Abstract
Neuropathic pain is the most difficult type of pain to control, and patients lose their motivation for the purposive pursuit with a decrease in their quality of life. Using a functional magnetic resonance imaging analysis, we demonstrated that blood oxygenation level-dependent signal intensity was increased in the ipsilateral nucleus accumbens (N.Acc.) following peripheral nerve injury. microRNAs are small, noncoding RNA molecules that direct the post-transcriptional suppression of gene expression, and play an important role in regulating synaptic plasticity. In this study, we found that sciatic nerve ligation induced a drastic decrease in the expression of miR200b and miR429 in N.Acc. neurons. The expression of DNA methyltransferase 3a (DNMT3a), which is the one of the predicted targets of miR200b/429, was significantly increased in the limbic forebrain including N.Acc. at 7 d after sciatic nerve ligation. Double-immunolabeling with antibodies specific to DNMT3a and NR1 showed that DNMT3a-immunoreactivity in the N.Acc. was located in NR1-labeled neurons, indicating that increased DNMT3a proteins were dominantly expressed in postsynaptic neurons in the N.Acc. area under a neuropathic pain-like state. The results of these analyses provide new insight into an epigenetic modification that is accompanied by a dramatic decrease in miR200b and miR429 along with the dysfunction of “mesolimbic motivation/valuation circuitry” under a neuropathic pain-like state. These phenomena may result in an increase in DNMT3a in neurons of the N.Acc. under neuropathic pain, which leads to the long-term transcription-silencing of several genes.
Introduction
Pain is a multidimensional experience with sensory-discriminative and motivational-affective components (Price, 2000; Narita et al., 2006). In the putative emotional circuit, both dopaminergic projections from the VTA to the nucleus accumbens (N.Acc.) and glutamatergic inputs to the N.Acc. from the amygdala, hippocampus, and prefrontal cortex participate in appetitive behaviors instructed by conditioned cues (Ambroggi et al., 2008; Carlezon and Thomas, 2009).
Recent human brain imaging studies have examined differences in activity in the N.Acc. in response to heat stimuli between controls and patients with chronic pain, and have revealed that the N.Acc. plays a role in predicting the value of a noxious stimulus and its offset, and in the consequent changes in the motivational state (Baliki et al., 2010). Thus, based on these reports of comorbidity between chronic pain and emotional disorders, it is possible that these disease states are linked. Nevertheless, the circuitry involved in emotion and motivation in response to chronic pain stimuli has not been fully explored.
There is a growing body of evidence that epigenetic mechanisms are molecular substrates that mediate long-lived changes in the brain. It has been considered that microRNAS (miRNAs), which are small, noncoding RNA molecules, play important roles as key modulators of post-transcriptional gene expression (Smalheiser and Lugli, 2009). In addition, several lines of evidence suggest that miRNAs are closely related to the regulation of synaptic plasticity (John et al., 2004; Vo et al., 2005; Wu and Xie, 2006; Klein et al., 2007; Nomura et al., 2008). These findings raise intriguing possibilities that miRNA-modulating cellular events along with epigenetic modifications may be associated with neural plasticity and neuronal adaptive responses in mesolimbic motivation/valuation circuitry under neuropathic pain. An important development in our understanding of the molecular processes that produce neural plasticity is multiplex analysis of gene-expression. Microarray studies are likely to be useful tools for this purpose. However, to date, few if any studies have used this approach in animal models under a persistent neuropathic pain-state. Therefore, we analyzed the miRNA profiles in the brain regions related to mesolimbic motivation/valuation circuitry in a neuropathic pain model using a miRNA microarray. Furthermore, we investigated whether changes in miRNA are associated with predicted changes in gene expression of candidate targets in these regions under neuropathic pain.
Materials and Methods
Animals.
The present study was conducted in accordance with the Guiding Principles for the Care and Use of Laboratory Animals, Hoshi University, as adopted by the Committee on Animal Research of Hoshi University, which is accredited by the Ministry of Education, Culture, Sports, Science and Technology of Japan. This study was approved by the Animal Research Committee of Hoshi University. Male C57BL/6J mice (6–8 weeks old) (Tokyo Laboratory Animals Science Co., Ltd.) were housed with ad libitum access to food and water and maintained on a 12 h light/dark cycle.
Neuropathic pain.
