Abstract
Establishment of proper connectivity between peripheral sensory neurons and their central targets is required for an animal to sense and respond to various external stimuli. Dorsal root ganglion (DRG) neurons convey sensory signals of different modalities via their axon projections to distinct laminae in the dorsal horn of the spinal cord. In this study, we found that c-Maf was expressed predominantly in the interneurons of laminae III/IV, which primarily receive inputs from mechanoreceptive DRG neurons. In the DRG, c-Maf+ neurons also coexpressed neurofilament-200, a marker for the medium- and large-diameter myelinated afferents that transmit non-noxious information. Furthermore, mouse embryos deficient in c-Maf displayed abnormal development of dorsal horn laminae III/IV neurons, as revealed by the marked reduction in the expression of several marker genes for these neurons, including those for transcription factors MafA and Rora, GABAA receptor subunit α5, and neuropeptide cholecystokinin. In addition, among the four major subpopulations of DRG neurons marked by expression of TrkA, TrkB, TrkC, and MafA/GFRα2/Ret, c-Maf was required selectively for the proper differentiation of MafA+/Ret+/GFRα2+ low-threshold mechanoreceptors (LTMs). Last, we found that the central and peripheral projections of mechanoreceptive DRG neurons were compromised in c-Maf deletion mice. Together, our results indicate that c-Maf is required for the proper development of MafA+/Ret+/GFRα2+ LTMs in the DRG, their afferent projections in the dorsal horn and Pacinian corpuscles, as well as neurons in laminae III/IV of the spinal cord.
Introduction
The accurate somatic perception requires proper connections between peripheral sensory neurons and their target neurons in the dorsal spinal cord. The dorsal horn of the mature spinal cord can be defined anatomically into five discrete parallel layers (laminae) (Rexed, 1952; Brown, 1981), with subsets of primary afferent fibers targeting spinal neurons within discrete laminae (Willis and Coggeshall, 1991). Afferents of nociceptors sensing pain and temperature mainly project to laminae I/II. Afferents for sensing innocuous mechanoreceptor signals such as texture, shape, vibration, and pressure project predominantly to internal laminae (III, IV, V). Afferents for sensing proprioceptive signals project through the dorsal horn to the ventrally located motor neurons (Brown, 1981; Willis and Coggeshall, 1991). Much progress has been made in understanding the development of dorsal horn neurons (Caspary and Anderson, 2003; Helms and Johnson, 2003; Fitzgerald, 2005; Ma, 2006). However, the molecular mechanism that controls the development of laminae III/IV neurons in dorsal horn remains poorly understood.
The proper differentiation of sensory neurons in the dorsal root ganglia (DRGs) is also important in establishing precise neural circuits for somatic sensation. Developmental studies have begun to define transcription factors that underlie the differentiation of diverse neuronal types of somatic sensory neurons (Marmigère and Ernfors, 2007; Liu and Ma, 2011; Reed-Geaghan and Maricich, 2011). At early stages of embryonic development, neurogenin1 (Ngn1) is required for the formation of TrkA-expressing sensory precursors, whereas Ngn2 is required for the generation of TrkB- or TrkC-expressing precursors (Ma et al., 1999). Transcription factors of the Runx family regulate further specification of these classes of sensory neurons during embryonic DRG development: Runx3 and Runx1 control the gene expression programs relevant to proprioceptive and nociceptive sensory neuron development and differentiation, respectively (Chen et al., 2006a,b; Kramer et al., 2006; Marmigère et al., 2006; Nakamura et al., 2008). Furthermore, transcription factor Shox2 (short stature homeobox2) is required for proper development of TrkB-expressing mechanosensory neurons (Scott et al., 2011). Last, three studies have found that Ret signaling was required for the specification of MafA+/Ret+/GFRα2+ low-threshold mechanoreceptor (LTMs) (Bourane et al., 2009; Luo et al., 2009; Honma et al., 2010). The finding that the phenotypes of central and peripheral projections of LTMs in MafA deletion mice do not recapitulate those observed in Ret mutants indicated that an unknown transcription factor may control the differentiation of MafA+/Ret+/GFRα2+ LTMs (Bourane et al., 2009).
The family of large Maf proteins, consisting of MafA, MafB, c-Maf, and Nrl, are bZIP (basic leucine-zipper) transcription factors of the AP-1 superfamily, and they act as key regulators of terminal differentiation in many tissues (Blank and Andrews, 1997; Kataoka, 2007; Yang and Cvekl, 2007). However, c-Maf's role in the nervous system is largely unknown. Here we showed that c-Maf was expressed in laminae III/IV neurons of the dorsal spinal cord and MafA+/Ret+/GFRα2+ LTMs in the DRGs. Deletion of c-Maf caused aberrant central and peripheral projections of the latter DRG neurons and the development of their target neurons in laminae III/IV of the spinal dorsal horn.
Materials and Methods
Mouse strains.
The generation of Tlx1, Tlx3, Lbx1, and Tlx3-cre mutant mice has been described previously (Roberts et al., 1994; Gross et al., 2000; Shirasawa et al., 2000; Xu et al., 2008). Ptf1acre mutant mice, which have Cre-recombinase in place of the Ptf1a protein-coding region, were kindly donated by Dr. Christopher Wright (Vanderbilt University, Nashville, TN) and obtained from the Mutant Mouse Regional Resource Centers (MMRRC) (Kawaguchi et al., 2002). Tau-nLacZ reporter line was kindly provided by Dr. Silvia Arber (University of Basel, Basel, Switzerland) (Hippenmeyer et al., 2005). Male Tlx3-cre knock-in mice were crossed with female Tau-nLacZ reporter mice to fate map the Tlx3-expressing neurons. MafA conditional knock-out mice were kindly donated by Dr. Roland Stein (Vanderbilt University, Nashville, TN) and obtained from MMRRC. Female EIIacre mice (Lakso et al., 1996) were crossed with male MafA conditional knock-out mice to generate the complete MafA deletion mice. c-Maf mutant mice were kindly donated by Dr. Greg Barsh (Stanford University, Palo Alto, CA) and obtained from the Jackson Laboratory (Ring et al., 2000). In all timed mating using both male and female mice, the morning that vaginal plugs were observed was designated as E0.5. Genotyping for Tlx1, Tlx3, Lbx1, and Ptf1a mutant mice was done as described previously (Gross et al., 2000; Qian et al., 2002; Huang et al., 2008). Tlx3-Cre mice were identified as described previously for detecting the presence of Cre in the mutant allele (Huang et al., 2008). Genotyping primers for identifying MafA and c-Maf mutant mice were provided by MMRRC and the Jackson Laboratory, respectively. All animal procedures are contained in protocols reviewed and approved by the Animal Care Committee at the Institute of Neuroscience, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences.
In situ hybridization and immunostaining.
In situ hybridization experiments were performed as described previously (Huang et al., 2010). The following mouse in situ probes were amplified with the following gene-specific sets of PCR primers and cDNA templates prepared from P0 mouse brain/spinal cord: MafA (NM_194350, 0.72 kb); c-Maf (NM_001025577, 0.83 kb); Rora (NM_013646, 0.89 kb); Rorb (NM_146095, 0.92 kb); Gfra2 (NM_008115, 0.97 kb); Netrin 1 (NM_008744, 1.01 kb); Ebf2 (NM_010095, 0.67 kb); Prrxl1 (DRG11) (NM_001001796, 0.66 kb); and Gabra5 (NM_176942, 0.73 kb). CCK (NM_031161, 0.47 kb) was amplified with cDNA prepared from differentiated P19 embryonic carcinoma cells. Other in situ probes were described previously (Qian et al., 2002; Cheng et al., 2004; Gray et al., 2004; Huang et al., 2008) or obtained from Dr. Qiufu Ma (Dana-Farber Cancer Institute, Harvard Medical School, Boston, MA).
Immunostaining was performed as described previously (Huang et al., 2010). The following antibodies were used: rabbit anti-c-Maf (1:500, Bethyl Laboratories); rabbit anti-MafA (1:50, Bethyl Laboratories); rabbit anti-Pax2 (1:50, Zymed); mouse anti-NF200 (neurofilament-200) (1:1000, Sigma); mouse anti-NeuN (1:1000, Millipore); rabbit anti-S100 (1:400, Dako); rabbit anti-TrkA (1:100, Advanced Targeting Systems); guinea-pig anti-VGLUT1 (1:100, Millipore); rabbit anti-parvalbumin (1:100, Swant); goat anti-Ret (1:25, R&D Systems); and chicken anti-β-gal (1:250, Abcam). Double staining combining the in situ hybridization and the fluorescent immunostaining was performed as described previously (Huang et al., 2008) with the proteinase K was used as 1 μg/ml.
TUNEL staining on cryostat sections.
Apoptotic cells in the developing spinal cord and DRG were analyzed by the TUNEL assay using the ApopTag-plus Fluorescein In Situ Apoptosis Detection Kit (Millipore Bioscience Research Reagents). Cell staining was performed according to the manufacturer's protocols.
Cell counting.
Lumbar DRGs from three pairs of control and mutant embryos (12 μm) were hybridized with various molecular markers. Positive cells with clear morphology in DRG were counted, and the values were presented as mean ± SEM. The differences in values were considered to be significant at p < 0.05 by Student's t test.
Quantification of central projections in the dorsal horn.
Sections (16 μm) from spinal cord at the lower cervical and upper thoracic level were used for the quantification analysis. The positive fibers were framed and quantitated from ImageJ (version 1.43, NIH). We set the fluorescence intensity in controls to 100%. Three pairs of embryos were used, and for each embryo four to six sections were chosen. The differences in values were considered to be significant at p < 0.05 by Student's t test.
