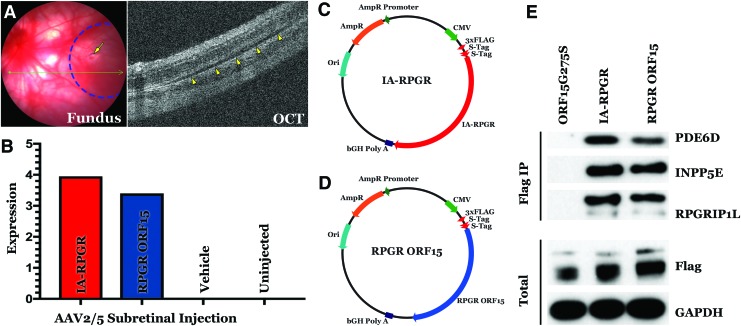
To determine if the IA-RPGR vector was capable of driving RPGR transcription in vivo, a series of subretinal injection experiments were performed in wild-type Sprague Dawley rats (Fig. 3A). Quantitative RT-PCR analysis detected similar levels of vector-driven transcript in eyes injected with either the IA-RPGR or RPGR-ORF15 vectors (Fig. 3B). Since the IA-RPGR vector is based on a naturally occurring transcript present in normally sighted people and has only been slightly modified with synonymous codon variants limited to the repetitive region of the gene, we expected that most of the important RPGR binding interactions would be unaffected by these modifications. To test this hypothesis, FLAG-S-tagged expression constructs (FLAG-S-tagged IA-RPGR, FLAG-S-tagged RPGR ORF15, and FLAG-S-tagged RPGR ORF15G275S [mutant binding control]) were generated and overexpressed in HEK293T cells (Fig. 3C, D). As shown in Fig. 3E, IA-RPGR-derived protein was found to bind PDE6D, INPP5E, and RPGRIP1L at levels similar to the RPGR ORF15-expressing construct. The specificity of these associations was confirmed by demonstrating that a construct driving a mutant allele (RPGR ORF15G275S—creation of a serine at position 275 results in generation of a known disease-causing mutation)25 failed to show any interactions with either of these binding partners (Fig. 3E). Collectively, these findings suggest that the newly developed IA-RPGR AAV vector is able to drive functional RPGR transcript and protein production and therefore seems suitable for clinical gene augmentation.
Figure 3.
Functional analysis of AAV2/5 IA-RPGR. (A) Fundus photograph (left) and OCT (right) of Sprague Dawley rat retina following subretinal injection. In the fundus photo, the dotted line demarcates the subretinal bleb area, and the arrow indicates the retinotomy site. In the OCT image, the arrowheads indicate the subretinal bleb (note dark fluid-filled space beneath the photoreceptor outer segments). (B) Quantitative RT-PCR analysis demonstrating expression of both IA-RPGR and RPGR ORF15 transcripts following subretinal injection in wild-type Sprague Dawley rats. (C, D) Maps of FLAG-tagged IA-RPGR and RPGR ORF15 plasmids. (E) Immunoprecipitation using FLAG-tagged RPGR ORF15G275S (mutant control), IA-RPGR, and RPGR ORF15 demonstrating that IA-RPGR-derived protein retains the ability to bind PDE6D, INPP5E, and RPGRIP1L. Anti-FLAG and anti-GAPDH antibodies were used as controls. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; OCT, optical coherence tomography; RT, reverse transcription. Color images are available online.
