Abstract
Temporal restriction of feeding can entrain circadian behavioral and physiological rhythms in mammals. These changes in biological rhythms are postulated to be brought about by a putative food-entrainable oscillator (FEO) that is independent of the suprachiasmatic nucleus (SCN). However, the anatomical substrates and molecular machinery of FEO remain elusive. We report here that mice with a nervous system-specific deletion of Bmal1, an essential clock component, had a marked deficit in entrainment of locomotor activity by periodic feeding, accompanied by reduced food intake and subsequent loss of body weight. These mice exhibited a nearly normal light-entrainable activity rhythm driven by the SCN, because deletion of the Bmal1 gene in the SCN was only partial. These findings suggest that an SCN-independent FEO in the nervous system requires Bmal1 and plays a critical role in adaptation of circadian locomotor activity and food intake to periodic feeding.
Introduction
When food availability is restricted to a single period scheduled at a fixed time of the day [restricted feeding (RF)], animals adapt to this condition within a few days by feeding during the period of food availability and increasing food-seeking activity in the preceding hours [food-anticipatory activity (FAA)] (Davidson, 2006). Such anticipatory behavior is often accompanied by an increase in body temperature, adrenal corticosterone secretion, and gastrointestinal motility. These RF-induced biological rhythms persist even when the suprachiasmatic nucleus (SCN) is ablated, indicating the existence of a circadian oscillator that is separate from and independent of the SCN. However, the anatomical substrate(s) and molecular machinery of the so-called “food-entrainable oscillator (FEO)” remains to be unknown. Moreover, despite the well characterized circadian nature of FAA, the involvement of canonical clock genes remains controversial (Challet et al., 2009). For example, one study demonstrated that arrhythmic Bmal1−/− mice also lack food-entrainable circadian rhythms (Fuller et al., 2008), while another two studies reported normal FAA of the same mouse line (Pendergast et al., 2009; Storch and Weitz, 2009).
These contradictory results may stem from the facts that Bmal1−/− mice appear particularly vulnerable to restricted feeding schedules and exhibit markedly reduced mobility, likely due in part to impaired glucose homeostasis (Rudic et al., 2004; Lamia et al., 2008; Marcheva et al., 2010) and the defects of joints and skeletal muscles in these mice (Bunger et al., 2005; Kondratov et al., 2006; McDearmon et al., 2006). These confounding factors make it difficult to evaluate food-entrainable circadian rhythms in Bmal1−/− mice. Additionally, even whether FAA exhibited by these mice is truly circadian has not been confirmed.
Therefore, we examined the food-entrainable activity rhythm of mice lacking Bmal1 specifically in the nervous system to elucidate whether Bmal1, an essential molecular component of circadian clocks, is indispensable or not for FAA, as well as to test whether an FEO resides in the nervous system.
Materials and Methods
Animals.
Bmal1wt/fl mice (Storch et al., 2007) and Nes-Cre transgenic mice (Tronche et al., 1999) were obtained from the Jackson Laboratory. These mice were backcrossed with C57BL/6J at least seven and six times, respectively. Bmal1nl allele was generated by crossing Bmal1wt/fl mice with Synapsin I-Cre transgenic mice (provided by Dr. Jamey D. Marth of University of California, Santa Barbara, CA), in which Cre is expressed in male germline cells in addition to neurons (Rempe et al., 2006). Male mice, aged 11–19 weeks, were used. All experimental procedures involving animals were approved by the appropriate institutional animal care and use committee of Kanazawa University.
Behavioral studies.
Spontaneous locomotor activity and food intake were recorded by infrared motion sensors to monitor general cage activity (O'Hara). Actogram and χ2 periodogram analyses were performed using ClockLab (Actimetrics). RF experiments were performed according to the method of Storch and Weitz (2009). Regular chow was provided in a feeder attached on the side of the cage.
In situ hybridization.
In situ hybridization was performed as described previously (Mieda et al., 2006). Per1 and Per2 expression levels were quantified using “Color Range” command of Photoshop (Adobe) as follows. First, the typical color of in situ hybridization staining (dark blue) was selected in a representative image. Then the number of pixels displaying colors similar to the selected color was counted in the region of interest for each image. Calculated values were expressed as percentage of the mean value from control mice.
Quantification of Cre-mediated recombination of Bmal1fl allele.