Results
Expression of MafA and c-Maf genes in developing spinal cord
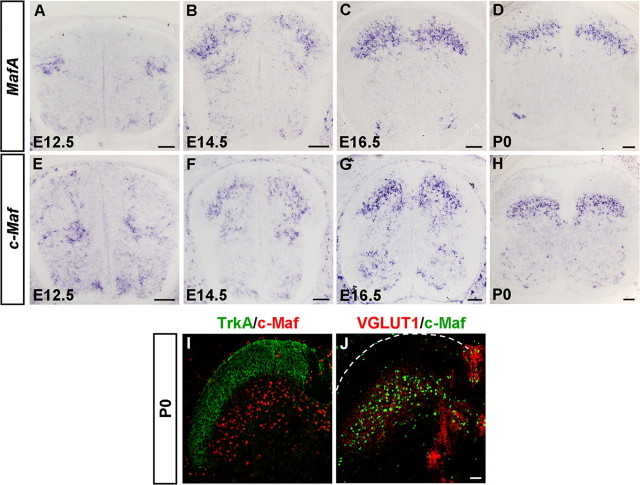
To study the dynamic expression of transcription factors MafA and c-Maf in the developing dorsal spinal cord, we performed in situ hybridization experiments. In wild-type embryos, MafA and c-Maf were expressed in the dorsal spinal cord, beginning around E11.5–E12.5 (Fig. 1A,E; and data not shown). At E12.5, MafA was expressed in the lateral region of the dorsal horn and c-Maf was initially expressed in cells adjacent to the ventricular zone of the dorsal spinal cord (Fig. 1A,E). Over the next few days, obvious expression of MafA and c-Maf was observed in laminae III/IV of the dorsal horn (Fig. 1B–D,F–H). It is noteworthy that the expression pattern of MafA was similar to that of c-Maf, except that some MafA+ cells were scattered in the superficial laminae at P0 (Fig. 1D,H).
Figure 1.
Expression of MafA and c-Maf in the developing mouse spinal cord. A–H, In situ hybridization was performed on transverse sections of the spinal cord at various developmental stages using MafA and c-Maf genes as the probes. MafA- and c-Maf-expressing cells first emerged at E12.5 in the dorsal spinal cord (A, E). From E14.5 to P0, most MafA- and c-Maf-expressing cells were enriched in the laminae III/IV (B–D, F–H). I, J, Double immunostaining of c-Maf with TrkA (I) and VGLUT1 (J) was performed on sections of P0 spinal cord. c-Maf+ cells were found enriched in laminae III/IV (mechanoreceptive input). Scale bars: A–H, 100 μm; I, J, 50 μm.
To determine which laminae c-Maf+ cells occupy, we performed double-staining experiments in the spinal cord of newborn mice, when laminae I/II and laminae III/IV are distinguishable. Previous studies have shown that central nociceptive projections (TrkA+) are mostly restricted to the superficial laminae I/II, and LTM afferents (VGLUT1+) mainly form synapses on interneurons in laminae III/IV of the dorsal horn (Todd et al., 2003; Alvarez et al., 2004). As expected, c-Maf+ cells were enriched in laminae III/IV, where VGLUT1+ afferents were found (Fig. 1I,J). These findings on the expression of MafA and c-Maf in the developing dorsal horn are in line with previous reports (Sakai et al., 1997; Li et al., 2006; Lecoin et al., 2010).
To further characterize the c-Maf+ and MafA+ neurons in the dorsal horn, we performed double staining of c-Maf and MafA with cell type-selective markers. Two major classes of dorsal interneurons have been defined by the expression of homeodomain transcription factor Lbx1. First, the Lbx1− early-born DI1–3 Class A neurons, and second, the Lbx1+ Class B neurons, including early-born DI4–6 neurons and late-born dILA and dILB neurons (Gross et al., 2002; Müller et al., 2002). In E12.5 dorsal horn, c-Maf was mainly expressed in DI1 and DI3 neurons (Fig. 2A,B,E,F), which are marked by Barhl1 and Isl1, respectively (Bermingham et al., 2001; Gross et al., 2002; Müller et al., 2002). In contrast, c-Maf+ neurons rarely expressed the DI2 marker Foxd3 (Gross et al., 2002) (Fig. 2C,D). Interestingly, a small subset of c-Maf+ cells was also Lbx1+ Class B neurons and coexpressed Lmx1b or Pax2 (Fig. 2G,H; and data not shown). MafA was largely colocalized with Pax2, which marks DI4, DI6, and dILA neurons (Fig. 2I,J). Moreover, in situ hybridization results showed that MafA expression was abolished in the dorsal cord of Lbx1 mutant mice at E12.5 (Fig. 2K,L), indicating a Class B fate of MafA+ neurons (Gross et al., 2002; Müller et al., 2002). We found that many Class B neurons coexpressed c-Maf and MafA in the dorsal horn at E14.5, and both c-Maf and MafA were allocated in Tlx3-derived excitatory neurons and Pax2+ inhibitory neurons at P0 (Fig. 2M–T; and data not shown).
Figure 2.
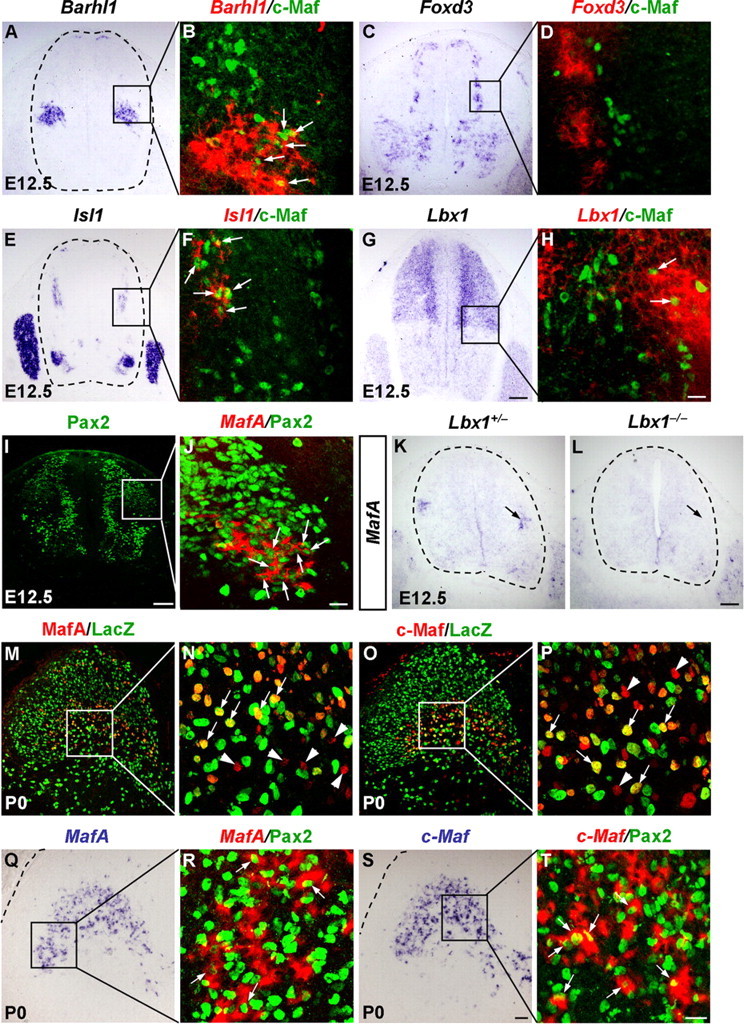
Characterization of c-Maf+ and MafA+ neurons in the dorsal spinal cord. A–J, Pseudo-color double staining of nuclear c-Maf protein (B, D, F, H, green) and Pax2 protein (J, green) with Barhl1 mRNA (B, red), Foxd3 mRNA (D, red), Isl1 mRNA (F, red), Lbx1 mRNA (H, red), or MafA mRNA (J, red) are shown in the boxed areas of A, C, E, G, and I, respectively. B, D, F, H, J, Confocal images, in which bright-field in situ hybridization signals were converted into red pseudo-fluorescent signals. Note c-Maf+ cells coexpressed Barhl1, Isl1, or Lbx1 in the dorsal horn at E12.5 (B, F, and H, arrows), and MafA+ cells coexpressed Pax2 (J, arrows). K, L, In situ hybridization was performed on spinal cord of control and Lbx1−/− mice at E12.5 with MafA as the probe. M–P, Double staining of the nLacZ protein (M–P, green) plus MafA (N, red), or c-Maf (P, red) protein was performed on sections through spinal cord of Tlx3-cre;Tau-nLacZ mice at P0. N and P are higher magnification of the boxed areas in M and O, respectively. Neurons coexpressing nLacZ and MafA (N, arrows) or c-Maf (P, arrows) appear yellow. Note that some neurons just express MafA or c-Maf (N, P, arrowheads). Q–T, Pseudo-color double staining of the nuclear Pax2 protein (R, T, green) with MafA mRNA (R, red) or c-Maf mRNA (T, red) are shown in the boxed areas of Q and S. Note that some MafA+ or c-Maf+ cells coexpressed Pax2 (R and T, arrows). Scale bars: (in L) A, C, E, G, I, K, L, 100 μm; (in S) M, O, Q, S, 50 μm; (in H, J, T) B, D, F, H, J, N, P, R, T, 20 μm.
Developmental defects in spinal dorsal horn of c-Maf −/− embryos
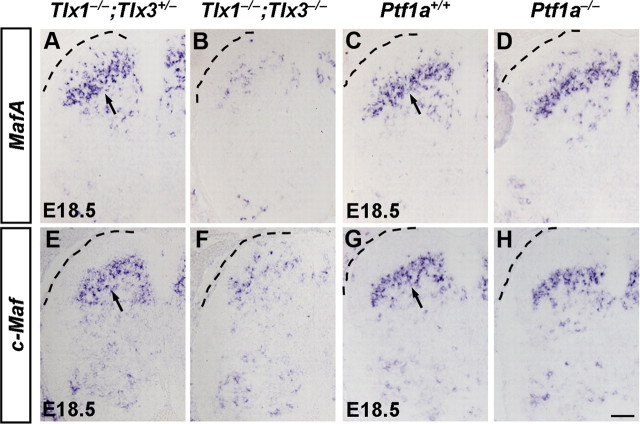
We have demonstrated previously that transcription factors Tlx1 and Tlx3 specify the glutamatergic cell fate and control a set of downstream targets including transcription factors, peptides, and transmitter receptors in the dorsal horn (Qian et al., 2002; Cheng et al., 2004; Cheng et al., 2005; Xu et al., 2008) (data not shown). The specific expression patterns of MafA and c-Maf prompted us to investigate whether MafA and c-Maf were involved in a transcriptional cascade to control the expression of some target genes of Tlx1 and Tlx3 in the developing dorsal spinal cord. In this study, we found indeed that Tlx1 and Tlx3 were required for the proper expression of a large portion of MafA and c-Maf in the dorsal horn at E18.5 (Fig. 3A,B,E,F). Meanwhile, double staining of MafA or c-Maf with Pax2 indicated that most of the remained Tlx1/3-independent MafA+ and c-Maf+ neurons at E18.5 were those Pax2+ inhibitory neurons (data not shown). In contrast, transcription factor Ptf1a, which specifies the GABAergic cell fate in dorsal spinal cord (Glasgow et al., 2005), was dispensable for the expression of MafA and c-Maf (Fig. 3C,D,G,H). Nevertheless, marked decrease of the expression of Pax2 in Ptf1a deletion mice at E18.5 suggested that Ptf1a plays a role in the development of MafA+;Pax2+ or c-Maf+;Pax2+ neurons (data not shown).
