Gram-negative bacteria are naturally resistant to many antibiotics because their surface is covered by the glycolipid LPS. Newly synthesized LPS is transported across the cell envelope by the multiprotein Lpt machinery, which includes LptB2FGC, an unusual ABC transporter that extracts LPS from the inner membrane. Like in other ABC transporters, the LptB2FGC transport cycle is driven by the cyclical conformational changes that a cytoplasmic, dimeric ATPase, LptB, undergoes when binding and hydrolyzing ATP. How these conformational changes are controlled in ABC transporters is poorly understood. Here, we identified two lethal changes in LptB that, when combined, remarkably restore wild-type transport function. Biochemical studies revealed that the two changes affect different steps in the transport cycle, having opposing, lethal effects on LptB’s dimerization cycle. Our work provides mechanistic details about the LptB2FGC extractor that could be used to develop Lpt inhibitors that would overcome the innate antibiotic resistance of Gram-negative bacteria.
KEYWORDS: ABC transporters, ATPase, cell envelope, lipopolysaccharide, outer membrane
ABSTRACT
ATP-binding cassette (ABC) transporters constitute a large family of proteins present in all domains of life. They are powered by dynamic ATPases that harness energy from binding and hydrolyzing ATP through a cycle that involves the closing and reopening of their two ATP-binding domains. The LptB2FGC exporter is an essential ABC transporter that assembles lipopolysaccharides (LPS) on the surface of Gram-negative bacteria to form a permeability barrier against many antibiotics. LptB2FGC extracts newly synthesized LPS molecules from the inner membrane and powers their transport across the periplasm and through the outer membrane. How LptB2FGC functions remains poorly understood. Here, we show that the C-terminal domain of the dimeric LptB ATPase is essential for LPS transport in Escherichia coli. Specific changes in the C-terminal domain of LptB cause LPS transport defects that can be repaired by intragenic suppressors altering the ATP-binding domains. Surprisingly, we found that each of two lethal changes in the ATP-binding and C-terminal domains of LptB, when present in combined form, suppressed the defects associated with the other to restore LPS transport to wild-type levels both in vivo and in vitro. We present biochemical evidence explaining the effect that each of these mutations has on LptB function and how the observed cosuppression results from the opposing lethal effects these changes have on the dimerization state of the LptB ATPase. We therefore propose that these sites modulate the closing and reopening of the LptB dimer, providing insight into how the LptB2FGC transporter cycles to export LPS to the cell surface and how to inhibit this essential envelope biogenesis process.
INTRODUCTION
The outer membrane (OM) of most Gram-negative bacteria is covered with lipopolysaccharide (LPS) molecules that create a permeability barrier against many antimicrobials (1). Since LPS biogenesis is also essential for the viability of important pathogens (2, 3), it is an attractive target for developing new molecules that might kill Gram-negative bacteria or might at least permeabilize their OM to antibiotics that otherwise cannot efficiently cross the LPS barrier. LPS biogenesis encompasses three main pathways: (i) LPS biosynthesis starting in the cytoplasm and ending in the inner membrane (IM), (ii) transport of precursors across the IM, and (iii) transport of fully synthesized LPS molecules from the IM to the OM (4). Compounds that target each of these three processes have been reported previously and are being developed as novel antibiotics that prevent LPS assembly at the cell surface (reviewed in reference 5). Excitingly, a peptidomimetic that inhibits LPS transport from the IM to the OM in Pseudomonas aeruginosa is undergoing clinical trials (6–8).
Transport of LPS from the IM to the OM relies on Lpt (LPS transport) proteins LptA to LptG, which form a transenvelope machine that likely functions similarly to a Pez candy dispenser (Fig. 1A) (reviewed in reference 9). The IM Lpt components continuously extract newly synthesized LPS molecules from the bilayer and place them in the Lpt periplasmic bridge, creating a stream of LPS along the Lpt machine that is pushed from the base of the transporter at the IM toward the OM (9–11). Mechanistic details of this intermembrane LPS transporter remain largely unknown, but it is clear that it is powered by LptB2FGC, an unusual ABC transporter localized at the IM (12–14). Like other ABC transporters, LptB2FGC contains two nucleotide-binding domains (NBDs; LptB2) that function as a single ATPase that interacts with and powers two transmembrane domains (TMDs; LptF and LptG), which transport the LPS substrate (10, 15). However, unlike most ABC transporters, LptB2FGC does not translocate its substrate across the membrane; instead, it extracts LPS from the IM and places it onto a periplasmic protein bridge. Recent structural and biochemical studies have revealed unique structural features and key functional details of this transporter (16, 17). The six transmembrane helices of each of the two TMDs, LptF and LptG, associate with the single transmembrane helix of another protein, LptC, to form a cavity in the IM. LPS enters the cavity formed by the transmembrane helices of LptFGC in an ATP-independent manner. The cavity then undergoes a collapse that “squeezes” or “pumps” LPS out and places it onto the Lpt periplasmic bridge. The periplasmic domains of LptFGC each adopt a β-jelly roll fold, and those in LptF and LptC interact to provide a continuous path that LPS follows after exiting the LptFG cavity (Fig. 1A).
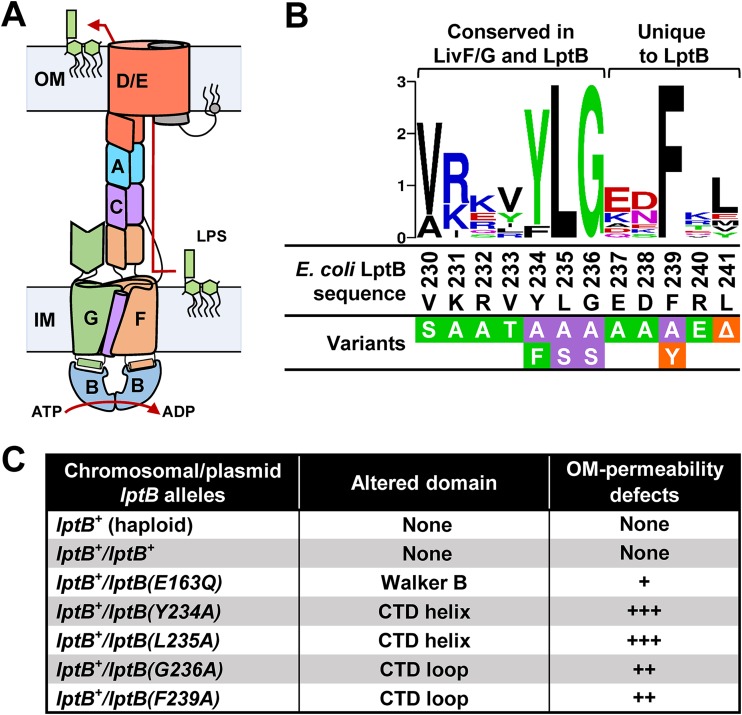
FIG 1.
The CTD of LptB is conserved and essential for function. (A) Architecture of the Lpt transenvelope complex. (B) Sequence logo of the CTD of LptB generated with WebLogo software (weblogo.berkeley.edu) using an alignment of LptB homologs (44). Regions conserved in LivF/G homologs and unique to LptB homologs are marked above the logo. See alignment of LivF/G and LptB in Fig. S2. Below the logo, the sequence of the CTD from E. coli LptBWT (residues 230 to 241) is shown in black. A summary of the results of functional analysis of variants with single amino acid substitutions in the CTD and encoded in pET23/42-LptB is shown. Substitutions conferring no defects in strains carrying a chromosomal ΔlptB deletion are shown in green boxes, those conferring a partial loss of function are shown in orange boxes, and those resulting in a total loss of function are shown in purple boxes. (C) Total-loss-of-function lptB alleles encoded in the pET23/42 plasmid confer dominant-negative effects, as determined by their ability to increase the OM permeability of merodiploid strains carrying the native chromosomal wild-type lptB allele (lptB+). The relative increase in OM permeability, which is indicative of LPS transport defects, is represented with plus signs (+). The catalytically defective LptB variant carrying the E163Q substitution in the Walker B domain was included for reference. Refer to Data set S1 for complete data and details.
Sequence alignment of LptB and the ATPases of branched-chain amino acid transporters. Clustal W alignment of LptB homologs with closely related branched-chain amino acid transporter ATPases LivF/G shows that they share homology at their CTDs. The protein family PF12399 sequence is a conserved sequence found in both protein families and ending in a characteristic YLG sequence (1). The CTD of LptB has additional conserved residues not found in PF12399. Colors correspond to the percent identity color scheme in Jalview (2). Light to dark purple coloring indicates 50% to 100% identity, respectively, and white indicates <50% identity. The protein sequences aligned were those of LivF (WP_000416891.1), LivG (WP_136807488.1), and LptB (WP_000224099.1) from E. coli K-12 substrain MG1655 and LptB homologs from Geobacter uraniireducens Rfr and Burkholderia cenocepacia (WP_011939814.1 and WP_124633598.1, respectively). Download FIG S2, PDF file, 0.4 MB (429.3KB, pdf) .
Copyright © 2019 Simpson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(Page 2) Loss-of-function substitutions at the CTD of LptB confer dominant-negative phenotypes. (Page 3) OM permeability defects of haploid strains with mutations in lptB that disrupt the interaction between the switch helix and CTD loop. (Page 4) Genetic interactions between changes in R144 and R198 and alterations to the CTD of LptB. (Page 5) Genetic interactions between changes in Walker A residue T45 and alterations to the CTD of LptB. (Page 6) Strains used in this study. (Page 9) Primers used in this study. Download Data Set S1, PDF file, 0.3 MB (306.8KB, pdf) .
Copyright © 2019 Simpson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ABC transporters use ATP binding and hydrolysis to cycle between conformations that promote substrate binding, substrate translocation, and resetting of the transporter to its ground state (18–22). ATP binding drives the formation of a closed NBD dimer that sandwiches two ATP molecules, and then hydrolysis promotes dimer opening to allow ADP/ATP exchange. Since NBDs are physically connected to TMDs, the nucleotide-driven dynamics of the NBDs cause the TMDs to cycle between states. In some transporters, ATP binding directs substrate transport, while ATP hydrolysis and subsequent release of ADP and inorganic phosphate (Pi) reset them to their ground state; in others, ATP hydrolysis powers substrate transport and the subsequent release of ADP and Pi resets them (19). Here, we discovered a novel C-terminal domain (CTD) in LptB that is crucial for LPS transport in Escherichia coli. Changes to this domain reduce ATP hydrolysis and prevent proper cycling of LptB. Our genetic, biochemical, and structural work provides insight into the LPS transport cycle by elucidating how different sites in LptB affect the function of this ATPase (summarized in Table 1).
TABLE 1.
Relevant domains and LptB variants explored in this study
| Domain altered | Domain function (reference[s]) | LptB variant(s) exploreda |
|---|---|---|
| Near/in Walker A | ATP binding (18, 29) | G33A/C, L35Q, T43S, T45A |
| Walker B | ATP hydrolysis (15, 18) | E163Q |
| D-loop helix | ATPase dimerization (41) | K177A/E, E199A |
| Signature helix | ATP binding (this study) | R144H¸ R144A/F/Q, R145A, I148S/T, S172A, D175A, I176T |
| Switch helix | Interaction with CTD loop (this study) | R198A/E |
| CTD helix | Unknown essential role (this study) | Y234A, L235A |
| CTD loop | ATPase dimerization and turnover (this study) | F239A, G236A, L241Δ |
| Extensions added to CTD loop | ATPase dimerization and turnover (this study) | LptB1, LptB-EHis8, LptB-R, LptB-RHis8 |
Variants that add extended sequences or tags to the CTD of LptB are indicated with the allele names shown in Fig. S1. Variants with amino acid substitutions are indicated with the substitution. The most extensively studied variants are shown in bold.