Genomic DNA was extracted from tissues dissected from sections prepared for in situ hybridization. Bmal1 and β-actin genomic DNA fragments were amplified and quantified by real-time PCR using a LightCycler 480 (Roche). Primers for Bmal1 were designed to amplify the Bmal1wt and Bmal1fl alleles but not the Bmal1nl allele. Copy number of Bmal1 alleles was normalized to that of β-actin gene. The primer sequences and Universal ProbeLibrary Probes (Roche) were as follows: Bmal1 upstream primer, 5′-TTTTGGGTGCTGGGTTATTC-3′; Bmal1 downstream primer, 5′-CAGGTTTGGTGGATGTGGAT-3′; Bmal1 probe, #69; β-actin upstream primer, 5′-AGCCATGTACGTAGCCATCC-3′; β-actin downstream primer, 5′-TCACAATGCCTGTGGTACG-3′; β-actin probe, #56.
Results
We generated mice lacking the Bmal1 gene specifically in the nervous system by breeding mice with a conditional Bmal1 allele (Bmal1fl) (Storch et al., 2007) with mice carrying a Cre recombinase transgene under control of the Nestin promoter, which acts selectively in the central and peripheral nervous systems (Nes-Cre) (Tronche et al., 1999). To improve the efficiency of deletion of Bmal1fl alleles, we generated Bmal1fl/nl;Nes-Cre mice, hereafter designated N-Bmal1−/− mice, which carry one flox (fl) and one null (nl) allele so that Cre-mediated inactivation of only one allele is sufficient to delete Bmal1 in a given cell. Data were also collected from littermates with the genotypes Bmal1fl/nl and Bmal1wt/fl;Nes-Cre (wt). Since these two genotypes behaved similarly, both were grouped together and referred to as control mice.
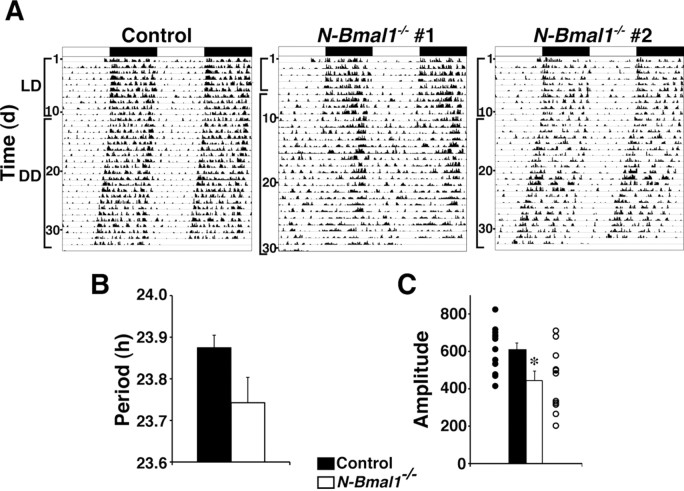
N-Bmal1−/− mice were not behaviorally arrhythmic under constant dark (DD) condition (Fig. 1A,C). Among 11 mice examined, 4 initially maintained daily pattern of spontaneous locomotor activity when they were released into DD, then their rhythmicity gradually attenuated, although they still preserved significant circadian periodicity during the 3 weeks in DD (N-Bmal1−/− #1). The remaining 7 appeared almost normal (N-Bmal1−/− #2). The mean free-running period during the last 10 d in DD did not differ between control and N-Bmal1−/− mice (Fig. 1B), whereas the mean amplitude of circadian periodicity during the same period was slightly but significantly lower in N-Bmal1−/− mice compared with controls (Fig. 1C). Considering the pivotal role of SCN in the regulation of light-entrainable circadian rhythms, these results suggest that most N-Bmal1−/− mice have residual cells in the SCN that retain a Bmal1fl allele.
Figure 1.
SCN-driven light-entrainable activity rhythm is only slightly attenuated in N-Bmal1−/− mice. A, Representative locomotor activity of one control and two N-Bmal1−/− mice. Animals were initially housed in LD conditions and then transferred to DD. Figure shows typical examples of activity rhythm of N-Bmal1−/− mice. Timing of LD cycle is indicated by horizontal bar above each record; dark bars indicate lights off, and open bars indicate lights on. B, C, Periodogram estimates of free-running period (B) and circadian amplitude (C, power from periodogram analyses) of control (black bars) and N-Bmal1−/− mice (white bars) during the last 10 d in DD. In C, value of each individual mouse is dot-plotted. *p < 0.05 by unpaired Student's t test. Values are mean ± SEM (n = 12 for control, n = 11 for N-Bmal1−/− mice).