Figure 3.
Expression of MafA and c-Maf in Tlx1−/−;Tlx3−/− and Ptf1a−/− mice. A–H, In situ hybridization was performed on lumbar spinal cords of control, Tlx1−/−;Tlx3−/−, and Ptf1a−/− mice at E18.5 with MafA or c-Maf as the probes. The expression of MafA and c-Maf were reduced in Tlx1−/−;Tlx3−/− mice (A, B, E, F), while no apparent changes were observed in Ptf1a−/− mice compared with control mice (C, D, G, H). Scale bar: (in H) A–H, 50 μm.
We then examined whether MafA and c-Maf were required for the proper development of dorsal spinal cord by mapping the expression of neuronal proteins. Using in situ hybridization, we identified four genes that were controlled by c-Maf: MafA, which encodes transcription factor MafA; Gabra5, which encodes GABAA receptor subunit α5; CCK, which encodes neuropeptide cholecystokinin; and Rora, which encodes transcription factor RAR-related orphan receptor α (Rora). In c-Maf−/− embryos at stage E18.5, MafA expression was markedly reduced in the dorsal spinal cord, and the high level expression of Gabra5 in the laminae III/IV was essentially eliminated (Fig. 4A,B,E,F). We also found that the expression of another two genes, CCK and Rora, was reduced ∼40% in the cervical spinal cord (Fig. 4I,J,M,N). The double-staining experiments showed that many neurons expressing MafA, Gabra5, CCK, and Rora also coexpressed c-Maf (Fig. 4C,D,G,H,K,L,O,P). These results indicated that c-Maf was required for the proper development of neurons in laminae III/IV of the dorsal horn. In contrast, MafA was dispensable for the expression of c-Maf and the above-mentioned neuronal marker genes examined in the developing spinal cord (Fig. 5A–H). In situ hybridization results also revealed the persistent expressions of c-Maf and its downstream genes in the dorsal horn at P30 (data not shown). Thus, c-Maf and its target genes may play some physiological roles in adult mice. The early lethality of c-Maf−/− mice at P0 warrants the generation and analysis of the c-Maf conditional knock-out mice.
Figure 4.
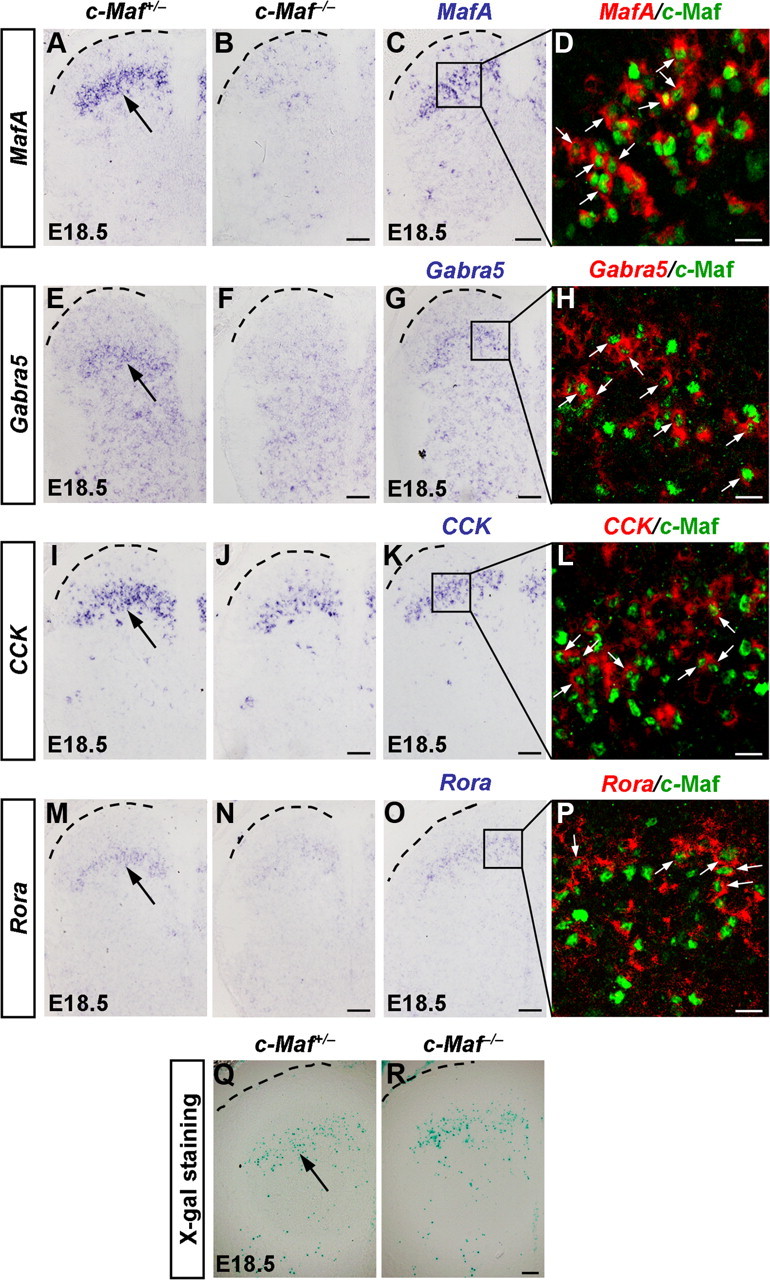
Reduced expression of MafA, Gabra5, CCK, and Rora in the dorsal horn of c-Maf−/− mice. A, B, E, F, I, J, M, N, In situ hybridization was performed with MafA (A, B), Gabra5 (E, F), CCK (I, J), and Rora (M, N) probes on sections of cervical/thoracic spinal cord in c-Maf+/− and c-Maf−/− mice at E18.5. Note a reduction of MafA, high-level Gabra5, CCK, and Rora expression in laminae III/IV neurons of the mutant spinal cord. C, D, G, H, K, L, O, P, Pseudo-color double staining of nuclear c-Maf protein (D, H, L, P, green) with MafA mRNA (D, red), Gabra5 mRNA (H, red), CCK mRNA (L, red), or Rora mRNA (P, red) are shown in the boxed areas of C, G, K, and O. D, H, L, and P are confocal images, in which bright-field in situ hybridization signals were converted into red pseudo-fluorescent signals. Many cells coexpressed c-Maf and MafA, Gabra5, CCK, or Rora in the dorsal horn at E18.5 (D, H, L, P, arrows). Q, R, X-gal staining was performed on sections of cervical/thoracic spinal cord in c-Maf+/− (Q) and c-Maf−/− (R) mice at E18.5. Scale bars: (in B, C, F, G, J, K, N, O, R) A–C, E–G, I–K, M–O, Q, R, 50 μm; D, H, L, P, 20 μm.
Figure 5.
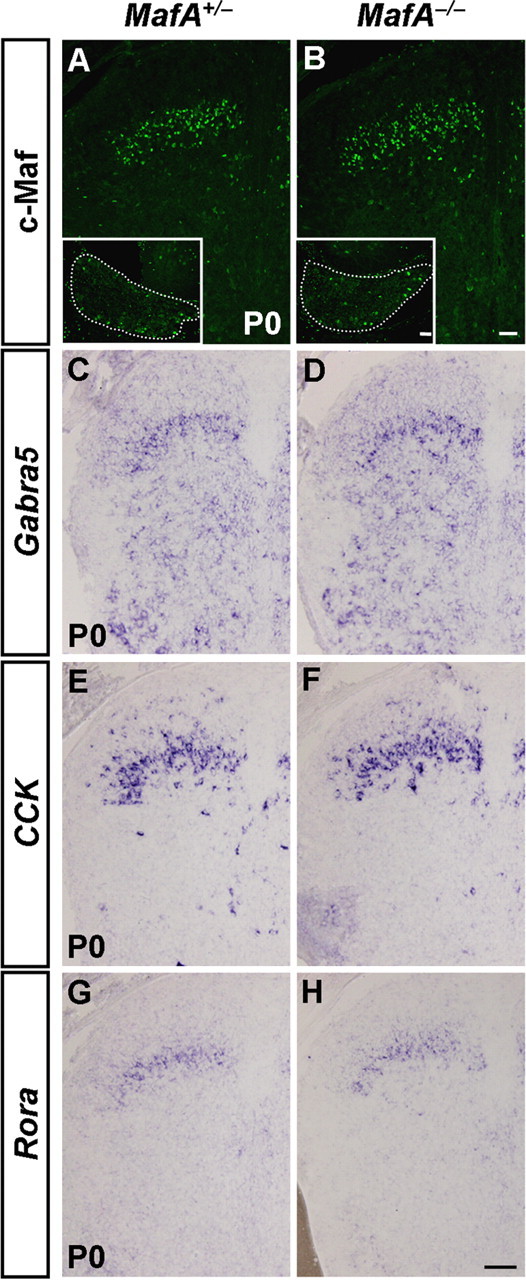
Expression of c-Maf and its target genes in the dorsal horn of MafA−/− mice. A, B, Immunostaining of c-Maf was performed on sections of spinal cord in MafA+/− and MafA−/− mice at P0. Insets show the anti-c-Maf immunofluorescence signal in the DRG of control and MafA deletion mice. Note comparable expression of c-Maf in both the dorsal horn and the DRG in MafA−/− versus control mice. C–H, In situ hybridization was performed with Gabra5 (C, D), CCK (E, F), and Rora (G, H) probes on sections of cervical/thoracic spinal cord in MafA+/− and MafA−/− mice at P0. No apparent difference was observed in MafA mutant and control mice. Scale bars: (in B, H) A–H, 50 μm; (in B, inset), A, B, insets, 50 μm.
