Abstract
Hindbrain dorsal interneurons that comprise the rhombic lip relay sensory information and coordinate motor outputs. The progenitor dA1 subgroup of interneurons, which is formed along the dorsal-most region of the caudal rhombic lip, gives rise to the cochlear and precerebellar nuclei. These centers project sensory inputs toward upper-brain regions. The fundamental role of dA1 interneurons in the assembly and function of these brainstem nuclei is well characterized. However, the precise en route axonal patterns and synaptic targets of dA1 interneurons are not clear as of yet. Novel genetic tools were used to label dA1 neurons and trace their axonal trajectories and synaptic connections at various stages of chick embryos. Using dA1-specific enhancers, two contralateral ascending axonal projection patterns were identified; one derived from rhombomeres 6–7 that elongated in the dorsal funiculus, while the other originated from rhombomeres 2–5 and extended in the lateral funiculus. Targets of dA1 axons were followed at later stages using PiggyBac-mediated DNA transposition. dA1 axons were found to project and form synapses in the auditory nuclei and cerebellum. Investigation of mechanisms that regulate the patterns of dA1 axons revealed a fundamental role of Lim-homeodomain (HD) proteins. Switch in the expression of the specific dA1 Lim-HD proteins Lhx2/9 into Lhx1, which is typically expressed in dB1 interneurons, modified dA1 axonal patterns to project along the routes of dB1 subgroup. Together, the results of this research provided new tools and knowledge to the assembly of trajectories and connectivity of hindbrain dA1 interneurons and of molecular mechanisms that control these patterns.
Introduction
The brainstem controls basic functions including respiration, consciousness, hearing, and motor coordination. Maintaining these functions is achieved by communicating between central/peripheral nervous systems via ascending/descending reticular networks. Brainstem centers, including cranial nerve, respiration, auditory, and precerebellar nuclei, process sensory/motor information to mediate these high-order activities (Altman and Bayer, 1997; Wang and Zoghbi, 2001; Ryugo and Parks, 2003; Milsom et al., 2004; Oertel and Young, 2004; Guthrie, 2007).
During early development, the brainstem precursor, the hindbrain, is subdivided along its anteroposterior (AP) axis into rhombomeres (r; Lumsden and Krumlauf, 1996). This segmentation assigns discrete neuronal groups that later generate the brainstem nuclei and neuronal networks in sync with their spatial coordinates (Moens and Prince, 2002; Lumsden, 2004). The link between hindbrain segmentation and the formation of brainstem circuitry is obscure.
The rhombic lip (RL) defines the dorsal hindbrain and is divided along the AP axis into upper (r1) and lower (r2–r8) domains (Hatten and Heintz, 1995; Alder et al., 1999). The upper RL generates cerebellar granule cells that form the external and internal granular layers of the cerebellum (EGL, IGL, respectively; Ben-Arie et al., 1997; Köster and Fraser, 2001; Machold and Fishell, 2005). The lower RL comprises cochlear nuclei (CN), through which auditory information is processed and relayed to the inferior colliculi and thalamus, and multiple precerebellar nuclei (PCN) that relay peripheral sensation to the cerebellum through mossy fiber (MF), or climbing fiber neurons (Altman and Bayer, 1980, 1987a,b,c,d; Cambronero and Puelles, 2000; Rodriguez and Dymecki, 2000; Díaz et al., 2003; Ryugo and Parks, 2003).
The basic helix-loop-helix (bHLH) transcription factor Atoh1 is expressed in the dorsal-most RL in a subset of interneuron progenitors, termed dA1 (Akazawa et al., 1995; Ben-Arie et al., 1997; Wang et al., 2005). Fate-mapping and mutant analyses revealed the fundamental role of Atoh1+/dA1 cells in the generation and organization of the EGL, CN, and PCN centers (Ben-Arie et al., 1997; Bermingham et al., 2001; Landsberg et al., 2005; Machold and Fishell, 2005; Wang et al., 2005; Farago et al., 2006; Fujiyama et al., 2009). However, knowledge is partial regarding the precise en route axonal patterns and targets of dA1 neurons.
This study aimed at elucidating the axonal projection patterns and targets of chick dA1 neurons. Using dA1-specific enhancer elements to drive neuronal, axonal, and synaptic reporter genes, and the PiggyBac DNA-mediated transposition system, dA1 axonal trajectories were decoded and found to project along two ascending contralateral trajectories, each derived from a distinct hindbrain level. Tracing these axons revealed target regions and synaptic connections in the medulla, cerebellum, and midbrain. To reveal molecular mechanisms that control dA1 axonal patterning, alteration of the Lim homeodomain (Lim-HD) code was performed in dA1 neurons by switching the dA1-Lim-HD proteins Lhx2/9 into the dB1-interneuron Lim-HD protein Lhx1. This change resulted in an axonal patterning switch from dA1 to dB1-like patterns. Together, the results of this study provide new tools and insights to the neuronal circuits of hindbrain dA1 interneurons and suggest a mechanism that controls their axonal projection.
Materials and Methods
In ovo electroporations.
Fertile Loman chick eggs were incubated for 2.5 d at 38°C until reaching stage 15 Hamburger Hamilton (HH). DNA solution (5 μg/μl, diluted in PBS) was injected into the lumen of the hindbrain together with fast-green (0.1%) to visualize injection site. Electroporation was performed with bent L-shaped gold electrodes (1 mm diameter) in a parallel holder (BTX, Harvard Apparatus) that were positioned in the caudal hindbrain, anode leftwards. Four 45 ms pulses of 25 V and pulse intervals of 300 ms were applied using BTX 3000 electroporator (Harvard Apparatus). Embryos were incubated for 1–11 d before analysis.
Plasmids.
Two enhancer elements, Atoh1 and edI1 (Helms et al., 2000; Pennacchio et al., 2006; Avraham et al., 2009) were cloned upstream to Cre-recombinase sequence and electroporated along with conditional reporter plasmids, in which a transcriptional floxed STOP cassette was inserted between the CAGG enhancer/promoter module and the nuclear GFP gene (pCAGG-LoxP-STOP-LoxP-nGFP), cytoplasmic-GFP gene (pCAGG-LoxP-STOP-LoxP-cGFP; Avraham et al., 2009, 2010a,b) or myristoylated-GFP (pCAGG-LoxP-STOP-LoxP-mGFP). For simultaneously assessing the electroporated region and the specificity of the enhancer elements, an alternating reporter plasmid was used in which a floxed mCherry gene was inserted between the CAGG enhancer/promoter module and the GFP sequence (pCAGG-LoxP-mCherry-LoxP-GFP; Avraham et al., 2009). For ectopic expression of Lhx1 in dA1 neurons, Lhx1 gene was inserted between floxed STOP module and a taumyc reporter (pCAGG-LoxP-STOP-LoxP-Lhx1-IRES-taumyc; Avraham et al., 2009). For tracking the axonal pathways of dB1 neurons, a plasmid containing the enhancer of Ptf1a, which drives expression of GFP (Meredith et al., 2009), was used. For integration of the reporter gene into the chick genome, the PiggyBac (PB) DNA-transposition method was applied (Wang et al., 2010), in which a Cre-conditional reporter cassette (mGFP) was cloned between the two PB arms (PB-CAG-LoxP-STOP-LoxP-mGFP-PB) and electroporated along with the Atoh1/edI1 enhancer plasmids and the Pbase transposase (Sanger Institute). For tracing synaptic connections, two conditional reporter plasmids containing the synaptic tracers SV2-GFP (PB-CAG-LoxP-STOP-LoxP-SV2-GFP-PB) or Synaptobrevin-GFP (PB-CAG-LoxP-STOP-LoxP-synB-GFP-PB; Alsina et al., 2001; Leal-Ortiz et al., 2008), were electroporated into the embryonic hindbrain along with the Atoh1/edI1 enhancers and the Pbase transposase. Schematic description of plasmids is provided in each figure.
In situ hybridization and immunofluorescence.
Whole-mount in situ hybridization was performed as described earlier (Weisinger et al., 2008), using a specific probe for chicken Atoh1 homolog (Cath1). Probe was labeled with digoxigenin-UTP and detected using 1:2000 alkaline phosphatase-coupled antibody followed by NBT/BCIP staining (Roche).
Whole-mount immunofluorescence was performed in embryos fixed overnight (ON) at 4°C in 4% paraformaldehyde (PFA)/0.1 M phosphate buffer, washed with PBS and incubated in PBS with 0.1% Tween20 (PBT)/ 5% goat serum for 2 h, before incubation for ON with the following antibodies: Pax2 (1:50, Abcam), Brn3a and 3A10 (both 1:50, Developmental Studies Hybridoma Bank), Myc (1:500, Santa Cruz Biotechnology). Following PBT washes, anti-rabbit or anti-mouse Alexa 488 and anti-rabbit Alexa 594 (all 1:400, Invitrogen) were added for ON. Immunofluorescence in frozen sections was performed on embryos fixed ON as described above and incubated in 30% sucrose/PBS for ON. Embryos were embedded in optimal cutting temperature solution. Cryostat sections (12 μm) were collected and incubated in PBT/5% goat serum for 2 h before incubation for ON at 4°C with the following antibodies: rabbit polyclonal Lhx2/9 (1:100, provided by Dr. T. Jessell, Columbia University, New York, NY), rabbit polyclonal Atoh1 (1:100, provided by Dr. J. Johnson, Southwestern Medical Center, Dallas, TX), mouse monoclonal Lmx1b, Brn3a, Lhx1/5, Tlx3, En-1, Islt1, Synaptotagmin, SV-2 (1:100, Developmental Studies Hybridoma Bank), rabbit polyclonal Pax2 (1:100, Abcam), Olig3 (1:400, provided by Dr. H. Takebayashy, Kumamoto University, Kumamoto, Japan), Axonin-1 (1:500, provided by Dr. E. Stoeckli, University of Zurich, Zurich, Switzerland), Zic1 (1:400, provided by Dr. R. Segal, Dana-Farber Cancer Institute, Boston, MA) and calbindin 28KD (1:80, Swant). Following PBS washes, anti-rabbit or anti-mouse Alexa 488 and anti-rabbit Alexa 594 (all 1:400, Invitrogen) were added for 2 h. All embryos were visualized under Nikon E400 microscope with a digital camera (DP70, Olympus), or by confocal analysis (Leica CTR 4000).
Cell counts.
Quantification of cells that conditionally express nGFP and thus represent dA1 cells was performed by calculating the number of nGFP+-cells that coexpress a related neuronal marker (Lhx2/9, Atoh1, Lhx1/5, Pax2, Lmx1b), out of the total nGFP+-cells. Quantification of dA1 cells that ectopically express Lhx1-taumyc was performed by counting the overlapping between Lhx1-taumyc-expressing cells and cells that express the various neuronal markers, out of the total Lhx1-taumyc-expressing cells. Quantification of dB1-GFP+ cells was performed by calculating the number of Ptf1a-GFP-expressing cells that coexpress a related neuronal marker (Lhx1/5, Pax2, Lmx1b), out of the total GFP+ cells. Numbers and percentages are taken from a section that represents >10 sections from three different embryos demonstrating similar percentages.
Results
Distribution of dorsal interneuron subtypes in the chick hindbrain
The localization of distinct dorsal interneuron (dI) subtypes in the murine hindbrain has been shown (Liu et al., 2008; Storm et al., 2009). These groups were divided into class A (dA) and class B (dB) neurons based on their distinct positions and gene expression; dA neurons are located in the dorsal alar plate of the hindbrain and express the bHLH gene Olig3, whereas dBs are more ventral and express the homeobox gene Lbx1. Each of the two groups is further subdivided along the dorsal-ventral (DV) axis based on the expression of multiple markers. Distribution of the markers also differs along the ventricular/mantel zone, reflecting the progenitor or mitotically inactive state of each neuron, respectively. Based on this information, we have used cell fate markers for mapping the different subtypes of hindbrain interneurons in the chick hindbrain. Immunofluorescence or in situ hybridization was performed in whole-mounted hindbrains as well as in transverse sections of E3 embryos (stage 18HH) at the levels of r4–r5. The position of class A and B hindbrain interneurons, from dorsal to ventral, is as follows: dA1-Lhx2/9+/Brn3a+/Olig3+ (Fig. 1A–E); dA3-Olig3+/Brn3a+/ Tlx3+/Lmx1b+ (Fig. 1B–D,H,I); dA4-Lhx1/5+(Fig. 1F,H); dB1-Lhx1/5+/Pax2+ (Fig. 1E–I,J); dB3-Brn3a+/Lmx1b+/Tlx3+ (Fig. 1C,D,I and data not shown); dB4-Lhx1/5+/Pax2+ (Fig. 1E–J). A summary of these results is provided (Fig. 1N), and shows the division of the hindbrain into multiple dA/dB subgroups along the DV axis, which is similar to that of the mice (Liu et al., 2008; Storm et al., 2009). Notably, dA2 (Lhx1+/ Foxd3+) subpopulation exists only from r7 and caudally (data not shown) and is thus not mentioned in the scheme. Moreover, in mice, dB1 (Lhx1/5+/Pax2+) subgroup also expresses Ptf1a (Meredith et al., 2009). We found that a Ptf1a-specific enhancer is expressed in chick dB1 subgroup (see Fig. 7K,L), and is therefore included in the scheme. dB2 (Lbx1+/phox2b+) subgroup is also included in the scheme between the dB1 and the dB3 domains (Sieber et al., 2007; Storm et al., 2009), although it could not be directly detected due to lack of chick antibodies. Pax6 and Islet1 are expressed in hindbrain motor neurons (Fig. 1G,J; Osumi et al., 1997), and are excluded from all dA/dB neurons.
