Abstract
Kinesin heavy chain (Khc) is crucially required for axonal transport and khc mutants show axonal swellings and paralysis. Here, we demonstrate that in Drosophila khc is equally important in glial cells. Glial-specific downregulation of khc by RNA interference suppresses neuronal excitability and results in spastic flies. The specificity of the phenotype was verified by interspecies rescue experiments and further mutant analyses. Khc is mostly required in the subperineurial glia forming the blood–brain barrier. Following glial-specific knockdown, peripheral nerves are swollen with maldistributed mitochondria. To better understand khc function, we determined Khc-dependent Rab proteins in glia and present evidence that Neurexin IV, a well known blood–brain barrier constituent, is one of the relevant cargo proteins. Our work shows that the role of Khc for neuronal excitability must be considered in the light of its necessity for directed transport in glia.
Introduction
The interaction of neurons and glia is crucial for nervous system function in manifold ways. Glia insulates and supports neurons, facilitates and modulates electrical conductance, and influences synaptic transmission (Barres, 2008; Nave and Trapp, 2008; Zlokovic, 2008). In Drosophila, different glial cell types can be distinguished which exert specific functions (Edwards and Meinertzhagen, 2010). The outermost layer of the entire nervous system is build by the perineurial and subperineurial glia. They set up the blood–brain barrier (BBB) regulating the uptake of ions and metabolites into the nervous system (Stork et al., 2008). The paracellular tightness of the BBB is assured by pleated septate junctions (pSJs), that resemble the septate-like junctions at the paranodes of vertebrates (Rodrigues et al., 2011). A central component of both septate and septate-like junctions is Neurexin IV/Caspr. In the absence of Neurexin IV (NrxIV), pSJs disassemble and small solutes enter the brain via paracellular flow, which results in the death of the embryo (Baumgartner et al., 1996).
In Drosophila, systematic gene silencing in only a few selected cells or cell types became possible with the generation of genome-wide collections of transgenes that express dsRNA (Dietzl et al., 2007). Using these collections, we performed an RNAi screen to identify new gene functions required in a cell-autonomous manner for glial development and function.
Here, we report that Kinesin heavy chain (Khc) functions in glia to control integrity of the BBB, which in turn is required for normal neuronal activity. Khc, a homodimeric mechanoenzyme, is needed for anterograde movement of cargoes such as membrane vesicles or mitochondria along microtubules (Hirokawa et al., 2010). The vertebrate homologs of Drosophila Khc are the KIF5 proteins. KIF5A and KIF5C are predominantly expressed in neuronal cells (Kanai et al., 2000; Dathe et al., 2004; Jaulin et al., 2007). KIF5B is more broadly expressed in the nervous system and is also found in glia (Kanai et al., 2000), but no glial-specific requirements have yet been reported. In contrast, glial functions were shown for other Kinesins. KIF13B transports the scaffolding protein Dlg1 to membrane remodeling sites during myelin formation in Schwann cells (Bolis et al., 2009). Zebrafish Kif1b was identified in a genetic screen for myelin basic protein (mbp) mRNA mislocalization (Lyons et al., 2009).
The linkage of Khc to different cargoes is mediated by a number of specific adaptor proteins (Hirokawa et al., 2010). The Milton protein is required for Khc-dependent transport of mitochondria (Stowers et al., 2002; Cox and Spradling, 2006; Glater et al., 2006; Pilling et al., 2006; Rice and Gelfand, 2006). The association of Khc with its cargoes is in turn regulated by different small G-proteins of the Rab family, which decorate the different vesicle pools (Simons and Zerial, 1993; Zerial and McBride, 2001). Here, we report that glial cell-specific knockdown of khc results in spastic flies. This not only unravels a so far unknown function of Khc in glial cells, but also highlights the essential contribution of glia for neuronal function.
Materials and Methods
Fly strains and genetics.
All crosses were performed on standard food at 25°C. A list of the 5000 screened fly strains is available upon request. The RNAi screen was performed using repo4.3Gal4;repoGal4/TM6 (Sepp and Auld, 1999; Lee and Jones, 2005). All RNAi lines were obtained from National Institute of Genetics Fly Stock Center (NIGfly) for screening and the Vienna Drosophila RNAi Center (VDRC). Khc8, khc6, and Df(2R)Jp6 were used for khc mutant analyzes (provided by B. Saxton, University of California, Santa Cruz, CA). For rescue experiments we generated phiC31-based transgenes in the landing sites 58A, 68E and 86Fb (Bischof et al., 2007). The different Rab-YFP, Rab-GFP and mitoGFP lines were obtained from the Bloomington Stock Center.
RNAi screen and the island assay.
Three to five males carrying a UAS-IR construct (inverted repeat; Dietzl et al., 2007) were crossed to five repo4.3Gal4/repo4.3Gal4;repoGal4/TM6 virgins. Crosses were kept at 25°C for 14 d. If gene suppression did not result in lethality, 25 RNAi-expressing flies of either sex were kept one additional week at 25°C and subsequently tested in the island assay for locomotion defects. The island assay consists of a 25 × 35 cm2 watery soap bath (soap is added to drown flies that touch the water). In the middle of the soap bath is a 10 × 10 cm2 island. Adult locomotion was evaluated regarding the following parameters: the time the flies needed to vanish, flight abilities, and hyper- or hypoactivity. If flies showed an abnormal locomotion, they were rescreened two additional times in a double-blind manner. Following videotaping, flies remaining on the platform were counted manually on the computer screen. At least three independent experiments were performed.
Larval locomotion analysis.
Wandering third instar larvae of either sex were tracked for 1 min [with 2 frames per second (fps)] on a 2% agarose plate. The tracking was performed in groups of 10 larvae at 25°C and controlled humidity (65%). For each genotype we tracked 130 larvae. To acclimatize, the larvae were kept at the tracking conditions for 5 min before starting the experiment. The migratory paths were recorded using Image Pro Plus (Media Cybernetics). The larval migration distance between two consecutive time steps was calculated by the tracking program and the set of all distance values of a single larva defines the movement pattern of an animal. All movement parameters of all larvae in a test group were then represented in an n-dimensional feature space, with each dimension representing a single movement between two consecutive time frames. Subsequently, we projected this n-dimensional feature space into two dimensions using multidimensional scaling (MDS) (Duda et al., 2001) using a Matlab programming environment. This resulted in a graphical representation of every single larval movement pattern in a single point in a 2D plot and allows us to visualize and determine clusters of similar locomotion patterns. In the resulting graph, larvae that behaved in a more similar way to each other are grouped more closely together than those differing very much from each other in their locomotion pattern for the analyzed parameter.
Intracellular recordings.
For electrophysiology, third instar larvae were dissected in a clear Sylgard dish containing chilled HL3.1 (Feng et al., 2004). In this saline solution, preparations typically remained viable for 1–2 h at room temperature. Each larva was positioned dorsal side up, and 0.1 mm insect pins were placed in the head and tail to make open fillets. Fat bodies, digestive organs, and brain lobes were removed. In tightly pinned preparations, this reduces locomotor rhythms but leaves motor neuron cell bodies, axons, and nerve terminals intact. We targeted larval muscle 6 (abdominal segment A6) for all intracellular recordings. Recordings were made with sharp glass electrodes (Harvard Apparatus GC100F-10; 1.0 mm outside diameter × 0.58 mm) prepared using a Flaming/Brown micropipette puller (Sutter Instruments Co. Model P-97) and filled with 3 m KCl. The electrodes and headstage were maneuvered with a micromanipulator. Voltage signals were amplified with an Axoclamp 900A amplifier. Data were digitized and analyzed using a Digidata 1440A data acquisition system (Axon CNS Instruments). We measured excitatory junction potentials (EJPs) by attaching a suction electrode to segmental nerves cut close to the CNS and delivering 0.3 ms duration voltage shocks with a model ISO-STIM 01D isolated pulse stimulator (voltage stimulator). The peak-to-trough amplitude of the response was measured after taking mean response of 10 pulses of voltage. When different genotypes were being tested, the unpaired t test was used to calculate significance. A value of p < 0.01 was taken to be significant.
Immunohistochemistry.
Anti-Khc antibody was obtained from Cytoskeleton, β3-tubulin (β3-tub) from D. Buttgereit and R. Renkawitz. Repo (Developmental Studies Hybridoma bank, Iowa City, IA), anti-GFP (Invitrogen), and anti-HRP Cy5 (Dianova) antisera were used according to the manufacturer's instructions. Fixation and treatment of tissues for immunohistochemistry was performed according to standard procedures. Fluorescently labeled specimens were analyzed using a Zeiss 510 LSM or a Zeiss 710 LSM laser scanning microscope; orthogonal sections were taken using the Zeiss image browser.