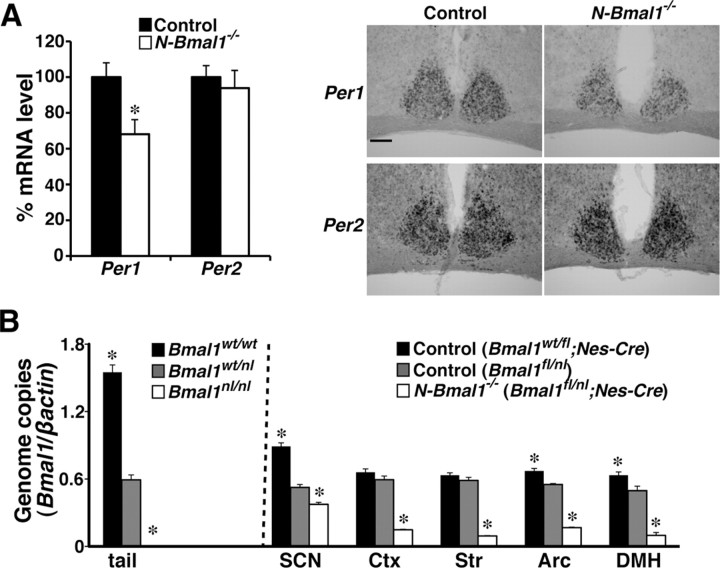
We then used in situ hybridization to evaluate the state of the molecular clock in the SCN of N-Bmal1−/− mice (Fig. 2A). We found that Per1 mRNA level in the SCN of N-Bmal1−/− mice at circadian time (CT)8, when its expression is peak, on the first day in DD was significantly decreased but only by 30.2% compared with that of control mice. Per2 mRNA level in the SCN was not significantly different between two groups. To estimate the efficiency of Cre-mediated recombination, we further quantified Bmal1wt and Bmal1fl alleles in genomic DNA extracted from control and N-Bmal1−/− mice, by real-time PCR (Fig. 2B). As expected, in the SCN, 71.3% of Bmal1fl alleles remained nonrecombined in N-Bmal1−/− (Bmal1fl/nl;Nes-Cre) mice compared with Bmal1fl/nl control mice. These results suggest that ∼70% of SCN cells retain a Bmal1fl allele in N-Bmal1−/− mice, which is likely to underlie the fact that N-Bmal1−/− mice showed merely slight impairment of circadian rhythmicity in DD.
Figure 2.
Bmal1 function is mostly retained in SCN of N-Bmal1−/− mice. A, Per1 and Per2 mRNA expression levels in SCN of control mice (black bar) and N-Bmal1−/− mice (white bar) at CT8 determined by in situ hybridization. *p < 0.05 by unpaired Student's t test. Values are mean ± SEM (n = 5). Representative images are shown on the right. Scale bar, 200 μm. B, Efficiency of Cre-mediated recombination of Bmal1fl allele. Amounts of Bmal1wt and Bmal1fl alleles were quantified by real-time PCR and normalized to those of β-actin gene. A pilot analysis of genomic DNA extracted from tails (shown in leftmost field) of Bmal1w/w (black bar), Bmal1wt/nl (gray bar), and Bmal1nl/nl (white bar) mice confirmed the specificity and quantitative performance of amplification. Genomic DNA extracted from SCN, cerebral cortex, striatum, arcuate hypothalamic nucleus (Arc), and DMH of Bmal1wt/fl;Nes-Cre (black bars) and Bmal1fl/nl (gray bars) control mice and N-Bmal1−/− mice (white bars) was subjected to similar quantitative PCR. *p < 0.05 by unpaired Student's t test vs Bmal1wt/nl mice (tail) or Bmal1fl/nl mice (brain tissues). Values are mean ± SEM (n = 3 for tail analysis, n = 4 for Bmal1wt/fl;Nes-Cre mice, n = 3 for Bmal1fl/nl mice, n = 6 for N-Bmal1−/− mice). Ctx, Cerebral cortex; Str, striatum.
Compared with the SCN, Cre-mediated recombination of the Bmal1fl allele was more efficient in other regions of the brain examined (Fig. 2B). As compared with Bmal1fl/nl control mice, 15∼30% of Bmal1fl alleles remained nonrecombined in N-Bmal1−/− mice in the cerebral cortex, striatum, arcuate hypothalamic nucleus, and dorsomedial hypothalamic nucleus (DMH). Thus, the efficiency of Cre-mediated recombination of the Bmal1fl allele varies among brain regions in N-Bmal1−/− mice. Since the FEO has been supposed to reside outside the SCN, where deletion of Bmal1fl alleles may be close to complete, we decided to further examine food-entrainable activity rhythm of N-Bmal1−/− mice.
