Abstract
Axotomized central neurons of most invertebrate species demonstrate a strong regenerative capacity, and as such may provide valuable molecular insights and new tools to promote axonal regeneration in injured mammalian neurons. In this study, we identified a novel molluscan protein, caltubin, ubiquitously expressed in central neurons of Lymnaea stagnalis and locally synthesized in regenerating neurites. Reduction of caltubin levels by gene silencing inhibits the outgrowth and regenerative ability of adult Lymnaea neurons and decreases local α- and β-tubulin levels in neurites. Caltubin binds to α- and/or β-tubulin in both Lymnaea and rodent neurons. Expression of caltubin in PC12 cells and mouse cortical neurons promotes NGF-induced axonal outgrowth and attenuates axonal retraction after injury. This is the first study illustrating that a xenoprotein can enhance outgrowth and prevent degeneration of injured mammalian neurons. These results may open up new avenues in molecular repair strategies through the insertion of molecular components of invertebrate regenerative pathways into mammalian neurons.
Introduction
Whereas adult mammalian peripheral neurons retain the capacity to regenerate injured axons, central mammalian neurons are severely limited in their regenerative growth capacity. The inability of central neurons to regenerate injured axons is due to both extrinsic (Richardson et al., 1980) and intrinsic factors (Sun and He, 2010). Extracellular inhibitory molecules [e.g., Nogo, myelin associated glycoprotein, and OMgp (oligodendrocyte myelin glycoprotein)] limit regeneration in the mature CNS (Mukhopadhyay et al., 1994; Goldberg and Barres, 2000; Wang et al., 2002). However, genetic deletion of these inhibitory molecules is insufficient for successful axonal regeneration (Lee et al., 2010). Manipulation of intrinsic neuronal processes can overcome these extrinsic inhibitory signals and promote axon regeneration both in vitro (Qiu et al., 2002; MacGillavry et al., 2009) and in vivo (Gao et al., 2004; Steward et al., 2008). Thus, understanding neuron-intrinsic mechanisms of axon regeneration is pivotal in the development of treatments for traumatic brain and spinal cord injuries, and neurodegenerative disorders.
In contrast to mature mammalian neurons, adult central neurons of some lower vertebrates and most invertebrates spontaneously regenerate following axonal injury (Ferretti et al., 2003; Mladinic et al., 2009). For instance, adult neurons of the freshwater snail Lymnaea stagnalis have the capacity to regenerate their injured axons and reform cell-type specific synapses, both in vitro (Syed et al., 1990; Feng et al., 2000; Spencer et al., 2000) and in vivo (Koert et al., 2001; Lee and Syed, 2004). A promising avenue to promote axon regeneration in the mammalian CNS may, therefore, be to identify unique proteins that mediate the intrinsic regenerative response in lower organisms and to test the growth-promoting properties of these proteins in the mammalian neurons, where they are not normally expressed. Invertebrate neurons and peripheral mammalian neurons share the capacity to locally synthesize proteins in axons and growth cones, and the identification of locally synthesized proteins has provided valuable insights into mechanisms of axon outgrowth and regeneration (Twiss and van Minnen, 2006). Approximately 100–200 local transcripts have been reported in isolated neurites of various neuronal preparations [e.g., in Aplysia sensory neurons (Moccia et al., 2003; Moroz et al., 2006), in squid axoplasm (Capano et al., 1987; Gioio et al., 1994), and in vertebrate axons (Hengst and Jaffrey, 2007; Willis et al., 2007)]. Many of the local transcripts encode cytoskeletal proteins (e.g., microtubule, microfilament, and intermediate filament proteins) (Kaplan et al., 1992; Eng et al., 1999; Vogelaar et al., 2009) or proteins that regulate cytoskeletal dynamics (e.g., Rho and β-thymosin) (Campbell and Holt, 2001; Verma et al., 2005; Wu et al., 2005; van Kesteren et al., 2006), suggesting that regulation of the cytoskeleton through local protein synthesis potentially serves as a conserved mechanism underlying axonal outgrowth and regeneration. Previously, we identified 15 axonally localized mRNAs in Lymnaea pedal A (PeA) neurons (van Kesteren et al., 2006). Of these transcripts, one encoded a novel putative calcium-binding protein, caltubin, with no apparent mammalian orthologs or homologs. Here, we present evidence that local synthesis of caltubin promotes axonal regeneration in Lymnaea PeA neurons by regulating tubulin level. Expression of caltubin in rat PC12 cells and mouse central neurons attenuates axonal degeneration following injury through a conserved interaction with β-tubulin.
Materials and Methods
Animals.
Freshwater snails, L. stagnalis (a hermaphrodite), were kept in water at 20°C under a 12 h light/dark cycle and fed green leaf lettuce twice a week (Gardzinski et al., 2007; Hui et al., 2007). Adult snails with shell lengths of 15–20 mm were used. All experiments were performed according to the guidelines of the Animal Care Committee of the University of Toronto.
Primary cell culture and cell isolation.
Snails were anesthetized with 10% (v/v) Listerine for 10 min as previously described (Syed et al., 1990; Hui et al., 2007). Central ganglia were dissected in snail saline containing the following (in mm): 51.3 NaCl, 1.7 KCl, 4.1 CaCl2, 1.5 MgCl2, pH was adjusted to 7.9 with 1 m HEPES/NaOH, and incubated with 3 mg/ml trypsin (type III; Sigma-Aldrich) for 20 min. Neurons were isolated and placed in poly-l-lysine-coated culture dishes and then maintained in conditioned medium (CM) (Feng et al., 1997) at room temperature up to a week. Anisomycin (A9789; Sigma-Aldrich), a protein synthesis inhibitor that acts by inhibiting peptidyl transferase activity, was added to culture media at various concentrations as previously described (Feng et al., 1997; Roche et al., 2009).
In situ hybridization.
Cultured neurons were fixed in 1% paraformaldehyde/1% acetic acid and permeabilized with 0.5% NP-40. Ganglia were fixed in 1% paraformaldehyde/1% acetic acid and embedded in paraffin, and 7 μm sections were adhered to 0.5% gelatin/0.5% chromalum-coated slides. Digoxigenin-labeled run-off sense and antisense RNA were synthesized from linearized pBluescript plasmids using T3 or T7 RNA polymerase and a dioxigenin-UTP labeling mixture (both from Roche Diagnostics). In situ hybridizations were performed as described previously (Smit et al., 1996).
Real-time quantitative PCR.
PeA neurons were cultured (8–10 cells per dish) for 2 d. RNA was isolated from the transacted neurites and from the corresponding somata separately. After removal of DNA by DNase I treatment, the RNA was random primed with 300 pmol of random hexanucleotides and reverse transcribed into cDNA. Quantitative PCR (qPCR) was performed in triplicate on each cDNA sample using an ABI PRISM 7700 Sequence Detection System (Applied Biosystems) with SYBR Green as the reporter dye. All reactions were performed according to the instructions of the manufacturer. The following primers were used: caltubin (forward, 5′-TTGTAACAGAGGCAGAGCTG-3′; reverse, 5′-AGTTTCCCGTCCTTGTTCG-3′), thymosin (forward, 5′-CTGTGAGCCATCAAGCTGTTG-3′; reverse, 5′-GGTGTACATGGCATCCGATTT-3′), β-tubulin (forward, 5′-AGCCATCCTTCTTGGGTATG-3′; reverse, 5′-AGTTTCCCGTCCTTGTTCG-3′), β-actin (forward, 5′-AGCCATCCTTCTTGGGTATG-3′; reverse, 5′-ATACCTGGGAACATGGTGGT-3′), and mitochondrial 16S rRNA (forward, 5′-ACCTTGACTGTGCTAAGGTAGCATAA-3′; reverse, 5′-CAGTTCTTCCCTATTAATCCGTTCAT-3′). Overall cDNA expression levels per sample were normalized to the expression of 16S rRNA. The relative expression level of the genes of interest was calculated by ΔΔCT methods.
Double-stranded siRNA production and delivery.
Caltubin-specific 27-mer siRNAs were designed using SciTools RNAi Design software (Integrated DNA Technologies). TriFECTa control RNA was used as a control in our experiments (Integrated DNA Technologies). The siRNA sequences are as follows: caltubin siRNA1 (LCa1), 5′-GAAACUUUCACUUGAUGAAUUCAAG-3′; caltubin siRNA3 (LCa3), 5′-CCACGUUCGACGAGGUCAAGAACTA-3′; caltubin siRNA6 (LCa6), 5′-UGGACACUGUCCAGGGUUAUUACAT-3′; caltubin siRNA10 (LCa10), 5′-UUAAUGAUGCCAUUGGUUAGAUATA-3′; TriFECTa control, 5′-UCACAAGGGAGAGAAAGAGAGGAAGGA-3′. Unless otherwise specified, the caltubin siRNA used in the experiments was LCa3.
For whole-animal RNA knockdown experiments, snails were anesthetized with 10% (v/v) Listerine, and 2 μl of 20 μm control siRNA or caltubin siRNA was injected into the head, above the central ganglia using a microliter syringe (Hamilton Company) (Fei et al., 2007; Hui et al., 2007; Lu and Feng, 2011). Ganglia were removed 48 h after injection, and RNA or protein was extracted. The mRNA and protein levels of caltubin were measured using semiquantitative RT-PCR and Western blotting, respectively, to confirm knockdown. For cultured cells, siRNA (7 nm) was directly applied to culture medium, as described previously (Hui et al., 2007; Lu and Feng, 2011).
Neurite outgrowth assays and neurite injury in Lymnaea neurons.