Life imaging of third instar larval peripheral nerves.
Crosses or stocks for life imaging experiments were set up on blue food (Carolina Biological Supply Company), which reduces autofluorescence of the gut system. Third instar larvae were anesthetized using an isoflurane bath for up to 10 s. Subsequently, they were washed with PBS and transferred to a coverslip. Wax spacers helped to gently squeeze the larva to improve accessibility of peripheral nerves. Imaging was performed with the Zeiss LSM 5 Live DuoScan for up to 30 min. Only data from larvae that recovered from anesthesia were evaluated.
Transgenes.
phiC31-based transgenes were made. First, pENTR clones containing the entire Drosophila melanogaster khc coding sequence from a cDNA, which was obtained by reverse transcription of isolated mRNA from late embryonic stages (Primers: Primer_CACC_KhcforI: CACCaagatctccgccctgaag; khc-rev_delta_STOP_53: CGAGTTGACAGGATTAACCTGGGCC; Primer3_Khc-revSTOPI: TTCAGCTCCGCTTTAAACTAAACTA). For the interspecies rescue we generated pENTR clones containing the full-length D. ananassae khc coding sequence we obtained using a fosmid (stock number 50412985) from Drosophila Genetic Resource Center (amplification Primers: Ana_full_Pr3_For caccATGTCCGCGGAACGCga; Ana_full_Pr3_rev: gatCTACGAGTTGACAGCATTAGCCgg). To generate base pair changes in the khcana rescue construct the kit QuikChange XL Site-Directed Mutagenesis (Stratagene) was used according to the manufacturer's instructions (site-directed mutagenesis primer for pENTR_Khc_ana_g159a_c309t: melTOana_muta_g159a_sense: agctcaaggagcaggtaatggagcaggaagaac; melTOana_muta_g159a_antisense: gttcttcctgctccattacctgctccttgagct; melTOana_muta_c309t_sense: gttggccgttaactacgatcagaaatctcaggaaatc; melTOana_muta_c309t_antisense: gatttcctgagatttctgatcgtagttaacggccaac). By subsequent Gateway cloning the clones were recombined into the pUAST-attB-rfA, pUAS-attB-EGFP-rfA, and pUAST-attB-rfA-EFGP vector. Full-length untagged versions of khcmel and khcana as well as C- and N-terminal GFP-tagged khcmel constructs were generated. The khcana construct was inserted into the 58A and 68E landing sites. khcmel constructs were inserted in the 68E, 86Fa and 86Fb landing sites.
Results
Identification of glial-specific gene functions
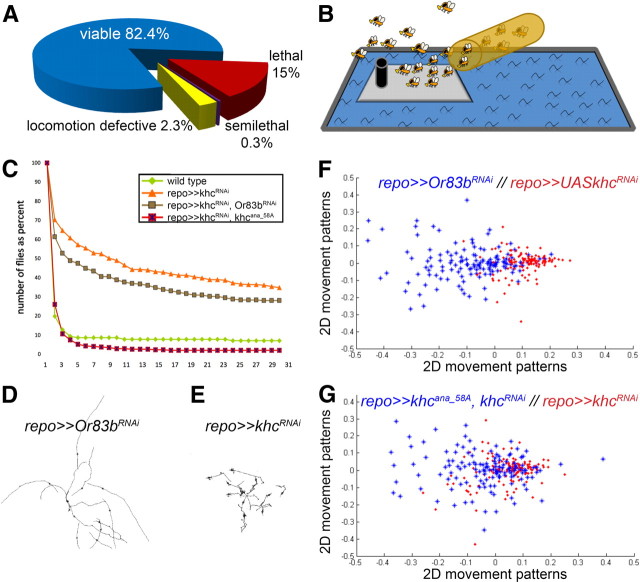
To search for genes required in glial cells, we used a glial cell-specific Gal4 driver and the collections of UAS-hairpinDNA transgenes provided by the VDRC and NIGfly stock centers to perform an RNAi-based screen (Dietzl et al., 2007). We selected ∼5000 putative candidate genes encoding transmembrane proteins, transcription factors as well as transport and cytoskeleton-associated proteins. For an exclusive glial knockdown of the respective gene functions, we used the panglial driver repoGal4 (Xiong et al., 1994; Sepp and Auld, 1999; Lee and Jones, 2005); see Materials and Methods). As a first read-out, we screened for RNAi-induced lethality. Silencing of ∼15% of the genes resulted in larval or pupal lethality (Fig. 1A) and their analysis will be presented elsewhere. In a second approach, we tested the requirement of the remaining genes for adult locomotion defects using a novel testing system: the island assay (Fig. 1B).
Figure 1.
A glial cell-specific RNAi screen reveals glial functions of kinesin. A, Summary of RNAi screen. A total of 5273 UAS-dsRNA elements were expressed in all glial cells. In 15% of the crosses glial-specific silencing resulted in lethality, in 0.3% we noted very weak flies (classified as semilethal), in 2.3% of the tested genes locomotion defects were noted. In the remaining 82.4% of the crosses no obvious defects were detected. B, Schematic view of the island assay. Seven-day-old flies expressing the different dsRNA constructs in all glial cells were released on the testing arena. The following locomotion defects were scored: jumper, sitter, and spastic. C, Elution profile of different flies released on the testing platform in seconds. The different genotypes are indicated. D, E, Crawling paths of 10 third instar larvae tracked for 1 min at two frames per second, the different genotypes are indicated. The glial suppression of khc (E) induces sluggish larval locomotion patterns compared with the RNAi mock control (D). F, G, Data analysis of the distance parameter during larval crawling using MDS (see Materials and Methods). The different genotypes of the larvae are indicated by red or blue color. A total of 100 animals were tracked for each genotype. The 2D movement patterns were plotted of all animals in the different test groups. Clustering of points in the graph indicates animals with similar movement patterns. F, Larvae expressing khcdsRNA in all glial cells clearly differ from wild-type larvae. G, Concomitant expression of khcana_58A rescues the larval crawling phenotype.
Fast and efficient screening for locomotion defects
The island assay allows high-throughput screening for different locomotion phenotypes. Flies expressing RNAi in glial cells were selected 2 weeks after egg-laying. Subsequently, 25 flies of either sex were kept for one additional week on standard food at 25°C before they were released on the testing area consisting of a 10 cm2 island surrounded by soap water (Fig. 1B). Wild-type flies placed on the island flew away within 2 s. Of the 4377 lines tested in the island assay, only 124 lines (2.3% of the total number screened) showed alterations in their locomotion pattern (data available upon request). The time it takes to clear the platform was determined (Fig. 1C).
Three distinct abnormal phenotypes were observed. The most frequent phenotypic class comprises genes whose silencing results in jumping but otherwise flightless flies (e.g., CG16771 encoding a predicted transmembrane alkaline phosphatase). The time to clear the island is only slightly longer compared with wild type, but flies simply bounced off into the surrounding water. A second class comprises genes whose knockdown results in generally lethargic flies and the testing platform is not cleared (e.g., CG2275, which encodes the transcription factor DJun).
Finally, glial-specific knockdown of only two genes resulted in similar, highly penetrant spastic locomotion phenotypes: khc and β3-tub (Movies 1, 2). The time window adult flies need to leave the platforms extends dramatically (Fig. 1C). To better quantify locomotion patterns we used a larval crawling assay. The migratory tracks of third instar wandering larvae were documented (>100 animals for each genotype) and analyzed using Image Pro plus (Fig. 1D,E). When we compared wild-type larvae with larvae in which khc function was downregulated in all glial cells, MDS analysis (see Materials and Methods) demonstrated a clear deviation of the locomotion pattern (Fig. 1F,G). Thus, khc is required in glial cells to assure proper locomotion.
Movie 1.
Downregulation of khc in glial cells induces a spastic locomotion defect. Escaper flies expressing khcdsRNA in glial cells remain on the island platform and are unable to fly.
Movie 2.
Glial-specific suppression of β3-tub leads to similar locomotion defects as the knockdown of khc. Specific downregulation of β3-tub expression in glial cells leads to a spastic locomotion defect.