Figure 1.
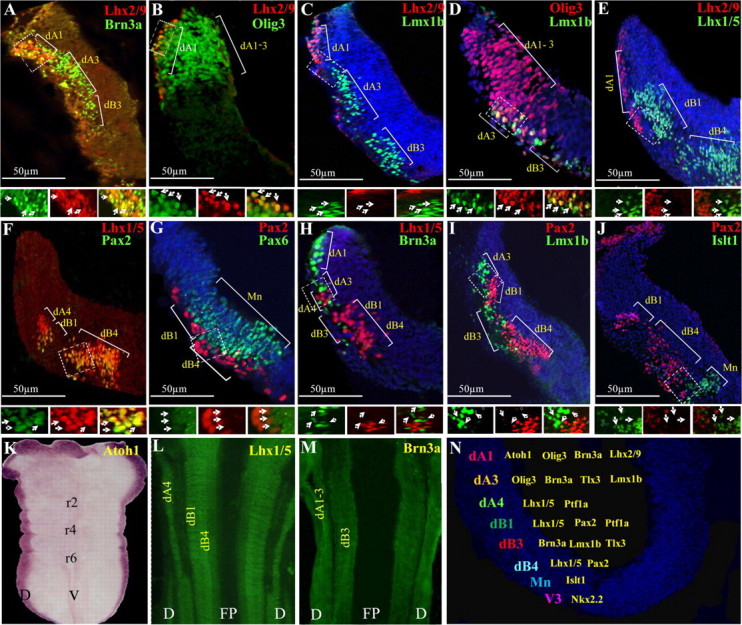
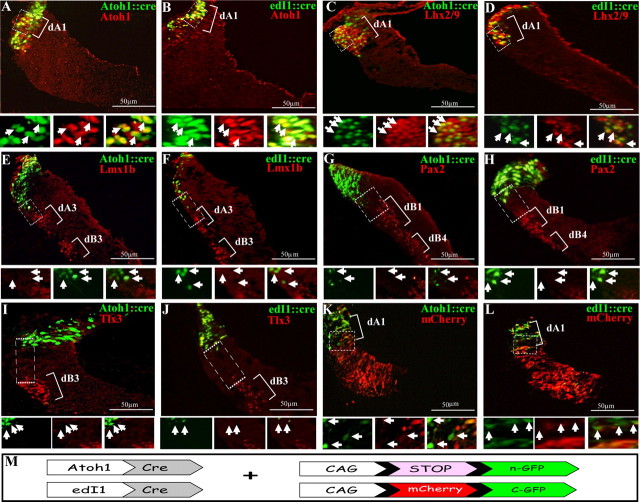
The distribution of dorsal neuronal subpopulations in the hindbrain. A–J, Double-labeled immunofluorescence was performed on transverse-sections of E3 chick hindbrains at the level of rhombomere 4. The localization of each dA/dB neuronal subgroup is marked. The boxed areas are represented as enlargements in their different channels at the bottom of each panel. The arrows point at representative neurons in all channels. K–M, Flat-mounted views of E3 hindbrains following in situ hybridization (K) or immunofluorescence (L, M) staining. N, Summary of the distribution of the dorsal neuronal subtypes in the DV axis of r4. In all panels, antibodies and probes are indicated in their respective colors and scale bars are marked. D, dorsal; r, rhombomere; FP, floor plate; V, ventral.
Figure 7.
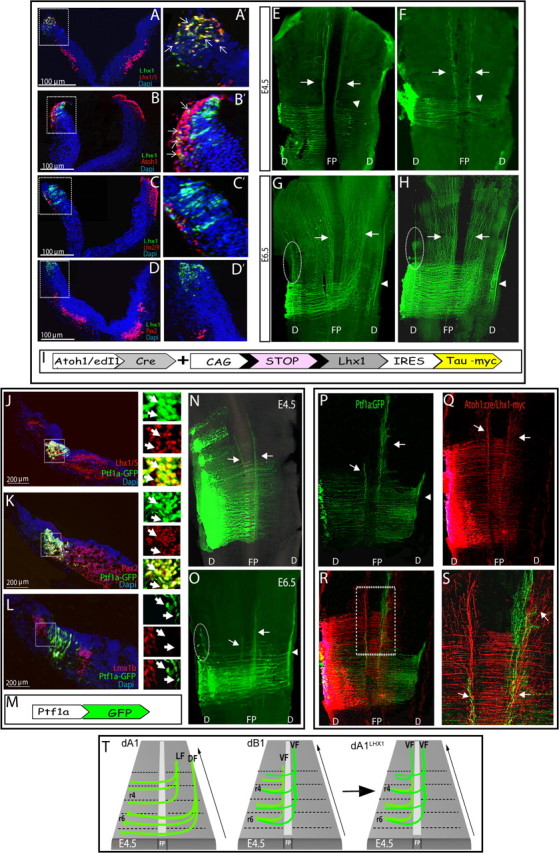
Alterations in dA1-axonal patterns by modification of the Lim-HD code. A–H, E2.5 hindbrains were electroporated with Atoh1 enhancer::Cre (A–E,G), or edI1 enhancer::Cre (F,H), and conditional Lhx1-taumyc plasmids. A–D, Transverse sections of E3.5 hindbrains examined for the distribution of dorsal interneurons, using antibodies against Lhx1/5 (A), Atoh1 (B), Lhx2/9 (C), and Pax2 (D). A′–D′, Enlargement views of the boxed areas in A–D to show the electroporated regions of the hindbrain. Arrows point at representative neurons that coexpress taumyc and the examined interneuron marker. Scale bars are indicated. E–H, Flat-mounted views of E4.5 (E, F) or E6.5 (G, H) hindbrains that were electroporated with showing axons of dA1 neurons expressing Lhx1. Arrows indicate ipsilateral and contralateral axons that ascend adjacent to the midline, arrowheads indicate axons that continue to grow laterally after crossing the midline and turn rostrally, and circled areas indicate dorsal ipsilateral ascending tract. I, A scheme of the plasmid used in A–H; Expression of Lhx1 and taumyc (pCAGG-LoxP-STOP-LoxP-Lhx1-IRES-taumyc) are driven by Atoh1::Cre or edI1::Cre. J–O, E2.5 hindbrains were electroporated with Ptf1a::GFP-expressing plasmid. J–L, Transverse sections of E3.5 hindbrains stained with antibodies against Lhx1/5 (J), Pax2 (K) and Lmx1b (L). Boxed areas in J–L are represented in their different channels in magnified views to the right of each image, respectively. Arrows point at representative Ptf1a::GFP-expressing neurons. Scale bars are indicated. N, O, Flat-mounted views of E4.5 (N) and E6.5 (O) hindbrains showing axonal trajectories of neurons expressing Ptf1a::GFP. M, A scheme of the plasmid used in J–O; expression of GFP is driven by Ptf1a enhancer. P–S, Flat-mounted views of E5 hindbrains coelectroporated with Ptf1a::GFP (P) and Atoh-cre::conditional Lhx1-taumyc (Q) plasmids (shown in I, M). R, A merged image of P, Q demonstrating colocalization of axons shown in P, Q. S, Magnified view of the boxed area in R. T, A schematic summary representing either the typical dA1 axonal projections, the typical dB1 axonal projections, or the dA1Lhx1 axonal tracts in E4.5 hindbrain. In all images, stages, antibodies, and plasmids are indicated. In all sections, scale bars are indicated. DF, dorsal funiculus; LF, lateral funiculus; VF, ventral funiculus; FP, floor plate; D, dorsal; r, rhombomere.
AP arrangement of chick hindbrain interneurons was next analyzed in whole-mounted preparations. The Atoh1 gene is expressed in a restricted pattern along the dorsal-most portion of the entire hindbrain (Fig. 1K). Furthermore, staining with Lhx1/5 and Brn3a demonstrates a homogenous expression pattern that refers to dA4/dB1/dB4 (Fig. 1L) and dA1/dA3/dB3 (Fig. 1M), respectively. These data confirm that the dA/dB neuronal subgroups are similarly expressed along the AP axis of the hindbrain while distinguished along the DV axis.
Labeling dA1 interneurons using specific enhancer elements
Genetic fate mapping using enhancer elements has been used in the murine hindbrain to define multiple interneuronal subtypes and to trace their migration routes in the brainstem (Helms and Johnson, 1998; Wang et al., 2005; Farago et al., 2006; Rose et al., 2009b; Renier et al., 2010). However, information regarding the detailed patterns and targets of their axonal projections is limited. We have previously used transient-transgenic chick screen of highly conserved human noncoding sequences and identified numerous enhancer elements that are expressed in dI1-dI3 of the chick spinal cord (Pennacchio et al., 2006; Visel et al., 2007; Avraham et al., 2009, 2010a,b). Unilateral electroporation of these elements enabled us to direct the expression of a reporter gene in specific neurons in a spatio-temporal controlled manner, and to follow or manipulate axonal projections on both sides of spinal cord (Reeber et al., 2008; Avraham et al., 2009, 2010b). This strategy circumvented the difficulty of tracing axonal projections in mice, where germ-line insertion of enhancer elements drives robust bilateral symmetric labeling of axons that may lead to masking of ascending/descending and ipsilateral/contralateral axonal tracts.
To trace hindbrain dA1 interneurons, we used two enhancer elements that have previously been characterized as specific for spinal dI1 neurons (Helms et al., 2000; Lumpkin et al., 2003; Reeber et al., 2008; Avraham et al., 2009) and analyzed their expression in the chick hindbrain. The tested enhancers were the mouse Atoh1 (Helms et al., 2000; Ebert et al., 2003; Lai et al., 2011) and edI1 enhancer element (Avraham et al., 2009). These elements were cloned upstream to Cre recombinase and coelectroporated at E2.5 along with a Cre-dependent nuclear Green Fluorescent Protein (nGFP) reporter plasmid (pCAGG-LoxP-Stop-LoxP-nGFP; Fig. 2M). To examine whether the Cre/LoxP-conditional expression system labels dA1 neurons specifically, hindbrains were stained at E3.5 with multiple neuronal markers (Fig. 2A–J). Expression of nGFP, driven by either Atoh1 or edI1 enhancer::Cre, was restricted to dA1 neurons, as indicated by the overlapping expression with Atoh1 and Lhx2/9 (Fig. 2A–D) and by the segregation from Lmx1B+/Tlx3+/Pax2+-neurons that mark dA3/dB3 and dB1/dB4 subpopulations, respectively (Fig. 2E–J). Quantification of these results confirmed that the vast majority of the nGFP+ neurons express Atoh1 or Lhx2/9 while no nGFP expression is evident in more ventral subpopulations (Table 1). Based on their position along the ventricular-marginal axis, the nGFP+/Lhx2/9− cells are likely to be the progenitors of dA1 that do not yet express Lhx2/9, while the nGFP+/Atoh− cells are postmitotic dA1 that downregulated Atoh1 expression. As an additional control to assess the efficiency of the electroporation and specificity of the enhancers, mcherry/GFP alternating conditional plasmid was used (Avraham et al., 2009), together with the enhancer::Cre constructs. Electroporated embryos showed GFP expression in dA1 neurons while mcherry was evident more ventrally in the non-dA1 neurons (Fig. 2K,L). Together, these data demonstrate the specificity of the Atoh1 and edI1 enhancers to dA1 interneurons.
Figure 2.
Labeling of dA1 neuronal subgroup using Atoh1/edI1 enhancer elements. Cross-section views of hindbrains at the level of r4–r5 that were electroporated with Atoh1::Cre (A, C, E, G, I, K) or edI1::Cre (B, D, F, H, J, L) plasmids, along with a conditional nuclear GFP reporter (A–J) or conditional alternating mCherry/GFP (K, L) plasmids. Sections in A–J were stained with Atoh1, Lhx2/9, Lmx1b, Pax2, or Tlx3 antibodies. In all images, dA/dB subgroups are marked. Higher-magnification views of the boxed areas are represented below each panel in different channels, and arrows indicate the same cells in all channels. Plasmids and antibodies are indicated in their respective colors and scale bars are indicated. M, A scheme of the Cre/loxP-based expression systems used in the experiments. A nuclear GFP (n-GFP) is flanked by two loxP sites (pCAGG-LoxP-STOP-LoxP-nGFP), or a floxed mCherry gene is inserted between the CAGG enhancer/promoter module and the GFP gene (CAGG-loxP-mCherry-loxP-GFP). The expression of GFP in dA1 neurons is driven by Atoh1::Cre or edI1::Cre.