PeA neurons were cultured in CM for 15 h to allow initiation of outgrowth, and cells with initial neurite lengths between 40 and 60 μm were used. Caltubin siRNA (7 nm) or control siRNA (7 nm) was then applied to culture medium (designated as t = 0) and neurite outgrowth was monitored for a period of 30 h using an Olympus inverted microscope (CK X41) with an Olympus C5050 digital camera. Images were analyzed using ImageTool 3.1 software. The net changes in the neurite lengths were measured at various time points following siRNA application (t = 0). Where there were multiple neurites for a given cell, an average length of all neurites belonging to each cell was calculated and counted as one sample. Neurite length was measured from the edge of the soma to the tip of the growth cone.
For neurite injury, PeA cells were cultured in CM for a period of 20 h to allow for neurite outgrowth, and cells with initial neurite lengths between 70 and 90 μm were used. The neurites were severed using glass micropipettes and then immediately treated with either caltubin siRNA (7 nm) or control siRNA (7 nm) (designated as t = 0). Neurite measurements were performed on the proximal (soma-attached) and distal (free) ends of the transected neurites at various time points, as described above.
Immunocytochemistry and confocal microscopy.
Antibodies against caltubin were raised in rat against synthetic peptides corresponding to the N-terminal (MERAFEDVRSQHRD) and C-terminal (DGKLSLDEFKTLYSP) regions of the protein. Cultured PeA neurons were fixed, permeabilized, and incubated with specific primary antibodies (NCS-1: 1:300, BIOMOL, catalog #BML-NL3750-0100; β-tubulin: 1:300, Sigma-Aldrich, catalog #T0198; α-tubulin: 1:300, Sigma-Aldrich, catalog #T6074; β-actin: 1:300, Sigma-Aldrich, catalog #A1978; caltubin: 1:200), as described previously (Gardzinski et al., 2007; Hui et al., 2007). Cells were then incubated with their respective secondary antibody (488 goat anti-rat, Millipore Bioscience Research Reagents; Alexa Fluor 568 goat anti-mouse, Invitrogen; Alexa Fluor 633 goat anti-rabbit, Invitrogen) for 2 h at room temperature. When cells were incubated with secondary antibodies only, no signal was observed under these conditions. Unless otherwise specified, immunocytochemistry of PeA neurons was conducted 6–8 h following siRNA treatment.
Confocal images were acquired using a TCS SL laser confocal microscope (Leica confocal software, version 2.5; build 1347; Leica Microsystems), as described previously (Gardzinski et al., 2007; Hui et al., 2007). Each Z-plane was 0.5 μm. Background fluorescence was determined in three regions not containing cells throughout the entire z-stack. All siRNA-treated cells were imaged using the same magnification and laser settings. To quantify antibody staining, the mean amplitude fluorescence intensities (in arbitrary units) were averaged from sections of the z-stack for each region of interest, and NCS-1 was used as a control against which microfilament intensities were measured. Each bar represents the average protein level from all cells under that condition.
Protein colocalization analysis.
The analysis was conducted to quantify distribution of the proteins labeled with different fluorescence markers and to measure the degree of overlap of the fluorescence signals within the same pixel. Colocalization suggests a high probability of two proteins co-occurring in close proximity. Confocal images of cells double-stained with antibodies for proteins of interest were scanned at a resolution of 1024 × 1024 pixels in the selected regions of interest (ROIs) in the neurites and growth cones of PeA cells, using a 63× oil-immersion lens (Leica Microsystems). Under a zoom of 1, the scan field for each image is 238 × 238 μm2, and thus each pixel is 0.054 μm2. Growth cone and neurite were scanned at a zoom of 2. Each fluorescence signal was imaged separately with corresponding channels, and protein colocalization was estimated based on the degree of overlay between the fluorescence intensities, as measured through analysis of each pixel in the ROI, without pixel saturation. For each image, an ROI was selected in the neurite, growth cone, and an area outside the cell to be used as the control. Using the intensity correlation analysis, a Pearson correlation coefficient was calculated using ImageJ software (http://rsb.info.nih.gov/ij/), where a coefficient close to 1.0 indicates close colocalization, and 0 indicates low probability for colocalization [also see Li et al. (2004) and Colocalization Module 2007, BiolImaging and Optics].
Western blotting.
Protein samples were prepared from snail ganglia, mouse brain, or PC12 cells expressing GFP, caltubin-GFP, or caltubin-myc, in RIPA buffer (50 mm Tris-Cl, pH 7.6, 150 mm NaCl, 2 mm EDTA, 1 mm PMSF plus 1% Igepal CA-630, 0.5% sodium deoxycholate, 1% Triton X-100) with a protease inhibitor mixture (5 μl/100 mg tissue; Sigma-Aldrich), as described previously (Fei et al., 2007; Hui et al., 2007). The samples were centrifuged at 4°C at 13,000 rpm for 15 min. Supernatant was extracted and protein concentrations were measured (Bio-Rad). Membranes were incubated with specific primary antibody against caltubin (1:500), β-tubulin (1:1000), α-tubulin (1:6000; Sigma-Aldrich), GFP (1:1000; Ab6673; Abcam), myc-tag (1:500; 05724; Upstate), or β-actin (1:10,000), overnight at 4°C. The membrane was then incubated with appropriate horseradish peroxidase-conjugated secondary antibody accordingly (1:10,000; Jackson ImmunoResearch Laboratories) for 1 h at room temperature. Antibody-labeled protein bands were visualized using enhanced chemiluminescent reagents (PerkinElmer) and analyzed by exposure to film (HyBlot CL).
Coimmunoprecipitation.
As described previously (Fei et al., 2007), snail ganglion protein (300 μg) or PC12 cell protein (250 μg) was incubated with Protein A/G Plus-Agarose beads (Santa Cruz Biotechnology) for 30 min on ice. The cell lysates were centrifuged at 10,000 × g for 3 min. The supernatants were collected and then incubated with caltubin primary antibody overnight at 4°C. The protein mix was incubated with Protein A/G Plus-Agarose beads and agitated for 2 h on ice and then centrifuged again at 10,000 × g for 1 min. A sample of supernatant was collected for Western blotting to detect the level of the unbound proteins. The pellet was washed with lysis buffer and centrifuged at 500 × g for 1 min, repeated five times. The pellet was placed in 40 μl of Laemmli buffer (eBioscience; with 50 mm DTT). After vortex, the pellet was boiled for 10 min, and then centrifuged at 10,000 × g for 3 min. The supernatant was collected for Western blotting.
Protein affinity purification (pull-down).
For affinity pull-down experiments, GST-caltubin fusion protein was prepared as described previously (Jarvis et al., 2002). Full-length caltubin cDNA was amplified by PCR and cloned into pGEX4T-1. The construct was resequenced to confirm appropriate insertion sites and the absence of spurious PCR generated nucleotide errors. Expression of the transformed plasmid in BL21 (DE3) Bacteria (C2527I; New England Biolabs) was induced by IPTG (isopropyl β-d-1-thiogalactopyranoside) (I5502; Sigma-Aldrich). GST-fusion protein was collected from bacterial lysate and purified using glutathione-Sepharose 4B beads as described by the manufacturer (17-0756-01; GE Healthcare). For affinity purification experiments, the solubilized protein extracts (300 μg of protein) were incubated overnight at 4°C with glutathione-Sepharose beads (GE Healthcare) bound to the indicated GST-fusion proteins (50 μg). Beads were washed three times with 600 μl of PBS containing 0.2% Triton X-100, and bound proteins were eluted with glutathione elution buffer. Eluates were incubated in sample buffer and subjected to 10% SDS-PAGE for Western blot analysis.
PC12 cell culture and transfection.
Rat pheochromocytoma PC12 cells were a kind gift from Dr. Shuzo Sugita (University Health Network, University of Toronto, Toronto, Ontario, Canada), who originally obtained the cells from Dr. Thomas Martin (University of Wisconsin, Madison, WI). The cells were maintained on 35 mm poly-l-lysine-coated dishes in a 37°C maintained in a 9% CO2 incubator in DMEM (HyClone) supplemented with 5% bovine calf serum (HyClone), 5% donor equine (HyClone), 200 U/ml penicillin, 0.2 mg/ml streptomycin, and 0.25 μg/ml Amphotericin B.
PC12 cells were transfected with caltubin-GFP or GFP plasmid (1–3 μg/ml) using Lipofectamine 2000 (Invitrogen). Six hours following transfection, 50 ng/ml nerve growth factor (NGF) (Invitrogen) was added to the culture medium (DMEM). For neurite outgrowth experiments, cells were maintained in culture for 7 d (DIV7). For the neurite injury experiment, neurite crush was induced on day 5 (DIV5) and cells were monitored for 30 h after injury.
Mouse cortical neuron culture and transfection.
All procedures were performed following the animal welfare guidelines at the University of Toronto and were approved by the institutional animal care and use committee. Cortical cultures were prepared from embryonic day 18 mice following the published protocols (Araki et al., 2001; Hares et al., 2011) with modification. Neurons were then dissociated and plated in low density (100,000/ml) on coverslips coated with poly-d-lysine (70–150 kDa; Invitrogen). Cells were maintained in Neurobasal medium with 2% B27 supplement, 1× Penn/Strep, and 2 mm l-glutamine (all from Invitrogen) at 37°C with 5% CO2. Neurons were transfected with caltubin-GFP or GFP plasmid (1–3 μg/ml) using Lipofectamine 2000 (Invitrogen). For neurite outgrowth experiments, cells were transfected on day 1 in culture (DIV1) and maintained for 3 d. For the neurite injury experiment, cells were transfected on DIV3 and neurite were injured on DIV5.
Statistics.