Altering the CTD of LptB results in functional defects. (A) The addition of CT extensions to LptB disrupts activity to differing degrees. A summary of functional analyses for various lptB haploid strains is presented. In investigating why some CT-tagged LptB variants could support growth while others could not, we noticed that the two types significantly differed at residue 242, which is located in the linker between the last residue (L241) of wild-type LptB and the tag. The lethal chromosomal lptB-His allele encodes a positively charged arginine, while the complementing plasmidborne lptB-EHis8 and chromosomal lptB1 alleles encode a negatively charged glutamate and a neutral isoleucine, respectively. To examine if the addition of an arginine after the native L241 residue affected LptB function on its own or in the context of the His tag, we constructed plasmids bearing lptB alleles encoding LptB followed by only an arginine (lptB-R) or an arginine followed by a polyhistidine tag (lptB-RHis8) and compared their function to that of wild-type lptB (lptB+) and lptB-EHis8 genes. The lptB-R and lptB-EHis8 alleles complemented chromosomal ΔlptB, but the resulting haploid strains were sensitive to hydrophobic antibiotics, indicating that these strains, like an lptB1 strain, are defective in LPS transport. In contrast, the lptB-RHis8 allele could not complement chromosomal ΔlptB despite resulting in the production of as much protein as an lptB-EHis8 strain (see below). Thus, adding an arginine or oligopeptide to the CTD of LptB causes partial loss-of-function defects, while the combination of an arginine and oligopeptide is lethal. (B) LptB immunoblot comparing strains that chromosomally encode LptBWT, labeled 754, or LptB1. (C) LptB immunoblot of samples from haploid strains bearing LptB CTD variants on the pET23/42-LptB plasmid. “WT” refers to haploid strain NR2101, which produces LptBWT from pET23/42-LptB. (D) LptB immunoblot of samples from merodiploid strains producing LptB CTD variants from the pET23/42-LptB plasmid and LptBWT from the native lptB locus. “754” refers to NR754, the wild-type strain with lptB+ at the native locus. “WT” refers to strain NR2583, which produces LptBWT from both the chromosome and pET23/42-LptB. Data are representative of results from at least three independent experiments. (E) Ability of CTD lptB alleles to complement a chromosomal ΔlptB allele in rich (LB) medium. (F) OM permeability of haploid strains producing LptB CTD variants was determined by disc diffusion assay. All alleles, except lptB+ in NR754 and lptB1, which are located on the chromosome, are borne on the pET23/42-LptB plasmid. Increased sensitivity was reproducibly observed for the strains shown and was indicative of LPS transport defects compared to wild-type lptB+ strain NR2101, while those not shown (containing changes V230S, K231A, R232A, V233T, E237A, D238A, F239Y, and R240A) behaved like wild-type strain NR2101. Numbers represent the diameter (in millimeters) of the zone of inhibition or that of partial growth (parentheses) around a 6-mm-diameter disc containing an antibiotic. Download FIG S1, PDF file, 0.3 MB (267.8KB, pdf) .
Copyright © 2019 Simpson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RESULTS
The CTD of LptB is a unique domain essential for LPS transport.
The LptB ATPase contains domains that are conserved among NBDs of ABC transporters and that are essential for LPS transport and viability in E. coli (13, 15). Here, we discovered an essential, unique domain in LptB while working with a C-terminally tagged variant. Previous studies showed that C-terminally His-tagged LptB proteins hydrolyze ATP in vitro (15, 23–26). One of them, LptB-EHis8, when encoded by a gene borne in a plasmid, supports growth of E. coli cells lacking chromosomal lptB (15). However, we found that the resulting lptB-EHis8 haploid cells are sensitive to hydrophobic antibiotics, a hallmark of LPS transport defects (see Fig. S1 in the supplemental material). Furthermore, we could not replace the native lptB gene with a chromosomal lptB-His allele. Instead, the only haploid strain that we isolated carried an altered chromosomal lptB allele (named lptB1) that we assume suppresses the lethality of the sought-after lptB-His mutant. This lptB1 allele, which resulted from a frameshift mutation, encodes full-length LptB with a 34-residue C-terminal (CT) extension and is partially functional since haploid lptB1 cells are sensitive to hydrophobic antibiotics (Fig. S1A) (27). We also found that the identity of the first amino acid of the CT extensions determines their impact on LptB function, explaining why lptB1, but not lptB-His, can substitute for chromosomal lptB (Fig. S1).
Since our results suggested a functional role for the CTD of LptB, we next characterized in vivo the function of LptB variants containing single substitutions in their CTD. We focused on the region encompassing residues 230 to 241 because it is fully conserved only in LptB orthologs and partially conserved in ATPases of closely related branched-chain amino acid transporters (Fig. S2; see also Text S1 in the supplemental material). We found that whereas substitutions at less-conserved positions did not affect function, those at the highly conserved residues Y234, L235, G236, and F239 abolished their ability to complement the loss of native lptB (Fig. 1B; see also Fig. S1E). These nonfunctional LptB variants were stable and exhibited dominant-negative effects, indicating they titrated wild-type LptB (LptBWT) and/or its partners LptFG into nonfunctional machinery (Fig. 1C; see also Data set S1 in the supplemental material). In fact, the dominance of the CTD mutant alleles was stronger than that of the catalytically dead lptB(E163Q) allele, suggesting that complexes containing LptBWT-LptBE163Q heterodimers might still be somewhat functional whereas those containing LptBWT and CTD-defective LptB variants might not. Together, these data demonstrate that the CTD of LptB is essential for LPS transport and suggest that the CTD might have a trans effect across monomers in the LptB dimer.
Supplemental references for Fig. S2 and Data set S1. Download Text S1, PDF file, 0.1 MB (114.4KB, pdf) .
Copyright © 2019 Simpson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The CTD of LptB forms critical interactions with the Walker A and switch helix domains.
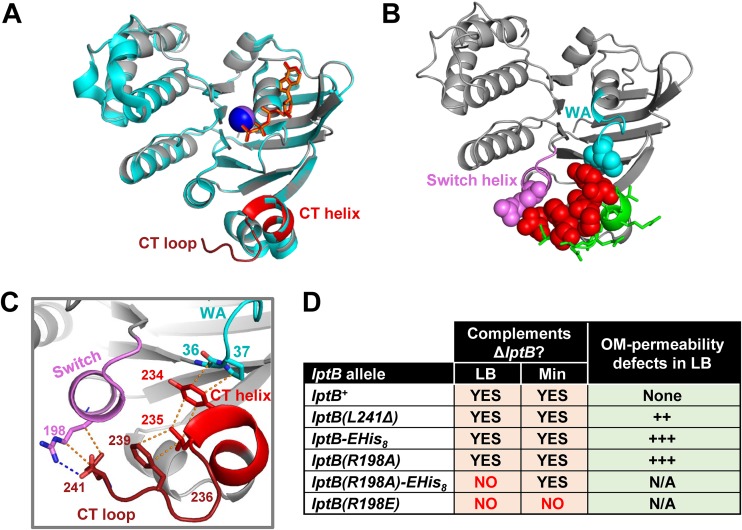
Previously reported structures derived from CT-tagged LptB variants failed to fully resolve the CTD, suggesting that it might be flexible when tagged (15, 25, 26, 28). Therefore, to gain structural insight into the role of the wild-type CTD of LptB, we attempted to crystallize ADP- and ATP-bound E. coli LptB proteins with N-terminal His tags after confirming that they do not affect function. To trap the ATP-bound form, we used the catalytically dead LptBE163Q protein as was done with CT-tagged LptBE163Q-EHis8 as previously described (15). We did not obtain a structure of ADP-bound His8-LptB, but we solved a 1.96-Å structure of ATP-bound His8-LptBE163Q. The new structure resembled that of LptBE163Q-EHis8, except that it included a fully resolved CTD (Fig. 2A; see also Fig. S3). The CTD is located at the LptB dimer interface below the ATP-binding sites, and it consists of a short helix (residues V230 to L235) followed by a turn and a loop (residues G236 to L241) (Fig. 2A). In ABC transporters, NBDs assemble in a head-to-tail conformation to form two ATP-binding sites, each composed of the Walker A and Walker B motifs of one monomer and the signature motif of the other monomer (18, 29). Within each LptB monomer, CTD-helix residue Y234 interacts with Walker-A invariant residue G36 and residue P37. In addition, CTD-helix residue L235 forms van der Waal interactions with CTD-loop residue F239 (Fig. 2B and C). Our in vivo data revealed that these interactions and placement of these residues are important for function, since altering them by substituting residues Y234, L235, G236, and F239 with alanine rendered LptB nonfunctional (Fig. 1; see also Fig. 2), while conservative substitutions (LptBY234F and LptBF239Y) that should have maintained their interactions had only a minor impact on function (Fig. S1E and F).
FIG 2.
The CTD of LptB makes critical contact with the Walker A and switch helix. (A) Alignment (generated with align function in PyMol; root mean square [RMS] = 0.183) of ATP-bound structures of His8-LptBE163Q (PDB 6MBN; gray) and LptB-EHis8E163Q (PDB 4P33; cyan). The CTD of His8-LptBE163Q is colored red. In PDB 6MBN, the Na+ atom is shown as a blue sphere and ATP as orange sticks. In 4P33, the Na+ atom is shown as a purple sphere and ATP as red sticks. (B) Cartoon representation of ATP-bound LptB structure (PDB 6MBN). CTD residues that, when substituted as in the Fig. 1 legend, resulted in no defects are shown as green sticks; those that caused total or partial loss of function are shown as red spheres. Functionally important CTD residues place the CTD helix and CTD loop such that they can interact with switch helix (violet) residue R198 (violet spheres) and Walker A (cyan) residues G36 and P37 (cyan spheres). (C) Detailed interactions of the functionally important CTD (red) residues shown in panel B. Dotted lines represent polar (blue) and nonpolar (orange) interactions. Y234 interacts with G36 and P37 in the Walker A motif (cyan); Y234, L235, and F239 stack together through hydrophobic interactions; G236 is likely critical for creating a turn that both facilitates stacking of L235 and F239 and properly orients L241; L241 interacts with R198 in the switch helix that follows the switch domain. (D) Functional characterization of mutant alleles encoding changes that disrupt the interaction between position R198 of the switch helix and the CTD loop. Table data indicate the ability of lptB alleles to complement a chromosomal ΔlptB allele in rich (LB) and minimal (Min) media. Haploid strains carrying alleles that complemented in LB were tested for increased OM permeability to antibiotics by disc diffusion assay. The relative increase in OM permeability is indicated with plus signs (+). Refer to Data set S1 for data and details.
The CTD of LptB mediates intra- and intermonomer contacts through interactions with the switch helix. (A) Data collection and refinement statistics of PDB 6MBN structure of the ATP-bound His8-LptBE163Q dimer. (B) Cartoon representation of the ATP-bound His8-LptBE163Q dimer (PDB 6MBN) showing that the carboxylic acid of the last residue in LptB, L241, forms ionic pairs (blue dotted lines) with residue R198 in the switch helix both within each monomer and across monomers. A bottom view of the LptB dimer is presented in which LptB monomers are colored cyan (monomer A) and light gray (monomer B), the switch helix containing R198 is colored pink, and the CTD is colored red. R198 and L241 are shown as sticks. (C) Detailed view of the interactions between residues R198 and L241. LptB monomers are colored cyan (monomer A) and light gray (monomer B). Download FIG S3, PDF file, 0.6 MB (662.5KB, pdf) .
Copyright © 2019 Simpson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The new ATP-bound structure also revealed a functional interaction between the carboxylate of CTD-loop residue L241, which is the last residue of LptB, and residue R198 from both monomers (Fig. 2B and C; see also Fig. S3). Residue R198 is located in the switch helix that follows the switch motif (or H loop) (18, 29). In LptB, the switch motif contains the essential histidine (H195) that interacts with the γ-phosphate of ATP and undergoes a large conformational change upon ATP hydrolysis to possibly allow the exit of Pi (15). Our previous results suggested that the R198-L241 interaction was functionally relevant, since haploid mutants producing LptB-R (in which the arginine added after residue L241 would likely cause a charge clash with R198) or CT-tagged LptB (in which the tag removes the carboxylate of L241 and likely interferes by steric hindrance) were partially defective in LPS transport (Fig. S1; see also Data set S1). To directly probe the relevance of this interaction, we constructed an LptB variant lacking L241 and found that haploid cells producing LptBΔL241 were viable but were partially defective in LPS transport (Fig. S1E and F). Furthermore, we tested if alterations to R198 also resulted in LPS transport defects. Complementation and antibiotic-sensitivity analyses showed that lptB(R198E) was a total-loss-of-function allele and that lptB(R198A) was a partial-loss-of-function allele whose function could be further compromised by altering the CTD of LptBR198A by adding a CT tag, as the lptB(R198A)-EHis8 haploid strain was viable in minimal medium but not in LB (Fig. 2D; see also Data set S1). This conditional lethality reflects severely compromised LPS transport that supports growth under slow-growth conditions (i.e., minimal medium) but not under fast-growth conditions (i.e., LB) (30). Together, the data from our structure-function analyses suggest that the essential function of the CTD of LptB depends on the proper placement of the CTD helix (V230 to L235) and CTD loop (G236 to L241) through intramonomer interactions with G36 and P37 in the Walker A domain and with R198 in the switch helix. In addition, the interactions between L241 and R198 across dimers (Fig. S3) might be functionally relevant and responsible for the aforementioned strong dominance of nonfunctional CTD variants and their trans effect in LptBWT-LptBCTD heterodimers (Fig. 1C).
Changes in the ATP-binding domains suppress defects caused by alterations to the CTD of LptB.
To further understand the function of the CTD of LptB, we selected for suppressors of lptB1, which encodes a partially functional LptB variant with a 34-residue CT extension (Fig. S1). Suppressors selected on vancomycin or novobiocin plates carried intragenic mutations in lptB1 and behaved in two distinct ways (previously described [27]): suppressors with substitutions G33A/C, L35Q, T43S, T45A, I148T, or S243Stop in LptB1 exhibited wild-type resistance to all antibiotics tested, while a suppressor with substitution R144H in LptB1 was resistant only to novobiocin (Fig. 3; see also Fig. S4). We do not further discuss the lptB1(S243stop) suppressor, which truncated the 34-residue CT extension to a single isoleucine, supporting our previous conclusion that the addition of an oligopeptide to the CTD negatively affects LptB function.