Several observations suggested that reduction of the above-mentioned genes in the dorsal spinal cord of c-Maf−/− mice were not due to neuronal loss. First, the TUNEL assay failed to show an increase in cell death in the dorsal horn of c-Maf−/− mice at the three different stages examined: E14.5, E16.5, and P0 (data not shown). Second, using X-gal staining, by replacing the C-terminal 351 aa of c-Maf with a lacZ gene in the c-Maf deletion mice, we found no obvious difference between laminae III/IV neurons of c-Maf−/− mice versus control mice (Fig. 4Q,R). Third, there was no apparent morphological defect in c-Maf mutants, as indicated by the expression pattern of the pan-neuronal marker NeuN (data not shown).
Despite the above-mentioned defects in the dorsal horn, we were unable to identify obvious alterations in the expression of several other neuronal markers in the dorsal spinal cord of c-Maf−/− embryos at E14.5 and E18.5. These include transcription factors Lmx1b, Prrxl1 (DRG11), Pbx3, Rorb, Ebf1, and Ebf2, and the axon guidance molecular Netrin-1, Sema3C (data not shown).
Expression of c-Maf in medium- and large-diameter DRG neurons
We also found that c-Maf was expressed in developing sensory neurons of DRGs from early embryonic stages to P0 (Fig. 6A–F). The early c-Maf+ cells were detectable from E10.5, and they were neurons, as revealed by the colocalization of c-Maf with NeuN, a marker of postmitotic neurons (Fig. 6A–C). Persistent expression of c-Maf was detected at later stages (Fig. 6D–F; and data not shown).
Figure 6.
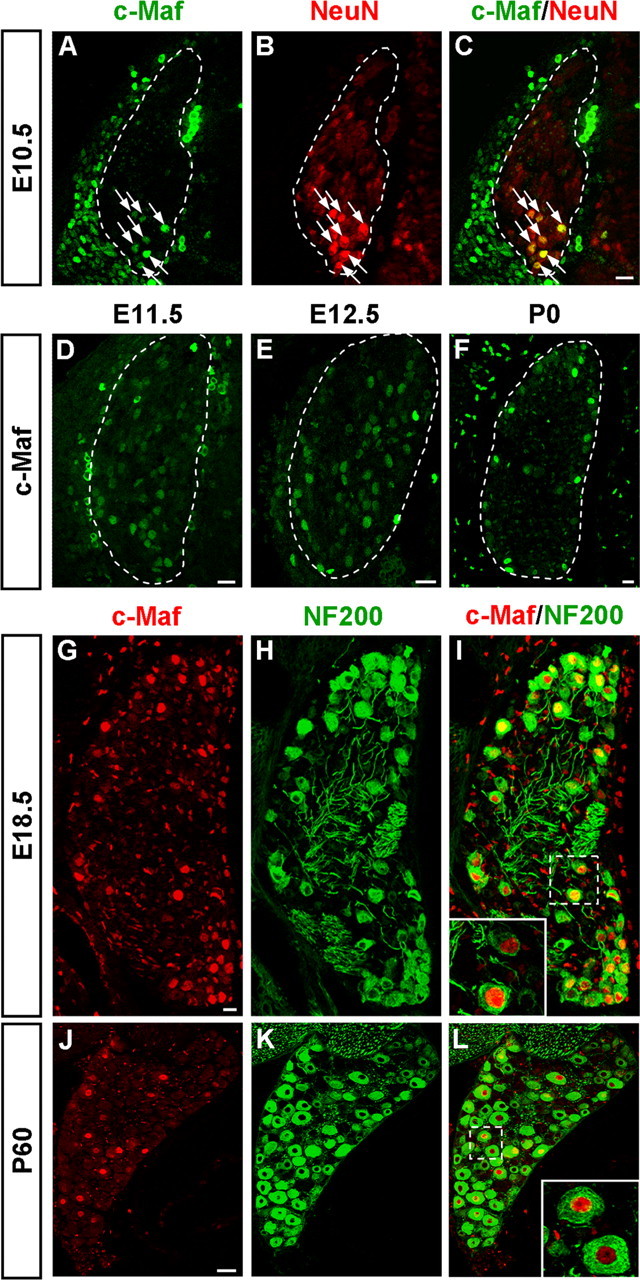
Expression of c-Maf in the developing DRG. A–F, Immunostaining of c-Maf was performed on sections of DRGs at E10.5 (A), E11.5 (D), E12.5 (E), and P0 (F). Cells expressing c-Maf appeared at around E10.5 and were found at all subsequent stages. c-Maf protein was found in neurons as shown by colocalization with the neuronal marker NeuN (A–C). G–L, c-Maf+ cells were medium- and large-diameter NF200+ DRG neurons. Double staining of c-Maf protein (G, J, red) and NF200 protein (H, K, green) was performed on sections of E18.5 (I) and P60 (L) DRGs. All c-Maf+ neurons coexpressed NF200; thus, the c-Maf+ neuronal population were medium- and large-diameter DRG neurons. Scale bars: A–I, 20 μm; J–L, 50 μm.
To determine which classes of DRG neurons express c-Maf, we performed double-staining experiments with c-Maf and molecular markers for DRG subpopulations. Neurons of medium and large diameter in the postnatal DRG are generally myelinated afferents characterized by the expression of NF200. By double labeling, we found that nearly all c-Maf+ cells were colocalized with NF200 at E18.5 and P60 (Fig. 6G–L). In contrast, almost none of the c-Maf+ cells were found to bind to isolectin B4 (marker for small-diameter nonpeptidergic nociceptors), and occasionally c-Maf+ cells were found to express calcitonin gene-related peptide (marker for small-diameter peptidergic nociceptors) (data not shown). Thus, we identified that c-Maf+ cells were early-born, and medium- and large-diameter DRG neurons.
To further explore the myelinated afferent subclass to which c-Maf+ cells belong, we performed double-labeling experiments that combined in situ hybridization for neurotrophin receptors TrkA, TrkB, and TrkC, as well as parvalbumin, together with immunostaining for c-Maf on sections of DRGs at E18.5. We found that ∼36% c-Maf+ neurons coexpressed TrkB, and ∼40% c-Maf+ neurons coexpressed TrkC (Fig. 7A,B). As a matter of fact, TrkB+ and TrkC+ subsets of c-Maf+ neurons represent two distinct populations of c-Maf+ neurons (Bourane et al., 2009; and data not shown). However, only neurons expressing low levels of parvalbumin coexpressed c-Maf, and a small population of c-Maf+ neurons coexpressed TrkA (data not shown).
Figure 7.
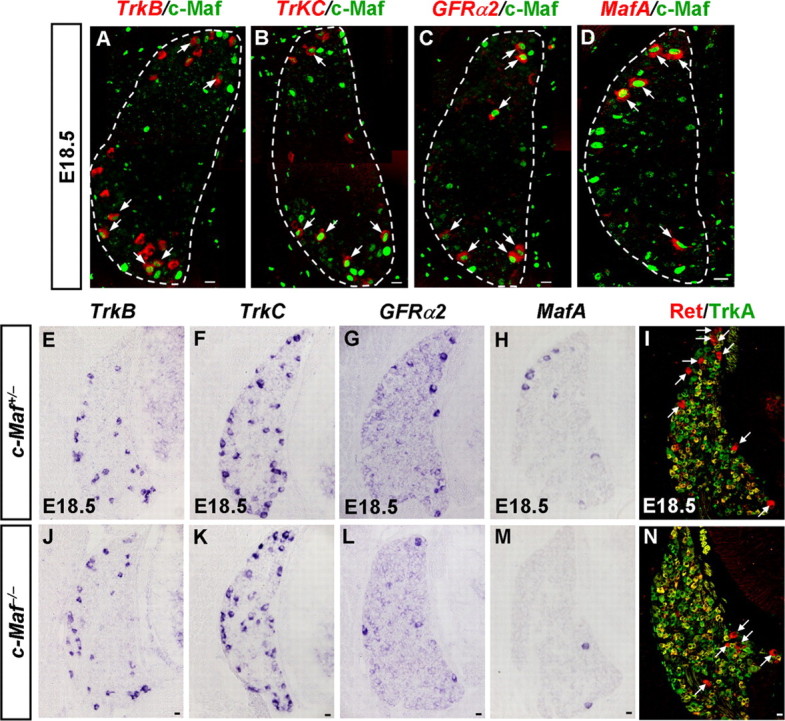
Defective differentiation of MafA+/Ret+/GFRα+ LTMs in c-Maf deletion mice. A–D, Coexpression studies of c-Maf with DRG neuronal markers at E18.5. Pseudo-color double staining of nuclear c-Maf protein (A–D, green) with TrkB mRNA (A, red), TrkC mRNA (B, red), GFRα2 mRNA (C, red), or MafA mRNA (D, red) was performed on DRG at E18.5. c-Maf+ neurons partially coexpressed TrkB or TrkC (A, B, arrows). Note that the majority of GFRα2+ and MafA+ neurons coexpressed c-Maf (C, D, arrows). E–N, In situ hybridization was performed on sections of DRGs from c-Maf+/− and c-Maf−/− mice at E18.5 with TrkB (E, J), TrkC (F, K), GFRα2 (G, L), and MafA (H, M) as the probes. Double staining of Ret (I and N, red) and TrkA (I and N, green) was performed on sections of DRGs from c-Maf+/− and c-Maf−/− mice at E18.5. Examples of large-diameter Ret+/TrkA− neurons are indicated by white arrows. Note that significant reductions of ∼55%, 70%, and 50%, respectively, in the numbers of GFRα2+, MafA+, and Ret+/TrkA− neurons were observed in c-Maf−/− mice. p < 0.001. Scale bar, 20 μm.
To assess whether the loss of c-Maf affected the expression of neurotrophin receptors in DRG neurons, we examined the expression of TrkA, TrkB, and TrkC in c-Maf−/− mice at E18.5. In situ hybridization results showed that the expression of TrkA, TrkB, and TrkC was comparable in c-Maf−/− mice versus control mice (Fig. 7E,F,J,K; and data not shown).
Together, these results suggested that c-Maf+ neurons were early-born myelinated afferents, many of which coexpressed the neurotrophin receptors TrkB or TrkC. The transcription factor c-Maf was not required for the expression of Trk receptors.