Specificity of the khc RNAi phenotype
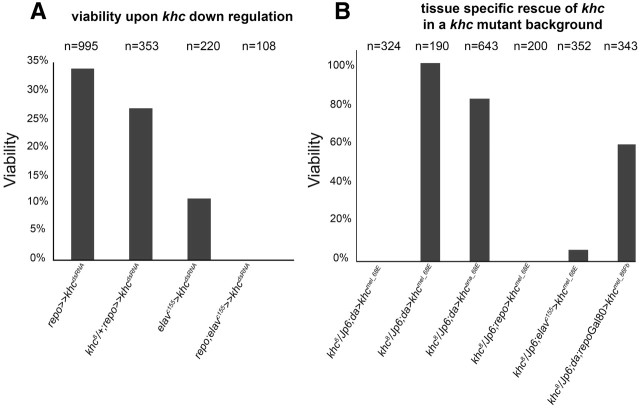
An RNAi-induced phenotype can be due to silencing of the respective gene or it can be due to off-target effects (Birmingham et al., 2006; Kulkarni et al., 2006; Dietzl et al., 2007). To test between these possibilities we silenced khc in a heterozygous mutant khc8 background. Under these conditions, we indeed observed an enhanced severity of the phenotype indicating that the khc knockdown by RNAi is not complete and supporting the notion that we identified a glial-specific khc phenotype (Fig. 2A). Silencing of khc in neurons using the elavGal4 driver significantly reduces viability, however, only concomitant silencing of khc in both, neurons and glial cells, causes lethality (Fig. 2A).
Figure 2.
Khc is required in glial cells. Downregulation of khc in specific tissues (A) or reexpression of khc in distinct cell type in khc mutant background (B). The expected number of rescued or silenced flies was compared with the number of actually eclosed non-balancer flies. A, khc downregulation in glial cells (using repo4.3Gal4;repoGal4) in wild-type and heterozygous khc8 mutant background. Note that heterozygous mutants show a decreased viability rate. No viable flies were obtained upon simultaneous suppression of khc in neurons and glial cells. B, Ubiquitous expression of khc in a khc6/Df(2R)Jp6 mutant background using daGal4 and khcmel_68E did not rescue lethality. In contrast, ubiquitous reexpression of khcmel_68E or khcana_58A in the khc8/Df(2R)Jp6 mutant rescued viability. Exclusive reexpression of khcmel_68E in glial cells (using repoGal4) did not rescue to viability. In contrast reexpression of khc in neurons (using elavc155Gal4) lead to a slight rescuing effect. However, reexpression in all cells but glia using a combination of daGal4 and repoGal80, results in a viability rate of 59%.
In the next step we verified the khc RNAi-induced phenotypes using an interspecies rescue approach (Kondo et al., 2009; Langer et al., 2010). khc is evolutionary very well conserved. Thus, we used a khc full-length cDNA from Drosophila ananassae, which over the entire coding region shares ∼85% DNA sequence identity with the D. melanogaster khc sequence (data not shown). Within the mRNA fragment, that is targeted by the used RNAi construct, two stretches of perfect sequence homology (21 and 17 nt long) were changed by site-directed mutagenesis to improve rescue abilities. A corresponding UAS-full-length construct was subsequently inserted into three different locations on the second and the third chromosome using phiC31-mediated transgenesis (Bischof et al., 2007).
Upon combining RNAi and rescue transgenes in glial cells, the locomotion defects were rescued (Figs. 1C,F,G; 2, data not shown). To exclude that the rescue is due to the presence of an additional UAS-element, which binds and thus titrates Gal4, we tested the effect of a control UAS-construct (UAS-Or83bdsRNA) in the khcdsRNA background. This did not diminish the neuronal phenotype, which further confirms the specificity of the RNAi construct (Fig. 1).
Khc is not exclusively required in neurons
In the past, different khc alleles have been analyzed (Saxton et al., 1991). Homozygous mutant animals display paralyzed abdominal segments and a so-called tail-up phenotype, which had been attributed to neuronal transport defects (Saxton et al., 1991). In consequence, khc mutants die during second and late third larval stages.
To further confirm the role of Khc in glial cells, we performed tissue-specific rescue experiments in a khc mutant background. We generated several UAS-based full-length D. melanogaster khc constructs (Khcmel), which we expressed in two different khc mutant alleles (khc8/Df(2R)Jp6 and khc6/Df(2R)Jp6). An untagged and an N-terminal GFP-tagged Khc were both equally active, whereas a C-terminal GFP-tagged Khc construct is not able to rescue the khc mutant phenotype. Unexpectedly, ubiquitous expression of Khcmel (untagged and N-terminal GFP-tagged) using daGal4 fully rescued the lethal phenotype associated with the null allele khc8 but was not able to rescue the khc6 phenotype. This indicates that khc6 may be an antimorphic mutation (Fig. 2B). We therefore continued our work with the null allele khc8.
To further determine the role of Khc in glial cells, we used two different rescue approaches in the khc8 mutant background. First, we restored Khc specifically in neurons using elavGal4. Second, we generated animals, which expressed khc in all cells except the glia. For this we used the ubiquitous activator strain daGal4 in combination with a glial-specific expressed Gal4 repressor Gal80 (repoGal80). These expression paradigms significantly shifted viability rates toward 6% and 59%, respectively (Fig. 2B). In summary, these data clearly demonstrate that Khc is required in glial cells.
Kinesin heavy chain is expressed in peripheral glial cells
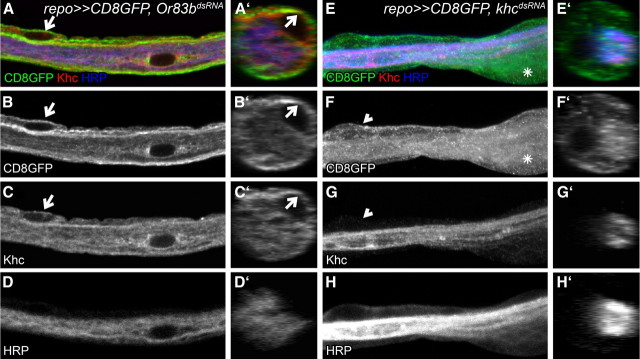
Recently, members of the KIF superfamily have been associated with functions in Schwann cells and oligodendrocytes during development (Bolis et al., 2009; Lyons et al., 2009). In Drosophila, the role of khc has been mostly studied in the context of fast axonal transport and no glial function has been reported so far (Saxton et al., 1991; Hurd and Saxton, 1996; Hurd et al., 1996). Using Khc-specific antibodies, we first tested whether Khc is expressed by glial cells. Within peripheral nerves, clear distinctions between neuronal and glial cell types can be made. Analysis of cross sections of peripheral nerves revealed an even distribution of Khc in neurons and glial cells (Fig. 3A,C). The glial expression is lost following glial-specific knockdown, demonstrating the specificity of the RNAi construct (Fig. 3E,G). Additionally, we noted a changed morphology of peripheral nerves, which upon panglial knockdown of khc often appear swollen (compare Fig. 3B,F asterisk). Of 64 nerves, three nerves showed four bulges, 10 nerves showed three bulges, 21 nerves showed two bulges, 21 nerves showed one bulge and nine nerves showed no bulging. In general the number of bulges correlates with the length of the nerve. These phenotypes are specifically associated with loss of khc function in glial cells and could be completely rescued by panglial expression of an RNAi insensitive khcana construct.
Figure 3.
Khc is expressed by glial cells and specifically silenced by glial expression of khcdsRNA. A–H, Confocal sections of peripheral nerves imaged with the same settings. Orthogonal sections shown in A′–H′ were generated using the Zen Software (Zeiss). Neuronal membranes are stained in blue using an anti-HRP-antibody (D, D′, H, H′), Khc is shown in red (C, C′, G, G′) and glial membranes are marked in green (repo≫CD8GFP; B, B′, F, F′). A–D′, Control nerves with the genotype repo≫CD8GFP, Or83bdsRNA. Note that Khc is expressed by neurons and glial cells (arrows). E–H′, Glial-specific expression of khcdsRNA (repo≫CD8GFP, khcdsRNA). Khc expression is specifically downregulated in glial cells (arrowhead). Note the swellings of peripheral glial induced upon khc knockdown (asterisk).
khc is required in the subperineurial glia for normal neuronal function
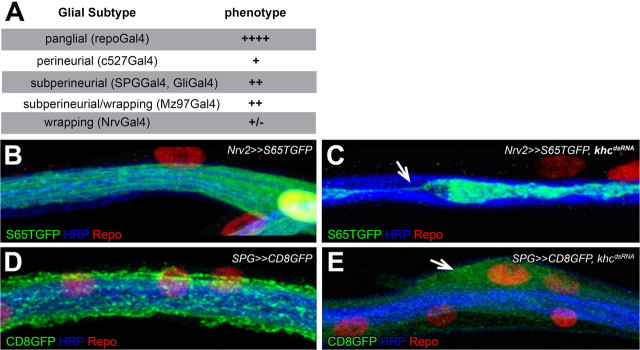
Within the Drosophila nervous system five different glial cell types can be described. The surface glial cells, perineurial and subperineurial glia, establish the blood–brain barrier, the wrapping glial cells insulate the individual axons to allow electrical conductance, the astrocyte-like glia likely modulates synaptic functions and the cortex glia participates in the compartmentalization of the nervous system (Pereanu et al., 2005; Freeman and Doherty, 2006; Stork et al., 2008; Doherty et al., 2009). To determine whether the spastic locomotion can be associated to a specific glial class, we analyzed adult locomotion following subtype-specific suppression of Khc. Only silencing of khc in the subperineurial glial cells resulted in locomotion defects but none of the flies showed the spastic phenotype (Fig. 4A). Half of the flies managed to directly fly away, similar to wild-type flies, whereas the other half remained on the island displaying a rather sluggish behavior.