We produced a partial sciatic nerve ligation model by tying a tight ligature with 8–0 silk suture ∼1/3 to 1/2 the diameter of the sciatic nerve on the right side (ipsilateral side) of mice under anesthesia with 3% isoflurane as described previously (Malmberg and Basbaum, 1998). In sham-operated mice, the nerve was exposed without ligation.
Heat stimulation.
Contact heat stimulation was applied using a custom-made, computer-controlled Peltier heating and cooling device. Peltier elements with a surface measuring 8.3 × 8.3 mm were fixed to the right hindpaw. A block design with two conditions (42−43°C heat stimulus, rest) was applied with stimulus blocks of 40 s and rest blocks of 60 s.
Functional imaging.
Experiments were performed with a Unity Inova spectrometer (Varian), which was interfaced to a 9.4-T/31 cm horizontal bore magnet equipped with actively shielded gradients capable of 300 mT/m in a risetime of 500 s (Magnex Scientific). During the measurements, mice were lightly anesthetized with isoflurane (0.5–1%), and heat stimuli were applied to the right hindpaw following the block design. A continuous fMRI scanning protocol was used to study changes in brain signal intensity using T2-weighted blood oxygenation level-dependent (BOLD) contrast.
A functional series was acquired using the echoplanar imaging technique (matrix = 64 × 64, TR = 2000 ms, TE = 35 ms, 2 acquisitions, slice thickness = 1 mm, field of view = 25.6 × 25.6 mm2). Anatomical scans with a high spatial resolution were collected using a fast spin echo pulse sequence (matrix = 256 × 256, TR = 2000 ms, TE = 45 ms, slice thickness = 1 mm, field of view = 25.6 × 25.6 mm2).
Data analysis was performed using the FEAT (http://www.fmrib.ox.ac.uk) software package. Z (Gaussianized T/F) statistic images were thresholded using clusters determined by Z > 2.3 and a (corrected) cluster significance threshold of p = 0.05. Regions of interest (ROIs) were manually selected and statistical analyses were performed using NIH ImageJ. ROIs were drawn according to an atlas of the mouse brain (Franklin and Paxinos, 1997). The BOLD signal intensity values in each ROI were extracted and expressed as percentages of the corresponding average BOLD signals in the first rest period (baseline).
Brain sample preparation for in vitro analysis.
As previously described (Narita et al., 2001), after the killing of mice and removal of whole brain, thick coronal sections of the brain, including the limbic forebrain, which included N.Acc. (0.1–2.0 mm from bregma), frontal cortex (0.1–2.0 mm from bregma), hippocampus (−0.9 to −2.8 mm from bregma) or amygdala (−0.9 to −2.8 mm from bregma), was initially dissected using Brain Blocker (Neuroscience Inc.) on an ice-cold glass plate. Then, each brain region was extracted from coronal sections. The decision of brain position was conducted according to an atlas of the mouse brain (Franklin and Paxinos, 1997).
TaqMan miRNA arrays.
Total RNA, including miRNAs, was extracted from the limbic forebrain including N.Acc., hippocampus and amygdala of mice using the mirVana miRNA Isolation Kit (Applied Biosystems Inc.). For miRNA cDNA synthesis, RNA was reverse-transcribed using the TaqMan miRNA Reverse Transcription (RT) kit (Applied Biosystems Inc.) in combination with the stem-loop Megaplex primer pool (Applied Biosystems Inc.). Cycling conditions were as follows: 40 cycles of 16°C for 2 min, 42°C for 1 min and 50°C for 1 s, followed by 85°C for 5 min. miRNA expression profiles were acquired using a TaqMan rodent microRNA array v2.0 A (Applied Biosystems Inc.). Quantitative RT-PCR was performed at Life Technologies Japan Ltd. using an Applied Biosystems 7900HT Fast Real-Time PCR system. Cycling conditions were as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Cycle threshold (Ct) values were calculated using SDS2.2.2 with an automatic baseline and a threshold of 0.2. Normalized expression (NE) was calculated as NE = 2ΔΔCt, where Ct is the threshold cycle for detecting fluorescence. The data were normalized to snoRNA202.
Quantification of miRNAs with a real-time PCR.