Table 1.
Quantification of the results obtained on neuronal markers by expression of the multiple constructs in dA1 cells
| Reporter plasmid | Marker | Plasmid-expressing cells (n) | Marker-expressing cells (n) | Overlapping (%) |
|---|---|---|---|---|
| a. Atoh1::Cre-nGFP | Lhx2/9 | 67 | 75 | 59/67 (88%) |
| b. Atoh1::Cre-nGFP | Atoh1 | 30 | 95 | 27/30 (90%) |
| c. Atoh1::Cre-nGFP | Pax2 | 82 | 158 | 0 |
| d. Atoh1::Cre-nGFP | Lmx1b | 59 | 41 | 0 |
| e. edI1::Cre-nGFP | Lhx2/9 | 52 | 65 | 48/52 (92%) |
| f. edI1::Cre-nGFP | Atoh1 | 29 | 52 | 27/29 (93%) |
| g. edI1::Cre-nGFP | Pax2 | 54 | 23 | 0 |
| h. edI1::Cre-nGFP | Lmx1b | 29 | 47 | 0 |
| i. Atoh1::Cre-Lhx1-IRES-taumyc | Lhx1/5 | 52 | 98 | 45/52 (87%) |
| j. Atoh1::Cre-Lhx1-IRES-taumyc | Lhx2/9 | 51 | 18 | 0 |
| k. Atoh1::Cre-Lhx1-IRES-taumyc | Atoh1 | 50 | 52 | 9/50 (18%) |
| l. Atoh1::Cre-Lhx1-IRES-taumyc | Pax2 | 73 | 117 | 0 |
| m. Ptf1a-GFP | Lhx1/5 | 81 | 388 | 73/81 (90%) |
| n. Ptf1a-GFP | Pax2 | 72 | 367 | 66/72 (91%) |
| o. Ptf1a-GFP | Lmx1b | 44 | 48 | 0 |
Mapping axonal projections of dA1 interneurons
Earlier studies revealed the presence of several longitudinal axonal tracts in the hindbrain that arise from ipsilateral or contralateral neurons (Lumsden and Keynes, 1989; Clarke and Lumsden, 1993). Yet, the link between the precise genetic identities of these neurons, especially those arising from dorsal subpopulations, and the individual axonal tracts that they provide, is unclear. Here we set to study the axonal patterns of dA1 neurons by labeling the cells with cytoplasmic-GFP (cGFP) under the control of each of the two dA1 enhancers (Fig. 3N; Avraham et al., 2009). Plasmids were coelectroporated at E2.5 at the level of r3–r7 and embryos were let to develop for further 2–4 d. Analyzing E4.5–E5 embryos revealed similar projection patterns of axonal trajectories for both enhancers (Fig. 3A–D). Two main axonal projection pathways were demonstrated, both crossing the midline and turning rostrally at the contralateral side; one projection originated from the caudal hindbrain and elongated longitudinally in the dorsal funiculus (DF; Fig. 3A–D, arrowheads), while the other arises from the more anterior hindbrain and elongated at the lateral funiculus (LF; Fig. 3A–D, arrows). In all experiments, the DF was more apparent compared with the LF, which seemed less condensed. To analyze whether these tracts originate from exclusive rhombomeric domains, hindbrains were costained with the neurofilament marker 3A10, which labels hindbrain axons that tend to accumulate at rhombomere borders (Lumsden and Keynes, 1989), and therefore allows the demonstration of repetitive segments. A sharp border between the two longitudinal paths of the dA1 axons was found (Fig. 3A,B, dotted lines). Axons derived from dA1 localized at r6–r7 ascended exclusively into the DF whereas axons derived from r2–r5 elongated only in the LF. E5.5 and E6 hindbrains revealed the same axonal patterns that further extended toward the upper brain (Fig. 3E–H), with the dorsal tract still being more noticeable than the lateral. At E6.5 the DF further extended rostrally and seemed to turn toward the developing cerebellar plate (Fig. 3I,J, arrowheads). Flat-mounted hindbrains of these embryos show defasciculation of axons from the main longitudinal tract at these sites (Fig. 3K–L′).
Figure 3.
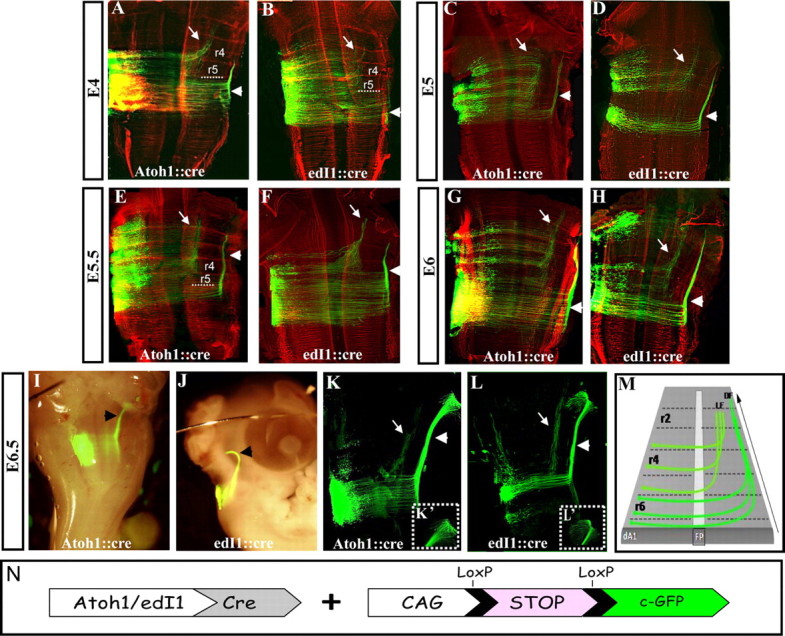
Axonal projections of dA1 neurons at E4–E6.5. Flat-mounted (A–H, K, L) or whole-mounted (I, J) views of hindbrains that were coelectroporated at E2.5 with Atoh1/edI1 enhancer::Cre and conditional cytoplasmic GFP (cGFP) reporter plasmids and inspected for axonal projections (in green) at E4 (A, B), E5 (C, D), E5.5 (E, F), E6 (G, H) and E6.5 (I–L). Images in A–H were immunolabeled with the neurofilament marker 3A10 (in red) to mark rhombomere borders. Boxed areas in K′ and L′ represent higher-magnification views of the defasciculating axons in K, L. M, Schematic summary of dA1 axonal projections along the hindbrain. In all images, white arrows indicate the LF that originates from dA1 cells at r2–r5, arrowheads indicate the DF that originates from dA1 cells at r6–r7, stages and plasmids are indicated. r, Rhombomere; LF, lateral funiculus; DF, dorsal funiculus; FP, floor plate. N, A scheme of the Cre/loxP-based expression system used. cGFP is flanked by two LoxP sites (pCAGG-LoxP-STOP-LoxP-cGFP). The expression of cGFP in dA1 neurons is driven by Atoh1::Cre or edI1::Cre.
We next aimed to inspect the axonal patterns and the destinations of dA1 neurons at later-embryonic stages (E7.5–13.5), in which the transiently expressed GFP fades away. Thus, the PiggyBac (PB) transposition method was applied (Figs. 4I, Wang et al., 2009). A reporter Cre-conditional-mGFP cassette cloned between the two PB arms (PB-CAG-LoxP-STOP-LoxP-mGFP-PB) was electroporated at E2.5 along with the Atoh1/edI1::Cre enhancers and the PiggyBac transposase vector (CAG-PBase). In this strategy, the electroporated Cre-conditional mGFP is integrated into the chick chromosomes and consequently the LoxP/STOP cassette is removed only in dA1 neurons, enabling prolonged expression of the reporter in dA1 cells (Lu et al., 2009). Flat-mounted E7.5 hindbrains show dA1-derived axons ascending in the contralateral side of the neural tube in dorsal or lateral paths and defasciculating at the rostral end of the DF (Fig. 4A,B, arrows and dotted box). These patterns are in agreement with the results obtained with the transiently expressed Cre/Lox plasmids (Fig. 3), confirming the specificity of the PB plasmids in labeling dA1 neurons. Notably, in addition to the two main trajectories, some ipsilateral tracts are evident at this stage (Fig. 4A,B, arrowheads). Such tracts were also found at this stage using the transient plasmid system, although the GFP level of expression was less intense compared with the PB system (Fig. 3K,L). Moreover, few axons seemed to project caudally at the contralateral side of the hindbrain (Fig. 4A,B, dotted circles). The additional tracts observed at later stages raise the possibility of late-born dA1 neurons that may provide these axonal projections. As a negative control, electroporation with the mGFP-conditional reporter and transposase plasmids but without the Atoh1/edI1 enhancer::Cre vector was performed and did not reveal any staining in the hindbrain (data not shown).
Figure 4.
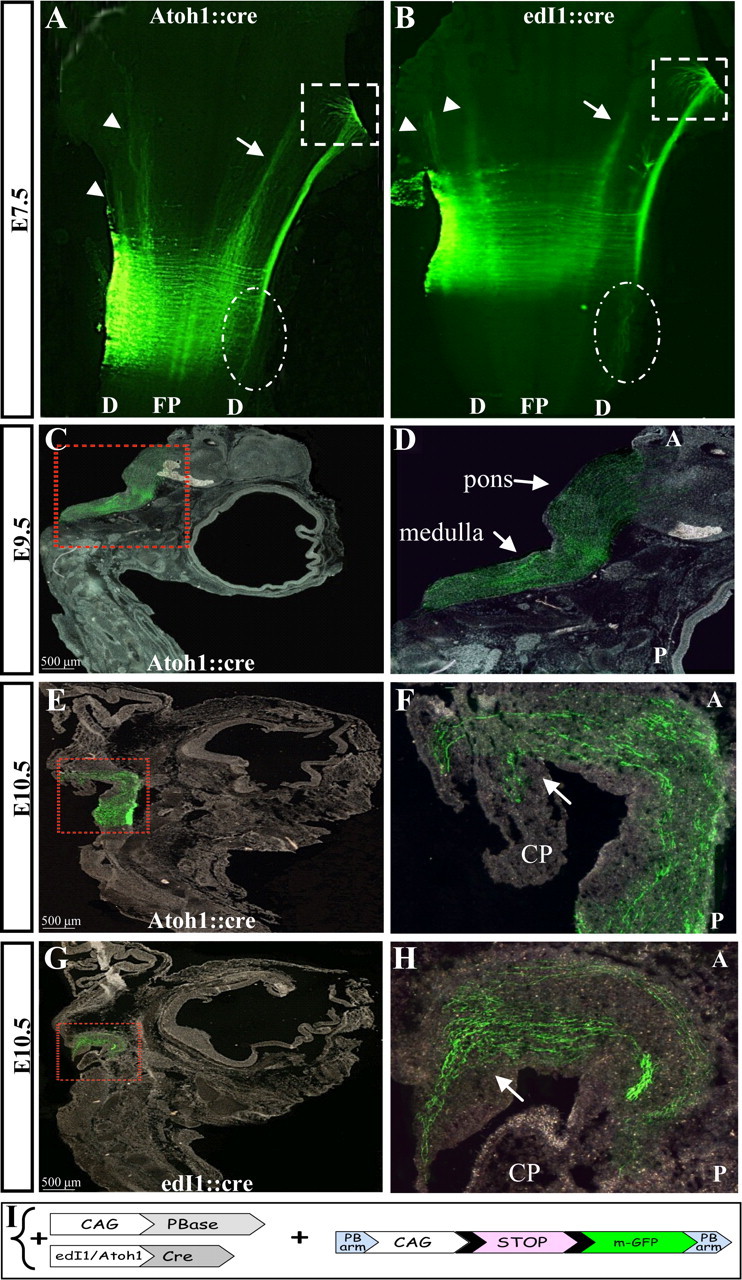
Axonal projections of dA1 interneurons at E7.5–10.5. Flat mounted views (A, B) and sagittal sections (C–H) of embryos that were electroporated with the PiggyBac system to enable the incorporation of the conditional myristoylated GFP (mGFP) into dA1 cell genome. Axons were inspected at E7.5 (A, B), E9.5 (C, D) and E10.5 (E–H). A, B, arrows indicate the ascending contralateral medial tract, arrowheads indicate ipsilateral tracts, boxed areas represent the fasciculating axons of the ascending contralateral lateral tract and circled areas indicate descending contralateral axons. C–H, boxed areas in C, E, G represent higher-magnification views in D, F, H, respectively. Arrows indicate ascending axons in the medulla, pons, and cerebellar plate. In all images, stages and plasmids are indicated. A, anterior; P, posterior; D, dorsal; FP, floor plate; CP, cerebellar plate. I, A scheme of the constructs in which mGFP cassette is flanked by two PB arms (PB-LoxP-STOP-LoxP-mGFP-PB). The integration of the reporter cassette into the genome and the expression of mGFP in dA1 neurons is driven by CAG::PBase and Atoh1::Cre or edI1::Cre.