Figure 4.
Khc is required in the subperineurial glia. A, We used the island assay to identify in which glial subtype khc function is required for proper locomotion. khc was downregulated in all glia (repo4.3Gal4, repoGal4), in the perineurial glia (c527Gal4), in the subperineurial glia (SPGGal4 or GliGal4), in subsets of wrapping and subperineurial glia (Mz97Gal4) and exclusively in wrapping glia (Nrv2Gal4, Nrv2Gal4). khc is needed in the subperineurial glia to allow wild-type behavior. B–E, Peripheral nerves of Drosophila third instar larvae. Repo staining marks glial cell nuclei (red), neuronal membranes are shown in blue. Glial membranes are labeled following glial-specific expression of GFP. The different genotypes are indicated. Loss of khc leads to a significant reduction of membrane protrusions in the wrapping glia (C, arrow) and to nerve bulges in subperineurial glia (E, arrow).
To address how khc affects glial differentiation, we analyzed glial morphology in the PNS of third instar larvae. Upon downregulation of khc in the wrapping glia, glial differentiation is severely affected. Wrapping glial cells do not engulf the entire length of the axonal fascicle but no nerve bulges were found (Fig. 4B,C; in 13 larvae with 156 nerves scored we found only three cells that appeared with normal wrapping glial morphology). In contrast, we noted bulged nerves upon subperineurial khc down-regulation similar to what we had seen following a panglial knockdown of khc (Figs. 3, 4D,E). Thus, Khc appears to be important in all glial cells for proper differentiation, but only downregulation in all glial cells or in subperineurial glia could induce the locomotion defects in adult flies.
To directly determine the neuronal consequences of glial khc function we measured evoked responses following short stimulation of peripheral nerves in third instar larval preparations (Fig. 5). In a fillet preparation, the A6 abdominal nerve was taken up by a suction electrode after detaching all the nerves from the ventral nerve cord. We determined the evoked responses at muscle fiber number 6 in all preparations. Silencing of khc in all glial cells or only in the subperineurial glial cells (using repoGal4 or gliotactinGal4) resulted in dramatically reduced amplitudes of evoked EJPs following voltage stimulus (p < 0.001) (Fig. 5C–E). No such reduction in the neuronal response was noted following silencing of khc in the wrapping glia (Fig. 5F). No changes were observed in the miniature EJPs amplitudes in any of the genotype (data not shown) suggesting the normal spontaneous firing in all the cases and neurons were healthy. The neuronal evoked response defects observed with khc RNAi could be rescued by expression of the khcana_58A construct but not by adding an additional UAS-construct as control (UASOr83b, Fig. 5G,H). Concomitantly, the latency was also reduced following glial-specific silencing of khc function (Fig. 5B). Similar results were obtained in a preparation where A6 nerve was left intact and attached to the ventral nerve cord and the current was injected in the CNS. In this scenario the action potential had to travel approximately twice as long before reaching the neuromuscular junction (NMJ). The amplitude of the EJPs was unchanged but the latency is almost doubled (data not shown). In conclusion, khc is needed in the subperineurial glia to establish neuronal excitability.
Figure 5.
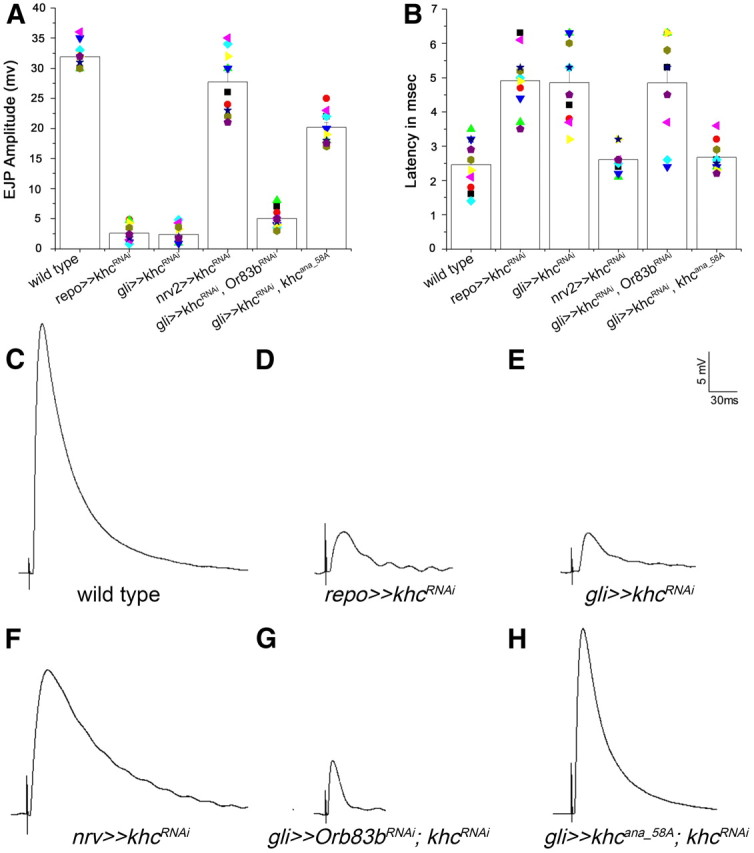
Glial khc knockdown affects neuronal activity. A, Comparison of evoked EJPs upon silencing khc in glial cells. Amplitudes of evoked EJPs after activating the A6 motor neuron at muscle 6 in response to a 0.3 ms voltage stimulus were determined. B, The latency of the muscle response is depicted. Glial reduction of khc results in delayed responses. C, Example of a wild-type response. D, E, Responses of silencing khc using dsRNA expression in all glial cells (repo≫khcRNAi) and specifically in the subperineurial glial layer (gli≫khcRNAi) were similar in amplitudes and duration to spontaneous EJPs. Control larvae showed much higher amplitudes (p < 0.001). Minis or spontaneous EJP responses were normal and similar in all the genotypes tested. F, No significance difference was observed in nrv2Gal4, UASkhdsRNA animals with control responses (p > 0.05). G, H, Coexpression of khcana_58A and khcdsRNA but not of Or83bdsRNA and khcdsRNA rescued the neuronal responses (p < 0.01). Pooled data are presented as means ± SE.
Mitochondrial defects in glia are not linked to khc-dependent locomotion deficits
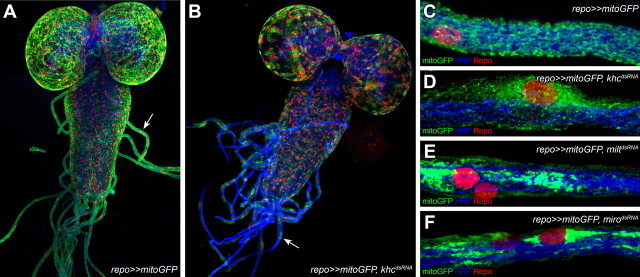
Kinesin is the major motor protein organizing anterograde transport along microtubules. Loss of khc leads to traffic jams of Khc-dependent cargoes in neurons causing axonal swellings (Hurd and Saxton, 1996). Similarly, we observed bulged peripheral glial cells upon glial-specific suppression of khc (Figs. 3, 4, 6).
Figure 6.