Mice were sacrificed by decapitation at 7 d after sciatic nerve ligation. Individual TaqMan assays (Applied Biosystems) were used to analyze the expression of the following mature mouse miRNAs. Total RNA (10 ng) extracted from the limbic forebrain including N.Acc., frontal cortex, hippocampus and amygdala of mice was used in the RT reaction and the transcribed cDNA was then used for subsequent PCR amplification using TaqMan 2X Universal PCR Master Mix, No AmpErase UNG (Applied Biosystems). Real-time PCR was conducted at 95°C for 10 min, followed by 45 cycles of 95°C for 15 s and 60°C for 1 min. The threshold cycle (Ct) method was used to determine the relative quantity of each miRNA.
miRNA target prediction.
miRNA-targeting predictions were queried in three target prediction databases: TargetScan (http://www.targetscan.org), PITA (http://genie.weizmann.ac.il/pubs/mir07), and DIANAmicroT (http://diana.pcbi.upenn.edu/cgi-bin/micro_t.cgi) with default parameters (for PITA, the no-flanking top table was used; see the website for details).
Western blotting.
Mice were killed by decapitation at 7 d after sciatic nerve ligation. Membrane or nucleus proteins were extracted from the limbic forebrain including N.Acc., hippocampus and amygdala of mice using a proteoExtract subcellular proteome extraction kit (Calbiochem). An aliquot of tissue sample was diluted with an equal volume of electrophoresis sample buffer (Protein Gel Loading Dye-2X, Amresco) and separated by SDS-polyacrylamide gradient gel, and transferred to nitrocellulose membranes. Immunodetection was performed using rabbit antibody to DNA methyltransferase 3a (DNMT3a; 1:1000, Cell Signaling Technology), Zinc finger E-box binding homeobox 2 (ZEB2; 1:3000, ProSci Inc., Poway, CA), EPS8 (1:1000, Abcam), methyl CpG binding protein 2 (MeCP2; 1:1000, Abcam), 25 kDa synaptosome-associated protein (SNAP25, 1;10,000, Cell Signaling Technology), neuronal membrane glycoprotein M6-a (Gpm6a, 1:6000, Abcam), fibrillarin (1:2000, Abcam) and goat antibody to NMDAζ1 (NR1, 1:2000, Santa Cruz Biotechnology) as primary antibodies, and antibody to rabbit or goat HRP (1:10,000, Southern Biotechnology Associates) as the secondary antibody. The antigen-antibody peroxidase complex was finally detected by enhanced chemiluminescence (Pierce) according to the manufacturer's instructions and visualized by exposure to GE Healthcare Hyperfilm (GE Healthcare Life Sciences).
Immunohistochemistry.
Seven days after nerve ligation, mice were deeply anesthetized with 3% isoflurane and intracardially perfusion-fixed with 4% paraformaldehyde in 0.1 m PBS (PBS, pH 7.4). After perfusion, the brain thick coronal sections including the N.Acc. were rapidly dissected and processed further, as described previously (Narita et al., 2010).
The sections were blocked in 10% normal horse serum (NHS) in 0.01 m PBS for 1 h at room temperature. For double-staining, goat anti-NR1 (1:1000, Santa Cruz Biotechnology) and rabbit anti-DNMT3a (1:1000, Abcam) were visualized using species-specific Alexa-conjugated secondary antibodies (Invitrogen).
Statistical data analysis.
Data are expressed as the mean with SEM. Two-way ANOVAs with independent and repeated measures, as well as planned comparisons or Student's t tests, were used as appropriate for the experimental design. All statistical analyses were performed with Prism version 5.0a (GraphPad Software).
Results
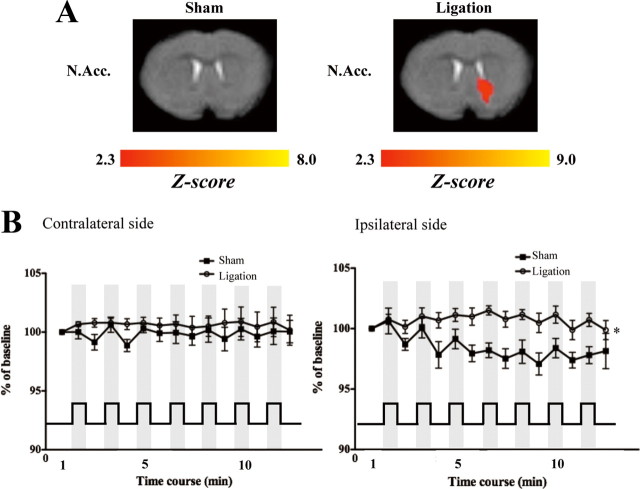
As shown in Figure 1A, the application of heat stimuli by Peltier elements caused positive signal activity in the ipsilateral N.Acc. of nerve-ligated mice, but not sham-operated mice. The BOLD signal was not changed by heat stimuli in the N.Acc. on the contralateral side after either sciatic nerve ligation or sham operation (Fig. 1B). With repeated applications of heat stimuli, decreased signal was noted in the ipsilateral N.Acc. of sham-operated mice, while signal was maintained in that of nerve-ligated mice (F(14,154) = 2.733, p < 0.05, sham-operated mice vs nerve-ligated mice).