Daily total activity in 12 h light/dark (LD) condition was not significantly different between control and N-Bmal1−/− mice (control mice, 19,327 ± 2374 counts, n = 8; N-Bmal1−/− mice, 13,963 ± 2322 counts, n = 6; p = 0.141 by unpaired Student's t test). Similarly, body weight was comparable between control and N-Bmal1−/− mice at 15 weeks of age (control mice, 26.7 ± 0.9 g, n = 11; N-Bmal1−/− mice, 25.4 ± 1.0 g, n = 6; p = 0.367 by unpaired Student's t test). Thus, N-Bmal1−/− mice mostly avoided the drawbacks of Bmal1−/− mice when studying food-entrainable circadian rhythms, such as extreme hypoactivity and growth retardation.
In accordance with the method of Storch and Weitz (2009), we adopted a gentle RF paradigm, which is reported to avoid vulnerability of Bmal1−/− mice to a sudden reduction in the duration of food availability. Consistently, no N-Bmal1−/− mice, as well as control mice, died or became sluggish in the current study. Locomotor activity in control mice started to exhibit a significant increase during the 2 h preceding food availability (FAA) on day 4 under RF, which reached a plateau on day 8 (Fig. 3A,B). In clear contrast, acquisition of FAA was markedly delayed in N-Bmal1−/− mice. They did not exhibit significant FAA until day 13 (Fig. 3A,B). Emergence of FAA was associated with a reduction of daily activity in the dark period, which was also delayed in N-Bmal1−/− mice compared with control mice (Fig. 3C). Taking these findings together, entrainment of circadian activity rhythm by feeding was markedly attenuated in N-Bmal1−/− mice compared with controls, suggesting that Bmal1 expression in the nervous system is critical for prompt and efficient entrainment of activity rhythm by feeding.
Figure 3.
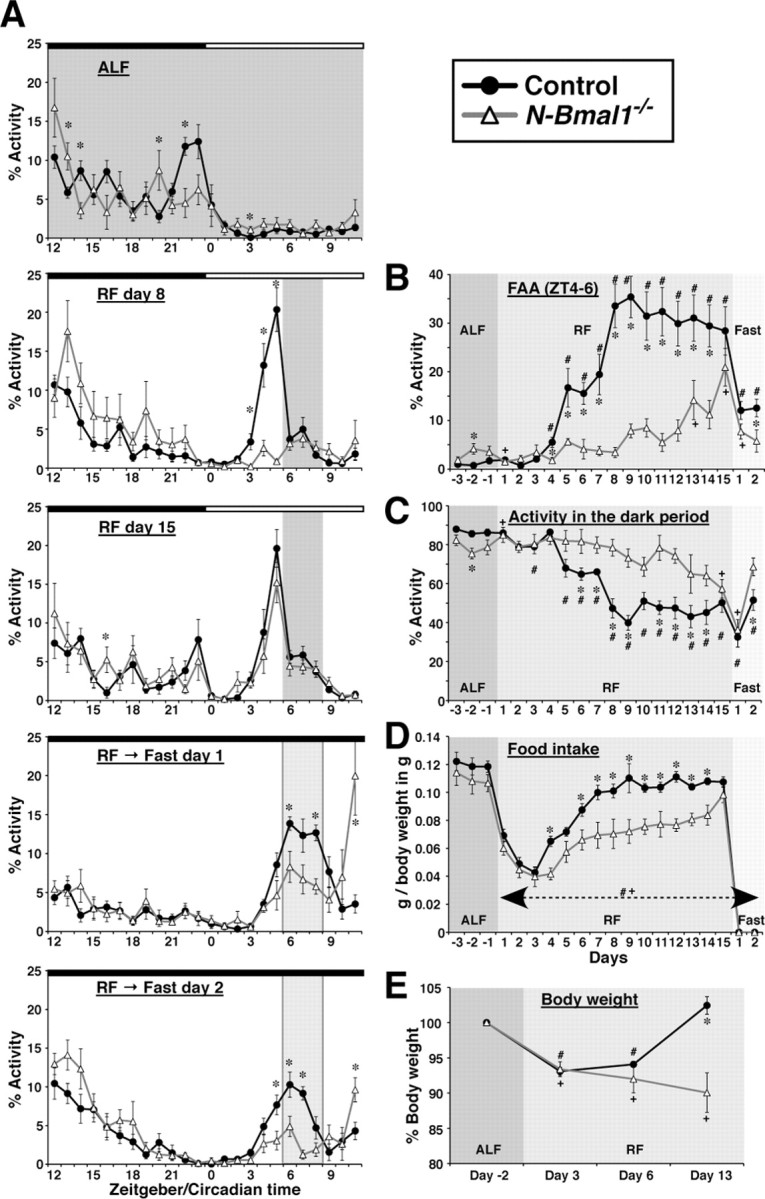
Entrainment of circadian activity rhythm and food intake by periodic feeding is markedly delayed and attenuated in N-Bmal1−/− mice. A, Hourly plots of locomotor activity of control and N-Bmal1−/− mice under ALF or on the indicated day under RF or fasting following RF. Activity counts are expressed as percentage of daily total. Note that FAA during fasting days appears to be reduced substantially compared with day 15 under RF due to this normalization. The period of food availability and expected period of food availability under fasting are shaded in dark and light gray, respectively. Black and white bars on top of each graph represent times of lights off and on, respectively. B, Time course of development of FAA in control and N-Bmal1−/− mice. The daily percentage of locomotor activity allocated to a 2 h time interval, zeitgeber time (ZT)4–6, is shown. C, Time course of reduction of activity during dark period. D, Time course of change in daily food intake. Data shown are normalized to body