Distribution of glial mitochondria is Khc dependent. Confocal images of third instar larval peripheral nerves. Glial nuclei are red (Repo), neurons are blue (HRP). The distribution of glial mitochondria (marked with repo≫mitoGFP) has been analyzed in the following genotypes: repo≫mitoGFP (A, C), repo≫mitoGFP, khcdsRNA (B, D), repo≫mitoGFP, miltdsRNA (E), and repo≫mitoGFP, mirodsRNA (F). Note the similar misdistribution upon glial-specific knockdown of khc and milt and miro, which encode adapter proteins linking Khc to its cargo.
One of the cargoes transported by Khc are mitochondria (Pilling et al., 2006). In wild-type glial cells, mitochondria marked by UAS-mitoGFP form an intricate but immobile network extending evenly throughout the entire cell (Fig. 6A,C). Following khc knockdown, mitochondria are missing over large stretches of the glia and accumulate in the perinuclear domain. Additionally, the formation of the mitochondrial mesh is completely disrupted (Fig. 6B,D). To analyze whether the remaining mitochondria are still functional, we used a JC-1 assay, which allows determination of the mitochondrial membrane potential (Cossarizza et al., 1993). No differences between the wild-type and the khc knockdown flies were observed, indicating that the energy balance is not grossly affected. To further test the relevance of the mitochondrial misdistribution phenotype for the locomotion phenotype, we silenced miro and milton, which encode well established adaptor proteins linking Khc to mitochondria (Stowers et al., 2002; Cox and Spradling, 2006; Glater et al., 2006). In both cases, we found a similar disruption in the mitochondrial distribution but did not observe any locomotion defects (Fig. 6E,F; >10 larvae with all nerves showing similar mitochondrial defects). Similarly, related morphological mitochondrial phenotypes but no locomotion defects were induced upon glial knockdown of Marf and pink1, genes which are involved in mitochondrial fusion or fission (Deng et al., 2008, data not shown). Thus, we assume that impairments of the glial mitochondrial mesh have no obvious relevance for survival or locomotion.
Proper locomotion requires Khc-dependent vesicle transport in glia
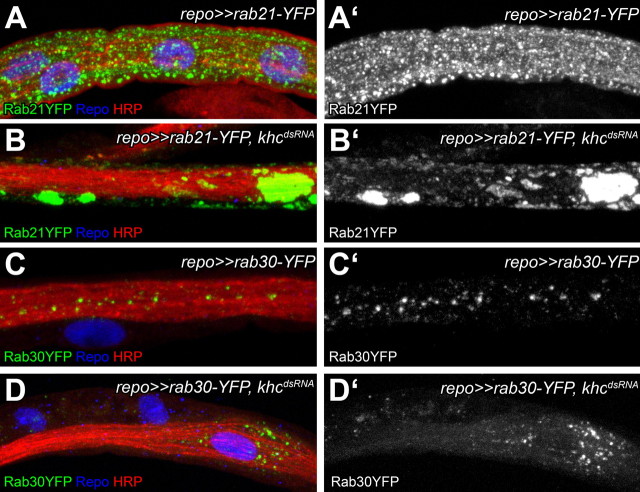
In addition to mitochondria, Khc is known to transport various vesicles along axons. Vesicle targeting and definition is largely controlled by small GTPases of the Rab superfamily. We tested different Rab proteins for a Khc-dependent motility in wild-type glial cells. From a panel of 16 different GFP-labeled Rab proteins (Zhang et al., 2007) we identified several Rab proteins as motile in glial cells in a life imaging approach (Movies 3, 4). Subsequently, we analyzed whether the motility of Rab-positive vesicles depends on Khc activity. Silencing of khc in glial cells resulted in the complete loss of motility and to a highly abnormal distribution of Rab21-YFP and Rab30-YFP (Fig. 7A–D, n > 70 nerves; Movies 5, 6; n = 8). Moreover, panglial-specific silencing of Rab21 also resulted in locomotion deficits. This demonstrates that Khc-dependent transport of specific Rab-positive vesicles is needed for proper glial function and, in consequence, for neuronal network activity.
Figure 7.
Rab21-YFP and Rab30YFP-positive vesicles are maldistributed upon glial-specific khc knockdown. Peripheral nerves of dissected Drosophila third instar larvae. Neuronal membranes are in red (anti-HRP staining), glial nuclei are shown in blue (anti-Repo staining). Rab21-YFP-positive vesicles (green, A, B) and Rab30-YFP-positive vesicles (C, D). A, C, Rab21-YFP and Rab30-YFP-positive vesicles are evenly distributed over the entire peripheral nerve of larvae. B, D, This pattern is highly disrupted upon glial-specific khc suppression.
Movie 3.
Rab21-YFP-positive vesicles are motile in wrapping glial cells. Third instar larvae expressing Rab21-YFP in wrapping glia (nrv2Gal4, UAS-Rab21-YFP) were imaged for 4 min using the LSM 5 Live Duo microscope. Every 3 s a new image stack was recorded.
Movie 4.
Rab30-YFP-positive vesicles are motile in all glial cells. Third instar larvae expressing Rab30-YFP in all glial cells (repo4.3Gal4, repoGal4, UAS-Rab30-YFP) were imaged for 17 min using the LSM 5 Live Duo microscope. Every 3 s a new image stack was recorded.
Movie 5.
Rab21-YFP motility is significantly reduced upon khc downregulation in wrapping glial cells. Third instar larvae expressing Rab21-YFP in wrapping glial cells in the background of khcdsRNA (nrv2Gal4, UAS-Rab21-YFP, USA-khcdsRNA) were imaged for 11 min using the LSM 5 Live Duo microscope. Every 3 s a new image stack was recorded.
Movie 6.
Rab30-YFP motility is significantly reduced upon khc downregulation in all glial cells. Third instar larvae expressing Rab30-YFP in all glial cells in the background of khcdsRNA (repo4.3Gal4, repoGal4, UAS-Rab30-YFP USA-khcdsRNA) were imaged for 3 min using the LSM 5 Live Duo microscope. Every 3 s a new image stack was recorded.
The distribution of Neurexin IV depends on Khc
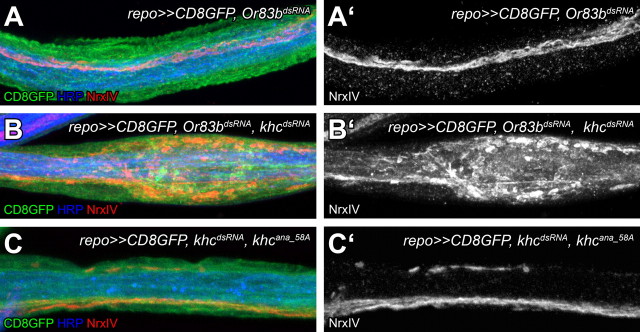
Suppression of khc either in all glia or only in subperineurial glia induces locomotion defects and leads to swollen peripheral nerves. In addition to impaired glial transport, a defective blood–brain barrier might be responsible for the bulged peripheral nerve phenotype. NrxIV is a component of pSJs that are necessary for the establishment and maintenance of the blood–brain barrier (Stork et al., 2008). In wild-type larvae, NrxIV is expressed as a thin line along peripheral nerves at the position of the pSJ (Fig. 8A). Upon khc knockdown in glial cells, a mislocalization of NrxIV can be observed at the bulged regions of peripheral nerves in 84% of the nerve bulges analyzed (Fig. 8B; n = 44). This misdistribution was restored upon the simultaneous expression of the Khcana_58A interspecies rescue construct (Fig. 8C). Interestingly, we could also observe a slight mislocalization of NrxIV upon glial knockdown of rab30. These findings are consistent with the notion that Rab30 primes NrxIV-containing vesicles for Khc-dependent transport to the forming septate junctions.
Figure 8.
NrxIV is abnormally localized in bulged regions of the peripheral nerves following khc suppression. Confocal images of dissected peripheral nerves of Drosophila third instar larvae are shown. Neurons are shown in blue (anti-HRP staining), the pleated septate junctions are shown in red (anti-NrxIV staining), and glial membranes are marked in green (repo≫CD8GFP). Peripheral nerves with the following genotypes were analyzed: repo≫CD8GFP, Or83bdsRNA (A, A′), repo≫CD8GFP, Or83bdsRNA khcdsRNA (B, B′), and repo≫CD8GFP, khcana_58A, khcdsRNA (C, C′). The stripe-like pattern along the nerve of NrxIV, which is highly disrupted following khc suppression in glial cells, can be restored upon the expression of the interspecies rescue construct.