Figure 1.
Change in BOLD signal intensity following the application of heat stimuli in the ipsilateral N.Acc. at 7 d after nerve ligation. A, Representative activation maps (Z statistic images) correspond to composite images of nerve-ligated or sham-operated mice. B, BOLD signals averaged for each block period from the contralateral or ipsilateral N.Acc. The bottom line shows the time course of the heat stimulus applied to the right hindpaw. The data are expressed as percentages of the corresponding average BOLD signals in the first rest period with SEM of 6–7 mice. F(14,154) = 2.733; *p < 0.05, sham-operated mice vs nerve-ligated mice.
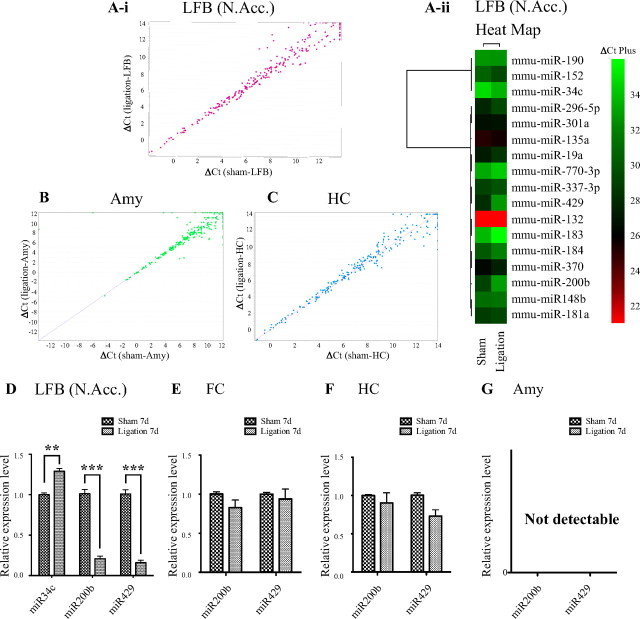
Expression levels of the 384 miRNAs spotted on microarrays from sham-operated and nerve ligated mice were analyzed with correlation plots (Fig. 2). This analysis revealed a number of miRNAs that were up- and downregulated. To confirm results of the microarray analysis, real-time PCR analysis was conducted in the limbic forebrain including the N.Acc. at 7 d after sciatic nerve ligation. The expression levels of miR200b (Sham, 1.01 ± 0.05; Ligation, 0.21 ± 0.03; n = 9, p < 0.001) and miR429 (Sham, 1.01 ± 0.05; Ligation, 0.16 ± 0.03; n = 9, p < 0.001) were drastically decreased, whereas that of miR34c was significantly increased (Sham, 1.00 ± 0.02; Ligation, 1.29 ± 0.03; n = 3, p < 0.01) in the limbic forebrain including the N.Acc. of nerve-ligated mice compared with sham-operated mice (Fig. 2D). On the other hand, there were no changes in the expression of miR200b or miR429 in the frontal cortex (miR200b; Sham, 1.00 ± 0.03; Ligation, 0.83 ± 0.10, miR429; Sham, 1.00 ± 0.02; Ligation, 0.94 ± 0.13, n = 9, Fig. 2E) and hippocampus (miR200b; Sham, 1.00 ± 0.01; Ligation, 0.90 ± 0.13, miR429; Sham, 1.00 ± 0.03; Ligation, 0.73 ± 0.08, n = 9, Fig. 2F) after sciatic nerve ligation. The expression of miR200b and miR429 were not detected in the amygdala (Fig. 2G).