After confirming the efficient and specific expression of the chromosomally integrated mGFP in dA1 cells, the dorsal ascending axonal route was traced. Embryos were electroporated at E2.5 at the caudal hindbrain (r6–r7) and analyzed by sagittal-sections (Fig. 4C–H). At E9.5, dA1 axons seemed to extensively elongate in the medulla and pons (Fig. 4C,D). At E10.5 axons extended more rostrally and turned dorsally toward the cerebellar plate (Fig. 4E–H). As PCN neurons, as well as subsets of CN neurons, project to the cerebellum (Mason, 1986; Hatten and Heintz, 1995; Bermingham et al., 2001; Díaz et al., 2003; Doucet and Ryugo, 2003; Cicirata et al., 2005; Dipietrantonio and Dymecki, 2009; Rose et al., 2009a), we then wished to inspect the dA1 axonal routes in detail within the cerebellum. The cerebellum was dissected out from E9.5-E13.5 embryos and stained for the zinc finger transcription factor Zic1, which marks the EGL and IGL, and with the Ca2+-binding protein Calbindin, which marks the Purkinje layer, to delineate the various cerebellar domains (Baimbridge et al., 1992; Aruga et al., 1994, 2002; Grinberg et al., 2004). Analysis of the cerebellum at E9.5 shows a broad staining of Zic1-expressing neurons, which is likely to refer to the EGL progenitors before their organization in the external layer of the cerebellum (Machold and Fishell, 2005; Morales and Hatten, 2006). Notably, many GFP+ axons are evident at these sites (Fig. 5A,B). At E11.5, more dA1 axons accumulate within the cerebellum and the expression of Zic1+ cells becomes more-restricted to the EGL (Fig. 5C,D). At E12.5, dA1 axons accumulate further in the EGL, demarcated by Zic1+ cells. Many axons are still seen in inner cerebellar positions at this stage. Some of these domains also express Zic1 (Fig. 5E,F), which reflects inner granular neurons (Aruga et al., 1994), suggesting the presence of dA1 axons also in the IGL. Staining with calbindin at E13.5 revealed axons that reached the Purkinje layer (Fig. 5G,H), as well as multiple axons in inner cerebellar positions, similar to the results shown in Figure 5, D and F. The distribution of dA1 axons at multiple cerebellar layers may either represent axons that are constantly extending from inner to outer cerebellar domains, or axons that form connections with multiple cerebellar neurons in the EGL, Purkinje, IGL, and deep cerebellar (DC) layers.
Figure 5.
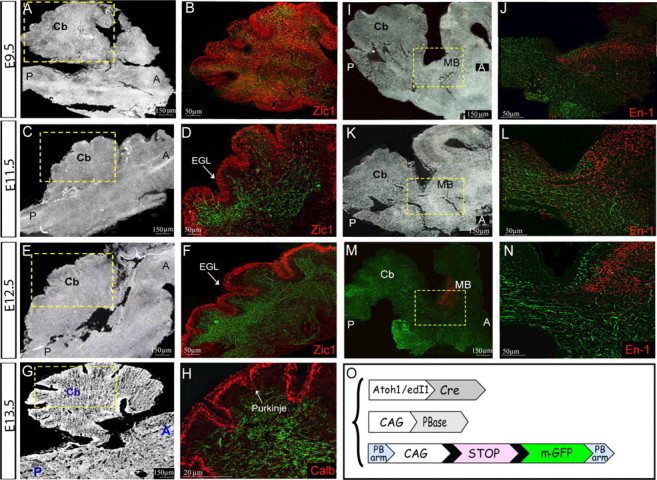
dA1 axonal projections in the cerebellum and midbrain. Sagittal sections of the brainstem, cerebellum and midbrain regions at E9.5 (A, B, I, J), E11.5 (C, D, K, L), E12.5 (E, F, M, N) and E13.5 (G, H) of embryos that were electroporated with the PiggyBac system to label dA1 axons. Boxed areas in A, C, E, G, I, K, M are shown as enlargements in panels B, D, F, H, J, L, N, respectively. A, C, E, G, I, K, Bright-field images of sections. B, D, F, H, J, L, N, Fluorescent images demonstrating axonal tracts (green) and Zic1 (B, D, F), Calbindin (H), and En-1 (J, L, N) immunolabeling (red). Stages, antibodies, anatomical axes, and landmarks are indicated in all panels. Scale bars are indicated. Cb, cerebellum; MB, midbrain; EGL, external granular layer; Calb, calbindin; A; anterior, P; posterior. O, A scheme of the constructs; mGFP-LoxP cassette is flanked by two PB arms. The integration of the reporter cassette into the genome and the expression of mGFP in dA1 neurons is driven by CAG::PBase and Atoh1::Cre/EdI1::Cre.
A main subset of cochlear nuclei axons elongates toward the mesencephalon in the lateral leminiscus to innervate the inferior colliculus (Cant, 1992; Ryugo et al., 2003). Therefore, we wished to inspect whether dA1 axons can also be found in the mesencephalon in our system. Embryos were stained with En-1, to mark the midbrain/hindbrain border (Wurst et al., 1994). At E9.5, dA1 axons have reached the anterior hindbrain toward the midbrain (Fig. 5I,J), whereas at E11.5 and 12.5, dA1 axons further extended and elongated along the posterior embryonic midbrain (Fig. 5K–N). Altogether, these data show that dA1 neurons, which originate from the caudal-most regions of the RL, extend axons into several domains: anterior hindbrain, multiple cerebellar layers, and the midbrain.
Synaptic targets of dA1 interneurons
An important question raised by these observations is whether the ascending dA1 axons merely extend through these domains or whether they form synaptic connections with target neurons at these positions. E2.5 embryos were electroporated with SV2-GFP or synaptobrevin-GFP (SynB-GFP) synaptic reporters (Alsina et al., 2001; Leal-Ortiz et al., 2008) along with Atoh1/edI1 enhancers and the PB transposase (Fig. 6A), and allowed to develop until E13.5. This method enables the expression of the GFP reporter in the presynaptic vesicles. As the two enhancers provided similar results, we present here data obtained by the Atoh1-enhancer. The brainstem and cerebellum were sectioned and stained with the synaptic markers synaptotagmin or SV2 (Gardzinski et al., 2007; Nowack et al., 2010), as well as with various neuronal markers, to allow the detection of dA1-synaptic target sites in correlation to other neurons along the embryonic brain.
Figure 6.
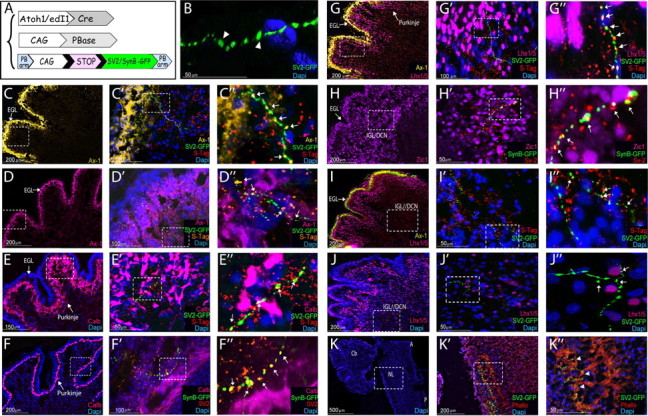
Synaptic targets of dA1 interneurons. A, A scheme of the constructs; a synaptic SV2-GFP or synaptobrevin-GFP (SynB) reporter cassette is flanked between two PB arms (PB-LoxP-STOP-LoxP-SV2-GFP-PB; PB-LoxP-STOP-LoxP-SynB-GFP-PB). The integration of the reporter cassette into the genome and the expression of SV2-GFP/SynB-GFP in dA1 neurons is driven by CAG::PBase and Atoh1::Cre. C–K, Sagittal sections of E13.5 cerebellum (C–J″) and medulla/pons (K–K″) from embryos electroporated at E2.5 with the PiggyBac system to label synaptic vesicles of dA1 axons. B, Confocal 3D imaging of SV2-GFP+ synapses in the cerebellum. For all images, each marker is indicated in a different color, higher-magnification views of the boxed areas in the left panels are presented at their respective right panels. C″–J″ represent digital magnifications of boxed areas in C′–J′. Arrowheads indicate SV2-GFP labeled synapses (green), and arrows indicate SV2-GFP/SynB-GFP labeled synapses coexpressing the synaptic markers synaptotagmin/SV2 (yellow). Scale bars are indicated. Cb, cerebellum; DCN, deep cerebellar neurons; EGL, external granular layer; IGL, internal granular layer; Phallo, phalloidin; NL, nuclear laminaris S-Tag, synaptotagmin; Ax-1; axonin 1; Calb, calbindin; A, anterior; P, posterior.
The expression of the SV2-GFP/SynB-GFP cassette in the synapses revealed a punctuated and succeeding staining of GFP+ particles (Fig. 6B–J″), which is typical of presynaptic vesicles (Alsina et al., 2001; Leal-Ortiz et al., 2008). Staining with the presynaptic markers synaptotagmin or SV2 highly resembled the characteristic synaptic morphology observed by the GFP+ synapses (Fig. 6C″–I″). In many instances, SV2-GFP+/SynB-GFP+ synapses were also colabeled with the presynaptic markers (Fig. 6C″–I″, arrows). These results validate the specificity of our GFP labeling method to detect dA1-presynaptic vesicles in the cerebellum. Notably, GFP+ synapses that were not costained with the presynaptic markers synaptotagmin/SV2 were also frequently detected. This finding may result from the former expression of SV2-GFP during the maturation steps of the presynaptic vesicle, compared with synaptotagmin, which is induced by SV2 and acts in the functional synapse at the cone termini (Chang and Südhof, 2009; Morgans et al., 2009; Yao et al., 2010; Bragina et al., 2011).
We next delineated the synaptic targets of dA1 interneurons in different cerebellar layers. Staining with the neuronal cell adhesion molecule Axonin-1 (Ax-1), which marks the EGL (Wolfer et al., 1998; Baeriswyl and Stoeckli, 2008), showed a close association between SV2-GFP+ vesicles and Ax-1+ neurons (Fig. 6C–D″). Staining with calbindin and Lhx1/5 was used to mark the nascent cerebellar Purkinje precursors (Baimbridge et al., 1992; Morales and Hatten, 2006). This staining revealed SV2-GFP+/SynB-GFP+ synapses also in the Purkinje layer of the cerebellum (Fig. 6E–G″). Furthermore, dA1 neurons also formed synapses in other, more inner positions, some of which are adjacent to Lhx1/5 as well as to Zic1-expressing cells (Fig. 6H–J″), which represent inner granular and deep cerebellar neurons (Aruga et al., 1994; Morales and Hatten, 2006). Together, these observations show for the first time that caudally originating dA1 axons form synaptic connections with external granular cells, Purkinje neurons and DC/IGL neurons, in agreement with previous reports (Mason et al., 1997; Kalinovsky et al., 2011).
Synaptic labeling of dA1 interneurons was also evident in a defined longitudinal position in the lateral medulla (Fig. 6K–K″). This region represents the nucleus laminaris (NL; L. Puelles, personal communication), which serves as part of the auditory complex in birds (analogous to the medial superior olive of mammals), and encodes sound localization. This finding is in agreement with previous descriptive data in which the NL was shown to be innervated by ipsilateral or contralateral axons of the posterior cochlear excitatory nuclei, the nucleus magnocellularis (NM; Rubel and Parks, 1988; Marín and Puelles, 1995; Díaz et al., 2003; Ryugo et al., 2003; Hendricks et al., 2006). The finding that dA1-derived synapses are mostly observed in the dorsal column of NL (Fig. 6K″), suggests that these connections develop either from an ipsilateral dA1 tract that begins to be apparent at E6.5 (Figs. 3K,L, 4A,B), or from previously unidentified contralateral derived dA1 neurons that can be detected by our labeling tools.