Defective differentiation of MafA+/Ret+/GFRα2+ LTMs in c-Maf−/− mice
Recent studies identified that a population of LTM neurons selectively expresses the transcription factor MafA, the early wave of Ret tyrosine kinase receptor, and Ret coreceptor GFRα2 (MafA+/Ret+/GFRα2+). These MafA+/Ret+/GFRα2+ LTMs were further identified as prospective rapid-adapting (RA) mechanoreceptors, including Meissner corpuscles, Pacinian corpuscles, and longitudinal lanceolate endings (Bourane et al., 2009; Luo et al., 2009; Honma et al., 2010). The identification of MafA, early wave of Ret, and GFRα2 as markers for LTMs allowed us to analyze the embryonic differentiation of MafA+/Ret+/GFRα2+ LTMs. By performing double-staining experiments that combined in situ hybridization and immunostaining, we found that the majority of GFRα2+ (86 ± 2%) and MafA+ (92 ± 5%) cells coexpressed c-Maf at E18.5 (Fig. 7C,D).
The marked reduction of MafA expression in laminae III/IV neurons of the dorsal horn in c-Maf deletion mice prompted us to examine whether MafA lies downstream of c-Maf in developing DRG neurons as well. In situ hybridization results showed that this was indeed the case. Because the expression of MafA, the early wave of Ret (Ret+/TrkA−), and Ret coreceptor GFRα2 all can be used to indicate the development of the MafA+/Ret+/GFRα2+ LTMs (Bourane et al., 2009; Luo et al., 2009; Honma et al., 2010), we quantified the numbers of cells expressing GFRα2, MafA, and early Ret (Ret+/TrkA−) in the DRG of control and c-Maf deletion mice at E18.5. We found that a reduction of 55%, 70%, and 50% in the number of GFRα2+, MafA+, and Ret+/TrkA− DRG neurons occurred in c-Maf−/− mice at E18.5 (Fig. 7G–I,L–N). Further TUNEL assay showed that there was no increased cell death in the DRG of c-Maf−/− embryos at E14.5, E16.5, and E18.5 (data not shown). Furthermore, we found that there was comparable expression of TrkB, TrkC, and S100, a marker for medium- and large-sized primary sensory neurons with myelinated axons (Ichikawa et al., 1997), in DRGs between c-Maf−/− mice and control mice (Fig. 7E,F,J,K; and data not shown). Thus, the downregulation of GFRα2, MafA, and early Ret (Ret+/TrkA−) expression in c-Maf−/− deletion mice may not be ascribed to cell death.
Aberrant central projections of MafA+/Ret+/GFRα2+ LTM afferents in c-Maf−/− mice
The defective development of neurons in the laminae III/IV of spinal dorsal horn and MafA+/Ret+/GFRα2+ LTMs of c-Maf−/− mice led us to ask whether the connection between them was also affected. Mechanoreceptors send collateral branches to deep dorsal horn neurons to form direct monosynaptic connections (Brown, 1981). We investigated the expression of VGLUT1 because it labels synapses of the central projections of mechanoreceptors and proprioceptors (Todd et al., 2003; Alvarez et al., 2004). Immunostaining of VGLUT1 was performed on spinal cord from c-Maf deletion mice and control mice. Because Ret expression in the laminae III/IV can be used for labeling MafA+/Ret+/GFRα2+ LTM projections (Bourane et al., 2009; Luo et al., 2009; Honma et al., 2010), we performed immunostaining of Ret in c-Maf deletion mice as well. Immunostaining of TrkA and parvalbumin, the molecular markers for labeling projections of nociceptors and proprioceptors, was performed on spinal cord sections adjacent to those used for VGLUT1 staining. We found that VGLUT1 expression was reduced in the laminae III/IV, particularly in the dorsolateral region, but was retained in the intermediate and ventral gray matter of the spinal cord of c-Maf-deficient mice at E18.5 (Fig. 8A,B). Quantification of VGLUT1 staining intensity in laminae III/IV revealed that the ratio of the staining intensity between the dorsolateral region and the intermediate region reduced by ∼38% in c-Maf deletion mice compared with that in the control mice (p < 0.001). As a control, the VGLUT1 staining signals in DRGs were indistinguishable between control and c-Maf mutant mice (67 ± 1 in control mice vs 65 ± 4 in c-Maf−/− mice) (Fig. 8A,B, insets). Similarly for Ret expression (Fig. 8C,D), Ret staining intensity in the laminae III/IV showed that the ratio of the staining intensity between the dorsolateral and intermediate regions was reduced by 33% in c-Maf deletion mice (p < 0.001). In line with the reduction of VGLUT1 expression in the laminae III/IV, the intensity of S100 staining decreased within layers III/IV of the spinal cord in c-Maf−/− mice (data not shown). In contrast, TrkA+ cutaneous nociceptive afferents in the laminae I/II were comparable in c-Maf−/− and c-Maf+/− mice (Fig. 8E,F). Consistent with the preservation of VGLUT1 expression in the intermediate and ventral spinal cord in c-Maf−/−-null mice, parvalbumin+ proprioceptive afferents were largely unaffected in c-Maf deletion mice (Fig. 8G,H). Meanwhile, the expression of VGLUT1, Ret, TrkA, and parvalbumin was not significantly different between MafA−/− and control mice (Fig. 9A–H).
Figure 8.
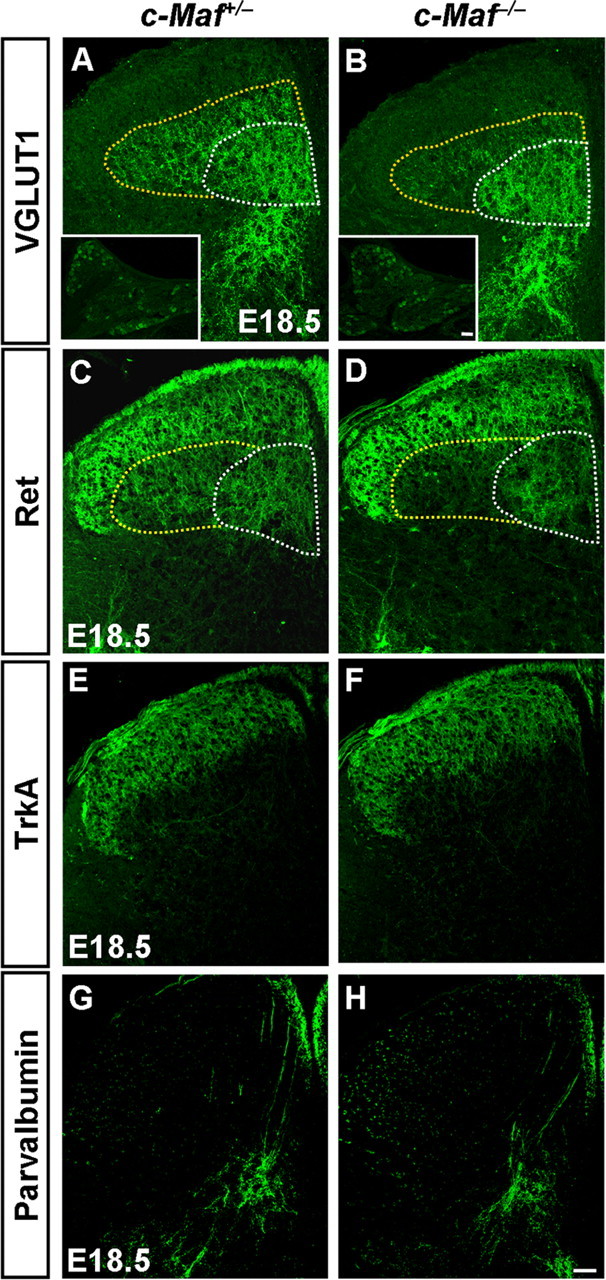
Aberrant projections of mechanoreceptive neurons in the deep dorsal horn of c-Maf deletion mice. A–D, VGLUT1 and Ret immunostaining in E18.5 mice revealed that mechanoreceptive central afferents projecting into the deep dorsal horn were reduced in c-Maf−/− mice (A, B, C, D, outlined by the dotted line). Quantification showed that the ratio of the staining intensity of the dorsolateral region outlined by the yellow dotted line compared with that of the intermediate region outlined by the white dotted line was reduced by ∼38% for VGLUT1 or 33% for Ret in c-Maf−/− mice versus control mice. p < 0.001. Insets show the anti-VGLUT1 immunofluorescence signal in the DRGs of control and c-Maf−/− mice. E–H, TrkA+ nociceptive afferents (E–F) and parvalbumin+ proprioceptive afferents (G–H) were largely unchanged in c-Maf+/− and c-Maf−/− mice. Scale bar, 50 μm.
Figure 9.
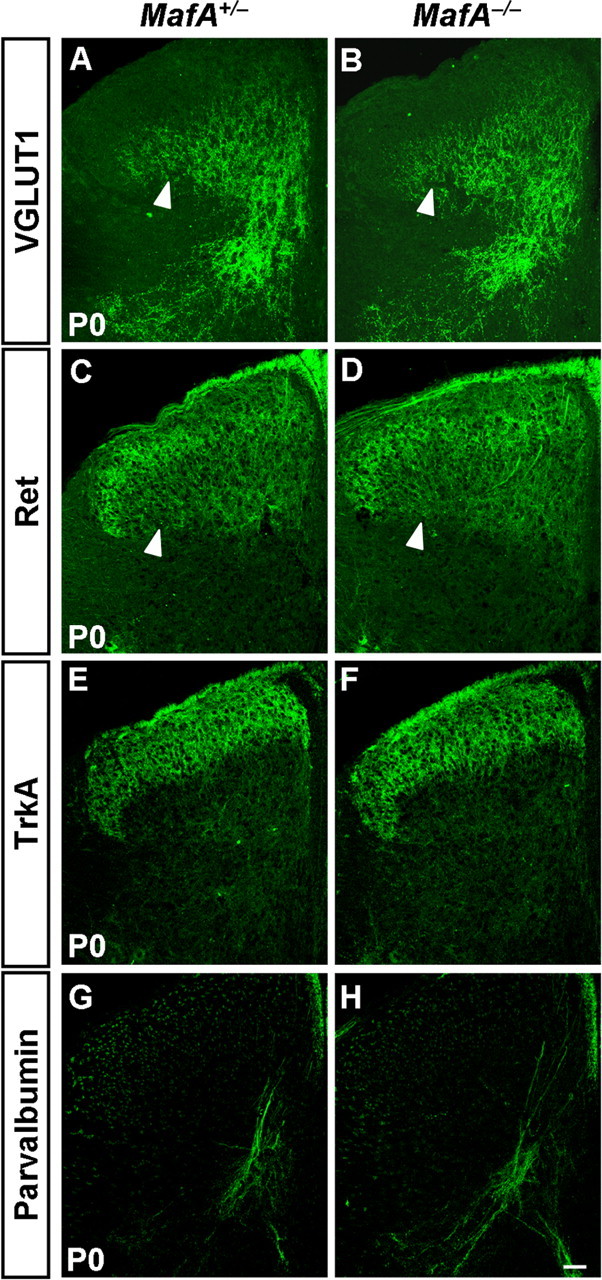
Normal projections of mechanoreceptive neurons in the deep dorsal horn of MafA deletion mice. A–H, Immunostaining of VGLUT1 (A, B), Ret (C, D), TrkA (E, F), and parvalbumin (G, H) was performed on sections of spinal cord in MafA+/− and MafA−/− mice at P0. No apparent differences of VGLUT1 and Ret staining were seen between control and MafA mutant mice (A–D, arrowheads). Scale bar, 50 μm.