Figure 2.
miRNA expression analysis using microarrays. Pearson's correlation scatter plot of miRNA levels in sham-operated and nerve-ligated mice at 7 d after surgery in the limbic forebrain (LFB) including the N.Acc. (r = 0.9865) (A-i), amygdala (Amy) (r = 0.8625) (B), and hippocampus (HC) (r = 0.9868) (C). A-ii, Heat map of 17 miRNAs that are differentially expressed in the LFB including the N.Acc. of nerve-ligated mice compared with that in sham-operated mice. Each miRNA is represented as a single bar based on its Ct value and the color coding represents a gradient from green (negative and lowest Ct values) to red (positive and highest Ct values). D–G, Quantitative analysis of miR34c, miR200b and miR429 expression in the LFB including the N.Acc. D, frontal cortex (FC) (E), hippocampus (HC) (F), and amygdala (Amy) (G) of mice at 7 d after nerve ligation. Each column represents the mean ± SEM of 3 samples. **p < 0.01, ***p < 0.001, sham-operated mice vs nerve-ligated mice.
We next investigated the changes in the expression of predicted miR200b/429 cluster targeted proteins in the limbic forebrain including the N.Acc. at 7 d after sciatic nerve ligation. The nucleus protein levels of ZEB2, MeCP2 and Eps8 (ZEB2; Sham, 100.0 ± 4.482; Ligation, 99.44 ± 3.696, MeCP2; Sham, 100.0 ± 2.966; Ligation, 103.5 ± 0.8380, Eps8; Sham, 100.0 ± 5.819; Ligation, 105.2 ± 6.436, n = 6, Fig. 3A–i,ii,iv) and the membrane-bound levels of SNAP25 and Gpm6a (SNAP25; Sham, 100.0 ± 4.871; Ligation, 106.7 ± 5.013, Gpm6a; Sham, 100.0 ± 3.674; Ligation, 107.7 ± 4.581, n = 6, Fig. 3A–v,vi) were not affected by nerve ligation, whereas the nucleus protein level of DNMT3a was significantly increased in the limbic forebrain including the N.Acc. of nerve-ligated mice compared with that of sham-operated mice. (Sham, 100.0 ± 8.242; Ligation, 132.4 ± 3.882, n = 6, p < 0.01, Fig. 3A-iii). In contrast, we demonstrated that partial sciatic nerve ligation had not detectable effect on the levels of DNMT3a expression in the frontal cortex or amygdala area of mice after nerve ligation (the frontal cortex; Sham, 100.0 ± 3.543, Ligation, 92.47 ± 3.105, amygdala; Sham, 100.0 ± 4.030, Ligation, 109.0 ± 3.203, n = 3, Fig. 3B–i,ii). To demonstrate the localization of DNMT3a protein, we performed an immunohistochemical analysis. DNMT3a-IR was detected in the N.Acc. of sham-operated mice and nerve-ligated mice. In nerve-ligated mice, DNMT3a-IR in the N.Acc. was increased compared with that in sham-operated mice at 7 d (Fig. 3C–i,ii). In an enlarged image, double-immunolabeling with antibodies specific to DNMT3a and NR1 showed that DNMT3a-IR in the N.Acc. was detected in the cytoplasm/nucleus of NR1-labeled neurons (Fig. 3D-iii).
Figure 3.
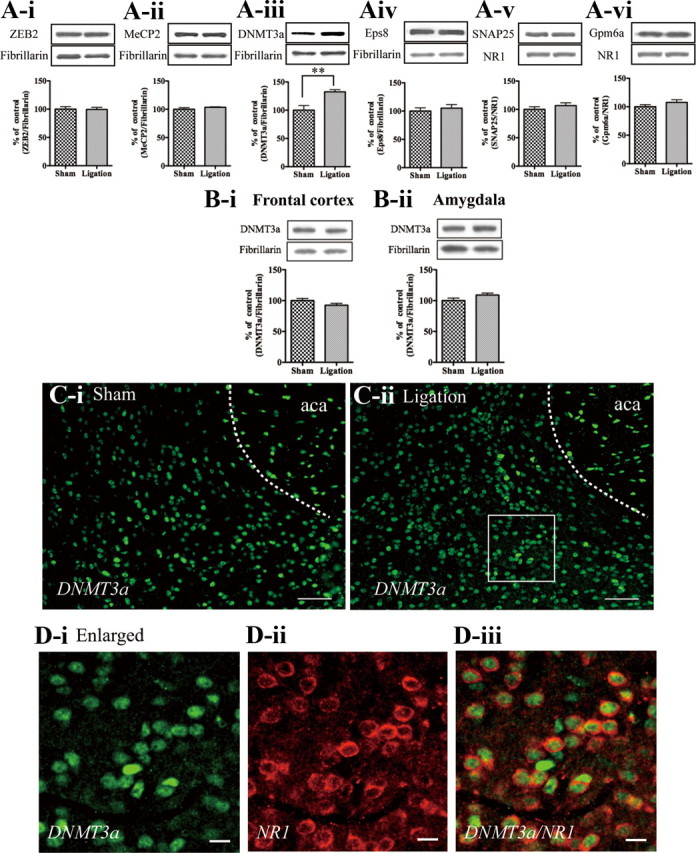
Changes in the expression of predicted miR200b/429 cluster targeted proteins at 7 d after sciatic nerve ligation. A, The nucleus protein levels of ZEB2 (A-i), MeCP2 (A-ii), DNMT3a (A-iii), and Eps8 (A-iv), and the membrane-bound levels of SNAP25 (A-v) and Gpm6a (A-vi), in the LFB including the N.Acc. at 7 d after sciatic nerve ligation. B, The nucleus protein levels of DNMT3a in the frontal cortex (B-i) and amygdala (B-ii) at 7 d after sciatic nerve ligation. Each column represents the mean ± SEM of 3 samples. **p < 0.01, sham-operated mice vs nerve-ligated mice. C, D, Immunofluorescent staining for DNMT3a-IR (C, D-i) and NR1-IR (D-ii) in the N.Acc. of sham-operated and nerve-ligated mice. Scale bars: (C-i, -ii) 50 μm, (D-i, -ii, -iii) 10 μm.