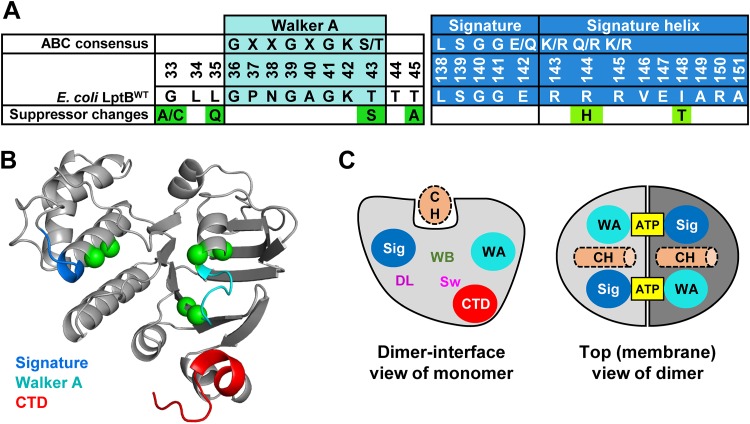
FIG 3.
Changes to the ATP-binding sites suppress defects in the CTD of LptB. (A) Consensus sequences for the Walker A motif, signature motif, and signature helix of the ABC transporter family (PF00005) (56). Below each consensus sequence, the E. coli LptBWT sequence and changes identified in suppressors are shown. (B) Cartoon representation of the structure of an His8-LptBE163Q monomer showing the location of the suppressors of lptB1 (represented as green spheres). The Walker A motif (cyan), signature helix (marine), and CTD (red) are colored in the structure. (C) The cartoon on the left shows the dimer interface view of an LptB monomer (light gray). The locations of the Walker A motif (WA; cyan) and signature (Sig; marine) motifs and of the CTD are marked. The locations of additional motifs important for function of ABC transporters are also shown as follows: “WB” marks the Walker B motif, “DL” the D loop, and “Sw” the switch loop. The Walker A and B motifs, together with the signature motif, constitute each ATP-binding site; the signature motif and D loop are thought to coordinate NBD-TMD coupling, and the switch loop is important for catalysis and possibly for release of Pi after hydrolysis. The structural groove on the top of each LptB monomer accommodates the coupling helix (CH, dotted light orange) of either LptF or LptG. The cartoon on the right shows the top view of the LptB dimer, where monomers appear in light and dark gray. Each ATP molecule is sandwiched between the Walker A motif and the signature motif of each monomer. The coupling helices (CH) are shown as dotted light orange cylinders.
Structure-function analysis of lptB1 suppressors. (A and B) OM permeability assay (A) and LptB immunoblots (B) of chromosomally encoded lptB1 and suppressors performed as described in the Fig. S1 legend and in Materials and Methods. “lptB+” refers to strain NR754. (C) LptB immunoblot of samples from haploid strains producing LptB variants from pET23/42LptB derivatives performed as described in the Fig. S1 legend. (D) OM permeability assay of haploid cells carrying pET23/42LptB-EHis8 derivatives with the substitutions we found as suppressors of lptB1. Assays were performed as described in in the Fig. S1 legend. Data are representative of results from at least three independent experiments. Download FIG S4, PDF file, 0.2 MB (232.4KB, pdf) .
Copyright © 2019 Simpson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The remaining changes suppressing LptB1 localized to the Walker A and signature motif regions, which mainly constitute the two halves of each ATP-binding domain (Fig. 3; see also Fig. S5) (18). Changes localized in or flanking the Walker A motif (G33A/C, L35Q, T43S, T45A) also suppressed defects conferred by the lptB-EHis8 allele (Fig. S4D). The remaining suppressors (I148T and R144H) mapped to the signature helix that follows the signature motif, the other half of the ATP-binding domain (Fig. 3) (31–35). The I148T substitution also suppressed lptB1 and lptB-EHis8 (Fig. S4). In contrast, as we reported earlier, the R144H substitution conferred resistance only to novobiocin, a known DNA gyrase inhibitor, because this antibiotic interacts with LptB at the LptFG interface to increase Lpt function (27). The fact that the combination of novobiocin and the R144H substitution suppressed lptB1 and lptB-EHis8 might suggest that the R144H change is a weak suppressor that needs the additional effect of novobiocin to fully suppress the CTD defect; however, as we describe below, this is not the case. Taking the results together, these suppressors revealed that the function of the CTD of LptB is related to ATP binding and/or hydrolysis.
Location of lptB1 suppressors. (A and B) Cartoon representations of the structures of an His8-LptBE163Q dimer (A) and monomer (B) showing the location of the suppressors of lptB1 (represented as forest green spheres). The Walker A domain (cyan), signature motif (marine), and CTD (red) are colored in the X-ray structure. ATP is shown in yellow sticks. The location of additional motifs important for function of ABC transporters is also shown on the cartoons on the left. “WB” marks the Walker B motif, “DL” the D loop, “Sw” the switch loop, and “CH” a coupling helix of either LptF or LptG. Numbers in green shown in panel B represent the number of the residue altered in the suppressors. Download FIG S5, PDF file, 0.6 MB (584KB, pdf) .
Copyright © 2019 Simpson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Defects caused by the R144H substitution and disruptions to the CTD loop-R198 interactions suppress one another.
We next investigated the effect that the aforementioned suppressor mutations had in an otherwise wild-type lptB allele. Neither the introduction of I148T nor the changes in the Walker A region had an effect, except that some substitutions caused a reduction in LptB levels (Fig. S4). These substitutions also decreased LptB1 levels, further suggesting they suppress lptB1 by conferring a functional change. When we introduced the R144H change into wild-type lptB, we generated a haploid lptB(R144H) strain that could grow in minimal medium but, surprisingly, not in LB (Fig. 4A). This conditional lethality is characteristic of severe Lpt defects (30), indicating that residue R144 is critical for LptB function. We considered that residue R144 might be functionally important because it forms polar interactions with the backbones of residues P84, D162, and Q163 (Fig. 4B), which are part of two motifs that are essential in ABC transporters. Residue P84 is in the Q loop, which is important for NBD-TMD communication; residues D162 and Q163 are in the Walker B motif, which is essential for ATP hydrolysis, with E163 being the catalytic glutamate in wild-type LptB (15, 18). Furthermore, the polar character is conserved at this position (R, K, or Q) in other ABC transporters (Fig. 3A). To investigate the relevance of these interactions, we substituted R144 with nonpolar alanine or phenylalanine, which abolished function, and with glutamine, which resulted in conditional lethality (Fig. 4A). These results suggested that polar contacts of R144 with the Walker B domain and/or the Q loop are critical for LptB function, implying that R144 might be important for ATP binding and/or hydrolysis and its coupling for the function of the TMDs.
FIG 4.
Specific changes to R144 and the CTD suppress each other. (A) Ability of variants with changes in R144 introduced in either LptBWT or LptB-EHis8 to complement a ΔlptB allele in LB and in M63 minimal media supplemented with glucose (Min). (B) Interactions of R144 (marine) of ATP-bound His8-LptBE163Q (PDB 6MBN) with the backbone of P84 of the Q-loop domain, D162, and Q163 in the Walker B domain (in wild-type LptB, E163 is the catalytic residue). (C) Cosuppression between the R144H and changes to R198 and the CTD loop. Mutant alleles were assayed for their ability to complement a chromosomal ΔlptB allele in LB. Haploid strains carrying alleles that resulted in complementation in LB were tested for increased OM permeability to antibiotics by disc diffusion assay. The relative levels of increases in OM permeability are indicated with plus signs (+). Refer to Data set S1 for complete data and details.
The inability of haploid cells producing untagged LptBR144H to grow in LB was surprising because those producing CT-altered LptB1R144H are viable in this medium. This disparity in results suggested that the CTD extension in LptB1 fixes defects conferred by the R144H substitution. Nevertheless, this fix must not be complete, as OM permeability defects of lptB1(R144H) can be further suppressed by the addition of novobiocin (27). Since these data suggested that the R144H substitution and the CT extension in LptB1 suppressed each other, we tested if this cosuppression was specific to LptB1 or was applicable to other CTD alterations. We found that introducing different changes to the CTD of LptBR144H also resulted in cosuppression (Fig. 4; see also Data set S1). Strikingly, whereas the lptB(G236A), lptB(F239A), and lptB(R144H) haploid strains were nonviable in LB, the lptB(R144H/G236A) and lptB(R144H/F239A) double mutant strains grew well in LB. In fact, the lptB(R144H/F239A) strain behaved similarly to the wild type. Thus, combining R144H, a severely compromising change in the signature helix, with F239A, a lethal change in the CTD, remarkably resulted in a double mutant that was phenotypically wild type.
We wondered whether the other suppressors of lptB1 could also suppress the lethal mutations in the CTD loop. We found that Walker A substitution T45A, like R144H, could suppress the lethal effects of the F239A change but that the resulting lptB(T45A/F239A) double mutant was more sensitive to antibiotics than the wild-type-like lptB(R144H/F239A) mutant (Data set S1). Notwithstanding, the cosuppression between either an R144H or T45A substitution and alterations to the CTD loop (residues G236 to L241) was not applicable to defects in the CTD helix, as neither an R144H or T45A change could rescue the lethality of the lptB(Y234A) and lptB(L235A) alleles. Thus, changes to the ATP-binding sites specifically suppress alterations to the CTD loop but not to the CTD helix. This suggests that alterations to the CTD helix, in addition to misplacing the CTD loop, cause an additional lethal defect.
On the basis of the ATP-bound LptB structure (Fig. 2), we predicted that the G236A and F239A substitutions, as well as the addition of the His tag, would each result in misplacement of the CTD loop and thereby in disruption of its interaction with switch-helix residue R198. We therefore expected that the cosuppression between the R144H and CTD loop alterations was caused by the breaking of the CTD loop-R198 interaction and that changes to R198 would also suppress the effects of the R144H change. Indeed, the otherwise lethal R198E and conditionally lethal R144H substitutions suppressed each other (Fig. 4C; see also Data set S1). Thus, the lethal defects conferred by the R144H change were suppressed by interfering with the interaction between the CTD loop and residue R198, and vice versa. This strong cosuppression indicates that the function of signature-helix residue R144 is related to that of the interaction between the CTD loop and switch-helix residue R198. From the unexpected cosuppression of these substitutions, we infer that the individual changes affect the ATP cycle in opposite ways such that altering each function individually causes severe defects in LPS transport but these alterations compensate for each other when combined.
It is also worth noting that (i) the CT tags that we used were less defective than the lethal F239A change, (ii) F239A and R144H fully suppressed each other’s severe defects, and (iii) the cosuppression between the less-defective CT extensions in LptB1 or LptB-EHis8 and R144H was only partial and was capable of being further improved by novobiocin. Therefore, it appears as if novobiocin somehow further increases the levels of defects caused by the CT extensions of LptB1 or LptB-EHis8 such that their combined defects can better suppress those conferred by the R144H substitution. These observations also support the aforementioned proposal that the R144H and CTD substitutions affect the ATP cycle in opposite ways that compensate for each other when combined.
A network of interactions connects the function of the signature helix and CTD of LptB.
Our genetic data revealed a functional link between the signature helix and the R198-CTD interaction. This connection would require a long-range network of interactions because these domains are not in close proximity (Fig. 5A). Upon examining the His8-LptBE163Q structure, we predicted that the D-loop helix, which follows the D loop, mediates interactions between the signature helix (which contains R144) and the switch helix (which contains R198) (Fig. 5). The D loop is an essential motif in ABC transporters that is thought to coordinate NBD-TMD coupling and NBD dimerization (22, 36). We reasoned that if this network were relevant to coordinating the functions of the signature helix and the R198-CTD loop interaction, disrupting interactions along the network should then result in genetic interactions similar to those described for mutations that alter R144, R198, and the CTD loop. Indeed, this idea was supported by our earlier finding that an I148T change, which should alter one of the interactions along this putative network, suppressed the CTD-loop defective lptB1 allele (Fig. 3; see also Fig. 5).
