Abstract
Food contains complex blends of structurally diverse bitter compounds that trigger bitterness through activation of one or more of the ∼25 human TAS2 bitter taste receptors. It remains unsolved, however, whether the perceived bitterness of binary bitter-compound mixtures can be considered an additive function of all bitter-inducing chemicals in the mouth, suggesting that little mutual interaction takes place among bitter substances or if mixture suppression and synergism occurs. Here we report on two natural sesquiterpene lactones from edible plants, which stimulate distinct sets of hTAS2Rs in transfected cells. Both chemicals also robustly inhibit different but overlapping subsets of agonist-activated hTAS2Rs. These findings demonstrate that mixtures of bitter compounds, because they normally occur in human foodstuff, likely elicit bitter perception in a complex and not in a merely additive manner. An unexpected implication of this discovery is that, during evolution, the naturally occurring bitter taste receptor antagonists have shaped some of the pharmacological properties of the receptors, such as overlapping recognition profiles and breadth of tuning.
Introduction
Numerous potentially noxious chemicals, synthetic, natural, or generated during food processing and aging, evoke bitter taste (Belitz and Wieser, 1985; Schieberle and Hofmann, 2003; DuBois et al., 2008). Given the rich presence of bitter substances in edible plants and other foodstuff, several of them are usually present in the mouth simultaneously when we eat (Belitz and Wieser, 1985; Stewart et al., 1996; Hofmann, 2009). Moreover, the bitter blends can combine with stimuli of other taste qualities to evoke complex flavor perceptions, including mixture suppression and synergistic effects (Bartoshuk, 1975; Drewnowski, 2001). For instance, sodium ions and some sweeteners can suppress the bitterness of various compounds in binary mixtures (Lawless, 1979; Kroeze and Bartoshuk, 1985; Schiffman et al., 1985; Calviño et al., 1990; Calviño and Garrido, 1991; Schifferstein and Frijters, 1993; Frijters and Schifferstein, 1994; Schiffman et al., 1994; Breslin and Beauchamp, 1995, 1997; Stevens, 1995; Prescott et al., 2001). Moreover, synergistic interactions occur between some sweeteners (Kamen, 1959; Stone and Oliver, 1969; Bartoshuk and Cleveland, 1977; Frank et al., 1989; Ayya and Lawless, 1992; Schifferstein, 1995, 1996; Schiffman et al., 1995). Conversely, it is unclear whether the perceived bitterness of binary bitter-compound mixtures is, in general, merely an additive function of the total bitter-inducing chemicals in the mouth, suggesting a lack of mutual interactions among bitter substances (Keast and Breslin, 2003; Keast et al., 2003) or, alternatively, whether bitter-compound mixtures exert suppression and/or synergistic effects.
Compounds that activate the G-protein-coupled receptors of the TAS2R family comprising ∼25 members in humans trigger bitterness perception (Zhang et al., 2003; Meyerhof, 2005; Mueller et al., 2005; Chandrashekar et al., 2006; Behrens and Meyerhof, 2009). To date, cognate bitter agonists for 20 hTAS2Rs have been identified (Meyerhof et al., 2010). TAS2Rs differ greatly in their tuning breadth, ranging from extreme promiscuity to pronounced selectivity (Meyerhof et al., 2010). Three hTAS2Rs, hTAS2R10, hTAS2RR14, and hTAS2RR46, display wide molecular receptive ranges and, together, detected ∼50% of all tested bitter compounds. The molecular receptive ranges of the TAS2Rs and the existence of numerous allelic variants of TAS2Rs likely account for people's and animal's ability to detect countless chemicals as bitter (Behrens et al., 2004; Brockhoff et al., 2007; Behrens and Meyerhof, 2009; Kuhn et al., 2010; Meyerhof et al., 2010).
One of the bitter taste receptors, hTAS2R46, is exquisitely sensitive to numerous sesquiterpene lactones (STLs), as well as to clerodane and labdane diterpenoids, strychnine, and denatonium (Brockhoff et al., 2007). However, several STLs that are structurally closely related to agonistic STLs failed to activate hTAS2R46. We now investigated the interaction of some of these natural STLs with hTAS2R46 and other hTAS2Rs in transfected cells. Our experiments demonstrate an unexpected level of complexity in the interaction of bitter compounds with their receptors. These findings may provide novel insights in the ability of humans to perceive the bitterness of complex mixtures, because they are usually presented in food and culinary preparations.
Materials and Methods
Taste active compounds.
Andrographolide, aristolochic acids, brucine, chloramphenicol, chloroquine diphosphate salt, colchicine, cromolyn, denatonium benzoate, denatonium saccharide, epigallocatechin gallate, ofloxacin, phenylthiocarbamide, picrotoxinin, quinine sulfate, d-salicin, and strychnine have been purchased in the highest purity from Sigma-Aldrich. Absinthin, 3β-hydroxypelenolide (3HP) from mountain wormwood (Artemisia absinthium L.), trans-isocohumulone, and cohumulone have been available from previous studies (Brockhoff et al., 2007; Intelmann et al., 2009). Costunolide has been purchased from LGC Standarts.
3β-Hydroxydihydrocostunolide (3HDC; artabin) has been isolated from sea wormwood, Artemisia arborescens L. as follows. Dried aerial parts (leaves and flowers, 500 g) were extracted with acetone at room temperature (2 × 2.5 L). Removal of the solvent left a black gum (6.8 g, 3.4%) that was dissolved in acetone and filtered over a bed of RP18-silica gel (50 g). The filtrate was evaporated, and the yellowish residue (5.1 g) was purified by gravity column chromatography on silica gel (100 g, petroleum ether-EtOAc gradient, from 9:1 to 5:5). Fractions eluted with petroleum ether-EtOAc at 7:3 afforded 0.95 g of an amorphous ∼1:4 mixture of matricin and 3β-hydroxydihydrocostunolide. The latter was obtained in pure form after acidic degradation of matricin to its 3,4-dehydro derivative. To this purpose, 320 mg of the mixture was dissolved in chloroform and irradiated with an immersion lamp to photolize chloroform at 254 nm and generate in situ HCl. The course of the reaction was checked by thin-layer chromatography on silica gel (petroleum ether-EtOAc at 7:3 as eluant; Rf matricin and 3β-hydroxydihydrocostunolide = 0.19; Rf 3,4-dehyd matricin = 0.40). After 6 h, the reaction was worked up by evaporation and purification by gravity column chromatography on silica gel (petroleum ether-EtOAc at 5:5) yielded 59 mg of 3β-hydroxydihydrocostunolide (0.036% from the dried plant material) as a gummy amorphous solid, identified by comparison with the published spectroscopic data (Akhmedov et al., 1970).
Functional expression of hTAS2Rs.