Discussion
In a recent study of 1,000 consecutive families seen by a single physician in our inherited eye disease clinic, we found that mutations in RPGR were the third most common cause of retinal degeneration, present in 4.8% of the probands in this cohort.1 The ability to accurately estimate the prevalence of RPGR-associated disease was hindered in past because sequencing the low complexity region of ORF15 is extraordinarily difficult using conventional PCR-based Sanger sequencing alone. To accurately identify disease-causing variants, a number of laboratories including ours have adopted a strategy that employs cloning of gel-purified PCR products using primers flanking the low complexity region. Given the difficulty in sequencing this region, it is not entirely surprising that past attempts to clone RPGR ORF15 cDNA have been complicated by an excessively high mutation rate.16,18 We hypothesized that the accumulation of mutations during cDNA replication is related to the propensity of the low complexity purine-rich region of ORF15 to adopt non-B DNA conformations (e.g., triplex DNA). Although we do not fully understand the function of this region at the DNA or RNA levels, we do know that maintaining the protein sequence of RPGR ORF15 is functionally critical. Specifically, the Glu-Gly-rich repetitive region of the protein is heavily glutamylated by Tubulin Tyrosine Ligase Like-5 (TTLL5).26 Deletion of TTLL5 in mice results in loss of RPGR glutamylation and subsequent retinal degeneration.26
Our goal in this study was to develop a gene augmentation vector for the treatment of RPGR-associated X-linked RP that is molecularly stable and that retains the exact protein sequence encoded by the native RPGR ORF15 transcript. As the repetitive ORF15 region has been shown to vary in length, to construct our gene augmentation vector we began by identifying the smallest naturally occurring ORF15 sequence in normally sighted individuals. By starting with the shortest repetitive region possible, the number of synonymous variations required to stabilize the cDNA was kept to a minimum. Next, we used Triplex,22 a bioinformatic tool, to predict which regions are most prone to the formation of triplex DNA and in turn responsible for conferring plasmid instability during replication. By taking this approach, we were able to generate a stable RPGR construct that contained the least amount of deviation from the reference sequence. This construct was used to robustly produce AAV vector that was capable of producing normal RPGR protein in vivo. The next step will be to generate a clinical-grade vector under current good manufacturing practice (cGMP) conditions and evaluate systemic toxicity and tumorigenicity in normal animals. This process can be reliably performed using wild-type rats, which, unlike mice, have eyes of sufficient size for subretinal injection of a human equivalent dose of vector (i.e., 10 μL). Individual rats also have sufficient quantities of blood for complete analysis of hematology and clinical chemistry.
Among genes with cDNAs that will fit into AAV, RPGR is the most common cause of inherited retinal disease. We have devised a molecularly stable therapeutic construct for gene augmentation therapy of this serious disorder and are now ready to proceed with cGMP manufacturing and preclinical evaluation of the clinical-grade product.
Acknowledgments
We would like to thank the University of Iowa, Institute for Vision Research, the NIH (grant 5T32GM007337), and Research to Prevent Blindness for providing funding for this project. In addition, support was received from the NHLBI-funded National Gene Vector Biorepository at Indiana University (contract no. 75N92019D00018).
Author Disclosure
No competing financial interests exist.
References
- 1. Stone EM, Andorf JL, Whitmore SS, et al. . Clinically focused molecular investigation of 1000 consecutive families with inherited retinal disease. Ophthalmology 2017;124:1314–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Meindl A, Dry K, Herrmann K, et al. . A gene (RPGR) with homology to the RCC1 guanine nucleotide exchange factor is mutated in X-linked retinitis pigmentosa (RP3). Nat Genet 1996;13:35–42 [DOI] [PubMed] [Google Scholar]
- 3. Roepman R, Van Duijnhoven G, Rosenberg T, et al. . Positional cloning of the gene for X-linked retinitis pigmentosa 3: homology with the guanine-nucleotide-exchange factor RCC1. Hum Mol Genet 1996;5:1035–1041 [DOI] [PubMed] [Google Scholar]
- 4. Vervoort R, Lennon A, Bird AC, et al. . Mutational hot spot within a new RPGR exon in X-linked retinitis pigmentosa. Nat Genet 2000;25:462–466 [DOI] [PubMed] [Google Scholar]