Alteration in dA1-axonal trajectories upon change of the Lim-HD code
Lim-HD proteins control multiple aspects of neuronal differentiation, such as subtype identity and axonal guidance (Sharma et al., 1998; Kania et al., 2000; Shirasaki and Pfaff, 2002; Wilson et al., 2008). Previous studies in the spinal cord demonstrated that alteration of the Lim-HD code of dI1 neurons by ectopic expression of the dI2-protein Lhx1, modifies their projection patterns (Avraham et al., 2009). To begin understanding whether the Lim-HD code has a similar role in patterning the axonal trajectories of hindbrain dA1 neurons, we misexpressed Lhx1 (which is typically expressed in dA4, dB1, and dB4 subgroups, Fig. 1E,F,H,L) in dA1 cells. As reciprocal cross-repression between Lhx1/5 and Lhx2/9 was demonstrated in the spinal cord, we first examined whether forced expression of Lhx1 in dA1 neurons affects their Lim-HD code. E2.5 embryos were coelectroporated with edI1/Atoh1::Cre and a conditional Lhx1 (pCAGG-LoxP-STOP-LoxP-Lhx1-IRES-taumyc) plasmids (Fig. 7I), to ectopically express Lhx1 and taumyc in dA1 neurons (dA1Lhx1). Cross sections of E3.5 embryos were stained with Lhx1/5, Lhx2/9, Atoh1, Pax2, and taumyc antibodies to assess the distribution of these neuronal markers. As expected, taumyc+ cells were observed in the dorsal hindbrain and coexpressed Lhx1/5 in dA1 cells (Fig. 7A, Table 1). Concomitantly, expression of Lhx2/9 was lost from the Lhx1/taumyc+ cells at the manipulated side of the dorsal hindbrain, whereas Lhx2/9 was evident at the control contralateral dA1 neurons (Fig. 7C). This result indicated that forced Lhx1 expression represses the expression of Lhx2/9 in dA1 neurons (Table 1), similar to the results obtained in the spinal cord (Avraham et al., 2009). In contrast, neither Atoh1, which marks dA1 progenitor cells, nor Pax2, which is expressed along with Lhx1/5 in dB1 and dB4 cells, were downregulated or upregulated, respectively, in dA1Lhx1 neurons (Fig. 7B,D, Table 1), indicating that ectopic Lhx1 in dA1 cells is sufficient to repress Lhx2/9 expression without altering their cell fate acquisition.
Using this tool, we then assessed whether Lhx1 expression in dA1 cells will impact their axonal projection patterns. E2.5 embryos were electroporated as before (Fig. 7I) and analyzed at E4.5 for dA1Lhx1 axons. A clear change in dA1Lhx1 axonal trajectories was found; two main longitudinal trajectories appeared, one ipsilateral and the other contralateral, both ascending adjacent to the floor plate at the ventral funiculi (VF; Fig. 7E,F, arrows). Moreover, some axons seemed to project more laterally after crossing the midline, but did not converge into a bundle at this stage (Fig. 7E,F, arrowheads). These patterns are in sharp contrast with the typical dA1 axonal patterns, in which two commissural lateral projections, at the LF and DF, are evident (Fig. 3). At E6.5, the two ventral axonal bundles were still apparent with more axons in each path, although the bundles seemed less compact (Fig. 7G,H, arrows). Furthermore, axons that projected laterally after crossing the midline at E4.5 seemed now to fasciculate into a lateral tract that extended rostrally (Fig. 7G,H, arrowheads). An ipsilateral ascending bundle at the DF also began to be apparent (Fig. 7G,H, circles). This experiment shows that Lim-code switch in dA1 neurons from Lhx2/9 to Lhx1 is capable of modifying their axonal paths, and suggests that the Lim-HD code may play a key role in controlling the axonal patterning of dA1 interneurons in the hindbrain.
Do the altered projections of dA1Lhx1 neurons correspond to axonal trajectories of another subtype of hindbrain interneurons that endogenously expresses Lhx1? Recently, Ptf1a-enhancer element was found to be specifically expressed in mice spinal cord dI4 (Lhx1/5+/Pax2/+Ptf1a+) and hindbrain Lhx1/5/+/Pax2+ subgroups (Meredith et al., 2009). Thus, we examined whether the expression of the Ptf1a-enhancer element in our system will be restricted to a certain Lhx1+ subgroup in the hindbrain. E2.5 embryos were electroporated with Ptf1a::GFP plasmid (Fig. 7M) and stained a day later with Lhx1/5, Lmx1b, and Pax2 antibodies. Ptf1a::GFP+ cells were restricted to dB1 neurons, as indicated by the overlapping expression with Lhx1/5 and Pax2 (Fig. 7J,K) and the segregation from Lmx1B+ areas, which mark the flanking dA3/dB3 subpopulations (Fig. 7L). Quantification of these results indicated that 90% of the Ptf1a-GFP+ neurons express Lhx1/5/Pax2 while no expression was evident in adjacent subpopulations (Table 1), further confirming the specificity of the Ptf1a enhancer in dB1 neurons. Based on their position along the ventricular-marginal axis, the remaining 10% of GFP+/Lhx1−/Pax2− cells are likely to represent dB1 progenitors that have not yet upregulated the expression of Lhx1 and Pax2.
We next inspected which axonal projection patterns will be derived by the Ptf1a::GFP-expressing neurons and compared them to those formed by dA1Lhx1 neurons. Embryos were electroporated at E2.5, using either the Ptf1a::GFP plasmid or a conditional Cre/GFP expression system (in which the Ptf1a-enhancer was cloned upstream to Cre recombinase and coelectroporated with a Cre-conditional GFP plasmid) and analyzed at E4.5. The presented data were obtained from the Ptf1a::GFP vector, as both types of plasmids provided similar results (Fig. 7N, and data not shown). Two main axonal trajectories of Ptf1a::GFP+ neurons were evident; one ipsilateral and the other contralateral, both ascending adjacent to the floor plate at the VF (Fig. 7N, arrows). Notably, some axons seemed to project more laterally after crossing the midline, without converging into a bundle at this stage. At E6.5 the two VFs were still apparent (Fig. 7O, arrows). Yet, additional patterns were formed, in which axons projected further laterally after crossing the midline and turned longitudinally in a dorsal ascending tract (DF; Fig. 7O, arrowhead), or turned into a dorsal ipsilateral ascending bundle (Fig. 7O, circle). In all experiments, the contralateral VF was more apparent than the ipsilateral VF, which seemed less condensed. These results show that the axonal patterns projected by the dB1 (Ptf1a::GFP+) subgroup (Fig. 7N,O) are highly similar to those observed by the dA1Lhx1-expressing neurons (Fig. 7E–G). To further confirm that dA1Lhx1 axonal patterns were shifted into dB1-like trajectories, we simultaneously labeled the two axonal subpopulations in the same embryo. E2.5 hindbrains were electroporated using Ptf1a::GFP vector to trace dB1 axons (Fig. 7P) and Atoh1::cre + CAGG-LoxP-stop-LoxP-Lhx1-IRES-taumyc plasmids to trace dA1LhX1 axons (Fig. 7Q). E5 embryos clearly showed colocalization of dA1lhx1 and dB1 axons in the same ipsilateral and contralateral paths (Fig. 7P–S). This experiment, together with the data shown in Figure 7E–H, N, and O, confirmed that the trajectories of dA1lhx axons were altered into dB1-like funiculi (Fig. 7T), demonstrating the ability of Lhx1 in shifting the typical axonal patterns of dA1 interneurons into a dB1-like patterns, without affecting dA1 cell-fate acquisition. This finding suggests a central role of the Lim-HD code in regulating axonal projections of hindbrain interneurons.
Discussion
Along the cRL, dA1 interneurons generate the CN and PCN and participate in establishing proprioceptive, interoceptive, auditory, and arousal circuitries. Multiple studies provided essential knowledge regarding the role of Atoh1 in these processes (Marín and Puelles, 1995; Cambronero and Puelles, 2000; Bermingham et al., 2001; Machold and Fishell, 2005; Wang et al., 2005; Rose et al., 2009a). Here we combined genetic and morphological tools to follow the axonal patterns of chick dA1-interneurons, to decipher their targets and connections, and to investigate the role of the Lim-HD code in patterning dA1-axonal paths.
Axonal patterns, targets, and connectivity of dA1-interneurons
We have previously used enhancer elements to drive a conditional expression of reporter genes in the chick spinal cord, and uncovered novel interneuron–axonal pathways (Avraham et al., 2009, 2010b). Here we used this strategy in the chick hindbrain and demonstrated for the first time the ability to target dA1-neurons and axons. Two main ascending contralateral trajectories were found: one derives from r6–r7 (dA1r6–7) and extends along a defined DF to predominantly populate the hindbrain, cerebellum, and midbrain, whereas the other originates from r2–r5 (dA1r2–5) and extends in a LF in a less-confined pattern. Synaptic connections of the dA1r6–7 axons were evident in the hindbrain and cerebellum.
Classical cell-tracing and grafting experiments in the chick have previously identified several ascending/descending axonal funiculi (Guthrie and Lumsden, 1992; Clarke and Lumsden, 1993; Díaz et al., 2003; Marín and Puelles, 1995). By retrograde/anterograde labeling, neurons were traced in the hindbrain alar/basal plate and projection sites were demonstrated, such as in the cerebellum. Other classical-labeling studies in adult rat brainstem also revealed projections of the PCN to the cerebellum (for review, see Cicirata et al., 2005). Together, these seminal studies revealed the segmental repetition, axonal directionalities, and routes of hindbrain neurons. Yet, certain neurons, axonal tracts, and projections were overlooked by these approaches, and are demonstrated here. Moreover, molecular identities of the interneurons that projected into the reported routes were unclear. Our genetic tools enabled the tracing of two dA1-derived tracts, supporting some of the previously reported funiculi, and providing new knowledge regarding interneuronal identities and trajectories that have not been accurately presented before. For instance, DiI labeling of caudal hindbrain commissures, identified as second order vestibular neurons (cC-VC), demonstrated their axonal turning in the DF (Díaz et al., 1998; Zhu et al., 2006). However, the genetic characteristics of cC-VC, their detailed localization in relation to other interneurons, and the progression of their axons, were not shown. Our data suggest this path to be contributed by dA1r6–7 cells. Furthermore, our constitutive-labeling strategy enabled us to trace the growth and connectivity of the DF at advanced stages, linking for the first time a subset of hindbrain interneurons with their axonal projections and targets.
Genetic strategies in mice extended the classical chick studies and revealed that dA1/Atoh1+ neurons along the cRL assemble the ventral/dorsal CN and PCN (Wang et al., 2005; Okada et al., 2007; Fu et al., 2009, 2011; Rose et al., 2009b). Application of intersectional/subtractive fate-map approaches further identified the contribution of rhombomere-specific interneurons to these nuclei (Farago et al., 2006; Maricich et al., 2009). For instance, r2–r5-derived Atoh1+ cells contribute neurons of the CN, whereas Atoh1+ from r6–r8 generates multiple precerebellar MF nuclei. These studies revealed the spatio-temporal development of mammalian brainstem centers and Atoh1 role in these assemblies. Yet, data describing the precise axonal patterns of each dA1 subgroup are partial, mostly because germ-line transgenesis results in general labeling of Atoh1+ neurons, complicating the ability to trace unilateral projections. Unilateral gene transfer by in utero electroporation is the only means to visualize hindbrain axonal projections in mice. Few such studies were previously performed to label PCN, such as the pontine-gray nuclei (PGN; Okada et al., 2007; Dipietrantonio and Dymecki, 2009). These analyses demonstrated that the majority of PGN axons extend contralaterally to the cerebellum, whereas fewer extend ipsilaterally. Yet, these methods used nonconditional GFP expression, as opposed to our strategy in which a precise GFP labeling of dA1-derived axons is provided. Moreover, the GFP transfer was performed in a restricted time point (i.e., E14.5) and axonal projections were analyzed only at one, much advanced, stage (i.e., P8; Okada et al., 2007; Dipietrantonio and Dymecki, 2009). Thus, questions regarding how dA1 neurons project axons at earlier stages and what the progression dynamics are from the hindbrain toward the cerebellum remained open. Our results corroborate and expand the mice studies by showing at much earlier chick stages contralateral dA1 axonal trajectories that progress in the DF toward the cerebellum, and the initiation of a dorsal ipsilateral tract at later stages. Hence, our genetic targeting of dA1 neurons in a unilateral and spatially restricted manner in chicks enables us to trace axonal trajectories on a stage-by-stage basis that are harder to visualize in mammals.