weight. E, Time course of change in body weight. Data are expressed as percentage of body weight under ALF. *p < 0.05 control vs N-Bmal1−/− mice at each time point by unpaired Student's t test; #p < 0.05 vs mean value under ALF in control mice; +p < 0.05 vs mean value under ALF in N-Bmal1−/− mice by paired Student's t test. For B–E, effects of genotype and time point, as well as interaction of genotype and time point, were all significant (p < 0.05) by two-way repeated-measures ANOVA. Values are mean ± SEM (n = 8 for control mice, closed circles; n = 6 for N-Bmal1−/− mice, open triangles).
To evaluate the circadian nature of daily rhythmicity induced by RF, we further recorded the activity of mice for 2 d, immediately following 15 d of RF, under food deprivation and constant darkness, where the effects of both light and feeding as zeitgebers were eliminated (Fig. 3A). On these days, control mice showed increased activity during the FAA period, sustained increased activity during the expected feeding period, and then reduced activity to the level in the pre-FAA period. Although N-Bmal1−/− mice also exhibited an increase in activity during both the food anticipation period and expected feeding period, the increases were significantly attenuated compared with those in control mice, especially on Fast day 2 (Fig. 3A,B). More importantly, in contrast to control mice, N-Bmal1−/− mice showed a further increase in activity after the expected feeding period (Fig. 3A). Activity counts during the 3 h after food availability (CT9–12) was 37.8 ± 9.0% and 163.7 ± 40.6% of those during the expected feeding period (CT6–9) in control and N-Bmal1−/− mice, respectively, on Fast day 1, and 40.6 ± 14.1% and 224.3 ± 60.3%, respectively, on Fast day 2. These results suggest that the apparent FAA in N-Bmal1−/− mice, whose acquisition was delayed under RF compared with controls, is driven largely by noncircadian mechanisms.
Daily food intake did not differ significantly between control and N-Bmal1−/− mice under ad libitum feeding (ALF) (Fig. 3D). Under RF until day 3, there was a similar reduction in food intake to ∼30% of ALF intake (Fig. 3D). This result suggests that control and N-Bmal1−/− mice respond similarly to acute fasting and have comparable desire for food. However, while control mice rapidly restored their food intake to reach 80∼90% of ALF intake on day 7 to day 15, N-Bmal1−/− mice failed to efficiently restore their food intake and ate ∼30% less than control mice until day 15 (Fig. 3D). Accordingly, when control mice had restored body weight to the state under ALF on day 13, N-Bmal1−/− mice continued to lose body weight (Fig. 3E). These results suggest that Bmal1 expression in the nervous system is essential for metabolic aspects of adaptation to RF.