Data are presented as the mean ± SEM. Statistical analysis was performed using SigmaStat 3.0 (Jandel Scientific). The significance of the differences among mean values for multiple experimental groups was tested using one-way ANOVA (Holm–Sidak post hoc comparison). Differences were considered significant when p < 0.05.
Results
Caltubin transcript and protein are expressed in neurites and growth cones of cultured PeA neurons
We first cloned the full-length cDNA of caltubin (Fig. 1A). Sequencing of a full-length caltubin cDNA predicted a 19 kDa protein with four putative calcium-binding EF-hand motifs (Fig. 1A). Sequence alignment with other calcium binding proteins that show highest sequence similarity with caltubin (i.e., the calmodulins and calcineurins) reveals very low overall sequence identity, implicating caltubin as a novel type of putative EF-hand calcium-binding protein (Fig. 1A,B).
Figure 1.

Caltubin is a novel EF-hand-containing protein ubiquitously expressed in CNS neurons of L. stagnalis. A, The full-length sequence of caltubin cDNA predicts a 179 aa protein containing four putative calcium-binding EF-hand motifs (red boxes). GenBank ID numbers are as follows: Neurocalcin Delta: Bt (Bos taurus), 5821829; Hs (Homo sapiens), 1196122; Dr (Danio rerio), 9473279; Dm (Drosophila melanogaster) (isoform B), 2724552; Calmodulin: Ac (Aplysia caligornica), 2130764; Dm (isoform B), 7303487; Hs, 825635; Os (Oryza sativa) (L32), 7532814; Caltubin: Ls (Lymnaea stagnalis). B, Phylogenetic tree of the proteins listed in A. In situ hybridization with a caltubin antisense RNA probe (C1) revealed presence of caltubin mRNA in somata and in neurites (arrows) in the ganglionic neuropil (arrow); or with sense control RNA probes (C2). D, In situ hybridizations of cultured PeA neurons showing differential localization of caltubin mRNA in cell soma and neurites labeled by the antisense RNA probe, but not the sense probe (negative control, inset). Caltubin mRNA clearly accumulates at specific regions within the neurite, as indicated with arrows. Scale bars: D, 20 μm; right inset, 20 μm; left inset, 10 μm. E, Quantification of caltubin mRNA level in neurites. PeA neurons were cultured for 2 d (E1, a representative neuron) and the cell soma was removed (E2), allowing mRNA to be isolated from somata and from neurites separately. E3, Transcript levels of caltubin, β-thymosin (Tmsb), and β-tubulin (Tubb) measured by qPCR analysis are higher in neurites than somata. All mRNA levels were first normalized against 16S rRNA levels, and neuritic mRNA levels were then divided by somatic mRNA levels. The data are presented as mean ± SEM (n). *p < 0.05. F, Western blot of total ganglionic protein lysate showing that the N-terminal caltubin antibody recognizes a single protein with a molecular weight of 20 kDa corresponding to the predicted molecular weight of caltubin. G, A representative confocal image of a cultured PeA cell labeled with the N-terminal caltubin antibody. Scale bar, 20 μm.
To study the distribution of caltubin transcript in Lymnaea neurons, we performed in situ hybridization with a caltubin antisense RNA probe, both on pedal ganglia sections and on cultured PeA neurons. In ganglia sections, abundant expression of caltubin mRNA was detected by the caltubin antisense probe in neuronal cell bodies (dark regions) as well as in neurites in the neuropil (arrows) (Fig. 1C1), whereas the sense control RNA probe showed negative in these experiments (Fig. 1C2), indicating the specificity of the RNA probe. In cultured PeA neurons, a strong presence of caltubin mRNA was detected in neuronal somata, and in localized regions of neurites and growth cones by caltubin antisense probe (Fig. 1D), but not by caltubin sense probe (Fig. 1D, right inset). The high-power image (Fig. 1D, left inset) showed that caltubin transcript was unevenly distributed in neurites and accumulated in specific regions (arrows). Real-time qPCR analysis confirmed that caltubin mRNA is expressed in isolated neurites in culture (Fig. 1E). After 2 d in culture (Fig. 1E1), the cell bodies were removed (Fig. 1E2) and mRNA was extracted from the cell bodies and from neurites separately. Real-time qPCR analysis was performed to measure the transcript levels of caltubin and two other known neuritic transcripts, β-thymosin and β-tubulin (van Kesteren et al., 2006), as positive controls. All mRNA levels were first normalized to the level of 16S rRNA, and neuritic mRNA levels were then divided by somatic mRNA levels. We found that caltubin mRNA level was approximately sixfold higher in the neurites than in the cell somata (Fig. 1E3). The neuritic/somatic expression ratio of caltubin mRNA was higher than that of β-thymosin and β-tubulin (the positive controls). These data further confirm that caltubin mRNA is indeed present at high levels in neurites. To study neuronal caltubin protein expression, we generated antibodies against the C terminus and N terminus of the protein. Immunoblotting showed that all antibodies recognized a single protein band with an apparent molecular weight of ∼20 kDa corresponding to the predicted molecular weight of caltubin (Fig. 1F). Confocal immunofluorescence imaging revealed caltubin immunoreactivity in the soma, neurites, and growth cones of cultured PeD1 neurons (Fig. 1G). The presence of both caltubin transcript and protein in neurites and growth cones of cultured adult PeA neurons raises the possibility that local synthesis of caltubin in these distal regions may play a role in regulating neurite development and/or regeneration in vitro.
Expression and local synthesis of caltubin are required for neurite outgrowth and regeneration of cultured adult PeA neurons
We next used an RNA interference approach to study the role of caltubin in cultured PeA neurons. Previous studies demonstrated that short siRNAs can be used for efficient gene silencing in cultured Lymnaea neurons (Hui et al., 2007). We first showed that caltubin-specific siRNAs effectively knocked down caltubin mRNA (Fig. 2A,B) and protein levels, both in intact ganglia in vivo (Fig. 2C,D) and in cultured PeA neurons in vitro (Fig. 2E,F) 6 h after siRNA application. Three siRNAs (LCa1, LCa3, and LCa10) targeting three different regions of the caltubin gene were tested and all resulted in a significant decrease in protein levels (Fig. 2C,D). Caltubin levels measured in cultured PeA neurons progressively decreased over time in caltubin siRNA (LCa3) treatment cells, but not in control siRNA-treated cells (Fig. 2G,H). Caltubin knockdown reached 30% reduction at 3 h (0 h: 1.92 ± 0.09, n = 11; 1.5 h: 1.74 ± 0.07, n = 11; 3 h: 1.19 ± 0.13, n = 9; p < 0.05) and 50% reduction at 6 h (0.81 ± 0.06; n = 10; p < 0.05), which was similar to that observed at 24 h.
Figure 2.

Caltubin siRNA treatment significantly reduces expression of both caltubin transcript and protein in vivo and in vitro. Relative expression levels of ganglial mRNA (A, B) or protein (C, D) of caltubin against β-actin were measured 24 h following injection of 2 μl of solution containing saline, control siRNA, or caltubin siRNA (LCa1, LCa3, or LCa10) into the snail central ganglia. A, C, Representative gel image and Western blot. B, D, Quantification of caltubin mRNA and protein levels. Caltubin siRNA treatment, but not control siRNA treatment, significantly reduced the expression of caltubin mRNA (41 ± 1.3%; n = 3) and protein (54 ± 1.6%; n = 3). E, Representative confocal images of caltubin and β-actin double-labeled PeA cells cultured in the presence of caltubin or control siRNA. F, Summary of caltubin to β-actin ratios in soma, neurites, and growths of cultured PeA cells treated with control or caltubin siRNA, showing a significant reduction in caltubin levels specifically in caltubin siRNA-treated cells. Soma: Control siRNA, n = 11; caltubin siRNA, n = 10. Neurites: Control siRNA, n = 12; caltubin siRNA, n = 10. Growth cones: Control siRNA, n = 12; caltubin siRNA, n = 11. G, H, Time course of caltubin siRNA treatment and caltubin protein expression. G, Representative confocal images of immunocytochemical staining of PeA cells at various time points following caltubin siRNA (LCa 3) treatment. Scale bar, 20 μm. H, A significant reduction in relative caltubin protein levels was already observed at 3 h [1.19 ± 0.13 (n = 9; p < 0.05)] and 6 h [0.81 ± 0.06 (n = 10; p < 0.05)] following siRNA treatment, compared with 0 h [1.92 ± 0.94 (n = 11)]. All the data are presented as mean ± SEM (n). *p < 0.05.
To elucidate the function of caltubin in neurite outgrowth, we compared neurite extension of cultured PeA neurons in the presence of either caltubin siRNA or control siRNA. PeA neurons were cultured for 15 h to allow initiation of outgrowth, and cells with an initial outgrowth of 40–60 μm were used for further analysis. Caltubin siRNA or control siRNA were added in the culture medium at t = 0, and neurite outgrowth was monitored for 30 h thereafter. We observed a large difference in the outgrowth between the two groups over this period. Neurites of control siRNA-treated cells grew 268.9 ± 40 μm (n = 17) on average, which was significantly longer than neurites of caltubin siRNA group (19 ± 7 μm; n = 26; p < 0.05) (Fig. 3A,B). These results indicate that caltubin siRNA treatment severely disrupts neurite extension of cultured adult PeA neurons. These findings led us to hypothesize that caltubin might be involved in neurite regeneration.
Figure 3.