Bitter taste receptor cDNA was cloned into pcDNA5/FRT (Invitrogen) or pEAK10 (Edge Biosystems). We used the same receptor sequences as reported by Meyerhof et al. (2010) with the exception of hTAS2R9 (GenBank accession number AF227135.1; cSNP, rs3741845; dbSNP, c.560 T > C) and hTAS2R30 (withdrawn gene symbol hTAS2R47; GenBank accession number AF494233.1). Nomenclature of hTAS2R genes was applied according to the HUGO Human Gene Nomenclature Committee. To improve plasma membrane targeting of the receptors in the heterologous expression system, they were fused to the first 45 aa of the rat somatostatin receptor subtype 3 at their N termini. This domain was shown to contain a plasma membrane targeting sequence that suffices to guide heterologously expressed GPCRs to the cell membrane (Ammon et al., 2002). The hTAS2R coding regions have been extended at their 3′ ends by a sequence coding for the glycoprotein D epitope of the Herpes simplex virus to enable immunological detection. For functional experiments, constructs were transiently transfected into HEK293T cells stably expressing the chimeric G-protein subunit Gα16gust44 (Ueda et al., 2003). Twenty-four hours after seeding into 96-well plates, cells were transfected with 150 ng of DNA per well mixed with 300 nl of Lipofectamine2000 (Invitrogen). Cells transfected with empty vector served as negative control. After 24 h, cells were loaded with Fluo4-AM for 1 h and subsequently washed triply using C1 solution (in mm: 130 NaCl, 5 KCl, 10 HEPES, pH 7.4, 2 CaCl2, and 10 glucose). Test substances were solved in dimethylsulfoxide (DMSO) if necessary and further diluted in C1. Changes in intracellular calcium levels during automatic application of test substances were measured at 510 nm in a Fluorometric Imaging Plate Reader (FLIPR; Molecular Devices) after excitation at 488 nm. To control for cell number and vitality, somatostatin-14 (SST14; 100 nm) was applied subsequent to the test substance application. Some bitter compounds were limited in their usable concentration because of unspecific stimulation of mock-transfected cells. In the absence of appropriate substitutions, these substances were used at the highest applicable concentration that did not elicit unspecific signals. For inhibition experiments, agonist and putative antagonist were premixed. The cellular response during agonist and antagonist coapplication was then compared to the sole application of the agonist. Experiments were performed in duplicates and repeated twice.
Analysis of calcium imaging experiments.
After measurement, the recorded calcium levels of mock cells were subtracted from receptor-transfected cells using the software FLIPR384 (Molecular Devices). Net signals were exported to SigmaPlot9 and normalized to background fluorescence (ΔF/F). Wells that received the same transfection and stimulus were averaged. For concentration–response and inhibition–response functions, ΔF/F was plotted against concentration of the test substance. By nonlinear regression of the plots to the function f(y) = (a − d)/(1 + (x/EC50)nh) + d, where a corresponds to the signal maximum, d to the minimum, x to the test substance concentration, and nh to the Hill coefficient, and half-maximal effective concentrations of agonists (EC50) were calculated. Half-maximal inhibitory concentrations of antagonists (IC50) were determined using the function f(x) = (a − b)/(1 + (x/IC50)nh) + b, where b is the signal minimum. For statistical analysis of the results, a one-way ANOVA at 5% α-risk level was applied, followed by a Bonferroni's post hoc test (GraphPad Prism).
Results
3β-Hydroxydihydrocostunolide and 3β-hydroxypelenolide activate several hTAS2Rs
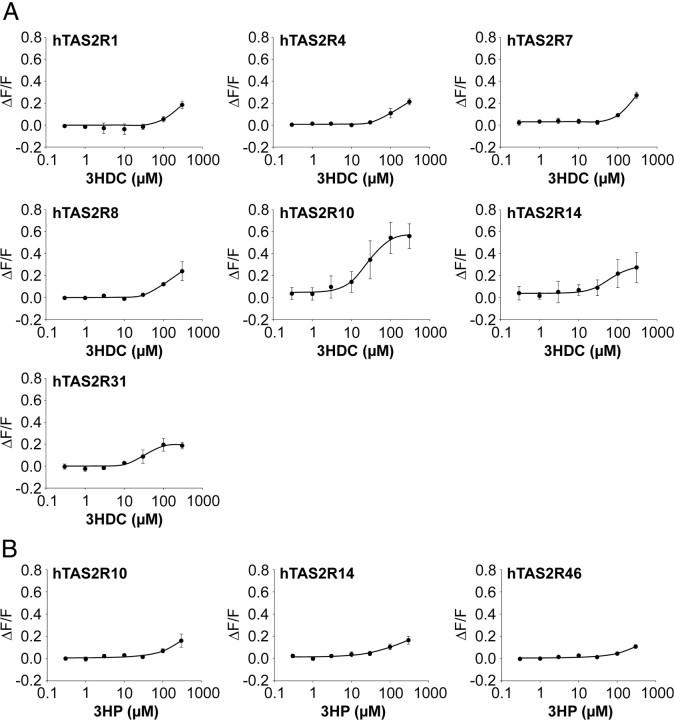
Recently, we have identified hTAS2R46 as bitter taste receptor with an exceptionally broad molecular receptive range being sensitive to numerous natural and synthetic chemicals, including various STLs (Brockhoff et al., 2007; Meyerhof et al., 2010). However, some STLs failed to activate hTAS2R46, although they showed only minor structural differences compared to the numerous STLs that stimulated the receptor. Because STLs are generally described as potent bitter compounds, we reasoned that STLs that did not activate hTAS2R46 could activate other hTAS2Rs. We therefore selected 3HDC and 3HP (Fig. 1B), which did not activate hTAS2R46 in our previous study but are structurally very similar to potent hTAS2R46 activators, such as costunolide and absinthin (Fig. 1A), to evaluate whether they act as cognate agonists of any of the other 24 hTAS2Rs. At a test concentration of 100 μm, 3HDC stimulated calcium responses from cells previously transfected with DNA for hTAS2R1, hTAS2R4, hTAS2R7, hTAS2R8, hTAS2R10, hTAS2R14, and hTAS2R31 (Fig. 1C). Marginal calcium responses of hTAS2R3- and hTAS2R39-expressing cells were not verified in replicate experiments. 3HP activated just three receptors, hTAS2R10, hTAS2R14 and, very weakly, hTAS2R46 (Fig. 1D). This weak stimulatory activity of 3HP on hTAS2R46-expressing cells was not obvious in our recent study (Brockhoff et al., 2007), probably because the previous preparation was less pure than the present one. Figure 1 also demonstrates that the costunolide derivative activated not only more TAS2Rs but also resulted in higher signal amplitudes than the pelenolide derivative. Thus, the data demonstrate that both substances activate subsets of the human TAS2Rs. This observation was validated by recording concentration–response curves of cells expressing the relevant TAS2R constructs during stimulation with 3HDC or 3HP (Fig. 2). Our data revealed that only hTAS2R10 and hTAS2R31 responded to 3HDC at concentrations lower than 100 μm (Fig. 2A). The substance exerted comparable potency on both receptors (hTAS2R10, EC50 = 27.8 ± 13.6 μm; hTAS2R31, EC50 = 24.0 ± 2.2 μm; n = 3) but a clearly higher efficacy on hTAS2R10 (maximum signal amplitude, hTAS2R10, ΔF/F = 0.56 ± 0.11; hTAS2R31, ΔF/F = 0.19 ± 0.06; n = 3). At the other receptors, TAS2R1, hTAS2R4, hTAS2R7, hTAS2R8, and hTAS2R14, 3HDC acted only as a weak partial agonist. The compound 3HP did not induce cellular responses below 100 μm, and its low potency at the tested receptors prevented the determination of EC50 values (Fig. 2B).
Figure 1.