Discussion
In the previous study, we have demonstrated that the persistent painful state only for the ipsilateral side caused by partial sciatic nerve ligation at 7 d after surgery in mice (Narita et al., 2006, Niikura et al., 2010). Under these conditions, the heat stimuli caused positive signal activity in the ipsilateral N.Acc. of nerve-ligated mice. The BOLD signal intensity in the sham-operated group was gradually decreased during repeated heat stimulus, while the basal signal was maintained in that of nerve-ligated mice.
These results raise questions regarding the mechanisms that induce the maintained signals in the N.Acc. after sciatic nerve ligation. There is a growing body of evidence that epigenetic mechanisms are likely to be used to maintain cell memory and to maintain and strengthen synaptic connections that maintain long-term changes in behavior and emotion (Colvis et al., 2005; Tsankova et al., 2007). Thus, the concept of the post-transcriptional suppression of gene expression addresses a key challenge in neurobiology of determining how stable changes in chronic pain are induced in neurons or glia to produce long-lasting emotional disorders. They silence the expression of target genes by reducing mRNA stability and translation. The production of miRNAs is controlled via developmental and environmental cues, and these small molecules, in concert with classical transcriptional regulators, amplify changes in neuronal maturation, dendrite morphogenesis, and synaptogenesis (John et al., 2004; Schratt et al., 2006; Klein et al., 2007; Nomura et al., 2008; Wayman et al., 2008). In the present study using miRNAs microarrays and real-time PCR, miR200b and miR429 were dramatically decreased in the limbic forebrain including the N.Acc., but not in the frontal cortex or hippocampus, of nerve-ligated mice. The expression of miR200b and miR429 was not detected in the amygdala. Furthermore, we demonstrated that the psychological stress, which was generated by the communication box test, had no effect on levels of miR200b/429 in the limbic forebrain including N.Acc. of mice (data not shown). These findings suggest that the dramatically decreased in the expression of miR200b/429 in the N.Acc. may be directly associated with sciatic nerve injury.
We next used 3 algorithms to identify the predicted targets for the miR209/429 cluster. This analysis revealed 363 genes in common. Many of the targets, such as ZEB2, MeCP2, DNMT3a, Eps8, Gpm6a and SNAP25, were mainly accompanied by cell morphology and epigenetic modifications. In Western blot analysis, the nucleus protein level of DNMT3a was significantly increased in the limbic forebrain including the N.Acc. of nerve-ligated mice compared with that in sham-operated mice. In agreement with the findings of present miRNA microarrays and real-time PCR analysis, partial sciatic nerve ligation had no effect on the protein level of DNMT3a in the frontal cortex or amygdala area of mice even after nerve ligation. We therefore hypothesize that a decrease in miRNA200b/429 cluster that binds to 3′-UTR of DNMT3a mRNA in the cytoplasm of the N.Acc. cells may lead to the enhancement of translational process of DNMT3a mRNA.