In both avian and mammals, the lower cRL (r6–r8) generates the precerebellar MF nuclei. In avian, this region also generates the posterior CN (nucleus magnocellularis), whereas the more anterior nucleus angularis (NA) counterpart, arises from r3–r6. In contrast, all mammalian CN progenitors arise from the auditory lip at r2–r5 (Marín and Puelles, 1995; Ryugo and Parks, 2003; Hendricks et al., 2006). Mice fate-map studies discovered dA1 interneurons from lower and upper cRL to generate the PCN and CN (Wang et al., 2005; Farago et al., 2006; Maricich et al., 2009; Rose et al., 2009b). Although likely, it is yet unclear whether dA1s assemble these centers in avian. Here we show dA1r6–7 neurons, which project into the DF, extend in the upper hindbrain, midbrain, and cerebellum and form synapses. However, we cannot distinguish between axons that originated from auditory (NM, posterior NA) or precerebellar progenitors. One possibility is that both subpopulations send axons into the DF and separate rostrally to project to different targets. This is supported by the synapses found both at the brainstem and cerebellum; although cerebellar synapses most probably originate from dA1-axons that extend from PCN, synapses in the NL are likely to represent dA1-axons originating from ipsilateral or contralateral NM (Marín and Puelles, 1995; Díaz et al., 2003). Another possibility is that early or later-born dA1 interneurons, which were previously reported in mice (Wang et al., 2005), also exist in avian (Cambronero and Puelles, 2000), and establish different trajectories at distinct times. This would suggest that the DF observed in our experiments originates from a subset of posterior dA1s, and is in agreement with our findings of additional tracts that appear at later stages, that may represent later-born dA1s. Assessing between these possibilities requires advanced tracing tools to label distinct dA1 subgroups (i.e., CN/PCN) or early versus later-born neurons. Such answers will highlight the conservation or difference between mammalian and avian cRL development.
How connectivity is established between brainstem neurons is not well understood. Previous mice data suggested that cRL-derived Atoh1+ cells project to Atoh1+ neurons in the cerebellum (Bermingham et al., 2001). Our findings of synapses in the EGL/IGL/DCN, formed by dA1r6–7 axons, confirm the brainstem Atoh1+ circuitry, since these layers are largely derived from Atoh1+ cells (Wang et al., 2005). Intriguingly, synapses were also observed in the Purkinje layer. This finding supports recent data in which PGN axons were found to project to Purkinje neurons in mice (Kalinovsky et al., 2011). Future work is required to unravel whether distinct dA1-subtypes in the cRL form the differential cerebellar connectivity, and whether the synapses observed are transient in character or stable. Moreover, it is still unclear whether Atoh1+-circuitry is also observed in the medulla, as the genetic characteristic of chick NL is unknown.
At r2–r5 hindbrain levels, dA1 interneurons project into the LF. This tract can be observed also at later stages (E12), where it reaches the pons and extends to the midbrain (data not shown). Based on previous reports, these axons may derive from the more anterior CN, the NA, which encompasses r2–r6 and projects toward the midbrain, thalamus, and cortex (Marín and Puelles, 1995; Díaz et al., 2003).
Possible cues that regulate dA1-axonal projection patterns
We found that dA1 progenitors give rise to two subpopulations; dA1r6–7 and dA1r2–5, which differ in their axonal patterns. Common cues should guide both groups' axons across the floor plate, while diverged cues should determine their longitudinal choice-point turning. Lim-HD proteins are involved in regulating neuronal cell fate and axonal guidance (Hobert and Westphal, 2000). In the spinal cord, alterations in Lim-HD code modified interneuron axonal projections without changing cell fates (Bachy et al., 2001; Pillai et al., 2007; Wilson et al., 2008; Avraham et al., 2009; Palmesino et al., 2010). In the hindbrain, Lhx2/9 proteins are expressed in dA1r6–7 and dA1r2–5 neurons. Here we show that alteration of dA1 Lim-HD code from Lhx2/9 to Lhx1 is sufficient to misdirect the patterns of dA1-axons toward dB1-like patterns, without affecting their cell-fate acquisition. These findings support the previous studies and suggest for the first time a role for Lim-HD code in patterning hindbrain axons. Intriguingly, dB1 (ptf1a+) interneurons are GABAergic/inhibitory, whereas dA1 (Atoh1+) interneurons are glutamatergic/excitatory. Whether the shift of dA1Lhx1 axons toward a dB1-like pattern would affect their latter neurotransmitter phenotype and synaptic targets remains to be elucidated.
While Lhx2/9 are uniformly expressed in all dA1-neurons, their axons form different ascending trajectories after crossing the midline. Hence, other transcription factors should cooperate with Lim-HD proteins in patterning their growth. Hox code was shown to control axonal pathfinding in the spinal cord and hindbrain (Dasen et al., 2005; Geisen et al., 2008; Narita and Rijli, 2009; Watari-Goshima and Chisaka, 2011). It is possible that the diverse dA1 axonal tracts relay on different Hox genes. Candidates are HoxA3/B2/B3/D3, which display differential expression levels in correlation with r5/r6 border. Another candidate is Zic1, which was found to drive the axonal projection choice of MF (PGN) nuclei toward the ipsilateral or contralateral cerebellum (Dipietrantonio and Dymecki, 2009). It will be important to explore the roles and cross talks between these transcription factors in patterning dA1 axonal paths.
Extracellular or membrane-bound attractive/repulsive cues are also critical to guide axons (Tessier-Lavigne and Goodman, 1996; Guthrie, 1997; Rodriguez et al., 2005; Dickson and Gilestro, 2006; Zhu et al., 2006). Wnts, Shh, and Robo/Slit are central in guiding commissural axons (including Atoh1+ neurons), before and after crossing the floor plate (Lyuksyutova et al., 2003; Bourikas et al., 2005; Howell et al., 2007; Renier et al., 2010). Hindbrain MF neurons were shown to require Robo/Slit as well as adhesion molecules (TAG-1, cadherins) for their normal soma or axonal migration along the DV axis (Backer et al., 2002; Marillat et al., 2004; Kawauchi et al., 2006; Taniguchi et al., 2006; Wilson et al., 2008; Renier et al., 2010). Moreover, Eph/Ephrin signaling was found to guide hindbrain CN and cC-VC axons (Zhu et al., 2006; Hsieh et al., 2010). The dorsal position of cC-VC neurons, the expression of Atoh1, and the commissural axonal projection followed by turning into the DF all implicate cC-VC neurons as being dA1r6–7 neurons. Thus, the different turning points of dA1r6–7 and dA1r2–5 might be determined by Eph/Ephrin cues. Interestingly, the expression of these guiding cues is regulated by Hox, Lim-HD, and Zic genes (García-Frigola et al., 2008; Geisen et al., 2008; Wilson et al., 2008). Thus, elucidating the network between the transcriptional codes and the guiding cues in governing dA1 axonal trajectories is the focus of future studies.
Footnotes
D.S.-D. was supported by grants from the Niedersachsen-Israeli Research Cooperation Program, The National Institute for Psychobiology in Israel, and the Israel Science Foundation (Grant 161/07); A. Klar was supported by grants from Israel Science Foundation (Grant 229/09), the Legacy Heritage Biomedical Science Partnership Program of the Israel Science Foundation (Grant 1930/08), Center of Excellence–Legacy Heritage Biomedical Science Partnership (Grant 1803/10), and the Israel Ministry of Health. We thank Tom Jessell, Hirohide Takebayashy, Esther Stoeckli, and Rosalind Segal for providing antibodies against Lhx2/9, Olig3, Axonin1, and Zic1 proteins, respectively. We also thank Jane Johnson for the Ptf1a::GFP plasmid, Susana Cohen-Cory for the Synaptobrevin-GFP plasmid, Noam Ziv for the SV2-GFP plasmid, and Xiaozhong Wang and the Sanger Institute for providing the PiggyBac system. We thank Amit Lilo for assisting in the illustrations and Irit Shoval for her help with the confocal analysis.
References
- Akazawa C, Ishibashi M, Shimizu C, Nakanishi S, Kageyama R. A mammalian helix-loop-helix factor structurally related to the product of Drosophila proneural gene atonal is a positive transcriptional regulator expressed in the developing nervous system. J Biol Chem. 1995;270:8730–8738. doi: 10.1074/jbc.270.15.8730. [DOI] [PubMed] [Google Scholar]
- Alder J, Lee KJ, Jessell TM, Hatten ME. Generation of cerebellar granule neurons in vivo by transplantation of BMP-treated neural progenitor cells. Nat Neurosci. 1999;2:535–540. doi: 10.1038/9189. [DOI] [PubMed] [Google Scholar]
- Alsina B, Vu T, Cohen-Cory S. Visualizing synapse formation in arborizing optic axons in vivo: dynamics and modulation by BDNF. Nat Neurosci. 2001;4:1093–1101. doi: 10.1038/nn735. [DOI] [PubMed] [Google Scholar]
- Altman J, Bayer SA. Development of the brain stem in the rat. II. Thymidine-radiographic study of the time of origin of neurons of the upper medulla, excluding the vestibular and auditory nuclei. J Comp Neurol. 1980;194:37–56. doi: 10.1002/cne.901940103. [DOI] [PubMed] [Google Scholar]
- Altman J, Bayer SA. Development of the precerebellar nuclei in the rat: I. The precerebellar neuroepithelium of the rhombencephalon. J Comp Neurol. 1987a;257:477–489. doi: 10.1002/cne.902570402. [DOI] [PubMed] [Google Scholar]
- Altman J, Bayer SA. Development of the precerebellar nuclei in the rat: II. The intramural olivary migratory stream and the neurogenetic organization of the inferior olive. J Comp Neurol. 1987b;257:490–512. doi: 10.1002/cne.902570403. [DOI] [PubMed] [Google Scholar]
- Altman J, Bayer SA. Development of the precerebellar nuclei in the rat: III. The posterior precerebellar extramural migratory stream and the lateral reticular and external cuneate nuclei. J Comp Neurol. 1987c;257:513–528. doi: 10.1002/cne.902570404. [DOI] [PubMed] [Google Scholar]
- Altman J, Bayer SA. Development of the precerebellar nuclei in the rat: IV. The anterior precerebellar extramural migratory stream and the nucleus reticularis tegmenti pontis and the basal pontine gray. J Comp Neurol. 1987d;257:529–552. doi: 10.1002/cne.902570405. [DOI] [PubMed] [Google Scholar]
- Altman J, Bayer SA. Development of the cerebellar system: in relation to its evolution, structure, and functions. Boca Raton, FL: CRC; 1997. [Google Scholar]
- Aruga J, Yokota N, Hashimoto M, Furuichi T, Fukuda M, Mikoshiba K. A novel zinc finger protein, zic, is involved in neurogenesis, especially in the cell lineage of cerebellar granule cells. J Neurochem. 1994;63:1880–1890. doi: 10.1046/j.1471-4159.1994.63051880.x. [DOI] [PubMed] [Google Scholar]
- Aruga J, Inoue T, Hoshino J, Mikoshiba K. Zic2 controls cerebellar development in cooperation with Zic1. J Neurosci. 2002;22:218–225. doi: 10.1523/JNEUROSCI.22-01-00218.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avraham O, Hadas Y, Vald L, Zisman S, Schejter A, Visel A, Klar A. Transcriptional control of axonal guidance and sorting in dorsal interneurons by the Lim-HD proteins Lhx9 and Lhx1. Neural Dev. 2009;4:21. doi: 10.1186/1749-8104-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avraham O, Hadas Y, Vald L, Hong S, Song MR, Klar A. Motor and dorsal root ganglion axons serve as choice points for the ipsilateral turning of dI3 axons. J Neurosci. 2010a;30:15546–15557. doi: 10.1523/JNEUROSCI.2380-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avraham O, Zisman S, Hadas Y, Vald L, Klar A. Deciphering axonal pathways of genetically defined groups of neurons in the chick neural tube utilizing in ovo electroporation. J Vis Exp. 2010b doi: 10.3791/1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachy I, Vernier P, Retaux S. The LIM-homeodomain gene family in the developing Xenopus brain: conservation and divergences with the mouse related to the evolution of the forebrain. J Neurosci. 2001;21:7620–7629. doi: 10.1523/JNEUROSCI.21-19-07620.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backer S, Sakurai T, Grumet M, Sotelo C, Bloch-Gallego E. Nr-CAM and TAG-1 are expressed in distinct populations of developing precerebellar and cerebellar neurons. Neuroscience. 2002;113:743–748. doi: 10.1016/s0306-4522(02)00221-x. [DOI] [PubMed] [Google Scholar]