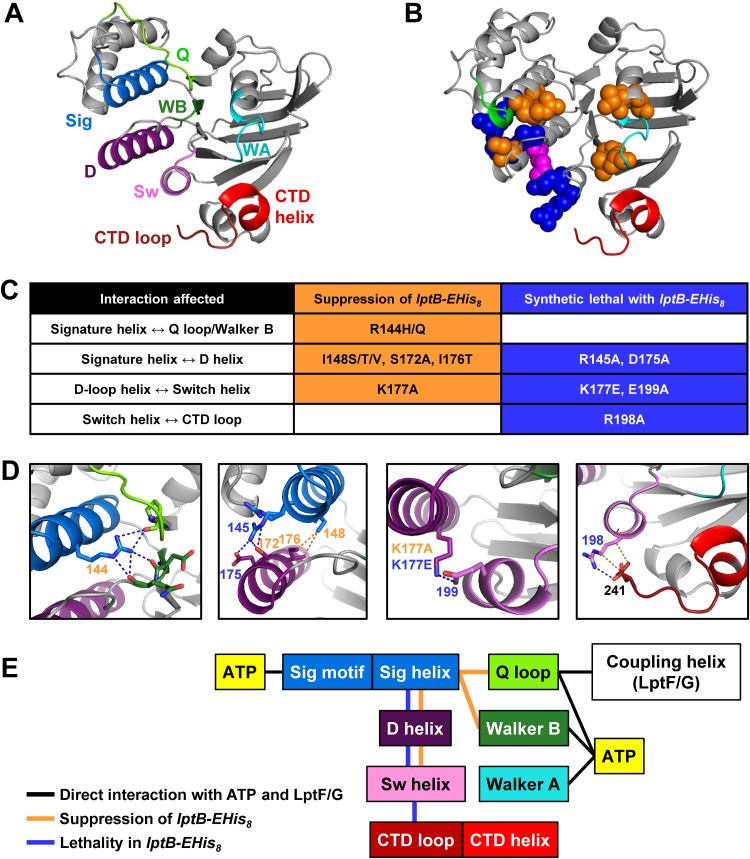
FIG 5.
A network of helices connects the functions of the signature helix and the CTD of LptB. (A) Cartoon view of the structure of a monomer of ATP-bound His8-LptBE163Q (PDB 6MBN). Relevant domains are the Q loop (Q; chartreuse), signature domain and helix (Sig; blue), Walker B domain (WB; forest green), Walker A domain (WA; cyan), D-loop helix (D; dark purple), switch helix (Sw; violet), CTD helix (red), and CTD loop (brick red). (B) Interactions observed in the ATP-bound crystal structure of His8-LptBE163Q (dimer interface view of PDB 6MBN). Spheres correspond to residues that, when substituted, resulted in suppression (in orange) or synthetic lethality (in blue) or both depending on the substitution (pink). The Walker A and signature motifs are colored in cyan and green, respectively, and the CTD is shown in red. (C) Summary of substitutions that suppress (in orange) or cause lethality (in blue) when introduced into LptB-EHis8 (complete data are shown in Fig. 4; see also Fig. S6B and C). (D) Expanded views of structures of LptB (PDB 6MBN) with residues depicted as sticks and polar (blue) or nonpolar (orange) interactions depicted as dotted lines. Residue numbers are colored according to whether substitutions shown in panel C led to suppression (orange) or lethality (blue). (E) Schematics showing key domains and relevant physical and genetic interactions. Direct interactions between LptB domains and ATP or the coupling helices of LptF/G are marked with black lines. Orange lines represent interactions that, when altered, led to suppression of defects conferred by lptB-EHis8. Blue lines represent interactions that, when changed, led to synthetic lethality with lptB-EHis8.
Synergistic effects of mutations combined with lptB-EHis8 identifying a network of functionally related residues. (A) Sequence logo (generated as described in the Fig. 1D legend and Materials and Methods) of the conservation of residues in the signature, D-loop, and switch helices (locations of each are marked with thick lines). Below the logo, the sequence of E. coli LptBWT is shown in black and a summary of the results of functional analysis of single amino acid substitutions designed to disrupt the observed interactions between R144 and the CTD is presented. Substitutions in pET23/42-LptB conferring no defects in strains carrying a chromosomal ΔlptB deletion are shown in green, those conferring partial loss of function are shown in orange, and those resulting in a total loss of function are shown in purple. (B) Ability of mutations that alter the indicated residue in either pET23/42LptB or pET23/42LptB-EHis8 to complement a ΔlptB allele in the indicated media as described in the Fig. S1A legend. (C) OM permeability of haploid strains carrying lptB mutations that exhibit effects synergistic with those of lptB-EHis8. Disc diffusion assays were performed on LB plates. Numbers indicate diameter (in millimeters) of the zone of total growth inhibition or that of partial growth (in parentheses). (D) LptB immunoblot of haploid lptB-EHis8 strains carrying pET23/42-LptB-EHis8 grown in LB. (E) LptB immunoblot of samples from haploid lptB strains carrying pET23/42-LptB grown in minimal medium to allow growth of strains carrying conditionally lethal alleles. “WT” refers to NR2101, which produces LptBWT from pET23/42-LptB. (F) LptB immunoblot of samples from merodiploid strains carrying the wild-type lptB allele and mutant lptB alleles carried in pET23/42-LptB grown in LB. “754” refers to haploid strain NR754, the wild-type strain with chromosomal lptB+ and “WT” to NR2583, which produces LptBWT from both the chromosome and pET23/42-LptB. (G) LptB immunoblot of samples from merodiploid strains carrying the chromosomal wild-type lptB allele and mutant lptB-EHis8 alleles encoded in pET23/42-LptB-EHis8 grown in LB. “754” refers to haploid strain NR754; “WT” refers to NR1872, which produces LptBWT from the chromosome and LptB-EHis8 from pET23/42-LptB-EHis8. Data are representative of results from at least three independent experiments. Download FIG S6, PDF file, 0.3 MB (276.8KB, pdf) .
Copyright © 2019 Simpson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To explore the relevance of this putative network, we introduced mutations expected to disrupt contacts between the signature, D-loop, and switch helices and tested their effects on wild-type and CT-loop-defective (lptB-EHis8) strains (Fig. 5; see also Fig. S6). We found that substitutions to networking residues in the signature (R144, R145, and I148), D-loop (D175 and K177) and switch (R198 and E199) helices caused LPS transport defects (Fig. S6). Moreover, these mutations either suppressed or were synthetically defective in lptB-EHis8, depending on the position or manner in which a position was altered (Fig. 5; see also Fig. S6). Thus, substitutions within the interaction network mimicked either the R144H change or those that break the R198-CTD interaction. Together, these data revealed a network of residues that functionally connects the signature helix and CTD loop of LptB and suggest that, depending on the manner in which the network is altered, LptB may favor one of two conformations or states.
Decoupling of ATP hydrolysis and LPS extraction in complexes containing LptBR144H/F239A.
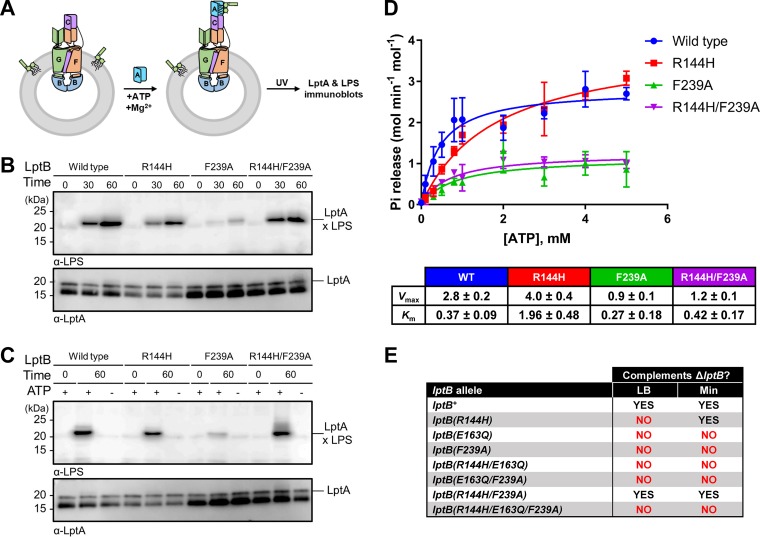
To further investigate the effect of altering the signature helix and CTD of LptB on LPS extraction and ATPase activity, we used an in vitro system with reconstituted LptB2FGC complexes (11). We prepared proteoliposomes containing LPS and different LptB2FGC complexes and compared the abilities of those complexes to transfer LPS to their soluble periplasmic partner LptA after extracting the glycolipid from the bilayer. For this, we used purified LptAI36pBpa, which contains the UV-cross-linkable amino acid para-benzoyl phenylalanine (pBPA) at a position that interacts with LPS (10). If LPS were extracted and transferred to LptAI36pBpa, exposure to UV would result in LPS-LptA adducts that could be detected using LPS immunoblotting (Fig. 6A). We found that, as previously reported (11), complexes containing LptBWT transferred LPS to LptA in a time-dependent manner under conditions of saturating ATP concentrations (Fig. 6B). Compared to wild-type complexes, those containing LptBR144H displayed some reduction in LPS extraction, while those with LptBF239A were highly defective (Fig. 6B). In contrast, and as expected from their in vivo wild-type-like behavior, complexes containing LptBR144H/F239A resembled wild-type complexes in their ability to extract LPS. For all complexes tested, LPS extraction required ATP (Fig. 6C). Furthermore, as previously reported (11), transfer of LPS to LptA required ATP hydrolysis, as shown by the lack of transport by complexes containing the catalytically dead LptBE163Q (Fig. S7A).
FIG 6.
Decoupling of the ATPase activity of LptB and LPS extraction in mutant LptB2FGC complexes. (A) In vitro assay monitoring release of LPS from proteoliposomes containing LptB2FGC complexes to LptAI36pBpa by UV cross-linking and detection of LPS-LptAI36pBpa adducts by LPS immunoblotting. Refer to Materials and Methods for details. (B) Defects in the signature helix and CTD of LptB affect LPS release. LPS released from proteoliposomes containing LptB2FGC complexes with various LptB variants were UV-cross-linked to LptAI36pBpa, and the resulting LPS-LptAI36pBpa adducts (marked LptA × LPS) were detected by LPS immunoblotting (top panel). Time (min) data indicate the incubation period of the LPS release assay in the presence of ATP. LptA immunoblots shown confirm total LptA levels (bottom panel). (C) The release of LPS shown in panel B was dependent on the presence of ATP. Time (min) data indicate the incubation period of the LPS release assay in the presence (+) or absence (−) of ATP. Immunoblots were performed as described for panel B. Data in panels B and C are representative of results from three independent biological replicates. (D) ATPase activity was monitored in the presence of various concentrations of ATP in the in vitro assay for LPS release from proteoliposomes containing LptB2FGC complexes with various LptB variants. Data represent averages and standard deviations of results determined using three different proteoliposome preparations for each LptB variant and a molybdate assay to monitor Pi release. (E) ATP hydrolysis is required for the cosuppression of defects conferred by the R144H and F239A substitutions in LptB, since introduction of the Walker B substitution E163Q into LptBR144H/F239A rendered the otherwise fully functional protein unable to complement the loss of chromosomal lptB.
ATP hydrolysis is required for LPS release from proteoliposomes. (A) Complexes containing the catalytically dead LptBE163Q variant are defective in extracting LPS from liposomes. An assay monitoring LPS release from liposomes containing LptB2FGC complexes with LptBWT and catalytically dead LptBE163Q variants was performed. Released LPS was UV-crosslinked to LptAI36pBpa, and the resulting LptAI36pBpa x LPS adducts were detected by LPS immunoblotting (top panel). Assays were initiated by the addition of ATP and incubation at 30°C for the indicated amount of time prior to crosslinking. LptA immunoblots are shown that confirm total LptA levels (bottom panel). Data are representative of results from three independent biological replicates. (B) In vitro assays for release of LPS from proteoliposomes containing LptB2CFG complexes with different LptB variants were performed in the presence of either ATP or nonhydrolyzable AMPPNP. After 60 min, LPS was cross-linked to LptAI36pBpa and the resulting LPS-LptAI36pBpa adducts (marked “LptA × LPS”) were detected by LPS immunoblot (top panel). Total levels of LptAI36pBpa in the assay were confirmed by LptA immunoblotting (bottom panel). We also did not detect LPS transport by complexes containing LptBWT or LptBR144H/F239A in using the nonhydrolyzable ATP analogs α,β-methylene ATP (ApCpp), 2-chloroadenosine β, γ-methylene ATP (AppCp), ATP-γS, and AppNH2. Download FIG S7, PDF file, 0.3 MB (342.6KB, pdf) .
Copyright © 2019 Simpson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Although the severe defect in LPS extraction displayed by complexes containing LptBF239A correlated with its inability to complement in vivo, we were surprised that those containing LptBR144H showed only a moderate defect, given that lptB(R144H) is a conditionally lethal allele. Since the aforementioned experiments were conducted under ATP-saturating conditions, we next investigated the ability of LptB2FGC complexes to bind and hydrolyze ATP in the presence of differing concentrations of ATP (Fig. 6D). Complexes containing LptBR144H yielded Vmax values similar to those yielded by wild-type complexes, but their Km values were higher, indicating that the binding affinity of LptBR144H for ATP is reduced. Notably, unlike the levels seen with wild-type complexes, the Km of LptBR144H complexes is above typical cellular ATP concentrations (37, 38). In contrast, the Km values of LptBF239A and LptBR144H/F239A complexes were similar to those of wild-type complexes, but their Vmax values were reduced by approximately half, indicating a defect in catalysis.