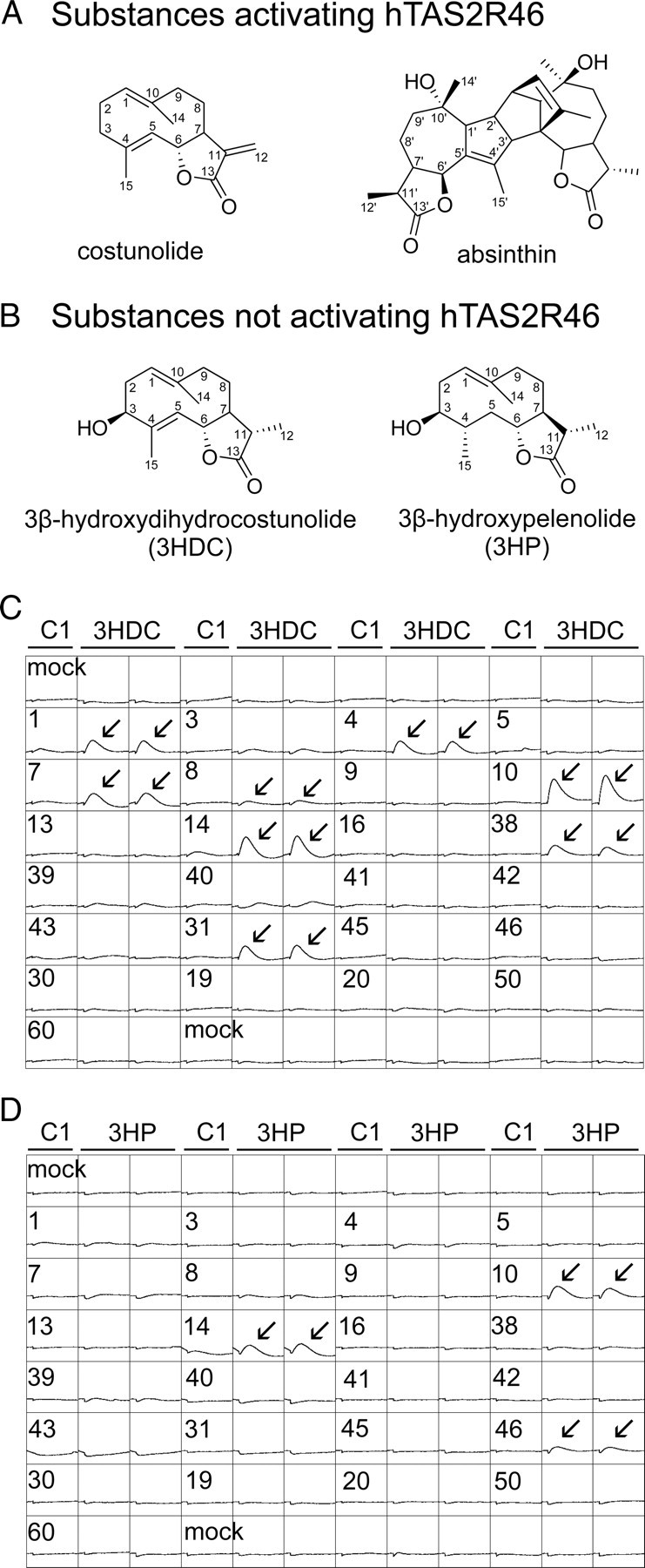
Chemical structure of substances activating hTAS2R46 (A) and substances not activating hTAS2R46 (B). The nomenclature of the carbon atoms is indicated. C, Activation of hTAS2R-expressing cells during bath application of 3HDC (100 μm). D, Activation of hTAS2R-expressing cells during bath application of 3HP (100 μm). HEK293T cells were transfected with cDNA for the indicated hTAS2Rs or empty vector (mock). Calcium signals elicited by the stimuli were recorded in the FLIPR. Arrows point to positive responses that were verified by replicates (n = 3). Apparent activation of hTAS2R38 by 3HDC is attributable to DMSO content of the test substance solution (Meyerhof et al., 2010). Scale: y, 15,000 counts; x, 8 min.
Figure 2.
Concentration–response functions of HEK293T cells transfected with hTAS2Rs to 3HDC (A) and 3HP (B). Cells were transfected with hTAS2R cDNA or empty vector. After 24 h, cells were loaded with Fluo4-AM and washed with C1 solution. Intracellular calcium levels were measured during bath application of test substances using a Fluorometric Imaging Plate Reader. Calcium signals were normalized to background fluorescence (ΔF/F). Concentration–response functions and EC50 values were calculated using SigmaPlot. Data were collected from three independent experiments.
Natural STLs as antagonists for hTAS2R46
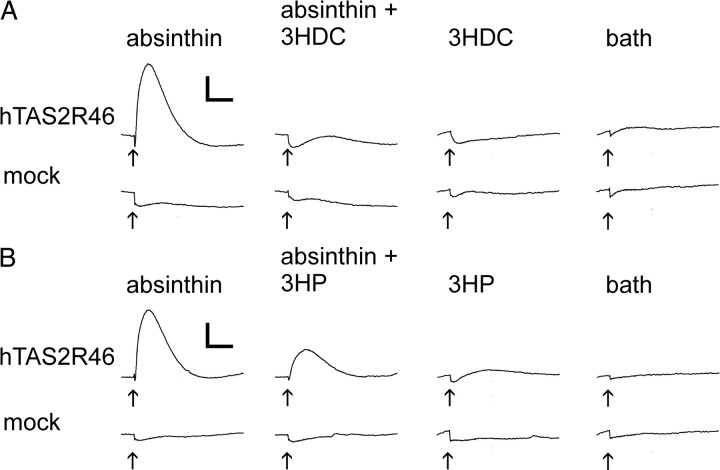
The close structural relationship between STLs that activate hTAS2R46 and STLs that fail to do so suggest that the latter compounds might fit as well into the binding pocket of hTAS2R46 but do not switch the receptor in the active conformation, thus acting as competitive antagonists. To evaluate this hypothesis, we stimulated hTAS2R46-transfected cells with the STL agonist absinthin in the absence or presence of 3HDC and 3HP, STLs that failed to activate this receptor (Fig. 3). Clearly, the responses to absinthin were substantially diminished by 3HP and completely blocked by 3HDC at a concentration of 100 μm, identifying these compounds as natural herbal bitter blockers.
Figure 3.
Inhibition of absinthin-induced calcium responses in hTAS2R46-expressing cells by 3HDC and 3HP. HEK293T Gα16gust44 cells previously transfected with hTAS2R46 cDNA or empty vector (mock) were loaded with Fluo4-AM. We recorded cytosolic calcium levels in these cells before and after bath application (arrows) of 3HDC (A; 100 μm) and 3HP (B; 100 μm) mixed with absinthin in a FLIPR. We applied absinthin at a concentration of 100 μm, which corresponds to the EC90 of hTAS2R46 (Brockhoff et al., 2007). Scale: y = 2000 counts; x = 2 min.