- Baeriswyl T, Stoeckli ET. Axonin-1/TAG-1 is required for pathfinding of granule cell axons in the developing cerebellum. Neural Dev. 2008;3:7. doi: 10.1186/1749-8104-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baimbridge KG, Celio MR, Rogers JH. Calcium-binding proteins in the nervous system. Trends Neurosci. 1992;15:303–308. doi: 10.1016/0166-2236(92)90081-i. [DOI] [PubMed] [Google Scholar]
- Ben-Arie N, Bellen HJ, Armstrong DL, McCall AE, Gordadze PR, Guo Q, Matzuk MM, Zoghbi HY. Math1 is essential for genesis of cerebellar granule neurons. Nature. 1997;390:169–172. doi: 10.1038/36579. [DOI] [PubMed] [Google Scholar]
- Bermingham NA, Hassan BA, Wang VY, Fernandez M, Banfi S, Bellen HJ, Fritzsch B, Zoghbi HY. Proprioceptor pathway development is dependent on Math1. Neuron. 2001;30:411–422. doi: 10.1016/s0896-6273(01)00305-1. [DOI] [PubMed] [Google Scholar]
- Bourikas D, Pekarik V, Baeriswyl T, Grunditz A, Sadhu R, Nardó M, Stoeckli ET. Sonic hedgehog guides commissural axons along the longitudinal axis of the spinal cord. Nat Neurosci. 2005;8:297–304. doi: 10.1038/nn1396. [DOI] [PubMed] [Google Scholar]
- Bragina L, Fattorini G, Giovedí S, Melone M, Bosco F, Benfenati F, Conti F. Analysis of synaptotagmin, SV2, and Rab3 expression in cortical glutamatergic and GABAergic axon terminals. Front Cell Neurosci. 2011;5:32. doi: 10.3389/fncel.2011.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambronero F, Puelles L. Rostrocaudal nuclear relationships in the avian medulla oblongata: a fate map with quail chick chimeras. J Comp Neurol. 2000;427:522–545. doi: 10.1002/1096-9861(20001127)427:4<522::aid-cne3>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Cant NB. The cochlear nucleus: neuronal types and their synaptic organization. In: Webster DB, Proper AN, Fay RR, editors. The mammalian auditory pathway: neuroanatomy. New York: Springer; 1992. pp. 66–116. [Google Scholar]
- Chang WP, Südhof TC. SV2 renders primed synaptic vesicles competent for Ca2+-induced exocytosis. J Neurosci. 2009;29:883–897. doi: 10.1523/JNEUROSCI.4521-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicirata F, Zappalà A, Serapide MF, Parenti R, Pantò MR, Paz C. Different pontine projections to the two sides of the cerebellum. Brain Res Rev. 2005;49:280–294. doi: 10.1016/j.brainresrev.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Clarke JD, Lumsden A. Segmental repetition of neuronal phenotype sets in the chick embryo hindbrain. Development. 1993;118:151–162. doi: 10.1242/dev.118.1.151. [DOI] [PubMed] [Google Scholar]
- Dasen JS, Tice BC, Brenner-Morton S, Jessell TM. A Hox regulatory network establishes motor neuron pool identity and target-muscle connectivity. Cell. 2005;123:477–491. doi: 10.1016/j.cell.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Díaz C, Puelles L, Marín F, Glover JC. The relationship between rhombomeres and vestibular neuron populations as assessed in quail-chicken chimeras. Dev Biol. 1998;202:14–28. doi: 10.1006/dbio.1998.8986. [DOI] [PubMed] [Google Scholar]
- Díaz C, Glover JC, Puelles L, Bjaalie JG. The relationship between hodological and cytoarchitectonic organization in the vestibular complex of the 11-day chicken embryo. J Comp Neurol. 2003;457:87–105. doi: 10.1002/cne.10528. [DOI] [PubMed] [Google Scholar]
- Dickson BJ, Gilestro GF. Regulation of commissural axon pathfinding by slit and its Robo receptors. Annu Rev Cell Dev Biol. 2006;22:651–675. doi: 10.1146/annurev.cellbio.21.090704.151234. [DOI] [PubMed] [Google Scholar]
- Dipietrantonio HJ, Dymecki SM. Zic1 levels regulate mossy fiber neuron position and axon laterality choice in the ventral brain stem. Neuroscience. 2009;162:560–573. doi: 10.1016/j.neuroscience.2009.02.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet JR, Ryugo DK. Axonal pathways to the lateral superior olive labeled with biotinylated dextran amine injections in the dorsal cochlear nucleus of rats. J Comp Neurol. 2003;461:452–465. doi: 10.1002/cne.10722. [DOI] [PubMed] [Google Scholar]
- Ebert PJ, Timmer JR, Nakada Y, Helms AW, Parab PB, Liu Y, Hunsaker TL, Johnson JE. Zic1 represses Math1 expression via interactions with the Math1 enhancer and modulation of Math1 autoregulation. Development. 2003;130:1949–1959. doi: 10.1242/dev.00419. [DOI] [PubMed] [Google Scholar]
- Farago AF, Awatramani RB, Dymecki SM. Assembly of the brainstem cochlear nuclear complex is revealed by intersectional and subtractive genetic fate maps. Neuron. 2006;50:205–218. doi: 10.1016/j.neuron.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Fu Y, Tvrdik P, Makki N, Palombi O, Machold R, Paxinos G, Watson C. The precerebellar linear nucleus in the mouse defined by connections, immunohistochemistry, and gene expression. Brain Res. 2009;1271:49–59. doi: 10.1016/j.brainres.2009.02.068. [DOI] [PubMed] [Google Scholar]
- Fu Y, Tvrdik P, Makki N, Paxinos G, Watson C. Precerebellar cell groups in the hindbrain of the mouse defined by retrograde tracing and correlated with cumulative Wnt1-cre genetic labeling. Cerebellum. 2011;10:570–584. doi: 10.1007/s12311-011-0266-1. [DOI] [PubMed] [Google Scholar]
- Fujiyama T, Yamada M, Terao M, Terashima T, Hioki H, Inoue YU, Inoue T, Masuyama N, Obata K, Yanagawa Y, Kawaguchi Y, Nabeshima Y, Hoshino M. Inhibitory and excitatory subtypes of cochlear nucleus neurons are defined by distinct bHLH transcription factors, Ptf1a and Atoh1. Development. 2009;136:2049–2058. doi: 10.1242/dev.033480. [DOI] [PubMed] [Google Scholar]
- García-Frigola C, Carreres MI, Vegar C, Mason C, Herrera E. Zic2 promotes axonal divergence at the optic chiasm midline by EphB1-dependent and -independent mechanisms. Development. 2008;135:1833–1841. doi: 10.1242/dev.020693. [DOI] [PubMed] [Google Scholar]
- Gardzinski P, Lee DW, Fei GH, Hui K, Huang GJ, Sun HS, Feng ZP. The role of synaptotagmin I C2A calcium-binding domain in synaptic vesicle clustering during synapse formation. J Physiol. 2007;581:75–90. doi: 10.1113/jphysiol.2006.127472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisen MJ, Di Meglio T, Pasqualetti M, Ducret S, Brunet JF, Chedotal A, Rijli FM. Hox paralog group 2 genes control the migration of mouse pontine neurons through slit-robo signaling. PLoS Biol. 2008;6:e142. doi: 10.1371/journal.pbio.0060142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinberg I, Northrup H, Ardinger H, Prasad C, Dobyns WB, Millen KJ. Heterozygous deletion of the linked genes ZIC1 and ZIC4 is involved in Dandy-Walker malformation. Nat Genet. 2004;36:1053–1055. doi: 10.1038/ng1420. [DOI] [PubMed] [Google Scholar]
- Guthrie S. Axon guidance: netrin receptors are revealed. Curr Biol. 1997;7:R6–R9. doi: 10.1016/s0960-9822(06)00007-8. [DOI] [PubMed] [Google Scholar]
- Guthrie S. Patterning and axon guidance of cranial motor neurons. Nat Rev Neurosci. 2007;8:859–871. doi: 10.1038/nrn2254. [DOI] [PubMed] [Google Scholar]
- Guthrie S, Lumsden A. Motor neuron pathfinding following rhombomere reversals in the chick embryo hindbrain. Development. 1992;114:663–673. doi: 10.1242/dev.114.3.663. [DOI] [PubMed] [Google Scholar]
- Hatten ME, Heintz N. Mechanisms of neural patterning and specification in the developing cerebellum. Annu Rev Neurosci. 1995;18:385–408. doi: 10.1146/annurev.ne.18.030195.002125. [DOI] [PubMed] [Google Scholar]
- Helms AW, Johnson JE. Progenitors of dorsal commissural interneurons are defined by MATH1 expression. Development. 1998;125:919–928. doi: 10.1242/dev.125.5.919. [DOI] [PubMed] [Google Scholar]
- Helms AW, Abney AL, Ben-Arie N, Zoghbi HY, Johnson JE. Autoregulation and multiple enhancers control Math1 expression in the developing nervous system. Development. 2000;127:1185–1196. doi: 10.1242/dev.127.6.1185. [DOI] [PubMed] [Google Scholar]
- Hendricks SJ, Rubel EW, Nishi R. Formation of the avian nucleus magnocellularis from the auditory anlage. J Comp Neurol. 2006;498:433–442. doi: 10.1002/cne.21031. [DOI] [PubMed] [Google Scholar]
- Hobert O, Westphal H. Functions of LIM-homeobox genes. Trends Genet. 2000;16:75–83. doi: 10.1016/s0168-9525(99)01883-1. [DOI] [PubMed] [Google Scholar]
- Howell DM, Morgan WJ, Jarjour AA, Spirou GA, Berrebi AS, Kennedy TE, Mathers PH. Molecular guidance cues necessary for axon pathfinding from the ventral cochlear nucleus. J Comp Neurol. 2007;504:533–549. doi: 10.1002/cne.21443. [DOI] [PubMed] [Google Scholar]
- Hsieh CY, Nakamura PA, Luk SO, Miko IJ, Henkemeyer M, Cramer KS. Ephrin-B reverse signaling is required for formation of strictly contralateral auditory brainstem pathways. J Neurosci. 2010;30:9840–9849. doi: 10.1523/JNEUROSCI.0386-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinovsky A, Boukhtouche F, Blazeski R, Bornmann C, Suzuki N, Mason CA, Scheiffele P. Development of axon-target specificity of ponto-cerebellar afferents. PLoS Biol. 2011;9:e1001013. doi: 10.1371/journal.pbio.1001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kania A, Johnson RL, Jessell TM. Coordinate roles for LIM homeobox genes in directing the dorsoventral trajectory of motor axons in the vertebrate limb. Cell. 2000;102:161–173. doi: 10.1016/s0092-8674(00)00022-2. [DOI] [PubMed] [Google Scholar]
- Kawauchi D, Taniguchi H, Watanabe H, Saito T, Murakami F. Direct visualization of nucleogenesis by precerebellar neurons: involvement of ventricle-directed, radial fibre-associated migration. Development. 2006;133:1113–1123. doi: 10.1242/dev.02283. [DOI] [PubMed] [Google Scholar]
- Köster RW, Fraser SE. Direct imaging of in vivo neuronal migration in the developing cerebellum. Curr Biol. 2001;11:1858–1863. doi: 10.1016/s0960-9822(01)00585-1. [DOI] [PubMed] [Google Scholar]
- Lai HC, Klisch TJ, Roberts R, Zoghbi HY, Johnson JE. In vivo neuronal subtype-specific targets of Atoh1 (Math1) in dorsal spinal cord. J Neurosci. 2011;31:10859–10871. doi: 10.1523/JNEUROSCI.0445-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landsberg RL, Awatramani RB, Hunter NL, Farago AF, Dipietrantonio HJ, Rodriguez CI, Dymecki SM. Hindbrain rhombic lip is comprised of discrete progenitor cell populations allocated by Pax6. Neuron. 2005;48:933–947. doi: 10.1016/j.neuron.2005.11.031. [DOI] [PubMed] [Google Scholar]
- Leal-Ortiz S, Waites CL, Terry-Lorenzo R, Zamorano P, Gundelfinger ED, Garner CC. Piccolo modulation of Synapsin1a dynamics regulates synaptic vesicle exocytosis. J Cell Biol. 2008;181:831–846. doi: 10.1083/jcb.200711167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Li H, Hu X, Yu L, Liu H, Han R, Colella R, Mower GD, Chen Y, Qiu M. Control of precerebellar neuron development by Olig3 bHLH transcription factor. J Neurosci. 2008;28:10124–10133. doi: 10.1523/JNEUROSCI.3769-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Lin C, Wang X. PiggyBac transgenic strategies in the developing chicken spinal cord. Nucleic Acids Res. 2009;37:e141. doi: 10.1093/nar/gkp686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumpkin EA, Collisson T, Parab P, Omer-Abdalla A, Haeberle H, Chen P, Doetzlhofer A, White P, Groves A, Segil N, Johnson JE. Math1-driven GFP expression in the developing nervous system of transgenic mice. Gene Expr Patterns. 2003;3:389–395. doi: 10.1016/s1567-133x(03)00089-9. [DOI] [PubMed] [Google Scholar]