Knockdown of caltubin mRNA inhibits elongation of intact neurites and regeneration of injured neurites. A, Representative phase-contrast images of a PeA neuron after control siRNA (top) or caltubin siRNA application (bottom). B, Quantification of the net changes in neurite lengths after siRNA treatment. C, Representative images of a transected PeA neuron after control siRNA (top) or caltubin siRNA application (bottom). The arrows indicate the site of injury. Quantification of net changes in the proximal (D) and distal (E) neurite lengths after siRNA treatment. F, Three different caltubin siRNAs (LCa3, LCa1, LCa10) all block neurite growth of intact, severed proximal, and severed distal neurites. siRNAs were added immediately following neurite transection. The net length of intact (untransected), and the proximal and the distal ends of transected neurites were measured at 30 h after siRNA treatment. *p < 0.05. Scale bars: A, C, 20 μm. All data in B, D, E, and F are presented as mean ± SEM.
To test the potential role of caltubin in neurite regeneration of PeA neurons, we transected neurites of cultured PeA cells, and then measured the growth capacity of both the proximal (soma-attached) neurite and the distal (disconnected) neurite (Fig. 3C) following treatment with either control siRNA or caltubin siRNA. Specifically, PeA neurons were maintained in culture for 20 h, and cells containing neurites with lengths of ∼70 μm were used. Neurites were severed using a glass micropipette and immediately treated with either control siRNA or caltubin siRNA. Neurite measurements were performed over a period of 30 h following injury (Fig. 3C). As expected, the growth of injured neurites was severely inhibited by caltubin siRNA. For proximal injured neurites, a net elongation of 168 ± 35 μm (n = 7) was observed in control siRNA condition; in contrast, a net retraction of 23 ± 4 μm (n = 6) was observed under the caltubin siRNA condition over the same 30 h period (p < 0.05; Fig. 3D). These data indicate that caltubin is required for postinjury regeneration of soma attached PeA neurites in culture. Similar to proximal neurites, the distal neurites of transected PeA neurons treated with control siRNA also showed a net increase in neurite length (133 ± 5 μm; n = 10) over a 30 h period, whereas the distal neurites of cells treated with caltubin siRNA experienced a net retraction of 73 ± 17 μm (n = 18) over the same period (p < 0.05; Fig. 3E). It is worth noting that all the transected neurites were treated with siRNA after the injury, and the differences observed in the isolated (distal) neurites between control and caltubin siRNA treatments over 30 h were likely a result of locally synthesized proteins, which were suppressed by caltubin siRNA. To confirm the specificity of our caltubin knockdown, cells were treated for 30 h with each of three caltubin siRNAs targeting different regions of the transcript. We found that all three siRNAs consistently reduced the outgrowth ability of the intact neurites and caused substantial retraction in the injured neurites (Fig. 3F). These observations indicate that caltubin is an important protein for normal neurite development, and we therefore decided to address the possible mechanisms by which caltubin might affect neurite outgrowth.
Caltubin regulates β-tubulin but not β-actin protein levels in PeA neurites
Neurite outgrowth and maintenance are highly dependent on the proper assembly of actin and tubulin (Campenot et al., 2003), and can be regulated by calcium-binding proteins (Hui et al., 2007). We investigated the possibility that caltubin affects neurite outgrowth and regeneration by regulating actin or tubulin expression. We conducted immunocytochemical triple labeling of caltubin, neuronal calcium sensor 1 (NCS-1), and β-actin or β-tubulin in cultured PeA cells treated with either control siRNA or caltubin siRNA. NCS-1 was used as a reference protein. We found that the protein expression levels of β-actin and NCS-1 in the soma, neurites, and growth cones of PeA neurons were not significantly different between control siRNA (n = 11, 11, 12)- and caltubin siRNA (n = 10, 10, 11)-treated cells (Fig. 4A,B), whereas the relative level of caltubin to NCS-1 was significantly reduced (Fig. 4C). These data are consistent with our immunoblot analysis, which showed that global β-actin levels did not vary with caltubin siRNA treatment (Fig. 2). In contrast, the relative β-tubulin protein levels in caltubin siRNA-treated cells were significantly reduced by 45 ± 2.7% (n = 11; p < 0.05) in the soma, 34 ± 5.8% (n = 6; p < 0.05) in the neurite, and 30 ± 9.7% (n = 6; p < 0.05) in growth cones compared with control siRNA-treated cells (n = 12, 12, 14) (Fig. 4D,E), corresponding with a significant decrease in caltubin level (Fig. 4F). Together, these data demonstrate that caltubin has a differential effect on the expression of cytoskeletal proteins. Knockdown of caltubin specifically reduced β-tubulin protein levels, without affecting β-actin protein levels, suggesting that the effect of caltubin on neurite outgrowth and regeneration may be, at least in part, through the regulation of β-tubulin levels.
Figure 4.
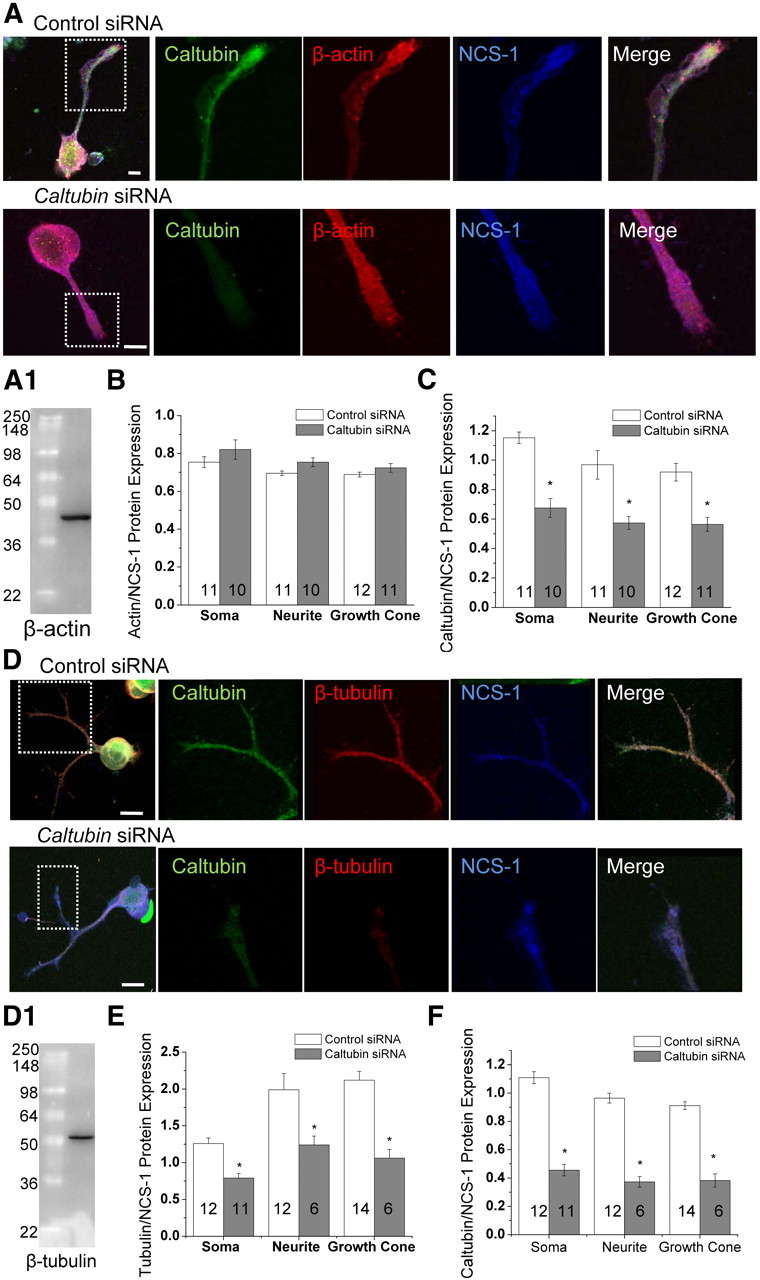
Knockdown of caltubin reduces β-tubulin protein levels, but not β-actin or NCS-1 protein levels. A, Representative confocal immunofluorescence images obtained from PeA cells treated with control or caltubin siRNA. Caltubin (green), β-actin (red), and NCS-1 (blue) are shown. A1, Western blot showing a single protein band labeled by the β-actin antibody used in A. Summary of the relative levels of β-actin (B) or caltubin (C) against NCS-1. D, Representatives of triple stainings with caltubin (green), β-tubulin (red), and NCS-1 (blue) in cells treated with control siRNA or caltubin siRNA. D1, Western blot showing a single protein band labeled by the β-tubulin antibody used in D. Summary of the relative protein expression levels of β-tubulin (E) or caltubin (F) against NCS-1. Scale bar, 20 μm. The data are presented as mean ± SEM. *p < 0.05.
Neurite outgrowth of PeA cells requires local synthesis of caltubin
Three lines of evidence suggested that caltubin might be synthesized locally in growth cones: (1) caltubin transcripts were present in neurites (Fig. 1D,E3), (2) caltubin siRNA reduced the caltubin level in growth cones (Fig. 2F), and (3) isolated (distal) neurites treated with caltubin siRNA after the injury showed retraction, in contrast to those treated with control siRNA, which showed elongation (Fig. 3E). Because the distal neurites no longer received proteins transported from cell somata, the proteins required for outgrowth of the isolated neurites were likely synthesized locally. Caltubin siRNA reduced caltubin level in growth cones (Fig. 2), which may be a result of interruption of the local synthesis of caltubin. To test this hypothesis, we measured the relative levels of caltubin over NCS-1 (as a control) in transected neurites of the PeA neurons either immediately or 3 h following injury (Fig. 5A). The levels of caltubin in both distal (Fig. 5B) and proximal (Fig. 5C) neurites increased significantly within 3 h following injury (distal: 0 h, 0.56 ± 0.05, n = 8; 3 h, 0.71 ± 0.05, n = 7; p < 0.05; proximal: 0 h, 0.45 ± 0.03, n = 8; 3 h, 0.72 ± 0.05, n = 7; p < 0.05). The increase in caltubin levels in distal neurites supports the hypothesis that the pro-regenerative effects of caltubin are, at least in part, derived through local synthesis of caltubin.