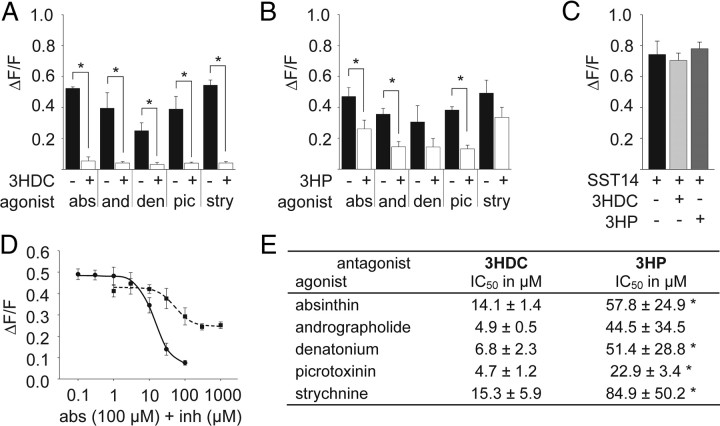
Next, we investigated whether the inhibitory effects of the two compounds on hTAS2R46 were specific for absinthin or apply to other agonists as well. Therefore, we challenged hTAS2R46-expressing cells with mixtures of either of the two compounds with EC90 concentrations of the agonists andrographolide, denatonium benzoate, picrotoxinin, or strychnine (Fig. 4A,B). The chosen agonists are structurally diverse and belong to different chemical classes but have been shown recently to fit into the same binding pocket of hTAS2R46 (Brockhoff et al., 2010). Picrotoxinin is an STL with two lactone rings, andrographolide, a diterpene lactone, strychnine, an indole alkaloid, and denatonium, a synthetic quaternary ammonium salt. As demonstrated in Figure 4A, 3HDC completely blocked the response of hTAS2R46-expressing cells to all tested agonists at a concentration of 100 μm. In contrast, the 3HP at the same concentration attenuated receptor activation only partially (Fig. 4B). The data suggest that the two inhibitors act generally to block the responses of hTAS2R46 elicited by all agonists. To assess the specificity of the inhibitory effects for the recombinant bitter receptor, we monitored the responses of hTAS2R46-expressing cells after application of SST14 in the absence and presence of the two inhibitors. The neuropeptide SST14 stimulates an endogenous somatostatin receptor in HEK293T cells, a member of the GPCR family (Behrens et al., 2004; Kuhn et al., 2004). Figure 4C reveals that the SST14-mediated calcium signals were not affected by 3HDC or 3HP, clearly demonstrating that the inhibitors act specifically on the recombinant bitter taste receptor.
Figure 4.
Inhibitory properties of 3HDC and 3HP. A, B, Calcium responses of hTAS2R46-expressing cells after stimulation with various agonists are inhibited by coapplication of 100 μm 3HDC or 3HP. Agonists were applied at their EC90 concentrations (abs, absinthin at 100 μm; and, andrographolide at 30 μm; den, denatonium benzoate at 300 μm; pic, picrotoxinin at 300 μm; stry, strychnine at 3 μm). C, Calcium responses of HEK293T Gα16gust44 cells after application of 100 nm somatostatin-14 in the absence or presence of 100 μm 3HDC or 3HP. D, Inhibition response curves of the effects of 3HDC (circles, solid line) or 3HP (squares, dotted line) on the absinthin-elevated intracellular calcium levels in hTAS2R46-expressing cells. E, IC50 values of 3HDC and 3HP for blocking hTAS2R46 responses to various agonists. Agonists were applied at their EC90 concentrations of activating hTAS2R46 (absinthin, 100 μm; andrographolide, 30 μm; denatonium benzoate, 300 μm; picrotoxinin, 300 μm; strychnine, 3 μm).
To compare the potency of both compounds to inhibit responses of hTAS2R46, we determined their concentration–response functions and calculated their half-maximal inhibitory concentrations (IC50). Figure 4, D and E, reveals that 3HDC was generally more potent than 3HP to block agonist-mediated responses of hTAS2R46. The former chemical exhibits significantly lower IC50 values than the latter for inhibiting hTAS2R46 responses to all tested agonists except andrographolide. In this case, the same trend is clearly seen but did not reach significance (Fig. 4E). The observed IC50 values did not only depend on the antagonists but also on the agonists. Although we used all hTAS2R46 activators approximately at their EC90 concentrations, the IC50 values of the two inhibitors varied twofold to threefold. Nevertheless, both antagonists act on hTAS2R46 approximately in a similar concentration range as its agonists [EC50 strychnine = 0.43 ± 0.02 μm; EC50 absinthin = 9.9 ± 0.3 μm; EC50 picrotoxinin = 70.0 ± 5.2 μm (Brockhoff et al., 2007)]. It is also worth noting that only 3HDC blocks the receptor response entirely, whereas receptor inhibition reaches a maximum of only 50% at a saturating concentration of ∼300 μm 3HP. The remaining receptor activity could be attributable to the comparatively low efficiency of 3HP to block hTAS2R46 stimulation and/or its weak agonistic property (Figs. 1D, 2B, 3B). Thus, 3HP may be considered as a weak partial agonist rather than an antagonist of hTAS2R46.
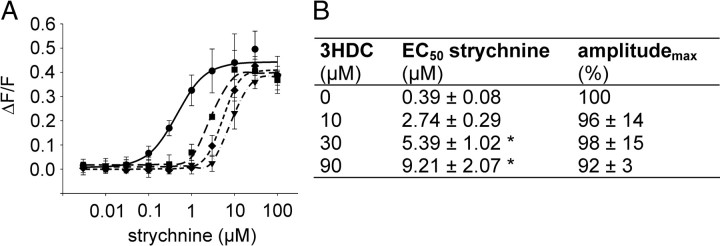
To investigate the type of antagonism of 3HDC, we established concentration–response functions using the most potent hTAS2R46 agonist, strychnine (Brockhoff et al., 2007), in the presence of various concentrations of the inhibitor. Figure 5A demonstrates that 3HDC shifts the concentration–response function of strychnine to higher concentrations, significantly increasing its EC50 value (Fig. 5B). However, the maximal signal amplitudes remained constant, suggesting that 3HDC acts as a surmountable antagonist (Gaddum et al., 1955) to block strychnine responses of hTAS2R46.
Figure 5.
Surmountable inhibition of strychnine-induced stimulation of hTAS2R46-expressing cells by 3HDC. A, Calcium responses of hTAS2R46-expressing cells to strychnine in the absence (circles, solid line) or presence of various concentrations of 3HDC (10 μm, squares, long dashed line; 30 μm, diamonds, short dashed line; 90 μm, triangles, dash-dotted line.). B, EC50 values and maximum amplitudes of the effects of strychnine on hTAS2R46-expressing cells in the absence or presence of various concentrations of 3HDC.
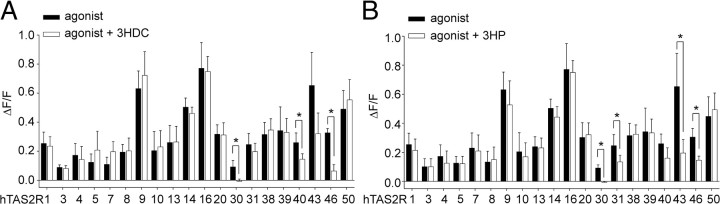
To further examine the inhibitory potential of the two STLs, we stimulated all 20 hTAS2Rs deorphaned so far with EC90 concentrations of their cognate agonists (Table 1) in the absence and presence of the costunolide and pelenolide derivatives (Fig. 6). Because of their low potency, the agonists for hTAS2R5 and hTAS2R7, denatonium saccharide and quinine sulfate, respectively, were used at their maximal applicable concentration. Coapplication of 3HDC with the respective receptor agonists led to a significant reduction of agonist-induced receptor responses in hTAS2R30-, hTAS2R40-, and hTAS2R46-expressing cells. The strongest inhibition was seen with hTAS2R30 and the least with hTAS2R40. A trend was also seen in the case of hTAS2R43, although inhibition did not reach significance (p = 0.100). Similarly, 3HP reduced the agonist-mediated calcium transients in cells expressing hTAS2R30, hTAS2R31, hTAS2R43, and hTAS2R46 significantly and in cells expressing hTAS2R40 by trend (p = 0.087). Only hTAS2R30 was entirely blocked by both antagonists. The other hTAS2Rs were less sensitive; inhibition ranged from 48% (hTAS2R43) to 59% (hTAS2R40) for 3HDC and from 30% (hTAS2R43) to 61% (hTAS2R40) for 3HP.