- 5. Hong D-H, Pawlyk B, Sokolov M, et al. . RPGR isoforms in photoreceptor connecting cilia and the transitional zone of motile cilia. Invest Ophthalmol Vis Sci 2003;44:2413–2421 [DOI] [PubMed] [Google Scholar]
- 6. Raghupathy RK, Gautier P, Soares DC, et al. . Evolutionary characterization of the retinitis pigmentosa GTPase regulator gene. Invest Ophthalmol Vis Sci 2015;56:6255–6264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wright RN, Hong D-H, Perkins B. RpgrORF15 connects to the usher protein network through direct interactions with multiple whirlin isoforms. Invest Ophthalmol Vis Sci 2012;53:1519–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Megaw R, Abu-Arafeh H, Jungnickel M, et al. . Gelsolin dysfunction causes photoreceptor loss in induced pluripotent cell and animal retinitis pigmentosa models. Nat Commun 2017;8:271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee J-J, Seo S. PDE6D binds to the C-terminus of RPGR in a prenylation-dependent manner. EMBO Rep 2015;16:1581–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rachel RA, Li T, Swaroop A. Photoreceptor sensory cilia and ciliopathies: focus on CEP290, RPGR and their interacting proteins. Cilia 2012;1:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Megaw RD, Soares DC, Wright AF. RPGR: its role in photoreceptor physiology, human disease, and future therapies. Exp Eye Res 2015;138:32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shu X, Fry AM, Tulloch B, et al. . RPGR ORF15 isoform co-localizes with RPGRIP1 at centrioles and basal bodies and interacts with nucleophosmin. Hum Mol Genet 2005;14:1183–1197 [DOI] [PubMed] [Google Scholar]
- 13. Khanna H, Hurd TW, Lillo C, et al. . RPGR-ORF15, which is mutated in retinitis pigmentosa, associates with SMC1, SMC3, and microtubule transport proteins. J Biol Chem 2005;280:33580–33587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hong D-H, Pawlyk BS, Adamian M, et al. . A single, abbreviated RPGR-ORF15 variant reconstitutes RPGR function in vivo. Invest Ophthalmol Vis Sci 2005;46:435–441 [DOI] [PubMed] [Google Scholar]
- 15. Beltran WA, Cideciyan AV, Lewin AS, et al. . Gene therapy rescues photoreceptor blindness in dogs and paves the way for treating human X-linked retinitis pigmentosa. Proc Natl Acad Sci USA 2012;109:2132–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pawlyk BS, Bulgakov OV, Sun X, et al. . Photoreceptor rescue by an abbreviated human RPGR gene in a murine model of X-linked retinitis pigmentosa. Gene Ther 2016;23:196–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fischer MD, McClements ME, Martinez-Fernandez de la Camara C, et al. . Codon-optimized RPGR improves stability and efficacy of AAV8 gene therapy in two mouse models of X-linked retinitis pigmentosa. Mol Ther J Am Soc Gene Ther 2017;25:1854–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Deng W-T, Dyka FM, Dinculescu A, et al. . Stability and safety of an AAV vector for treating RPGR-ORF15 X-linked retinitis pigmentosa. Hum Gene Ther 2015;26:593–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang G, Vasquez KM. Naturally occurring H-DNA-forming sequences are mutagenic in mammalian cells. Proc Natl Acad Sci USA 2004;101:13448–13453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Davis TL, Firulli AB, Kinniburgh AJ. Ribonucleoprotein and protein factors bind to an H-DNA-forming c-myc DNA element: possible regulators of the c-myc gene. Proc Natl Acad Sci USA 1989;86:9682–9686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wu Z, Hiriyanna S, Qian H, et al. . A long-term efficacy study of gene replacement therapy for RPGR-associated retinal degeneration. Hum Mol Genet 2015;24:3956–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hon J, Martínek T, Rajdl K, et al. . Triplex: an R/Bioconductor package for identification and visualization of potential intramolecular triplex patterns in DNA sequences. Bioinforma 2013;29:1900–1901 [DOI] [PubMed] [Google Scholar]
- 23. Zolotukhin S, Byrne BJ, Mason E, et al. . Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther 1999;6:973–985 [DOI] [PubMed] [Google Scholar]
- 24. Parfitt DA, Lane A, Ramsden CM, et al. . Identification and correction of mechanisms underlying inherited blindness in human iPSC-derived optic cups. Cell Stem Cell 2016;18:769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Banin E, Mizrahi-Meissonnier L, Neis R, et al. . A non-ancestral RPGR missense mutation in families with either recessive or semi-dominant X-linked retinitis pigmentosa. Am J Med Genet A 2007;143A:1150–1158 [DOI] [PubMed] [Google Scholar]
- 26. Sun X, Park JH, Gumerson J, et al. . Loss of RPGR glutamylation underlies the pathogenic mechanism of retinal dystrophy caused by TTLL5 mutations. Proc Natl Acad Sci USA 2016;113:E2925–E2934 [DOI] [PMC free article] [PubMed] [Google Scholar]