Discussion
It is well established that the modulation of neuronal functionality depends on the ability of glial cells to provide metabolic support, regulate neurotransmitter homeostasis and influence the electrical conductance. To decipher these processes we have performed a large-scale RNAi screen and silenced ∼5000 genes specifically in glial cells. Here, we present an unexpected glial function of Drosophila khc in regulating neuronal activity. The specificity of the phenotype was ascertained by interspecies rescue studies and cell type-specific rescue of the khc mutant. Furthermore, the fact that glial-specific knockdown of β3-tubulin results in identical neuronal phenotypes demonstrates the functional relevance of Khc-dependent transport along microtubules.
Drosophila Khc was identified more than 20 years ago (Saxton et al., 1988, 1991; Yang et al., 1988). In khc mutant larvae, vesicles, lysosomes, and mitochondria accumulate in axons. This results in swollen axons and eventually in larval paralysis (Saxton et al., 1991; Gho et al., 1992; Hurd and Saxton, 1996). Here we have generated a number of UAS-driven khc constructs to examine cell type-specific requirements of Khc. We used two different khc alleles, khc6 and khc8 (Saxton et al., 1991), in trans to a genomic deficiency to eliminate background effects. Surprisingly, ubiquitous expression of Khc is not able to rescue the hypomorphic allele khc6. In contrast the null mutation khc8 can be completely rescued by Khc expression. khc6 contains an alteration in coil 2 of the stalk domain, a region implemented to be important for Kinesin light chain and cargo linkage (Pilling et al., 2006). Thus, khc6 may induce some antimorphic functions. But the introduction of a genomic rescue construct of khc (P-khc+) has been described as being able to rescue khc6-associated lethality, which points to very high levels of endogenous khc expression (Flybase; Saxton et al., 1991).
Using a null mutant background (khc8/Df(2R)Jp6), we were able to perform cell type-specific rescue experiments. Following expression of khc in neurons using elavGal4 only 6% of the expected flies survived. These flies were very sluggish and died within 6 d. To analyze glial-specific requirements of Khc, we compared the ubiquitous Gal4-dependent rescue with and without a glial cell-specific Gal4 repressor (repoGal80). We noted a significant difference, as the repressor reduced viability down to ∼60%, which further supports the crucial role of Khc in glial cells.
Glial-specific knockdown of khc revealed an adult phenotype, which resembled a hyperexcitation phenotype. khc knockdown flies have the ability to walk. However, to initiate flight, flies need to jump (Card and Dickinson, 2008a,b). Upon khc knockdown, flies fail to perform this jump but rather directly start with a wing beat. Such flies then turn on their backs or spin over the floor, which then results in the characteristic popcorn-like movements (Movie 1). Thus, khc knockdown delays or prevents the jumping response.
In larvae, glial knockdown of khc results in reduced and delayed evoked responses at the muscle. Concomitantly, we noted swellings of the peripheral nerves. In the majority of the swellings (and only in the swellings) we found a mislocalization of the septate junction protein Neurexin IV. This indicates that knockdown of khc results in an opening of the blood–brain barrier. In turn, this results in influx of potassium into the nervous system. Such influx is known to induce changes in axonal physiology and conductance velocity (Baumgartner et al., 1996; Ueda et al., 2008). Recently the role of focal adhesion kinase 56 (fak56) in Drosophila nerves was reported (Ueda et al., 2008). Focal adhesion kinase acts downstream of integrin receptors, which are also known to be expressed by Drosophila glial cells (Murakami et al., 2007; Xie and Auld, 2011). Interestingly, loss of fak56 function in the subperineurial glial cells (which constitute the blood–brain barrier) lowers axonal conductance in larvae and also renders the larvae sluggish pointing toward the notion that adhesion is required for a full establishment of the blood–brain barrier (Ueda et al., 2008).
The observed suppression of EJP could in principle also be due to presynaptic defects or defects in the glutamate receptors expressed by the muscle. However, in contrast to vertebrates, where terminal Schwann cells cover the NMJ, the NMJ of Drosophila larvae grows into the muscle cell, which forms a so called subsynaptic reticulum around the NMJ. In addition, only few (subperineurial) glial processes invade parts of the NMJ in a highly dynamic fashion and where they participate in the removal of presynaptic debris (Jia et al., 1993; Sepp et al., 2000; Fuentes-Medel et al., 2009). To further discriminate between a possible synaptic or an axonal defect we compared the latency of EJP suppression by injecting the current in the nerve 200–300 μm distant from the NMJ or in the ventral nerve cord 1000 μm distant from the NMJ. In this setting the latency doubled suggesting that axonal conductance is impaired upon glial khc knockdown. Therefore, we anticipate that the suppression of the EJPs caused by glial reduction of khc function is unlikely due to synaptic defects.
Moreover, if khc would modulate synaptic activities, it would be acting in adult flies. However, when we silenced khc function only during adult stages using the TARGET system (McGuire et al., 2003, 2004), we noted no abnormal locomotion, which demonstrated that Khc is needed during development and not during adult stages.
Several Rab proteins were identified as highly motile in glia and their motility was significantly reduced upon khc suppression. Rab30 has been identified as a downstream target of JNK signaling, which can control kinesin cargo linkage (Horiuchi et al., 2007; Thomas et al., 2009). In addition, JNK-interacting proteins (JIPs) link Kinesin with membrane vesicles and are involved in Khc activation and cargo release (Verhey et al., 2001; Taru et al., 2002; Horiuchi et al., 2005; Blasius et al., 2007; Koushika, 2008). For vertebrate Rab21 a possible role in targeted Integrin trafficking from and to the cleavage furrow during cell division and an involvement in the endocytic pathway have been described (Simpson et al., 2004; Simpson and Jones, 2005; Pellinen et al., 2006, 2008; Burgo et al., 2009).
The swollen nerves are likely due to a defective blood–brain barrier. The integrity of septate junctions for normal locomotion has also been suggested by the analysis of mutants affecting several septate junction components (Strigini et al., 2006). At the end of embryogenesis, lachesin, neurexin IV, and gliotactin mutant embryos are hyperactive before becoming immobile. Possibly, the influx of potassium and sodium ions into the nervous system affects conductivity. Interestingly, panglial downregulation of the Na-K-Cl cotransporter Ncc69 or the serine/threonine kinase Fray (Leiserson et al., 2000, 2011) also results in locomotion defects characterized by jumping flies (data not shown). Fray and Ncc69 act in the subperineurial glia but no affect on neuronal conductance had been determined in peripheral nerves (Leiserson et al., 2011; Leiserson and Keshishian, 2011). Thus, the disruption of septate junctions as seen following knockdown of khc likely affects ion homeostasis more dramatically.
Drosophila NrxIV is highly mislocalized upon glial khc suppression. Other examples of a role for kinesin in cell polarization have been described. KIF5B and KIF5C are involved in polarized transport of certain apically distributed proteins in cultured mammalian epithelial cells (Jaulin et al., 2007; Astanina and Jacob, 2010). Additionally, KIF13B has been shown to directly transport proteins like Dlg1 to myelinating sites in Schwann cells (Bolis et al., 2009). Thus, a similar function might be performed by Drosophila Khc in subperineurial glial cells and Khc might be involved in the directed transport of transmembrane proteins to septate junctions.
Footnotes
This work was supported through grants of the Deutsche Forschungsgemeinschaft to C.K. (SFB629) and the European Community through FP7/2007–2013 under Grant Agreement HEALTH-F2–2008-201535. We thank B. Saxton, D. Buttgereit, R. Renkawitz-Pohl, and T. Schwarz for flies and antibodies. We thank R. Dierkes and C. Kühn for help during initial screening, N. Otto for generating the UAS-khcana_58A construct and the generation of GFP-tagged UAS-Khc construct, and N. Otto and G. Grün for help in generating the UAS-Khc flies, and R. Baines, E. Knoche, and S. Sigrist for much help during the initial electrophysiology experiments. Antibodies were provided by Developmental Studies Hybridoma Bank (Iowa City, IA) and Cytoskeleton, Inc. RNAi flies were obtained from National Institute of Genetics Fly Stock Center, Kyoto and the Vienna Drosophila RNAi Center. All general fly stocks were provided by the Bloomington Stock Center. cDNA of D. ananassae was received through the Drosophila Genomics Resource Center. We are thankful to FlyBase for providing information. We are grateful to H. Aberle, B. Goellner, A. Püschel, and S. Sigrist for discussion and comments on the manuscript and all members of the Klämbt laboratory for support throughout the project. S.T. acknowledges a predoctoral fellowship of the Boehringer Ingelheim Foundation.