Table 1.
Specific hTAS2R agonists used to assess the putative antagonistic effect of 3HDC and 3HP
| Receptor | Agonist | Concentration (μm) |
|---|---|---|
| hTAS2R1 | Trans-isocohumulone | 30 |
| hTAS2R3 | Chloroquine | 1300 |
| hTAS2R4 | Colchicine | 3000 |
| hTAS2R5 | Denatonium saccharide | 3000a |
| hTAS2R7 | Quinine sulfate | 10a |
| hTAS2R8 | Chloramphenicol | 1000 |
| hTAS2R9 | Ofloxacin | 6000 |
| hTAS2R10 | Strychnine | 90 |
| hTAS2R13 | Denatonium benzoate | 3000a |
| hTAS2R14 | Aristolochic acids | 3 |
| hTAS2R16 | D-Salicin | 3000 |
| hTAS2R20 | Cromolyn | 1000 |
| hTAS2R30 | Brucine | 100 |
| hTAS2R31 | Aristolochic acids | 3 |
| hTAS2R38 | Phenylthiocarbamide | 30 |
| hTAS2R39 | Epigallocatechin gallate | 30 |
| hTAS2R40 | Cohumulone | 0.3 |
| hTAS2R43 | Aristolochic acids | 0.3 |
| hTAS2R46 | Picrotoxinin | 300 |
| hTAS2R50 | Andrographolide | 100 |
Receptors hTAS2R41, hTAS2R42, hTAS2R45, hTAS2R19, and hTAS2R60 were not analyzed because there is no cognate agonist known so far. Agonists recently identified (Dotson et al., 2008; Intelmann et al., 2009; Meyerhof et al., 2010; Slack et al., 2010) were applied at EC90 concentration.
aBecause of low potency, agonists for hTAS2R5, hTAS2R7, and hTAS2R13 were used at their maximum applicable concentration that did not cause unspecific fluorescence signals in control cells.
Figure 6.
Inhibition of agonist-induced fluorescence in hTAS2R-expressing cells by 3HDC (A) or 3HP (B). Cells were stimulated by either sole application of a specific agonist (black bars; see Table 1) or mixtures of specific stimuli with 3HDC or 3HP, respectively (white bars, 100 μm) (n = 3). Significant depression of the agonist response by the inhibitors is indicated by asterisks (p < 0.05).
Discussion
Human TAS2R46 is a very broadly tuned receptor that can be considered a general sensor for bitter STL, including absinthin from wormwood, the archetypal non-alkaloid bitter agent. However, some structurally similar STL surprisingly did not activate hTAS2R46 (Brockhoff et al., 2007). We have now found that two of these compounds, 3HDC and 3HP, although they stimulate various other hTAS2Rs, inhibit activation of hTAS2R46 by a host of agonists, including not only STL but also structurally unrelated chemicals, such as strychnine and denatonium benzoate. Remarkably, both compounds occur in wormwood, one of the bitterest plants known and an unusual source for bitter inhibitors. Whereas 3HDC is a potent antagonist of hTAS2R46, completely abolishing receptor activity, 3HP functions as a weak partial agonist, blocking hTAS2R46 activity only by ∼50%. Thus, an important discovery of this work is the demonstration that natural compounds can simultaneously act as both agonists and antagonists of bitter receptors. The ability of the two compounds to block hTAS2R46 activation by structurally dissimilar agonists further supports the existence of only one binding site in hTAS2Rs (Biarnés et al., 2010; Brockhoff et al., 2010; Sakurai et al., 2010).
Besides hTAS2R46, 3HDC also inhibits hTAS2R30 and hTAS2R40, and 3HP hTAS2R30, hTAS2R31, and hTAS2R43. With the exception of hTAS2R40, the inhibited receptors show close sequence relationship, attributable to recent gene amplifications in the primate lineage (Shi et al., 2003), likely explaining the common sensitivity of the receptors to these inhibitory STLs. This assumption is further supported by TAS2R40, which is less related to the above-mentioned hTAS2Rs and responds weakest to 3HDC and even less to 3HP. Interestingly, hTAS2R31, hTAS2R40, and hTAS2R43 are not only blocked by 3HDC and 3HP but also by the small synthetic molecule, 4-(2,2,3-trimethylcyclopentyl)butanoic acid (GIV3727), which in addition diminished responses mediated by hTAS2R4, hTAS2R7, and hTAS2R20 (Slack et al., 2010). Thus, the two STL inhibitors and GIV3727, despite a lack of obvious structural similarity, block a partly overlapping set of TAS2Rs. This indicates that bitter taste receptor antagonists, just like the bitter agonists, interact with several receptors, and, conversely, a given receptor is sensitive to several antagonists. Thus, TAS2Rs vary in their molecular receptive range for antagonists. Therefore, our data predict that some TAS2Rs would be particularly prone to inhibition because their molecular receptive range allows them to bind more antagonists. For instance, several compounds of distinct chemical classes selectively reduce the bitter off-taste of the artificial sweeteners saccharin and acesulfam K (Ley, 2008), suggesting that the cognate bitter receptors hTAS2R8, hTAS2R31, and hTAS2R43 (Kuhn et al., 2004; Pronin et al., 2007; Roudnitzky et al., 2011) are subject to pronounced receptor antagonism.
The ambivalent effects of 3HDC and 3HP on TAS2Rs raise the question of whether these properties are unique or shared by other bitter substances. We often observed that compounds expected to activate a hTAS2R under study actually failed to do so (Bufe et al., 2002, 2005; Brockhoff et al., 2007; Intelmann et al., 2009). These compounds include glucopyranosides as activators of hTAS2R16, thioamides, carbamides, or isothiocyanates as activators of hTAS2R38, and sesquiterpene lactones as activators of hTAS2R46. Moreover, in a screening approach with 104 bitter compounds, we failed to assign receptors to 22 compounds (Meyerhof et al., 2010). This failure could be attributable to the specificities of the cell-based receptor assay. Alternatively, these compounds may represent agonists for the remaining orphan receptors, which, like hTAS2R38 (Kim et al., 2003; Bufe et al., 2005), could exist as taster and non-taster variants. However, some of these 22 compounds could bind to TAS2Rs but are unable to activate them or could even block them.