- Lumsden A. Segmentation and compartition in the early avian hindbrain. Mech Dev. 2004;121:1081–1088. doi: 10.1016/j.mod.2004.04.018. [DOI] [PubMed] [Google Scholar]
- Lumsden A, Keynes R. Segmental patterns of neuronal development in the chick hindbrain. Nature. 1989;337:424–428. doi: 10.1038/337424a0. [DOI] [PubMed] [Google Scholar]
- Lumsden A, Krumlauf R. Patterning the vertebrate neuraxis. Science. 1996;274:1109–1115. doi: 10.1126/science.274.5290.1109. [DOI] [PubMed] [Google Scholar]
- Lyuksyutova AI, Lu CC, Milanesio N, King LA, Guo N, Wang Y, Nathans J, Tessier-Lavigne M, Zou Y. Anterior-posterior guidance of commissural axons by Wnt-frizzled signaling. Science. 2003;302:1984–1988. doi: 10.1126/science.1089610. [DOI] [PubMed] [Google Scholar]
- Machold R, Fishell G. Math1 is expressed in temporally discrete pools of cerebellar rhombic-lip neural progenitors. Neuron. 2005;48:17–24. doi: 10.1016/j.neuron.2005.08.028. [DOI] [PubMed] [Google Scholar]
- Maricich SM, Xia A, Mathes EL, Wang VY, Oghalai JS, Fritzsch B, Zoghbi HY. Atoh1-lineal neurons are required for hearing and for the survival of neurons in the spiral ganglion and brainstem accessory auditory nuclei. J Neurosci. 2009;29:11123–11133. doi: 10.1523/JNEUROSCI.2232-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marillat V, Sabatier C, Failli V, Matsunaga E, Sotelo C, Tessier-Lavigne M, Chédotal A. The slit receptor Rig-1/Robo3 controls midline crossing by hindbrain precerebellar neurons and axons. Neuron. 2004;43:69–79. doi: 10.1016/j.neuron.2004.06.018. [DOI] [PubMed] [Google Scholar]
- Marín F, Puelles L. Morphological fate of rhombomeres in quail/chick chimeras: a segmental analysis of hindbrain nuclei. Eur J Neurosci. 1995;7:1714–1738. doi: 10.1111/j.1460-9568.1995.tb00693.x. [DOI] [PubMed] [Google Scholar]
- Mason CA. Axon development in mouse cerebellum: embryonic axon forms and expression of synapsin I. Neuroscience. 1986;19:1319–1333. doi: 10.1016/0306-4522(86)90146-6. [DOI] [PubMed] [Google Scholar]
- Mason CA, Morrison ME, Ward MS, Zhang Q, Baird DH. Axon-target interactions in the developing cerebellum. Perspect Dev Neurobiol. 1997;5:69–82. [PubMed] [Google Scholar]
- Meredith DM, Masui T, Swift GH, MacDonald RJ, Johnson JE. Multiple transcriptional mechanisms control Ptf1a levels during neural development including autoregulation by the PTF1-J complex. J Neurosci. 2009;29:11139–11148. doi: 10.1523/JNEUROSCI.2303-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milsom WK, Chatburn J, Zimmer MB. Pontine influences on respiratory control in ectothermic and heterothermic vertebrates. Respir Physiol Neurobiol. 2004;143:263–280. doi: 10.1016/j.resp.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Moens CB, Prince VE. Constructing the hindbrain: insights from the zebrafish. Dev Dyn. 2002;224:1–17. doi: 10.1002/dvdy.10086. [DOI] [PubMed] [Google Scholar]
- Morales D, Hatten ME. Molecular markers of neuronal progenitors in the embryonic cerebellar anlage. J Neurosci. 2006;26:12226–12236. doi: 10.1523/JNEUROSCI.3493-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgans CW, Kensel-Hammes P, Hurley JB, Burton K, Idzerda R, McKnight GS, Bajjalieh SM. Loss of the Synaptic Vesicle Protein SV2B results in reduced neurotransmission and altered synaptic vesicle protein expression in the retina. PLoS One. 2009;4:e5230. doi: 10.1371/journal.pone.0005230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita Y, Rijli FM. Hox genes in neural patterning and circuit formation in the mouse hindbrain. Curr Top Dev Biol. 2009;88:139–167. doi: 10.1016/S0070-2153(09)88005-8. [DOI] [PubMed] [Google Scholar]
- Nowack A, Yao J, Custer KL, Bajjalieh SM. SV2 regulates neurotransmitter release via multiple mechanisms. Am J Physiol Cell Physiol. 2010;299:C960–C967. doi: 10.1152/ajpcell.00259.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oertel D, Young ED. What's a cerebellar circuit doing in the auditory system? Trends Neurosci. 2004;27:104–110. doi: 10.1016/j.tins.2003.12.001. [DOI] [PubMed] [Google Scholar]
- Okada T, Keino-Masu K, Masu M. Migration and nucleogenesis of mouse precerebellar neurons visualized by in utero electroporation of a green fluorescent protein gene. Neurosci Res. 2007;57:40–49. doi: 10.1016/j.neures.2006.09.010. [DOI] [PubMed] [Google Scholar]
- Osumi N, Hirota A, Ohuchi H, Nakafuku M, Iimura T, Kuratani S, Fujiwara M, Noji S, Eto K. Pax-6 is involved in the specification of hindbrain motor neuron subtype. Development. 1997;124:2961–2972. doi: 10.1242/dev.124.15.2961. [DOI] [PubMed] [Google Scholar]
- Palmesino E, Rousso DL, Kao TJ, Klar A, Laufer E, Uemura O, Okamoto H, Novitch BG, Kania A. Foxp1 and lhx1 coordinate motor neuron migration with axon trajectory choice by gating Reelin signalling. PLoS Biol. 2010;8:e1000446. doi: 10.1371/journal.pbio.1000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, Plajzer-Frick I, Akiyama J, De Val S, Afzal V, Black BL, Couronne O, Eisen MB, Visel A, Rubin EM. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- Pillai A, Mansouri A, Behringer R, Westphal H, Goulding M. Lhx1 and Lhx5 maintain the inhibitory-neurotransmitter status of interneurons in the dorsal spinal cord. Development. 2007;134:357–366. doi: 10.1242/dev.02717. [DOI] [PubMed] [Google Scholar]
- Reeber SL, Sakai N, Nakada Y, Dumas J, Dobrenis K, Johnson JE, Kaprielian Z. Manipulating Robo expression in vivo perturbs commissural axon pathfinding in the chick spinal cord. J Neurosci. 2008;28:8698–8708. doi: 10.1523/JNEUROSCI.1479-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renier N, Schonewille M, Giraudet F, Badura A, Tessier-Lavigne M, Avan P, De Zeeuw CI, Chédotal A. Genetic dissection of the function of hindbrain axonal commissures. PLoS Biol. 2010;8:e1000325. doi: 10.1371/journal.pbio.1000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez CI, Dymecki SM. Origin of the precerebellar system. Neuron. 2000;27:475–486. doi: 10.1016/s0896-6273(00)00059-3. [DOI] [PubMed] [Google Scholar]
- Rodriguez J, Esteve P, Weinl C, Ruiz JM, Fermin Y, Trousse F, Dwivedy A, Holt C, Bovolenta P. SFRP1 regulates the growth of retinal ganglion cell axons through the Fz2 receptor. Nat Neurosci. 2005;8:1301–1309. doi: 10.1038/nn1547. [DOI] [PubMed] [Google Scholar]
- Rose MF, Ahmad KA, Thaller C, Zoghbi HY. Excitatory neurons of the proprioceptive, interoceptive, and arousal hindbrain networks share a developmental requirement for Math1. Proc Natl Acad Sci U S A. 2009a;106:22462–22467. doi: 10.1073/pnas.0911579106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MF, Ren J, Ahmad KA, Chao HT, Klisch TJ, Flora A, Greer JJ, Zoghbi HY. Math1 is essential for the development of hindbrain neurons critical for perinatal breathing. Neuron. 2009b;64:341–354. doi: 10.1016/j.neuron.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubel EW, Parks TN. Organization and development of the avian brainstem auditory system. In: Edelman GM, Gall WE, Cowan WM, editors. Auditory function. New York: Wiley; 1988. pp. 3–92. [Google Scholar]
- Ryugo DK, Parks TN. Primary innervation of the avian and mammalian cochlear nucleus. Brain Res Bull. 2003;60:435–456. doi: 10.1016/s0361-9230(03)00049-2. [DOI] [PubMed] [Google Scholar]
- Ryugo DK, Haenggeli CA, Doucet JR. Multimodal inputs to the granule cell domain of the cochlear nucleus. Exp Brain Res. 2003;153:477–485. doi: 10.1007/s00221-003-1605-3. [DOI] [PubMed] [Google Scholar]
- Sharma K, Sheng HZ, Lettieri K, Li H, Karavanov A, Potter S, Westphal H, Pfaff SL. LIM homeodomain factors Lhx3 and Lhx4 assign subtype identities for motor neurons. Cell. 1998;95:817–828. doi: 10.1016/s0092-8674(00)81704-3. [DOI] [PubMed] [Google Scholar]
- Shirasaki R, Pfaff SL. Transcriptional codes and the control of neuronal identity. Annu Rev Neurosci. 2002;25:251–281. doi: 10.1146/annurev.neuro.25.112701.142916. [DOI] [PubMed] [Google Scholar]
- Sieber MA, Storm R, Martinez-de-la-Torre M, Müller T, Wende H, Reuter K, Vasyutina E, Birchmeier C. Lbx1 acts as a selector gene in the fate determination of somatosensory and viscerosensory relay neurons in the hindbrain. J Neurosci. 2007;27:4902–4909. doi: 10.1523/JNEUROSCI.0717-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm R, Cholewa-Waclaw J, Reuter K, Bröhl D, Sieber M, Treier M, Müller T, Birchmeier C. The bHLH transcription factor Olig3 marks the dorsal neuroepithelium of the hindbrain and is essential for the development of brainstem nuclei. Development. 2009;136:295–305. doi: 10.1242/dev.027193. [DOI] [PubMed] [Google Scholar]
- Taniguchi H, Kawauchi D, Nishida K, Murakami F. Classic cadherins regulate tangential migration of precerebellar neurons in the caudal hindbrain. Development. 2006;133:1923–1931. doi: 10.1242/dev.02354. [DOI] [PubMed] [Google Scholar]
- Tessier-Lavigne M, Goodman CS. The molecular biology of axon guidance. Science. 1996;274:1123–1133. doi: 10.1126/science.274.5290.1123. [DOI] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser-a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35:D88–D92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Miller ED, Simmons GS, Miller TA, Tabashnik BE, Park Y. piggyBac-like elements in the pink bollworm, Pectinophora gossypiella. Insect Mol Biol. 2010;19:177–184. doi: 10.1111/j.1365-2583.2009.00964.x. [DOI] [PubMed] [Google Scholar]
- Wang VY, Zoghbi HY. Genetic regulation of cerebellar development. Nat Rev Neurosci. 2001;2:484–491. doi: 10.1038/35081558. [DOI] [PubMed] [Google Scholar]
- Wang VY, Rose MF, Zoghbi HY. Math1 expression redefines the rhombic lip derivatives and reveals novel lineages within the brainstem and cerebellum. Neuron. 2005;48:31–43. doi: 10.1016/j.neuron.2005.08.024. [DOI] [PubMed] [Google Scholar]
- Wang W, Bradley A, Huang Y. A piggyBac transposon-based genome-wide library of insertionally mutated Blm-deficient murine ES cells. Genome Res. 2009;19:667–673. doi: 10.1101/gr.085621.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watari-Goshima N, Chisaka O. Chicken HOXA3 gene: its expression pattern and role in branchial nerve precursor cell migration. Int J Biol Sci. 2011;7:87–101. doi: 10.7150/ijbs.7.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisinger K, Wilkinson DG, Sela-Donenfeld D. Inhibition of BMPs by follistatin is required for FGF3 expression and segmental patterning of the hindbrain. Dev Biol. 2008;324:213–225. doi: 10.1016/j.ydbio.2008.09.005. [DOI] [PubMed] [Google Scholar]
- Wilson SI, Shafer B, Lee KJ, Dodd J. A molecular program for contralateral trajectory: Rig-1 control by LIM homeodomain transcription factors. Neuron. 2008;59:413–424. doi: 10.1016/j.neuron.2008.07.020. [DOI] [PubMed] [Google Scholar]
- Wolfer DP, Giger RJ, Stagliar M, Sonderegger P, Lipp HP. Expression of the axon growth-related neural adhesion molecule TAG-1/axonin-1 in the adult mouse brain. Anat Embryol (Berl) 1998;197:177–185. doi: 10.1007/s004290050129. [DOI] [PubMed] [Google Scholar]
- Wurst W, Auerbach AB, Joyner AL. Multiple developmental defects in Engrailed-1 mutant mice: an early mid-hindbrain deletion and patterning defects in forelimbs and sternum. Development. 1994;120:2065–2075. doi: 10.1242/dev.120.7.2065. [DOI] [PubMed] [Google Scholar]
- Yao J, Nowack A, Kensel-Hammes P, Gardner RG, Bajjalieh SM. Cotrafficking of SV2 and synaptotagmin at the synapse. J Neurosci. 2010;30:5569–5578. doi: 10.1523/JNEUROSCI.4781-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Guthrie S, Murakami F. Ephrin A/EphA controls the rostral turning polarity of a lateral commissural tract in chick hindbrain. Development. 2006;133:3837–3846. doi: 10.1242/dev.02564. [DOI] [PubMed] [Google Scholar]