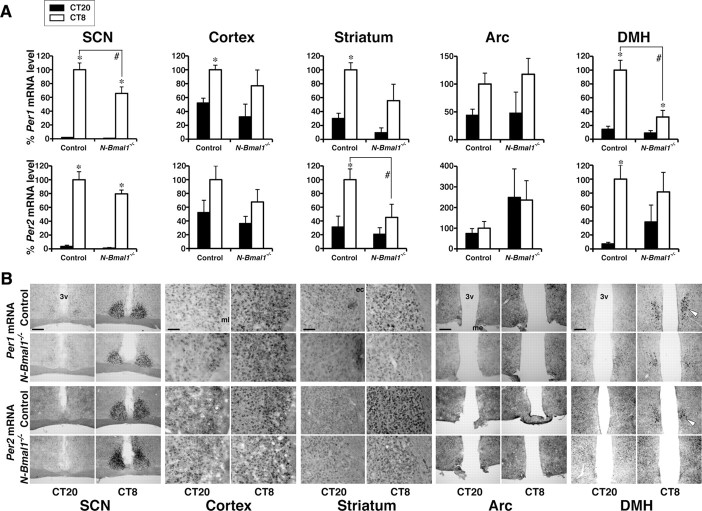
We next evaluated the states of the molecular clocks in the brain of N-Bmal1−/− mice by in situ hybridization. Previous studies demonstrated that RF entrains and shifts the circadian expression of clock genes in several non-SCN regions of the brain and peripheral tissues, whereas rhythmicity in the SCN remains phase-locked to the external light/dark cycle (Damiola et al., 2000; Stokkan et al., 2001; Wakamatsu et al., 2001; Mieda et al., 2006; Moriya et al., 2009). Thus, we next studied Per1 and Per2 mRNA expression rhythms entrained by feeding in several brain regions of N-Bmal1−/− and control mice using in situ hybridization. Mice were housed under RF for 7 d in LD condition, and then fasted on the following day in DD. We collected brains on the fasting day at CT20 and CT8, when Per1 and Per2 mRNA expression is reported to show trough and peak levels, respectively, in many brain areas. As previously reported, Per1 and Per2 mRNA levels were higher at CT8 than at CT20 in the cerebral cortex, striatum, and the compact part of the DMH of control mice under this condition (Fig. 4). These diurnal patterns of expression were observed also in N-Bmal1−/− mice. However, diurnal oscillation of Per expression in the brain was attenuated in N-Bmal1−/− mice compared with that in control mice, especially in the striatum and the compact part of the DMH. These results suggest that food-entrainment of Per1 and Per2 expression rhythms are attenuated, at least in several regions, in the brain of N-Bmal1−/− mice, which may be responsible for the deficit in food-entrainble circadian rhythms of these mice.
Figure 4.
Food-entrainable rhythms of Per1 and Per2 mRNA expression are attenuated in the brain of N-Bmal1−/− mice. A, Per1 and Per2 mRNA levels. Brains from control mice (black bars) and N-Bmal1−/− mice (white bars) were collected at CT20 and CT8 on the first day of food deprivation in DD following 7 d under RF in LD. Per1 and Per2 mRNA levels were determined by in situ hybridization. Data are expressed as percentage of mRNA level at CT8 in each region in control mice. *p < 0.05 for effect of sampling time point in each genotype and #p < 0.05 for effect of genotype by two-way factorial ANOVA followed by post hoc unpaired Student's t test. Values are mean ± SEM (n = 7 and 5 for control and N-Bmal1−/− mice, respectively at CT8, n = 4 and 3 at CT20). B, Representative images. Expression in the compact part of the DMH is indicated by white arrowhead. ec, External capsule; me, medial eminence; ml, molecular layer of the cerebral cortex; 3v, third ventricle. Scale bars, 200 μm.
Discussion
In the present study, we have shown that Bmal1 expression in the nervous system is necessary for prompt entrainment of circadian activity rhythm by temporally restricted feeding. Our observations further suggest that the locus of FEO(s), or at least those regulating locomotor activity and food intake, is in the nervous system outside the SCN, which complements classical experiments showing that lesions of the SCN do not abolish food-entrainable circadian rhythms. In addition, the absence of FEO in the nervous system impairs adaptation of food intake and regulation of body weight under RF, underscoring the physiological importance of the FEO and food-entrainable circadian rhythms to temporally coordinate food-seeking activity and food consumption.
Although they failed to show entrainment of locomotor activity to feeding in a prompt manner, N-Bmal1−/− mice still exhibited significant FAA after 2 weeks of RF. FAA in N-Bmal1−/− mice, as well as feeding-entrained rhythms of Per1 and Per2 mRNA in their brain, might be driven by residual cells in the nervous system having nonrecombined functional Bmal1fl alleles (i.e., somatic mosaicism), by Bmal2 (a paralog of Bmal1) (Shi et al., 2010), and/or by Bmal1 outside the nervous system. Circadian mechanisms without canonical clock machinery may be also involved (Hastings et al., 2008). This delayed component of FAA, however, seems to include noncircadian mechanisms, since N-Bmal1−/− mice responded to food deprivation following RF with a further increase in locomotor activity after the expected feeding period, in clear contrast to the response of control mice to the same condition. The noncircadian nature of FAA exhibited by Bmal1−/− mice was also manifested clearly by the fact that it can be entrained to periodic food availability with periods shorter than the circadian range, such as 15 h, to which FAA of wild-type mice cannot be entrained (W. Nakamura, personal communication).