Strikingly, as seen for the antagonistic molecules presented here, several antagonists of odorant receptors exhibit structural similarities with odorants acting at the same receptor (Oka et al., 2004a,b). The structural similarity between agonist and antagonist likely provides the basis for competitive binding to the ligand binding pocket of the corresponding odorant receptor, which becomes evident by a competitive mode of inhibition (Oka et al., 2004a). The molecular difference between the agonist and the antagonist, conversely, then may account for the differential ability to allow the receptor to adopt an active conformation. An example for such closely related agonist–antagonist pairs was identified by a screen for antagonistic molecules for the mouse odorant receptor mOR-EG (Oka et al., 2004a), whose canonical agonist is eugenol, a small molecule present in clove oil (Kajiya et al., 2001). Whereas eugenol and its derivative methyl-eugenol potently activate mOR-EG, the stereoisomer methyl-isoeugenol represents a powerful inhibitor (Oka et al., 2004a). Later, the molecular characteristics of agonists and antagonists and their interaction with particular amino acid residues of mOR-EG were investigated in detail (Katada et al., 2005). Intriguingly, it was demonstrated that a serine residue in transmembrane domain 3 of mOR-EG was involved in hydrogen bond formation with an oxygen group connected to the C4 atom of both the agonist and antagonist, whereas a phenylalanine residue present in transmembrane domain 6 facing the side chains connected to the C1 atoms of agonists and antagonists was involved in the differential receptor responses observed. It is therefore tempting to speculate that molecular differences among the agonists and antagonists shown in Figure 1, A and B, e.g., the hydroxyl groups at C4 positions of both inhibitors, may negatively interfere with bitter receptor activation. Along this line, another observation made in the olfactory system, that the differential activation of odorant receptors by some enantiomers, e.g., (+) and (−) carvone (Friedman and Miller, 1971), may have important implications for the future search for additional bitter taste receptor antagonists. Because olfactory quality coding relies on the generation of differential activity patterns in the olfactory bulb (Buck, 2004), enantiomers that evoke distinct odors such as (+) carvone smelling like caraway and (−) carvone smelling like spearmint, respectively, must activate, at least in part, different subsets of odorant receptors. The different activation pattern of such substances could arise in part from competitive inhibition mechanisms, which may apply to bitter-tasting enantiomers as well.
The existence of natural bitter blockers could hamper the crucial detection of poisonous compounds in food and prevent efficient protection from oral intoxication. From an evolutionary point of view, the development of a large heterogeneous group of TAS2R bitter sensors with binding pockets of distinct molecular constitution was seemingly necessary to minimize failure of detecting bitter toxins mixed with analogs that block bitter taste receptors. Although bitter substances can stimulate up to 15 different hTAS2Rs (Meyerhof et al., 2010), so far no compound is known that interacts with all 25 hTAS2Rs. It is therefore likely that hTAS2R antagonists, like their agonists, are unable to interact with all human bitter taste receptors. However, bitter food contains usually a complex blend of different bitter taste-active compounds. The activation of some and the simultaneous inhibition of other hTAS2Rs could diminish or prevent the perception of bitter compounds in mixture, affecting the perceived bitterness of certain foods. Daily life experiences support this assumption. For instance, intensely bitter honey from the strawberry tree (Arbutus unedo) and the chestnut tree, two prolific producers of bitter phenolics, are traditionally consumed with fermented cheeses, such as Roquefort, that have already a bitter note because of the presence of certain peptides. As a result, the bitterness of both foods is diminished, a well-known culinary experience (Weinzweig, 2003). Likewise, in a comprehensive psychophysical study testing binary mixtures of eight well known bitter tastants, Keast et al. (2003) identified that, whereas the majority of substances exhibited additive effects, urea suppressed the bitter intensity if mixed with other substances in a compound and concentration-dependent manner. These phenomena are reminiscent of mixture suppression, a phenomenon that has been observed frequently in the olfactory system and that results from the simultaneous presence of more than one odorant (Laing and Willcox, 1987). Mixture suppression is induced by odorants that inhibit olfactory receptors and thereby olfactory sensory neurons (Sanhueza et al., 2000; Oka et al., 2004a,b; Ache and Young, 2005; Ukhanov et al., 2010). Thus, the ability of volatiles to activate and inhibit olfactory receptors contributes to the combinatorial coding in the smell system and creates the odor of the source of the volatile.
In very much the same way, the ambivalent effect of bitter compounds, such as 3HDC and 3HP, on hTAS2Rs proposes that bitter compounds in our diets mutually influence taste perception, exerting a unique sensation of the ingested food. Thus, the co-occurrence of bitter agonists and antagonists in the same food is intriguing and might have shaped the complexity of the bitter receptor system, whose aim is the detection of poisonous dietary chemicals in the “noisy” sensory environment of food, in which hundreds of compounds occur, and not in reductionistically simplified experimental systems of single agents dissolved in buffer systems.
Footnotes
This work was supported by German Research Foundation Grant Me 1024/2-3 (W.M.).
References
- Ache BW, Young JM. Olfaction: diverse species, conserved principles. Neuron. 2005;48:417–430. doi: 10.1016/j.neuron.2005.10.022. [DOI] [PubMed] [Google Scholar]
- Akhmedov IS, Kasymov SZ, Sidyakin GP. Artabin: a new lactone from Artemisia absinthium. Chem Nat Compounds. 1970;6:634. [Google Scholar]
- Ammon C, Schäfer J, Kreuzer OJ, Meyerhof W. Presence of a plasma membrane targeting sequence in the amino-terminal region of the rat somatostatin receptor 3. Arch Physiol Biochem. 2002;110:137–145. doi: 10.1076/apab.110.1.137.908. [DOI] [PubMed] [Google Scholar]
- Ayya N, Lawless HT. Quantitative and qualitative evaluation of high-intensity sweeteners and sweetener mixtures. Chem Senses. 1992;17:245–259. [Google Scholar]
- Bartoshuk LM. Taste mixtures: is mixture suppression related to compression? Physiol Behav. 1975;14:643–649. doi: 10.1016/0031-9384(75)90193-6. [DOI] [PubMed] [Google Scholar]