Figure 5.

Injury increases caltubin protein levels and blocking protein translation reduces caltubin levels in the distal portion of transected neurites. A, Representative confocal immunofluorescence images obtained from transected PeA neuron fixed either immediately following injury (top panel) or following a 3 h delay (bottom panel). Relative levels of caltubin in distal (B) and proximal (C) portions of neurite measured immediately and 3 h after injury. D, Representative confocal immunofluorescence images obtained from injured PeA cells 30 h after treatment with either control siRNA (top panel) or control siRNA plus 11.3 μg/ml anisomycin (bottom panel). Cells were treated immediately following transection. E, Summary of the expression levels of caltubin, β-tubulin, and NCS-1 in the distal neurites with or without anisomycin treatment in presence of control siRNA. F, Net changes in neurites following anisomycin treatments. Anisomycin (8.3 or 11.3 μg/ml) was added in the culture dish immediately after transection. The lengths of the intact neurites and of proximal and distal ends of transected neurites were measured at 30 h after the treatment. Shown are caltubin (green), β-tubulin (red), and NCS-1 (blue). Scale bar, 20 μm. All the data are presented as mean ± SEM. *p < 0.05.
To confirm that transected neurites indeed synthesize caltubin locally, we compared the expression of caltubin, β-tubulin, and NCS-1 in distal regions of transected neurites, in the presence or absence of a protein translation inhibitor, anisomycin (11.3 μg/ml) (Fig. 5D). We found that both caltubin and β-tubulin protein levels, but not NCS-1 protein levels, were significantly reduced in distal neurites 30 h following anisomycin treatment compared with the control (Fig. 5E). Outgrowth of the intact neurites and the severed neurites following transection was suppressed and a retraction of severed neurites was observed in the presence of anisomycin (Fig. 5F). These observations further support the notion that neurite outgrowth/regeneration in cultured Lymnaea PeA neurons requires local translation of caltubin in the distal regions of neurites.
Caltubin colocalizes and interacts with tubulin but not β-actin in Lymnaea neurons
To further explore the mechanism of β-tubulin regulation by caltubin, we first tested the degree of spatial colocalization between caltubin and β-tubulin using confocal immunofluorescence intensity correlation analysis. Protein colocalization suggests a high probability of two proteins co-occurring in close proximity. Colocalization was evaluated by measuring the level of overlap between signal intensities of the corresponding fluorescent channels over the selected pixels in randomly selected regions of PeA neurites and growth cones. PeA cells previously treated with control siRNA were double-labeled for caltubin and either β-tubulin, β-actin, or NCS-1. Colocalization of proteins in neurites and/or growth cones was estimated based on the Pearson correlation coefficient: a coefficient (Rr) close to 1 indicates colocalization, while a coefficient close to 0 indicates no colocalization (Li et al., 2004). Our data show that caltubin is colocalized with β-tubulin in neurites and growth cones (neurites: Rr = 0.67 ± 0.08, n = 6; growth cones: Rr = 0.75 ± 0.10, n = 6; Fig. 6A). In contrast, a much lower correlation was found between caltubin and β-actin (Rr = 0.10–0.20; Fig. 6B) and between caltubin and NCS-1 (Rr = 0.18–0.30), indicating that there was a low probability for colocalization between caltubin and β-actin or NCS-1. Protein colocalization only implies a high probability of two proteins occurring together in a specific region. Thus, to further determine whether the colocalized proteins physically interact, we performed a series of immunoprecipitation (IP) or affinity pull-down analyses. We found that caltubin bound to β-tubulin (Fig. 6C), but not to β-actin (Fig. 6C) or the negative control IPs (Fig. 6C). Interestingly, we also observed caltubin binding to α-tubulin (Fig. 6D). To further investigate whether caltubin also regulates α-tubulin levels in growth cones, we used triple immunocytochemical staining (Fig. 6E1). Similar to β-tubulin, relative α-tubulin protein levels in growth cones of caltubin siRNA-treated cells were significantly reduced (0.53 ± 0.05; n = 8) compared with control siRNA-treated cells (1.14 ± 0.13; n = 8; p < 0.05) (Fig. 6E2, top panel) and corresponded with a significant decrease in caltubin levels (caltubin siRNA: 0.18 ± 0.024, n = 8; control siRNA: 0.47 ± 0.042, n = 8; p < 0.05) (Fig. 6E2, bottom panel). Together, these results indicate that caltubin colocalizes and interacts with tubulins in Lymnaea neurons, and point to a direct mechanism in the regulation of microtubule proteins by caltubin.
Figure 6.

Caltubin is colocalized with and binds to tubulins, but not to β-actin, in PeA neurons. Representative confocal images of double immunocytochemical staining for caltubin and either β-tubulin (A) or β-actin (B). The correlations between the local staining intensities of the two proteins was measured in neurites, in growth cones, and in the background, and indicated by Pearson coefficient values (Rr). Scale bars, 20 μm. C, Representative Western blots showing that β-tubulin, but not β-actin, coimmunoprecipitates with caltubin. Both β-tubulin and β-actin are detected in input (extracted Lymnaea ganglionic protein) as well as in supernatant fractions; however, only β-tubulin is detected in the caltubin IP fraction, indicating specific coimmunoprecipitation between β-tubulin and caltubin. D, Representative Western blot of α-tubulin after affinity precipitation revealed a single band in the input and in presence of GST-caltubin, but not with GST alone. E, Knockdown of caltubin reduces α-tubulin levels. E1, Representative confocal immunofluorescence images obtained from PeA neurons treated with control or caltubin siRNA. Caltubin (green), β-actin (red), and NCS-1 (blue) are shown. Scale bar, 7 μm. E2, Summary of the relative levels of caltubin (top panel) or α-tubulin (bottom panel) against NCS-1, from PeA neurons treated with control siRNA (n = 8) or caltubin siRNA (n = 8). All the data are presented as mean ± SEM. *p < 0.05.
Caltubin interacts with β-tubulin, promotes axonal elongation, and attenuates neurite retraction of PC12 cells
Since tubulins are highly conserved (Burns and Surridge, 1990), we asked whether the mechanism of β-tubulin regulation by caltubin might translate to mammalian cells. We first tested the potential binding between caltubin and β-tubulin in rat PC12 neurons by conducting coimmunoprecipitation experiments on cultured PC12 cells, which had been transfected with caltubin-myc, caltubin-eGFP, or eGFP only (each construct at 3 μg/ml) and allowed a period of 3 d for growth. Our Western blot data show that β-tubulin was present in all three input samples (Fig. 7A1); however, the relative levels of β-tubulin in caltubin-expressing cells over GFP-expressing cells significantly increased (caltubin-GFP: 1.17 ± 0.07; caltubin-myc: 1.12 ± 0.04; n = 4; p < 0.05) (Fig. 7A2). In immunoprecipitated samples, β-tubulin was detected in protein complexes obtained from PC12 cells transfected with caltubin-eGFP and caltubin-myc, but not from cells transfected with only eGFP (Fig. 7A1). To confirm these findings, we measured the level of unbound β-tubulin remaining in the supernatant collected following the immunoprecipitation assay. We found that the level of unbound β-tubulin remaining in the supernatant of the eGFP-transfected cells was higher than that in the supernatant of the caltubin-eGFP and caltubin-myc transfected cells, consistent with our IP data. These experiments indicate that, in mammalian cells, the ability of caltubin to bind to β-tubulin is conserved.
Figure 7.

Caltubin expression in rat PC12 cells promotes axonal elongation and attenuates axonal degeneration. A1, Representative immunoblots of β-tubulin in cultured PC12 cells transfected with caltubin-GFP, caltubin-myc, or eGFP only (each construct at 3 μg/ml) and immunoprecipitated (IP) for caltubin. β-Tubulin was detected in all three input samples (lysates), while coimmunoprecipitated β-tubulin was only detected from caltubin-eGFP or caltubin-myc transfected cells. A2, Summary of relative levels of β-tubulin in PC12 cells expressing caltubin-GFP or caltubin-myc. The dashed line indicates β-tubulin level in PC12 cells expressing GFP only. B, Representative images of PC12 cells expressing caltubin-eGFP or eGFP only. Scale bar, 20 μm. C, Summary of NGF-dependent axonal growth of PC12 cells expressing eGFP, or caltubin-eGFP measured over a period of 6 d after transfection. D, Time course of the postinjury changes in axon lengths of PC12 cells expressing eGFP or caltubin-eGFP. Cells were transfected with either caltubin-eGFP (3 μg/ml) or eGFP only. Injury was induced by cutting off the growth cone on day 5 after transfection. All the data are presented as mean ± SEM. *p < 0.05.
Next, we tested whether caltubin can influence axonal elongation in PC12 cells. Cells were transfected with either 1, 2, or 3 μg/ml caltubin-eGFP construct or 3 μg/ml eGFP control construct (Fig. 7B), and NGF was added on the first day following transfection. Consistent with previous reports, PC12 cells extended processes following NGF exposure (Greene and Tischler, 1976) and were capable of axonal elongation for up to 10 d in culture (Fujii et al., 1982). Overexpression of caltubin-eGFP significantly increased axon extension compared with the eGFP-only controls, in a time-dependent and dose-dependent fashion (Fig. 7C). Specifically, following transfection, the axons of control cells expressing eGFP extended over time in culture (3 d: 47.34 ± 5.1 μm, n = 19; 5 d: 56.31 ± 5.9 μm, n = 34; 6 d: 85.4 ± 5.1 μm, n = 21). The axons of cells treated with caltubin-eGFP (1 μg of construct) extended significantly longer (3 d: 77 ± 9.1 μm, n = 29; 5 d: 79 ± 5.8 μm, n = 32; 6 d: 116.4 ± 9.3 μm, n = 26; p < 0.05), compared with the controls. The increase in axonal extension was also observed at higher caltubin-eGFP concentrations (2 and 3 μg) (Fig. 7B,C).