It is well known that animals respond to food deprivation by increased locomotor activity (Yamanaka et al., 2003), and this fasting-induced hyperactivity is likely to be involved in the noncircadian mechanisms of FAA. This view is also supported by our observation that N-Bmal1−/− mice were losing body weight when they started to exhibit FAA. Since fasting-induced hyperactivity should critically depend on the internal status of energy balance and Bmal1−/− mice demonstrate impaired glucose homeostasis (Rudic et al., 2004; Lamia et al., 2008; Marcheva et al., 2010) and a considerable decrease of adipose tissue (Kondratov et al., 2006), FAA observed in these mice may be a direct response to fasting that is enhanced compared with wild-type mice, as discussed by Pendergast et al. (2009). Possible contributions of homeostatic and other cues to FAA have been also argued thoroughly by Fuller et al. (2009).
These findings suggest that at least two independent elements are likely to be involved in the expression of FAA; (1) A clock gene-dependent mechanism, which resides in the nervous system, but independent of the SCN, (2) A clock gene-independent mechanism, which could involve a machinery that responds to fasting and other homeostatic information, and/or neural mechanisms involved in subjective time perception.
Mammalian circadian organization at the whole-organism level has been thought to be multioscillatory in nature; multiple circadian clocks exist in almost all neural and non-neural tissues and are entrained to the master clock in the SCN under ALF. Thus, reciprocal interactions among multiple clocks throughout the body including the SCN seem very important for determining temporal patterns of animals' behavior and physiology. Importantly, suppressive effects of SCN-derived signals on FAA have been suggested (Acosta-Galvan et al., 2011). In Bmal1−/− mice, the absence of all circadian clocks throughout the body may enhance the emergence of FAA regulated by noncircadian mechanisms. In contrast, N-Bmal1−/− mice retain circadian clocks in peripheral tissues and the SCN, which may force the noncircadian FAA to play only a minor role in the adaptation of activity rhythm to RF. Thus, as discussed by Pendergast et al. (2009), Bmal1 may be an important molecular component of the wild-type FEO, while in the absence of Bmal1 the mechanism regulating the expression of FAA becomes an interval timer.
In conclusion, Bmal1-containing circadian clocks in the nervous system, which reside outside the SCN, are important for adaptation of not only activity rhythm but also food intake and body weight regulation to periodic feeding. Our observations justify approaches based on canonical clock genes to find anatomical substrates of the FEO. In addition, N-Bmal1−/− mice will provide an excellent model to study the roles of FEO in the nervous system in the regulation of appetite and physiology of peripheral tissues.
Footnotes
This study was supported in part by a Career Development Award from the Human Frontier Science Program, grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan, and the Cabinet Office, Government of Japan through its “Funding Program for Next Generation World-Leading Researchers.” We thank S. Sawada and E. Hasegawa for technical assistance, J. D. Marth for Synapsin I-Cre transgenic mice, and W. Nakamura for sharing his unpublished observations.