- Bartoshuk LM, Cleveland CT. Mixtures of substances with similar tastes. A test of a psychophysical model of taste mixture interactions. Sens Processes. 1977;1:177–186. [PubMed] [Google Scholar]
- Behrens M, Meyerhof W. Mammalian bitter taste perception. Results Probl Cell Differ. 2009;47:203–220. doi: 10.1007/400_2008_5. [DOI] [PubMed] [Google Scholar]
- Behrens M, Brockhoff A, Kuhn C, Bufe B, Winnig M, Meyerhof W. The human taste receptor hTAS2R14 responds to a variety of different bitter compounds. Biochem Biophys Res Commun. 2004;319:479–485. doi: 10.1016/j.bbrc.2004.05.019. [DOI] [PubMed] [Google Scholar]
- Belitz H, Wieser H. Bitter compounds: occurrence and structure-activity relationships. Food Rev Int. 1985;1:271–354. [Google Scholar]
- Biarnés X, Marchiori A, Giorgetti A, Lanzara C, Gasparini P, Carloni P, Born S, Brockhoff A, Behrens M, Meyerhof W. Insights into the binding of Phenyltiocarbamide (PTC) agonist to its target human TAS2R38 bitter receptor. PLoS One. 2010;5:e12394. doi: 10.1371/journal.pone.0012394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslin PA, Beauchamp GK. Suppression of bitterness by sodium: variation among bitter taste stimuli. Chem Senses. 1995;20:609–623. doi: 10.1093/chemse/20.6.609. [DOI] [PubMed] [Google Scholar]
- Breslin PA, Beauchamp GK. Salt enhances flavour by suppressing bitterness. Nature. 1997;387:563. doi: 10.1038/42388. [DOI] [PubMed] [Google Scholar]
- Brockhoff A, Behrens M, Massarotti A, Appendino G, Meyerhof W. Broad tuning of the human bitter taste receptor hTAS2R46 to various sesquiterpene lactones, clerodane and labdane diterpenoids, strychnine, and denatonium. J Agric Food Chem. 2007;55:6236–6243. doi: 10.1021/jf070503p. [DOI] [PubMed] [Google Scholar]
- Brockhoff A, Behrens M, Niv MY, Meyerhof W. Structural requirements of bitter taste receptor activation. Proc Natl Acad Sci U S A. 2010;107:11110–11115. doi: 10.1073/pnas.0913862107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck LB. Olfactory receptors and odor coding in mammals. Nutr Rev. 2004;62:S184–S188. doi: 10.1111/j.1753-4887.2004.tb00097.x. discussion S224–S241. [DOI] [PubMed] [Google Scholar]
- Bufe B, Hofmann T, Krautwurst D, Raguse JD, Meyerhof W. The human TAS2R16 receptor mediates bitter taste in response to beta-glucopyranosides. Nat Genet. 2002;32:397–401. doi: 10.1038/ng1014. [DOI] [PubMed] [Google Scholar]
- Bufe B, Breslin PA, Kuhn C, Reed DR, Tharp CD, Slack JP, Kim UK, Drayna D, Meyerhof W. The molecular basis of individual differences in phenylthiocarbamide and propylthiouracil bitterness perception. Curr Biol. 2005;15:322–327. doi: 10.1016/j.cub.2005.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calviño AM, Garrido D. Spatial and temporal suppressive behavior of bitter-sweet mixtures. Percept Mot Skills. 1991;73:1216. doi: 10.2466/pms.1991.73.3f.1216. [DOI] [PubMed] [Google Scholar]
- Calviño AM, García-Medina MR, Cometto-Muniz JE. Interactions in caffeine–sucrose and coffee–sucrose mixtures: evidence of taste and flavor suppression. Chem Senses. 1990;15:505–519. [Google Scholar]
- Chandrashekar J, Hoon MA, Ryba NJ, Zuker CS. The receptors and cells for mammalian taste. Nature. 2006;444:288–294. doi: 10.1038/nature05401. [DOI] [PubMed] [Google Scholar]
- Dotson CD, Zhang L, Xu H, Shin YK, Vigues S, Ott SH, Elson AE, Choi HJ, Shaw H, Egan JM, Mitchell BD, Li X, Steinle NI, Munger SD. Bitter taste receptors influence glucose homeostasis. PLoS One. 2008;3:e3974. doi: 10.1371/journal.pone.0003974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewnowski A. The science and complexity of bitter taste. Nutr Rev. 2001;59:163–169. doi: 10.1111/j.1753-4887.2001.tb07007.x. [DOI] [PubMed] [Google Scholar]
- DuBois GE, DeSimone JA, Lyall V. Chemistry of gustatory stimuli. In: Firestein S, Beauchamp GK, editors. Olfaction and taste. Amsterdam: Elsevier; 2008. pp. 27–74. [Google Scholar]
- Frank RA, Mize SJS, Carter R. An assessment of binary mixture interactions for nine sweeteners. Chem Senses. 1989;14:621–632. [Google Scholar]
- Friedman L, Miller JG. Odor incongruity and chirality. Science. 1971;172:1044–1046. doi: 10.1126/science.172.3987.1044. [DOI] [PubMed] [Google Scholar]
- Frijters JE, Schifferstein HN. Perceptual interactions in mixtures containing bitter tasting substances. Physiol Behav. 1994;56:1243–1249. doi: 10.1016/0031-9384(94)90372-7. [DOI] [PubMed] [Google Scholar]
- Gaddum JH, Hameed KA, Hathway DE, Stephens FF. Quantitative studies of antagonists for 5-hydroxytryptamine. Q J Exp Physiol Cogn Med Sci. 1955;40:49–74. doi: 10.1113/expphysiol.1955.sp001097. [DOI] [PubMed] [Google Scholar]
- Hofmann T. Identification of the key bitter compounds in our daily diet is a prerequisite for the understanding of the hTAS2R gene polymorphisms affecting food choice. Ann N Y Acad Sci. 2009;1170:116–125. doi: 10.1111/j.1749-6632.2009.03914.x. [DOI] [PubMed] [Google Scholar]
- Intelmann D, Batram C, Kuhn C, Haseleu G, Meyerhof W, Hofmann T. Three TAS2R bitter taste receptors mediate the psychophysical responses to bitter compounds of hops (Humulus lupulus L.) and beer. Chem Percept. 2009;2:118–132. [Google Scholar]
- Kajiya K, Inaki K, Tanaka M, Haga T, Kataoka H, Touhara K. Molecular bases of odor discrimination: reconstitution of olfactory receptors that recognize overlapping sets of odorants. J Neurosci. 2001;21:6018–6025. doi: 10.1523/JNEUROSCI.21-16-06018.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamen JM. Interaction of sucrose and calcium cyclamate on perceived intensity of sweetness. Food Res. 1959;24:279–282. [Google Scholar]
- Katada S, Hirokawa T, Oka Y, Suwa M, Touhara K. Structural basis for a broad but selective ligand spectrum of a mouse olfactory receptor: mapping the odorant-binding site. J Neurosci. 2005;25:1806–1815. doi: 10.1523/JNEUROSCI.4723-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keast RSJ, Breslin PAS. An overview of binary taste–taste interactions. Food Qual Prefer. 2003;14:111–124. [Google Scholar]
- Keast RS, Bournazel MM, Breslin PA. A psychophysical investigation of binary bitter-compound interactions. Chem Senses. 2003;28:301–313. doi: 10.1093/chemse/28.4.301. [DOI] [PubMed] [Google Scholar]
- Kim UK, Jorgenson E, Coon H, Leppert M, Risch N, Drayna D. Positional cloning of the human quantitative trait locus underlying taste sensitivity to phenylthiocarbamide. Science. 2003;299:1221–1225. doi: 10.1126/science.1080190. [DOI] [PubMed] [Google Scholar]
- Kroeze JH, Bartoshuk LM. Bitterness suppression as revealed by split-tongue taste stimulation in humans. Physiol Behav. 1985;35:779–783. doi: 10.1016/0031-9384(85)90412-3. [DOI] [PubMed] [Google Scholar]