It is thus likely that the increased DNMT3a proteins can be subsequently translocated to the nucleus where it modulates expression of a number of genes. Double-immunolabeling with antibodies specific to DNMT3a and NR1 showed that DNMT3a-IR in the N.Acc. was detected in the cytoplasm/nucleus of NR1-labeled neurons, indicating that increased DNMT3a proteins were dominantly expressed in postsynaptic neurons in the N.Acc. area under a neuropathic pain-like state.
It has been well recognized that DNMT3a is an important DNA methyltransferase that is expressed in postmitotic neurons. DNA methylation can induce long-term transcriptional-silencing by directly interfering with transcription factor binding. Recent studies have implicated the misregulation of DNA methylation and DNMTs in cognitive disorders such as schizophrenia, Rett syndrome, and Fragile X mental retardation (Sutcliffe et al., 1992; Amir et al., 1999; Veldic et al., 2004). Behavioral studies have suggested that DNA methylation is required for hippocampal-dependent memory formation (Miller and Sweatt, 2007; Feng et al., 2010). On the other hand, it has been reported that N.Acc.-specific manipulations that block DNA methylation potentiate cocaine reward and exert antidepressant-like effects, whereas N.Acc.-specific DNMT3a overexpression attenuates cocaine reward and is prodepressant (LaPlant et al., 2010). In the previous studies, we clearly demonstrated that sciatic nerve ligation caused a dramatic decrease in a morphine-induced conditioned place preference associated with dysfunction of the mesolimbic dopaminergic circuitry in rodents (Narita et al., 2006, Niikura et al., 2010). Together with the present findings, these data support the idea that increased DNMT3a associated with decreased miRNA 200b/429 clusters in the N.Acc. of mice with peripheral nerve injury may produce negative emotions along with dysfunction of the “mesolimbic motivation/valuation circuitry.” In fact, we found in our recent studies that mice with the same sciatic nerve injury used in the present study displayed sleep disturbance (Narita et al., 2011) and anxiety (Narita et al., 2006). The exact mechanisms driving changes in gene expression in the N.Acc following nerve injury should be further examined, in particular, those initiating the changes. Nevertheless, the changes in the N.Acc observed in the present study support the suggestion that persistent nociceptive stimuli, such as that associated with peripheral nerve injury may induce negative emotions.
Footnotes
This work was supported in part by grants from the Ministry of Health, Labor and Welfare, and the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- Ambroggi F, Ishikawa A, Fields HL, Nicola SM. Basolateral amygdala neurons facilitate reward-seeking behavior by exciting nucleus accumbens neurons. Neuron. 2008;59:648–661. doi: 10.1016/j.neuron.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Baliki MN, Geha PY, Fields HL, Apkarian AV. Predicting value of pain and analgesia: nucleus accumbens response to noxious stimuli changes in the presence of chronic pain. Neuron. 2010;66:149–160. doi: 10.1016/j.neuron.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlezon WA, Jr, Thomas MJ. Biological substrates of reward and aversion: a nucleus accumbens activity hypothesis. Neuropharmacology. 2009;56(Suppl 1):122–132. doi: 10.1016/j.neuropharm.2008.06.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvis CM, Pollock JD, Goodman RH, Impey S, Dunn J, Mandel G, Champagne FA, Mayford M, Korzus E, Kumar A, Renthal W, Theobald DE, Nestler EJ. Epigenetic mechanisms and gene networks in the nervous system. J Neurosci. 2005;25:10379–10389. doi: 10.1523/JNEUROSCI.4119-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, Silva AJ, Fan G. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KBJ, Paxinos G. The mouse brain in stereotaxic coordinates. Ed 1. San Diego: Academic; 1997. [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein ME, Lioy DT, Ma L, Impey S, Mandel G, Goodman RH. Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat Neurosci. 2007;10:1513–1514. doi: 10.1038/nn2010. [DOI] [PubMed] [Google Scholar]