References
- Astanina K, Jacob R. KIF5C, a kinesin motor involved in apical trafficking of MDCK cells. Cell Mol Life Sci. 2010;67:1331–1342. doi: 10.1007/s00018-009-0253-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barres BA. The mystery and magic of glia: a perspective on their roles in health and disease. Neuron. 2008;60:430–440. doi: 10.1016/j.neuron.2008.10.013. [DOI] [PubMed] [Google Scholar]
- Baumgartner S, Littleton JT, Broadie K, Bhat MA, Harbecke R, Lengyel JA, Chiquet-Ehrismann R, Prokop A, Bellen HJ. A Drosophila neurexin is required for septate junction and blood-nerve barrier formation and function. Cell. 1996;87:1059–1068. doi: 10.1016/s0092-8674(00)81800-0. [DOI] [PubMed] [Google Scholar]
- Birmingham A, Anderson EM, Reynolds A, Ilsley-Tyree D, Leake D, Fedorov Y, Baskerville S, Maksimova E, Robinson K, Karpilow J, Marshall WS, Khvorova A. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat Methods. 2006;3:199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci U S A. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasius TL, Cai D, Jih GT, Toret CP, Verhey KJ. Two binding partners cooperate to activate the molecular motor Kinesin-1. J Cell Biol. 2007;176:11–17. doi: 10.1083/jcb.200605099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolis A, Coviello S, Visigalli I, Taveggia C, Bachi A, Chishti AH, Hanada T, Quattrini A, Previtali SC, Biffi A, Bolino A. Dlg1, Sec8, and Mtmr2 regulate membrane homeostasis in Schwann cell myelination. J Neurosci. 2009;29:8858–8870. doi: 10.1523/JNEUROSCI.1423-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgo A, Sotirakis E, Simmler MC, Verraes A, Chamot C, Simpson JC, Lanzetti L, Proux-Gillardeaux V, Galli T. Role of Varp, a Rab21 exchange factor and TI-VAMP/VAMP7 partner, in neurite growth. EMBO Rep. 2009;10:1117–1124. doi: 10.1038/embor.2009.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Card G, Dickinson M. Performance trade-offs in the flight initiation of Drosophila. J Exp Biol. 2008a;211:341–353. doi: 10.1242/jeb.012682. [DOI] [PubMed] [Google Scholar]
- Card G, Dickinson MH. Visually mediated motor planning in the escape response of Drosophila. Curr Biol. 2008b;18:1300–1307. doi: 10.1016/j.cub.2008.07.094. [DOI] [PubMed] [Google Scholar]
- Cossarizza A, Baccarani-Contri M, Kalashnikova G, Franceschi C. A new method for the cytofluorimetric analysis of mitochondrial membrane potential using the J-aggregate forming lipophilic cation 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolcarbocyanine iodide (JC-1) Biochem Biophys Res Commun. 1993;197:40–45. doi: 10.1006/bbrc.1993.2438. [DOI] [PubMed] [Google Scholar]
- Cox RT, Spradling AC. Milton controls the early acquisition of mitochondria by Drosophila oocytes. Development. 2006;133:3371–3377. doi: 10.1242/dev.02514. [DOI] [PubMed] [Google Scholar]
- Dathe V, Prols F, Brand-Saberi B. Expression of kinesin kif5c during chick development. Anat Embryol (Berl) 2004;207:475–480. doi: 10.1007/s00429-003-0370-1. [DOI] [PubMed] [Google Scholar]
- Deng H, Dodson MW, Huang H, Guo M. The Parkinson's disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc Natl Acad Sci U S A. 2008;105:14503–14508. doi: 10.1073/pnas.0803998105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Doherty J, Logan MA, Taşdemir OE, Freeman MR. Ensheathing glia function as phagocytes in the adult Drosophila brain. J Neurosci. 2009;29:4768–4781. doi: 10.1523/JNEUROSCI.5951-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda RO, Hart PE, Stork DG. Pattern classification. Ed 2. New York: Wiley; 2001. [Google Scholar]
- Edwards TN, Meinertzhagen IA. The functional organisation of glia in the adult brain of Drosophila and other insects. Prog Neurobiol. 2010;90:471–497. doi: 10.1016/j.pneurobio.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Ueda A, Wu CF. A modified minimal hemolymph-like solution, HL3.1, for physiological recordings at the neuromuscular junctions of normal and mutant Drosophila larvae. J Neurogenet. 2004;18:377–402. doi: 10.1080/01677060490894522. [DOI] [PubMed] [Google Scholar]
- Freeman MR, Doherty J. Glial cell biology in Drosophila and vertebrates. Trends Neurosci. 2006;29:82–90. doi: 10.1016/j.tins.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Fuentes-Medel Y, Logan MA, Ashley J, Ataman B, Budnik V, Freeman MR. Glia and muscle sculpt neuromuscular arbors by engulfing destabilized synaptic boutons and shed presynaptic debris. PLoS Biol. 2009;7:e1000184. doi: 10.1371/journal.pbio.1000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gho M, McDonald K, Ganetzky B, Saxton WM. Effects of kinesin mutations on neuronal functions. Science. 1992;258:313–316. doi: 10.1126/science.1384131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glater EE, Megeath LJ, Stowers RS, Schwarz TL. Axonal transport of mitochondria requires milton to recruit kinesin heavy chain and is light chain independent. J Cell Biol. 2006;173:545–557. doi: 10.1083/jcb.200601067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Niwa S, Tanaka Y. Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron. 2010;68:610–638. doi: 10.1016/j.neuron.2010.09.039. [DOI] [PubMed] [Google Scholar]
- Horiuchi D, Barkus RV, Pilling AD, Gassman A, Saxton WM. APLIP1, a kinesin binding JIP-1/JNK scaffold protein, influences the axonal transport of both vesicles and mitochondria in Drosophila. Curr Biol. 2005;15:2137–2141. doi: 10.1016/j.cub.2005.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi D, Collins CA, Bhat P, Barkus RV, Diantonio A, Saxton WM. Control of a kinesin-cargo linkage mechanism by JNK pathway kinases. Curr Biol. 2007;17:1313–1317. doi: 10.1016/j.cub.2007.06.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd DD, Saxton WM. Kinesin mutations cause motor neuron disease phenotypes by disrupting fast axonal transport in Drosophila. Genetics. 1996;144:1075–1085. doi: 10.1093/genetics/144.3.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd DD, Stern M, Saxton WM. Mutation of the axonal transport motor kinesin enhances paralytic and suppresses Shaker in Drosophila. Genetics. 1996;142:195–204. doi: 10.1093/genetics/142.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaulin F, Xue X, Rodriguez-Boulan E, Kreitzer G. Polarization-dependent selective transport to the apical membrane by KIF5B in MDCK cells. Dev Cell. 2007;13:511–522. doi: 10.1016/j.devcel.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia XX, Gorczyca M, Budnik V. Ultrastructure of neuromuscular junctions in Drosophila: comparison of wild type and mutants with increased excitability. J Neurobiol. 1993;24:1025–1044. doi: 10.1002/neu.480240804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Okada Y, Tanaka Y, Harada A, Terada S, Hirokawa N. KIF5C, a novel neuronal kinesin enriched in motor neurons. J Neurosci. 2000;20:6374–6384. doi: 10.1523/JNEUROSCI.20-17-06374.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S, Booker M, Perrimon N. Cross-species RNAi rescue platform in Drosophila melanogaster. Genetics. 2009;183:1165–1173. doi: 10.1534/genetics.109.106567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koushika SP. “JIP”ing along the axon: the complex roles of JIPs in axonal transport. Bioessays. 2008;30:10–14. doi: 10.1002/bies.20695. [DOI] [PubMed] [Google Scholar]
- Kulkarni MM, Booker M, Silver SJ, Friedman A, Hong P, Perrimon N, Mathey-Prevot B. Evidence of off-target effects associated with long dsRNAs in Drosophila melanogaster cell-based assays. Nat Methods. 2006;3:833–838. doi: 10.1038/nmeth935. [DOI] [PubMed] [Google Scholar]