These results indicated that c-Maf was required selectively for mechanoreceptive neurons to project properly in the deep layers of spinal dorsal horn, and was dispensable for the invasion of proprioceptive and nociceptive afferents into the dorsal horn.
Impaired development of Pacinian corpuscles in c-Maf−/− mice
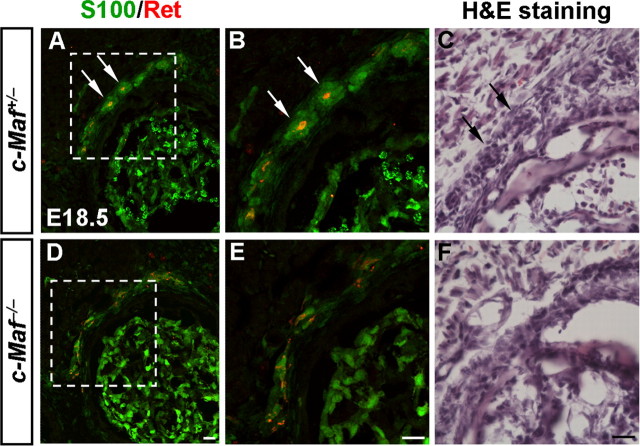
Since the central projections of mechanoreceptive neurons were defective in c-Maf−/− mice, we examined whether the peripheral innervation of MafA+/Ret+/GFRα2+ LTMs was affected as well. Recent studies demonstrate that early-born Ret+ sensory neurons are RA mechanoreceptors, whose peripheral nerve terminals are associated with Meissner corpuscles, longitudinal lanceolate endings, and Pacinian corpuscles (Bourane et al., 2009; Luo et al., 2009; Honma et al., 2010). Pacinian corpuscles are the only LTM peripheral endings, majority of which (91.7%) are innervated by early Ret+ fibers, and are completely absent in the periosteum membrane of the fibula of Ret deletion mice at P14 (Luo et al., 2009). Since a reduction of 50% in the number of early Ret+ (Ret+/TrkA−) DRG neurons occurred in c-Maf deletion mice at E18.5 (Fig. 7I,N), we have examined whether the development of Pacinian corpuscles was abnormal in c-Maf−/− mice. Indeed, we found that Pacinian corpuscles were underdeveloped in the periosteum of the fibula of c-Maf−/− mice at E18.5, as revealed by double staining of S100 and Ret (Fig. 10A,B,D,E) and hematoxylin and eosin (H&E) staining (Fig. 10C,F).
Figure 10.
Defective development of Pacinian corpuscles in c-Maf deletion mice. A, B, D, E, Pacinian corpuscles (arrows) in the periosteum of the fibula of control (A, B) and c-Maf−/− (D, E) mice at E18.5 were visualized by double staining of S100 and Ret. Note the underdevelopment of Pacinian corpuscles in the mutant mice. B and E are higher magnification of the boxed areas in A and D, respectively. H&E staining of Pacinian corpuscles was performed in E18.5 control (C, arrows) and c-Maf−/− (F) mice. H&E staining is used here to rule out the potential confounding issue of decreased S100 and Ret expression in c-Maf deletion mice. Scale bar, 20 μm.
Discussion
Much progress has been made in elucidating the genetic program underlying the diversity of neurons in the DRGs and dorsal spinal cord. However, little is known about the identity of transcription factors that control the development of MafA+/Ret+/GFRα2+ LTMs in DRG and laminae III/IV neurons in the dorsal horn. In this study, we found that transcription factor c-Maf was expressed in MafA+/Ret+/GFRα2+ LTMs and laminae III/IV neurons of dorsal horn. Deletion of c-Maf resulted in abnormal development of the laminae III/IV neurons of the spinal dorsal horn, MafA+/Ret+/GFRα2+ LTMs in the DRGs, and the central and peripheral projections of these DRG neurons.
Transcriptional control of development of dorsal horn neurons
Much progress has been made in understanding of the development of dorsal horn neurons in the past decade (Caspary and Anderson, 2003; Helms and Johnson, 2003; Fitzgerald, 2005; Ma, 2006). A set of transcription factors, including Gsx1/2, Tlx1/3, Ptf1a, and Lbx1, has been found to specify the glutamatergic versus GABAergic cell fates in the dorsal horn (Gross et al., 2002; Müller et al., 2002; Cheng et al., 2004, 2005; Glasgow et al., 2005; Mizuguchi et al., 2006). Furthermore, some additional transcription factors control the specification and the migration of subsets of dorsal horn neurons, and are required for the generation of normal pain and itch sensation (Chen et al., 2001; Ding et al., 2004; Holstege et al., 2008; Rottkamp et al., 2008; Rebelo et al., 2010; Ross et al., 2010). How neurons in laminae III/IV of the dorsal horn are specified during development remains largely unknown. c-Maf was identified as one of two transcription factors whose expression is restricted to the laminae III/IV (Li et al., 2006). We found that c-Maf controlled the development of a subset of laminae III/IV neurons. In the laminae III/IV of dorsal spinal cord, the expression of MafA and three other neuronal markers (Gabra5, CCK, and Rora) was reduced in c-Maf mutant mice (Fig. 4A–P), whereas the expression of c-Maf and its target genes (Gabra5, CCK, and Rora) remained largely unaffected in MafA deletion mice (Fig. 5). This indicates that MafA lies downstream of c-Maf in the dorsal horn and loss of MafA may account for some of the phenotypes of c-Maf mutants. Because c-Maf knock-out mice die within a few hours after birth, it will be of interest to generate c-Maf conditionally deficient mice and investigate whether the somatosensation is affected when c-Maf is deleted in the dorsal horn. Studies in the past century have suggested that the perception of specific sensory modalities might be best explained by the population-coding (also called selectivity) hypothesis (Ma, 2010). One important step in sensory biology is to better delineate the neuronal subtypes within the dorsal horn and characterize their physiological functions (Basbaum et al., 2009; Patel and Dong, 2010; Todd, 2010; Liu and Ma, 2011).
c-Maf is required for development of MafA+/Ret+/GFRα2+ LTMs
Recent studies have shown that MafA+/Ret+/GFRα2+ LTMs can be identified shortly after DRG genesis by the unique expression of the transcription factor MafA, the Ret receptor and coreceptor GFRα2 (Bourane et al., 2009; Luo et al., 2009). Ret was required for the proper differentiation of MafA+/Ret+/GFRα2+ LTMs and their central and peripheral projections (Bourane et al., 2009; Luo et al., 2009). Defective development of MafA+/Ret+/GFRα2+ LTMs was observed in MafA mutant mice as well (Bourane et al., 2009). It was proposed that a part of the “MafA/Ret-only” population lost Ret and gained TrkB. In contrast, the central and peripheral projections of MafA+/Ret+/GFRα2+ LTMs in MafA mutant mice remain largely unaffected (Bourane et al., 2009). The marked reduction of MafA expression in Ret mutants indicates that MafA acts downstream of Ret signaling to control the expression of neurotrophic factor receptors within the MafA+/Ret+/GFRα2+ LTMs (Bourane et al., 2009).
Here we have identified c-Maf as a transcription factor that is required for the proper development of MafA+/Ret+/GFRα2+ LTMs. The initial development of MafA+/Ret+/GFRα2+ LTMs was largely normal in c-Maf−/− mice, as revealed by comparable expression of Ret, MafA, and GFRα2 in c-Maf−/− and control mice at E12.5 (data not shown). However, marked reduction of GFRα2, MafA, and early Ret (Ret+/TrkA−) expression (Fig. 7G–I,L–N) accompanied by reduced innervation of these neurons in the laminae III/IV of the dorsal horn were observed in c-Maf−/− mice at E18.5 (Fig. 8A–D). We have examined peripheral innervation of MafA+/Ret+/GFRα2+ LTMs in the c-Maf deletion mice as well. The c-Maf−/− mice die within a few hours of birth, when the peripheral axons have reached their targets, but have not yet fully differentiated into specialized sensory endings (Albuerne et al., 2000; Hasegawa et al., 2007). Nevertheless, we found that Pacinian corpuscles were underdeveloped in c-Maf deletion mice (Fig. 10). Reduction of MafA and the early expression of Ret (Ret+/TrkA−) was observed in DRG neurons of c-Maf deletion mice (Fig. 7H,I,M,N), whereas c-Maf expression was comparable in DRG neurons of Ret or MafA mutant mice (Fig. 5A,B, insets; and data not shown), indicating that early Ret and MafA lie downstream of c-Maf to control the development of MafA+/Ret+/GFRα2+ LTMs. It will be of interest to further examine whether loss of Ret accounts for the peripheral innervation defects in c-Maf deletion mice.
In this study, we found that a reduction of 70% in the number of MafA+ DRG neurons occurred in c-Maf−/− mice at E18.5 (Fig. 7H,M). Our observation that TrkB expression was comparable between wild-type controls and c-Maf mutant mice is different from the increased TrkB expression observed in MafA mutant cells (Bourane et al., 2009). The reason for this difference is not clear. The comparable TrkB expression may be ascribed to the residual MafA expression in c-Maf deletion mice. On the other hand, introduction of a better reporter gene to label the c-Maf mutant cells will clarify whether TrkB expression is altered when c-Maf is deleted.