- LaPlant Q, Vialou V, Covington HE, 3rd, Dumitriu D, Feng J, Warren BL, Maze I, Dietz DM, Watts EL, Iñiguez SD, Koo JW, Mouzon E, Renthal W, Hollis F, Wang H, Noonan MA, Ren Y, Eisch AJ, Bolaños CA, Kabbaj M, et al. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat Neurosci. 2010;13:1137–1143. doi: 10.1038/nn.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmberg AB, Basbaum AI. Partial sciatic nerve injury in the mouse as a model of neuropathic pain: behavioral and neuroanatomical correlates. Pain. 1998;76:215–222. doi: 10.1016/s0304-3959(98)00045-1. [DOI] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Narita M, Mizoguchi H, Narita M, Nagase H, Suzuki T, Tseng LF. Involvement of spinal protein kinase Cgamma in the attenuation of opioid mu-receptor-mediated G-protein activation after chronic intrathecal administration of [D-Ala2,N-MePhe4,Gly-Ol(5)]enkephalin. J Neurosci. 2001;21:3715–3720. doi: 10.1523/JNEUROSCI.21-11-03715.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita M, Kuzumaki N, Narita M, Kaneko C, Hareyama N, Miyatake M, Shindo K, Miyoshi K, Nakajima M, Nagumo Y, Sato F, Wachi H, Seyama Y, Suzuki T. Chronic pain-induced emotional dysfunction is associated with astrogliosis due to cortical delta-opioid receptor dysfunction. J Neurochem. 2006;97:1369–1378. doi: 10.1111/j.1471-4159.2006.03824.x. [DOI] [PubMed] [Google Scholar]
- Narita M, Matsushima Y, Niikura K, Narita M, Takagi S, Nakahara K, Kurahashi K, Abe M, Saeki M, Asato M, Imai S, Ikeda K, Kuzumaki N, Suzuki T. Implication of dopaminergic projection from the ventral tegmental area to the anterior cingulate cortex in mu-opioid-induced place preference. Addict Biol. 2010;15:434–447. doi: 10.1111/j.1369-1600.2010.00249.x. [DOI] [PubMed] [Google Scholar]
- Narita M, Niikura K, Nanjo-Niikura K, Narita M, Furuya M, Yamashita A, Saeki M, Matsushima Y, Imai S, Shimizu T, Asato M, Kuzumaki N, Okutsu D, Miyoshi K, Suzuki M, Tsukiyama Y, Konno M, Yomiya K, Matoba M, Suzuki T. Sleep disturbances in a neuropathic pain-like condition in the mouse are associated with altered GABAergic transmission in the cingulate cortex. Pain. 2011;152:1358–1372. doi: 10.1016/j.pain.2011.02.016. [DOI] [PubMed] [Google Scholar]
- Niikura K, Narita M, Butelman ER, Kreek MJ, Suzuki T. Neuropathic and chronic pain stimuli downregulate central mu-opioid and dopaminergic transmission. Trends Pharmacol Sci. 2010;31:299–305. doi: 10.1016/j.tips.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Nomura T, Kimura M, Horii T, Morita S, Soejima H, Kudo S, Hatada I. MeCP2-dependent repression of an imprinted miR-184 released by depolarization. Hum Mol Genet. 2008;17:1192–1199. doi: 10.1093/hmg/ddn011. [DOI] [PubMed] [Google Scholar]
- Price DD. Psychological and neural mechanisms of the affective dimension of pain. Science. 2000;288:1769–1772. doi: 10.1126/science.288.5472.1769. [DOI] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Smalheiser NR, Lugli G. microRNA regulation of synaptic plasticity. Neuromolecular Med. 2009;11:133–140. doi: 10.1007/s12017-009-8065-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe JS, Nelson DL, Zhang F, Pieretti M, Caskey CT, Saxe D, Warren ST. DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum Mol Genet. 1992;1:397–400. doi: 10.1093/hmg/1.6.397. [DOI] [PubMed] [Google Scholar]
- Tsankova N, Renthal W, Kumar A, Nestler EJ. Epigenetic regulation in psychiatric disorders. Nat Rev Neurosci. 2007;8:355–367. doi: 10.1038/nrn2132. [DOI] [PubMed] [Google Scholar]
- Veldic M, Caruncho HJ, Liu WS, Davis J, Satta R, Grayson DR, Guidotti A, Costa E. DNA-methyltransferase 1 mRNA is selectively overexpressed in telencephalic GABAergic interneurons of schizophrenia brains. Proc Natl Acad Sci U S A. 2004;101:348–353. doi: 10.1073/pnas.2637013100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayman GA, Davare M, Ando H, Fortin D, Varlamova O, Cheng HY, Marks D, Obrietan K, Soderling TR, Goodman RH, Impey S. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc Natl Acad Sci U S A. 2008;105:9093–9098. doi: 10.1073/pnas.0803072105. [DOI] [PMC free article] [PubMed] [Google Scholar]