To test whether caltubin can affect axonal degeneration following transection in PC12 neurons, we expressed caltubin-eGFP (3 μg/ml) or eGFP alone and allowed processes to grow for a period of 5 d. Cells with initial axons of ∼70 μm were identified and marked for time-lapse imaging. Axons were injured by cutting off the growth cone using a glass micropipette, and axonal behavior was monitored over various time points following the injury. At 45 min after the procedure, injured axons of cells transfected with eGFP alone showed a significant net retraction of 20.72 ± 1.94 μm (n = 19), whereas axons of cells transfected with caltubin-eGFP showed a net retraction of only 12.89 ± 3.45 μm (n = 15) (p < 0.05; Fig. 8D). The axons in the eGFP-only group continued to retract over the next 24 h (total net retraction of 31.62 ± 3.9 μm; n = 19), whereas axons expressing caltubin-eGFP remained stable over the same period of time (total net retraction of 12.62 ± 6.4 μm; n = 15) (p < 0.05; Fig. 8D). Together, these results demonstrate that caltubin can interact with endogenous mammalian β-tubulin to promote NGF-induced outgrowth and prevent retraction of neuron-like processes following injury in mammalian cells.
Figure 8.

Caltubin expression in mouse primary cortical neurons promotes axonal elongation and attenuates axonal retraction. Representative Western blots of mouse brain β-tubulin (A) or α-tubulin (B) after affinity precipitation revealed a single band in the presence of GST-caltubin, but not with GST alone. Input (mouse brain tissue-extracted protein) in each experiment serves as the control for indicated proteins. C, Summary of NGF-induced axonal growth of mouse primary cortical cells expressing either eGFP, or caltubin-eGFP at DIV3 (transfection at DIV1). D, Representative images of cultured cortical neurons expressing eGFP (top panel) or caltubin-eGFP (bottom panel). Neurons were injured at DIV5, 3 d following transfection. Injury was induced by a cut through the growth cone of cultured cells using a glass micropipette, and the dashed line indicates the site and the direction of injury. Scale bar, 20 μm. E, Time course of postinjury changes in axon lengths of cortical neurons expressing eGFP or caltubin-eGFP. All the data are presented as mean ± SEM. *p < 0.05.
Caltubin interacts with tubulin, promotes axonal elongation, and attenuates axonal retraction of cultured mouse cortical neurons
To confirm that caltubin indeed has a conserved role in mammalian neurons, we tested whether caltubin binds to tubulin and promotes neurite outgrowth in mouse cortical neurons. Our affinity pull-down assays showed that GST-caltubin fusion protein, but not GST, precipitated with both β- (Fig. 8A) and α-tubulin (Fig. 8B), indicating that caltubin binds to mouse microtubule proteins. We then expressed caltubin-eGFP in neonatal cortical neurons on the first day in culture (DIV1) and measured neurite lengths of neurons expressing either eGFP or caltubin-eGFP at DIV3. Neurons expressing caltubin-eGFP had significantly longer average neurite lengths (186 ± 11 μm; n = 24) compared with neurons expressing eGFP only (115 ± 6, n = 31; p < 0.05) (Fig. 8C), indicating that the snail protein caltubin enhances the growth capability of the mouse neurons. To test whether caltubin also has an effect on neurite regeneration/degeneration, we transfected neurons at DIV3 and cut through the growth cone of one of the neurites at DIV5. We then measured the net changes in neurite length for a period of 8 h following injury (Fig. 8D,E). We observed that cells expressing caltubin-eGFP were able to maintain their neurite length following injury over the 8 h period (8 h: −5.11 ± 7.4 μm, n = 24), whereas those expressing eGFP only showed neurite retraction (8 h: −58.38 ± 10.1 μm, n = 25; p < 0.05) (Fig. 8D). These data confirm our findings in PC12 cells and indicate that caltubin serves to promote neurite outgrowth and attenuate neuronal retraction in mammalian neurons, where it is normally not expressed.
Discussion
In this study, we report that the neurite growth-promoting and degeneration-attenuating effects of a novel Lymnaea protein, named caltubin, can be transferred to rodent neurons. Expression of caltubin in rat PC12 cells or mouse cortical neurons effectively enhanced outgrowth and attenuated axonal degeneration following transection. These findings implicate caltubin as a novel molecular player in a conserved cellular mechanism of neurite outgrowth and highlight its potential as a possible therapeutic tool in nerve regeneration.
Local protein synthesis is one of the crucial mechanisms regulating neurite outgrowth, growth cone behavior, and axonal regeneration (Gumy et al., 2010). Whereas the exact downstream mechanisms by which local protein synthesis regulates axon growth remain unclear, most mRNAs that are identified in axonal growth cones appear to be structural proteins in both invertebrates (Plunet et al., 2002; Gioio et al., 2004) and in vertebrates (Willis et al., 2005; Hengst et al., 2009), and translational regulation of these proteins occurs during axon regeneration (Twiss et al., 2000). For instance, stimulus-driven axonal growth is dependent on local translation of the cytoskeleton regulator PAR (Hengst et al., 2009), and inhibition of local translation of the actin regulating protein β-thymosin enhances rates of neurite outgrowth (van Kesteren et al., 2006). In this study, we provided multiple lines of evidence that local protein synthesis of caltubin is required for neurite regeneration through regulation of β-tubulin level. First, caltubin mRNA and protein are highly expressed in growth cones of snail neurons. Second, local caltubin levels in isolated neurites can be reduced by targeted siRNA knockdown or anisomycin treatment. Third, reduction of local caltubin levels in isolated neurites suppresses neurite extension and regeneration following injury in cultured CNS neurons. Fourth, caltubin protein level increased in distal neurites 3 h after transection, indicating the potential de novo synthesis of proteins in these regions. Finally, caltubin colocalizes and physically interacts with tubulin, and a reduction of local levels of caltubin by targeted siRNA knockdown leads to a decrease in local tubulin levels. Together, our findings indicate that local synthesis of de novo proteins is essential for neurite regeneration during this period of time, and that locally synthesized caltubin promotes regenerative neurite outgrowth.
The cytoskeleton is an essential element for cell architecture and neurite structure. Most of the local transcripts are cytoskeletal proteins (e.g., microtubule, microfilament, and intermediate filament proteins) (Kaplan et al., 1992; Eng et al., 1999; Vogelaar et al., 2009), or proteins that regulate cytoskeletal dynamics (e.g., Rho and β-thymosin) (Campbell et al., 2001; Verma et al., 2005; van Kesteren et al., 2006). Stabilization and continuous restructuring of the cytoskeleton plays an important role in the processes of neurite formation and regeneration, allowing growth cone formation, vesicle accumulation, and neurite elongation (Spira et al., 2001; Witte et al., 2008). We found that caltubin colocalizes with and binds to both α- and β-tubulin. Thus, the neurite growth-promoting effects of caltubin are likely through local regulation of tubulin. Caltubin knockdown decreased local α- and β-tubulin protein levels, but did not affect β-actin or NCS-1 levels, indicating that caltubin specifically regulates tubulin levels to achieve its effects on promoting neurite outgrowth and regeneration. Microtubules are comprised of αβ-tubulin heterodimers (Westermann and Weber, 2003), and polymerization of microtubules stabilizes the cytoskeleton (Nakano et al., 2010), leading to maintenance of neurite integrity (Karima et al., 2010), and promotion of neurite outgrowth (Tanaka et al., 1995) and regeneration (Ertürk et al., 2007). While the mechanisms by which caltubin may regulate tubulin levels remain unclear, we speculate that binding of caltubin may cause overall stabilization of tubulin polymers, which in turn enhances neurite elongation. Similarly, the ring finger protein ZNRF1, encoded by a nerve injury-induced neuronal gene Nin283 (Araki et al., 2001), interacts with β-tubulin to enhance neurite-like elongation (Yoshida et al., 2009), suggesting that other tubulin-binding proteins may also regulate axonal elongation following injury. Knockdown of caltubin may destabilize microtubule proteins, leading to neurite retraction. Alternatively, caltubin may regulate local translation or degradation of tubulin in the axon. While further investigation is required for understanding how exactly caltubin regulates tubulin levels, our data clearly indicate that in Lymnaea neurons caltubin binding to tubulin is a prerequisite for efficient axon growth and regeneration.
Vertebrate and invertebrate growth cones and axons share many common mechanisms of neuronal growth, including organization of the microtubule cytoskeleton, response to axon guidance molecules, and dynamic changes in intracellular calcium signaling (Dent et al., 2003). Many of the crucial proteins involved in neurite outgrowth and regeneration, including tubulin, are highly conserved across species (Burns and Surridge, 1990). Yet vertebrate and invertebrate adult central neurons differ in their regenerative capacity after injury: in most vertebrates, central neurons seldom regenerate injured neurites, whereas injury to invertebrate central neurons results in a strong regenerative response. Taking advantage of the high conservation between tubulins across species, we tested whether caltubin, which has no obvious sequence orthologs in mammals, might also promote neurite outgrowth and regeneration in mammalian cells. We show that caltubin not only binds to endogenous tubulin but also promotes NGF-induced axonal outgrowth in PC12 cells and cultured mouse cortical neurons, and reduces rates of axonal retraction. Thus, our data suggest that caltubin might be one of the critical molecules essential for the robust intrinsic ability to regenerate in snail neurons, and that some of its downstream mechanisms are conserved in mammalian central neurons. Indeed, our findings show that the molecular interactions and cellular functions of caltubin are conserved in invertebrate and vertebrate neurons, leading to a promising new avenue for using an invertebrate protein as a potential therapeutic target to remodel neuronal properties favoring axonal regeneration and/or recovery from neural injury and neurodegenerative disorders.