References
- Acosta-Galvan G, Yi CX, van der Vliet J, Jhamandas JH, Panula P, Angeles-Castellanos M, Del Carmen Basualdo M, Escobar C, Buijs RM. Interaction between hypothalamic dorsomedial nucleus and the suprachiasmatic nucleus determines intensity of food anticipatory behavior. Proc Natl Acad Sci U S A. 2011;108:5813–5818. doi: 10.1073/pnas.1015551108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunger MK, Walisser JA, Sullivan R, Manley PA, Moran SM, Kalscheur VL, Colman RJ, Bradfield CA. Progressive arthropathy in mice with a targeted disruption of the Mop3/Bmal-1 locus. Genesis. 2005;41:122–132. doi: 10.1002/gene.20102. [DOI] [PubMed] [Google Scholar]
- Challet E, Mendoza J, Dardente H, Pévet P. Neurogenetics of food anticipation. Eur J Neurosci. 2009;30:1676–1687. doi: 10.1111/j.1460-9568.2009.06962.x. [DOI] [PubMed] [Google Scholar]
- Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson AJ. Search for the feeding-entrainable circadian oscillator: a complex proposition. Am J Physiol Regul Integr Comp Physiol. 2006;290:R1524–R1526. doi: 10.1152/ajpregu.00073.2006. [DOI] [PubMed] [Google Scholar]
- Fuller PM, Lu J, Saper CB. Differential rescue of light- and food-entrainable circadian rhythms. Science. 2008;320:1074–1077. doi: 10.1126/science.1153277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller PM, Lu J, Saper CB. Standards of evidence in chronobiology: a response. J Circadian Rhythms. 2009;7:9. doi: 10.1186/1740-3391-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings MH, Maywood ES, O'Neill JS. Cellular circadian pacemaking and the role of cytosolic rhythms. Curr Biol. 2008;18:R805–R815. doi: 10.1016/j.cub.2008.07.021. [DOI] [PubMed] [Google Scholar]
- Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev. 2006;20:1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci U S A. 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH, Ivanova G, Omura C, Mo S, Vitaterna MH, Lopez JP, Philipson LH, Bradfield CA, Crosby SD, JeBailey L, Wang X, Takahashi JS, Bass J. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466:627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDearmon EL, Patel KN, Ko CH, Walisser JA, Schook AC, Chong JL, Wilsbacher LD, Song EJ, Hong HK, Bradfield CA, Takahashi JS. Dissecting the functions of the mammalian clock protein BMAL1 by tissue-specific rescue in mice. Science. 2006;314:1304–1308. doi: 10.1126/science.1132430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieda M, Williams SC, Richardson JA, Tanaka K, Yanagisawa M. The dorsomedial hypothalamic nucleus as a putative food-entrainable circadian pacemaker. Proc Natl Acad Sci U S A. 2006;103:12150–12155. doi: 10.1073/pnas.0604189103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya T, Aida R, Kudo T, Akiyama M, Doi M, Hayasaka N, Nakahata N, Mistlberger R, Okamura H, Shibata S. The dorsomedial hypothalamic nucleus is not necessary for food-anticipatory circadian rhythms of behavior, temperature or clock gene expression in mice. Eur J Neurosci. 2009;29:1447–1460. doi: 10.1111/j.1460-9568.2009.06697.x. [DOI] [PubMed] [Google Scholar]
- Pendergast JS, Nakamura W, Friday RC, Hatanaka F, Takumi T, Yamazaki S. Robust food anticipatory activity in Bmal1-deficient mice. PLoS One. 2009;4:e4860. doi: 10.1371/journal.pone.0004860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rempe D, Vangeison G, Hamilton J, Li Y, Jepson M, Federoff HJ. Synapsin I Cre transgene expression in male mice produces germline recombination in progeny. Genesis. 2006;44:44–49. doi: 10.1002/gene.20183. [DOI] [PubMed] [Google Scholar]
- Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB, Fitzgerald GA. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S, Hida A, McGuinness OP, Wasserman DH, Yamazaki S, Johnson CH. Circadian clock gene Bmal1 is not essential; functional replacement with its paralog, Bmal2. Curr Biol. 2010;20:316–321. doi: 10.1016/j.cub.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science. 2001;291:490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- Storch KF, Weitz CJ. Daily rhythms of food-anticipatory behavioral activity do not require the known circadian clock. Proc Natl Acad Sci U S A. 2009;106:6808–6813. doi: 10.1073/pnas.0902063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storch KF, Paz C, Signorovitch J, Raviola E, Pawlyk B, Li T, Weitz CJ. Intrinsic circadian clock of the mammalian retina: importance for retinal processing of visual information. Cell. 2007;130:730–741. doi: 10.1016/j.cell.2007.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronche F, Kellendonk C, Kretz O, Gass P, Anlag K, Orban PC, Bock R, Klein R, Schütz G. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet. 1999;23:99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- Wakamatsu H, Yoshinobu Y, Aida R, Moriya T, Akiyama M, Shibata S. Restricted-feeding-induced anticipatory activity rhythm is associated with a phase-shift of the expression of mPer1 and mPer2 mRNA in the cerebral cortex and hippocampus but not in the suprachiasmatic nucleus of mice. Eur J Neurosci. 2001;13:1190–1196. doi: 10.1046/j.0953-816x.2001.01483.x. [DOI] [PubMed] [Google Scholar]
- Yamanaka A, Beuckmann CT, Willie JT, Hara J, Tsujino N, Mieda M, Tominaga M, Yagami K, Sugiyama F, Goto K, Yanagisawa M, Sakurai T. Hypothalamic orexin neurons regulate arousal according to energy balance in mice. Neuron. 2003;38:701–713. doi: 10.1016/s0896-6273(03)00331-3. [DOI] [PubMed] [Google Scholar]