Given that LptBF239A and LptBR144H/F239A complexes were similarly defective in ATP hydrolysis and that lptB(F239A) cells are nonviable whereas lptB(R144H/F239A) cells are wild-type-like, we tested whether the R144H substitution could bypass the need for LptB to hydrolyze ATP. To test this in vivo, we generated new mutant lptB alleles in which the E163Q change, which abolishes ATP hydrolysis (11, 15), was combined with R144H and R144H/F239A substitutions. None of these alleles complemented the loss of native lptB (Fig. 6E). Furthermore, in vitro, LptBR144H/F239A complexes could not extract LPS from liposomes in the presence of various nonhydrolyzable ATP analogs (Fig. S7B). Therefore, in order to function, wild-type-like LptBR144H/F239A hydrolyzes about half the amount of ATP as LptBWT.
Our genetic data demonstrate that the R144H and F239A substitutions have opposite, compensating effects on LptB function. The biochemical data show that the R144H substitution in the signature helix decreases the ability of LptB to bind ATP, while the F239A substitution in the CTD reduces its ability to hydrolyze ATP, but that combining the two substitutions suppresses defects in LPS transport by fixing the ATP binding defect without restoring the defect in ATP hydrolysis.
DISCUSSION
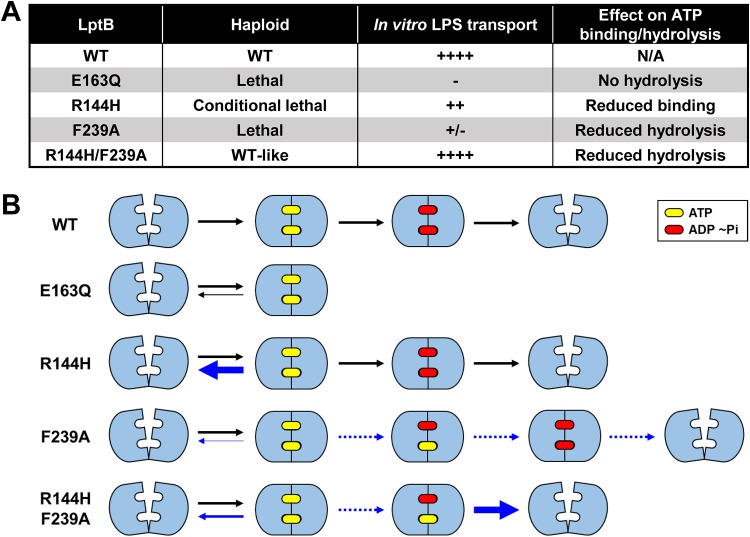
We have found a pair of lethal mutations in lptB that individually impair two distinct steps of the ATP hydrolysis cycle but that together restore wild-type levels of LPS transport and cellular growth. Biochemical experiments showed that, individually, one of these changes (R144H) results in impaired ATP binding, as judged by an increase in Km, while the other (F239A) causes impaired turnover, as judged by a decrease in Vmax. Restoration of LPS transport in complexes containing both changes (LptBR144H/F239A) requires rescue only of ATP-binding activity (Km) and not of ATP hydrolysis activity (Vmax). The simplest explanation of these results is that LPS transport by wild-type LptB2FGC is normally driven by the defective step that becomes suppressed in LptBR144H/F239A complexes, namely, ATP binding. If ATP hydrolysis determined LPS extraction, LptBR144H/F239A complexes should still be defective in LPS transport because they remain defective in ATP hydrolysis. We therefore propose that ATP binding triggers the collapse of the LptFG cavity that moves LPS to the periplasmic bridge, while ATP hydrolysis is needed to return the LptB2FGC transporter to the ground state to initiate another LPS extraction.
The restoration of LPS transport in LptBR144H/F239A must be driven by the addition of opposing defects caused by each change. LptBR144H has impaired ATP binding such that it is easier to open the ATP-bound dimer (see bold blue arrow in Fig. 7B). It follows that the F239A change can fix (suppress) the defect conferred by the R144H change because it stabilizes the conformation in which the LptB dimer is closed around ATP (see thin blue arrow in Fig. 7B). This conclusion is supported by the kinetic characterization of LptBF239A. Moreover, the fact that the low Vmax of LptBF239A was not suppressed by the R144H change implies that this catalytic defect (see dotted blue arrows in Fig. 7B) cannot be caused simply by the increased stabilization of the ATP-bound state; instead, LptBF239A must have an additional yet-to-be determined defect. Nevertheless, the lowered dimer stability conferred by the R144H change allows LptBR144H/F239A to reopen after hydrolysis of a single ATP molecule (see bold blue arrow for double mutant in Fig. 7B). It is formally possible that the double LptBR144H/F239A mutant complex eliminated the requirement of ATP hydrolysis for LPS transport; however, all attempts to observe transport with nonhydrolyzable ATP analogues or in the presence of the catalytically dead E163Q change were unsuccessful.
FIG 7.
Model for the effects of the R144H and F239A changes on LptB dimerization. (A) Summary of relevant phenotypes to consider for the model shown in panel B. In the absence of ATP, the wild-type LptB (WT) dimers are in the open-dimer state. Upon binding to ATP, the LptB dimer closes; ATP hydrolysis at both ATP-binding sites is then needed to open the dimer such that the transporter returns to its ground state and initiates another transport cycle. The LptBE163Q variant can bind ATP, but it cannot hydrolyze it. We propose that a complex containing LptBE163Q can undergo only one round of transport and stalls in a closed-dimer state. The LptBR144H variant has lower affinity for ATP, disfavoring the closed-dimer state, and therefore shows an increased tendency to reopen (indicated by reverse blue arrow); consequently, transporters with LptBR144H do not function efficiently, and haploid lptB(R144H) mutants are viable only under conditions of slow growth. In contrast, LptBF239A has a defect that favors the closed-dimer state (thinner blue reverse arrow) and decreases the rate of the subsequent ATP hydrolysis reactions such that hydrolysis occurs at only one site during a time span that would normally allow it to occur at both sites. Because hydrolysis of both ATP molecules is required to open the LptB dimer and because LptBF239A favors the closed-dimer state (dotted blue lines), the resetting of the transporter is highly defective; consequently, complexes with LptBF239A are defective in both ATP hydrolysis and LPS transport. In the LptBR144H/F239A complexes, the opposite effects of the R144H and F239A substitutions on the stability of the nucleotide-bound states compensate for each other. As a result, the instability of the ATP-bound closed-dimer state caused by R144H is somewhat lowered by the higher stability of the closed-dimer state induced by the F239A change (thinner blue arrow); in addition, the miscoordination of the hydrolysis activities at the two ATP-binding sites and the resulting difficulty in opening an ATP-ADP dimer caused by the F239A substitution are compensated for by the lower stability of the closed-dimer state caused by the R144H change. Thus, the R144H/F239A combination allows the complexes to reset to the open-dimer state without fixing the defect in the ATP hydrolysis rate such that the double mutant complexes hydrolyze less ATP when transporting LPS.
Further, upon being tested for dominance in merodiploid strains, production of LptBF239A (or other total-loss-of-function CTD-defective variants) in the presence of wild-type LptB resulted in greater defects in LPS transport than coproduction of the catalytically dead LptBE163Q and wild-type LptB. This difference implies that CTD defects cause a worse defect than simply that of killing catalytic activity. The simplest explanation is that, while LptBE163Q cannot hydrolyze ATP, mixed LptBWT-LptBE163Q complexes can turn over (i.e., open the dimer) at a higher rate than LptBWT-LptBF239A complexes. This difference in the levels of dominance of the defective variants illustrates that LptB2FGC can, in certain circumstances, turn over with defective ATP hydrolysis and that LptBF239A cannot be defective only in ATP hydrolysis. Indeed, these results are consistent with the idea that the F239A change results in an overly stable dimer that is defective in turnover, even in mixed LptBWT-LptBF239A complexes.
Our studies have identified interdomain interactions that functionally connect the distant signature helix and the CTD in LptB. Key features identified to be important here are conserved in other ABC transporters. For example, residue Q140 of MalK, the NBD of the MalK2FG maltose importer, is equivalent to R144 in LptB, and substitutions at Q140 in MalK result in nonfunctional transporters owing to the presence of dimers that are trapped in either an open or a closed conformation (39, 40). In addition, we found that the D-loop helix is central in the network that functionally links the signature helix containing R144 and the CTD loop containing F239 in LptB. In the TAP (transporter associated with antigen processing) ABC transporter, a change in the D loop was shown to decrease affinity of the NBD dimer (36), while its D-loop helix was also demonstrated to play a critical role in the dimerization of the NBDs (41). Additional studies will be needed to better understand how the network of functional interactions that we have uncovered couples the movement of LptFG with the dimerization state of LptB, as well as how conserved their role is across ABC transporters.
MATERIALS AND METHODS
Strains and growth conditions.
All strains are listed in Data set S1 in the supplemental material and were derived, unless otherwise indicated, from NR754, an ara+ derivative of MC4100 (42, 43). Cells were grown with aeration at 37°C in LB or M63 minimal media with 0.2% (wt/vol) glucose. Plates were prepared with 1.5% (wt/vol) agar, and top agar was prepared with 0.75% (wt/vol) agar. When applicable, the medium was prepared with the addition of ampicillin (125 μg/ml), isopropyl-β-d-1-thiogalactopyranoside (IPTG; 0.16 mM), kanamycin (30 μg/ml), novobiocin (5 or 33 μg/ml), tetracycline (25 μg/ml), vancomycin (75 μg/ml), or X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; 33 μg/ml).
Mutant allele construction.
Mutant alleles of lptB were constructed in pET23/42LptB by site-directed mutagenesis (SDM) PCR and confirmed by DNA sequencing with the primer T7 seq (15). SDM PCR was performed with PfuTurbo DNA polymerase and primers designed per the protocol specified for QuikChange (Agilent Technologies, Inc., Santa Clara, CA). All primers are listed in Data set S1. PCR products were then digested with DpnI (New England Biolabs, Ipswich, MA) and electroporated into DH5α competent cells.
Functional analysis of mutant alleles.
Functionality of lpt alleles was assessed as previously described (44). Briefly, pET23/42LptB-carried mutant alleles were transformed into strain NR2050 carrying a chromosomal lptB deletion and a wild-type copy of lptB on segregation-defective plasmid pRC7 (15, 45). Transformants were selected on LB containing ampicillin, IPTG, and X-Gal. Without selective pressure, the pRC7-derived plasmid is easily lost, and that loss is monitored by detection of the presence of an encoded lacZ gene (45). Mutant alleles that could sustain cell viability lost the pRC7-derived plasmid in the resulting strain and had white colonies (LacZ−). In contrast, loss-of-function alleles yielded only blue colonies (LacZ+) because the strain required the pRC7-carried copy of lptB. Functionality was checked on LB and glucose minimal solid media. Mutant alleles that were unable to complement deletion of chromosomal lptB on minimal medium were transformed into NR754 to generate lptB merodiploid strains and to assess dominance.
OM permeability assays.
Sensitivity to hydrophobic antibiotics (bacitracin, novobiocin, erythromycin, and rifampin) was used to assess OM permeability of strains that were haploid or merodiploid with respect to lptB with pET2342LptB-encoded mutant alleles. Disc diffusion assays were performed as described previously (15).
Immunoblotting of LptB variants.
Strains were grown overnight in glucose minimal medium or LB broth as indicated, and samples were normalized for differences in cell density. The resulting samples were subjected to electrophoresis on 12% SDS polyacrylamide gels. Proteins were transferred to polyvinylidene difluoride (PVDF) membranes (Roche Diagnostics, Basel, Switzerland) and probed with anti-LptB antisera (1:10,000 dilution) followed by anti-rabbit horseradish peroxidase (HRP) antibodies (GE Amersham, Chicago, IL) (1:10,000 dilution). Clarity Western ECL substrate (Bio-Rad, Hercules, CA) was used to develop signal and a ChemiDoc XRS+ system, and ImageLab 5.2.1 software (Bio-Rad) was used for imaging.
Suppressor analysis of lptB1.
Several cultures of NR1768, which contains the lptB1 allele, were grown overnight in LB as previously described (27). From each culture, a 100-μl volume was plated onto LB agar containing either novobiocin (33 μg/ml) or vancomycin (75 μg/ml). A kanamycin resistance cassette immediately downstream of lptB1 was used to test for linkage to lptB1 by P1vir transduction. When the suppressor phenotype was 98% to 100% linked to the kanamycin resistance cassette, this suggested the suppressor mutations were located in lptB1. PCR was performed to amplify the chromosomal locus with primers 5LptB77up and 3LptB50down. The resulting PCR products were sequenced. Each of the suppressors described here had a single-base-pair missense mutation in lptB, including the following: a G98C mutation resulting in a G33A substitution, a G97T mutation resulting in a G33C substitution, a T104A mutation resulting in an L35Q substitution, a C128G mutation resulting in a T43S substitution, a A133G mutation resulting in a T45A substitution, a G431A mutation resulting in an R144H substitution, a T443C mutation resulting in an I148T substitution, and a C728A mutation resulting in a stop codon after codon 242.
Construction of pET28b(+)-His8TEV-LptB(E163Q).
pCDFduet-His6LptB(E163Q)-LptFG (15) was modified to lengthen the His tag and to include a tobacco etch virus (TEV) protease cleavage site by use of the primers His8TEV-LptB-fwd and His8TEV-LptB-rev (Data set S1). Restriction enzymes NcoI and EcoRI were used to transfer the gene encoding His8TEV-LptB(E163Q) into pET28b+.