Transcriptional matching in control of somatosensory circuit formation
Specific transcription factors were found to be expressed in sensory neurons and their central target neurons. The transcription factor Phox2b is expressed in the peripheral afferent visceral pathways (carotid body and visceral sensory ganglia), the central projection site the nucleus of the solitary tract and associated area postrema, and is required for their differentiation (Pattyn et al., 1997; Dauger et al., 2003). The paired homeodomain gene Prrxl1 (DRG11) is expressed in both primary sensory neurons and secondary somatic dorsal horn neurons (Saito et al., 1995) and is required for the projection of the former to the latter (Chen et al., 2001). The Ets-domain transcription factors, such as Er81 and Pea3, are coordinately expressed by subsets of muscle afferent sensory neurons and the motor neurons they innervate in the chick spinal cord, and Er81 is essential for the formation of such connections (Lin et al., 1998; Arber et al., 2000). Here we found that c-Maf was selectively expressed in laminae III/IV neurons of the dorsal horn and also in NF200+ DRG sensory neurons. We showed in this report that deletion of c-Maf resulted in the abnormal development of laminae III/IV and LTM neurons, and LTM projections in the spinal dorsal horn. However, it remains unclear whether the projection defect reflects an intrinsic role for c-Maf in sensory neurons, laminae III/IV neurons, or both. Generation of the c-Maf conditional knock-out mice should help to clarify this issue.
Footnotes
This work was supported by the 973 Program of The Ministry of Science and Technology of China (Grant 2011CBA00400), the “Strategic Priority Research Program” of the Chinese Academy of Sciences (Grant XDA01020306), and the 973 Program Grant 2007CB947102. We are grateful to Dr. M-m. Poo for comments on the manuscript; Drs. S. Shirasawa and S. Korsmeyer for Tlx1 and Tlx3-null mice; M. Goulding for Lbx1 mutant mice; C. Wright and the MMRRC for Ptf1acre mutant mice; S. Arber for the Tau-nLacZ reporter mice; and Q. Ma for the Tlx3-cre mice.
References
- Albuerne M, De Lavallina J, Esteban I, Naves FJ, Silos-Santiago I, Vega JA. Development of Meissner-like and Pacinian sensory corpuscles in the mouse demonstrated with specific markers for corpuscular constituents. Anat Rec. 2000;258:235–242. doi: 10.1002/(SICI)1097-0185(20000301)258:3<235::AID-AR2>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Alvarez FJ, Villalba RM, Zerda R, Schneider SP. Vesicular glutamate transporters in the spinal cord, with special reference to sensory primary afferent synapses. J Comp Neurol. 2004;472:257–280. doi: 10.1002/cne.20012. [DOI] [PubMed] [Google Scholar]
- Arber S, Ladle DR, Lin JH, Frank E, Jessell TM. ETS gene Er81 controls the formation of functional connections between group Ia sensory afferents and motor neurons. Cell. 2000;101:485–498. doi: 10.1016/s0092-8674(00)80859-4. [DOI] [PubMed] [Google Scholar]
- Basbaum AI, Bautista DM, Scherrer G, Julius D. Cellular and molecular mechanisms of pain. Cell. 2009;139:267–284. doi: 10.1016/j.cell.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham NA, Hassan BA, Wang VY, Fernandez M, Banfi S, Bellen HJ, Fritzsch B, Zoghbi HY. Proprioceptor pathway development is dependent on Math1. Neuron. 2001;30:411–422. doi: 10.1016/s0896-6273(01)00305-1. [DOI] [PubMed] [Google Scholar]
- Blank V, Andrews NC. The Maf transcription factors: regulators of differentiation. Trends Biochem Sci. 1997;22:437–441. doi: 10.1016/s0968-0004(97)01105-5. [DOI] [PubMed] [Google Scholar]
- Bourane S, Garces A, Venteo S, Pattyn A, Hubert T, Fichard A, Puech S, Boukhaddaoui H, Baudet C, Takahashi S, Valmier J, Carroll P. Low-threshold mechanoreceptor subtypes selectively express MafA and are specified by Ret signaling. Neuron. 2009;64:857–870. doi: 10.1016/j.neuron.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Brown A, editor. Organization in the spinal cord. New York: Springer; 1981. [Google Scholar]
- Caspary T, Anderson KV. Patterning cell types in the dorsal spinal cord: what the mouse mutants say. Nat Rev Neurosci. 2003;4:289–297. doi: 10.1038/nrn1073. [DOI] [PubMed] [Google Scholar]
- Chen AI, de Nooij JC, Jessell TM. Graded activity of transcription factor Runx3 specifies the laminar termination pattern of sensory axons in the developing spinal cord. Neuron. 2006a;49:395–408. doi: 10.1016/j.neuron.2005.12.028. [DOI] [PubMed] [Google Scholar]
- Chen CL, Broom DC, Liu Y, de Nooij JC, Li Z, Cen C, Samad OA, Jessell TM, Woolf CJ, Ma Q. Runx1 determines nociceptive sensory neuron phenotype and is required for thermal and neuropathic pain. Neuron. 2006b;49:365–377. doi: 10.1016/j.neuron.2005.10.036. [DOI] [PubMed] [Google Scholar]
- Chen ZF, Rebelo S, White F, Malmberg AB, Baba H, Lima D, Woolf CJ, Basbaum AI, Anderson DJ. The paired homeodomain protein DRG11 is required for the projection of cutaneous sensory afferent fibers to the dorsal spinal cord. Neuron. 2001;31:59–73. doi: 10.1016/s0896-6273(01)00341-5. [DOI] [PubMed] [Google Scholar]
- Cheng L, Arata A, Mizuguchi R, Qian Y, Karunaratne A, Gray PA, Arata S, Shirasawa S, Bouchard M, Luo P, Chen CL, Busslinger M, Goulding M, Onimaru H, Ma Q. Tlx3 and Tlx1 are post-mitotic selector genes determining glutamatergic over GABAergic cell fates. Nat Neurosci. 2004;7:510–517. doi: 10.1038/nn1221. [DOI] [PubMed] [Google Scholar]
- Cheng L, Samad OA, Xu Y, Mizuguchi R, Luo P, Shirasawa S, Goulding M, Ma Q. Lbx1 and Tlx3 are opposing switches in determining GABAergic versus glutamatergic transmitter phenotypes. Nat Neurosci. 2005;8:1510–1515. doi: 10.1038/nn1569. [DOI] [PubMed] [Google Scholar]
- Dauger S, Pattyn A, Lofaso F, Gaultier C, Goridis C, Gallego J, Brunet JF. Phox2b controls the development of peripheral chemoreceptors and afferent visceral pathways. Development. 2003;130:6635–6642. doi: 10.1242/dev.00866. [DOI] [PubMed] [Google Scholar]
- Ding YQ, Yin J, Kania A, Zhao ZQ, Johnson RL, Chen ZF. Lmx1b controls the differentiation and migration of the superficial dorsal horn neurons of the spinal cord. Development. 2004;131:3693–3703. doi: 10.1242/dev.01250. [DOI] [PubMed] [Google Scholar]
- Fitzgerald M. The development of nociceptive circuits. Nat Rev Neurosci. 2005;6:507–520. doi: 10.1038/nrn1701. [DOI] [PubMed] [Google Scholar]
- Glasgow SM, Henke RM, Macdonald RJ, Wright CV, Johnson JE. Ptf1a determines GABAergic over glutamatergic neuronal cell fate in the spinal cord dorsal horn. Development. 2005;132:5461–5469. doi: 10.1242/dev.02167. [DOI] [PubMed] [Google Scholar]
- Gray PA, Fu H, Luo P, Zhao Q, Yu J, Ferrari A, Tenzen T, Yuk DI, Tsung EF, Cai Z, Alberta JA, Cheng LP, Liu Y, Stenman JM, Valerius MT, Billings N, Kim HA, Greenberg ME, McMahon AP, Rowitch DH, et al. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science. 2004;306:2255–2257. doi: 10.1126/science.1104935. [DOI] [PubMed] [Google Scholar]
- Gross MK, Moran-Rivard L, Velasquez T, Nakatsu MN, Jagla K, Goulding M. Lbx1 is required for muscle precursor migration along a lateral pathway into the limb. Development. 2000;127:413–424. doi: 10.1242/dev.127.2.413. [DOI] [PubMed] [Google Scholar]
- Gross MK, Dottori M, Goulding M. Lbx1 specifies somatosensory association interneurons in the dorsal spinal cord. Neuron. 2002;34:535–549. doi: 10.1016/s0896-6273(02)00690-6. [DOI] [PubMed] [Google Scholar]
- Hasegawa H, Abbott S, Han BX, Qi Y, Wang F. Analyzing somatosensory axon projections with the sensory neuron-specific Advillin gene. J Neurosci. 2007;27:14404–14414. doi: 10.1523/JNEUROSCI.4908-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms AW, Johnson JE. Specification of dorsal spinal cord interneurons. Curr Opin Neurobiol. 2003;13:42–49. doi: 10.1016/s0959-4388(03)00010-2. [DOI] [PubMed] [Google Scholar]
- Hippenmeyer S, Vrieseling E, Sigrist M, Portmann T, Laengle C, Ladle DR, Arber S. A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biol. 2005;3:e159. doi: 10.1371/journal.pbio.0030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege JC, de Graaff W, Hossaini M, Cano SC, Jaarsma D, van den Akker E, Deschamps J. Loss of Hoxb8 alters spinal dorsal laminae and sensory responses in mice. Proc Natl Acad Sci U S A. 2008;105:6338–6343. doi: 10.1073/pnas.0802176105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma Y, Kawano M, Kohsaka S, Ogawa M. Axonal projections of mechanoreceptive dorsal root ganglion neurons depend on Ret. Development. 2010;137:2319–2328. doi: 10.1242/dev.046995. [DOI] [PubMed] [Google Scholar]
- Huang M, Huang T, Xiang Y, Xie Z, Chen Y, Yan R, Xu J, Cheng L. Ptf1a, Lbx1 and Pax2 coordinate glycinergic and peptidergic transmitter phenotypes in dorsal spinal inhibitory neurons. Dev Biol. 2008;322:394–405. doi: 10.1016/j.ydbio.2008.06.031. [DOI] [PubMed] [Google Scholar]