Footnotes
This work was supported by Canadian Institutes of Health Research Operating Grant CIHR MOP151437 (Z.-P.F.) and also a start-up fund (H.-S.S.) and a Discovery grant from Natural Sciences and Engineering Research Council of Canada (NSERC) (RGPIN402733, H.-S.S.). N.N. is the recipient of an NSERC Canada Graduate Scholarship–PhD studentship, and T.Z.L. is the recipient of a NSERC Postgraduate Scholarship–PhD studentship. Z.-P.F. holds a New Investigator Award from the Heart and Stroke Foundation of Canada. We thank Qing Li for her technical assistance and Dr. Milton Charlton for his critical comments on this manuscript.
References
- Araki T, Nagarajan R, Milbrandt J. Identification of genes induced in peripheral nerve after injury. Expression profiling and novel gene discovery. J Biol Chem. 2001;276:34131–34141. doi: 10.1074/jbc.M104271200. [DOI] [PubMed] [Google Scholar]
- Burns RG, Surridge C. Analysis of beta-tubulin sequences reveals highly conserved, coordinated amino acid substitutions. Evidence that these “hot spots” are directly involved in the conformational change required for dynamic instability. FEBS Lett. 1990;271:1–8. doi: 10.1016/0014-5793(90)80359-q. [DOI] [PubMed] [Google Scholar]
- Campbell DS, Holt CE. Chemotropic responses of retinal growth cones mediated by rapid local protein synthesis and degradation. Neuron. 2001;32:1013–1026. doi: 10.1016/s0896-6273(01)00551-7. [DOI] [PubMed] [Google Scholar]
- Campbell DS, Regan AG, Lopez JS, Tannahill D, Harris WA, Holt CE. Semaphorin 3A elicits stage-dependent collapse, turning, and branching in Xenopus retinal growth cones. J Neurosci. 2001;21:8538–8547. doi: 10.1523/JNEUROSCI.21-21-08538.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campenot RB, Soin J, Blacker M, Lund K, Eng H, MacInnis BL. Block of slow axonal transport and axonal growth by brefeldin A in compartmented cultures of rat sympathetic neurons. Neuropharmacology. 2003;44:1107–1117. doi: 10.1016/s0028-3908(03)00042-x. [DOI] [PubMed] [Google Scholar]
- Capano CP, Giuditta A, Castigli E, Kaplan BB. Occurrence and sequence complexity of polyadenylated RNA in squid axoplasm. J Neurochem. 1987;49:698–704. doi: 10.1111/j.1471-4159.1987.tb00950.x. [DOI] [PubMed] [Google Scholar]
- Dent EW, Tang F, Kalil K. Axon guidance by growth cones and branches: common cytoskeletal and signaling mechanisms. Neuroscientist. 2003;9:343–353. doi: 10.1177/1073858403252683. [DOI] [PubMed] [Google Scholar]
- Eng H, Lund K, Campenot RB. Synthesis of β-tubulin, actin, and other proteins in axons of sympathetic neurons in compartmented cultures. J Neurosci. 1999;19:1–9. doi: 10.1523/JNEUROSCI.19-01-00001.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertürk A, Hellal F, Enes J, Bradke F. Disorganized microtubules underlie the formation of retraction bulbs and the failure of axonal regeneration. J Neurosci. 2007;27:9169–9180. doi: 10.1523/JNEUROSCI.0612-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei G, Guo C, Sun HS, Feng ZP. Chronic hypoxia stress-induced differential modulation of heat-shock protein 70 and presynaptic proteins. J Neurochem. 2007;100:50–61. doi: 10.1111/j.1471-4159.2006.04194.x. [DOI] [PubMed] [Google Scholar]
- Feng ZP, Klumperman J, Lukowiak K, Syed NI. In vitro synaptogenesis between the somata of identified Lymnaea neurons requires protein synthesis but not extrinsic growth factors or substrate adhesion molecules. J Neurosci. 1997;17:7839–7849. doi: 10.1523/JNEUROSCI.17-20-07839.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng ZP, Hasan SU, Lukowiak K, Syed NI. Target cell contact suppresses neurite outgrowth from soma-soma paired Lymnaea neurons. J Neurobiol. 2000;42:357–369. doi: 10.1002/(sici)1097-4695(20000215)42:3<357::aid-neu7>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Ferretti P, Zhang F, O'Neill P. Changes in spinal cord regenerative ability through phylogenesis and development: lessons to be learnt. Dev Dyn. 2003;226:245–256. doi: 10.1002/dvdy.10226. [DOI] [PubMed] [Google Scholar]
- Fujii DK, Massoglia SL, Savion N, Gospodarowicz D. Neurite outgrowth and protein synthesis by PC12 cells as a function of substratum and nerve growth factor. J Neurosci. 1982;2:1157–1175. doi: 10.1523/JNEUROSCI.02-08-01157.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Deng K, Hou J, Bryson JB, Barco A, Nikulina E, Spencer T, Mellado W, Kandel ER, Filbin MT. Activated CREB is sufficient to overcome inhibitors in myelin and promote spinal axon regeneration in vivo. Neuron. 2004;44:609–621. doi: 10.1016/j.neuron.2004.10.030. [DOI] [PubMed] [Google Scholar]
- Gardzinski P, Lee DW, Fei GH, Hui K, Huang GJ, Sun HS, Feng ZP. The role of synaptotagmin I C2A calcium-binding domain in synaptic vesicle clustering during synapse formation. J Physiol. 2007;581:75–90. doi: 10.1113/jphysiol.2006.127472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gioio AE, Chun JT, Crispino M, Capano CP, Giuditta A, Kaplan BB. Kinesin mRNA is present in the squid giant axon. J Neurochem. 1994;63:13–18. doi: 10.1046/j.1471-4159.1994.63010013.x. [DOI] [PubMed] [Google Scholar]
- Gioio AE, Lavina ZS, Jurkovicova D, Zhang H, Eyman M, Giuditta A, Kaplan BB. Nerve terminals of squid photoreceptor neurons contain a heterogeneous population of mRNAs and translate a transfected reporter mRNA. Eur J Neurosci. 2004;20:865–872. doi: 10.1111/j.1460-9568.2004.03538.x. [DOI] [PubMed] [Google Scholar]
- Goldberg JL, Barres BA. Nogo in nerve regeneration. Nature. 2000;403:369–370. doi: 10.1038/35000309. [DOI] [PubMed] [Google Scholar]
- Greene LA, Tischler AS. Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc Natl Acad Sci U S A. 1976;73:2424–2428. doi: 10.1073/pnas.73.7.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumy LF, Tan CL, Fawcett JW. The role of local protein synthesis and degradation in axon regeneration. Exp Neurol. 2010;223:28–37. doi: 10.1016/j.expneurol.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hares K, Kemp K, Gray E, Scolding N, Wilkins A. Neurofilament dot blot assays: novel means of assessing axon viability in culture. J Neurosci Methods. 2011;198:195–203. doi: 10.1016/j.jneumeth.2011.03.025. [DOI] [PubMed] [Google Scholar]
- Hengst U, Jaffrey SR. Function and translational regulation of mRNA in developing axons. Semin Cell Dev Biol. 2007;18:209–215. doi: 10.1016/j.semcdb.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengst U, Deglincerti A, Kim HJ, Jeon NL, Jaffrey SR. Axonal elongation triggered by stimulus-induced local translation of a polarity complex protein. Nat Cell Biol. 2009;11:1024–1030. doi: 10.1038/ncb1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui K, Fei GH, Saab BJ, Su J, Roder JC, Feng ZP. Neuronal calcium sensor-1 modulation of optimal calcium level for neurite outgrowth. Development. 2007;134:4479–4489. doi: 10.1242/dev.008979. [DOI] [PubMed] [Google Scholar]
- Jarvis SE, Barr W, Feng ZP, Hamid J, Zamponi GW. Molecular determinants of syntaxin 1 modulation of N-type calcium channels. J Biol Chem. 2002;277:44399–44407. doi: 10.1074/jbc.M206902200. [DOI] [PubMed] [Google Scholar]
- Kaplan BB, Gioio AE, Capano CP, Crispino M, Giuditta A. beta-Actin and beta-Tubulin are components of a heterogeneous mRNA population present in the squid giant axon. Mol Cell Neurosci. 1992;3:133–144. doi: 10.1016/1044-7431(92)90017-v. [DOI] [PubMed] [Google Scholar]
- Karima O, Riazi G, Yousefi R, Movahedi AA. The enhancement effect of beta-boswellic acid on hippocampal neurites outgrowth and branching (an in vitro study) Neurol Sci. 2010;31:315–320. doi: 10.1007/s10072-010-0220-x. [DOI] [PubMed] [Google Scholar]
- Koert CE, Spencer GE, van Minnen J, Li KW, Geraerts WP, Syed NI, Smit AB, van Kesteren RE. Functional implications of neurotransmitter expression during axonal regeneration: serotonin, but not peptides, auto-regulate axon growth of an identified central neuron. J Neurosci. 2001;21:5597–5606. doi: 10.1523/JNEUROSCI.21-15-05597.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JK, Geoffroy CG, Chan AF, Tolentino KE, Crawford MJ, Leal MA, Kang B, Zheng B. Assessing spinal axon regeneration and sprouting in Nogo-, MAG-, and OMgp-deficient mice. Neuron. 2010;66:663–670. doi: 10.1016/j.neuron.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TK, Syed NI. Transplantation and restoration of functional synapses between an identified neuron and its targets in the intact brain of Lymnaea stagnalis. Synapse. 2004;51:186–193. doi: 10.1002/syn.10295. [DOI] [PubMed] [Google Scholar]