Expression, purification, and crystallization of His-LptB(E163Q).
Overnight cultures of KRX E. coli containing pET28b(+)-His8TEV-LptB(E163Q) were diluted 1:100 into fresh LB (Miller) with 50 μg/ml kanamycin, grown at 37°C to an optical density at 600 nm (OD600) of approximately 1.0, and then cooled to 16°C, at which point expression was induced by addition of 0.05% (wt/vol) l-rhamnose monohydrate (Sigma-Aldrich, St. Louis, MO) and 100 μM IPTG. After 18 h of expression, cells were collected by centrifugation at 4,200 × g and 4°C for 20 min. Cells from a 1.5-liter culture were resuspended in 40 ml buffer A (150 mM NaCl, 20 mM Tris [pH 8], 20% [vol/vol] glycerol, 0.5 mM dithiothreitol [DTT]; Sigma-Aldrich), supplemented with 20 mM imidazole, 0.5% (wt/vol) n-dodecyl β-d-maltoside (DDM; Anatrace, Maumee, OH), 2 mM ATP (Sigma-Aldrich), 1 mM phenylmethylsulfonyl fluoride (PMSF; Sigma-Aldrich), 100 μg/ml lysozyme (Sigma-Aldrich), and 50 μg/ml DNase I (bovine; Sigma-Aldrich). Cells were lysed by 3 passages through an Emulsiflex C3 homogenizer (Avestin, Ottawa, CA) at 15,000 lb/in2. Unbroken cells were removed from the lysate by centrifugation at 12,000 × g and 4°C for 10 min, and then other insoluble debris was removed by ultracentrifugation at 100,000 × g and 4°C for 30 min. Nickel-nitrilotriacetic acid (Ni-NTA) affinity resin was preequilibrated with buffer A and 20 mM imidazole, and the clarified supernatant was rocked with the resin for 1 h at 4°C. After incubation and flowthrough of the clarified lysate, the resin was washed with 20 column volumes (cv) buffer A with 20 mM imidazole and then 15 cv buffer A with 40 mM imidazole. Protein was eluted with 3 cv buffer A with 200 mM imidazole and the eluate supplemented with 2 mM ATP and 1 mM EDTA (Sigma-Aldrich). The protein sample was concentrated using an Amicon 10-kDa-molecular-weight-cutoff centrifugation filter (EMD Millipore, Burlington, MA) and further purified by gel filtration using a Superdex 200 10/300 GL column (GE Healthcare, Chicago, Illinois) with a mixture consisting of 150 mM NaCl, 20 mM Tris (pH 8), 20% (vol/vol) glycerol, and 0.5 mM Tris(3-hydroxypropyl)phosphine (THP; EMD Millipore). Peak fractions containing purified protein were pooled, supplemented with 2 mM ATP and 1 mM EDTA, concentrated to 20 mg/ml, and then either flash frozen and stored at –80°C or diluted 1:1 with buffer containing 150 mM NaCl, 20 mM Tris (pH 8), 0.5 mM THP, 2 mM ATP, and 1 mM EDTA. The diluted protein samples (∼10 mg/ml) were used to set crystallization trials using previously published conditions (15). The best diffracting crystals appeared in hanging drops set with a 1:1 protein-to-reservoir ratio, with the reservoir containing 28% polyethylene glycol (PEG) 4000 (Sigma-Aldrich), 100 mM MES (morpholineethanesulfonic acid; Tocris Biosciences, Bristol, United Kingdom) (pH 6.75), and 600 mM NaCl. After 3 weeks of growth, crystals were harvested, cryoprotected with a mixture containing 28% PEG 4000, 100 mM MES (pH 6.75), 600 mM NaCl, 20% glycerol, 2 mM ATP, and 1 mM EDTA, and then flash-frozen in liquid nitrogen.
Data collection, data processing, and structure refinement.
X-ray diffraction data were collected at 0.97918 Å on beamline 24-ID-E at Argonne National Laboratory. The His8-LptBE163Q crystals belonged to space group C2221. Data were indexed and integrated in XDS (46) or iMosflm (47) and were then scaled in the CCP4 suite (48) program AIMLESS (49). Molecular replacement was performed using Phaser (50) with the ATP-bound LptBE163Q-EHis8 structure (PDB 4P33) (15). Manual building was performed using Coot (51), while automated refinement was performed in Phenix (52). Drops of His8-LptBE163Q were crystallized in the presence of ATP, and the structure of the inactive variant contained high electron density corresponding to ATP. This is consistent with previous studies of LptB (15, 27). All software was accessed through the SBGrid consortium (53). Data collection and refinement statistics are given in Fig. S3A in the supplemental material.
Purification of LptB2FGC complexes.
LptB2FGC complexes containing different LptB variants were purified as previously described, with slight modifications (54). Overnight cultures of BL21(λDE3) E. coli containing pCDFduet-LptB-LptFG (wild-type or mutant LptB, as applicable) and pET22/42-LptC-His7 were diluted 1:100 into LB containing carbenicillin and spectinomycin. Cells were grown at 37°C to an OD600 of ∼0.8 and were then cooled to 18°C. IPTG (200 μM) was added after 20 min, and cells were grown overnight. Cells were harvested by centrifugation (4,200 × g, 20 min), resuspended in lysis buffer (50 mM Tris [pH 7.4], 300 mM NaCl, 1 mM PMSF, 100 μg/ml lysozyme, 50 μg/ml DNase I), homogenized, and subjected to passage through an EmulsiFlex-C3 high-pressure cell disruptor 3 times. The cell lysate was centrifuged (10,000 × g, 10 min), and the supernatant was further centrifuged (100,000 × g, 45 min). The resulting pellets were resuspended and solubilized in solubilization buffer (20 mM Tris [pH 7.4], 300 mM NaCl, 10% glycerol, 5 mM MgCl2, 1% [wt/vol] DDM [Anatrace Maumee, OH], 50 μM PMSF, 15 mM imidazole, 2 mM ATP) and rocked at 4°C for 1 h. The mixture was centrifuged (100,000 × g, 30 min) and the supernatant was rocked with Ni-NTA Superflow resin (Qiagen) for 1 h. The resin was then washed with affinity buffer (20 mM Tris [pH 7.4], 300 mM NaCl, 10% glycerol, 0.01% DDM, 0.01% LMNG [lauryl maltose neopentyl glycol; Anatrace]) containing 35 mM imidazole. Protein was eluted with affinity buffer containing 200 mM imidazole, concentrated using a 100-kDa-molecular-weight-cutoff Amicon Ultra centrifugal filter (Millipore), and purified by size exclusion chromatography on a Superdex 200 increase column in buffer containing 20 mM Tris-HCl (pH 7.4), 300 mM NaCl, 0.005% (wt/vol) LMNG, and 10% glycerol. The His tag was cleaved by overnight incubation with restriction-grade thrombin (Sigma). The solution was spiked with 8 mM imidazole, and the uncleaved protein was removed by passage through Ni-NTA resin and benzamidine Sepharose. Purified protein was either used within 24 h or flash frozen and stored at –80°C.
Purification of LptAI36pBPA.
LptAI36pBPA was purified as previously described, with slight modifications (10). Briefly, Bl21 (λDE3) E. coli cells containing pSup-BpaRS-6TRN and pET22b-LptA(I36Am) were grown to an OD600 of ∼0.6 at 37°C in LB media containing 50 μg/ml carbenicillin, 30 μg/ml chloramphenicol, and 0.8 mM pBPA (BaChem, Bubendorf, Switzerland). Cells were then induced with 50 μM IPTG; allowed to grow for 2 h; harvested; resuspended in a mixture containing 50 mM Tris-HCl (pH 7.4), 250 mM sucrose, and 3 mM EDTA; incubated on ice for 30 min; and pelleted (6,000 × g, 10 min). The supernatant was supplemented with 1 mM PMSF and 10 mM imidazole and pelleted (100,000 × g, 30 min). The supernatant was incubated with Ni-NTA resin, which was then washed twice (20 column volumes of 20 mM Tris-HCl [pH 8.0], 150 mM NaCl, 10% [vol/vol] glycerol, and 20 mM imidazole). LptA was eluted twice (2.5 column volumes of wash buffer supplemented with an additional 180 mM imidazole), concentrated using a 10-Da-cutoff Amicon centrifugal concentrator (Millipore), flash frozen, and stored at –80°C until use.
Preparation of LptB2FGC liposomes.
Liposomes were prepared using a detergent dilution method as previously described (11). Aqueous E. coli polar lipid extract (Avanti Polar Lipids Inc.) (30 mg/ml) and aqueous LPS from E. coli EH100 (Ra mutant; Sigma) (2 mg/ml) were sonicated briefly for homogenization. A mixture of 20 mM Tris-HCl (pH 8.0), 150 mM NaCl, 7.5 mg/ml E. coli polar lipids, 0.5 mg/ml LPS, and 0.25% DDM was prepared and kept on ice for 10 min. Purified LptB2FGC was added to a final concentration of 0.86 μM, and the mixture was left on ice for 20 min. The mixture was diluted 100-fold with cold 20 mM Tris-HCl (pH 8.0) and 150 mM NaCl and kept on ice for 20 min. The proteoliposomes were pelleted (300,000 × g, 2 h, 4°C), resuspended in 20 mM Tris-HCl (pH 8.0) and 150 mM NaCl, diluted 100×, and centrifuged (300,000 × g, 2 h, 4°C). The pellets were resuspended in a mixture of 20 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 10% glycerol (250 μl per 100 μl of the original predilution solution), homogenized by sonication, flash frozen, and stored at –80°C until use.
LPS release assay.
The levels of release of LPS from proteoliposomes to LptA were measured as previously described, with slight modifications (11). Assays used 60% proteoliposomes (by volume) in a solution containing 50 mM Tris-HCl (pH 8.0), 500 mM NaCl, 10% glycerol, and 2 μM LptAI36pBPA. Reactions were initiated by the addition of ATP (or nonhydrolyzable ATP analogues ATP and AMP-PNP [adenylyl-imidodiphosphate] from Sigma, St. Louis, MO; all other analogues were from Jenna Bioscience, Jena, Germany) and MgCl2 (final concentrations of 5 mM and 2 mM, respectively) and proceeded at 30°C. Aliquots (33 μl) were removed from the reaction mixtures and irradiated with UV light (365 nm) on ice for 10 min using a B-100AP lamp (Fisher Scientific, Hampton, NH). Following UV irradiation, samples were added to a mixture consisting of 297 μl cold 20 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 0.2% DDM. To precipitate the protein, 330 μl 20% trichloroacetic acid (Sigma) was added, after which the precipitated protein was collected by centrifugation. The precipitates were resuspended in 50 μl 1:1 2× SDS-PAGE sample loading buffer supplemented with 2% β-mercaptoethanol–1 M Tris-HCl (pH 8.0) and boiled for 10 min, and proteins were separated using Tris-HCl 4% to 20% polyacrylamide gradient gels with Tris-glycine running buffer. Proteins were transferred onto Immun-Blot PVDF membranes (Bio-Rad). Mouse monoclonal antiserum against the LPS core (Hycult Biotechnology, Uden, The Netherlands), rabbit anti-LptA antisera (11), sheep anti-mouse horseradish peroxidase (HRP) conjugate secondary antibody (GE Amersham), and donkey anti-rabbit HRP conjugate secondary antibody (GE Amersham) were used for the immunoblots. Bands were visualized using ECL Prime Western blotting detection reagent (GE Amersham) and an Azure c400 imaging system.
ATPase assay.
ATPase assays were done using a modified molybdate method as previously reported (11). Assays used 60% proteoliposomes (by volume) in a mixture containing 50 mM Tris-HCl (pH 8.0), 500 mM NaCl, and 10% glycerol. Reactions were initiated by the addition of ATP and MgCl2 (final concentrations of 5 mM and 2 mM, respectively, unless otherwise indicated) and run at 30°C. Aliquots (25 μl) were taken at 0, 20, 40, and 60 min. Reactions were quenched with an equal volume of 12% sodium dodecyl sulfate (SDS). The amounts of Pi were determined using a colorimetric method, and potassium phosphate was used as a standard (55). Reagents were obtained from Sigma-Aldrich. After the addition of SDS, a mixture containing 50 μl of 30 mg/ml ascorbic acid, 0.5 N HCl, 5 mg/ml ammonium molybdate, and 6% SDS was added. The samples were incubated at room temperature for 7 min, and 75 μl of an aqueous solution containing 20 mg/ml sodium citrate tribasic dihydrate, 2 mg/ml sodium arsenite, and 2% (vol/vol) acetic acid was added. The absorbance at 850 nm was measured using a Spectramax Plus 384 plate reader (Molecular Devices, CA) after 20 min. Error bars indicate the standard deviations of the average rates measured over 3 biological replicates.
ATPase assays for measuring Vmax and Km.