- Huang T, Liu Y, Huang M, Zhao X, Cheng L. Wnt1-cre-mediated conditional loss of Dicer results in malformation of the midbrain and cerebellum and failure of neural crest and dopaminergic differentiation in mice. J Mol Cell Biol. 2010;2:152–163. doi: 10.1093/jmcb/mjq008. [DOI] [PubMed] [Google Scholar]
- Ichikawa H, Jacobowitz DM, Sugimoto T. S100 protein-immunoreactive primary sensory neurons in the trigeminal and dorsal root ganglia of the rat. Brain Res. 1997;748:253–257. doi: 10.1016/s0006-8993(96)01364-9. [DOI] [PubMed] [Google Scholar]
- Kataoka K. Multiple mechanisms and functions of maf transcription factors in the regulation of tissue-specific genes. J Biochem. 2007;141:775–781. doi: 10.1093/jb/mvm105. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, Cooper B, Gannon M, Ray M, MacDonald RJ, Wright CV. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat Genet. 2002;32:128–134. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- Kramer I, Sigrist M, de Nooij JC, Taniuchi I, Jessell TM, Arber S. A role for Runx transcription factor signaling in dorsal root ganglion sensory neuron diversification. Neuron. 2006;49:379–393. doi: 10.1016/j.neuron.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, Westphal H. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci U S A. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecoin L, Rocques N, El-Yakoubi W, Ben Achour S, Larcher M, Pouponnot C, Eychène A. MafA transcription factor identifies the early ret-expressing sensory neurons. Dev Neurobiol. 2010;70:485–497. doi: 10.1002/dneu.20790. [DOI] [PubMed] [Google Scholar]
- Li MZ, Wang JS, Jiang DJ, Xiang CX, Wang FY, Zhang KH, Williams PR, Chen ZF. Molecular mapping of developing dorsal horn-enriched genes by microarray and dorsal/ventral subtractive screening. Dev Biol. 2006;292:555–564. doi: 10.1016/j.ydbio.2006.01.033. [DOI] [PubMed] [Google Scholar]
- Lin JH, Saito T, Anderson DJ, Lance-Jones C, Jessell TM, Arber S. Functionally related motor neuron pool and muscle sensory afferent subtypes defined by coordinate ETS gene expression. Cell. 1998;95:393–407. doi: 10.1016/s0092-8674(00)81770-5. [DOI] [PubMed] [Google Scholar]
- Liu Y, Ma Q. Generation of somatic sensory neuron diversity and implications on sensory coding. Curr Opin Neurobiol. 2011;21:52–60. doi: 10.1016/j.conb.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Enomoto H, Rice FL, Milbrandt J, Ginty DD. Molecular identification of rapidly adapting mechanoreceptors and their developmental dependence on ret signaling. Neuron. 2009;64:841–856. doi: 10.1016/j.neuron.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q. Transcriptional regulation of neuronal phenotype in mammals. J Physiol. 2006;575:379–387. doi: 10.1113/jphysiol.2006.113449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q. Labeled lines meet and talk: population coding of somatic sensations. J Clin Invest. 2010;120:3773–3778. doi: 10.1172/JCI43426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Fode C, Guillemot F, Anderson DJ. Neurogenin1 and neurogenin2 control two distinct waves of neurogenesis in developing dorsal root ganglia. Genes Dev. 1999;13:1717–1728. doi: 10.1101/gad.13.13.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmigère F, Ernfors P. Specification and connectivity of neuronal subtypes in the sensory lineage. Nat Rev Neurosci. 2007;8:114–127. doi: 10.1038/nrn2057. [DOI] [PubMed] [Google Scholar]
- Marmigère F, Montelius A, Wegner M, Groner Y, Reichardt LF, Ernfors P. The Runx1/AML1 transcription factor selectively regulates development and survival of TrkA nociceptive sensory neurons. Nat Neurosci. 2006;9:180–187. doi: 10.1038/nn1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi R, Kriks S, Cordes R, Gossler A, Ma Q, Goulding M. Ascl1 and Gsh1/2 control inhibitory and excitatory cell fate in spinal sensory interneurons. Nat Neurosci. 2006;9:770–778. doi: 10.1038/nn1706. [DOI] [PubMed] [Google Scholar]
- Müller T, Brohmann H, Pierani A, Heppenstall PA, Lewin GR, Jessell TM, Birchmeier C. The homeodomain factor lbx1 distinguishes two major programs of neuronal differentiation in the dorsal spinal cord. Neuron. 2002;34:551–562. doi: 10.1016/s0896-6273(02)00689-x. [DOI] [PubMed] [Google Scholar]
- Nakamura S, Senzaki K, Yoshikawa M, Nishimura M, Inoue K, Ito Y, Ozaki S, Shiga T. Dynamic regulation of the expression of neurotrophin receptors by Runx3. Development. 2008;135:1703–1711. doi: 10.1242/dev.015248. [DOI] [PubMed] [Google Scholar]
- Patel KN, Dong X. An itch to be scratched. Neuron. 2010;68:334–339. doi: 10.1016/j.neuron.2010.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattyn A, Morin X, Cremer H, Goridis C, Brunet JF. Expression and interactions of the two closely related homeobox genes Phox2a and Phox2b during neurogenesis. Development. 1997;124:4065–4075. doi: 10.1242/dev.124.20.4065. [DOI] [PubMed] [Google Scholar]
- Qian Y, Shirasawa S, Chen CL, Cheng L, Ma Q. Proper development of relay somatic sensory neurons and D2/D4 interneurons requires homeobox genes Rnx/Tlx-3 and Tlx-1. Genes Dev. 2002;16:1220–1233. doi: 10.1101/gad.982802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebelo S, Reguenga C, Lopes C, Lima D. Prrxl1 is required for the generation of a subset of nociceptive glutamatergic superficial spinal dorsal horn neurons. Dev Dyn. 2010;239:1684–1694. doi: 10.1002/dvdy.22305. [DOI] [PubMed] [Google Scholar]
- Reed-Geaghan EG, Maricich SM. Peripheral somatosensation: a touch of genetics. Curr Opin Genet Dev. 2011;21:240–248. doi: 10.1016/j.gde.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexed B. The cytoarchitectonic organization of the spinal cord in the rat. J Comp Neurol. 1952;96:415–466. doi: 10.1002/cne.900960303. [DOI] [PubMed] [Google Scholar]
- Ring BZ, Cordes SP, Overbeek PA, Barsh GS. Regulation of mouse lens fiber cell development and differentiation by the Maf gene. Development. 2000;127:307–317. doi: 10.1242/dev.127.2.307. [DOI] [PubMed] [Google Scholar]
- Roberts CW, Shutter JR, Korsmeyer SJ. Hox11 controls the genesis of the spleen. Nature. 1994;368:747–749. doi: 10.1038/368747a0. [DOI] [PubMed] [Google Scholar]
- Ross SE, Mardinly AR, McCord AE, Zurawski J, Cohen S, Jung C, Hu L, Mok SI, Shah A, Savner EM, Tolias C, Corfas R, Chen S, Inquimbert P, Xu Y, McInnes RR, Rice FL, Corfas G, Ma Q, Woolf CJ, et al. Loss of inhibitory interneurons in the dorsal spinal cord and elevated itch in Bhlhb5 mutant mice. Neuron. 2010;65:886–898. doi: 10.1016/j.neuron.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottkamp CA, Lobur KJ, Wladyka CL, Lucky AK, O'Gorman S. Pbx3 is required for normal locomotion and dorsal horn development. Dev Biol. 2008;314:23–39. doi: 10.1016/j.ydbio.2007.10.046. [DOI] [PubMed] [Google Scholar]
- Saito T, Greenwood A, Sun Q, Anderson DJ. Identification by differential RT-PCR of a novel paired homeodomain protein specifically expressed in sensory neurons and a subset of their CNS targets. Mol Cell Neurosci. 1995;6:280–292. doi: 10.1006/mcne.1995.1022. [DOI] [PubMed] [Google Scholar]
- Sakai M, Imaki J, Yoshida K, Ogata A, Matsushima-Hibaya Y, Kuboki Y, Nishizawa M, Nishi S. Rat maf related genes: specific expression in chondrocytes, lens and spinal cord. Oncogene. 1997;14:745–750. doi: 10.1038/sj.onc.1200869. [DOI] [PubMed] [Google Scholar]
- Scott A, Hasegawa H, Sakurai K, Yaron A, Cobb J, Wang F. Transcription factor short stature homeobox 2 is required for proper development of tropomyosin-related kinase B-expressing mechanosensory neurons. J Neurosci. 2011;31:6741–6749. doi: 10.1523/JNEUROSCI.5883-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasawa S, Arata A, Onimaru H, Roth KA, Brown GA, Horning S, Arata S, Okumura K, Sasazuki T, Korsmeyer SJ. Rnx deficiency results in congenital central hypoventilation. Nat Genet. 2000;24:287–290. doi: 10.1038/73516. [DOI] [PubMed] [Google Scholar]
- Todd AJ. Neuronal circuitry for pain processing in the dorsal horn. Nat Rev Neurosci. 2010;11:823–836. doi: 10.1038/nrn2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd AJ, Hughes DI, Polgár E, Nagy GG, Mackie M, Ottersen OP, Maxwell DJ. The expression of vesicular glutamate transporters VGLUT1 and VGLUT2 in neurochemically defined axonal populations in the rat spinal cord with emphasis on the dorsal horn. Eur J Neurosci. 2003;17:13–27. doi: 10.1046/j.1460-9568.2003.02406.x. [DOI] [PubMed] [Google Scholar]
- Willis W, Coggeshall R, editors. Sensory mechanisms of the spinal cord. New York: Plenum; 1991. [Google Scholar]
- Xu Y, Lopes C, Qian Y, Liu Y, Cheng L, Goulding M, Turner EE, Lima D, Ma Q. Tlx1 and Tlx3 coordinate specification of dorsal horn pain-modulatory peptidergic neurons. J Neurosci. 2008;28:4037–4046. doi: 10.1523/JNEUROSCI.4126-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Cvekl A. Large Maf transcription factors: cousins of AP-1 proteins and important regulators of cellular differentiation. Einstein J Biol Med. 2007;23:2–11. doi: 10.23861/ejbm20072347. [DOI] [PMC free article] [PubMed] [Google Scholar]