- Langer CC, Ejsmont RK, Schönbauer C, Schnorrer F, Tomancak P. In vivo RNAi rescue in Drosophila melanogaster with genomic transgenes from Drosophila pseudoobscura. PLoS One. 2010;5:e8928. doi: 10.1371/journal.pone.0008928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BP, Jones BW. Transcriptional regulation of the Drosophila glial gene repo. Mech Dev. 2005;122:849–862. doi: 10.1016/j.mod.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Leiserson WM, Keshishian H. Maintenance and regulation of extracellular volume and the ion environment in Drosophila larval nerves. Glia. 2011 doi: 10.1002/glia.21132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiserson WM, Harkins EW, Keshishian H. Fray, a Drosophila serine/threonine kinase homologous to mammalian PASK, is required for axonal ensheathment. Neuron. 2000;28:793–806. doi: 10.1016/s0896-6273(00)00154-9. [DOI] [PubMed] [Google Scholar]
- Leiserson WM, Forbush B, Keshishian H. Drosophila glia use a conserved cotransporter mechanism to regulate extracellular volume. Glia. 2011;59:320–332. doi: 10.1002/glia.21103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons DA, Naylor SG, Scholze A, Talbot WS. Kif1b is essential for mRNA localization in oligodendrocytes and development of myelinated axons. Nat Genet. 2009;41:854–858. doi: 10.1038/ng.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- McGuire SE, Mao Z, Davis RL. Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci STKE. 2004;2004:pl6. doi: 10.1126/stke.2202004pl6. [DOI] [PubMed] [Google Scholar]
- Murakami S, Umetsu D, Maeyama Y, Sato M, Yoshida S, Tabata T. Focal adhesion kinase controls morphogenesis of the Drosophila optic stalk. Development. 2007;134:1539–1548. doi: 10.1242/dev.001529. [DOI] [PubMed] [Google Scholar]
- Nave KA, Trapp BD. Axon-glial signaling and the glial support of axon function. Annu Rev Neurosci. 2008;31:535–561. doi: 10.1146/annurev.neuro.30.051606.094309. [DOI] [PubMed] [Google Scholar]
- Pellinen T, Arjonen A, Vuoriluoto K, Kallio K, Fransen JA, Ivaska J. Small GTPase Rab21 regulates cell adhesion and controls endosomal traffic of beta1-integrins. J Cell Biol. 2006;173:767–780. doi: 10.1083/jcb.200509019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellinen T, Tuomi S, Arjonen A, Wolf M, Edgren H, Meyer H, Grosse R, Kitzing T, Rantala JK, Kallioniemi O, Fässler R, Kallio M, Ivaska J. Integrin trafficking regulated by Rab21 is necessary for cytokinesis. Dev Cell. 2008;15:371–385. doi: 10.1016/j.devcel.2008.08.001. [DOI] [PubMed] [Google Scholar]
- Pereanu W, Shy D, Hartenstein V. Morphogenesis and proliferation of the larval brain glia in Drosophila. Dev Biol. 2005;283:191–203. doi: 10.1016/j.ydbio.2005.04.024. [DOI] [PubMed] [Google Scholar]
- Pilling AD, Horiuchi D, Lively CM, Saxton WM. Kinesin-1 and Dynein are the primary motors for fast transport of mitochondria in Drosophila motor axons. Mol Biol Cell. 2006;17:2057–2068. doi: 10.1091/mbc.E05-06-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice SE, Gelfand VI. Paradigm lost: milton connects kinesin heavy chain to miro on mitochondria. J Cell Biol. 2006;173:459–461. doi: 10.1083/jcb.200604071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues F, Schmidt I, Klämbt C. Comparing peripheral glial cell differentiation in Drosophila and vertebrates. Cell Mol Life Sci. 2011;68:55–69. doi: 10.1007/s00018-010-0512-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton WM, Porter ME, Cohn SA, Scholey JM, Raff EC, McIntosh JR. Drosophila kinesin: characterization of microtubule motility and ATPase. Proc Natl Acad Sci U S A. 1988;85:1109–1113. doi: 10.1073/pnas.85.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton WM, Hicks J, Goldstein LS, Raff EC. Kinesin heavy chain is essential for viability and neuromuscular functions in Drosophila, but mutants show no defects in mitosis. Cell. 1991;64:1093–1102. doi: 10.1016/0092-8674(91)90264-y. [DOI] [PubMed] [Google Scholar]
- Sepp KJ, Auld VJ. Conversion of lacZ enhancer trap lines to GAL4 lines using targeted transposition in Drosophila melanogaster. Genetics. 1999;151:1093–1101. doi: 10.1093/genetics/151.3.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepp KJ, Schulte J, Auld VJ. Developmental dynamics of peripheral glia in Drosophila melanogaster. Glia. 2000;30:122–133. doi: 10.1002/(sici)1098-1136(200004)30:2<122::aid-glia2>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Simons K, Zerial M. Rab proteins and the road maps for intracellular transport. Neuron. 1993;11:789–799. doi: 10.1016/0896-6273(93)90109-5. [DOI] [PubMed] [Google Scholar]
- Simpson JC, Jones AT. Early endocytic Rabs: functional prediction to functional characterization. Biochem Soc Symp. 2005;(72):99–108. doi: 10.1042/bss0720099. [DOI] [PubMed] [Google Scholar]
- Simpson JC, Griffiths G, Wessling-Resnick M, Fransen JA, Bennett H, Jones AT. A role for the small GTPase Rab21 in the early endocytic pathway. J Cell Sci. 2004;117:6297–6311. doi: 10.1242/jcs.01560. [DOI] [PubMed] [Google Scholar]
- Stork T, Engelen D, Krudewig A, Silies M, Bainton RJ, Klämbt C. Organization and function of the blood–brain barrier in Drosophila. J Neurosci. 2008;28:587–597. doi: 10.1523/JNEUROSCI.4367-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowers RS, Megeath LJ, Górska-Andrzejak J, Meinertzhagen IA, Schwarz TL. Axonal transport of mitochondria to synapses depends on milton, a novel Drosophila protein. Neuron. 2002;36:1063–1077. doi: 10.1016/s0896-6273(02)01094-2. [DOI] [PubMed] [Google Scholar]
- Strigini M, Cantera R, Morin X, Bastiani MJ, Bate M, Karagogeos D. The IgLON protein Lachesin is required for the blood-brain barrier in Drosophila. Mol Cell Neurosci. 2006;32:91–101. doi: 10.1016/j.mcn.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Taru H, Kirino Y, Suzuki T. Differential roles of JIP scaffold proteins in the modulation of amyloid precursor protein metabolism. J Biol Chem. 2002;277:27567–27574. doi: 10.1074/jbc.M203713200. [DOI] [PubMed] [Google Scholar]
- Thomas C, Rousset R, Noselli S. JNK signalling influences intracellular trafficking during Drosophila morphogenesis through regulation of the novel target gene Rab30. Dev Biol. 2009;331:250–260. doi: 10.1016/j.ydbio.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Ueda A, Grabbe C, Lee J, Palmer RH, Wu CF. Mutation of Drosophila focal adhesion kinase induces bang-sensitive behavior and disrupts glial function, axonal conduction and synaptic transmission. Eur J Neurosci. 2008;27:2860–2870. doi: 10.1111/j.1460-9568.2008.06252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhey KJ, Meyer D, Deehan R, Blenis J, Schnapp BJ, Rapoport TA, Margolis B. Cargo of kinesin identified as JIP scaffolding proteins and associated signaling molecules. J Cell Biol. 2001;152:959–970. doi: 10.1083/jcb.152.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Auld VJ. Integrins are necessary for the development and maintenance of the glial layers in the Drosophila peripheral nerve. Development. 2011;138:3813–3822. doi: 10.1242/dev.064816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong WC, Okano H, Patel NH, Blendy JA, Montell C. repo encodes a glial-specific homeo domain protein required in the Drosophila nervous system. Genes Dev. 1994;8:981–994. doi: 10.1101/gad.8.8.981. [DOI] [PubMed] [Google Scholar]
- Yang JT, Saxton WM, Goldstein LS. Isolation and characterization of the gene encoding the heavy chain of Drosophila kinesin. Proc Natl Acad Sci U S A. 1988;85:1864–1868. doi: 10.1073/pnas.85.6.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- Zhang J, Schulze KL, Hiesinger PR, Suyama K, Wang S, Fish M, Acar M, Hoskins RA, Bellen HJ, Scott MP. Thirty-one flavors of Drosophila rab proteins. Genetics. 2007;176:1307–1322. doi: 10.1534/genetics.106.066761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlokovic BV. The blood-brain barrier in health and chronic neurodegenerative disorders. Neuron. 2008;57:178–201. doi: 10.1016/j.neuron.2008.01.003. [DOI] [PubMed] [Google Scholar]