Assays were performed as described above for the ATPase assays done under LPS release conditions with the following modifications. Assays included 30% proteoliposomes (by volume) in a mixture containing 50 mM Tris-HCl (pH 8.0), 500 mM NaCl, and 10% glycerol and were initiated by the addition of ATP and MgCl2 (the final concentration of ATP was adjusted as indicated; 2 mM final concentration of MgCl2) and run at 33°C. Aliquots (5 μl) were taken at 0, 30, 60, and 90 min. They were quenched with an equal volume of 12% sodium dodecyl sulfate (SDS). The amounts of Pi were determined using a published colorimetric method, and potassium phosphate was used as a standard as described above and previously (55). Error bars indicated the standard deviations of the rates measured over 3 biological replicates.
Data availability.
The atomic coordinates and structure factors have been deposited at the RCSB Protein Data Bank with accession number 6MBN.
ACKNOWLEDGMENTS
This work was supported by grants R01-GM100951 to N.R. and R01 GM066174 and R01 AI081059 to D.K. The content is solely our responsibility and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Citation Simpson BW, Pahil KS, Owens TW, Lundstedt EA, Davis RM, Kahne D, Ruiz N. 2019. Combining mutations that inhibit two distinct steps of the ATP hydrolysis cycle restores wild-type function in the lipopolysaccharide transporter and shows that ATP binding triggers transport. mBio 10:e01931-19. https://doi.org/10.1128/mBio.01931-19.
Contributor Information
Matthew R. Chapman, University of Michigan—Ann Arbor.
Alessandra Polissi, University of Milan.
Steven Rutherford, Genentech, Inc.
REFERENCES
- 1.Nikaido H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67:593–656. doi: 10.1128/mmbr.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang G, Meredith TC, Kahne D. 2013. On the essentiality of lipopolysaccharide to Gram-negative bacteria. Curr Opin Microbiol 16:779–785. doi: 10.1016/j.mib.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Powers MJ, Trent MS. 2018. Expanding the paradigm for the outer membrane: Acinetobacter baumannii in the absence of endotoxin. Mol Microbiol 107:47–56. doi: 10.1111/mmi.13872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertani B, Ruiz N. 1 August 2018, posting date. Function and biogenesis of lipopolysaccharides. EcoSal Plus 2018 doi: 10.1128/ecosalplus.ESP-0001-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simpson BW, Trent MS. 2019. Pushing the envelope: LPS modifications and their consequences. Nat Rev Microbiol 17:403–416. doi: 10.1038/s41579-019-0201-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andolina G, Bencze LC, Zerbe K, Muller M, Steinmann J, Kocherla H, Mondal M, Sobek J, Moehle K, Malojcic G, Wollscheid B, Robinson JA. 2018. A peptidomimetic antibiotic interacts with the periplasmic domain of LptD from Pseudomonas aeruginosa. ACS Chem Biol 13:666–675. doi: 10.1021/acschembio.7b00822. [DOI] [PubMed] [Google Scholar]
- 7.Robinson JA. 2019. Folded synthetic peptides and other molecules targeting outer membrane protein complexes in Gram-negative bacteria. Front Chem 7:45. doi: 10.3389/fchem.2019.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Srinivas N, Jetter P, Ueberbacher BJ, Werneburg M, Zerbe K, Steinmann J, Van der Meijden B, Bernardini F, Lederer A, Dias RL, Misson PE, Henze H, Zumbrunn J, Gombert FO, Obrecht D, Hunziker P, Schauer S, Ziegler U, Kach A, Eberl L, Riedel K, DeMarco SJ, Robinson JA. 2010. Peptidomimetic antibiotics target outer-membrane biogenesis in Pseudomonas aeruginosa. Science 327:1010–1013. doi: 10.1126/science.1182749. [DOI] [PubMed] [Google Scholar]
- 9.Okuda S, Sherman DJ, Silhavy TJ, Ruiz N, Kahne D. 2016. Lipopolysaccharide transport and assembly at the outer membrane: the PEZ model. Nat Rev Microbiol 14:337–345. doi: 10.1038/nrmicro.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okuda S, Freinkman E, Kahne D. 2012. Cytoplasmic ATP hydrolysis powers transport of lipopolysaccharide across the periplasm in E. coli. Science 338:1214–1217. doi: 10.1126/science.1228984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sherman DJ, Xie R, Taylor RJ, George AH, Okuda S, Foster PJ, Needleman DJ, Kahne D. 2018. Lipopolysaccharide is transported to the cell surface by a membrane-to-membrane protein bridge. Science 359:798–801. doi: 10.1126/science.aar1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruiz N, Gronenberg LS, Kahne D, Silhavy TJ. 2008. Identification of two inner-membrane proteins required for the transport of lipopolysaccharide to the outer membrane of Escherichia coli. Proc Natl Acad Sci U S A 105:5537–5542. doi: 10.1073/pnas.0801196105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sperandeo P, Cescutti R, Villa R, Di Benedetto C, Candia D, Deho G, Polissi A. 2007. Characterization of lptA and lptB, two essential genes implicated in lipopolysaccharide transport to the outer membrane of Escherichia coli. J Bacteriol 189:244–253. doi: 10.1128/JB.01126-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sperandeo P, Lau FK, Carpentieri A, De Castro C, Molinaro A, Deho G, Silhavy TJ, Polissi A. 2008. Functional analysis of the protein machinery required for transport of lipopolysaccharide to the outer membrane of Escherichia coli. J Bacteriol 190:4460–4469. doi: 10.1128/JB.00270-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sherman DJ, Lazarus MB, Murphy L, Liu C, Walker S, Ruiz N, Kahne D. 2014. Decoupling catalytic activity from biological function of the ATPase that powers lipopolysaccharide transport. Proc Natl Acad Sci U S A 111:4982–4987. doi: 10.1073/pnas.1323516111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Owens TW, Taylor RJ, Pahil KS, Bertani BR, Ruiz N, Kruse AC, Kahne D. 2019. Structural basis of unidirectional export of lipopolysaccharide to the cell surface. Nature 567:550–553. doi: 10.1038/s41586-019-1039-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, Orlando BJ, Liao M. 2019. Structural basis of lipopolysaccharide extraction by the LptB2FGC complex. Nature 567:486–490. doi: 10.1038/s41586-019-1025-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davidson AL, Dassa E, Orelle C, Chen J. 2008. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol Mol Biol Rev 72:317–364. doi: 10.1128/MMBR.00031-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Locher KP. 2016. Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat Struct Mol Biol 23:487–493. doi: 10.1038/nsmb.3216. [DOI] [PubMed] [Google Scholar]
- 20.Saurin W, Hofnung M, Dassa E. 1999. Getting in or out: early segregation between importers and exporters in the evolution of ATP-binding cassette (ABC) transporters. J Mol Evol 48:22–41. doi: 10.1007/PL00006442. [DOI] [PubMed] [Google Scholar]
- 21.Thomas C, Tampe R. 2018. Multifaceted structures and mechanisms of ABC transport systems in health and disease. Curr Opin Struct Biol 51:116–128. doi: 10.1016/j.sbi.2018.03.016. [DOI] [PubMed] [Google Scholar]
- 22.Szollosi D, Rose-Sperling D, Hellmich UA, Stockner T. 2018. Comparison of mechanistic transport cycle models of ABC exporters. Biochim Biophys Acta Biomembr 1860:818–832. doi: 10.1016/j.bbamem.2017.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Narita S, Tokuda H. 2009. Biochemical characterization of an ABC transporter LptBFGC complex required for the outer membrane sorting of lipopolysaccharides. FEBS Lett 583:2160–2164. doi: 10.1016/j.febslet.2009.05.051. [DOI] [PubMed] [Google Scholar]
- 24.Sherman DJ, Okuda S, Denny WA, Kahne D. 2013. Validation of inhibitors of an ABC transporter required to transport lipopolysaccharide to the cell surface in Escherichia coli. Bioorg Med Chem 21:4846–4851. doi: 10.1016/j.bmc.2013.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong H, Zhang Z, Tang X, Paterson NG, Dong C. 2017. Structural and functional insights into the lipopolysaccharide ABC transporter LptB(2)FG. Nat Commun 8:222. doi: 10.1038/s41467-017-00273-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo Q, Yang X, Yu S, Shi H, Wang K, Xiao L, Zhu G, Sun C, Li T, Li D, Zhang X, Zhou M, Huang Y. 2017. Structural basis for lipopolysaccharide extraction by ABC transporter LptB2FG. Nat Struct Mol Biol 24:469–474. doi: 10.1038/nsmb.3399. [DOI] [PubMed] [Google Scholar]
- 27.May JM, Owens TW, Mandler MD, Simpson BW, Lazarus MB, Sherman DJ, Davis RM, Okuda S, Massefski W, Ruiz N, Kahne D. 2017. The antibiotic novobiocin binds and activates the ATPase that powers lipopolysaccharide transport. J Am Chem Soc 139:17221–17224. doi: 10.1021/jacs.7b07736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z, Xiang Q, Zhu X, Dong H, He C, Wang H, Zhang Y, Wang W, Dong C. 2014. Structural and functional studies of conserved nucleotide-binding protein LptB in lipopolysaccharide transport. Biochem Biophys Res Commun 452:443–449. doi: 10.1016/j.bbrc.2014.08.094. [DOI] [PubMed] [Google Scholar]
- 29.Locher KP. 2009. Review. Structure and mechanism of ATP-binding cassette transporters. Philos Trans R Soc Lond B Biol Sci 364:239–245. doi: 10.1098/rstb.2008.0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yao Z, Davis RM, Kishony R, Kahne D, Ruiz N. 2012. Regulation of cell size in response to nutrient availability by fatty acid biosynthesis in Escherichia coli. Proc Natl Acad Sci U S A 109:E2561–E2568. doi: 10.1073/pnas.1209742109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bakos E, Klein I, Welker E, Szabó K, Müller M, Sarkadi B, Váradi A. 1997. Characterization of the human multidrug resistance protein containing mutations in the ATP-binding cassette signature region. Biochem J 323:777–783. doi: 10.1042/bj3230777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoof T, Demmer A, Hadam MR, Riordan JR, Tummler B. 1994. Cystic fibrosis-type mutational analysis in the ATP-binding cassette transporter signature of human P-glycoprotein MDR1. J Biol Chem 269:20575–20583. [PubMed] [Google Scholar]
- 33.Jones PM, George AM. 1999. Subunit interactions in ABC transporters: towards a functional architecture. FEMS Microbiol Lett 179:187–202. doi: 10.1111/j.1574-6968.1999.tb08727.x. [DOI] [PubMed] [Google Scholar]
- 34.Schmees G, Stein A, Hunke S, Landmesser H, Schneider E. 1999. Functional consequences of mutations in the conserved ‘signature sequence’ of the ATP-binding-cassette protein MalK. Eur J Biochem 266:420–430. doi: 10.1046/j.1432-1327.1999.00871.x. [DOI] [PubMed] [Google Scholar]
- 35.Shyamala V, Baichwal V, Beall E, Ames GF. 1991. Structure-function analysis of the histidine permease and comparison with cystic fibrosis mutations. J Biol Chem 266:18714–18719. [PubMed] [Google Scholar]
- 36.Grossmann N, Vakkasoglu AS, Hulpke S, Abele R, Gaudet R, Tampe R. 2014. Mechanistic determinants of the directionality and energetics of active export by a heterodimeric ABC transporter. Nat Commun 5:5419. doi: 10.1038/ncomms6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yaginuma H, Kawai S, Tabata KV, Tomiyama K, Kakizuka A, Komatsuzaki T, Noji H, Imamura H. 2015. Diversity in ATP concentrations in a single bacterial cell population revealed by quantitative single-cell imaging. Sci Rep 4:6522. doi: 10.1038/srep06522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schneider DA, Gourse RL. 2004. Relationship between growth rate and ATP concentration in Escherichia coli: a bioassay for available cellular ATP. J Biol Chem 279:8262–8268. doi: 10.1074/jbc.M311996200. [DOI] [PubMed] [Google Scholar]
- 39.Daus ML, Grote M, Muller P, Doebber M, Herrmann A, Steinhoff HJ, Dassa E, Schneider E. 2007. ATP-driven MalK dimer closure and reopening and conformational changes of the “EAA” motifs are crucial for function of the maltose ATP-binding cassette transporter (MalFGK2). J Biol Chem 282:22387–22396. doi: 10.1074/jbc.M701979200. [DOI] [PubMed] [Google Scholar]
- 40.Daus ML, Landmesser H, Schlosser A, Muller P, Herrmann A, Schneider E. 2006. ATP induces conformational changes of periplasmic loop regions of the maltose ATP-binding cassette transporter. J Biol Chem 281:3856–3865. doi: 10.1074/jbc.M511953200. [DOI] [PubMed] [Google Scholar]
- 41.Vakkasoglu AS, Srikant S, Gaudet R. 2017. D-helix influences dimerization of the ATP-binding cassette (ABC) transporter associated with antigen processing 1 (TAP1) nucleotide-binding domain. PLoS One 12:e0178238. doi: 10.1371/journal.pone.0178238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Casadaban MJ. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol 104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 43.Ruiz N, Wu T, Kahne D, Silhavy TJ. 2006. Probing the barrier function of the outer membrane with chemical conditionality. ACS Chem Biol 1:385–395. doi: 10.1021/cb600128v. [DOI] [PubMed] [Google Scholar]
- 44.Simpson BW, Owens TW, Orabella MJ, Davis RM, May JM, Trauger SA, Kahne D, Ruiz N. 2016. Identification of residues in the lipopolysaccharide ABC transporter that coordinate ATPase activity with extractor function. mBio 7:e01729-16. doi: 10.1128/mBio.01729-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Boer PA, Crossley RE, Rothfield LI. 1989. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell 56:641–649. doi: 10.1016/0092-8674(89)90586-2. [DOI] [PubMed] [Google Scholar]
- 46.Kabsch W. 2010. XDS. Acta Crystallogr D Biol Crystallogr 66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leslie AGW, Powell HR. 2007. Processing diffraction data with mosflm. In Read RJ, Sussman JL (eds), Evolving methods for macromolecular crystallography NATO Science Series, vol 245 Springer, Dordrecht. doi: 10.1007/978-1-4020-6316-9_4. [DOI] [Google Scholar]
- 48.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Evans PR, Murshudov GN. 2013. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr 69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morin A, Eisenbraun B, Key J, Sanschagrin PC, Timony MA, Ottaviano M, Sliz P. 2013. Collaboration gets the most out of software. Elife 2:e01456. doi: 10.7554/eLife.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mandler MD, Baidin V, Lee J, Pahil KS, Owens TW, Kahne D. 2018. Novobiocin enhances polymyxin activity by stimulating lipopolysaccharide transport. J Am Chem Soc 140:6749–6753. doi: 10.1021/jacs.8b02283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang G, Baidin V, Pahil KS, Moison E, Tomasek D, Ramadoss NS, Chatterjee AK, McNamara CW, Young TS, Schultz PG, Meredith TC, Kahne D. 2018. Cell-based screen for discovering lipopolysaccharide biogenesis inhibitors. Proc Natl Acad Sci U S A 115:6834–6839. doi: 10.1073/pnas.1804670115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, Bateman A. 2008. The Pfam protein families database. Nucleic Acids Res 36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence alignment of LptB and the ATPases of branched-chain amino acid transporters. Clustal W alignment of LptB homologs with closely related branched-chain amino acid transporter ATPases LivF/G shows that they share homology at their CTDs. The protein family PF12399 sequence is a conserved sequence found in both protein families and ending in a characteristic YLG sequence (1). The CTD of LptB has additional conserved residues not found in PF12399. Colors correspond to the percent identity color scheme in Jalview (2). Light to dark purple coloring indicates 50% to 100% identity, respectively, and white indicates <50% identity. The protein sequences aligned were those of LivF (WP_000416891.1), LivG (WP_136807488.1), and LptB (WP_000224099.1) from E. coli K-12 substrain MG1655 and LptB homologs from Geobacter uraniireducens Rfr and Burkholderia cenocepacia (WP_011939814.1 and WP_124633598.1, respectively). Download FIG S2, PDF file, 0.4 MB (429.3KB, pdf) .