- Kuhn C, Bufe B, Winnig M, Hofmann T, Frank O, Behrens M, Lewtschenko T, Slack JP, Ward CD, Meyerhof W. Bitter taste receptors for saccharin and acesulfame K. J Neurosci. 2004;24:10260–10265. doi: 10.1523/JNEUROSCI.1225-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn C, Bufe B, Batram C, Meyerhof W. Oligomerization of TAS2R bitter taste receptors. Chem Senses. 2010;35:395–406. doi: 10.1093/chemse/bjq027. [DOI] [PubMed] [Google Scholar]
- Laing DG, Willcox ME. An investigation of the mechanisms of odor suppression using physical and dichorhinic mixtures. Behav Brain Res. 1987;26:79–87. doi: 10.1016/0166-4328(87)90157-4. [DOI] [PubMed] [Google Scholar]
- Lawless HT. Evidence for neural inhibition in bittersweet taste mixtures. J Comp Physiol Psychol. 1979;93:538–547. doi: 10.1037/h0077582. [DOI] [PubMed] [Google Scholar]
- Ley JP. Masking bitter taste by molecules. Chem Percept. 2008;1:58–77. [Google Scholar]
- Meyerhof W. Elucidation of mammalian bitter taste. Rev Physiol Biochem Pharmacol. 2005;154:37–72. doi: 10.1007/s10254-005-0041-0. [DOI] [PubMed] [Google Scholar]
- Meyerhof W, Batram C, Kuhn C, Brockhoff A, Chudoba E, Bufe B, Appendino G, Behrens M. The molecular receptive ranges of human TAS2R bitter taste receptors. Chem Senses. 2010;35:157–170. doi: 10.1093/chemse/bjp092. [DOI] [PubMed] [Google Scholar]
- Mueller KL, Hoon MA, Erlenbach I, Chandrashekar J, Zuker CS, Ryba NJ. The receptors and coding logic for bitter taste. Nature. 2005;434:225–229. doi: 10.1038/nature03352. [DOI] [PubMed] [Google Scholar]
- Oka Y, Omura M, Kataoka H, Touhara K. Olfactory receptor antagonism between odorants. EMBO J. 2004a;23:120–126. doi: 10.1038/sj.emboj.7600032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka Y, Nakamura A, Watanabe H, Touhara K. An odorant derivative as an antagonist for an olfactory receptor. Chem Senses. 2004b;29:815–822. doi: 10.1093/chemse/bjh247. [DOI] [PubMed] [Google Scholar]
- Prescott J, Ripandelli N, Wakeling I. Binary taste mixture interactions in prop non-tasters, medium-tasters and super-tasters. Chem Senses. 2001;26:993–1003. doi: 10.1093/chemse/26.8.993. [DOI] [PubMed] [Google Scholar]
- Pronin AN, Xu H, Tang H, Zhang L, Li Q, Li X. Specific alleles of bitter receptor genes influence human sensitivity to the bitterness of aloin and saccharin. Curr Biol. 2007;17:1403–1408. doi: 10.1016/j.cub.2007.07.046. [DOI] [PubMed] [Google Scholar]
- Roudnitzky N, Bufe B, Thalmann S, Kuhn C, Gunn HC, Xing C, Crider BP, Behrens M, Meyerhof W, Wooding SP. Genomic, genetic and functional dissection of bitter taste responses to artificial sweeteners. Hum Mol Genet. 2011;20:3437–3449. doi: 10.1093/hmg/ddr252. [DOI] [PubMed] [Google Scholar]
- Sakurai T, Misaka T, Ishiguro M, Masuda K, Sugawara T, Ito K, Kobayashi T, Matsuo S, Ishimaru Y, Asakura T, Abe K. Characterization of the β-d-glucopyranoside binding site of the human bitter taste receptor hTAS2R16. J Biol Chem. 2010;285:28373–28378. doi: 10.1074/jbc.M110.144444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanhueza M, Schmachtenberg O, Bacigalupo J. Excitation, inhibition, and suppression by odors in isolated toad and rat olfactory receptor neurons. Am J Physiol Cell Physiol. 2000;279:C31–C39. doi: 10.1152/ajpcell.2000.279.1.C31. [DOI] [PubMed] [Google Scholar]
- Schieberle P, Hofmann T. Die molekulare Welt des Lebensmittelgenusses. Chemie unserer Zeit. 2003;6:388–401. [Google Scholar]
- Schifferstein HN. Prediction of sweetness intensity for equiratio aspartame/sucrose mixtures. Chem Senses. 1995;20:211–219. doi: 10.1093/chemse/20.2.211. [DOI] [PubMed] [Google Scholar]
- Schifferstein HN. An equiratio mixture model for non-additive components: a case study for aspartame/acesulfame-K mixtures. Chem Senses. 1996;21:1–11. doi: 10.1093/chemse/21.1.1. [DOI] [PubMed] [Google Scholar]
- Schifferstein HN, Frijters JE. Perceptual integration in heterogeneous taste percepts. J Exp Psychol Hum Percept Perform. 1993;19:661–675. doi: 10.1037//0096-1523.19.3.661. [DOI] [PubMed] [Google Scholar]
- Schiffman SS, Gill JM, Diaz C. Methyl xanthines enhance taste: evidence for modulation of taste by adenosine receptor. Pharmacol Biochem Behav. 1985;22:195–203. doi: 10.1016/0091-3057(85)90377-6. [DOI] [PubMed] [Google Scholar]
- Schiffman SS, Gatlin LA, Sattely-Miller EA, Graham BG, Heiman SA, Stagner WC, Erickson RP. The effect of sweeteners on bitter taste in young and elderly subjects. Brain Res Bull. 1994;35:189–204. doi: 10.1016/0361-9230(94)90122-8. [DOI] [PubMed] [Google Scholar]
- Schiffman SS, Booth BJ, Carr BT, Losee ML, Sattely-Miller EA, Graham BG. Investigation of synergism in binary mixtures of sweeteners. Brain Res Bull. 1995;38:105–120. doi: 10.1016/0361-9230(95)00062-j. [DOI] [PubMed] [Google Scholar]
- Shi P, Zhang J, Yang H, Zhang YP. Adaptive diversification of bitter taste receptor genes in mammalian evolution. Mol Biol Evol. 2003;20:805–814. doi: 10.1093/molbev/msg083. [DOI] [PubMed] [Google Scholar]
- Slack JP, Brockhoff A, Batram C, Menzel S, Sonnabend C, Born S, Galindo MM, Kohl S, Thalmann S, Ostopovici-Halip L, Simons CT, Ungureanu I, Duineveld K, Bologa CG, Behrens M, Furrer S, Oprea TI, Meyerhof W. Modulation of bitter taste perception by a small molecule hTAS2R antagonist. Curr Biol. 2010;20:1104–1109. doi: 10.1016/j.cub.2010.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens JC. Detection of heteroquality taste mixtures. Percept Psychophys. 1995;57:18–26. doi: 10.3758/bf03211846. [DOI] [PubMed] [Google Scholar]
- Stewart RE, Heck GL, Desimone JA. Taste-mixture suppression: functional dissection of cellular and paracellular origins. J Neurophysiol. 1996;75:2124–2128. doi: 10.1152/jn.1996.75.5.2124. [DOI] [PubMed] [Google Scholar]
- Stone H, Oliver SM. Measurement of relative sweetness of selected sweeteners on their sweetener mixtures. J Food Sci. 1969;34:215–222. [Google Scholar]
- Ueda T, Ugawa S, Yamamura H, Imaizumi Y, Shimada S. Functional interaction between T2R taste receptors and G-protein α subunits expressed in taste receptor cells. J Neurosci. 2003;23:7376–7380. doi: 10.1523/JNEUROSCI.23-19-07376.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukhanov K, Corey EA, Brunert D, Klasen K, Ache BW. Inhibitory odorant signaling in Mammalian olfactory receptor neurons. J Neurophysiol. 2010;103:1114–1122. doi: 10.1152/jn.00980.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinzweig A. New York: Houghton Mifflin Harcourt; 2003. Zingerman's guide to good eating: how to choose the best bread, cheeses, olive oil, pasta, chocolate, and much more. [Google Scholar]
- Zhang Y, Hoon MA, Chandrashekar J, Mueller KL, Cook B, Wu D, Zuker CS, Ryba NJ. Coding of sweet, bitter, and umami tastes: different receptor cells sharing similar signaling pathways. Cell. 2003;112:293–301. doi: 10.1016/s0092-8674(03)00071-0. [DOI] [PubMed] [Google Scholar]