- Li Q, Lau A, Morris TJ, Guo L, Fordyce CB, Stanley EF. A syntaxin 1, Gαo, and N-type calcium channel complex at a presynaptic nerve terminal: analysis by quantitative immunocolocalization. J Neurosci. 2004;24:4070–4081. doi: 10.1523/JNEUROSCI.0346-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu TZ, Feng ZP. A sodium leak current regulates pacemaker activity of adult central pattern generator neurons in Lymnaea stagnalis. PLoS One. 2011;6:e18745. doi: 10.1371/journal.pone.0018745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGillavry HD, Stam FJ, Sassen MM, Kegel L, Hendriks WT, Verhaagen J, Smit AB, van Kesteren RE. NFIL3 and cAMP response element-binding protein form a transcriptional feedforward loop that controls neuronal regeneration-associated gene expression. J Neurosci. 2009;29:15542–15550. doi: 10.1523/JNEUROSCI.3938-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mladinic M, Muller KJ, Nicholls JG. Central nervous system regeneration: from leech to opossum. J Physiol. 2009;587:2775–2782. doi: 10.1113/jphysiol.2009.169938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moccia R, Chen D, Lyles V, Kapuya E, E Y, Kalachikov S, Spahn CM, Frank J, Kandel ER, Barad M, Martin KC. An unbiased cDNA library prepared from isolated Aplysia sensory neuron processes is enriched for cytoskeletal and translational mRNAs. J Neurosci. 2003;23:9409–9417. doi: 10.1523/JNEUROSCI.23-28-09409.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroz LL, Edwards JR, Puthanveettil SV, Kohn AB, Ha T, Heyland A, Knudsen B, Sahni A, Yu F, Liu L, Jezzini S, Lovell P, Iannucculli W, Chen M, Nguyen T, Sheng H, Shaw R, Kalachikov S, Panchin YV, Farmerie W, et al. Neuronal transcriptome of Aplysia: neuronal compartments and circuitry. Cell. 2006;127:1453–1467. doi: 10.1016/j.cell.2006.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay G, Doherty P, Walsh FS, Crocker PR, Filbin MT. A novel role for myelin-associated glycoprotein as an inhibitor of axonal regeneration. Neuron. 1994;13:757–767. doi: 10.1016/0896-6273(94)90042-6. [DOI] [PubMed] [Google Scholar]
- Nakano A, Kato H, Watanabe T, Min KD, Yamazaki S, Asano Y, Seguchi O, Higo S, Shintani Y, Asanuma H, Asakura M, Minamino T, Kaibuchi K, Mochizuki N, Kitakaze M, Takashima S. AMPK controls the speed of microtubule polymerization and directional cell migration through CLIP-170 phosphorylation. Nat Cell Biol. 2010;12:583–590. doi: 10.1038/ncb2060. [DOI] [PubMed] [Google Scholar]
- Plunet W, Kwon BK, Tetzlaff W. Promoting axonal regeneration in the central nervous system by enhancing the cell body response to axotomy. J Neurosci Res. 2002;68:1–6. doi: 10.1002/jnr.10176. [DOI] [PubMed] [Google Scholar]
- Qiu J, Cai D, Dai H, McAtee M, Hoffman PN, Bregman BS, Filbin MT. Spinal axon regeneration induced by elevation of cyclic AMP. Neuron. 2002;34:895–903. doi: 10.1016/s0896-6273(02)00730-4. [DOI] [PubMed] [Google Scholar]
- Richardson PM, McGuinness UM, Aguayo AJ. Axons from CNS neurons regenerate into PNS grafts. Nature. 1980;284:264–265. doi: 10.1038/284264a0. [DOI] [PubMed] [Google Scholar]
- Roche FK, Marsick BM, Letourneau PC. Protein synthesis in distal axons is not required for growth cone responses to guidance cues. J Neurosci. 2009;29:638–652. doi: 10.1523/JNEUROSCI.3845-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AB, Spijker S, Van Minnen J, Burke JF, De Winter F, Van Elk R, Geraerts WP. Expression and characterization of molluscan insulin-related peptide VII from the mollusc Lymnaea stagnalis. Neuroscience. 1996;70:589–596. doi: 10.1016/0306-4522(95)00378-9. [DOI] [PubMed] [Google Scholar]
- Spencer GE, Syed NI, van Kesteren E, Lukowiak K, Geraerts WP, van Minnen J. Synthesis and functional integration of a neurotransmitter receptor in isolated invertebrate axons. J Neurobiol. 2000;44:72–81. doi: 10.1002/1097-4695(200007)44:1<72::aid-neu7>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Spira ME, Oren R, Dormann A, Ilouz N, Lev S. Calcium, protease activation, and cytoskeleton remodeling underlie growth cone formation and neuronal regeneration. Cell Mol Neurobiol. 2001;21:591–604. doi: 10.1023/A:1015135617557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O, Zheng B, Tessier-Lavigne M, Hofstadter M, Sharp K, Yee KM. Regenerative growth of corticospinal tract axons via the ventral column after spinal cord injury in mice. J Neurosci. 2008;28:6836–6847. doi: 10.1523/JNEUROSCI.5372-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, He Z. Neuronal intrinsic barriers for axon regeneration in the adult CNS. Curr Opin Neurobiol. 2010;20:510–518. doi: 10.1016/j.conb.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed NI, Bulloch AG, Lukowiak K. In vitro reconstruction of the respiratory central pattern generator of the mollusk Lymnaea. Science. 1990;250:282–285. doi: 10.1126/science.2218532. [DOI] [PubMed] [Google Scholar]
- Tanaka E, Ho T, Kirschner MW. The role of microtubule dynamics in growth cone motility and axonal growth. J Cell Biol. 1995;128:139–155. doi: 10.1083/jcb.128.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twiss JL, van Minnen J. New insights into neuronal regeneration: the role of axonal protein synthesis in pathfinding and axonal extension. J Neurotrauma. 2006;23:295–308. doi: 10.1089/neu.2006.23.295. [DOI] [PubMed] [Google Scholar]
- Twiss JL, Smith DS, Chang B, Shooter EM. Translational control of ribosomal protein L4 mRNA is required for rapid neurite regeneration. Neurobiol Dis. 2000;7:416–428. doi: 10.1006/nbdi.2000.0293. [DOI] [PubMed] [Google Scholar]
- van Kesteren RE, Carter C, Dissel HM, van Minnen J, Gouwenberg Y, Syed NI, Spencer GE, Smit AB. Local synthesis of actin-binding protein β-thymosin regulates neurite outgrowth. J Neurosci. 2006;26:152–157. doi: 10.1523/JNEUROSCI.4164-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma P, Chierzi S, Codd AM, Campbell DS, Meyer RL, Holt CE, Fawcett JW. Axonal protein synthesis and degradation are necessary for efficient growth cone regeneration. J Neurosci. 2005;25:331–342. doi: 10.1523/JNEUROSCI.3073-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelaar CF, Gervasi NM, Gumy LF, Story DJ, Raha-Chowdhury R, Leung KM, Holt CE, Fawcett JW. Axonal mRNAs: characterisation and role in the growth and regeneration of dorsal root ganglion axons and growth cones. Mol Cell Neurosci. 2009;42:102–115. doi: 10.1016/j.mcn.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Koprivica V, Kim JA, Sivasankaran R, Guo Y, Neve RL, He Z. Oligodendrocyte-myelin glycoprotein is a Nogo receptor ligand that inhibits neurite outgrowth. Nature. 2002;417:941–944. doi: 10.1038/nature00867. [DOI] [PubMed] [Google Scholar]
- Westermann S, Weber K. Post-translational modifications regulate microtubule function. Nat Rev Mol Cell Biol. 2003;4:938–947. doi: 10.1038/nrm1260. [DOI] [PubMed] [Google Scholar]
- Willis D, Li KW, Zheng JQ, Chang JH, Smit A, Kelly T, Merianda TT, Sylvester J, van Minnen J, Twiss JL. Differential transport and local translation of cytoskeletal, injury-response, and neurodegeneration protein mRNAs in axons. J Neurosci. 2005;25:778–791. doi: 10.1523/JNEUROSCI.4235-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis DE, van Niekerk EA, Sasaki Y, Mesngon M, Merianda TT, Williams GG, Kendall M, Smith DS, Bassell GJ, Twiss JL. Extracellular stimuli specifically regulate localized levels of individual neuronal mRNAs. J Cell Biol. 2007;178:965–980. doi: 10.1083/jcb.200703209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte H, Neukirchen D, Bradke F. Microtubule stabilization specifies initial neuronal polarization. J Cell Biol. 2008;180:619–632. doi: 10.1083/jcb.200707042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KY, Hengst U, Cox LJ, Macosko EZ, Jeromin A, Urquhart ER, Jaffrey SR. Local translation of RhoA regulates growth cone collapse. Nature. 2005;436:1020–1024. doi: 10.1038/nature03885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Watanabe M, Hatakeyama S. ZNRF1 interacts with tubulin and regulates cell morphogenesis. Biochem Biophys Res Commun. 2009;389:506–511. doi: 10.1016/j.bbrc.2009.09.011. [DOI] [PubMed] [Google Scholar]