Copyright © 2019 Simpson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(Page 2) Loss-of-function substitutions at the CTD of LptB confer dominant-negative phenotypes. (Page 3) OM permeability defects of haploid strains with mutations in lptB that disrupt the interaction between the switch helix and CTD loop. (Page 4) Genetic interactions between changes in R144 and R198 and alterations to the CTD of LptB. (Page 5) Genetic interactions between changes in Walker A residue T45 and alterations to the CTD of LptB. (Page 6) Strains used in this study. (Page 9) Primers used in this study. Download Data Set S1, PDF file, 0.3 MB (306.8KB, pdf) .
Copyright © 2019 Simpson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Altering the CTD of LptB results in functional defects. (A) The addition of CT extensions to LptB disrupts activity to differing degrees. A summary of functional analyses for various lptB haploid strains is presented. In investigating why some CT-tagged LptB variants could support growth while others could not, we noticed that the two types significantly differed at residue 242, which is located in the linker between the last residue (L241) of wild-type LptB and the tag. The lethal chromosomal lptB-His allele encodes a positively charged arginine, while the complementing plasmidborne lptB-EHis8 and chromosomal lptB1 alleles encode a negatively charged glutamate and a neutral isoleucine, respectively. To examine if the addition of an arginine after the native L241 residue affected LptB function on its own or in the context of the His tag, we constructed plasmids bearing lptB alleles encoding LptB followed by only an arginine (lptB-R) or an arginine followed by a polyhistidine tag (lptB-RHis8) and compared their function to that of wild-type lptB (lptB+) and lptB-EHis8 genes. The lptB-R and lptB-EHis8 alleles complemented chromosomal ΔlptB, but the resulting haploid strains were sensitive to hydrophobic antibiotics, indicating that these strains, like an lptB1 strain, are defective in LPS transport. In contrast, the lptB-RHis8 allele could not complement chromosomal ΔlptB despite resulting in the production of as much protein as an lptB-EHis8 strain (see below). Thus, adding an arginine or oligopeptide to the CTD of LptB causes partial loss-of-function defects, while the combination of an arginine and oligopeptide is lethal. (B) LptB immunoblot comparing strains that chromosomally encode LptBWT, labeled 754, or LptB1. (C) LptB immunoblot of samples from haploid strains bearing LptB CTD variants on the pET23/42-LptB plasmid. “WT” refers to haploid strain NR2101, which produces LptBWT from pET23/42-LptB. (D) LptB immunoblot of samples from merodiploid strains producing LptB CTD variants from the pET23/42-LptB plasmid and LptBWT from the native lptB locus. “754” refers to NR754, the wild-type strain with lptB+ at the native locus. “WT” refers to strain NR2583, which produces LptBWT from both the chromosome and pET23/42-LptB. Data are representative of results from at least three independent experiments. (E) Ability of CTD lptB alleles to complement a chromosomal ΔlptB allele in rich (LB) medium. (F) OM permeability of haploid strains producing LptB CTD variants was determined by disc diffusion assay. All alleles, except lptB+ in NR754 and lptB1, which are located on the chromosome, are borne on the pET23/42-LptB plasmid. Increased sensitivity was reproducibly observed for the strains shown and was indicative of LPS transport defects compared to wild-type lptB+ strain NR2101, while those not shown (containing changes V230S, K231A, R232A, V233T, E237A, D238A, F239Y, and R240A) behaved like wild-type strain NR2101. Numbers represent the diameter (in millimeters) of the zone of inhibition or that of partial growth (parentheses) around a 6-mm-diameter disc containing an antibiotic. Download FIG S1, PDF file, 0.3 MB (267.8KB, pdf) .
Copyright © 2019 Simpson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supplemental references for Fig. S2 and Data set S1. Download Text S1, PDF file, 0.1 MB (114.4KB, pdf) .
Copyright © 2019 Simpson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The CTD of LptB mediates intra- and intermonomer contacts through interactions with the switch helix. (A) Data collection and refinement statistics of PDB 6MBN structure of the ATP-bound His8-LptBE163Q dimer. (B) Cartoon representation of the ATP-bound His8-LptBE163Q dimer (PDB 6MBN) showing that the carboxylic acid of the last residue in LptB, L241, forms ionic pairs (blue dotted lines) with residue R198 in the switch helix both within each monomer and across monomers. A bottom view of the LptB dimer is presented in which LptB monomers are colored cyan (monomer A) and light gray (monomer B), the switch helix containing R198 is colored pink, and the CTD is colored red. R198 and L241 are shown as sticks. (C) Detailed view of the interactions between residues R198 and L241. LptB monomers are colored cyan (monomer A) and light gray (monomer B). Download FIG S3, PDF file, 0.6 MB (662.5KB, pdf) .
Copyright © 2019 Simpson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Structure-function analysis of lptB1 suppressors. (A and B) OM permeability assay (A) and LptB immunoblots (B) of chromosomally encoded lptB1 and suppressors performed as described in the Fig. S1 legend and in Materials and Methods. “lptB+” refers to strain NR754. (C) LptB immunoblot of samples from haploid strains producing LptB variants from pET23/42LptB derivatives performed as described in the Fig. S1 legend. (D) OM permeability assay of haploid cells carrying pET23/42LptB-EHis8 derivatives with the substitutions we found as suppressors of lptB1. Assays were performed as described in in the Fig. S1 legend. Data are representative of results from at least three independent experiments. Download FIG S4, PDF file, 0.2 MB (232.4KB, pdf) .
Copyright © 2019 Simpson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Location of lptB1 suppressors. (A and B) Cartoon representations of the structures of an His8-LptBE163Q dimer (A) and monomer (B) showing the location of the suppressors of lptB1 (represented as forest green spheres). The Walker A domain (cyan), signature motif (marine), and CTD (red) are colored in the X-ray structure. ATP is shown in yellow sticks. The location of additional motifs important for function of ABC transporters is also shown on the cartoons on the left. “WB” marks the Walker B motif, “DL” the D loop, “Sw” the switch loop, and “CH” a coupling helix of either LptF or LptG. Numbers in green shown in panel B represent the number of the residue altered in the suppressors. Download FIG S5, PDF file, 0.6 MB (584KB, pdf) .
Copyright © 2019 Simpson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Synergistic effects of mutations combined with lptB-EHis8 identifying a network of functionally related residues. (A) Sequence logo (generated as described in the Fig. 1D legend and Materials and Methods) of the conservation of residues in the signature, D-loop, and switch helices (locations of each are marked with thick lines). Below the logo, the sequence of E. coli LptBWT is shown in black and a summary of the results of functional analysis of single amino acid substitutions designed to disrupt the observed interactions between R144 and the CTD is presented. Substitutions in pET23/42-LptB conferring no defects in strains carrying a chromosomal ΔlptB deletion are shown in green, those conferring partial loss of function are shown in orange, and those resulting in a total loss of function are shown in purple. (B) Ability of mutations that alter the indicated residue in either pET23/42LptB or pET23/42LptB-EHis8 to complement a ΔlptB allele in the indicated media as described in the Fig. S1A legend. (C) OM permeability of haploid strains carrying lptB mutations that exhibit effects synergistic with those of lptB-EHis8. Disc diffusion assays were performed on LB plates. Numbers indicate diameter (in millimeters) of the zone of total growth inhibition or that of partial growth (in parentheses). (D) LptB immunoblot of haploid lptB-EHis8 strains carrying pET23/42-LptB-EHis8 grown in LB. (E) LptB immunoblot of samples from haploid lptB strains carrying pET23/42-LptB grown in minimal medium to allow growth of strains carrying conditionally lethal alleles. “WT” refers to NR2101, which produces LptBWT from pET23/42-LptB. (F) LptB immunoblot of samples from merodiploid strains carrying the wild-type lptB allele and mutant lptB alleles carried in pET23/42-LptB grown in LB. “754” refers to haploid strain NR754, the wild-type strain with chromosomal lptB+ and “WT” to NR2583, which produces LptBWT from both the chromosome and pET23/42-LptB. (G) LptB immunoblot of samples from merodiploid strains carrying the chromosomal wild-type lptB allele and mutant lptB-EHis8 alleles encoded in pET23/42-LptB-EHis8 grown in LB. “754” refers to haploid strain NR754; “WT” refers to NR1872, which produces LptBWT from the chromosome and LptB-EHis8 from pET23/42-LptB-EHis8. Data are representative of results from at least three independent experiments. Download FIG S6, PDF file, 0.3 MB (276.8KB, pdf) .
Copyright © 2019 Simpson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ATP hydrolysis is required for LPS release from proteoliposomes. (A) Complexes containing the catalytically dead LptBE163Q variant are defective in extracting LPS from liposomes. An assay monitoring LPS release from liposomes containing LptB2FGC complexes with LptBWT and catalytically dead LptBE163Q variants was performed. Released LPS was UV-crosslinked to LptAI36pBpa, and the resulting LptAI36pBpa x LPS adducts were detected by LPS immunoblotting (top panel). Assays were initiated by the addition of ATP and incubation at 30°C for the indicated amount of time prior to crosslinking. LptA immunoblots are shown that confirm total LptA levels (bottom panel). Data are representative of results from three independent biological replicates. (B) In vitro assays for release of LPS from proteoliposomes containing LptB2CFG complexes with different LptB variants were performed in the presence of either ATP or nonhydrolyzable AMPPNP. After 60 min, LPS was cross-linked to LptAI36pBpa and the resulting LPS-LptAI36pBpa adducts (marked “LptA × LPS”) were detected by LPS immunoblot (top panel). Total levels of LptAI36pBpa in the assay were confirmed by LptA immunoblotting (bottom panel). We also did not detect LPS transport by complexes containing LptBWT or LptBR144H/F239A in using the nonhydrolyzable ATP analogs α,β-methylene ATP (ApCpp), 2-chloroadenosine β, γ-methylene ATP (AppCp), ATP-γS, and AppNH2. Download FIG S7, PDF file, 0.3 MB (342.6KB, pdf) .
Copyright © 2019 Simpson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The atomic coordinates and structure factors have been deposited at the RCSB Protein Data Bank with accession number 6MBN.